Telomere replication is problematic, as the ends have to be appropriately processed to produce 3′ overhangs. Although an important aspect of telomere biology, little is known about the relevant steps. Wright and colleagues develop new approaches to monitor the sequential steps of end processing. They demonstrate that the rate of telomere shortening is determined by the final RNA primer position during lagging strand replication. The work provides detailed insight into the end replication problem and identifies the temporally separated steps that generate mature overhangs.
Keywords: telomere overhangs, DNA replication, Okazaki fragment RNA primer, aging, cancer
Abstract
Telomere overhangs are essential for telomere end protection and telomerase extension, but how telomere overhangs are generated is unknown. Leading daughter strands synthesized by conventional semiconservation DNA replication are initially blunt, while lagging daughter strands are shorter by at least the size of the final RNA primer, which is thought to be located at extreme chromosome ends. We developed a variety of new approaches to define the steps in the processing of these overhangs. We show that the final lagging RNA primer is not terminal but is randomly positioned ∼70–100 nucleotides from the ends and is not removed for more than an hour. This identifies an important intrinsic step in replicative aging. Telomeric termini are processed in two distinct phases. During the early phase, which occupies 1–2 h following replication of the duplex telomeric DNA, several steps occur on both leading and lagging daughters. Leading telomere processing remains incomplete until late S/G2, when the C-terminal nucleotide is specified—referred to as the late phase. These observations suggest the presence of previously unsuspected complexes and signaling events required for the replication of the ends of human chromosomes.
Telomeres are nucleoprotein structures at the ends of linear chromosomes that are essential for preventing chromosomal end-to-end fusions and degradation. Adult human telomeres contain ∼5–15 kb of hexameric DNA repeats (5′-TTAGGG-3′) ending with 3′ single-stranded G-rich overhangs ranging in size from ∼12 to 300 nucleotides (nt) (Makarov et al. 1997; Wright et al. 1997; Stewart et al. 2003; Zhao et al. 2008; Dai et al. 2010). In cancer and stem cells, this overhang provides the single-stranded substrate required for telomerase extension. This G overhang can be inserted into homologous double-stranded regions, resulting in a lasso-like telomere loop (t-loop) structure thought to prevent chromosome ends from being recognized as double-stranded breaks (Griffith et al. 1999).
The mechanism generating these overhangs has been sought for many years. Many DNA damage signaling factors such as ATM, ATR, the MRN complex, and their homologs in other species (Chai et al. 2006b; Goudsouzian et al. 2006; Denchi and de Lange 2007; Deng et al. 2009; Dimitrova and de Lange 2009); helicases (Tomita et al. 2004; Bonetti et al. 2009; Li et al. 2009); telomere-associated proteins (Garvik et al. 1995; Nugent et al. 1996; Grandin et al. 1997, 2001; van Steensel et al. 1998; Churikov et al. 2006; Wu et al. 2006; Song et al. 2008; Ballal et al. 2009; Price et al. 2010); and nucleases (Tomita et al. 2003; Tom and Greider 2010; Wu et al. 2010) have been found to affect overhang length. However, it is very difficult to interpret the mechanism of action of these factors without knowing the specific steps involved in overhang generation. Some of these factors may affect overhang length by influencing the action of telomerase (Chai et al. 2006b; Zhao et al. 2009), while others may affect additional overhang processing steps. Chromatin immunoprecipitation (ChIP) analysis has shown that the association of a variety of proteins at telomeres change at S/G2 in normal diploid cells (Verdun et al. 2005; Verdun and Karlseder 2006), suggesting that some processing events might occur at S/G2. However, the number, nature, and timing of the steps involved in producing an appropriately sized G-rich 3′ overhang following replication of the duplex telomeric DNA in the absence of telomerase remain unknown. How overhang maturation is coupled with duplex telomeric DNA replication also remains undefined. In particular, the differences in the biogenesis of leading versus lagging overhangs have not been characterized. It is generally assumed that the nuclease activity of C-strand resection is tightly associated with the replication machinery and is dependent on the alteration of telomere chromatin during replication fork progression.
The original hypothesis of the end replication problem by Watson (1972) and Olovnikov (1973) ∼40 years ago has traditionally been formulated as the inability to copy the 10–14 nt at the end of the chromosome after the final lagging Okazaki fragment RNA primer has been removed. This has led to the model presented in most modern textbooks: The final primer of Okazaki fragment synthesis is positioned at the very end of the chromosome. Short G overhangs have been observed in several organisms; e.g., ∼14 nt in ciliates (Klobutcher et al. 1981; Jacob et al. 2001) and ∼12–14 nt in Saccharomyces cerevisiae (Larrivee et al. 2004). This is approximately the size of the RNA primer laid down by polymerase α during lagging strand synthesis (Kunkel and Burgers 2008; Nick McElhinny et al. 2008). However, it is also possible that this final overhang size of 12–14 nt in S. cerevisiae (Larrivee et al. 2004) may not actually represent the site of the final Okazaki priming event because both leading and lagging strands have a transient >30-nt overhang that is later filled in by an unknown mechanism to generate the final overhang (Wellinger et al. 1992, 1993, 1996). Although in some organisms it may be possible for the conventional replication machinery to position the final lagging RNA primer at the very terminus, the presence and location of the final RNA primer has not yet been directly demonstrated in any organism.
In the absence of telomerase, the size of the overhangs represents a reduction in the length of C strands. In the absence of damage to telomeres, this determines the rate of telomere shortening and the amount that telomerase would have to extend to compensate and maintain telomere length (Zhao et al. 2009). In yeast, the rate of telomere shortening in the absence of telomerase (estimated at 3–5 base pairs [bp] per division) (Marcand et al. 1999) is close to what would be expected based on the observed 12- to 14-nt overhangs (Larrivee et al. 2004). Telomerase only adds ∼44 nt to ∼7% of the ends (Teixeira et al. 2004), for an average addition that matches the shortening rate. The rate of telomere shortening in cultured telomerase-negative human cells is 50–100 bp per division, which is also close to what would be expected when the average overhang in telomerase-negative cells is ∼60–70 nt (∼30 nt for the daughters of leading strand semiconservative replication [“leading daughters”] and ∼100 nt for the daughters of lagging strand semiconservative replication [“lagging daughters”]) (Zhao et al. 2008). In human cancer cells, telomerase adds ∼60 nt to every chromosome (Zhao et al. 2009), again closely matching the shortening rate. There are two alternate mechanisms thought to explain why the length of telomere loss per cell division in human cells is considerably longer than a simple result of the end replication problem upon removal of a terminal RNA primer. The first assumes that the RNA primer is positioned at the very terminal end and that both leading and lagging daughters are processed by nuclease resection on the C strands. An alternative model would be that the initial lagging overhang reflects the position of the final Okazaki RNA priming event as the replication complex runs off its linear template. The removal of the RNA primer could then generate an initial overhang that is almost the size of the fully processed overhang. Meanwhile, the initially blunt leading daughter strands resulting from semiconservative DNA replication would be processed by nucleases or helicases to generate mature overhangs (Sfeir et al. 2005). Uncovering the steps that regulate the processing of human telomeric overhangs is thus fundamental in understanding the roles of candidate proteins involved and potentially manipulating rates of replicative aging and telomerase action.
In the present study, combinations of modified and newly developed biochemical assays were designed to monitor the dynamics of overhang sizes, RNA primers on lagging daughters, and the C-terminal nucleotides during S phase. We found that lagging daughters had an almost mature overhang size very soon after replication of the duplex telomeric DNA and were capped by a terminal RNA primer for ∼1 h following replication prior to its removal, leaving a specified C-terminal nucleotide. In contrast, leading daughters only developed a mature size 1–2 h after duplex telomere replication and did not acquire a specific C-terminal sequence until S/G2. We present a model characterizing overhang processing in two distinct phases to summarize the complex series of steps by which human linear chromosomal ends are replicated.
Results
Mature overhang sizes are present by 2 h after replication
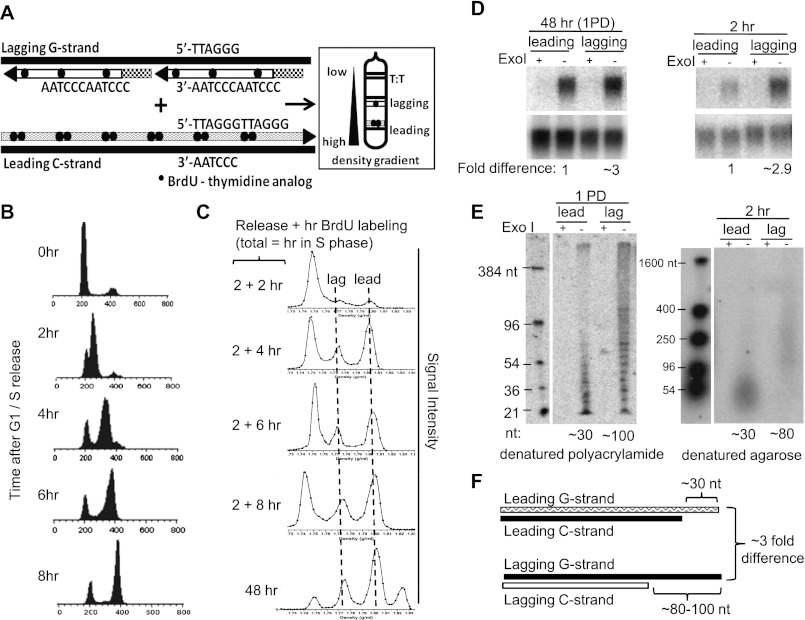
In order to establish the window within which mature overhang sizes were established, we examined the end structure in telomeres labeled for increasing times with 5-bromodeoxyuridine (BrdU). Normal human foreskin fibroblast BJ cells were synchronized at the G1/S interface with an efficiency of 65%–75% (Fig. 1B) and with an S phase of ∼6–8 h. Telomeres in BJ cells replicate throughout S phase, but there is a delay of ∼2 h before significant numbers of telomeres begin to replicate (Wright et al. 1999). Cells were first released into S phase in fresh medium for 2 h before treating with pulses of 100 μM BrdU (Fig. 1A). This procedure maximized the amount of BrdU incorporated into the newly synthesized telomeres during short periods. BrdU was differentially incorporated into daughter telomeres such that two BrdU molecules were incorporated per leading repeat (TTAGGG) versus only one BrdU per complementary lagging repeat (CCCTAA) (Fig. 1A). Leading and lagging daughter strands synthesized by conventional semiconservation DNA replication were then separated on cesium chloride (CsCl) gradients, where leading daughters banded at a heavier density than lagging daughters, with unreplicated telomeres having the lightest density. Aliquots of the gradient fractions were hybridized with a telomere-specific probe on a slot blot to differentiate leading versus lagging daughters from unreplicated telomeres. Cesium centrifugation of samples labeled for 2, 4, 6, and 8 h showed distinct leading versus lagging peaks, as expected (Fig. 1C). Cells labeled for one population doubling ([PD], ∼48 h) served as a positive control for fully processed overhang sizes. The additional fourth peak of the highest density observed at 48 h represented cells that had entered the second cell cycle and had incorporated BrdU on both leading and lagging daughter strands (Fig. 1C). The fractions corresponding to leading versus lagging telomeres were then pooled, purified, and analyzed.
Figure 1.
Overhangs approach mature sizes within 2 h of replication in BJ fibroblasts. (A) Design of the purification of leading versus lagging daughters using CsCl centrifugation. Leading strand replication produces the highest density, followed by lagging, while unreplicated DNA has the lowest density. (B,C) FACS analysis of BJ fibroblasts synchronized at G1/S and every 2 h upon release until S/G2 (B) and the corresponding separation of leading versus lagging daughters (C). Forty-eight hours represents the one PD control. (D) In-gel hybridization of telomeres for one PD (1PD; left) versus 2 h (right). The native gel was first hybridized to show the single-stranded overhang, then denatured and rehybridized to show the total telomeric signal. Overhang signals are normalized to the total genomic input (native/denatured). Both 2 h and one PD lagging overhangs have three times higher overhang signals as compared with that of the leading overhangs. (E) DSN of one PD on denatured polyacrylamide gel (1 PD; left) versus 2 h on alkaline agarose gel (right). Lagging daughter overhangs have three times the size of leading daughter overhangs when fully processed at one PD, and direct measurement establishes that both have approximately mature sizes after a 2-h density label. (F) Summary of the expected fold difference between leading versus lagging overhangs signals of both one PD (1PD) and 2 h obtained in D, and overhang length (in nucleotides) obtained in E.
Two independent approaches were used to quantify overhang lengths. Signals from single-stranded overhangs hybridized to a telomere probe under native conditions were normalized to the total telomere signal obtained after the dsDNA was denatured. This native in-gel hybridization approach measures relative changes in overhang abundance. Electrophoresis was stopped after telomeres had penetrated ∼1 cm into the gel, so that telomeres of various lengths did not separate significantly according to size but remained as a compact band that concentrated the very low overhang signal into a small area. Alternatively, duplex-specific nuclease (DSN), which digests double-stranded telomere DNA to <10 bp in length with no effect on single-stranded telomere G overhangs, was used for the quantitative determination of overhang sizes as short as 12 nt (Zhao et al. 2008). Due to the heterogeneity of overhangs, a wide range of sizes were distributed on a denaturing gel.
In control cells labeled with BrdU for one PD, in-gel hybridization showed that lagging overhangs were approximately threefold more intense than leading overhangs after normalizing the overhang signals (obtained from native hybridization) to the total genomic DNA input (obtained from hybridization upon denaturing the gel) (Fig. 1D,F). Consistently, average leading overhangs were ∼30 nt and lagging overhangs were ∼100 nt (thus, lagging overhangs were approximately threefold longer than that of leading) using DSN assays on a denaturing polyacrylamide gel (Fig. 1E,F). In both assays, a 3′ → 5′ exonuclease (ExoI) was used as a negative control to digest 3′ overhangs. These controls were consistent with our previous observations (Chai et al. 2006a; Zhao et al. 2008). Using the same analyses, we showed that leading and lagging overhangs were processed to nearly mature sizes within 2 h (0–2 h released, 2–4 h BrdU incorporation) (Fig. 1D–F). The DSN at 2 h was performed on an alkaline agarose gel, in which the decreased resolution makes it visually easier to estimate overhang sizes without formal quantitation. In summary, 2-h-labeled telomeres had almost mature overhang lengths, similar to that of the one-PD control.
Lagging but not leading overhangs have almost mature overhang size immediately following replication
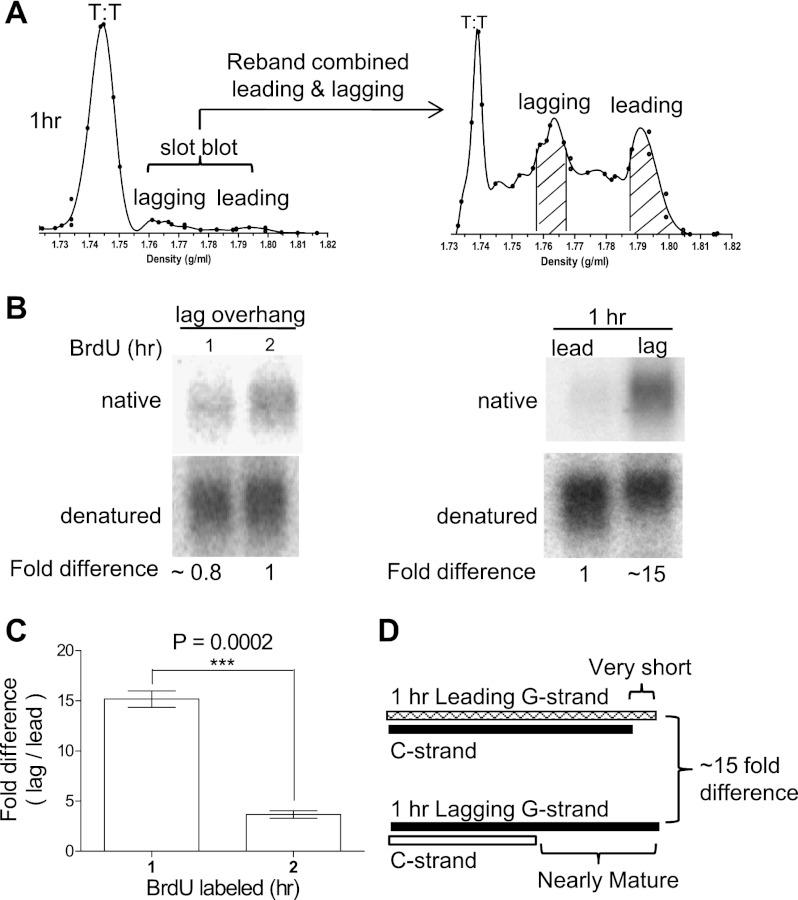
We then determined whether overhangs were processed to mature sizes immediately following semiconservative replication at the earliest time point we could analyze. Only fully replicated telomeres can be resolved into distinct peaks on a CsCl gradient, since partially replicated telomeres have densities ranging from that of unreplicated DNA to midway between the leading and lagging peaks (Supplemental Fig. S1). Five-hundred micrograms of genomic DNA from a 1-h BrdU label (0–2 h released, 2–3 h BrdU incorporation) generated only minimal leading and lagging peaks on a CsCl gradient centrifuguation. Leading and lagging peaks from two independent CsCl gradient centrifugations (∼1 mg of total genomic DNA) were thus combined and rebanded on a second CsCl gradient to better resolve the replicated leading and lagging daughter strands. In addition, a hypersensitive telomere probe was developed to maximize the sensitivity of overhang signal detection (see the Materials and Methods). In some cases, iododeoxyuridine (IdU), which has a higher density than BrdU, was used to increase resolution (data not shown). Distinct leading and lagging peaks were present within 1 h of BrdU labeling, indicating that some BJ telomeres had been completely replicated within 1 h (Fig. 2A). The 1-h-labeled leading and lagging daughters were purified, and overhang lengths were examined.
Figure 2.
Lagging daughter overhang size is almost mature following replication, but leading daughter overhang size maturation is delayed. (A) The small amount of semiconservative replicated telomeres observed following a 1-h label of BJ fibroblasts was somewhat contaminated by the trailing edge of unreplicated DNA. The lagging + leading fractions from the first CsCl gradient were combined and rebanded on a second CsCl gradient for higher resolution of fully replicated leading and lagging peaks (shaded area). (B) In-gel hybridization comparing relative overhang signals of lagging daughters at 1 h versus 2 h (left) and overhangs of leading versus lagging daughters at 1 h (right). Leading overhangs should be more dense and thus uncontaminated by semiconservative replication intermediates (Supplemental Fig. S1), and any contamination should have mature overhangs and contribute an overhang signal. The very faint signal thus cannot be explained by incomplete semiconservative replication. (C) Quantification of (lagging/leading) fold difference after normalizing to total genomic DNA input, comparing 1-h versus 2-h controls from three independent experiments using two-tailed Student's t-test with P < 0.001 (***) (P = 0.0002); 99% confidence level. Error bars represent SEM. (D) Summary of the results of 1-h leading versus lagging from B and C, illustrating that the overhangs of 1-h lagging daughters have an almost mature size but are ∼15-fold longer than the overhang sizes of 1-h leading strand daughters, which are still very short.
By in-gel hybridization, 1-h- and 2-h-labeled lagging daughter telomeres had similar overhang signals (0.8 vs. 1.0, normalized signals), indicating that lagging telomeres had almost mature overhang sizes at the earliest time point we could analyze following replication (Fig. 2B). Using 1-h lagging daughter overhang sizes as a reference point, we examined whether mature leading daughter overhang sizes were observed by 1 h of BrdU labeling. However, the overhang signal from 1-h leading daughters was barely detectable—only an average of ∼15-fold less intense than the almost mature 1-h lagging overhangs (Fig. 2C,D), far less than the roughly threefold difference found in the mature overhangs control (Fig. 1D–F). Leading overhangs were thus not processed to almost mature lengths until an additional hour had passed. Thus, while lagging daughter overhangs had close to mature overhang lengths after a 1-h label, it took an additional hour before leading daughters exhibited approximately mature sizes.
The terminal lagging Okazaki RNA primers remain for ∼1 h after replication on lagging daughters
We developed two independent assays to determine whether the final primer of Okazaki fragment synthesis was still present immediately following replication of the duplex telomeric DNA or whether nuclease processing had removed the final primer and potentially generated the long overhang on lagging daughters.
λ Exonuclease (λ exo) assay to monitor lagging terminal RNA primers
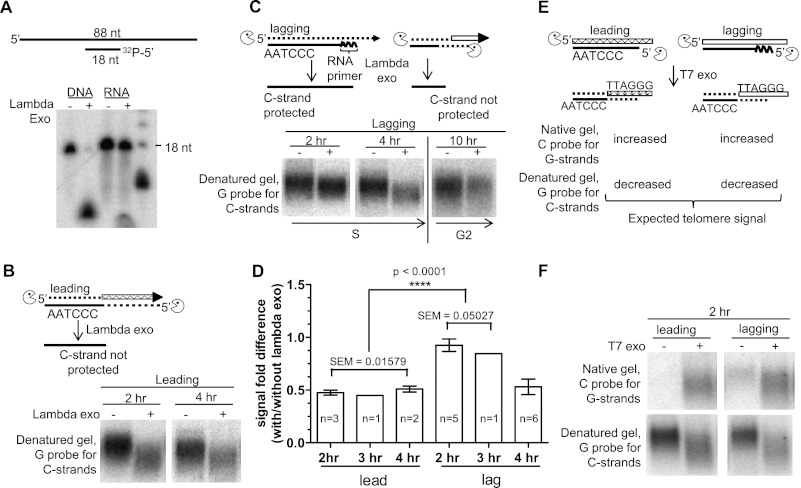
λ Exo is a highly processive 5′ → 3′ exonuclease that removes 5′ mononucleotides from dsDNA. Using model oligonucleotides, we first verified that λ exo was able to digest dsDNA but unable to digest RNA in RNA/DNA heteroduplexes. Eighteen-nucleotide DNA or RNA oligos were first 5′-end-labeled with 32P-γ ATP and annealed to an excess of complementary 88-nt oligos to generate either dsDNA or heteroduplex substrates with a 3′ single-stranded overhang (Fig. 3A). After λ exo digestion, the products were analyzed on a denaturing polyacrylamide gel to detect whether the 18-nt radiolabeled DNA or RNA had been digested. The 18-nt DNA oligonucleotide was entirely digested, confirming the sensitivity of dsDNA to λ exo treatment. However, the 18-nt RNA oligonucleotide in the RNA/DNA heteroduplex was resistant to λ exo digestion. λ Exo was unable to remove even the single 5′-labeled ribonucleotide in the DNA/RNA heteroduplex (Fig. 3A). This assay was then applied to telomeres, where purified lagging daughter strands should be resistant to λ exo digestion if the final RNA primer had not been removed.
Figure 3.
RNA primers remain on lagging C strands following replication in HeLa cells: λ exo assay. (A) In vitro validation that λ exo cannot digest RNA. Denaturing polyacrylamide gels show that a radioactive DNA oligonucleotide in duplex DNA is digested by λ exo, while the radioactive RNA oligonucleotide in an RNA/DNA heteroduplex is resistant to λ exo treatment. (B) Positive control: Leading 2- and 4-h HeLa telomeres are λ exo-sensitive. The C-strand signal is reduced in both size and intensity, as predicted, due to the absence of a final RNA primer on the leading daughter C strands. (C) Dynamics of the presence of a HeLa lagging terminal RNA primer across S phase. Lagging telomeres are λ exo-resistant at 2 h and become sensitive to λ exo at 4 h and 10 h following replication of the duplex telomeric DNA. (D) Multiple independent experiments were quantified by comparing +/− λ exo signal fold differences of lagging and leading strand daughter overhangs at 2, 3, and 4 h using two-tailed Student's t-test. The “n” within each bar represents the number of independent experiments. The values for each of the six independent experiments for leading strand daughters were used to calculate the SEM for that group. The six values for lagging strand daughters at 2 and 3 h were used to determine their SEM. P < 0.0001; 99% confidence level. Error bars represent SEM for those time points at which N > 1. (E) Predicted results of T7 exo, which digests 5′ mononucleotides of both dsDNA and dsRNA of RNA/RNA hybrids and thus serves as a positive control. Both leading and lagging daughters (even with intact RNA primer) should be sensitive to T7 exo and thus result in increased G-strand signal when hybridized to a C-rich probe under native conditions. Furthermore, after denaturation, a G-rich probe should yield a decreased C-strand signal. (F) As predicted above, 2-h leading and lagging daughter telomeres both show sensitivity to T7 exo.
Semiconservative replication intermediates will have densities on CsCl gradients ranging from slightly more dense than unreplicated DNA at the earliest times to half-way between the peaks of lagging and leading daughters just before replication is complete and the daughter strands separate (Supplemental Fig. S1). Although HeLa cells and BJ fibroblasts progress through S phase at approximately the same rate (Supplemental Fig. S2), the rate of telomere replication is much more rapid in HeLa cells, and fully replicated telomeres are present within 15 min of adding a density label (TT Chow, PG Smiraldo, Y Zhao, S Mak, JW Shay, and WE Wright, in prep.). The presence of semiconservative replication intermediates could potentially complicate the interpretation of λ exo assays due to the presence of the RNA primer on Okazaki fragments near the replication fork. HeLa cells rather than BJ fibroblasts were used for these experiments because after a 1- or 2-h label, the fraction of replication intermediates is too small to affect the interpretation of the results (Supplemental Fig. S3).
The C strand of leading daughters terminates in DNA (with no RNA primer immediately following duplex DNA replication) and was used as a positive control for the action of λ exo on telomeres. HeLa cell leading daughters labeled for 2, 3, and 4 h (0–2, 0–3, and 0–4 h BrdU incorporation upon release into S phase) showed the expected decrease in size and the expected 50% decrease in intensity of the C strand following λ exo digestion (Fig. 3B,D). However, the C strand of lagging daughters from HeLa cells labeled for 2 or 3 h immediately upon release into S (0–2 h or 0–3 h of BrdU incorporation upon release into S phase) were resistant to digestion. The lagging daughters became sensitive to λ exo after 4-h and 10-h labeling periods (0–4 h and 0–10 h of BrdU incorporation upon release into S phase), suggesting that the RNA primer initially protecting the C strands from digestion was removed by 4 h (Fig. 3C,D). Quantification demonstrated that the 2- and 3-h lagging daughters retained most of the C-strand signal, while the 4-h lagging daughters had an ∼50% C-strand signal reduction. Unpaired two-tailed t-test with 99% confidence indicated a significant difference between the combined 2-, 3-, and 4-h leading versus 2- and 3-h lagging signals, with a P-value of <0.0001 (SEM error bars in Fig. 3D). Meanwhile, 4-h lagging daughters became similar to the control leading daughters with an ∼50% C-strand signal (with no significant difference upon t-test analysis) (Fig. 3D). T7 exonuclease (T7 exo) digestion was used as an additional positive control.
T7 exo is a 5′ → 3′ exonuclease with low processivity that can degrade both DNA duplexes and RNA/DNA heteroduplexes. The 5′ end of both lagging daughters (with intact RNA primers) and leading daughters should be sensitive to T7 exo. 5′ End resection of the C strand should produce an increased G-overhang signal (C-probe hybridization). Meanwhile, G-probe hybridization upon denaturation should show the amount of C strands that had survived digestion (Fig. 3E). HeLa 2-h-labeled lagging and leading daughters both showed increased G-overhang and decreased C-strand signal upon T7 exo treatment (Fig. 3F), as expected for this positive control. These results indicate that something, presumably an RNA primer, was inhibiting λ exo but not T7 exo for ∼3 h following the completion of lagging strand replication.
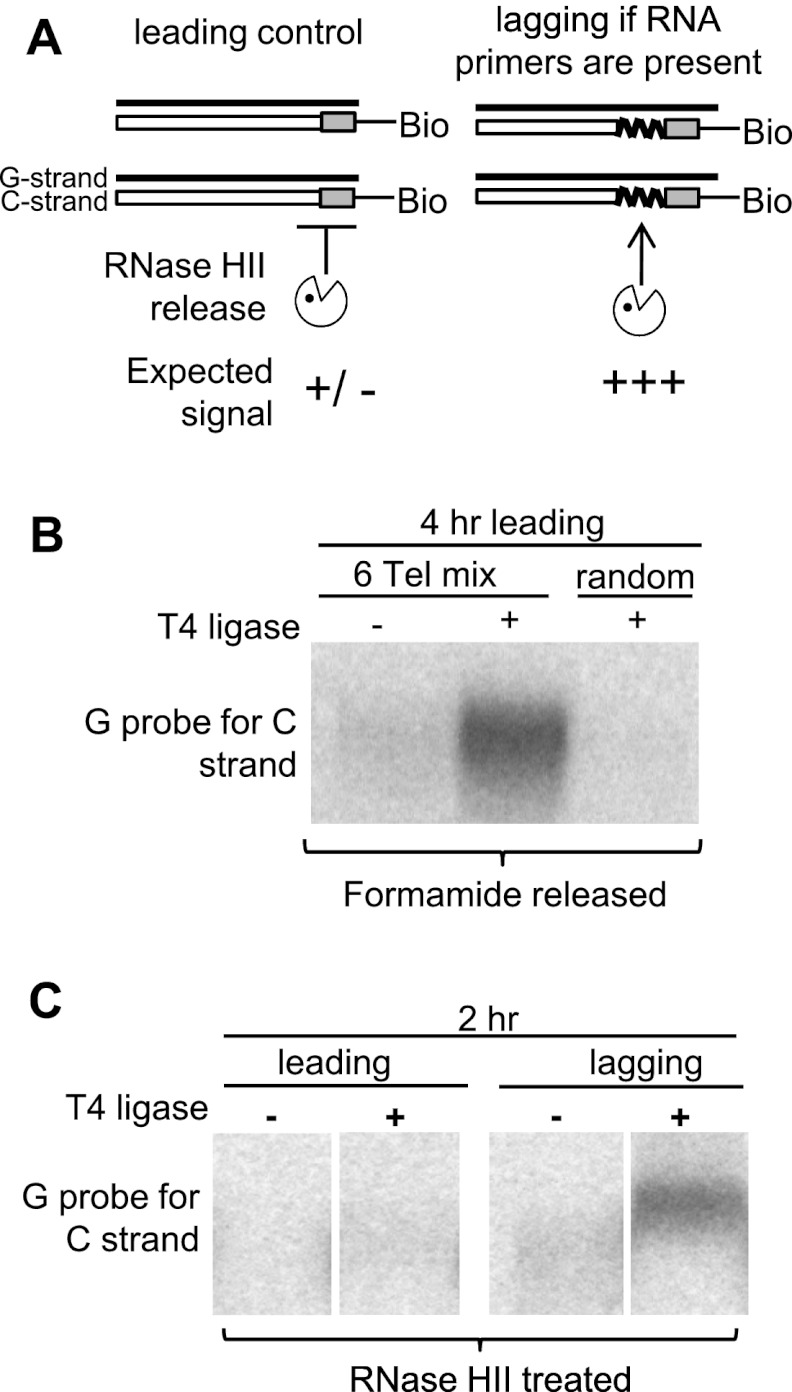
Pull-down assay to monitor lagging terminal RNA primers
The identification of RNA as the block to λ exo digestion was determined by the ability of RNase HII (an endonuclease that nicks heteroduplexes at multiple sites within the RNA) to release purified lagging C strands following ligation of biotinylated oligonucleotides to their 5′ ends (Fig. 4A). A mixture of six biotinylated telorette oligos (as used for single-telomere length analysis [STELA] in Figs. 5, 6) was ligated to either purified leading (control) or lagging 2-h-labeled HeLa DNA and retrieved using streptavidin-coated magnetic beads. RNase HII should release telomeres anchored to the beads via ligation to RNA but not those in which the telorette oligo is ligated directly to DNA (Fig. 4A). We first verified that the ligation was telomere-specific and ligase-dependent by ligating purified HeLa leading 4-h-labeled telomeres to either a mixture of the six telorette oligos or biotinylated random oligos (Fig. 4B). Telomeres retrieved from the streptavidin pull-down were then released using formamide. Only ligation with the telorette oligos yielded a C-strand signal upon release, and the signal was dependent on the inclusion of ligase (Fig. 4B). The experiment was then repeated using 2-h-labeled HeLa DNA (0–2 h of BrdU label upon release into S phase) and released with RNase HII rather than formamide (Fig. 4C). Leading control telomeres showed no detectable released telomeres, while lagging telomeres were liberated (Fig. 4C). This confirmed that RNA was still present on telomeric lagging daughters for a significant time following replication of the duplex telomeric DNA.
Figure 4.
RNA primers remain on lagging C strands following replication in HeLa: pull-down assay. (A) A mixture of biotinylated C-telorette oligos (gray-shaded squares—Bio) were ligated to either leading or lagging HeLa telomeres and pulled down with streptavidin beads followed by RNase HII treatment. Only ligated products containing RNA should be released by RNase HII. (B) Ligation is required and is specific to the six C-telorette oligos but not the random oligos. (C) Two-hour lagging daughters, but not leading daughters, are released by RNase HII. This establishes the presence of RNA at the C-strand termini of lagging strand daughters.
Figure 5.
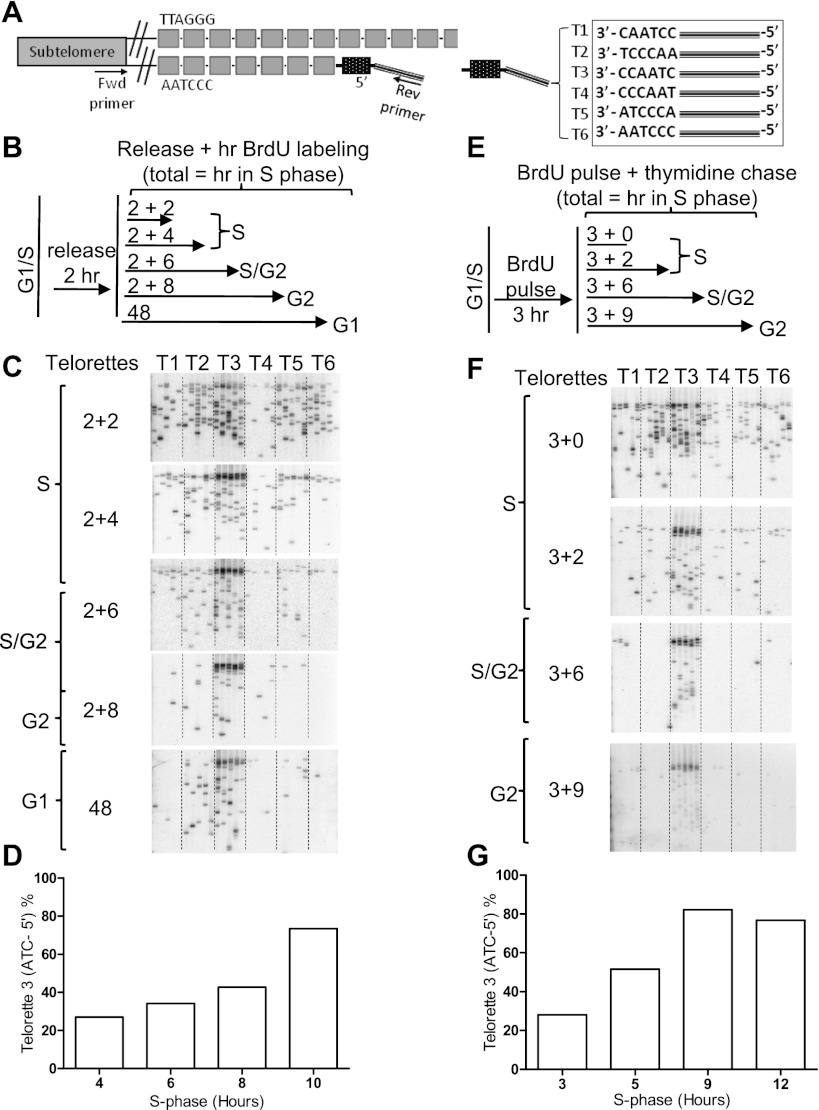
Leading daughter C-strand-terminal nucleotides are random until S/G2. (A) Individual C telorettes representing the six permutations of the hexameric C-rich repeat were ligated in separate reactions to the same amount of DNA and amplified using a forward Xp/Yp subtelomere-specific primer and a reverse teltail primer. Only the C telorette that annealed adjacent to the last base of the complementary strand would be ligated to the end of the telomere and thus produce a PCR product. Each amplified band represents ligation to an individual telomere. (B) Approach: Synchronized BJ fibroblasts released into S phase for 2 h were labeled for 2, 4, 6, 8, or 48 h. (C) T3 is the sequence that ligates to C strands ending in 3′-CCAATC-5′. Two-hour leading strands (2 h + 2 h in S phase) initially show randomized ends but become specified for CCAATC-5′ (T3) at S/G2. (D) Quantification of C showing a 73% specification for T3 at 10 h (2 + 8). (E) Approach: Synchronized BJ fibroblasts were pulsed for 3 h with BrdU and thymidine-chased for 0, 2, 6, 9 h. (F,G) Three-hour BrdU pulse results in randomized ends, as indicated by the PCR products in T1–T6 (28% T3 preference) that become specified at S/G2 (3 + 6 h) with 82% CCAATC-5′.
Figure 6.
Lagging C-strand-terminal nucleotides are rapidly specified. (A) Approach: Synchronized BJ fibroblasts were released into S phase for 2 h in fresh medium followed by BrdU labeling for 2, 4, 8 h, with 48 h as a positive control. (B) A 3′-CCAATC-5′ (T3) preference is evident at all time points. (C) Quantification of 3′-CCAATC-5′ specificity on lagging strand daughters ranges from 55% to 67%.
Leading terminal nucleotide is specified at S/G2
Fully processed overhangs exhibit a specific C-terminal nucleotide (Sfeir et al. 2005). We then asked when the processing of leading daughter C strands to produce this CCAATC-5′-terminal sequence occurred. This timing was monitored using a modified STELA (Fig. 5A; Baird et al. 2003; Sfeir et al. 2005), which involved a ligation step followed by PCR with oligonucleotides (Telorettes 1–6) representing the six possible permutations of the hexameric telomere repeat. Six independent samples (each containing a specific telorette) were first ligated to the C strand of DNA. PCR was then performed using a forward primer located at the Xp/Yp subtelomeric region and a reverse “teltail” primer complementary to a unique sequence at the 5′ end of the telorettes. Amplified PCR products were hybridized with subtelomeric probe such that each band represented the terminal nucleotide specificity of a single telomere. Quantification of the number of amplified PCR bands by individual telorette (T1–T6) defined the C-terminal nucleotide distribution across S phase. T3 corresponds to the mature specification of the 5′ end as 3′-CCAATC-5′. Two independent experimental designs were applied such that BJ cells were either BrdU-labeled for increasing periods of time starting at 2 h after release into S phase (i.e., 0–2 h of release in fresh medium followed by x h of BrdU incorporation) (Fig. 5B) or BrdU-labeled for 3 h at the time of release into S phase and chased with thymidine for increasing periods of time (i.e., 0–3 h of BrdU incorporation followed by x h of thymidine chase) (Fig. 5E). Replicated leading daughters were then isolated on CsCl gradients and analyzed. Both protocols indicated that C-strand 5′ ends of leading daughters were initially random (PCR products were observed across T1–T6)—although mature overhang sizes have already been generated—and were only processed so that most ended in CCAATC-5′ late in S phase or S/G2 (corresponding to the majority of the PCR products resulting from T3 reactions) (Fig. 5C,D,F,G). These results suggested that there was a delay in C-strand-terminal nucleotide specification between when leading overhang length approached mature sizes and the end of S/G2.
Lagging terminal nucleotide is specified rapidly
In contrast to leading ends, the terminal nucleotides of lagging daughters were specified within 2 h after replication (0–2-h released and 2–4 h of BrdU incorporation). BJ cells were labeled for increasing periods of time starting at 2 h after release into S phase (Fig. 6A). The sequence CCAATC-5′ was predominant even at the earliest times examined, at which the majority of the PCR products were detected with the T3 telorette (Fig. 6B,C).
Discussion
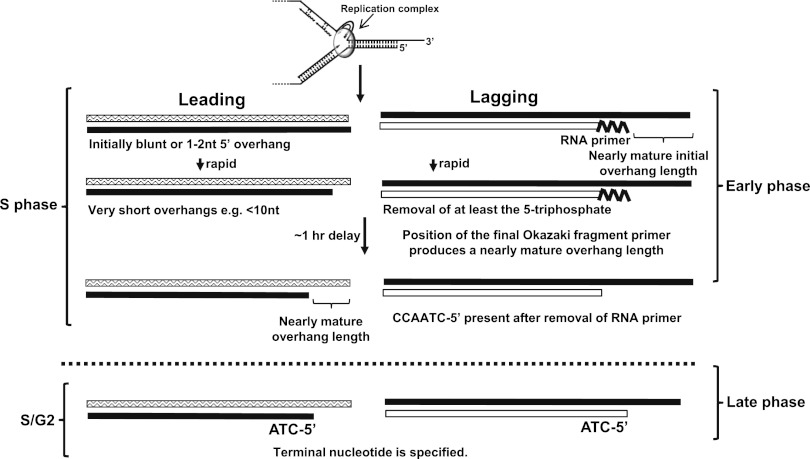
Telomeres require 3′ overhangs for chromosome end protection. The details of how these overhangs are generated in normal human cells are unknown. The results presented here provide important insights implicating several distinguishable steps in telomere end processing that occur in two distinct phases that are summarized in Figure 7. The early phase lasts several hours following the completion of basic semiconservative replication (the ability to physically separate the daughters of leading and lagging strand synthesis). The late phase occurs at S/G2. Since human telomeres replicate throughout S phase (Ten Hagen et al. 1990; Wright et al. 1999), there can be many hours intervening between the first and second phases of end processing.
Figure 7.
Model for telomerase-negative human telomere overhang processing. See the text for details.
We showed previously that telomerase extended leading strands at the shortest BrdU incorporation time (30-min pulse) that we could measure (Zhao et al. 2009). Telomerase requires at least 6 nt of single-stranded G-rich overhangs to function (Lai et al. 2001); thus, we infer that there is a very rapid processing event that generates an initial overhang. However, we were unable to detect meaningful overhang signals from leading strands in normal diploid cells after a 1-h label. Unless the timing is different in normal versus tumor cells or there is an additional rapid processing event that is specifically present in telomerase-positive tumor cells, this initial overhang must be very short. The nearly mature size of the leading overhang was only detected later, after an additional hour.
Eukaryotic Okazaki fragments are ∼100–200 nt in length (Sakabe and Okazaki 1966; Okazaki et al. 1968; Blumenthal and Clark 1977; Bielinsky and Gerbi 1999; Burgers 2009; Balakrishnan and Bambara 2011). If the replication apparatus simply ran off the end of the telomere while the position of the final RNA primer is random, then lagging daughters should have the final RNA priming event occur at approximately half of the average Okazaki fragment size (or 50–100 nt from the end). We observed that the final lagging RNA primer remained intact immediately following replication, with an overhang size of ∼80 nt at that time, consistent with the above prediction. This resolves a long-standing question about the mechanism of overhang generation at lagging daughter telomeres that is fundamental to our understanding of telomere shortening. Since the gap produced by removal of the RNA primer only contributes ∼10% to the size of the human overhang, the end replication problem in humans should now be revised to state that the conventional replication complex cannot copy the gap between the final RNA priming event and the extreme end of the chromosome. Therefore, the position of the final priming event of replication (rather than processing that degrades the C strand) is an important intrinsic step in the rate of telomere shortening in human cells. Other mechanisms, such as oxidative stress, may also contribute to the rate of shortening.
The first nucleotide incorporated during RNA primer synthesis contains a 5′ triphosphate. Neither T7 exonuclease nor T4 ligase should be able to function on an RNA containing a 5′ triphosphate (Kerr and Sadowski 1972; Engler and Richardson 1982), yet both of these enzymes are able to function on the RNA-capped DNA in our assays. We infer that there is a rapid initial processing of the RNA primer after replication that removes at least the triphosphates, followed by a delay of several hours before the rest of the RNA is removed in the early phase.
In telomerase-positive tumor cells, C-strand fill-in of the telomerase-extended G strands is delayed until S/G2 (Zhao et al. 2009). Normal diploid cells do not have extended G strands needing fill-in, but they also exhibit processing events during this second phase of replication of the duplex telomeric DNA. The C-rich strand-terminal nucleotides of leading daughter telomeres remain random until S/G2, when CCAATC-5′ specification is acquired. In contrast, lagging C strands exhibit specific ends once the RNA primer is removed during the early phase following replication. As leading daughters have DNA while lagging daughters have RNA at their C terminus, it is possible that entirely different mechanisms are used to generate C-strand end specificity on leading versus lagging daughters, reflecting the activation of separate enzymatic complexes. However, if the phenomenon of the presence of a transient overhang independent of telomerase in yeast (Wellinger et al. 1992, 1993, 1996; Dionne and Wellinger 1996) also exists in humans, a transient short resection of the leading daughter C strand could occur at S/G2 and be filled in by the same mechanism that fills in the extended overhangs in telomerase-positive cells. This mechanism presumably requires an RNA primer, possibly produced by polymerase α recruited by the Cst1–Stn1–Ten1 (CST) complex (Price et al. 2010). In this scenario, leading strands would acquire an RNA primer at their C-strand termini. Since leading daughters acquire an almost mature overhang size within ∼1 h of replication, any transient resection and fill-in at S/G2 would thus need to produce minimum net change in overhang size. It is possible that both leading and lagging daughters undergo a transient resection so that the final end specification of both C strands could occur at S/G2 by the same mechanism.
The telomeric G strand is always generated by leading strand synthesis and should end in GGTTAG, since 80%–85% of fully processed telomeres have C strands ending in the complementary sequence CCAATC-5′ (Sfeir et al. 2005). However, approximately equal amounts of G ends of the sequences GGTTAG, GGTTA, and GGTT were found in normal diploid cells (Sfeir et al. 2005). This is consistent with the concept that the replication apparatus often simply runs off and dissociates from the extreme chromosome termini prior to incorporating the final 1 or 2 nt (Sfeir et al. 2005). The initial result of leading strand synthesis thus probably has a 0-, 1-, or 2-nt 5′ overhang due to the difficulty of making fully blunt-ended products (Fig. 7).
Telomeres present many challenges to replication. It is unknown whether t-loops refold following replication or remain unfolded following replication until the end of S phase. The long delay in processing of both leading and lagging telomeric daughters suggests that telomeres remain in an unfolded state for at least 1–2 h following replication. The mechanism preventing the single-stranded overhang at telomeres from initiating an ATR-mediated DNA damage response during this time may be Pot1-mediated, but that remains to be determined. The repetitive nature of the telomeric repeat and the ability to form G quartets make telomeres sensitive to fork stalling (Hao and Tan 2002; Lipps and Rhodes 2009; Paeschke et al. 2011), and a variety of proteins are required to allow replication of duplex telomeric DNA to proceed (Crabbe et al. 2004; Sfeir et al. 2009). The series of separate steps that we presented here introduce another layer of choreography required to successfully reproduce the structures present prior to replication. This will now allow an analysis of the specific steps affected by different factors that influence overhang structure, and understanding the regulation of these steps may provide the opportunities for manipulation that can be beneficial to both cancer therapy and regenerative medicine.
Materials and methods
Cell culture and cell cycle synchronization
BJ foreskin fibroblasts were cultured at 37°C in 5% CO2 in high-glucose DMEM medium (Hyclone) containing 15% Cosmic calf serum (Hyclone). For synchronization, logarithmically growing cells were washed twice with 1× PBS, presynchronized with low serum (high-glucose DMEM with 0.1% fetal bovine serum, 20 mM HEPES) for 48 h, and then fed fresh medium containing 15% fetal bovine serum and 2 μg/mL aphidicolin (A.G. Scientific) for 24 h. Cells were then washed three times with 1× PBS, released into fresh medium for 2 h, and then labeled with 100 μM BrdU. For pulse-chase experiments, cells were washed twice with 1× PBS and chased with 100 μM thymidine.
HeLa cervical carcinoma cells were used for some experiments requiring short labeling times and a large number of cells. HeLa cells were cultured at 37°C in 5% CO2 in high-glucose DMEM medium with 10% Cosmic calf serum (Hyclone). Exponentially growing HeLa cells were synchronized with 2 mM thymidine (Sigma) for 19 h, washed three times with 1× PBS, and incubated with fresh medium for 9 h. Two millimolar thymidine was added again for 16 h. Cells were washed three times with 1× PBS and released into fresh medium with 100 μM BrdU for 0–10 h.
Genomic DNA isolation
Genomic DNA was purified by Qiagen Blood and Cell Culture Midi kit. Precipitated DNA was washed twice with 70% ethanol and suspended in 10 mM Tris-HCl (pH 8). DNA was dissolved overnight at 37°C.
Generation of telomere probes
Twenty-four-nucleotide probes containing six 32P-dC or six 32P-dG were synthesized as described (Herbert et al. 2003). A hypersensitive C probe was designed to incorporate 13 total radioactive nucleotides (nine32P-dC + four32P-dA). T3C3+3 (5′-TTTCCCTAA) was used instead of T3C3+9 (5′-TTTCCCTAACCCTAA) and annealed to GTU4 (5′-GGGUUAGGGUUAGGGUUAGGGAAA) for 1 min at 90°C, for 15 min at 20°C, and for 15 min at 17°C. The synthesis reaction (Herbert et al. 2003) was modified as follows: 3.1 μL of 8× Roche buffer M, 1 μL of annealed template oligo (1.7 pmol/μL), 1 μL of dTTP (1.25 mM stock, 50 μM final), 7 μL of 32P-dCTP (3000 Ci/mmol), 4 μL of 32P-dATP (3000 Ci/mmol), 7.9 μL of Millipore H2O, and 1 μL of Klenow (5 U/μL) were combined in a final volume of 25 μL. After extension for 30 min at 20°C and for 5 min at 95°C (to inactivate Klenow to prevent probe degradation upon uracil deglycosylase [UDG] treatment), the reaction was cooled to room temperature. UDG (0.5 μL; 1 U/μL) was added to degrade the GTU template, incubated for 15 min at 37°C, and then inactivated for 10 min at 95°C. Free isotopes were then removed using an Illustra Microspin G-25 column (GE Healthcare).
CsCl separation of leading and lagging telomeric daughters
CsCl gradient separation was performed as described (Zhao et al. 2011b) with modifications. In brief, 500 μg of purified genomic DNA was digested with HinfI and RsaI overnight in 250 μL and terminated by the addition of EDTA to 10 mM. DNA was mixed with CsCl solution (density of 1.79 g/mL with 5 mM Tris-HCl at pH 8, 2 mM EDTA) to obtain a final density of ∼1.760–1.770 g/mL and added to a polyallomer quick-seal centrifuge tube (Beckman). Samples were centrifuged at 55,000 rpm for 20 h at 25°C using a VTi-80 vertical rotor (Beckman). Fractions of the sample were collected, and aliquots were denatured and hybridized with a telomere-specific probe on a slot blot to identify the fractions that contained telomere DNA. The corresponding densities were obtained by measuring the refractive index. Leading DNA was located at a density of 1.790–1.800 g/mL, lagging DNA was located at a density of 1.760–1.770 g/mL, and unreplicated DNA was located at a density of 1.740–1.750 g/mL. Four to five fractions of each peak were pooled and desalted by agarose dialysis (rocking on a plug of 2% agarose made with 10 mM Tris-HCl at pH 8 in a 50-mL tube for 1 h at room temperature) followed by ethanol precipitation. DNA was then dissolved in 10 mM Tris-HCl (pH 8).
Telomere overhang analysis by in-gel hybridization
Five micrograms of purified leading or lagging DNA was briefly run into an 0.8% agarose gel in 1× TAE so that telomeres were not significantly separated by size and remained as a relatively tight band. DNA that had been digested with 10 U of exonuclease I (20 U/μL) (Epicentre Biotechnologies) in a 20-μL reaction for 1 h at 37°C and terminated by adding 0.5 μL of EDTA (0.5 M stock) served as a negative control. The gel was dried, prehybridized (6× SSC, 5× Denhardt's solution, 0.5% [w/v] SDS), and hybridized with C probe for G overhangs under native conditions. Upon obtaining the signal from the native hybridization, the gel was denatured (0.5 M NaOH, 1.5 M NaCl for 1 h), rinsed three times with distilled water, neutralized (0.5 M Tris-HCl at pH 8, 1.5 M NaCl for 30 min), and then hybridized again with the C probe to obtain the denatured signal for total telomere input.
Overhang analysis by DSN assay
DSN assay was performed as described (Zhao et al. 2008, 2011a). In brief, 2–5 μg of purified leading versus lagging was digested with 2 U of DSN (Evrogen) for 2 h at 37°C. For negative controls, 10 U of ExoI (Epicentre Biotechnologies) was added to the genomic DNA for 1 h at 37°C prior to DSN digestion. DSN digestion was stopped by adding EDTA to a final concentration of 25 mM. For denaturing polyacrylamide gels, a 1:1 ratio of deionized formamide was added. DNA was then heated for 5 min at 65°C and loaded on a 6% polyacrylamide gel with 8 M urea. The gel was run at 15 V/cm in 0.5× TBE until the bromophenyl blue dye front-migrated two thirds into the length of the gel. DNA was electro-transferred onto a Amersham Hybond-N+ membrane (GE Healthcare) with 0.5× TBE. The membrane was air-dried, UV-cross-linked, hybridized to a telomere-specific probe as described (Herbert et al. 2003), and exposed on a PhosphorImager (GE Healthcare). Alkaline agarose gels (1% agarose made with 50 mM NaOH, 1 mM EDTA) were run in 50 mM NaOH and 1 mM EDTA at 4°C with a low voltage (1–2 V/cm) until the dye migrated ∼6–8 cm. DNA was capillary-transferred to Amersham Hybond-XL (GE Healthcare) and processed as above. Average overhang sizes were calculated using the formula mean average length = ∑(Inti)/∑(Inti/MWi), where Inti is the signal intensity, and MWi is the molecular weight of the DNA at position i (Chai et al. 2006a; Zhao et al. 2008).
Modified STELA
STELA was performed as described (Sfeir et al. 2005) with modifications. Ligations were performed using each of the six C-telorette oligos (T1–T6) corresponding to six permutations of the hexameric telomere repeat. Five-hundred nanogram of purified leading or lagging daughters were incubated in a 10-μL reaction (1× T4 ligase buffer, 0.001 μM individual C telorettes, 50 U New England Biolabs T4 ligase) for 20 h at 35°C. Multiple PCR amplifications were performed with 0.5 ng of ligated DNA and 0.5 μM primers (XpYpE2 forward and C-teltail reverse primers) using Abgene Hi-Fidelity PCR Master 2× mix (AB-0792, Thermo Scientific) in a final volume of 25 μL for 28 cycles of 15 sec at 94°C, 30 sec at 65°C, and 15 min at 68°C. Amplified products were resolved on a 0.7% agarose with 1× TAE and capillary-transferred onto Amersham Hybond-N+ membrane overnight. Membranes were fixed for 2 h at 80°C, hybridized with a subtelomeric probe generated by PCR amplification with Xp/YpE2 and Xp/YpB2 primers, and 32P-labeled by random priming. The membrane was then exposed to a PhosphorImager screen.
λ Exo assay
Two micrograms of purified leading or lagging DNA was digested with 1 μL of λ exo (10 U/μL) (Epicenter Biotechnologies) for 1.5 h at 37°C. DNA was run on a 0.8% agarose gel in 1× TAE at 10 V/cm for 40 min, denatured with an alkaline solution containing 0.5 M NaOH and 1.5 M NaCl for 30 min, rinsed three times with distilled water, and dried at room temperature. The gel was then neutralized with 0.5 M Tris-HCl (pH 8) and 1.5 M NaCl for 30 min and hybridized overnight at 42°C with G-telomere probes to detect the C strands.
RNA primer pull-down assay
Prior to CsCl gradient centrifugation, genomic DNA was digested with RsaI and HaeIII, generating only blunt ends to minimize background. Two micrograms of purified leading or lagging DNA was ligated overnight at 16°C to a 0.5 μM (final concentration) mixture of the six telorette oligos (Sfeir et al. 2005) that were biotinylated. DNA was pulled down using Dynabeads kilobaseBINDER kit (Invitrogen) overnight at room temperature. DNA was released by 7.5 U of RNase HII (New England Biolabs) for 2 h. Control DNA was released using 95% formamide and 10 mM EDTA (pH 8.2) for 10 min at 90°C.
Acknowledgments
We thank Jinyong Kim, PhD, and Guido Stadler, PhD, for their insightful scientific discussions. This research was supported by BC083375 from the Department of Defense Predoctoral Cancer Research Program (T.T.C) and AG01228 from the National Institute of Aging (W.E.W).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187211.112.
References
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D 2003. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 [DOI] [PubMed] [Google Scholar]
- Balakrishnan L, Bambara RA 2011. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem 286: 6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballal RD, Saha T, Fan S, Haddad BR, Rosen EM 2009. BRCA1 localization to the telomere and its loss from the telomere in response to DNA damage. J Biol Chem 284: 36083–36098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA 1999. Chromosomal ARS1 has a single leading strand start site. Mol Cell 3: 477–486 [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Clark EJ 1977. Discrete sizes of replication intermediates in Drosophila cells. Cell 12: 183–189 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Burgers PM 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284: 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE 2006a. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE 2006b. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep 7: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Wei C, Price CM 2006. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol 26: 6971–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J 2004. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Dai X, Huang C, Bhusari A, Sampathi S, Schubert K, Chai W 2010. Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J 29: 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S 2009. Multiple roles for MRE11 at uncapped telomeres. Nature 460: 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T 2009. Cell cycle-dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of nonhomologous end joining (NHEJ) in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol 29: 5552–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci 93: 13902–13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler MJ, Richardson CC 1982. DNA ligases. In The enzymes (ed. PD Boyer), pp. 3–29. Academic Press, San Diego [Google Scholar]
- Garvik B, Carson M, Hartwell L 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsouzian LK, Tuzon CT, Zakian VA 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol Cell 24: 603–610 [DOI] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Hao YH, Tan Z 2002. The generation of long telomere overhangs in human cells: A model and its implication. Bioinformatics 18: 666–671 [DOI] [PubMed] [Google Scholar]
- Herbert BS, Shay JW, Wright WE 2003. Analysis of telomeres and telomerase. Current Protoc Cell Biol 18.6.1–18.6.20 doi: 10.1002/0471143030.cb1806s20 [DOI] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM 2001. G-overhang dynamics at Tetrahymena telomeres. EMBO J 20: 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr C, Sadowski PD 1972. Gene 6 exonuclease of bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem 247: 305–310 [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci 78: 3015–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM 2008. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol 18: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K 2001. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol 21: 990–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Reddy S, Comai L 2009. Sequence-specific processing of telomeric 3′ overhangs by the Werner syndrome protein exonuclease activity. Aging 1: 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D 2009. G-quadruplex structures: In vivo evidence and function. Trends Cell Biol 19: 414–422 [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J 18: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA 2008. Division of labor at the eukaryotic replication fork. Mol Cell 30: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V 1996. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci 59: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41: 181–190 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA 2011. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Okazaki R 1966. A unique property of the replicating region of chromosomal DNA. Biochim Biophys Acta 129: 651–654 [DOI] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE 2008. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Ben-Porath I, Carey VJ, O'Connor BF, Hahn WC, Weinberg RA 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat Genet 33: 492–496 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Gilbert DM, Willard HF, Cohen SN 1990. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol 10: 6348–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom HI, Greider CW 2010. A sequence-dependent exonuclease activity from Tetrahymena thermophila. BMC Biochem 11: 45 doi: 10.1186/1471-2091-11-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, et al. 2003. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Kibe T, Kang HY, Seo YS, Uritani M, Ushimaru T, Ueno M 2004. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol 24: 9557–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J 2006. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Watson JD 1972. Origin of concatemeric T7 DNA. Nat New Biol 239: 197–201 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA 1992. Use of non-denaturing Southern hybridization and two dimensional agarose gels to detect putative intermediates in telomere replication in Saccharomyces cerevisiae. Chromosoma 102: S150–S156 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA 1996. Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev 11: 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Liao ML, Shay JW 1999. Normal human telomeres are not late replicating. Exp Cell Res 251: 492–499 [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE 2008. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res 36: e14 doi: 10.1093/nar/gkm1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shay JW, Wright WE 2011a. Telomere G-overhang length measurement method 1: The DSN method. Methods Mol Biol 735: 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shay JW, Wright WE 2011b. Telomere terminal G/C strand synthesis: Measuring telomerase action and C-rich fill-in. Methods Mol Biol 735: 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]