Unlike other cell cycle regulators that are targeted for degradation to allow cell cycle progression, the retinoblastoma protein (pRb) is regulated by cyclic phosphorylation and dephosphorylation. In this issue of Genes & Development, Burke et al. describe how the structure of pRb is altered by phosphorylation at T373 or S608. These modifications cause specific conformational changes and alter pRb's interaction with E2F via two distinct mechanisms. The structures suggest that the panel of phosphorylation sites represents a versatile set of tools that are used to sculpt pRb in precise, but very different, ways.
Keywords: Retinoblastoma protein, cell cycle regulation, multisite phosphorylation, cyclin-dependent kinase, X-ray crystal structure, small-angle X-ray scattering (SAXS)
Abstract
In this issue of Genes & Development, Burke and colleagues (pp. 1156–1166) describe how the structure of retinoblastoma protein (pRb) is altered by phosphorylation at T373 or S608. These modifications cause specific conformational changes and alter pRb's interaction with E2F via two distinct mechanisms. The structures suggest that the panel of phosphorylation sites represents a versatile set of tools that are used to sculpt pRb in precise, but very different, ways.
At first glance, the retinoblastoma protein (pRb) presents a paradox: a seemingly potent inhibitor of cell proliferation that is present in the dividing cells of normal tissues. The explanation for this conundrum stems from the fact that the activity of pRb is controlled by cyclin-dependent kinases (Cdks). Unlike other cell cycle regulators that are targeted for degradation at key moments to allow cell cycle progression, pRb is a stable protein. Instead, cyclic phosphorylation and dephosphorylation cause pRb to oscillate between active and inactive states. The observation that pRb becomes heavily modified during the G1-to-S transition (Buchkovich et al. 1989; Chen et al. 1989; DeCaprio et al. 1989) was an important insight that came just a few years after the discovery that mutational inactivation of the retinoblastoma tumor susceptibility gene is rate-limiting in the genesis of retinoblastoma, a malignant childhood cancer (Friend et al. 1986). These changes in pRb phosphorylation led to the idea that the proliferation-suppressing activities of pRb are switched off each time a cell progresses through a division cycle. We now know that the phosphorylation of pRb during late G1 phase allows the activation of E2F-dependent transcription, a program of gene expression that controls genes that drive the cell cycle and are needed for DNA replication (Weinberg 1995). In many cancers, the regulation of pRb becomes unbalanced, and elevated Cdk activity prevents pRb from being an effective brake.
Understanding how phosphorylation alters pRb is central to understanding how pRb functions. Human pRb contains at least 16 potential Cdk phosphorylation sites, most of which are located in unstructured regions of the protein. It has long been known that the modification of pRb disrupts its ability to form protein/protein interactions, but structural studies are finally beginning to explain the basis for these effects. The results described by Burke et al. (2012) illustrate that phosphorylation does not simply cause a random loss of pRb structure, but drives pRb into specific conformations, with individual phosphorylation events disrupting different features of the protein.
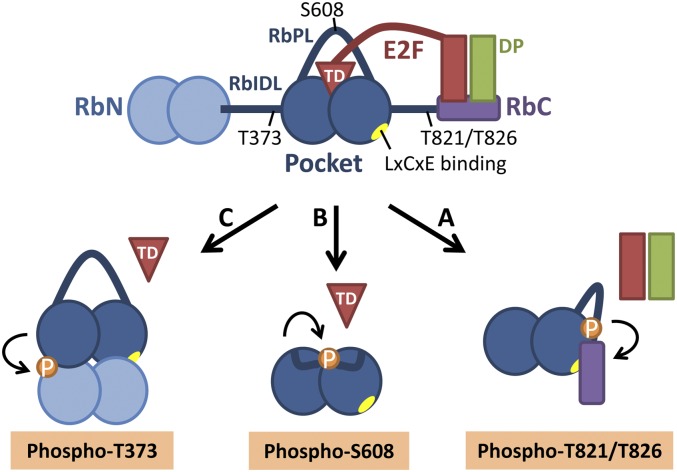
pRb regulates E2F activity in at least two ways: It binds directly to the transactivation domain of activator E2Fs and, in addition, uses a second site, the LxCxE-binding domain, to interact with transcriptional corepressors and recruit these to E2F target genes (for a recent review, see Chinnam and Goodrich 2011). The structures of individual pRb domains and their complexes with small E2F fragments have shed light on the pRb–E2F interaction. The central pocket domain of pRb consists of a tandem cyclin fold that interacts with the E2F transactivation domain (E2FTD) and is separated by the less-structured pocket domain loop (RbPL) (Fig. 1; Lee et al. 1998, 2002; Xiao et al. 2003). The pRb N-terminal domain (RbN) also contains a tandem cyclin fold and is connected with the pocket through an unstructured interdomain linker (RbIDL) (Hassler et al. 2007). The intrinsically disordered C-terminal tail of pRb binds the Marked Box domains of the E2F1–DP1 heterodimer, and this interaction is destabilized by the phosphorylation of C-terminal serines (S788/S795) or threonines (T821/T826) (Rubin et al. 2005). Despite this progress, the structural basis for the pRb phosphorylation-induced release of the E2FTD from the pRb pocket, a key feature of the pRb–E2F cell cycle switch, remained unclear.
Figure 1.
Cdk phosphorylation of discrete sites in pRb causes distinct conformational changes that inhibit pRb's binding to E2F. (A) Phosphorylation of T821/T826 stabilizes an interaction between the pRb C-terminal domain (RbC) and the pocket and may exclude the E2F–DP complex from RbC (Rubin et al. 2005). (B) Phosphorylation of S608 induces the RbPL to interact with the pRb pocket. This intramolecular interaction competitively blocks the binding of the E2FTD (Burke et al. 2012). (C) Phosphorylation of T373 within the RbIDL stabilizes the docking of the RbN against the pocket. This leads to a rotation of the pocket subdomains relative to one another and allosterically inhibits E2FTD binding to the pocket (Burke et al. 2012). (A,C) The LxCxE-binding cleft in the pocket is blocked by RbN–pocket docking or the RbC–pocket interaction (Rubin et al. 2005; Burke et al. 2012).
Previous studies had identified serine or threonine residues within pRb that are critical for the dissociation of the pRb–E2F complex (Knudsen and Wang 1996, 1997; Zarkowska and Mittnacht 1997; Brown et al. 1999; Lents et al. 2006). Cell-based assays had pointed to S608/S612 in the RbPL and T373 as crucial sites for growth arrest or repression of E2F transcription (Knudsen and Wang 1997; Lents et al. 2006; Gorges et al. 2008), but overall, the notion remained that cumulative pRb phosphorylation was necessary to abrogate the pRb–E2F interaction. This was clarified by a previous study by Burke et al. (2010) who used purified proteins in biophysical assays to show that multiple phosphorylation events on pRb (T356/T373, S608/S612, S788/S795, or T821/T826) could independently destabilize the pRb–E2F complex. These results already provided evidence for two distinct phosphorylation-induced conformational changes in pRb that occluded the E2FTD-binding site upon S608/S612 phosphorylation or required the RbN domain following T356/T373 phosphorylation, but the structural basis for these effects was not known.
In the current study, Burke et al. (2012) elucidate the structural features that are induced by the two distinct phosphorylation events in pRb. In order to observe the effect of the S608 phosphorylation, the investigators crystallized the pocket domain with a shortened RbPL and a phosphoserine mimetic S608E that was confirmed to inhibit E2FTD binding (Rb380–787Δ616–642/S608E/S612A/S780A). In the crystal structure, the residues 601–608 of the RbPL interact with the cleft between the pocket subdomains and partially occlude the E2FTD-binding site. Some RbPL–pocket contacts are analogous to E2FTD–pocket side chain interactions, while other contacts are not superimposable with E2FTD. Thus, RbPL phosphorylation at S608 orders the flexible loop so that it forms an intramolecular interaction with the pocket domain (Fig. 1). This interaction partially mimics and directly blocks E2FTD binding to the pocket.
A second structure described by Burke et al. (2012) is the first multidomain structure of pRb. To address the conformational changes that are induced by RbIDL T356/T373 phosphorylation and depend on RbN, the investigators crystallized a large phosphorylated Rb fragment that contains RbN, RbIDL, and the pocket but lacks the unstructured loops within the domains (Rb53–787,Δ245–267,Δ582–642). In this structure, phosphothreonine T373 stabilizes a closed conformation in which the RbN subdomains dock against the pocket subdomains on the opposite face as E2FTD (Fig. 1). This closed Rb conformation is consistent with previous evidence indicating that RbN can directly interact with the pocket (Hassler et al. 2007). Small-angle X-ray scattering suggests that pRb exists in an equilibrium between the open and closed conformation, which is shifted toward the closed state upon RbIDL phosphorylation. The shift to the closed conformation has great impact on the interaction surfaces of the pocket domain. The pocket subdomains are rotated relative to one another in the closed conformation. This opens the E2F-binding site between the subdomains so widely that E2F binding to the pocket is allosterically inhibited. Furthermore, the closed conformation prevents protein binding to the LxCxE-binding site of the pocket, but does not exclude the interaction between the pocket and the phosphorylated C terminus of pRb.
These results suggest that phosphorylation of S608 and T373 induces two distinct types of conformational changes in pRb structure. These changes share a common principle that unstructured pRb loops become more ordered and form intramolecular interactions with the pocket, but the changes inhibit E2FTD–pocket binding by remarkably discrete mechanisms. While the phosphorylated pocket loop competitively blocks the E2FTD binding, RbIDL phosphorylation stabilizes the allosteric shift of the pocket subdomains, which disables their hold on E2FTD. The distinct features of these conformational changes have interesting implications. The competitive inhibition of E2FTD binding explains how the formation of new pRb–E2F complexes can be blocked, and this may be important to fine-tune the levels of free E2F. The allosteric changes, however, allow the dissociation of preformed pRb–E2F complexes. It may be possible to exploit this mechanism therapeutically by stabilizing pRb–E2F complexes with small molecules that prevent RbN from docking to the pocket. Furthermore, the allosteric shift prevents protein binding to the pocket LxCxE-binding cleft, which has been reported to mediate the interactions of pRb to >25 different proteins, including chromatin-modifying proteins (Dick 2007). The loss of these interactions suggests that the allosteric mechanism not only releases the E2FTD from pRb, but also prevents pRb from binding to proteins that have been implicated in the pRb-mediated repression of E2F target genes.
These latest structures still leave questions to be answered. One caveat is that our current picture of pRb structure is still a montage that has been assembled from snapshots taken of different fragments of the pRb protein (Fig. 1). It is not certain how the structural changes caused by the various phosphorylation events impact one another in the context of the full-length protein. The roles played by several phosphorylation sites have yet to be identified, and it is likely that pRb will have additional surprises in store for us. Aside from the changes in phosphorylation, pRb is also methylated and acetylated on specific residues. These modifications occur in specific contexts and also seem likely to affect the activation or inactivation of specific properties of pRb.
Close to 200 different cellular proteins have been reported to physically associate with pRb (Morris and Dyson 2001; Goodrich 2006). This literature has lead to the idea that pRb is a multifunctional protein that interacts with different proteins in different contexts. The notion that individual phosphorylation events regulate specific elements of the protein structure supports the very appealing model that the phosphorylation of discrete sites may determine the sets of proteins that pRb targets and the types of functions that it can perform. Future studies are needed to assign specific Rb phosphorylation events to the gain or loss of interactions with its partners. If this model is correct, then it should be possible to identify natural biological contexts in which the phosphorylation of pRb on a specific site regulates a specific subset of pRb complexes, independent of the bulk phosphorylation events that occur during cell cycle progression. Structural studies may lead the way forward by unraveling the combinations of interaction surfaces that are available in distinct conformations of pRb.
Acknowledgments
We thank Michael Korenjak for helpful discussions on the manuscript. We also apologize to our colleagues whose work was not cited due to space limitations. A.H. and N.D. are supported by NIH grant CA64402 (to N.D.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.195552.112.
References
- Brown VD, Phillips RA, Gallie BL 1999. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol 19: 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich K, Duffy LA, Harlow E 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58: 1097–1105 [DOI] [PubMed] [Google Scholar]
- Burke JR, Deshong AJ, Pelton JG, Rubin SM 2010. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem 285: 16286–16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JR, Hura GL, Rubin SM 2012. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev (this issue). doi: 10.1101/gad.189837.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Scully P, Shew JY, Wang JY, Lee WH 1989. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell 58: 1193–1198 [DOI] [PubMed] [Google Scholar]
- Chinnam M, Goodrich DW 2011. RB1, development, and cancer. Curr Top Dev Biol 94: 129–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang CM, Livingston DM 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58: 1085–1095 [DOI] [PubMed] [Google Scholar]
- Dick FA 2007. Structure–function analysis of the retinoblastoma tumor suppressor protein—is the whole a sum of its parts?. Cell Div 2: 26 doi: 10.1186/1747-1028-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646 [DOI] [PubMed] [Google Scholar]
- Goodrich DW 2006. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 25: 5233–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges LL, Lents NH, Baldassare JJ 2008. The extreme COOH terminus of the retinoblastoma tumor suppressor protein pRb is required for phosphorylation on Thr-373 and activation of E2F. Am J Physiol Cell Physiol 295: C1151–C1160 doi: 10.1152/ajpcell.00300.2008 [DOI] [PubMed] [Google Scholar]
- Hassler M, Singh S, Yue WW, Luczynski M, Lakbir R, Sanchez-Sanchez F, Bader T, Pearl LH, Mittnacht S 2007. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol Cell 28: 371–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY 1996. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem 271: 8313–8320 [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Wang JY 1997. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol 17: 5771–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, Russo AA, Pavletich NP 1998. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391: 859–865 [DOI] [PubMed] [Google Scholar]
- Lee C, Chang JH, Lee HS, Cho Y 2002. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev 16: 3199–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lents NH, Gorges LL, Baldassare JJ 2006. Reverse mutational analysis reveals threonine-373 as a potentially sufficient phosphorylation site for inactivation of the retinoblastoma tumor suppressor protein (pRB). Cell Cycle 5: 1699–1707 [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ 2001. Retinoblastoma protein partners. Adv Cancer Res 82: 1–54 [DOI] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP 2005. Structure of the Rb C-terminal domain bound to E2F1–DP1: A mechanism for phosphorylation-induced E2F release. Cell 123: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Weinberg RA 1995. The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceno C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ 2003. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc Natl Acad Sci 100: 2363–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkowska T, Mittnacht S 1997. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem 272: 12738–12746 [DOI] [PubMed] [Google Scholar]