Melanoma is often considered one of the most aggressive and treatment-resistant human cancers. Aside from early surgical resection, no therapeutic modality has been found to afford a high likelihood of curative outcome. In this review by Fisher and colleagues, the authors discuss recent breakthroughs that range from new understandings of the molecular basis of melanoma all the way to new therapeutic strategies that produce unquestionable clinical benefit.
Keywords: BRAF, MITF pathway, melanoma
Abstract
Melanoma is often considered one of the most aggressive and treatment-resistant human cancers. It is a disease that, due to the presence of melanin pigment, was accurately diagnosed earlier than most other malignancies and that has been subjected to countless therapeutic strategies. Aside from early surgical resection, no therapeutic modality has been found to afford a high likelihood of curative outcome. However, discoveries reported in recent years have revealed a near avalanche of breakthroughs in the melanoma field—breakthroughs that span fundamental understanding of the molecular basis of the disease all the way to new therapeutic strategies that produce unquestionable clinical benefit. These discoveries have been born from the successful fruits of numerous researchers working in many—sometimes-related, although also distinct—biomedical disciplines. Discoveries of frequent mutations involving BRAF(V600E), developmental and oncogenic roles for the microphthalmia-associated transcription factor (MITF) pathway, clinical efficacy of BRAF-targeted small molecules, and emerging mechanisms underlying resistance to targeted therapeutics represent just a sample of the findings that have created a striking inflection in the quest for clinically meaningful progress in the melanoma field.
The disease
Melanomas can arise within any anatomic territory occupied by melanocytes. Although cutaneous melanoma, which develops from epidermal melanocytes of the skin, represents the most common site of origination, noncutaneous melanocytes such as those lining the choroidal layer of the eye, respiratory, gastrointestinal, and genitourinary mucosal surfaces, or the meninges do occasionally undergo malignant transformation, albeit at a low frequency.
Clinical morphologists have traditionally divided the cutaneous disease into several subgroups, including superficial spreading melanoma, nodular melanoma, acral lentiginous melanoma, and lentigo maligna melanoma (Fig. 1), and other uncommon variants such as desmoplastic melanoma and nevoid melanoma. Histological patterns have been well described, and microscopic features that correlate with clinical subgroups have been thoroughly codified. Perhaps one of the most unusual and tested aspects of melanoma physiology is the mathematical relationship between tumor thickness (i.e., diameter of tumor sphere) and outcome (Balch et al. 2009). Other features, including mitotic rate and ulceration, also play significant roles in determining prognosis. Despite decades of study, an understanding of the melanoma subsets that are destined to be lethal remains incomplete.
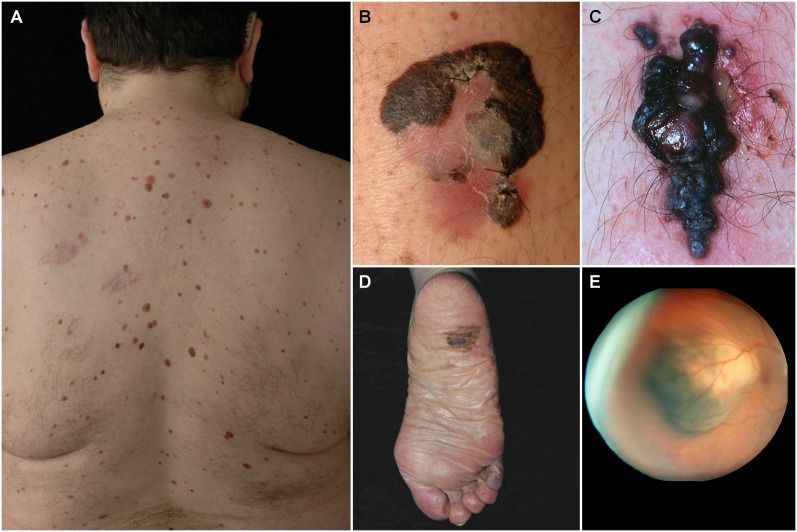
Figure 1.
Clinical disease. (A) A patient with multiple clinically atypical moles. Since ∼80% of all acquired nevi harbor BRAF(V600E) mutations, hundreds of BRAF-activating events occur in such nevi-prone individuals without progression to melanoma. It is thought that melanocyte growth arrest results from OIS. Common forms of cutaneouos melanoma include superficial spreading melanoma (B) and nodular melanoma (C). Both of these subtypes are associated with BRAF or NRAS mutations. (D) Acral lentiginous melanoma is the most common subtype among darker-skinned individuals and is more often associated with KIT aberrations. (E) Ocular melanoma is rarer than cutaneous melanoma and is not associated with BRAF, NRAS, or KIT changes, but rather with GNAQ or GNA11 alterations (picture courtesy of Dr. Ivana Kim, Massachusetts Eye and Ear Infirmary).
Despite recent therapeutic advances in management of advanced melanoma, several crucial biological questions remain, including: (1) What is the relationship between environmental exposures and melanoma risk? (2) Do biomarkers exist that may predict clinical behavior and thus guide therapies? (3) Which genomic alterations drive invasion, metastasis, and drug resistance? (4) Which molecular lesions underlie tumor maintenance? (5) Which aberrant pathways and targets are amenable to either preventative or therapeutic intervention?
Genetic loci and variants that confer melanoma risk
A family history of melanoma occurs in 10% of melanoma patients and confers an approximately twofold increase in melanoma risk (Gandini et al. 2005). One can argue that melanoma is fundamentally a genetic disease, since the range of heritable risk factors—from physical characteristics such as light complexion, an inability to tan, red hair, and blue eyes to the familial atypical mole/melanoma (FAMM) syndrome—are all determined by distinct genetic elements. Hereditary melanoma itself is often associated with (1) multiple cases of melanoma in several generations on one side of the family, (2) multiple primary melanomas in a given individual, and (3) early onset of disease. In this section, we review recent discoveries in melanoma predisposition and survey known risk loci, especially those uncovered through genome-wide association studies (GWAS).
High-risk melanoma loci
To date, the weight of evidence suggests that the retinoblastoma (RB) pathway, which serves to regulate the G1/S checkpoint, is uniquely vulnerable in melanoma susceptibility.
Cyclin-dependent kinase N2A (CDKN2A)
It has been recognized for decades that there are families with an increased occurrence of both melanoma and clinically atypical moles (i.e., dysplastic nevi) (Fig. 1A). Through the systematic collection of these kindreds worldwide, linkage analysis on melanoma families led to putative loci on chromosomes 1p36 (Bale et al. 1989) and 9p21 (Cannon-Albright et al. 1992). Within the 9p21 region, the p16 (now CDKN2A) gene quickly became an attractive candidate for the melanoma locus, since p16 was shown to be a potent cell cycle inhibitor through a direct negative interaction with CDK4 (Serrano et al. 1993). Hussussian et al. (1994) then demonstrated deleterious germline mutations in CDKN2A among a subset of melanoma-prone families that exhibited linkage to chromosome 9p21 markers, thereby establishing the first high-risk susceptibility locus in melanoma. Around this time, several groups also reported homozygous deletions and deleterious mutations of CDKN2A in a variety of cancer cell lines (Kamb et al. 1994; Nobori et al. 1994). Thus, within a span of a few years, CDKN2A catapulted into the center of cancer biology as a critical target of inactivation at both the germline and somatic levels.
The CDKN2A locus is composed of four exons and encodes for two distinct proteins through alternative splicing: p16INK4a and p14ARF (Fig. 2; for review, see Chin 2003); interestingly, both proteins are potent tumor suppressors with distinct but equally crucial roles in cell cycle and apoptosis regulation. p16INK4a binds to and inhibits CDK4/6, thereby preventing CDK4/6 from phosphorylating the RB protein (Koh et al. 1995). Since hyperphosphorylation of RB triggers the release of E2F1, a transcriptional inducer of S-phase genes, loss of p16INK4a encourages G1–S transition and re-entry into the cell cycle. On the other hand, p14ARF binds to human double minute-2 (HDM2) protein at its N terminus and promotes the rapid degradation of HDM2. Since HDM2 in turn ubiquitinates and condemns p53 to destruction, the net effect of p14ARF loss is a destabilization of p53 (Kamijo et al. 1998; Stott et al. 1998; Zhang et al. 1998). Thus, in true genetic economy, CDKN2A lesions eliminate both the RB and p53 pathways through loss of p16INK4a and p14ARF, respectively (Lin et al. 2008).
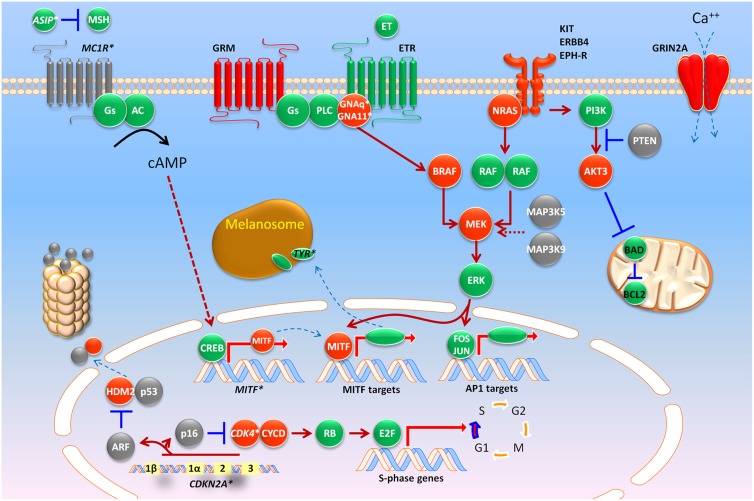
Figure 2.
A molecular map of melanoma. Heritable loci with risk alleles or SNPs are shown in italics with asterisks (e.g., CDKN2A*). Red and gray colors indicate somatic alterations that result in gain of function (i.e., oncogenes such as BRAF) or loss of function (i.e., tumor suppressor genes such as PTEN), respectively. See the text for abbreviations.
Most germline mutations that confer melanoma risk occur in exons 1α and 2, which suggests that p16INK4a is the preferentially targeted, and functionally dominant, component of CDKN2A. For p14ARF, rare deletions and a 16-base-pair (bp) insertion at exon 1β with a mutational hot spot at the exon 1β splice site (Hewitt et al. 2002; Harland et al. 2005b) have also been reported. The presence of exon 1β-specific alterations suggest that p14ARF is a bona fide melanoma susceptibility gene independent from p16. Even rarer deep intronic mutations of CDKN2A have also been described, although these account for very few cases worldwide (Harland et al. 2001, 2005a).
Several studies now have estimated the risk of developing cutaneous melanoma among CDKN2A mutation carriers. For familial cases, the overall penetrance has been calculated to be 30% by age 50, and 67% by age 80, although this risk is higher in sunnier climes. The risk of melanoma by age 50 reaches 13% in Europe, 50% in the United States, and 32% in Australia; by age 80, it is 58% in Europe, 76% in the United States, and 91% in Australia (Bishop et al. 2002). Among sporadic CDKN2A carriers, the risk appears lower: 14%, 24%, and 28% by ages 50, 70, and 80, respectively (Begg et al. 2005). Other coinherited modifiers (such as MC1R variants) (Demenais et al. 2010) may enhance risk, although the familial ascertainment itself may introduce bias and produce a higher calculated penetrance. Absolute melanoma risk among CDKN2A mutation carriers is clearly modulated by pedigree structure and environmental input.
Several animal models have provided biological proof of CDKN2A's involvement in carcinogenesis, particularly in melanoma. These models demonstrated formation of fibrosarcomas and lymphomas with high frequency in mice containing a targeted disruption of Cdkn2a (Serrano et al. 1996). Evidence for involvement of this locus in melanoma has been shown using Cdkn2a−/− mice with alleles containing activating mutations in the oncogenes HRAS (Chin et al. 1997) and NRAS (VanBrocklin et al. 2010). These mice were readily induced to form melanoma-like cutaneous tumors. Within this family of oncogenes, however, observations have revealed NRAS as the most frequently mutated family member in human tumors (Hocker and Tsao 2007). Further proof of Cdkn2a involvement is shown in a gene exposure model through ultraviolet (UV) irradiation of Cdkn2a−/− mice. In this model, in the absence of DNA excision repair (e.g., inactivation or loss of Xpc), UV irradiation of mice led to rapid formation of cutaneous melanomas. It is interesting to note that the melanomas induced in this background harbored activating mutations in KRAS (Yang et al. 2007). This model illustrates the importance of fidelity in DNA repair and that any UV-induced damage to the DNA without this repair machinery in place may result in activation of the RAS pathway, thereby leading to melanomagenesis.
CDK4
Recurrent mutations of CDK4, which lead to cell cycle progression, have been reported in the germline of melanoma-prone families and in tumors (Wolfel et al. 1995; Zuo et al. 1996; Soufir et al. 1998; Tsao et al. 1998a; Molven et al. 2005). These mutations were observed to occur at a conserved arginine residue (Arg 24) that abolished regulatory interactions with p16INK4a. Carcinogen treatment of a knock-in mouse model expressing the R24C mutation in CDK4 had a marked increase in potential to acquire melanomas after exposure. In concordance with these observations, some human melanomas have been known to amplify and/or mutate CDK4 (Muthusamy et al. 2006).
RB1
Hereditary retinoblastoma has been linked to inactivated copies of RB1 in the germline; however, such carriers have a fourfold to 80-fold elevated risk to develop melanoma (Draper et al. 1986; Sanders et al. 1989; Eng et al. 1993; Fletcher et al. 2004). Development of sarcomas in patients with loss of RB1 function decreases proportionately with decreased use of radiotherapy. However, the incidence of melanomas does not track with radiotherapy use, which is likely due to loss of heterozygosity (LOH) for the RB1 allele. This demonstrates the intricate linkages between the CDKN2A/CDK/RB pathways of tumor suppression in humans.
Low-to-moderate-risk melanoma loci
In contrast to high-risk alleles, low-to-moderate disease variants often lack familial clustering and dictate cancer-susceptible traits, such as skin color, rather than cancer itself. Although some of these variants are quite common in the general population, some risk-conferring alleles have minor allele frequencies (MAFs) below 1%. These low-to-moderate-risk loci are outlined in the following sections.
Determinants of pigmentation and melanoma risk
MC1R
A host of epidemiological studies long established a direct positive connection between light skin color and melanoma risk (for review, see Gandini et al. 2005). Among the plethora of genes known to regulate constitutive and facultative pigmentation, the melanocortin-1 receptor (MC1R) has emerged as one of the leading moderate-risk loci for melanoma susceptibility (for review, see Miller and Tsao 2010).
MC1R is a seven-transmembrane G-protein-coupled receptor that activates adenylate cyclase (Fig. 2) in response to α-MSH binding (Garcia-Borron et al. 2005). Subsequent increases in cAMP up-regulates the microphthalmia-associated transcription factor (MITF), which consequently induces the transcription of pigment synthetic genes and the production of eumelanin—the major source of UV attenuation in darkly pigmented skin. Germline variants of MC1R that disrupt this signaling cascade are present in ∼80% of individuals with red hair color (RHC), <20% of people with brown or black hair, and <4% of persons with a robust tanning response (Valverde et al. 1995). Association studies have found that the MC1R variants p.D84E, p.R151C, p.R160W, p.D294H, p.R142H, and p.I155T are strongly associated with the RHC phenotype (i.e., “R” variants), while the p.V60L, p.V92M, and p.R163Q variants seem to have weaker association with the RHC phenotype (i.e., “r” alleles) (Raimondi et al. 2008). A recent meta-analysis found that all but the p.V60L and p.V92M variants were associated with melanoma risk with odds ratios (ORs) ranging from 1.42-fold for p.R163Q to 2.45-fold for p.I155T (Raimondi et al. 2008). Interestingly, some variants are associated with melanoma risk but not pigmentary phenotypes, suggesting that MC1R may harbor subtle cancer effects that go beyond hair and skin color (Kennedy et al. 2001).
MITF
As discussed above, engagement of α-MSH and MC1R leads to increased cAMP and the induction of MITF by the cAMP response element-binding protein (CREB) (Fig. 2). Although MITF's role in melanoma risk was thought to be purely responsive to MSH signaling, it now appears that variants in MITF itself are also instructive of risk. Through a whole-genome sequencing effort, a novel germline MITF variant (p.E318K) was discovered in a patient with melanoma. Among families harboring this variant, linkage analysis generated a log-of-odds (LOD) score of 2.7, implicating MITF(E318K) as a potential risk variant. In a large Australian/United Kingdom case control series of 3940 melanoma patients and 4036 controls, the p.E318K variant was found to confer a 2.19-fold risk, which is the range calculated for MC1R variants, although the prevalence of the MITF(E318K) alteration is significantly lower (<1%) in the general white population compared with MC1R RHC variants. Functional analysis (see “MITF—Melanocyte Master Regulator” ) showed that the missense mutation at codon 318 abrogated a conserved SUMOylation site, thereby altering the transcription of several MITF targets, including hypoxia-inducible factor 1a (HIF1a), a regulatory event that appears to also confer renal cell carcinoma risk among MITF(E318K) carriers (Bertolotto et al. 2011; Yokoyama et al. 2011).
Lessons from GWAS
Multiple GWAS have also yielded several risk-associated single-nucleotide polymorphisms (SNPs) across the genome (for review, see Chatzinasiou et al. 2011). The most substantiated loci include MC1R (per allele OR = 1.83; 95% confidence interval [CI] = 1.44–2.32), ASIP (per allele OR = 1.35; 95% CI = 1.08–1.68), TYR (per allele OR = 1.34; 95% CI = 1.14–1.58), IRF4 (per allele OR = 0.80; 95% CI = 0.67–0.95), and SLC45A2 (per allele OR = 0.72; 95% CI = 0.44–1.18). Additional loci that have recently been found include loci at CASP8 (OR = 1.11; 95% CI = 1.06–1.18), CCND1 (OR = 1.07; 95% CI = 1.01–1.13), ATM (OR = 0.87; 95% CI = 0.81–0.94), MX2 (OR = 0.91; 95% CI = 0.86–0.96), and chromosome 1q21.3 (OR = 0.89; 95% CI = 0.85–0.95), including a region containing the ARNT and SETDB1 genes (Chatzinasiou et al. 2011; Macgregor et al. 2011). It is clear from careful correlative analysis that some of these SNPs define overlapping phenotypes such as sun sensitivity, propensity for nevus formation, and skin cancer risk, including melanoma and nonmelanoma skin cancers.
In summary, a harvest of novel low–moderate melanoma risk loci has now been discovered through candidate and systematic genome-wide approaches. At this point, it is unlikely that there are many undisclosed recurrent high-risk loci (e.g., CDKN2A). Rather, mutations in multiple high-penetrant genes or the presence of several moderate-risk alleles in a single kindred may explain the balance of strong melanoma pedigrees. On a population level, most of the attributable risk for melanoma may in fact result from an untoward conspiracy of moderate-risk SNPs.
Biological drivers and therapeutic targets in melanoma
Receptor tyrosine kinase (RTK) activation
Over the years, a considerable amount of evidence has accrued in support of the notion that RTKs may contribute importantly to melanoma biology. Whereas earlier studies focused primarily on receptor overexpression, recent genomic studies suggest that genetic dysregulation may also play a role in some cases. These advances raise the possibility that small-molecule therapeutics targeting certain RTKs may prove clinically useful against specific subsets of melanoma. An integrated diagram of signaling molecules and biological drivers in melanoma is shown in Figure 2.
KIT
The c-KIT gene encodes the RTK for stem cell factor (SCF). Although early reports described a sequential loss of c-KIT expression from benign to primary and metastatic melanomas (Montone et al. 1997; Shen et al. 2003; Zhu and Fitzpatrick 2006), reconstitution of the RTK in metastatic melanoma cells apparently restored sensitivity to SCF-induced apoptosis in vitro (S Huang et al. 1996). The role of c-KIT in melanoma biology had been uncertain until more recent genomic screens.
A survey of copy number imbalances in primary melanomas found the KIT locus (chromosome 4q11) to be amplified and/or mutated in 39% of mucosal, 36% of acral, and 28% of melanomas on chronically sun-damaged skin (CSD), respectively (Curtin et al. 2006). However, subsequent large series from Australia (Handolias et al. 2010) and China (Kong et al. 2011) revealed much lower rates of aberrations. It is notable that many of the activating KIT mutations described in melanomas overlap with those reported for gastrointestinal stromal tumors (GISTs) (Curtin et al. 2006; Antonescu et al. 2007; Rivera et al. 2008; Smalley et al. 2008; Ashida et al. 2009), suggesting that a similar trophic signaling pathway is shared between the two tumor types.
Given the success of imatinib mesylate in KIT-mutated GISTs, a pharmacological framework for targeting KIT-mutated melanomas became evident. Several early reports of dramatic responses using imatinib (Hodi et al. 2008; Lutzky et al. 2008) led to the development of two phase II (open label) trials of imatinib for KIT-mutated melanomas. In one trial (Carvajal et al. 2011), 28 patients (N = 25 evaluable) with metastatic KIT-altered melanomas were treated, and two complete responses (CRs; 94 and 95 wk), two durable partial responses (PRs; 53 and 89 wk), and two transient PRs (12 and 18 wk) were reported. The median progression-free survival (PFS) and overall survival (OS) were 12 wk and 46.3 wk, respectively. In terms of the genetics, 23.4% of the cases harbored KIT mutations and/or amplifications, with the most significant responses occurring in patients with c-KIT(K642E) or c-KIT(L576P) mutations and those with tumors that enriched for the presence of a KIT mutation (i.e., mutant:allele ratio >1). In the second trial (Guo et al. 2011), 43 patients with metastatic melanomas harboring KIT aberrations (i.e., exons 9, 11, 13, 17, and 18 mutations and/or copy number gains) were treated with imatinib. Partial responses were observed in 10 patients (23.3%), with disease stabilization observed in another 13 patients (30.2%), and progressive disease observed in an additional 20 patients (46.5%). The median PFS and OS were 3.5 mo and 14.0 mo, respectively, in this second trial. Genotype–phenotype correlations did not reveal any evident relationships between response and KIT mutations. It is clear that imatinib exhibited only modest effects, although other RTK inhibitors (e.g., sunitinib, nilotinib, and dasatanib) are being tested with hopes of being more efficacious.
Epidermal growth factor receptor (EGFR)
The gene encoding the EGFR is located on chromosome 7, which has long been known to undergo frequent polysomy in advanced melanoma (Koprowski et al. 1985; Bastian et al. 1998; Udart et al. 2001). Whereas enforced activation of EGFR may promote metastatic progression in cell line studies (de Wit et al. 1992; TS Huang et al. 1996), neither activating EGFR mutations nor focal EGFR amplifications have been observed in melanoma. Thus, most studies linking EGFR activation to melanoma biology have relied on protein expression or activation studies (Topcu-Yilmaz et al. 2010; Tworkoski et al. 2011). An assortment of in vitro studies has suggested that ectopic EGFR expression may enhance melanoma cell growth (Diaz et al. 2007; Ueno et al. 2008) and that pharmacological blockade of EGFR using small-molecule inhibitors or monoclonal antibodies may suppress growth in melanoma cell lines (Boone et al. 2011), either alone or in combination with other targeted agents (Ivanov and Hei 2005; Schicher et al. 2009). Also, preliminary results raise the possibility that EGFR activation may contribute to uveal melanoma pathogenesis in some instances (Wu et al. 2012). However, some studies found that the gefitinib GI50 values reached the 2–3 μM range and that melanoma cells continued to grow even at 10 μM gefitinib (Djerf et al. 2009). Together with the paucity of driver EGFR mutations in melanoma, such results have dampened preclinical enthusiasm for EGFR as a target in this malignancy. In fact, one recent study found that EGFR expression may modestly suppress melanoma growth in a B16 model (Diaz et al. 2009).
The limited clinical experience with EGFR inhibitors has been similarly disappointing. A phase II trial of the EGFR inhibitor gefitinib in 46 metastatic melanoma patients resulted in a median PFS of only 1.4 mo and a median OS of only 9.7 mo. During treatment, there were no reproducible changes in tumoral p-ERK1/2, p-AKT, and PAK1 and serum vascular endothelial growth factor (VEGF) and IL-8 levels (Patel et al. 2011). Thus, despite the aforementioned experimental evidence of EGFR's involvement in melanoma progression, there are scant clinical data to support single-agent anti-EGFR therapy.
Despite the relatively poor support for either EGFR dependency or monotherapy in melanoma, recent results raise the possibility that the combination of EGFR and RAF inhibitors might prove beneficial in some cases of de novo resistance to RAF inhibition. Bernards and colleagues (Prahallad et al. 2012) performed a synthetic-lethal RNAi screen in the setting of RAF inhibition in BRAF mutant colon cancer cells, which show limited sensitivity to RAF inhibitor monotherapy. They found that EGFR knockdown and pharmacological blockade were synergistic with vemurafenib in the suppression of BRAF mutant cells that were intrinsically resistant to vemurafenib alone. In this study, the addition of EGFR inhibition appeared to interdict a feedback up-regulation engendered by vemurafenib treatment. Although these results were seen in BRAF mutant colorectal cancer, they raise the intriguing possibility that at least some BRAF mutant melanomas that exhibit de novo resistance to RAF inhibition may be candidates for concomitant EGFR inhibition in future clinical trials.
MET
The RTK c-MET is normally activated by binding of its ligand, hepatocyte growth factor/scatter factor (HGF). Autocrine activation of HGF-MET has been described in melanoma progression (for review, see Vande Woude et al. 1997; Li et al. 2001). Although increased c-MET expression has been observed in metastatic melanoma (Natali et al. 1993), the MET locus also resides on chromosome 7, which commonly undergoes polysomy in this setting (as described above). As with EGFR, MET amplifications and activating point mutations thus far have not been described in melanoma. Thus, the evidence that MET activation constitutes a bona fide melanoma dependency remains scant. However, several lines of experimental evidence suggest that MET signaling may enhance melanoma growth and metastasis. For example, HGF exposure may promote increased melanocytic cell mobility (Damm et al. 2010), and HIF-1α may promote MET-dependent invasion and vasculogenic mimicry in melanoma cells (Comito et al. 2011). Thus, MET activation may serve to augment rather than “drive” melanomagenesis and progression per se.
A small-molecule inhibitor of c-MET (SU11274) is now available for potential clinical use. In one preclinical analysis, melanoma lines that exhibited constitutive c-MET stimulation in the absence of MET alterations underwent decreased proliferation and an increase in apoptosis when exposed to SU11274. In a xenograft model, the compound also demonstrated significant anti-tumor activity (Kenessey et al. 2010). Currently, the role of anti-c-MET treatment in melanoma remains a theoretical promise at best.
Ephrin receptors
Ephrin receptors constitute the largest family of RTKs in the kinome. Physiologically, ephrin ligands are membrane-bound, so forward (through Eph receptors) and reverse (through ephrin molecules) signaling can occur upon cell–cell interaction (Genander and Frisen 2010). One specific Eph member, EphA2, has been directly implicated in tumor formation. Published studies report an increased expression of EphA2 in multiple cancer types, including melanoma (Easty et al. 1999; Seftor et al. 2002; Hendrix et al. 2003; Kinch and Carles-Kinch 2003; Miyazaki et al. 2003; Herath et al. 2006; Genander and Frisen 2010). Furthermore, the EPHA2 locus maps to chromosomal region 1p36, which is a region frequently altered in melanoma (Sulman et al. 1997). Mechanistically, EphA2 appears to participate in cross-talk between other cancer signaling circuits, such as the RAS–phosphatidylinositol 3-kinase (PI3K)–AKT and RAS–MAPK pathways (Menges and McCance 2008). Recent investigations revealed that some melanomas are, in fact, “addicted” to EphA2 (Udayakumar et al. 2011). shRNA-mediated silencing of EphA2 led to a rapid apoptotic response along with tumor suppression in a xenograft model, while ectopic expression of EphA2 led to enhanced colony formation and migration. Interestingly, EphA2 appears to be essential for UV-mediated apoptosis, and acute introduction of EphA2 into normal and immortalized cells also elicits an apoptotic response (Zhang et al. 2008). These findings suggest that Eph receptors play a complex role in melanocytes and melanomas. It is possible that early death purges the genetically susceptible population, thereby leaving more aggressive tumor cells that come to depend on EphA2 signaling for sustenance; this is compatible with an “overdose/addiction” model whereby oncogene stress, which is a common physiological response, serves as a selection pressure for tumorigenic recruits.
Although mutagenic activation of EPHA2 is not commonly observed (Udayakumar et al. 2011), other Eph receptors have been shown to be mutated in melanoma. In particular, multiple lesions have been observed in EPHA6, EPHA10, EPHB1, EPHB2, and EPHB6 (Prickett et al. 2009). Given the various molecular systems that are impacted by these receptors, functional classification of the Eph receptors into oncogenes or tumor suppressors is not yet possible without further study.
ERBB4
In a sweeping analysis of the tyrosine kinome, Prickett et al. (2009) screened the coding exons of 86 protein tyrosine kinases and identified 99 nonsynonymous somatic mutations. Most prominent among these was ERBB4, which was mutated in 19% of melanoma cases. The missense mutations were oncogenic in several in vitro assays, such as NIH-3T3 transformation and soft agar growth; a recent genotyping effort (Dutton-Regester et al. 2012) points to a possible low-prevalence ERBB4(E452K) hot spot. Inhibition of ERBB4 by lapatinib also led to apoptosis in ERBB4-mutated cells. Given the broad-based nature of this kinome screen, it is unlikely that common forms of melanoma harbor high rates of recurrent activating mutations in any single RTK. It is possible that melanoma cells have bypassed the need for more upstream signaling, given the high rate of oncogenic changes in downstream molecules such as NRAS and BRAF (see below).
Glutamate receptor dysfunction
GRMs
Although RTKs represent attractive biological targets for oncogenic activation, other surface receptors may promote melanomagenesis in unsuspecting ways. It is intriguing that several neurotransmitter receptors have now been shown to participate in melanoma pathogenesis. Since melanocytes are derived embryologically from neural crest cells, it is possible that a shared mechanistic circuitry between neural and melanocyte descendants permits melanoma cells privileged access to neurophysiological molecules. The earliest hints that glutamate receptors contribute to melanoma formation were derived from mouse studies (Pollock et al. 2003a) in which aberrant Grm1 expression was associated with a melanoma-prone phenotype. GRM1 is expressed in human melanoma specimens but not benign melanocytic nevi. This surface molecule is a member of the metabotropic glutamate receptor family, which is comprised of G-protein-coupled receptors (GPCRs) that activate phospholipase C upon ligand binding. More recently, Choi et al. (2011) showed that mice in which metabotropic glutamate receptor 5 (mGluR5) expression was driven by the Trp1 promoter also developed murine melanomas with high penetrance. Furthermore, mGluR5 expression could be documented in human melanomas and may provide an oncogenic signal through ERK (Choi et al. 2011).
The role of metabotropic glutamate receptors remained circumstantial until mutations were uncovered in another metabotropic glutamate receptor, GRM3, during a GPCR family-wide screen of human melanoma specimens (Prickett et al. 2011). Four of these mutations were subjected to functional analysis (p.Gly561Glu, p.Ser610Leu, p.Glu767Lys, and p.Glu870Lys) and found to stimulate MEK1/2 in the presence of agonist and melanoma migration even in the absence of agonists. Melanoma cells with GRM3 variants were also more sensitive to MEK inhibition by AZD6244. On the surface, activated GRM3 appears to be an accessory to MAPK signaling in melanomas. However, since BRAF or NRAS is often mutated in melanoma cells (see below), the precise contribution of GRM3 to this pathway is unclear. It is known that even among BRAF-mutated melanomas, there is a range of primary sensitivity to BRAF and MEK inhibitors (Flaherty et al. 2010; Chapman et al. 2011). Thus, accessory MAPK signal flux through proteins such as GRM3 may play a compensatory role.
GRIN2A
Through an unbiased exome-wide sequencing effort, mutations in GRIN2A have also been discovered (Wei et al. 2011). Unlike GRM3, GRIN2A is an ionotropic glutamate-gated ion channel that binds N-methyl-D-aspartate (NMDA). This ligand-gated channel is permeable to cations, including Ca2+. Mutations in GRIN2A was found in 34 out of 135 melanoma samples (25.2%). Unlike GRM3, however, there were multiple missense and several nonsense mutations scattered along GRIN2A, suggesting that this gene is not a canonical oncogene in melanoma. Functional validation of GRIN2A as an oncogene has yet to be performed.
Small G proteins, including RAS
The RAS family of small G proteins serves to transduce signals triggered by extracellular growth factors (Ji et al. 2012a). Unlike other solid tumors, activating RAS mutations occur in a relatively small fraction of melanomas (∼10%–15%), with a higher frequency noted in amelanotic nodular melanoma subtypes (for review, see Chin et al. 1998). Among RAS genes, melanoma mutations are most common in NRAS, which is also thought to be mutated in the majority of congenital nevi (Papp et al. 1999) but rarely in dysplastic nevi (Albino et al. 1989; Jafari et al. 1995; Papp et al. 1999). HRAS mutation has been associated with Spitz nevi, based on both genomic amplifications and mutations (Bastian et al. 2000). KRAS and HRAS mutations have both been reported in ∼2% of melanomas (http://www.sanger.ac.uk/genetics/CGP/cosmic).
Interestingly, differences between HRAS and NRAS have also been noted in relation to the consequences of their transgenic targeting to melanocytes. Whereas activated HRAS together with loss-of-function mutations in Cdkn2a and/or Trp53 produce nonmetastatic melanomas in mice (Chin et al. 1997; Bardeesy et al. 2001; Sharpless et al. 2003), activated NRAS together with Cdkn2a deficiency produce melanomas with major metastatic propensity to both lymph nodes and distant sites (Ackermann et al. 2005).
Since oncogenic RAS proteins were among the first oncogenes described in humans, potent pharmacological inhibitors of RAS proteins have been a source of aggressive development. Farnesyl transferase inhibitors (FTIs; e.g., R115777/tibifarnib) were first deployed as selective RAS inhibitors, given the drug's ability to interfere with the requisite lipid modification of RAS (James et al. 1993); despite initial enthusiasm, FTIs have not fared well in clinical trials (Caponigro et al. 2003). One hypothesis for this failure is that FTIs impair other farnesylated proteins, which then lead to dose-limiting toxicities. Alternatively, RAS proteins may use geranylgeranyltransferases, thereby bypassing the block imposed by FTIs (Whyte et al. 1997). More recently, S-trans, trans-farnesylthiosalicylic acid (FTS) was developed to mimic the C-terminal farnesylcysteine (Weisz et al. 1999), thereby competing with the active, GTP-bound forms of RAS for specific binding sites on the cellular membrane (Aharonson et al. 1998). FTS appears to be effective in inhibiting melanoma growth both in vitro and in animal models (Jansen et al. 1999; Smalley and Eisen 2002). Clinical studies in pancreatic cancer also showed some possible survival benefits (Johnson et al. 2009; Laheru et al. 2009), although the efficacy of FTS as a single agent in melanoma awaits clinical testing.
GNAQ
The story behind GNAQ's involvement in melanoma unfolded through an exchange between developmental biology and cancer genetics. In a forward genetic screen, Barsh's laboratory (Van Raamsdonk et al. 2004) identified hypermorphic mutations in GNAQ and GNA11 as causative of diffuse hyperpigmentation and dermal melanocytosis in mice. Among comparable human lesions, blue nevi represent a benign proliferation of dermal melanocytes. Given the phenotypical overlap, analyses of GNAQ and GNA11 were performed in a collection of benign and malignant melanocytic tumors, and GNAQ mutations were found in 83% of blue nevi (N = 29), 50% of “malignant blue nevi” (N = 2), and 46% of uveal melanomas (N = 48) (Van Raamsdonk et al. 2009). Like other oncogenes, a single missense change (p.Q209L) accounts for all identified GNAQ mutations. This GNAQ variant fully activates the MAPK pathway and is oncogenic in both in vitro and in vivo assays. In a follow-up study (Van Raamsdonk et al. 2010), somatic mutations in exon 5 (affecting Q209) and exon 4 (affecting R183) of both GNA11 and GNAQ were seen in a mutually exclusive pattern. GNA11 mutations are present in 7% of blue nevi, 32% of primary uveal melanomas, and 57% of uveal melanoma metastases, while GNAQ alterations were present in 55% of blue nevi, 45% of uveal melanomas, and 22% of uveal melanoma metastases. Both GNAQ and GNA11 mutations activate the MAPK pathway. The epistatic relationship between GNAQ and GNA11 implies that both fulfill overlapping functions in melanocytes or that the presence of both mutations creates a synthetic-lethal condition. There is also early evidence that GNA11 alterations may be more predictive of metastases and thus may represent a potentially crucial therapeutic target (Van Raamsdonk et al. 2010).
Other members of the heterotrimeric G-protein family, which includes GNAQ and GNA11, have also been screened in unselected metastatic melanomas (Cardenas-Navia et al. 2010). No other recurrent alterations were detected, although the overall nonsynonymous somatic mutation rate was 17.5%.
BRAF activation—the heart of melanoma oncogenesis
The discovery of oncogenic BRAF mutations in melanoma (Davies et al. 2002) stands as one of the most powerful affirmations of the transformative potential of systematic cancer genome characterization. In hindsight, it is all the more remarkable that BRAF mutations were discovered by Stratton and colleagues (Davies et al. 2002) using a discovery set of only 15 tumor/normal pairs—only one of which was a melanoma sample! Since that seminal discovery, BRAF mutations—most commonly a valine-to-glutamic acid substitution at codon 600—have been observed in ∼50% of melanomas (Maldonado et al. 2003; Pollock et al. 2003b; Uribe et al. 2003; Daniotti et al. 2004; Kumar et al. 2004; Shinozaki et al. 2004; Libra et al. 2005) and to a lesser extent in other cancers (Ciampi and Nikiforov 2005; Young et al. 2005). BRAF mutations also occur at high frequencies (>80%) in melanocytic nevi (Pollock et al. 2003b; Yazdi et al. 2003; Kumar et al. 2004; Saldanha et al. 2004), suggesting that these somatic alterations occur early in melanomagenesis. Interestingly, individuals with germline BRAF mutations develop cardio–facio–cutaneous syndrome but do not exhibit increased cancer risk (Niihori et al. 2006; Rodriguez-Viciana et al. 2006)—nor do they harbor the V600E mutation that is so prevalent in melanoma, colorectal cancer, and thyroid cancer.
Although it is tempting to speculate that the BRAF(V600E) mutation is induced by UV damage, the T → A transversion that converts the valine to glutamic acid at amino acid 600 (V600E) is not part of the “classic” UV-induced mutational signature (Daya-Grosjean et al. 1995). Nonetheless, a durable epidemiological relationship between BRAF mutations and sun exposure has been noted. In particular, BRAF mutations are much more common in melanomas arising on intermittent sun-exposed (solar elastosis of arms, trunk, etc.) than in acral melanomas—which arise on less sun-exposed glabrous skin—and mucosal melanomas (e.g., gastrointestinal, vaginal origin, etc.) (Maldonado et al. 2003; Edwards et al. 2004; Curtin et al. 2005; Bauer et al. 2011). Strikingly, BRAF mutations are absent in uveal melanoma (Cohen et al. 2003; Cruz et al. 2003; Edmunds et al. 2003; Rimoldi et al. 2003; Weber et al. 2003). Conceivably, the substitution that undergirds the BRAF(V600E) mutation may reflect a secondary effect of UV damage, such as the generation of reactive oxygen species. Toward this end, recent results suggest that melanocytic cells may be deficient in repair of oxidative DNA damage (Wang et al. 2010). Alternatively, this event may arise as a result of “nonclassic” DNA lesions induced by UV (for review, see Besaratinia and Pfeifer 2008).
Given that melanocytic nevi rarely progress into melanoma, it stands to reason that BRAF(V600E)-induced checkpoint mechanisms may produce a senescence-like state in the absence of additional genetic or molecular events that promote tumorigenesis. Toward this end, congenital nevi stain positively for senescence-associated acidic β-galactosidase (SA-β-Gal) (Michaloglou et al. 2005), and BRAF(V600E) expression in primary human melanocytes induces cell cycle arrest. Thus, oncogene-induced senescence (OIS) appears to constrain progression of premalignant melanocytic lesions (Sharpless and DePinho 2005). Senescent melanocytes exhibit a mosaic p16-staining pattern, suggesting that melanocytic senescence is not invariably dependent on p16 up-regulation (Michaloglou et al. 2005).
While BRAF(V600E) mutation proved insufficient to transform human melanocytes by itself, multiple lines of evidence showed that dysregulated MAPK activation was necessary for melanoma cell viability in this setting. Suppression of oncogenic BRAF by RNAi-mediated knockdown resulted in markedly reduced cell growth, diminished ERK phosphorylation, and induction of apoptosis in some instances (Hingorani et al. 2003; Karasarides et al. 2004; Wellbrock et al. 2004). BRAF knockdown also reduced tumor formation in murine xenograft models (Hoeflich et al. 2006). Furthermore, selective small-molecule RAF inhibitors potently suppressed the growth of BRAF mutant melanoma cell lines but had little effect on melanoma cells that lacked these mutations (Joseph et al. 2010). Thus, the presence of BRAF mutations conferred a stringent tumor dependency on MAPK signaling in general and ectopic BRAF activation in particular.
Several groups have identified genes whose protein products may drive oncogenesis together with BRAF in melanoma. The master melanocyte regulator MITF was found to cooperate with BRAF in melanoma tumor formation in vitro (Garraway et al. 2005) and in vivo (Jane-Valbuena et al. 2010). In zebrafish, BRAF activation alone resulted in benign nevus formation, while malignant transformation requires concurrent loss of p53 (Patton et al. 2005). Expression of the BRAF(V600E) allele alone in TERT-immortalized RB–p53 mutant human melanocytes produced only junctional moles in a human/mouse skin graft model, in contrast to activated NRAS or PI3K p110a mutants, which generated invasive melanoma lesions (Chudnovsky et al. 2005). Two very similar Braf(V600E) murine models have emerged, confirming the tumorigenic potential of the mutated Braf allele (Dankort et al. 2009; Dhomen et al. 2009). While both models capitalize on a Tyr-cre-inducible “knock-in” of the oncogenic change (i.e., V600E), the phenotypic details differ in subtle but important ways. The Dankort model (Dankort et al. 2009) provided evidence for cooperativity between Braf activation and Pten loss—an observation first made in human melanoma cell lines (Tsao et al. 2004). Furthermore, they found that Braf(V600E) expression alone in skin melanocytes led to widespread benign melanocytic neoplasia but very few, if any, frank melanomas. This is consistent with the idea that BRAF activation alone results in senescence and nevi. In contrast, the Dhomen model (Dhomen et al. 2009) generated a 60%–70% rate of melanoma tumor formation with Braf(V600E) expression alone, although abrogation of Cdkn2a enhanced tumor multiplicity and shortened the latency period. Interestingly, loss of Cdkn2a did not constrain nevus development, suggesting that senescence from Braf(V600E) is independent of Cdkn2a. The animal models offer substantive proof that BRAF(V600E) mediates melanoma growth in various melanocytic systems.
Biochemical and biophysical studies have recently unraveled the mechanism by which RAF molecules become catalytically primed to phosphorylate MEK. RAF enzymatic activity appears to be triggered by a specific “side-to-side” mode of dimerization as either dimers between two RAF proteins or heterodimers with the RAF-related pseudo-kinase KSR (kinase suppressor of RAS). This side-to-side mechanism is essential for signaling by oncogenic BRAF mutants (Rajakulendran et al. 2009) and is also a critical determinant of resistance to anti-BRAF therapies (see below).
Sorafenib, a type II kinase inhibitor, was the first RAF inhibitor subjected to clinical trial. Sorafenib inhibits several kinases, including BRAF, CRAF, and the VEGF and PDGF RTKs (Wilhelm et al. 2004). After failing to demonstrate a survival benefit in a phase III trial of patients with advanced melanoma (Hauschild et al. 2009), sorafenib has now been largely abandoned in melanoma, certainly as a single agent. Whittaker et al. (2010) provided evidence against the dogma that sorafenib's effects were primarily against BRAF. They engineered cells with a BRAF “gatekeeper” mutation (i.e., p.T529N) that rendered the BRAF(T529N/V600E) cells resistant to sorafenib in vitro and yet sensitive to sorafenib in vivo, a paradox that does not exist with more selective BRAF inhibitors (SBIs) such as PLX4720 (see below). It is thus possible that sorafenib possesses BRAF-independent cellular effects and may also explain why sorafenib causes toxicity at a lower dose than needed for MEK inhibition (Whittaker et al. 2010).
SBIs are now available in both clinical and research settings. The first of these SBIs, vemurafenib, is 10-fold more selective for mutated BRAF compared with its wild-type counterpart (Tsai et al. 2008), although its effectiveness against CRAF (IC50 = 48 nM) in vitro is often overlooked (Poulikakos et al. 2010). The clinical efficacy of vemurafenib in the metastatic setting has now been thoroughly validated in clinical trials (Flaherty et al. 2009, 2010; Chapman et al. 2011). A total of 675 patients with documented BRAF(V600E) mutations enrolled in BRIM-3 (the pivotal phase III study). Patients received either dacarbazine or vemurafenib. The rates of OS were 84% in the vemurafenib (N = 336) group and 64% in the dacarbazine (N = 336) group (hazard ratio = 0.37; P < 0.001). This translated to an estimated median PFS of 1.6 mo in the dacarbazine group and 5.3 mo in the vemurafenib group. The compelling outcomes of BRIM-3 led to the approval of vemurafenib by the Food and Drug Administration (FDA) in 2011.
There are several other BRAF inhibitors that are currently undergoing clinical testing (Smalley and Flaherty 2009). GSK2118436 is another promising BRAF inhibitor (Kefford et al. 2010), with potential benefits for patients with brain metastasis. In wild-type BRAF tumors, CRAF may also be an effective therapeutic target (Garnett et al. 2005; Gray-Schopfer et al. 2005), especially since CRAF appears to play a key role in NRAS-mutated melanomas (Dumaz et al. 2006). Broader-spectrum pan-RAF inhibitors may be developed for NRAS-mutated and NRAS/BRAF dual wild-type tumors.
Despite vemurafenib's success as an SBI, several sentinel questions have also surfaced as a result of deeper investigations: (1) How do SBIs paradoxically stimulate MEK–ERK signaling in BRAF wild-type (e.g., RAS-mutated) cells? (2) How do BRAF-mutated cells eventually escape SBI suppression? As alluded to above, RAF activation involves a side-to-side dimerization, and two models that invoke this mechanism have emerged to answer the first question (Heidorn et al. 2010; Poulikakos et al. 2010).
In response to growth factor receptor engagement or in the face of an oncogenic mutation, RAS mobilizes to the plasma membrane and induces homodimers and heterodimers of BRAF and CRAF, which then leads to MEK phosphorylation and activation. In BRAF(V600E) cells, MEK is largely phosphorylated by the constitutively active BRAF, and consequently, an SBI abrogates nearly all downstream MEK–ERK signaling. So why is there paradoxical activation of MAPK signaling in BRAF wild-type cells? In one model, low concentrations of any RAF inhibitor leads to inactivation of a single monomer in the RAF dimer, thereby “transactivating” its partner RAF molecule and triggering MAPK signaling (Poulikakos et al. 2010); increasing levels of the RAF inhibitor inhibits both RAF partners, and all signaling is thus abrogated. In the Heidorn model (Heidorn et al. 2010), wild-type BRAF translocates to the cell membrane upon SBI binding, dimerizes with CRAF, and further stimulates CRAF signaling. A pan-RAF inhibitor suppresses both BRAF and CRAF, thereby effectively shutting down all MAPK signaling. Gatekeeper CRAF mutations [e.g., CRAF(T421N)] that interfere with the binding of RAF inhibitor to CRAF could restore signaling. The bottom line is that RAS-mutated cells are stimulated by a SBI (Fig. 3), which could explain poor response to vemurafenib among patients with BRAF wild-type tumors and the observed HRAS-driven squamous cell carcinomas (SCCs) that develop while on vemurafenib (Su et al. 2012) and other first-generation RAF inhibitors. Thus, the use of vemurafenib requires absolute genetic precision in order to avoid acceleration of disease and potentially untoward side effects.
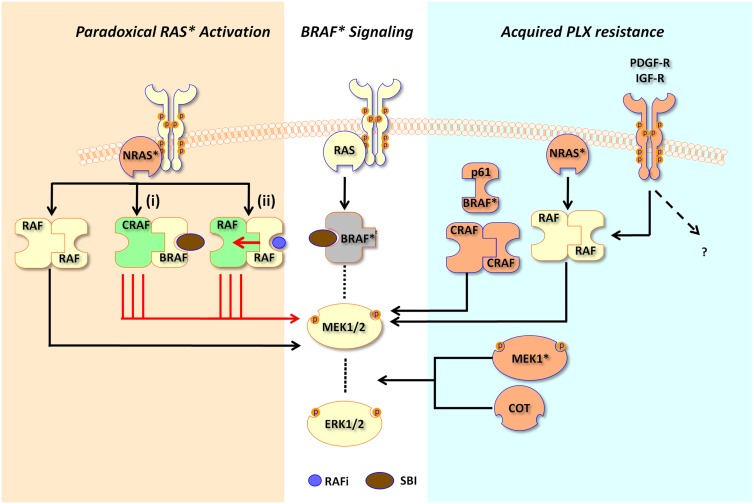
Figure 3.
Pharmacological mechanisms underlying selective BRAF inhibition. In cells dependent on BRAF(V600E) (BRAF*) signaling, binding of a SBI such as PLX4720/4032 leads to attenuation of downstream MEK1/2 and ERK1/2 signaling. In RAS*-dependent cells (tan background shading), MEK and ERK are paradoxically stimulated by RAF inhibitors. In one model (i), binding of SBIs to BRAF leads to binding between BRAF/CRAF and increased stimulation of CRAF by RAS*, since BRAF activity is inhibited by the SBI. In another model (ii), RAS* mediates dimerization between RAF partners; when one RAF molecule within the dimer is inhibited by a RAF inhibitor (RAFi), there is transactivation of the uninhibited partner, thereby stimulating downstream signaling. In cells with acquired SBI resistance, several mechanisms have been described. BRAF gene amplification, a BRAF(V600E) splice variant (p61BRAF*), MEK1 mutation (MEK1*), secondary NRAS activation (NRAS*), and stimulation of PDGF-R or IGF-R have all been observed in tumors samples. Overexpression of CRAF and COT1 has been shown to confer resistance in functional screens.
How do cells escape SBI inhibition? Multiple mechanisms have now come to light to explain vemurafenib resistance (Fig. 3). The acquisition of a new NRAS or MEK1 mutation or the up-regulation of PDGFRβ or IGFR results in reactivation of MAPK signaling and acquired resistance to vemurafenib (Nazarian et al. 2010; Villanueva 2010; Wagle et al. 2011). Ectopic expression of both CRAF and COT/TPL2/MAP3K8 was also associated with greater resistance to PLX4720, a tool compound of vemurafenib (Johannessen et al. 2010). Most recently, a 61-kDa variant form of BRAF(V600E) [p61BRAF(V600E)], which lacks the RAS-binding domain, was found to enhance RAF dimerization as compared with full-length BRAF(V600E) (Poulikakos et al. 2011). Ectopic p61BRAF(V600E) expression creates constitutive ERK signaling that is unaltered by the inhibitor. Splice variants that lack the RAS-binding domain were also detected in six out of 19 tumors from patients with acquired resistance to vemurafenib. BRAF amplification has also been recently reported to be associated with vemurafenib resistance (Shi et al. 2012). It is clear at this moment that vemurafenib creates a high selection pressure for survival, and any genetic or epigenetic mechanism that allows for reactivation or bypass of ERK signaling will likely induce resistance. One clinically untenable situation is the emergence of multiple distinct resistance mechanisms within different metastatic deposits, thereby rendering a uniform secondary therapeutic attack nearly impossible.
Downstream MAPK effectors
Recent whole-exome sequencing approaches have yielded other MAPK pathway components that are also mutated in addition to NRAS and BRAF. In the first study from Australia, investigators performed exome-wide sequencing of eight melanoma lines along with matched normal germline DNA. In this screen, isolated mutations in MAP3K5 and MAP3K9 were identified (Stark et al. 2011). A subsequently expanded screen found that eight out of 85 additional melanoma cell lines harbored nonsynonymous changes in MAP3K5, and 13 out of 85 cell lines had mutations in MAP3K9. Functional analysis of several MAPK3K5 alterations (p.Glu663Lys, c.G1987A, and p.Ile780Phe) and MAP3K9 changes (p.Trp333* and p.Lys171Ala) led to a loss of kinase function. In a parallel set of studies, these investigators also found that suppression of MAP3K9 activity also contributed to temozolomide resistance in melanoma. Undertaking a similar approach, a Swiss group (Nikolaev et al. 2011) also performed whole-exome sequencing of seven metastatic melanoma specimens and identified two samples with somatic mutations in MEK1 and MEK2. In a subsequent screen of MEK1 and MEK2 in 127 additional melanomas, 10 (8%) samples harbored mutations in either MAP2K1 or MAP2K2, although the presence of these mutations did not correlate with BRAF mutation status. Although it is possible that the MEK1/2 mutations activate ERK, the presence of these alterations in the face of oncogenic BRAF(V600E) lesions suggests that other signaling effects may be occurring. Initial evidence has also accrued to suggest that at least some MEK1/2 mutations may also confer resistance to RAF inhibition (Wagle et al. 2011).
Although MEK mutations have only recently been described in melanoma, MEK inhibition has long been recognized as an attractive therapeutic approach in tumors with BRAF(V600E) mutations. Before potent and selective RAF inhibitors were widely available, MEK inhibitors were found to exhibit exquisite potency against BRAF(V600E) melanomas (Solit et al. 2006). Accordingly, multiple small-molecule MEK inhibitors are in development. Early clinical trials of first-generation MEK inhibitors were confounded by suboptimal potency and pharmacodynamic parameters (e.g., CI-1040) (Rinehart et al. 2004). In hindsight, then, it is perhaps not surprising that these inhibitors showed only moderate effects in phase I trials of a limited number of patients with BRAF(V600E) melanomas (Adjei et al. 2008). However, new MEK inhibitors have shown clinical promise both as single agents and in combination with RAF inhibitors (Gray-Schopfer et al. 2007; Infante et al. 2010; Gilmartin et al. 2011). Thus, MEK inhibition may ultimately find a role in the treatment of BRAF mutant melanoma.
PTEN, negative regulator of the PI3K–AKT pathway
The PI3K pathway is often dysregulated in melanoma. The PTEN tumor suppressor gene encodes a lipid and protein phosphatase that regulates cell growth and survival through PI3K/AKT signaling. PTEN negatively regulates signal transduction that uses phosphatidylinositol phosphate (PIP3) as a cytosolic second messenger. Upon cell surface receptor (e.g., RTK) activation, growth factors augment intracellular PIP3 levels, which triggers downstream events that typically converge on phosphorylation of the serine/threonine kinase AKT. AKT is a well-known oncogene that sends many downstream signals to promote cell growth and survival. In melanoma, elevated phospho-AKT levels may correlate adversely with patient survival (Dai et al. 2005).
PTEN/PI3K pathway genomic alterations are considerably less prevalent than MAPK pathway alterations in melanoma; however, recent results may suggest a larger diversity of genetic events affecting this pathway—at least indirectly—than was initially appreciated. Since PI3K itself is rarely mutated in melanoma (Omholt et al. 2006) in contrast to other malignancies, the PTEN tumor suppressor gene has emerged as the dominant genetic target in this pathway. The PTEN locus is situated on chromosome 10q, which undergoes frequent hemizygous deletion and LOH in melanoma (Bastian 2003; Wu et al. 2003). In melanoma, allelic loss or altered expression of PTEN comprises 20% and 40% of melanoma tumors, respectively (Tsao et al. 1998b, 2003; Pollock et al. 2002; Mikhail et al. 2005; Slipicevic et al. 2005; Goel et al. 2006). Somatic point mutations and homozygous deletions are uncommon (Lin et al. 2008), although focal deletions affecting the PTEN locus may be more common than initially appreciated, based on recent genome sequencing studies (Berger et al. 2012). This same study, along with an earlier whole-genome study of a single melanoma cell line (Pleasance et al. 2010), also found recurrent MAGI2 mutations. MAGI2 encodes a protein known to interact with and stabilize PTEN (Tolkacheva et al. 2001; Vazquez et al. 2001); thus, MAGI2 disruption may offer an additional means of PTEN dysregulation in melanoma. More recently, Karreth et al. (2011) described a competitive endogenous RNA (ceRNA), designated ZEB2, which modulates PTEN levels and whose attenuation leads to increased PI3K/AKT pathway signaling. Phosphatase-independent function, such as direct binding and transactivation of p53 (Tang and Eng 2006), may also contribute to its melanoma-suppressive effects. Forced expression of PTEN in PTEN-deficient melanoma cells suppresses AKT phosphorylation, cell growth, and several other tumorigenic phenotypes (Robertson et al. 1998; Stewart et al. 2002; Stahl et al. 2003; for review, see Robertson 2005).
Several genetic studies have noted that hemizygous deletions spanning chromosome 10q (which includes the PTEN locus) occur with high frequency in BRAF mutant melanomas (Bastian 2003; Lin et al. 2008). In contrast, PTEN deletions are less common in NRAS mutant melanomas. These observations suggested that BRAF and PTEN may cooperate in melanoma tumorigenesis (Tsao et al. 2004). Indeed, this notion has been borne out in genetically engineered mouse models in which melanocytes were engineered to undergo PTEN inactivation in the setting of BRAF(V600E) expression (Dankort et al. 2009). The melanomas that form are often highly aggressive, but their growth can be inhibited by combined MEK/target of rapamycin (TOR) inhibition (Dankort et al. 2009). In one mouse model, the mechanism of metastasis appears to involve β-catenin signaling (Damsky et al. 2011). Together, these observations suggest that cooperation between dysregulated MAPK and PI3K signaling (governed by BRAF mutation and PTEN deficiency) may comprise an oncogenic driver for a substantial subset of melanomas.
Aberrant AKT activation can also promote melanocyte transformation (Chudnovsky et al. 2005), as predicted from the aforementioned studies. Some lines of evidence point to a possible role for AKT3 in this process. For example, copy gains spanning AKT3 have been observed in melanoma, and AKT3 activation may be common (Stahl et al. 2004). Similarly, point mutations in AKT3 have occasionally been reported (Davies et al. 2008). Despite these observations, the role of AKT as a melanoma oncogene remains incompletely understood. For example, AKT1 activation inhibits migration and invasion in some cancer models (Yoeli-Lerner et al. 2005), including MDA-MB435, a cell line previously annotated as a breast cancer model but subsequently found by genetic studies to be melanoma-derived (Ross et al. 2000; Garraway et al. 2005). Thus, while the general importance of PTEN/PI3K dysregulation in at least a subset of melanoma seems well-established, the contribution of AKT as a downstream effector remains a topic of active investigation.
There is substantial evidence that the mammalian TOR (mTOR) pathway acts as a major effector of the AKT oncogenic signal downstream from PI3K activation. Thus far, however, TOR inhibitors have proved mostly ineffective in melanoma, at least as single agents (Margolin et al. 2004; Xu et al. 2004). However, no trial has been completed that evaluates TOR inhibitors in combination with the selective RAF and MEK inhibitors that are either FDA-approved or in clinical trials. Conceivably, such trials might be further enhanced by the concomitant incorporation of genomic profiling to identify patients whose tumors harbor genetic alterations predicted to dysregulate PTEN/PI3K signaling.
Apoptosis regulators in melanoma
An abundance of mechanistic studies indicated that the capacity to undergo efficient apoptosis may predict sensitivity to anti-cancer therapeutics (Johnstone et al. 2002). The melanocyte lineage uses several mechanisms that appear to diminish the propensity to activate an apoptotic program. Synthesis of eumelanin provides UV protection. Yet melanocytes and melanoma cells exhibit greater protection against UV-induced apoptosis than adjacent keratinocytes, suggesting that additional survival signals may protect the melanocyte lineage against cell death (for review, see Soengas and Lowe 2003). Melanomas commonly resist induction of apoptosis (Glinsky et al. 1997) and appear to be protected by multiple mechanisms, which include activation of the MAPK and PI3K/AKT pathways (Wada and Penninger 2004; Kharas and Fruman 2005). In melanoma, the MAPK pathway antagonizes apoptosis via multiple mechanisms that include suppression of Smac/DIABLO release from mitochondria (Zhang et al. 2003), expression of anti-apoptotic BCL2 through MITF (McGill et al. 2002), suppression of the proapoptotic protein BAD (Eisenmann et al. 2003), and phosphorylation-sensitive degradation of proapoptotic BIM (Cartlidge et al. 2008). While a fully integrated understanding of apoptosis regulation remains to be completed for melanoma, it is likely that such information will be mechanistically informative regarding therapeutic efficacy and resistance for the new generation of melanoma treatments.
BCL2 has long been known to be expressed in both melanocytes and melanoma cells (Plettenberg et al. 1995). Its expression has been seen to be up-regulated by a variety of growth factors, including KIT ligand (SCF) (Zhai et al. 1996; von Willebrand et al. 2005), NRAS (Borner et al. 1999), and MITF (McGill et al. 2002). BCL2 expression has been correlated with several poor prognostic features that include presence of ulceration and patient survival (Leiter et al. 2000; Ilmonen et al. 2005). Genetic deletion of BCL2 in the mouse germline results in a striking hair-graying phenotype that was found to result from abrupt death of hair follicle melanocyte stem cells at approximately postnatal day 8 (Nishimura et al. 2005). Other anti-apoptotic BCL2 family members, such as BCL-XL and MCL1, may also contribute to melanoma survival and drug resistance (Tron et al. 1995; Selzer et al. 1998; Skvara et al. 2005).
Attempts to therapeutically target BCL2 were carried out through the use of Genasense (G3139; Genta, Inc.), which was an antisense oligonucleotide targeting BCL2 mRNA (Badros et al. 2005; Marcucci et al. 2005; O'Brien et al. 2005; Tolcher et al. 2005). Despite encouraging data from phase II studies in prostate cancer and myeloma (Badros et al. 2005; Tolcher et al. 2005), phase III data in advanced melanoma patients did not demonstrate a significant survival advantage, resulting in lack of FDA approval for melanoma (FDA summary report, http://www.fda.gov/ohrms/dockets/ac/04/slides/4037S1_02_FDA-Kane-Yang%20.ppt). While these data were not encouraging for the antisense targeting of BCL2, it remains plausible that BCL2 targeting may offer therapeutic benefit in subsets of melanoma patients or that targeting of BCL2 may be particularly advantageous in a combinatorial context with other agents for whom treatment resistance might be mediated by BCL2.
NEDD9, a melanoma metastasis gene identified by cross-species comparison
The NEDD9 gene was discovered as a metastasis gene for melanoma using a novel strategy that used genetically engineered mouse models in order to discover new oncogenes (Kim et al. 2006). A comparison of parental melanomas that were not metastatic, relative to a metastatic variant in an inducible system (Chin et al. 1999) using genome-wide high-resolution array comparative genomic hybridization, was employed and revealed genomic amplification that correlated with acquired metastatic potential. The genomic region in mice was found to be syntenic to human 6p24-25, a genomic region known to undergo gains in copy number within ∼36% of human metastatic, but not primary, melanomas (Bastian et al. 1998; Namiki et al. 2005). The 6p gain in human melanomas typically involves a vast genomic region, making identification of individual driver oncogenes extremely difficult. In contrast, the focal and recurrent amplifications seen in the mouse model permitted identification of the NEDD9 locus. At the protein level, NEDD9 expression was found to correlate with progression in human melanomas (Kim et al. 2006). Loss- and gain-of-function analyses further validated NEDD9's functional role in progression and metastasis. NEDD9 was seen to localize to dynamic focal contacts at the periphery of the cell, where it interacts with focal adhesion kinase to mediate invasive behavior. These data suggest the possibility that suppression of this signaling complex may inhibit progression from early melanoma to late stage disease. It will be of value to determine whether NEDD9 elevations are prognostically meaningful in predicting future disease progression.
GOLPH3—a new class of oncoprotein identified by genomics
Another approach toward gene discovery leveraged extensive comparative analyses of array genomic hybridization profiles from multiple tumor types with the goal of prioritizing common copy number alterations that are more likely to be oncogenically relevant (Scott et al. 2009). This analysis identified frequent amplifications at 5p13, and fluorescence in situ hybridization on tumor tissue microarrays (TMAs) confirmed significant amplification frequencies ranging from 24% to 56% in several tumor types that included breast, colorectal, and non-small-cell lung cancer, among others. Detailed mapping of focally amplified specimens delimited an informative minimal common region of gain comprised of four genes (GOLPH3, MTMR12, ZFR, and SUB1). Subsequent functional analyses of these genes identified GOLPH3, which encodes a highly conserved 34-kDa protein first identified through proteomic characterizations of the Golgi apparatus (Scott and Chin 2010), as one gene that is likely targeted in cancers with 5p13 gain. Parallel studies using Saccharomyces cerevisiae coupled with confirmatory biochemical and functional assays in human cell systems established that Golph3 physically interacts with the Vps35 subunit of the retromer protein-recycling complex (Scott et al. 2009). In addition, those studies also revealed that GOLPH3 expression modulates signaling through the mTOR, a serine/threonine protein kinase that is a critical oncogenic effector in the RTK–PI3K pathway that integrates input from multiple signaling pathways to control cell growth, proliferation, and survival (Guertin and Sabatini 2007).
Additional expression analyses indicated that 5p13 copy number correlated with increased phosphorylation of the mTOR substrate p70 S6 kinase in non-small-cell lung cancer specimens, which further linked Golph3 function to mTOR activity. When transplanted into mice, human tumor cell lines overexpressing GOLPH3 not only developed tumors faster than control, but also were significantly more sensitive to rapamycin, a potent inhibitor of mTOR (Sabatini 2006), providing pharmacological proof that the protumorigenic activity of GOLPH3 is mediated through mTOR signaling. These results also raise the possibility that GOLPH3 expression level or copy number status may serve as a predictive biomarker for sensitivity to mTOR inhibitors, especially for advanced cancers driven by aberrant RTK–PI3K signaling that remain refractory to standard chemotherapy regimens.
MITF—melanocyte master regulator
The identification of MITF as a melanoma oncogene came about through an integrated analysis of genomic copy number gain (amplification) with matched mRNA expression analysis (Garraway et al. 2005), while loss of function was already known to affect survival of the entire melanocyte lineage (Hodgkinson et al. 1993). In assessing MITF's potential role as a melanoma oncogene, it was also found that enforced overexpression of MITF participated actively in conferring a transformed phenotype when introduced together with BRAF(V600E) into immortalized human melanocytes (Garraway et al. 2005). Furthermore, survival analysis indicated that MITF amplification was associated with worsened 5-year survival.
Genomic sequencing of sporadic melanomas has provided evidence for the occurrence of scattered somatic point mutations within the MITF gene (Cronin et al. 2009), consistent with the possibility of their participating as oncogenic variants of MITF. While additional functional evidence for such activity will be of great interest, recently, a coding variant was discovered in studies seeking to identify familial melanoma genes. Two independent studies identified the same variant, MITF(E318K), which largely cosegregates with affected individuals in instances of familial melanoma, as well as a small fraction of sporadic melanomas and renal cell carcinomas (see above) (Bertolotto et al. 2011; Yokoyama et al. 2011). The point mutation occurs at a site that had been previously identified as a target for SUMO modification on MITF (Miller et al. 2005). Modification by SUMOylation on MITF had been shown to modestly suppress MITF activity on certain transcriptional target genes. Thus the MITF(E318K) variant was predicted to represent a gain-of-function allele for MITF, consistent with MITF's role as a melanoma oncogene. Interestingly, clinical analysis of affected individuals carrying the germline variant suggested a statistically significant incidence of nonblue eye color (Yokoyama et al. 2011). Since MITF is known to transcriptionally regulate pigment gene expression, for which stronger gene expression is predicted to produce darker pigmentation, these observations are further consistent with the MITF(E318K) variant representing gain-of-function biological activity for MITF. Thus far, the transcriptional targets of MITF that mediate its oncogenic activity, as distinct from its pigmentation activity, remain to be fully elucidated.
Multiple genes besides MITF are known to be essential to melanocyte lineage survival. Certain ones are known to function as upstream regulators of MITF expression. These include Pax3 and SOX10, factors whose germline mutation shares clinical pigmentary features with germline Mitf mutation in humans (for review, see Price and Fisher 2001). Additional regulators of MITF that are important for melanocyte survival are growth factor receptor genes and the pathways that they regulate. These factors include endothelin 3, endothelin receptor B, SCF, c-KIT, and components of their signaling pathways, which include RAS, RAF, GNAQ/11, and others. One biochemical consequence of MAPK pathway activation is the direct phosphorylation of MITF by ERK/MAPK on Ser 73, an effect that was shown to enhance molecular recruitment of the coactivator p300 to MITF (Hemesath et al. 1998; Price et al. 1998). Another consequence of this MITF phosphorylation is ubiquitination and proteolysis of MITF (Wu et al. 2000; Xu et al. 2000), an effect recently shown to be antagonized by the deubiquitinase USP13 (Zhao et al. 2011). Of note, bacterial artificial chromosomes containing Ser 73-to-alanine mutation of MITF were capable of rescuing coat color/pigmentation of Mitf-deficient mice (Bauer et al. 2009). It will be interesting to determine whether other biological activities of MITF, such as its roles in melanomagenesis, are affected by the MAPK phosphorylation at Ser 73. This question is important because a very high fraction of human melanomas contain mutations that produce constitutive overactivity of the MAPK pathway. MITF undergoes several other post-translational modifications, which include phosphorylation by p38 kinase (Mansky et al. 2002) and phosphorylation by glycogen synthase kinase 3b (Takeda et al. 2000). The MITF genomic organization is complex, in that a series of upstream promoters can be alternatively spliced onto common downstream exons, most of which contain common coding sequences. The most 3′ (downstream) of the promoter/exon 1 units is the melanocyte-specific M-MITF promoter (Fuse et al. 1996), whose expression is controlled by a combination of factors that include WNT, cAMP/CREB, SOX10, and PAX3.
MITF's DNA-binding domain is characterized by the basic helix–loop–helix/leucine zipper (bHLHzip) DNA-binding/dimerization motifs (Hemesath et al. 1994). This motif structure, while homologous to those found in the MYC oncoprotein family, confers heterodimerization capabilities with three other transcription factors—TFEB, TFE3, and TFEC (the MiT family)—but not with MYC oncoproteins. Genetic studies in mice by Steingrimsson et al. (2002) demonstrated genetic redundancy between MITF and TFE3 within the osteoclast lineage—a feature that provides genetic support to the concept that members of the MiT family share considerable biochemical activities in vivo.
Another MiT family-associated human malignancy is clear cell sarcoma (also referred to as melanoma of soft parts). This tumor expresses multiple melanocytic markers, is often pigmented (Granter et al. 2001), and has been known to contain a diagnostic translocation that fuses the EWS gene to the ATF1 gene (Fujimura et al. 1996). The resulting EWS–ATF1 fusion protein mimicks the activity of CREB, thus stimulating uncontrolled expression of MITF within clear cell sarcoma (Davis et al. 2006)—and imitating MITF's oncogenic role seen in melanoma.
A large list of transcriptional target genes for MITF has accrued in recent years. While a complete discussion of these genes is beyond the scope of this review, it is notable that numerous pigmentation-related genes appear to be direct transcriptional targets of MITF. From its initial molecular characterization, it was rapidly recognized that MITF protein binds and transcriptionally activates gene expression at consensus DNA-binding elements containing the consensus sequence CA[C/T]GTG, a sequence known to be present in the promoters of most or all pigmentation-related genes (Bentley et al. 1994; Hemesath et al. 1994; Yasumoto et al. 1994). Some of these melanocytic genes include Pmel17/silver/gp100 (which encodes the melanoma diagnostic epitope HMB45) (Halaban et al. 1996; Baxter and Pavan 2003; Du et al. 2003) and MelanA/Mart1 (Du et al. 2003). HMB45, Mart1, and MITF are now commonly used as immunohistochemical stains of the melanocytic lineage for melanoma diagnosis. Many additional MITF target genes have been identified, with some notable ones including Melastatin (TRPM1) (Miller et al. 2004; Zhiqi et al. 2004), AIM1 (ocular albinism 4 gene) (Du and Fisher 2002), Ocular albinism 1 gene (OA1) (Vetrini et al. 2004), VMD2 (Esumi et al. 2004), HIF1α (Busca et al. 2005), and Plasminogen activator inhibitor-1 (Murakami et al. 2006); a variety of mast cell genes including Prostaglandin D2; multiple mast cell proteases; adhesion molecules; and others (Ito et al. 2004; Morii et al. 2004; Takeda et al. 2006).
Another interesting set of MITF target genes includes those involved in cell cycle regulation. This list includes the CDK inhibitors (CDKi) p16INK4a (Loercher et al. 2005) and p21 (Carreira et al. 2005). In addition, the cell cycle kinase CDK2 was found to be a direct transcriptional target of MITF selectively within the melanocyte lineage (Du et al. 2004). The CDK2 genomic locus was found to reside directly adjacent to the melanocyte-specific pmel17/gp100 locus. Via an enhancer element containing the consensus MITF-binding sequence, MITF was seen to coregulate these genes, with CDK2 expression being uniquely regulated by MITF within melanocytes (its expression is constitutively regulated in other lineages). Slug has also been suggested to be a direct transcriptional target of MITF (Sanchez-Martin et al. 2002), a notable finding because Waardenburg syndrome type 2 patients were also identified to carry deletions of Slug (either germline or somatic). Slug has also been shown to contribute invasive behavior to melanocytes (Gupta et al. 2005). c-Met, the RTK activated by the ligand HGF, was also identified as a direct transcriptional target of MITF (McGill et al. 2006). It is anticipated that the coming years will provide a comprehensive picture of target genes regulated by MITF in melanocytes and melanoma cells through the use of genome-wide technologies such as ChIP-seq (chromatin immunoprecipitation in combination with deep sequencing) and RNA-seq (RNA sequencing) (Strub et al. 2011). Moreover, in all cases, it is virtually certain that additional transcription factors beyond MITF play similarly important roles in controlling target gene expression and perhaps function as partners with MITF to control specific biological pathways (such as pigmentation). A key mechanistic question involves the means by which MITF induces melanomagenesis on the one hand versus differentiation/pigmentation on the other. It is possible that such activities relate to altered protein levels, distinct post-translational modifications, or other contextual features within the melanocyte.
Cell cycle dysregulation as a melanoma therapeutic target
Deletion, mutation, or silencing of CDKN2A is common in melanoma (Chin 2003) and, in combination with evidence for CDK2 dependency (Du et al. 2004), suggests that the RB pathway should represent a potentially “druggable” dependency in melanoma cells. Activation of CDK2 and CDK4/6 results in RB hyperphosphorylation in melanoma cells (Halaban 1999). In the case of CDK2, there appears to be a lineage-selective melanoma dependency because multiple nonmelanomas and normal tissues are resistant to CDK2 suppression (Tetsu and McCormick 2003; Du et al. 2004). However, selective CDK2 antagonists have not yet appeared, and less-selective CDK2 inhibitors (such as flavopiridol, which antagonizes CDK2, CDK1, CDK4, and CDK7) are the types of small molecules that have been clinically tested for a variety of malignancies. Numerous additional CDKi are in various stages of development currently.
Circumventing p53 DNA damage signaling
Inactivation of the p53 pathway appears to be a rite of passage for virtually all tumor cells. While most human solid tumors neutralize this tumor-suppressive pathway at the level of p53 itself (e.g., mutations within the TP53-coding region), melanoma provides a notable exception to this rule. In a recent meta-analytical survey of 645 uncultured melanoma specimens, there were only 85 (13.2%) TP53 mutations reported. Interestingly, more than half of these (N = 43) were UV signature changes (Hocker and Tsao 2007), suggesting that TP53 is, in fact, a melanoma-promoting target of UV. With the recognition that p14ARF (an exon 1β-specific transcript of CDKN2A) down-regulates HDM2 (Kamijo et al. 1998; Pomerantz et al. 1998; Stott et al. 1998; Zhang et al. 1998), it became clear that functional inactivation of p53 could be achieved by the melanoma cell through CDKN2A/p14ARF loss. In fact, concomitant deletion of the CDKN2A locus does occur in the face of activating CDK4 mutations (Yang et al. 2005), suggesting that abrogation of the RB pathway through a CDK4-mediated mechanism is still insufficient and that secondary inactivation of p53 is needed and accomplished through abrogation of p14ARF. If direct TP53 mutagenesis and p14ARF represent two possible mechanisms for p53 pathway injury, one would expect HDM2 amplification as a third potential mechanism; indeed, HDM2 amplification has been described in 3%–5% of human melanomas (Muthusamy et al. 2006).
Since p53 is only rarely mutated but only functionally constrained by HDM2, restoration of p53 function through small-molecule antagonists of HDM2 represents a viable therapeutic approach. Ji et al. (2012b) recently demonstrated that melanomas can be suppressed by nutlin-3, a canonical HDM2 antagonist, in a TP53-dependent fashion. Moreover, synergy with MAPK pathway inhibition suggests that optimization may be possible with future development (Ji et al. 2012b).
Conclusion: future perspectives on melanoma
The recent approval of ipilimumab and vemurafenib marks 2011 as a historical year for melanoma therapeutics. However, rather than celebrating the extraordinary knowledge that has been uncovered in the last 20 years, perhaps it is also appropriate to conclude with the most pressing and challenging questions in melanoma today.
(1) Is there a melanoma-initiating cell? The existence of a rare melanoma stem cell population was suggested by early studies that identified a CD20-enriched subpopulation of cells from melanoma lines or metastases that grew as nonadherent spheroids when cultured in human embryonic stem cell-like medium and that could be induced to differentiate toward multiple lineages, including melanocyte, adipocyte, osteoblast, and chondrocyte, suggesting the presence of such subpopulations with the stem cell phenotype (Fang et al. 2005). Separately, Frank et al. (2005) isolated a small subpopulation of melanoma cells that were enriched for expression of both ABCB5 and CD133. In xenograft experiments, the melanoma-initiating cell frequencies for unsegregated and ABCB5+ populations were one in 1,090,336 versus one in 158,170, respectively, suggesting that the marker enriched for a more tumorigenic population of cells; i.e., stem cells. Furthermore, selective depletion of ABCB5+ melanoma cells using anti-ABCB5 antibodies also led to inhibition of tumor formation (Schatton et al. 2008). However, no less than a few months after this initial publication, Sean Morrison's group (Quintana et al. 2008) showed that, under the proper experimental context, up to one in four melanoma cells could initiate tumors, thereby challenging the notion that there is a defined cellular hierarchy in melanoma tumor initiation. Other putative markers of melanoma-initiating cells have also been proposed, including CD271 (Boiko et al. 2010). Amidst conflicting results, varied technical approaches, and nonuniform definitions of a “cancer stem cell,” this debate will likely continue.
(2) Why is resistance to anti-BRAF therapies so rampant? One of the greatest challenges in melanoma therapeutics is ironic juxtaposition of rapid and widespread response to vemurafenib with the often rapid and fatal relapses that occur months to years later. The systematic layer-by-layer unpeeling of mechanisms suggests that single-agent molecular approaches in melanoma may not be ultimately curative but that cocktails and combination targeted/immunotherapy will be needed.
(3) How are patients with BRAF wild-type and NRAS-mutated tumors to be treated? A tremendous amount of effort exists for optimizing anti-BRAF treatments despite a large need among patients with BRAF wild-type tumors. With a significant set of recurrent melanomas having been shown to harbor activating NRAS mutations, the next generation of melanoma treatments will have to contend with RAS-mediated reactivation of MAPK and non-MAPK mechanisms. Approaches including the targeting of MEK and the PI3K pathway have recently been incorporated into clinical trials and will undoubtedly be vigorously tested in the coming months.
(4) Is it possible to cripple “master regulators”? While traditionally considered nondruggable, the increasing recognition that transcription factors (e.g., MITF) may play oncogenic roles in melanoma has driven efforts to devise alternative approaches to target this protein class. One such approach involves the use of peptide to block transcription factor activities. In melanoma, ATF2 has been targeted in this fashion with impressive preclinical results (Bhoumik et al. 2002, 2004). These observations are particularly intriguing given evidence that ATF2 nuclear localization correlates with prognosis in human melanoma specimens (Berger et al. 2003). Small molecules that block transcription factor dimerization represents another approach and are currently being developed for the inhibition of MYC–MAX interaction (Prochownik and Vogt 2010). An additional strategy involves elucidating the critical downstream targets of oncogenic transcription factors, some of which may prove to be more amenable to small-molecule compound inhibition. Attempts to target the MITF pathway are being increasingly scrutinized, balancing the challenges of antagonizing a noncatalytic protein against the benefits of inhibiting a lineage-selective master survival factor.
(5) How do we maximally leverage the advances in immunotherapy with those in targeted therapeutics? In parallel evolution, progress in our understanding of tumor immunology and immunotherapy has led to equally exciting and novel approaches to melanoma treatment using immune-based modifiers (for review, see Mellman et al. 2011; Sharma et al. 2011). Although a full exploration of immunological treatments is beyond the scope of this review, there are scientific and clinical considerations to suggest combinations of molecularly based and immunologically driven approaches. For instance, suppression of BRAF(V600E) by PLX induces the expression of melanocyte differentiation antigens, raising the possibility that concurrent treatment with SBIs may in fact improve the efficacy of immunological targeting (Boni et al. 2010).
(6) The unprecedented propagation of melanoma genetics, genomics, and biological understanding justifies an optimistic view of its future therapeutic outlook. Undoubtedly, the road ahead will remain stunted with challenges and occasional setbacks; however, salient tumor dependencies that underlie definable melanoma subtypes have begun to emerge. In partnership with versatile in vitro and in vivo model systems, increasingly rigorous target validation (for maintenance) and rational drug development efforts, and judicious use of correlative “-omics” studies for robust biomarkers, melanoma promises to provide an instructive framework for progress against even the most brittle of human malignancies.
Acknowledgments
We extend our sincere apology to those colleagues whose studies were not cited in this review due to space constraints. Funding for this scholarly effort was supported in part by the NIH (K24 CA149202 to H.T., R01 AR043369 to D.F., and RO1 CA093947 to L.C.), the NIH Director's New Innovator Award to L.G., the Starr Cancer Consortium to L.G., the Melanoma Research Alliance (to all authors), the American Skin Association/Abby S. and Howard P. Milstein Award to H.T. and L.C., and the Sheldon and Dr. Miriam Adelson Medical Research Foundation to L.G. and D.F.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.191999.112.
References
- Ackermann J, Frutschi M, Kaloulis K, McKee T, Trumpp A, Beermann F 2005. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Res 65: 4005–4011 [DOI] [PubMed] [Google Scholar]
- Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, et al. 2008. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol 26: 2139–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharonson Z, Gana-Weisz M, Varsano T, Haklai R, Marciano D, Kloog Y 1998. Stringent structural requirements for anti-Ras activity of S-prenyl analogues. Biochim Biophys Acta 1406: 40–50 [DOI] [PubMed] [Google Scholar]
- Albino AP, Nanus DM, Mentle IR, Cordon-Cardo C, McNutt NS, Bressler J, Andreeff M 1989. Analysis of ras oncogenes in malignant melanoma and precursor lesions: Correlation of point mutations with differentiation phenotype. Oncogene 4: 1363–1374 [PubMed] [Google Scholar]
- Antonescu CR, Busam KJ, Francone TD, Wong GC, Guo T, Agaram NP, Besmer P, Jungbluth A, Gimbel M, Chen CT, et al. 2007. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer 121: 257–264 [DOI] [PubMed] [Google Scholar]
- Ashida A, Takata M, Murata H, Kido K, Saida T 2009. Pathological activation of KIT in metastatic tumors of acral and mucosal melanomas. Int J Cancer 124: 862–868 [DOI] [PubMed] [Google Scholar]
- Badros AZ, Goloubeva O, Rapoport AP, Ratterree B, Gahres N, Meisenberg B, Takebe N, Heyman M, Zwiebel J, Streicher H, et al. 2005. Phase II study of G3139, a Bcl-2 antisense oligonucleotide, in combination with dexamethasone and thalidomide in relapsed multiple myeloma patients. J Clin Oncol 23: 4089–4099 [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. 2009. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 27: 6199–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale SJ, Dracopoli NC, Tucker MA, Clark WH Jr, Fraser MC, Stanger BZ, Green P, Donis-Keller H, Housman DE, Greene MH 1989. Mapping the gene for hereditary cutaneous malignant melanoma-dysplastic nevus syndrome to chromosome 1p. N Engl J Med 320: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L 2001. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol 21: 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian BC 2003. Understanding the progression of melanocytic neoplasia using genomic analysis: From fields to cancer. Oncogene 22: 3081–3086 [DOI] [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D 1998. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 58: 2170–2175 [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Pinkel D 2000. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol 157: 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer GL, Praetorius C, Bergsteinsdottir K, Hallsson JH, Gisladottir BK, Schepsky A, Swing DA, O'Sullivan TN, Arnheiter H, Bismuth K, et al. 2009. The role of MITF phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue. Genetics 183: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Buttner P, Murali R, Okamoto I, Kolaitis NA, Landi MT, Scolyer RA, Bastian BC 2011. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res 24: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter LL, Pavan WJ 2003. Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns 3: 703–707 [DOI] [PubMed] [Google Scholar]
- Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Anton-Culver H, Zanetti R, et al. 2005. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 97: 1507–1515 [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Eisen T, Goding CR 1994. Melanocyte-specific expression of the human tyrosinase promoter: Activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 14: 7996–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AJ, Kluger HM, Li N, Kielhorn E, Halaban R, Ronai Z, Rimm DL 2003. Subcellular localization of activating transcription factor 2 in melanoma specimens predicts patient survival. Cancer Res 63: 8103–8107 [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Lissanu Deribe Y, Lawrence MS, Protopopov A, Ivanova E, Stransky N, Watson IR, Nickerson E, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature doi: 10.1038/nature11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. 2011. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 480: 94–98 [DOI] [PubMed] [Google Scholar]
- Besaratinia A, Pfeifer GP 2008. Sunlight ultraviolet irradiation and BRAF V600 mutagenesis in human melanoma. Hum Mutat 29: 983–991 [DOI] [PubMed] [Google Scholar]
- Bhoumik A, Huang TG, Ivanov V, Gangi L, Qiao RF, Woo SL, Chen SH, Ronai Z 2002. An ATF2-derived peptide sensitizes melanomas to apoptosis and inhibits their growth and metastasis. J Clin Invest 110: 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoumik A, Gangi L, Ronai Z 2004. Inhibition of melanoma growth and metastasis by ATF2-derived peptides. Cancer Res 64: 8222–8230 [DOI] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Paillerets BB, Chompret A, Ghiorzo P, Gruis N, Hansson J, et al. 2002. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 94: 894–903 [DOI] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, et al. 2010. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 466: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A, Cogdill AP, Dang P, Udayakumar D, Njauw CN, Sloss CM, Ferrone CR, Flaherty KT, Lawrence DP, Fisher DE, et al. 2010. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res 70: 5213–5219 [DOI] [PubMed] [Google Scholar]
- Boone B, Jacobs K, Ferdinande L, Taildeman J, Lambert J, Peeters M, Bracke M, Pauwels P, Brochez L 2011. EGFR in melanoma: Clinical significance and potential therapeutic target. J Cutan Pathol 38: 492–502 [DOI] [PubMed] [Google Scholar]
- Borner C, Schlagbauer Wadl H, Fellay I, Selzer E, Polterauer P, Jansen B 1999. Mutated N-ras upregulates Bcl-2 in human melanoma in vitro and in SCID mice. Melanoma Res 9: 347–350 [DOI] [PubMed] [Google Scholar]
- Busca R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribere L, Bertolotto C, et al. 2005. Hypoxia-inducible factor 1α is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J Cell Biol 170: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, Fountain JW, Hegi ME, Wiseman RW, Petyy EM, Bale AE, et al. 1992. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science 258: 1148–1152 [DOI] [PubMed] [Google Scholar]
- Caponigro F, Casale M, Bryce J 2003. Farnesyl transferase inhibitors in clinical development. Expert Opin Investig Drugs 12: 943–954 [DOI] [PubMed] [Google Scholar]
- Cardenas-Navia LI, Cruz P, Lin JC, Rosenberg SA, Samuels Y 2010. Novel somatic mutations in heterotrimeric G proteins in melanoma. Cancer Biol Ther 10: 33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433: 764–769 [DOI] [PubMed] [Google Scholar]
- Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, McMahon M 2008. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res 21: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, et al. 2011. KIT as a therapeutic target in metastatic melanoma. JAMA 305: 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzinasiou F, Lill CM, Kypreou K, Stefanaki I, Nicolaou V, Spyrou G, Evangelou E, Roehr JT, Kodela E, Katsambas A, et al. 2011. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst 103: 1227–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L 2003. The genetics of malignant melanoma: Lessons from mouse and man. Nat Rev Cancer 3: 559–570 [DOI] [PubMed] [Google Scholar]
- Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW II, DePinho RA 1997. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev 11: 2822–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Merlino G, DePinho RA 1998. Malignant melanoma: Modern black plague and genetic black box. Genes Dev 12: 3467–3481 [DOI] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, et al. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400: 468–472 [DOI] [PubMed] [Google Scholar]
- Choi KY, Chang K, Pickel JM, Badger JD II, Roche KW 2011. Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc Natl Acad Sci 108: 15219–15224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA 2005. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet 37: 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi R, Nikiforov YE 2005. Alterations of the BRAF gene in thyroid tumors. Endocr Pathol 16: 163–172 [DOI] [PubMed] [Google Scholar]
- Cohen Y, Goldenberg-Cohen N, Parrella P, Chowers I, Merbs SL, Pe'er J, Sidransky D 2003. Lack of BRAF mutation in primary uveal melanoma. Invest Ophthalmol Vis Sci 44: 2876–2878 [DOI] [PubMed] [Google Scholar]
- Comito G, Calvani M, Giannoni E, Bianchini F, Calorini L, Torre E, Migliore C, Giordano S, Chiarugi P 2011. HIF-1α stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Radic Biol Med 51: 893–904 [DOI] [PubMed] [Google Scholar]
- Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, Vemula S, Burrell AS, Agrawal NS, Lin JC, et al. 2009. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res 22: 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F III, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, Bainbridge T, Heinrich MC, Corless CL 2003. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res 63: 5761–5766 [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, et al. 2005. Distinct sets of genetic alterations in melanoma. N Engl J Med 353: 2135–2147 [DOI] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC 2006. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 24: 4340–4346 [DOI] [PubMed] [Google Scholar]
- Dai DL, Martinka M, Li G 2005. Prognostic significance of activated Akt expression in melanoma: A clinicopathologic study of 292 cases. J Clin Oncol 23: 1473–1482 [DOI] [PubMed] [Google Scholar]
- Damm S, Koefinger P, Stefan M, Wels C, Mehes G, Richtig E, Kerl H, Otte M, Schaider H 2010. HGF-promoted motility in primary human melanocytes depends on CD44v6 regulated via NF-κ B, Egr-1, and C/EBP-β. J Invest Dermatol 130: 1893–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, Taketo MM, Dankort D, Rimm DL, McMahon M, et al. 2011. β-Catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 20: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, et al. 2004. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene 23: 5968–5977 [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, You MJ, DePinho RA, McMahon M, Bosenberg M 2009. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 41: 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954 [DOI] [PubMed] [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB 2008. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer 99: 1265–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. 2006. Oncogenic MITF dysregulation in clear cell sarcoma: Defining the MiT family of human cancers. Cancer Cell 9: 473–484 [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean L, Dumaz N, Sarasin A 1995. The specificity of p53 mutation spectra in sunlight induced human cancers. J Photochem Photobiol B 28: 115–124 [DOI] [PubMed] [Google Scholar]
- Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, Kanetsky PA, Hayward NK, Gillanders E, Elder DE, et al. 2010. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: A GenoMEL study. J Natl Cancer Inst 102: 1568–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit PE, Moretti S, Koenders PG, Weterman MA, van Muijen GN, Gianotti B, Ruiter DJ 1992. Increasing epidermal growth factor receptor expression in human melanocytic tumor progression. J Invest Dermatol 99: 168–173 [DOI] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R 2009. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15: 294–303 [DOI] [PubMed] [Google Scholar]
- Diaz A, Suarez E, Blanco R, Badia T, Rivero D, Lopez-Requena A, Lopez A, Montero E 2007. Functional expression of human-epidermal-growth-factor receptor in a melanoma cell line. Biotechnol Appl Biochem 48: 21–27 [DOI] [PubMed] [Google Scholar]
- Diaz A, Suarez E, Blanco R, Lopez A, Montero E 2009. Epidermal growth factor receptor modulates the tumorigenic potential of melanoma. Front Biosci 14: 159–166 [DOI] [PubMed] [Google Scholar]
- Djerf EA, Trinks C, Abdiu A, Thunell LK, Hallbeck AL, Walz TM 2009. ErbB receptor tyrosine kinases contribute to proliferation of malignant melanoma cells: Inhibition by gefitinib (ZD1839). Melanoma Res 19: 156–166 [DOI] [PubMed] [Google Scholar]
- Draper GJ, Sanders BM, Kingston JE 1986. Second primary neoplasms in patients with retinoblastoma. Br J Cancer 53: 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Fisher DE 2002. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem 277: 402–406 [DOI] [PubMed] [Google Scholar]
- Du J, Miller AJ, Widlund HR, Horstmann MA, Ramaswamy S, Fisher DE 2003. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol 163: 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE 2004. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6: 565–576 [DOI] [PubMed] [Google Scholar]
- Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R 2006. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res 66: 9483–9491 [DOI] [PubMed] [Google Scholar]
- Dutton-Regester K, Irwin D, Hunt P, Aoude L, Tembe V, Pupo GM, Lanagan C, Carter CD, O'Connor L, O'Rourke M, et al. 2012. A high throughput panel for identifying clinically-relevant mutation profiles in melanoma. Mol Cancer Ther 11: 888–897 [DOI] [PubMed] [Google Scholar]
- Easty DJ, Hill SP, Hsu MY, Fallowfield ME, Florenes VA, Herlyn M, Bennett DC 1999. Up-regulation of ephrin-A1 during melanoma progression. Int J Cancer 84: 494–501 [DOI] [PubMed] [Google Scholar]
- Edmunds SC, Cree IA, Di Nicolantonio F, Hungerford JL, Hurren JS, Kelsell DP 2003. Absence of BRAF gene mutations in uveal melanomas in contrast to cutaneous melanomas. Br J Cancer 88: 1403–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RH, Ward MR, Wu H, Medina CA, Brose MS, Volpe P, Nussen-Lee S, Haupt HM, Martin AM, Herlyn M, et al. 2004. Absence of BRAF mutations in UV-protected mucosal melanomas. J Med Genet 41: 270–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann KM, VanBrocklin MW, Staffend NA, Kitchen SM, Koo HM 2003. Mitogen-activated protein kinase pathway-dependent tumor-specific survival signaling in melanoma cells through inactivation of the proapoptotic protein bad. Cancer Res 63: 8330–8337 [PubMed] [Google Scholar]
- Eng C, Li FP, Abramson DH, Ellsworth RM, Wong FL, Goldman MB, Seddon J, Tarbell N, Boice JD Jr 1993. Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst 85: 1121–1128 [DOI] [PubMed] [Google Scholar]
- Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ 2004. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem 279: 19064–19073 [DOI] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M 2005. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 65: 9328–9337 [DOI] [PubMed] [Google Scholar]
- Flaherty K, Puzanov I, Sosman J, Kim K, Ribas A, McArthur G, Lee RJ, Grippo JF, Nolop K, Chapman P 2009. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol 27: 15s, Abstract 9000 [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher O, Easton D, Anderson K, Gilham C, Jay M, Peto J 2004. Lifetime risks of common cancers among retinoblastoma survivors. J Natl Cancer Inst 96: 357–363 [DOI] [PubMed] [Google Scholar]
- Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W, Frank MH 2005. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res 65: 4320–4333 [DOI] [PubMed] [Google Scholar]
- Fujimura Y, Ohno T, Siddique H, Lee L, Rao VN, Reddy ES 1996. The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene 12: 159–167 [PubMed] [Google Scholar]
- Fuse N, Yasumoto K, Suzuki H, Takahashi K, Shibahara S 1996. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem Biophys Res Commun 219: 702–707 [DOI] [PubMed] [Google Scholar]
- Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, Boyle P, Melchi CF 2005. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 41: 2040–2059 [DOI] [PubMed] [Google Scholar]
- Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C 2005. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res 18: 393–410 [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Rana S, Paterson H, Barford D, Marais R 2005. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell 20: 963–969 [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122 [DOI] [PubMed] [Google Scholar]
- Genander M, Frisen J 2010. Ephrins and Eph receptors in stem cells and cancer. Curr Opin Cell Biol 22: 611–616 [DOI] [PubMed] [Google Scholar]
- Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, Rominger CM, Erskine S, Fisher KE, Yang J, et al. 2011. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 17: 989–1000 [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Glinsky VV, Ivanova AB, Hueser CJ 1997. Apoptosis and metastasis: Increased apoptosis resistance of metastatic cancer cells is associated with the profound deficiency of apoptosis execution mechanisms. Cancer Lett 115: 185–193 [DOI] [PubMed] [Google Scholar]
- Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG 2006. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol 126: 154–160 [DOI] [PubMed] [Google Scholar]
- Granter SR, Weilbaecher KN, Quigley C, Fletcher CD, Fisher DE 2001. Clear cell sarcoma shows immunoreactivity for microphthalmia transcription factor: Further evidence for melanocytic differentiation. Mod Pathol 14: 6–9 [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, da Rocha Dias S, Marais R 2005. The role of B-RAF in melanoma. Cancer Metastasis Rev 24: 165–183 [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer VC, Karasarides M, Hayward R, Marais R 2007. Tumor necrosis factor-α blocks apoptosis in melanoma cells when BRAF signaling is inhibited. Cancer Res 67: 122–129 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM 2007. Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Guo J, Si L, Kong Y, Flaherty KT, Xu X, Zhu Y, Corless CL, Li L, Li H, Sheng X, et al. 2011. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 29: 2904–2909 [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA 2005. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 37: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R 1999. Melanoma cell autonomous growth: The Rb/E2F pathway. Cancer Metastasis Rev 18: 333–343 [DOI] [PubMed] [Google Scholar]
- Halaban R, Bohm M, Dotto P, Moellmann G, Cheng E, Zhang Y 1996. Growth regulatory proteins that repress differentiation markers in melanocytes also downregulate the transcription factor microphthalmia. J Invest Dermatol 106: 1266–1272 [DOI] [PubMed] [Google Scholar]
- Handolias D, Salemi R, Murray W, Tan A, Liu W, Viros A, Dobrovic A, Kelly J, McArthur GA 2010. Mutations in KIT occur at low frequency in melanomas arising from anatomical sites associated with chronic and intermittent sun exposure. Pigment Cell Melanoma Res 23: 210–215 [DOI] [PubMed] [Google Scholar]
- Harland M, Mistry S, Bishop DT, Bishop JA 2001. A deep intronic mutation in CDKN2A is associated with disease in a subset of melanoma pedigrees. Hum Mol Genet 10: 2679–2686 [DOI] [PubMed] [Google Scholar]
- Harland M, Taylor CF, Bass S, Churchman M, Randerson-Moor JA, Holland EA, Mann GJ, Bishop DT, Newton Bishop JA 2005a. Intronic sequence variants of the CDKN2A gene in melanoma pedigrees. Genes Chromosomes Cancer 43: 128–136 [DOI] [PubMed] [Google Scholar]
- Harland M, Taylor CF, Chambers PA, Kukalizch K, Randerson-Moor JA, Gruis NA, de Snoo FA, ter Huurne JA, Goldstein AM, Tucker MA, et al. 2005b. A mutation hotspot at the p14ARF splice site. Oncogene 24: 4604–4608 [DOI] [PubMed] [Google Scholar]
- Hauschild A, Agarwala SS, Trefzer U, Hogg D, Robert C, Hersey P, Eggermont A, Grabbe S, Gonzalez R, Gille J, et al. 2009. Results of a phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J Clin Oncol 27: 2823–2830 [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. 2010. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 8: 2770–2780 [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE 1998. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature 391: 298–301 [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE 2003. Molecular plasticity of human melanoma cells. Oncogene 22: 3070–3075 [DOI] [PubMed] [Google Scholar]
- Herath NI, Spanevello MD, Sabesan S, Newton T, Cummings M, Duffy S, Lincoln D, Boyle G, Parsons PG, Boyd AW 2006. Over-expression of Eph and ephrin genes in advanced ovarian cancer: ephrin gene expression correlates with shortened survival. BMC Cancer 6: 144 doi: 10.1186/1471-2407-6-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C, Lee Wu C, Evans G, Howell A, Elles RG, Jordan R, Sloan P, Read AP, Thakker N 2002. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet 11: 1273–1279 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA 2003. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res 63: 5198–5202 [PubMed] [Google Scholar]
- Hocker T, Tsao H 2007. Ultraviolet radiation and melanoma: A systematic review and analysis of reported sequence variants. Hum Mutat 28: 578–588 [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix–loop–helix-zipper protein. Cell 74: 395–404 [DOI] [PubMed] [Google Scholar]
- Hodi FS, Friedlander P, Corless CL, Heinrich MC, Mac Rae S, Kruse A, Jagannathan J, Van den Abbeele AD, Velazquez EF, Demetri GD, et al. 2008. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol 26: 2046–2051 [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Gray DC, Eby MT, Tien JY, Wong L, Bower J, Gogineni A, Zha J, Cole MJ, Stern HM, et al. 2006. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res 66: 999–1006 [DOI] [PubMed] [Google Scholar]
- Huang S, Luca M, Gutman M, McConkey DJ, Langley KE, Lyman SD, Bar-Eli M 1996. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene 13: 2339–2347 [PubMed] [Google Scholar]
- Huang TS, Rauth S, Das Gupta TK 1996. Overexpression of EGF receptor is associated with spontaneous metastases of a human melanoma cell line in nude mice. Anticancer Res 16: 3557–3563 [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH Jr, Tucker MA, Dracopoli NC 1994. Germline p16 mutations in familial melanoma. Nat Genet 8: 15–21 [DOI] [PubMed] [Google Scholar]
- Ilmonen S, Hernberg M, Pyrhonen S, Tarkkanen J, Asko-Seljavaara S 2005. Ki-67, Bcl-2 and p53 expression in primary and metastatic melanoma. Melanoma Res 15: 375–381 [DOI] [PubMed] [Google Scholar]
- Infante JR, Fecher LA, Nallapareddy S, Gordon MS, Flaherty KT, Cox DS, DeMarini DJ, Morris SR, Burris HA, Messersmith WA 2010. Safety and efficacy results from the first-in-human study of the oral MEK 1/2 inhibitor GSK1120212. J Clin Oncol 28: 15s, Abstract 2503 [Google Scholar]
- Ito A, Koma Y, Watabe K, Jippo T, Wakayama T, Iseki S, Kitamura Y 2004. Contribution of the SgIGSF adhesion molecule to survival of cultured mast cells in vivo. Biochem Biophys Res Commun 319: 200–206 [DOI] [PubMed] [Google Scholar]
- Ivanov VN, Hei TK 2005. Combined treatment with EGFR inhibitors and arsenite upregulated apoptosis in human EGFR-positive melanomas: A role of suppression of the PI3K–AKT pathway. Oncogene 24: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari M, Papp T, Kirchner S, Diener U, Henschler D, Burg G, Schiffmann D 1995. Analysis of ras mutations in human melanocytic lesions: Activation of the ras gene seems to be associated with the nodular type of human malignant melanoma. J Cancer Res Clin Oncol 121: 23–30 [DOI] [PubMed] [Google Scholar]
- James GL, Goldstein JL, Brown MS, Rawson TE, Somers TC, McDowell RS, Crowley CW, Lucas BK, Levinson AD, Marsters JC Jr 1993. Benzodiazepine peptidomimetics: Potent inhibitors of Ras farnesylation in animal cells. Science 260: 1937–1942 [DOI] [PubMed] [Google Scholar]
- Jane-Valbuena J, Widlund HR, Perner S, Johnson LA, Dibner AC, Lin WM, Baker AC, Nazarian RM, Vijayendran KG, Sellers WR, et al. 2010. An oncogenic role for ETV1 in melanoma. Cancer Res 70: 2075–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen B, Schlagbauer-Wadl H, Kahr H, Heere-Ress E, Mayer BX, Eichler H, Pehamberger H, Gana-Weisz M, Ben-David E, Kloog Y, et al. 1999. Novel Ras antagonist blocks human melanoma growth. Proc Natl Acad Sci 96: 14019–14024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Flaherty KT, Tsao H 2012a. Targeting the RAS pathway in melanoma. Trends Mol Med 18: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H 2012b. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol 132: 356–364 [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. 2010. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Rizvi NA, Ginsberg MS, Miller VA, Kris MG, Pao W, Riely GJ 2009. A phase II trial of salirasib in patients with stage IIIB/IV lung adenocarcinoma enriched for KRAS mutations. J Clin Oncol 27: 15s, Abstract 8012 [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Ruefli AA, Lowe SW 2002. Apoptosis: A link between cancer genetics and chemotherapy. Cell 108: 153–164 [DOI] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. 2010. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci 107: 14903–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS III, Johnson BE, Skolnick MH 1994. A cell cycle regulator potentially involved in genesis of many tumor types. Science 264: 436–440 [DOI] [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci 95: 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, Ogilvie L, Hedley D, Martin J, Marshall CJ, et al. 2004. B-RAF is a therapeutic target in melanoma. Oncogene 23: 6292–6298 [DOI] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. 2011. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefford RAAH, Brown MP, Millward M, Infante JR, Long GV, Ouellet D, Curtis M, Lebowitz PF, Falchook GS 2010. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol 28: 15s, Abstract 8503 [Google Scholar]
- Kenessey I, Keszthelyi M, Kramer Z, Berta J, Adam A, Dobos J, Mildner M, Flachner B, Cseh S, Barna G, et al. 2010. Inhibition of c-Met with the specific small molecule tyrosine kinase inhibitor SU11274 decreases growth and metastasis formation of experimental human melanoma. Curr Cancer Drug Targets 10: 332–342 [DOI] [PubMed] [Google Scholar]
- Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN 2001. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol 117: 294–300 [DOI] [PubMed] [Google Scholar]
- Kharas MG, Fruman DA 2005. ABL oncogenes and phosphoinositide 3-kinase: Mechanism of activation and downstream effectors. Cancer Res 65: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. 2006. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 125: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Kinch MS, Carles-Kinch K 2003. Overexpression and functional alterations of the EphA2 tyrosine kinase in cancer. Clin Exp Metastasis 20: 59–68 [DOI] [PubMed] [Google Scholar]
- Koh J, Enders GH, Dynlacht BD, Harlow E 1995. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 375: 506–510 [DOI] [PubMed] [Google Scholar]
- Kong Y, Si L, Zhu Y, Xu X, Corless CL, Flaherty KT, Li L, Li H, Sheng X, Cui C, et al. 2011. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res 17: 1684–1691 [DOI] [PubMed] [Google Scholar]
- Koprowski H, Herlyn M, Balaban G, Parmiter A, Ross A, Nowell P 1985. Expression of the receptor for epidermal growth factor correlates with increased dosage of chromosome 7 in malignant melanoma. Somat Cell Mol Genet 11: 297–302 [DOI] [PubMed] [Google Scholar]
- Kumar R, Angelini S, Snellman E, Hemminki K 2004. BRAF mutations are common somatic events in melanocytic nevi. J Invest Dermatol 122: 342–348 [DOI] [PubMed] [Google Scholar]
- Laheru D, Rudek M, Taylor G, Goldsweig H, Rajeshkumar NV, Linden S, Angenendt M, Le D, Donehower R, Jimeno A, et al. 2009. Integrated development of S-trans, trans-farnesylthiosalicyclic acid (FTS, salirasib) in advanced pancreatic cancer. J Clin Oncol 27: 15s, Abstract 4529. [Google Scholar]
- Leiter U, Schmid RM, Kaskel P, Peter RU, Krahn G 2000. Antiapoptotic bcl-2 and bcl-xL in advanced malignant melanoma. Arch Dermatol Res 292: 225–232 [DOI] [PubMed] [Google Scholar]
- Li G, Schaider H, Satyamoorthy K, Hanakawa Y, Hashimoto K, Herlyn M 2001. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene 20: 8125–8135 [DOI] [PubMed] [Google Scholar]
- Libra M, Malaponte G, Navolanic PM, Gangemi P, Bevelacqua V, Proietti L, Bruni B, Stivala F, Mazzarino MC, Travali S, et al. 2005. Analysis of BRAF mutation in primary and metastatic melanoma. Cell Cycle 4: 1382–1384 [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. 2008. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res 68: 664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loercher AE, Tank EM, Delston RB, Harbour JW 2005. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol 168: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzky J, Bauer J, Bastian BC 2008. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res 21: 492–493 [DOI] [PubMed] [Google Scholar]
- Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, Schmid H, Holland EA, Duffy DL, Zhang M, et al. 2011. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet 43: 1114–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado JL, Fridlyand J, Patel H, Jain AN, Busam K, Kageshita T, Ono T, Albertson DG, Pinkel D, Bastian BC 2003. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst 95: 1878–1890 [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sankar U, Han J, Ostrowski MC 2002. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-κB ligand signaling. J Biol Chem 277: 11077–11083 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Stock W, Dai G, Klisovic RB, Liu S, Klisovic MI, Blum W, Kefauver C, Sher DA, Green M, et al. 2005. Phase I study of oblimersen sodium, an antisense to Bcl-2, in untreated older patients with acute myeloid leukemia: Pharmacokinetics, pharmacodynamics, and clinical activity. J Clin Oncol 23: 3404–3411 [DOI] [PubMed] [Google Scholar]
- Margolin KA, Longmate J, Baratta T, Synold T, Weber J, Gajewski T, Quirt I, Christensen S, Doroshow JH 2004. CCI-779 in metastatic melanoma: A phase II trial of the California Cancer Consortium. ASCO Annual Meeting. Abstract no. 7523 [DOI] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109: 707–718 [DOI] [PubMed] [Google Scholar]
- McGill GG, Haq R, Nishimura EK, Fisher DE 2006. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem 281: 10365–10373 [DOI] [PubMed] [Google Scholar]
- Mellman I, Coukos G, Dranoff G 2011. Cancer immunotherapy comes of age. Nature 480: 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges CW, McCance DJ 2008. Constitutive activation of the Raf–MAPK pathway causes negative feedback inhibition of Ras–PI3K–AKT and cellular arrest through the EphA2 receptor. Oncogene 27: 2934–2940 [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436: 720–724 [DOI] [PubMed] [Google Scholar]
- Mikhail M, Velazquez E, Shapiro R, Berman R, Pavlick A, Sorhaindo L, Spira J, Mir C, Panageas KS, Polsky D, et al. 2005. PTEN expression in melanoma: Relationship with patient survival, Bcl-2 expression, and proliferation. Clin Cancer Res 11: 5153–5157 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Tsao H 2010. New insights into pigmentary pathways and skin cancer. Br J Dermatol 162: 22–28 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE 2004. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res 64: 509–516 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE 2005. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem 280: 146–155 [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Kato H, Fukuchi M, Nakajima M, Kuwano H 2003. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int J Cancer 103: 657–663 [DOI] [PubMed] [Google Scholar]
- Molven A, Grimstvedt MB, Steine SJ, Harland M, Avril MF, Hayward NK, Akslen LA 2005. A large Norwegian family with inherited malignant melanoma, multiple atypical nevi, and CDK4 mutation. Genes Chromosomes Cancer 44: 10–18 [DOI] [PubMed] [Google Scholar]
- Montone KT, van Belle P, Elenitsas R, Elder DE 1997. Proto-oncogene c-kit expression in malignant melanoma: Protein loss with tumor progression. Mod Pathol 10: 939–944 [PubMed] [Google Scholar]
- Morii E, Oboki K, Ishihara K, Jippo T, Hirano T, Kitamura Y 2004. Roles of MITF for development of mast cells in mice: Effects on both precursors and tissue environments. Blood 104: 1656–1661 [DOI] [PubMed] [Google Scholar]
- Murakami M, Ikeda T, Saito T, Ogawa K, Nishino Y, Nakaya K, Funaba M 2006. Transcriptional regulation of plasminogen activator inhibitor-1 by transforming growth factor-β, activin A and microphthalmia-associated transcription factor. Cell Signal 18: 256–265 [DOI] [PubMed] [Google Scholar]
- Muthusamy V, Hobbs C, Nogueira C, Cordon-Cardo C, McKee PH, Chin L, Bosenberg MW 2006. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer 45: 447–454 [DOI] [PubMed] [Google Scholar]
- Namiki T, Yanagawa S, Izumo T, Ishikawa M, Tachibana M, Kawakami Y, Yokozeki H, Nishioka K, Kaneko Y 2005. Genomic alterations in primary cutaneous melanomas detected by metaphase comparative genomic hybridization with laser capture or manual microdissection: 6p gains may predict poor outcome. Cancer Genet Cytogenet 157: 1–11 [DOI] [PubMed] [Google Scholar]
- Natali PG, Nicotra MR, Di Renzo MF, Prat M, Bigotti A, Cavaliere R, Comoglio PM 1993. Expression of the c-Met/HGF receptor in human melanocytic neoplasms: Demonstration of the relationship to malignant melanoma tumour progression. Br J Cancer 68: 746–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, et al. 2006. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet 38: 294–296 [DOI] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, et al. 2011. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 44: 133–139 [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE 2005. Mechanisms of hair graying: Incomplete melanocyte stem cell maintenance in the niche. Science 307: 720–724 [DOI] [PubMed] [Google Scholar]
- Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA 1994. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368: 753–756 [DOI] [PubMed] [Google Scholar]
- O'Brien SM, Cunningham CC, Golenkov AK, Turkina AG, Novick SC, Rai KR 2005. Phase I to II multicenter study of oblimersen sodium, a Bcl-2 antisense oligonucleotide, in patients with advanced chronic lymphocytic leukemia. J Clin Oncol 23: 7697–7702 [DOI] [PubMed] [Google Scholar]
- Omholt K, Krockel D, Ringborg U, Hansson J 2006. Mutations of PIK3CA are rare in cutaneous melanoma. Melanoma Res 16: 197–200 [DOI] [PubMed] [Google Scholar]
- Papp T, Pemsel H, Zimmermann R, Bastrop R, Weiss DG, Schiffmann D 1999. Mutational analysis of the N-ras, p53, p16INK4a, CDK4, and MC1R genes in human congenital melanocytic naevi. J Med Genet 36: 610–614 [PMC free article] [PubMed] [Google Scholar]
- Patel SP, Kim KB, Papadopoulos NE, Hwu WJ, Hwu P, Prieto VG, Bar-Eli M, Zigler M, Dobroff A, Bronstein Y, et al. 2011. A phase II study of gefitinib in patients with metastatic melanoma. Melanoma Res 21: 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. 2005. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol 15: 249–254 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plettenberg A, Ballaun C, Pammer J, Mildner M, Strunk D, Weninger W, Tschachler E 1995. Human melanocytes and melanoma cells constitutively express the Bcl-2 proto-oncogene in situ and in cell culture. Am J Pathol 146: 651–659 [PMC free article] [PubMed] [Google Scholar]
- Pollock PM, Walker GJ, Glendening JM, Que Noy T, Bloch NC, Fountain JW, Hayward NK 2002. PTEN inactivation is rare in melanoma tumours but occurs frequently in melanoma cell lines. Melanoma Res 12: 565–575 [DOI] [PubMed] [Google Scholar]
- Pollock PM, Cohen-Solal K, Sood R, Namkoong J, Martino JJ, Koganti A, Zhu H, Robbins C, Makalowska I, Shin SS, et al. 2003a. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat Genet 34: 108–112 [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, et al. 2003b. High frequency of BRAF mutations in nevi. Nat Genet 33: 19–20 [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R 2012. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483: 100–103 [DOI] [PubMed] [Google Scholar]
- Price ER, Fisher DE 2001. Sensorineural deafness and pigmentation genes: Melanocytes and the Mitf transcriptional network. Neuron 30: 15–18 [DOI] [PubMed] [Google Scholar]
- Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem 273: 17983–17986 [DOI] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y 2009. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet 41: 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V, Gartner J, Jiang J, Cherukuri PF, Molinolo A, et al. 2011. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet 43: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik EV, Vogt PK 2010. Therapeutic targeting of Myc. Genes Cancer 1: 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ 2008. Efficient tumour formation by single human melanoma cells. Nature 456: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, Fargnoli MC 2008. MC1R variants, melanoma and red hair color phenotype: A meta-analysis. Int J Cancer 122: 2753–2760 [DOI] [PubMed] [Google Scholar]
- Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M 2009. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461: 542–545 [DOI] [PubMed] [Google Scholar]
- Rimoldi D, Salvi S, Lienard D, Lejeune FJ, Speiser D, Zografos L, Cerottini JC 2003. Lack of BRAF mutations in uveal melanoma. Cancer Res 63: 5712–5715 [PubMed] [Google Scholar]
- Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, Hamid O, Varterasian M, Asbury P, Kaldjian EP, et al. 2004. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol 22: 4456–4462 [DOI] [PubMed] [Google Scholar]
- Rivera RS, Nagatsuka H, Gunduz M, Cengiz B, Gunduz E, Siar CH, Tsujigiwa H, Tamamura R, Han KN, Nagai N 2008. C-kit protein expression correlated with activating mutations in KIT gene in oral mucosal melanoma. Virchows Arch 452: 27–32 [DOI] [PubMed] [Google Scholar]
- Robertson GP 2005. Functional and therapeutic significance of Akt deregulation in malignant melanoma. Cancer Metastasis Rev 24: 273–285 [DOI] [PubMed] [Google Scholar]
- Robertson GP, Furnari FB, Miele ME, Glendening MJ, Welch DR, Fountain JW, Lugo TG, Huang HJ, Cavenee WK 1998. In vitro loss of heterozygosity targets the PTEN/MMAC1 gene in melanoma. Proc Natl Acad Sci 95: 9418–9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA 2006. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311: 1287–1290 [DOI] [PubMed] [Google Scholar]
- Ross P, Hall L, Haff LA 2000. Quantitative approach to single-nucleotide polymorphism analysis using MALDI-TOF mass spectrometry. Biotechniques 29: 620–629 [DOI] [PubMed] [Google Scholar]
- Sabatini DM 2006. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer 6: 729–734 [DOI] [PubMed] [Google Scholar]
- Saldanha G, Purnell D, Fletcher A, Potter L, Gillies A, Pringle JH 2004. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int J Cancer 111: 705–710 [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I 2002. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet 11: 3231–3236 [DOI] [PubMed] [Google Scholar]
- Sanders BM, Jay M, Draper GJ, Roberts EM 1989. Non-ocular cancer in relatives of retinoblastoma patients. Br J Cancer 60: 358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. 2008. Identification of cells initiating human melanomas. Nature 451: 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicher N, Paulitschke V, Swoboda A, Kunstfeld R, Loewe R, Pilarski P, Pehamberger H, Hoeller C 2009. Erlotinib and bevacizumab have synergistic activity against melanoma. Clin Cancer Res 15: 3495–3502 [DOI] [PubMed] [Google Scholar]
- Scott KL, Chin L 2010. Signaling from the Golgi: Mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clin Cancer Res 16: 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. 2009. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459: 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seftor EA, Meltzer PS, Schatteman GC, Gruman LM, Hess AR, Kirschmann DA, Seftor RE, Hendrix MJ 2002. Expression of multiple molecular phenotypes by aggressive melanoma tumor cells: Role in vasculogenic mimicry. Crit Rev Oncol Hematol 44: 17–27 [DOI] [PubMed] [Google Scholar]
- Selzer E, Schlagbauer-Wadl H, Okamoto I, Pehamberger H, Potter R, Jansen B 1998. Expression of Bcl-2 family members in human melanocytes, in melanoma metastases and in melanoma cell lines. Melanoma Res 8: 197–203 [DOI] [PubMed] [Google Scholar]
- Serrano M, Hannon GJ, Beach D 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366: 704–707 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27–37 [DOI] [PubMed] [Google Scholar]
- Sharma P, Wagner K, Wolchok JD, Allison JP 2011. Novel cancer immunotherapy agents with survival benefit: Recent successes and next steps. Nat Rev Cancer 11: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA 2005. Cancer: Crime and punishment. Nature 436: 636–637 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Kannan K, Xu J, Bosenberg MW, Chin L 2003. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene 22: 5055–5059 [DOI] [PubMed] [Google Scholar]
- Shen SS, Zhang PS, Eton O, Prieto VG 2003. Analysis of protein tyrosine kinase expression in melanocytic lesions by tissue array. J Cutan Pathol 30: 539–547 [DOI] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. 2012. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun 3: 724 doi: 10.1038/ncomms1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki M, Fujimoto A, Morton DL, Hoon DS 2004. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res 10: 1753–1757 [DOI] [PubMed] [Google Scholar]
- Skvara H, Thallinger C, Wacheck V, Monia BP, Pehamberger H, Jansen B, Selzer E 2005. Mcl-1 blocks radiation-induced apoptosis and inhibits clonogenic cell death. Anticancer Res 25: 2697–2703 [PubMed] [Google Scholar]
- Slipicevic A, Holm R, Nguyen MT, Bohler PJ, Davidson B, Florenes VA 2005. Expression of activated Akt and PTEN in malignant melanomas: Relationship with clinical outcome. Am J Clin Pathol 124: 528–536 [DOI] [PubMed] [Google Scholar]
- Smalley KS, Eisen TG 2002. Farnesyl thiosalicylic acid inhibits the growth of melanoma cells through a combination of cytostatic and pro-apoptotic effects. Int J Cancer 98: 514–522 [DOI] [PubMed] [Google Scholar]
- Smalley KS, Flaherty KT 2009. Integrating BRAF/MEK inhibitors into combination therapy for melanoma. Br J Cancer 100: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley KS, Contractor R, Nguyen TK, Xiao M, Edwards R, Muthusamy V, King AJ, Flaherty KT, Bosenberg M, Herlyn M, et al. 2008. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res 68: 5743–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Lowe SW 2003. Apoptosis and melanoma chemoresistance. Oncogene 22: 3138–3151 [DOI] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. 2006. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, Stoppa-Lyonnet D, Benard J, Bressac-de Paillerets B 1998. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet 7: 209–216 [DOI] [PubMed] [Google Scholar]
- Stahl JM, Cheung M, Sharma A, Trivedi NR, Shanmugam S, Robertson GP 2003. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res 63: 2881–2890 [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP 2004. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res 64: 7002–7010 [DOI] [PubMed] [Google Scholar]
- Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, Chow D, Sereduk C, Niemi NM, Tang N, et al. 2011. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet 44: 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA 2002. Mitf and Tfe3, two members of the Mitf–Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci 99: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AL, Mhashilkar AM, Yang XH, Ekmekcioglu S, Saito Y, Sieger K, Schrock R, Onishi E, Swanson X, Mumm JB, et al. 2002. PI3 kinase blockade by Ad-PTEN inhibits invasion and induces apoptosis in RGP and metastatic melanoma cells. Mol Med 8: 451–461 [PMC free article] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J 17: 5001–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C, et al. 2011. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. 2012. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulman EP, Tang XX, Allen C, Biegel JA, Pleasure DE, Brodeur GM, Ikegaki N 1997. ECK, a human EPH-related gene, maps to 1p36.1, a common region of alteration in human cancers. Genomics 40: 371–374 [DOI] [PubMed] [Google Scholar]
- Takeda K, Takemoto C, Kobayashi I, Watanabe A, Nobukuni Y, Fisher DE, Tachibana M 2000. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Hum Mol Genet 9: 125–132 [DOI] [PubMed] [Google Scholar]
- Takeda K, Yokoyama S, Aburatani H, Masuda T, Han F, Yoshizawa M, Yamaki N, Yamamoto H, Eguchi N, Urade Y, et al. 2006. Lipocalin-type prostaglandin D synthase as a melanocyte marker regulated by MITF. Biochem Biophys Res Commun 339: 1098–1106 [DOI] [PubMed] [Google Scholar]
- Tang Y, Eng C 2006. PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res 66: 736–742 [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F 2003. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 3: 233–245 [DOI] [PubMed] [Google Scholar]
- Tolcher AW, Chi K, Kuhn J, Gleave M, Patnaik A, Takimoto C, Schwartz G, Thompson I, Berg K, D'Aloisio S, et al. 2005. A phase II, pharmacokinetic, and biological correlative study of oblimersen sodium and docetaxel in patients with hormone-refractory prostate cancer. Clin Cancer Res 11: 3854–3861 [DOI] [PubMed] [Google Scholar]
- Tolkacheva T, Boddapati M, Sanfiz A, Tsuchida K, Kimmelman AC, Chan AM 2001. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res 61: 4985–4989 [PubMed] [Google Scholar]
- Topcu-Yilmaz P, Kiratli H, Saglam A, Soylemezoglu F, Hascelik G 2010. Correlation of clinicopathological parameters with HGF, c-Met, EGFR, and IGF-1R expression in uveal melanoma. Melanoma Res 20: 126–132 [DOI] [PubMed] [Google Scholar]
- Tron VA, Krajewski S, Klein-Parker H, Li G, Ho VC, Reed JC 1995. Immunohistochemical analysis of Bcl-2 protein regulation in cutaneous melanoma. Am J Pathol 146: 643–650 [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al. 2008. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci 105: 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Benoit E, Sober AJ, Thiele C, Haluska FG 1998a. Novel mutations in the p16/CDKN2A binding region of the cyclin-dependent kinase-4 gene. Cancer Res 58: 109–113 [PubMed] [Google Scholar]
- Tsao H, Zhang X, Benoit E, Haluska FG 1998b. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene 16: 3397–3402 [DOI] [PubMed] [Google Scholar]
- Tsao H, Mihm MC Jr, Sheehan C 2003. PTEN expression in normal skin, acquired melanocytic nevi, and cutaneous melanoma. J Am Acad Dermatol 49: 865–872 [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG 2004. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol 122: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, Muthusamy V, Bosenberg M, Krauthammer M, Halaban R, Stern DF 2011. Phosphoproteomic screen identifies potential therapeutic targets in melanoma. Mol Cancer Res 9: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udart M, Utikal J, Krahn GM, Peter RU 2001. Chromosome 7 aneusomy. A marker for metastatic melanoma? Expression of the epidermal growth factor receptor gene and chromosome 7 aneusomy in nevi, primary malignant melanomas and metastases. Neoplasia 3: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayakumar D, Zhang G, Ji Z, Njauw CN, Mroz P, Tsao H 2011. Epha2 is a critical oncogene in melanoma. Oncogene 30: 4921–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Sakurai H, Tsunoda S, Choo MK, Matsuo M, Koizumi K, Saiki I 2008. Heregulin-induced activation of ErbB3 by EGFR tyrosine kinase activity promotes tumor growth and metastasis in melanoma cells. Int J Cancer 123: 340–347 [DOI] [PubMed] [Google Scholar]
- Uribe P, Wistuba II, Gonzalez S 2003. BRAF mutation: A frequent event in benign, atypical, and malignant melanocytic lesions of the skin. Am J Dermatopathol 25: 365–370 [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 11: 328–330 [DOI] [PubMed] [Google Scholar]
- VanBrocklin MW, Robinson JP, Lastwika KJ, Khoury JD, Holmen SL 2010. Targeted delivery of NRASQ61R and Cre-recombinase to post-natal melanocytes induces melanoma in Ink4a/Arflox/lox mice. Pigment Cell Melanoma Res 23: 531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Woude GF, Jeffers M, Cortner J, Alvord G, Tsarfaty I, Resau J 1997. Met-HGF/SF: Tumorigenesis, invasion and metastasis. Ciba Found Symp 212: 119–130 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS 2004. Effects of G-protein mutations on skin color. Nat Genet 36: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC 2009. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, et al. 2010. Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Grossman SR, Takahashi Y, Rokas MV, Nakamura N, Sellers WR 2001. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem 276: 48627–48630 [DOI] [PubMed] [Google Scholar]
- Vetrini F, Auricchio A, Du J, Angeletti B, Fisher DE, Ballabio A, Marigo V 2004. The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: Link between melanin synthesis and melanosome biogenesis. Mol Cell Biol 24: 6550–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J 2010. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18: 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Willebrand M, Kohler K, Alanko T, Laiho M, Saksela O 2005. FGF-2 blocks TGF-β1-mediated suppression of Bcl-2 in normal melanocytes. Exp Dermatol 14: 202–208 [DOI] [PubMed] [Google Scholar]
- Wada T, Penninger JM 2004. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23: 2838–2849 [DOI] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 29: 3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HT, Choi B, Tang MS 2010. Melanocytes are deficient in repair of oxidative DNA damage and UV-induced photoproducts. Proc Natl Acad Sci 107: 12180–12185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Hengge UR, Urbanik D, Markwart A, Mirmohammadsaegh A, Reichel MB, Wittekind C, Wiedemann P, Tannapfel A 2003. Absence of mutations of the BRAF gene and constitutive activation of extracellular-regulated kinase in malignant melanomas of the uvea. Lab Invest 83: 1771–1776 [DOI] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, et al. 2011. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 43: 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz B, Giehl K, Gana-Weisz M, Egozi Y, Ben-Baruch G, Marciano D, Gierschik P, Kloog Y 1999. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene 18: 2579–2588 [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D, Springer CJ, Marais R 2004. V599EB-RAF is an oncogene in melanocytes. Cancer Res 64: 2338–2342 [DOI] [PubMed] [Google Scholar]
- Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, Affolter A, Nourry A, Niculescu-Duvaz D, Springer C, et al. 2010. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med 2: 35ra41 doi: 10.1126/scitranslmed.3000758 [DOI] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK 1997. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272: 14459–14464 [DOI] [PubMed] [Google Scholar]
- Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al. 2004. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 64: 7099–7109 [DOI] [PubMed] [Google Scholar]
- Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde KH, Beach D 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 14: 301–312 [PMC free article] [PubMed] [Google Scholar]
- Wu H, Goel V, Haluska FG 2003. PTEN signaling pathways in melanoma. Oncogene 22: 3113–3122 [DOI] [PubMed] [Google Scholar]
- Wu X, Zhou J, Rogers AM, Janne PA, Benedettini E, Loda M, Hodi FS 2012. c-Met, epidermal growth factor receptor, and insulin-like growth factor-1 receptor are important for growth in uveal melanoma and independently contribute to migration and metastatic potential. Melanoma Res 22: 123–132 [DOI] [PubMed] [Google Scholar]
- Xu W, Gong L, Haddad MM, Bischof O, Campisi J, Yeh ET, Medrano EE 2000. Regulation of microphthalmia-associated transcription factor MITF protein levels by association with the ubiquitin-conjugating enzyme hUBC9. Exp Cell Res 255: 135–143 [DOI] [PubMed] [Google Scholar]
- Xu G, Zhang W, Bertram P, Zheng XF, McLeod H 2004. Pharmacogenomic profiling of the PI3K/PTEN–AKT-mTOR pathway in common human tumors. Int J Oncol 24: 893–900 [PubMed] [Google Scholar]
- Yang G, Rajadurai A, Tsao H 2005. Recurrent patterns of dual RB and p53 pathway inactivation in melanoma. J Invest Dermatol 125: 1242–1251 [DOI] [PubMed] [Google Scholar]
- Yang G, Curley D, Bosenberg MW, Tsao H 2007. Loss of xeroderma pigmentosum C (Xpc) enhances melanoma photocarcinogenesis in Ink4a–Arf-deficient mice. Cancer Res 67: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S 1994. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol 14: 8058–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi AS, Palmedo G, Flaig MJ, Puchta U, Reckwerth A, Rutten A, Mentzel T, Hugel H, Hantschke M, Schmid-Wendtner MH, et al. 2003. Mutations of the BRAF gene in benign and malignant melanocytic lesions. J Invest Dermatol 121: 1160–1162 [DOI] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A 2005. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell 20: 539–550 [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, et al. 2011. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 480: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Barker MA, Simms LA, Walsh MD, Biden KG, Buchanan D, Buttenshaw R, Whitehall VL, Arnold S, Jackson L, et al. 2005. Evidence for BRAF mutation and variable levels of microsatellite instability in a syndrome of familial colorectal cancer. Clin Gastroenterol Hepatol 3: 254–263 [DOI] [PubMed] [Google Scholar]
- Zhai S, Yaar M, Doyle SM, Gilchrest BA 1996. Nerve growth factor rescues pigment cells from ultraviolet-induced apoptosis by upregulating BCL-2 levels. Exp Cell Res 224: 335–343 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF–INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734 [DOI] [PubMed] [Google Scholar]
- Zhang XD, Borrow JM, Zhang XY, Nguyen T, Hersey P 2003. Activation of ERK1/2 protects melanoma cells from TRAIL-induced apoptosis by inhibiting Smac/DIABLO release from mitochondria. Oncogene 22: 2869–2881 [DOI] [PubMed] [Google Scholar]
- Zhang G, Njauw CN, Park JM, Naruse C, Asano M, Tsao H 2008. EphA2 is an essential mediator of UV radiation-induced apoptosis. Cancer Res 68: 1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE 2011. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun 2: 414 doi: 10.1038/ncomms1421 [DOI] [PubMed] [Google Scholar]
- Zhiqi S, Soltani MH, Bhat KM, Sangha N, Fang D, Hunter JJ, Setaluri V 2004. Human melastatin 1 (TRPM1) is regulated by MITF and produces multiple polypeptide isoforms in melanocytes and melanoma. Melanoma Res 14: 509–516 [DOI] [PubMed] [Google Scholar]
- Zhu YI, Fitzpatrick JE 2006. Expression of c-kit (CD117) in Spitz nevus and malignant melanoma. J Cutan Pathol 33: 33–37 [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward NK, Dracopoli NC 1996. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 12: 97–99 [DOI] [PubMed] [Google Scholar]