This study identifies human RNF4 as a key player in the DNA damage response. RNF4 is recruited to damage foci via interaction with sumoylated proteins and then functions as a ubiquitin E3 ligase to enhance the turnover of MDC1 and RPA. Turnover of the latter is required for RPA replacement by RAD51, allowing homologous recombination to ensue. RNF4 is thus identified as a crucial modulator of dynamics at DNA damage foci.
Keywords: RNF4, SUMO, ubiquitin–proteasome system (UPS), DNA double-strand breaks (DSBs), RPA, RAD51
Abstract
Protein ubiquitylation and sumoylation play key roles in regulating cellular responses to DNA double-strand breaks (DSBs). Here, we show that human RNF4, a small ubiquitin-like modifier (SUMO)-targeted ubiquitin E3 ligase, is recruited to DSBs in a manner requiring its SUMO interaction motifs, the SUMO E3 ligases PIAS1 and PIAS4, and various DSB-responsive proteins. Furthermore, we reveal that RNF4 depletion impairs ubiquitin adduct formation at DSB sites and causes persistent histone H2AX phosphorylation (γH2AX) associated with defective DSB repair, hypersensitivity toward DSB-inducing agents, and delayed recovery from radiation-induced cell cycle arrest. We establish that RNF4 regulates turnover of the DSB-responsive factors MDC1 and replication protein A (RPA) at DNA damage sites and that RNF4-depleted cells fail to effectively replace RPA by the homologous recombination factors BRCA2 and RAD51 on resected DNA. Consistent with previous data showing that RNF4 targets proteins to the proteasome, we show that the proteasome component PSMD4 is recruited to DNA damage sites in a manner requiring its ubiquitin-interacting domains, RNF4 and RNF8. Finally, we establish that PSMD4 binds MDC1 and RPA1 in a DNA damage-induced, RNF4-dependent manner and that PSMD4 depletion cause MDC1 and γH2AX persistence in irradiated cells. RNF4 thus operates as a DSB response factor at the crossroads between the SUMO and ubiquitin systems.
DNA double-strand breaks (DSBs) are generated by ionizing radiation (IR) and various DNA-damaging chemicals. If they are not repaired or are repaired incorrectly, DSBs result in cell death or genomic instability that can lead to immune deficiencies, neurodegeneration, premature aging, and cancer (Jackson and Bartek 2009; Ciccia and Elledge 2010). DSB formation triggers activation of the DNA damage response (DDR) protein kinases ATM, ATR, and DNA-PK, which phosphorylate many protein targets, including the histone variant H2AX (H2AFX). Phosphorylated H2AX (γH2AX) then mediates the accumulation of DDR proteins such as MDC1 (NFBD1), 53BP1 (TP53BP1), and BRCA1 into IR-induced foci (IRIF) that are thought to amplify DSB signaling and promote DSB repair (Downs et al. 2007; Bekker-Jensen and Mailand 2010; Ciccia and Elledge 2010; Polo and Jackson 2011).
DSBs are repaired by two principal pathways: nonhomologous end-joining (NHEJ) that is active throughout the cell cycle, and homologous recombination (HR) that is normally restricted to S and G2 cells (Hartlerode and Scully 2009; Pardo et al. 2009). HR but not NHEJ relies on the presence of a sister chromatid and also requires cell cycle-regulated 5′-to-3′ exonucleolytic processing (resection) of DNA ends that generates stretches of ssDNA. This ssDNA is bound by replication protein A (RPA), which is then replaced by RAD51 to produce a RAD51-ssDNA nucleoprotein filament, promoting DNA strand invasion and subsequent HR events. A key protein player in the essential step of replacing RPA with RAD51 is BRCA2, which directly binds RAD51 and promotes RAD51 loading onto RPA-coated ssDNA (Liu et al. 2010; Thorslund et al. 2010; Holloman 2011). Notably, RPA-coated ssDNA also leads to recruitment and activation of the checkpoint kinase ATR, which phosphorylates various targets, including the downstream checkpoint kinase CHK1 (Cortez et al. 2001; Zou and Elledge 2003; Cimprich and Cortez 2008). Recent work has revealed that resection is promoted by various factors, including human CtIP (RBBP8) (Sartori et al. 2007), and has indicated how resection is subject to cell cycle control (Sartori et al. 2007; Gravel et al. 2008; Nimonkar et al. 2008, 2011; Huertas 2010). However, it remains to be determined specifically how the activities of the various HR-promoting factors are controlled and precisely how other HR events, such as the transition from RPA-ssDNA to RAD51-ssDNA, are mediated.
The reversible, covalent attachment of ubiquitin and small ubiquitin-like modifier (SUMO) proteins to DDR factors is critical for effective DSB repair and signaling in eukaryotic cells (Bergink and Jentsch 2009; Al-Hakim et al. 2010; Bekker-Jensen and Mailand 2010; Morris 2010a,b). For example, ubiquitin conjugates and ubiquitin-conjugating enzymes such as UBC13 (UBE2N), BRCA1, RNF8, HERC2, RNF168, RAD18, and the Fanconi anemia (FA) protein complex accumulate at DSB sites in mammalian cells, and defects in these factors result in impaired DSB repair and signaling associated with hypersensitivity toward DNA-damaging agents (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Zhao et al. 2007; Bekker-Jensen et al. 2009; Doil et al. 2009; Huang et al. 2009; Stewart et al. 2009; Watanabe et al. 2009; Kee and D'Andrea 2010; Yang et al. 2010). While the ubiquitin–proteasome system (UPS) was originally described as the main protein turnover/degradation machinery of eukaryotic cells, it is now evident that it is also a major nondegradative regulator of various cellular processes, including the DDR (Kirkin and Dikic 2007; Bergink and Jentsch 2009; Motegi et al. 2009; Al-Hakim et al. 2010). Furthermore, it has been shown that proteasomes accumulate on damaged chromatin, suggesting that they may promote DDR signaling as well as DDR-dependent protein turnover and HR (Ustrell et al. 2002; Blickwedehl et al. 2007; Jacquemont and Taniguchi 2007; Murakawa et al. 2007; Shi et al. 2008; Motegi et al. 2009; Al-Hakim et al. 2010; Ben-Aroya et al. 2010; Levy-Barda et al. 2011). The kinetics and mechanisms governing proteasome accumulation at DNA damage regions and the functions and targets of proteasomes at such sites, however, remain largely obscure.
It was recently established that modification of the largest RPA subunit (RPA1) by SUMO2/3 promotes the interaction of RPA with RAD51 and HR-mediated DSB repair (Dou et al. 2010). Furthermore, we and others have established that SUMO conjugates and the SUMO-conjugating enzymes UBC9 (UBE2I), PIAS1 (protein inhibitor of activated STAT 1), and PIAS4 (PIASy) accumulate at DSB sites and are required for effective DSB signaling and repair (Galanty et al. 2009; Morris et al. 2009). This work also revealed that PIAS1- and PIAS4-mediated sumoylation promotes accumulation of 53BP1 and effective RNF8-, RNF168-, and BRCA1-dependent accumulation of ubiquitin conjugates at DSB sites and that PIAS1/4-mediated sumoylation directly promotes BRCA1 E3 ubiquitin ligase activity (Galanty et al. 2009; Morris et al. 2009). Thus, rather than operating separately, DDR–protein sumoylation and ubiquitylation are intimately interconnected.
An additional link between ubiquitylation and sumoylation has been established through work on the mammalian RNF4 protein and its yeast counterparts (Slx5/Slx8 in Saccharomyces cerevisiae and Slx8/Rfp1 or Rfp2 in Schizosaccharomyces pombe) (Prudden et al. 2007; Sun et al. 2007). These proteins act as SUMO-targeted ubiquitin E3 ligases (STUbLs), which recognize sumoylated or SUMO-like domain-containing protein targets. For example, sumoylation of the acute promyelocytic leukemia (PML) protein triggers its recognition by RNF4, which then mediates PML ubiquitylation and proteasome-dependent PML degradation (Lallemand-Breitenbach et al. 2008; Tatham et al. 2008; Weisshaar et al. 2008). Since various DDR components are sumoylated and because RNF4 counterparts in yeast are implicated in suppressing HR via an as-yet-undefined mechanism (Burgess et al. 2007; Kosoy et al. 2007; Prudden et al. 2007), we investigated whether human RNF4 functions in the DDR. Indeed, as described below, we show that RNF4 accumulates at DSBs, regulates protein turnover/exchange at these sites, and is required for effective DSB repair.
Results
SUMO-interacting motif (SIM)-dependent accumulation of RNF4 at DSB sites
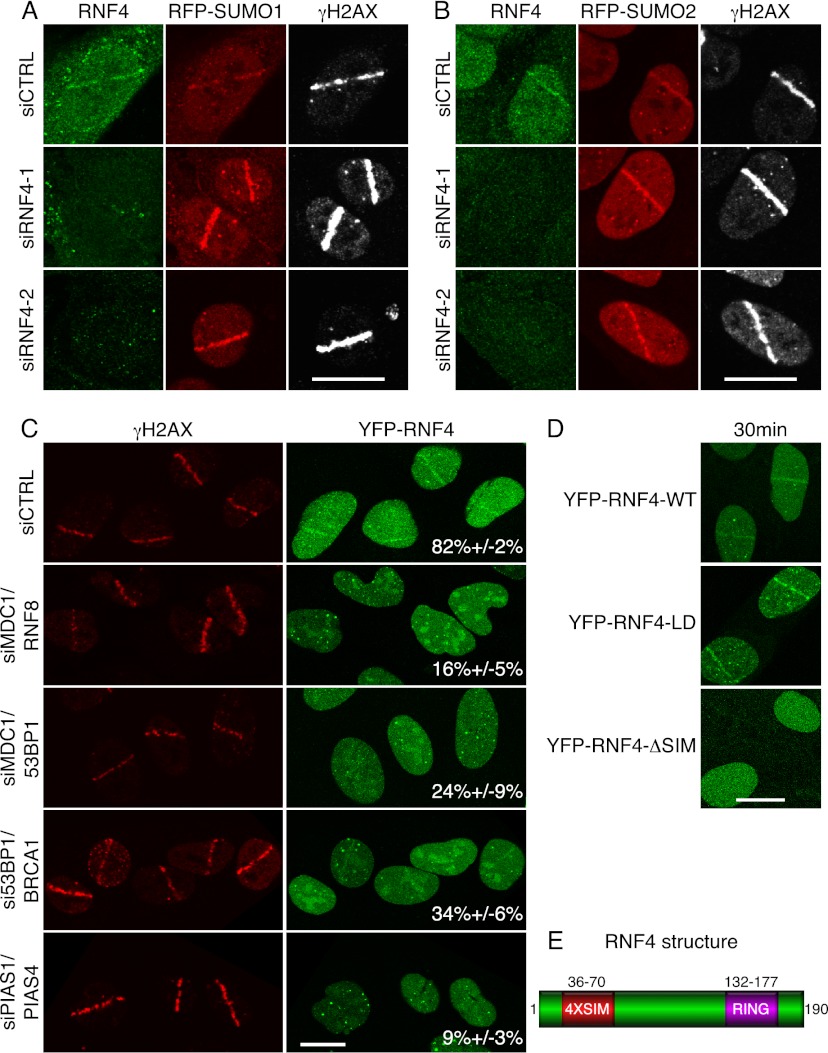
To investigate potential DDR functions for RNF4, we first determined whether it accumulates at DNA damage sites. Although we could not detect RNF4 recruitment to IRIF, by using both live-cell imaging and fixed-cell staining, we found that endogenous RNF4 (Fig. 1A,B) and recombinant RNF4 (Fig. 1C,D; Supplemental Fig. S1A,B) accumulated at DNA damage sites induced by laser micro-irradiation. RNF4 accrual was weakly detectable within 15 min after irradiation (data not shown) but became readily apparent after 30 min (Fig. 1D) and then persisted for up to several hours (note that Fig. 1A–C displays cells 2 h after laser micro-irradiation; Supplemental Fig. S1B; data not shown). This persistence of RNF4 recruitment within laser-micro-irradiated regions suggested that RNF4 might function at DSBs, which in many cases take several hours to repair. Consistent with this idea, RNF4 depletion by either of two distinct siRNAs enhanced the persistence of SUMO proteins and the well-defined DSB marker γH2AX at micro-irradiated sites (Fig. 1A,B). Furthermore, effective RNF4 accrual at DNA damage sites required the DSB-responsive proteins MDC1, RNF8, 53BP1, and BRCA1 (Fig. 1C; Supplemental Fig. S1A, numbers represent the proportion of cells displaying γH2AX recruitment that also displayed RNF4 recruitment), with RNF4 recruitment being most markedly diminished when several of these factors were codepleted (see Supplemental Fig. S1A for effects of individual factor depletions). These data therefore suggested that RNF4 might be targeted to DNA damage sites, most likely DSBs, by interactions with multiple DDR proteins.
Figure 1.
RNF4 accumulation at DNA damage sites requires its SIM domains, PIAS1, PIAS4, and DDR proteins. (A) U2OS cells stably expressing RFP-SUMO1 and transfected with the indicated siRNAs were laser-micro-irradiated, fixed after 2 h, and then analyzed by immunofluorescence. (B) As in A, but with cells expressing RFP-SUMO2. (C) Experiments with U2OS cells stably expressing YFP-RNF4 were performed as in A. For quantifications, numbers represent proportions of cells showing RNF4 accumulation out of γH2AX-positive cells ±SED (n > 100). Single depletions and their quantifications are presented in Supplemental Figure S1A. (D) U2OS cells stably expressing YFP-RNF4 wild-type (WT), LD, or SIM-deleted (ΔSIM) were micro-irradiated and live-imaged after 30 min. Additional time points are presented in Supplemental Figure S1B. For siRNA depletions, see Supplemental Figure S11, A and C. (E) Schematic of RNF4 structure. Bars: this and all other figures, 10 μm.
Because RNF4 contains SIMs (Fig. 1E) and since the SUMO E3 ligases PIAS1 and PIAS4 mediate SUMO accrual at DSBs (Galanty et al. 2009; Morris et al. 2009), we tested whether PIAS1/4 depletion affected RNF4 recruitment to laser-induced DNA damage. Indeed, RNF4 accrual was essentially abolished by codepleting PIAS1 and PIAS4 (Fig. 1C, bottom row; Supplemental Fig. S1A). Notably, efficient accumulation of RNF4 also required its four tandem SIMs but was not impaired by mutating its RING finger ubiquitin E3 ligase domain to yield an E3 ligase-dead (LD) protein (Fig. 1D; Supplemental Fig. S1B). Collectively, these findings indicated that PIAS-mediated sumoylation of DDR proteins at DSB sites mediates SIM-dependent recruitment of RNF4 to these regions.
RNF4 depletion impairs ubiquitin accrual but causes MDC1 and γH2AX persistence at DNA damage sites
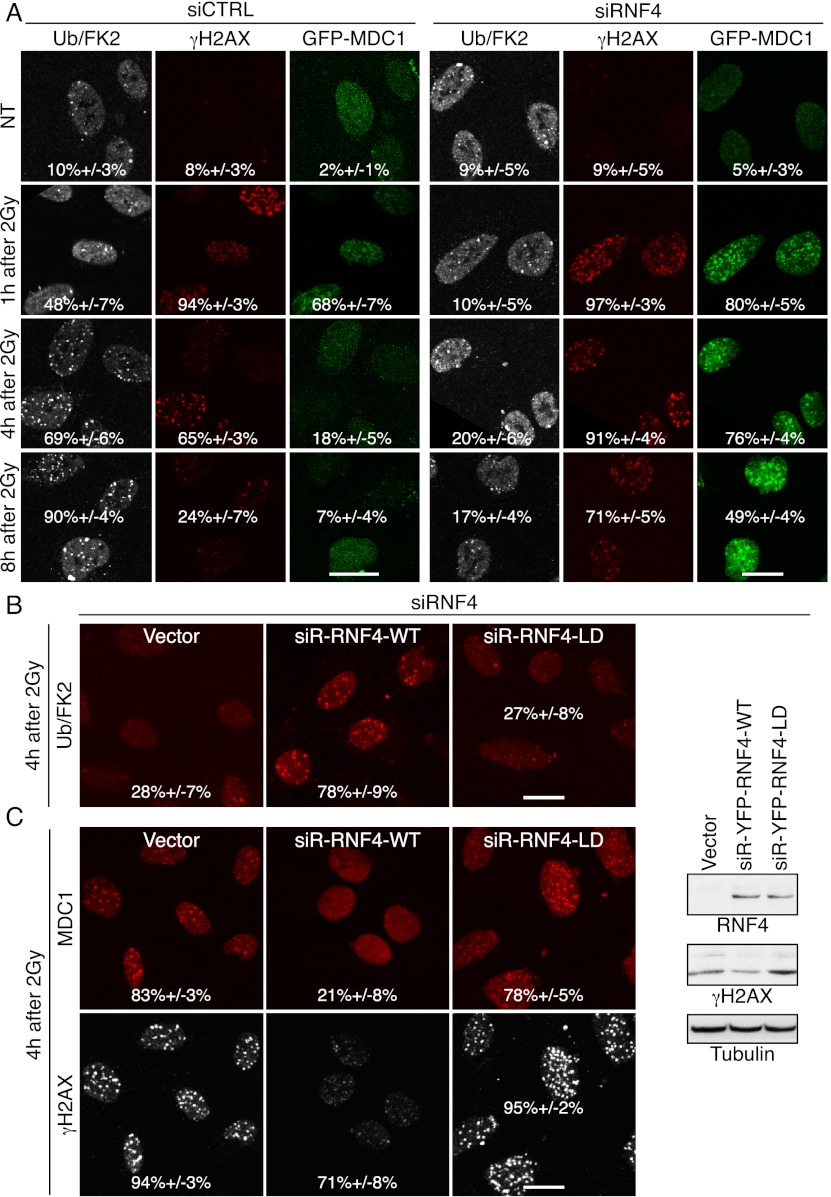
Previous work has shown that RNF4 functions as a ubiquitin E3 ligase in a manner that requires its RING finger domain (Hakli et al. 2004). Consistent with RNF4 functioning in this way at DNA damage sites, RNF4 depletion significantly reduced the production of ubiquitin adducts within IRIF, as detected by the FK2 antibody (Figs. 2A, 5E [below]). Moreover, complementation experiments with stable cell lines expressing siRNA-resistant RNF4 derivatives revealed that wild-type but not RING finger-mutated (LD) RNF4 corrected the impaired ubiquitin adduct staining caused by siRNA-mediated depletion of endogenous RNF4 (Fig. 2B; Supplemental Fig. S2A). In line with our data on laser-micro-irradiated cells (Fig. 1A,B), RNF4 depletion also caused the persistence of γH2AX staining in IRIF (Fig. 2A). Furthermore, RNF4 depletion also markedly affected the γH2AX-binding factor MDC1 (Stucki et al. 2005), whose IRIF staining was substantially enhanced in RNF4-depleted cells compared with control cells, particularly at later time points (Fig. 2A, numbers represent the proportion of cells displaying detectable IRIF). As these effects of RNF4 depletion on MDC1 and γH2AX were complemented by wild-type but not RING finger-mutated RNF4 (Fig. 2C; Supplemental Fig. S2B), these findings suggested that impaired RNF4-mediated ubiquitylation leads to γH2AX and MDC1 persistence at DSB sites, possibly reflecting defective DSB repair.
Figure 2.
RNF4 depletion causes persistence of MDC1 and γH2AX foci. (A) U2OS cells stably expressing GFP-MDC1 were transfected with the indicated siRNAs, exposed to 2 Gy of IR, fixed after the indicated times, and analyzed by immunofluorescence. Quantification numbers represent proportions of cells showing Ub/FK2, γH2AX, or MDC1 foci ±SED (n > 100). (B,C) Cells stably expressing siRNA-resistant (siR) rat YFP-RNF4 wild type (WT) or LD or vector only were transfected with RNF4 siRNAs, exposed to 2 Gy of IR, fixed 4 h later, and processed as in A. MDC1 detection was with an anti-pSDTD-MDC1 antibody (Chapman and Jackson 2008) that detects constitutively casein kinase 2 phosphorylated MDC1. Quantifications for Ub/FK2 (B) and MDC1 and γH2AX (C) were done as in A. (Right panel) Corresponding samples were collected for immunoblotting. For additional time points, siRNA depletions, and resistant clones, see Supplemental Figures S2, A and B, and S11, A and B.
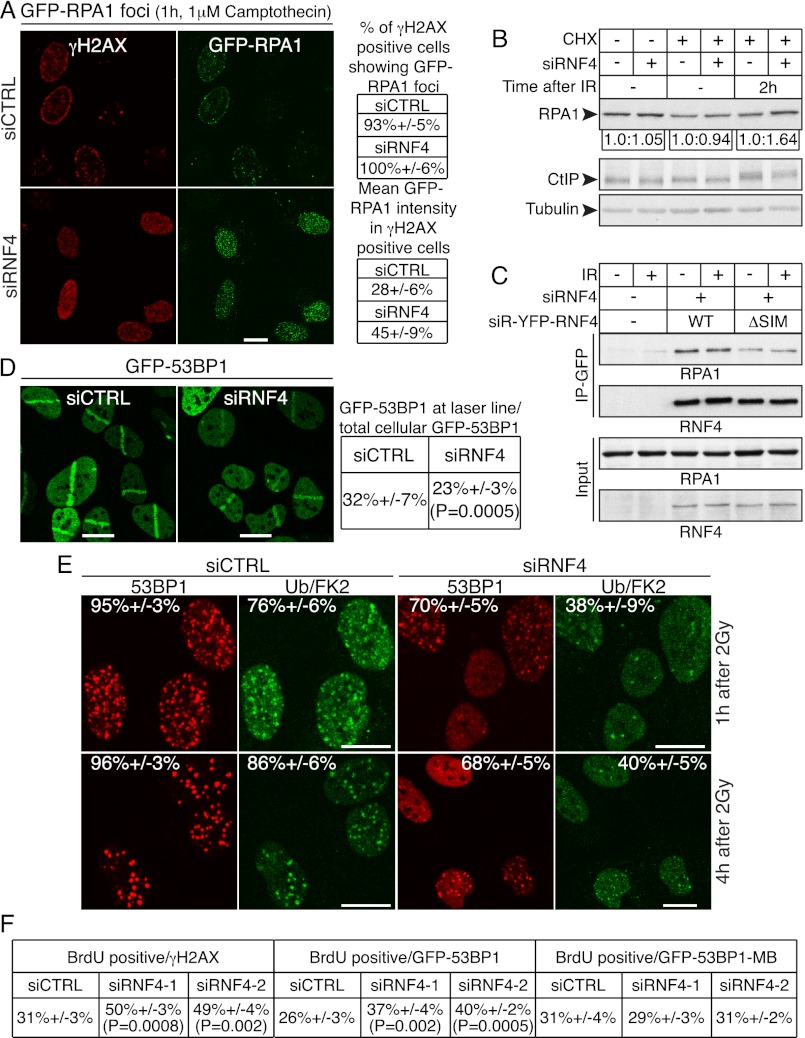
Figure 5.
RNF4 depletion causes hyperaccumulation of RPA and BrdU staining on damaged DNA and affects RPA levels following IR treatment. (A) U2OS cells stably expressing GFP-RPA1 (wild-type [WT]) were transfected with siRNAs and treated with 1 μM camptothecin for 1 h and then pre-extracted, fixed, and stained for γH2AX and DAPI (not shown); to the right, proportions of γH2AX-positive cells displaying GFP-RPA1 foci and the mean GFP-RPA1 fluorescence intensities are presented ±SED. Fluorescence intensity was calculated with Volocity 6.0 software; n > 100 cells per siRNA, accumulated over two independent experiments. (B) RNF4 regulates RPA1 turnover following IR treatment. U2OS cells were transfected with siRNAs. Forty-eight hours later, they were mock-treated or treated with 100 μg/mL cycloheximide (CHX) and/or 10 Gy of IR as indicated. Extracts were prepared 2 h following treatments, and samples were analyzed by 4%–12% gradient SDS-PAGE and immunoblotted as indicated. Quantifications of RPA1 were acquired with ImageJ software and normalized to tubulin. For each pair of samples (siCTRL and siRNF4), RPA1 levels in the siCTRL sample was set to 1.0, and RPA1 levels in the corresponding siRNF4 sample was normalized to the siCTRL value. (C) U2OS cells stably expressing GFP or siRNA-resistant (siR) YFP-RNF4 wild-type (WT) or SIM-deleted (ΔSIM) were transfected with siRNF4 (+). Forty-eight hours later, cells were mock-treated or treated with 10 Gy of IR and left to recover for 2 h. Samples were prepared with Benzonase (see the Materials and Methods), YFP-RNF4 immunoprecipitations were with GFP-Trap-A beads, and samples were analyzed by 4%–12% gradient SDS-PAGE and immunoblotting as indicated. Immunoprecipitation and detection of YFP-RNF4 derivatives were done with anti-GFP-Trap-A beads and GFP antibody, respectively. (D) Cells stably expressing GFP-53BP1 were transfected with RNF4 siRNA. Forty-eight hours later, they were laser-micro-irradiated and, 20 min after this, imaged, and 53BP1 accumulation was calculated (n = 24 independent measurements; error bars, ±SED). 53BP1 accumulation at DNA damage sites reached a steady-state level 20 min after micro-irradiation. For equations and calculations, see the Materials and Methods. (E) U2OS cells were transfected with siRNAs and treated with IR, fixed after the indicated time points, and then pre-extracted, fixed, and stained for 53BP1 and Ub/FK2. Quantification numbers represent proportions of cells showing 53BP1 or Ub/FK2 foci ±SED (n > 100). (F) Control U2OS cells and U2OS cells stably expressing GFP-53BP1 or GFP-53BP1 whose BRCT region was replaced by that of MDC1 (GFP-53BP1-MB) were transfected with siRNAs, laser-micro-irradiated, fixed after 2 h, and then analyzed by immunofluorescence as indicated. Numbers presented are cells displaying BrdU staining at DNA damage sites under native conditions as a readout of resection as proportions of cells displaying γH2AX or 53BP1 recruitment ±SED (n > 100); see Supplemental Figures S7–S9 for representative images and Supplemental Figure S11A for siRNA depletions.
RNF4 promotes DSB repair
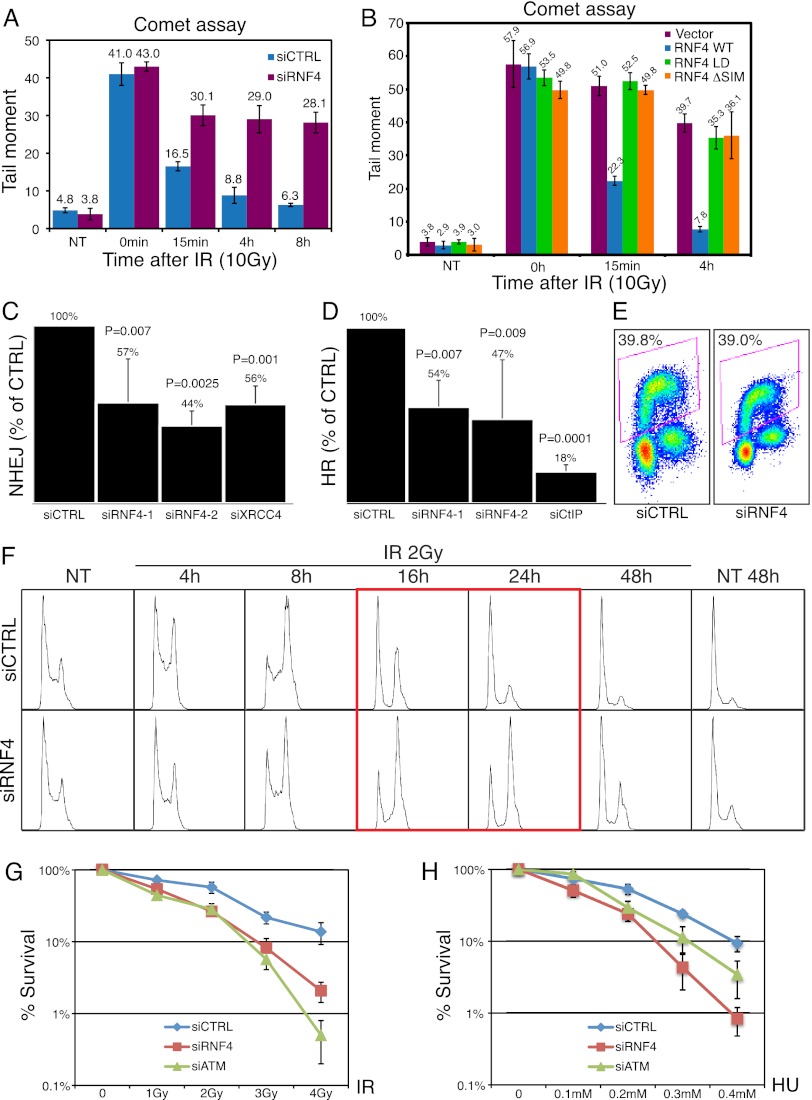
In accord with a model in which RNF4 promotes DNA repair, we found that RNF4 depletion reduced rates of chromosomal DSB repair, as measured by neutral comet assays (Fig. 3A; Supplemental Fig. S3). Furthermore, these effects of RNF4 depletion on DSB repair, as measured by comet assay, were complemented by reintroducing wild-type but not RING finger-mutated or SIM-deleted RNF4 into RNF4-depleted cells (Fig. 3B; Supplemental Fig. S4). To see which DSB repair pathways were affected by RNF4 depletion, we assessed DSB repair by NHEJ or HR individually. Thus, by using random plasmid integration into genomic DNA as a measure of NHEJ (Stucki et al. 2005), we found that this pathway was markedly impaired by RNF4 depletion (Fig. 3C, note that similar effects were observed with two different siRNAs targeting RNF4 and that depletion of the NHEJ component XRCC4 served as a positive control). Furthermore, RAD51-mediated repair of chromosomal DSBs by HR (Pierce et al. 2001) was also impaired by RNF4 depletion (Fig. 3D, depletion of the HR-promoting factor CtIP served as a positive control). Importantly, these effects of RNF4 depletion on HR did not reflect indirect effects on cell cycle progression because flow cytometry analyses revealed that cell cycle profiles of RNF4-depleted cell populations were essentially equivalent to those of control cells (Figs. 3F, 4B). Furthermore, RNF4-depleted cells progressed efficiently into and through S phase in the absence of DNA damage, as measured by quantifying incorporation of the thymidine analog ethynyl deoxyuridine (EdU) by pulse labeling (Fig. 3E).
Figure 3.
RNF4 promotes DSB repair, recovery from IR-induced G2 checkpoint arrest, and cell survival following genotoxic stress. (A) DSB repair is impaired in RNF4-depleted cells. U2OS cells were transfected with siRNAs and, 48 h following siRNA transfection, exposed to 10 Gy of IR, harvested at the indicated times, and subjected to neutral comet assays; tail moment quantifications are presented in the histogram (n > 150 measurements accumulated over three independent experiments; error bars, ±SED), (NT) Cells not treated with IR. Note that the total amount of DNA damage present in the cells immediately after irradiation was not significantly different between control and RNF4-depeleted cells. See Supplemental Figure S3 for representative images. (B) U2OS cells stably expressing vector only or siRNA-resistant YFP-RNF4 wild type, LD, or ΔSIM were transfected with siRNAs and, 48 h later, exposed to 10 Gy of IR, harvested at the indicated times, and subjected to neutral comet assays. Tail moment quantifications are presented. See Supplemental Figure S4 for representative images and expression levels of siRNA-resistant RNF4 derivatives. (C,D) Effects of RNF4 depletion on NHEJ (C) or HR-mediated gene conversion (D); error bars, ±SED. (E) S-phase index of control and RNF4-depleted cells as measured by pulse EdU incorporation. (F) Cell cycle profiles of irradiated and nonirradiated control and RNF4-depleted cells. U2OS cells were transfected with siRNAs and, 48 h following siRNA transfection, exposed to 2 Gy of IR, harvested at the indicated times, and subjected to flow cytometry (16- and 24-h time points following irradiation are boxed in red). (G,H) Effects of RNF4 depletion on colony formation following exposure to IR (G) or HU (H); error bars, ±SED; data represent four independent experiments. For siRNA depletions, see Supplemental Figure S11, A and C.
Figure 4.
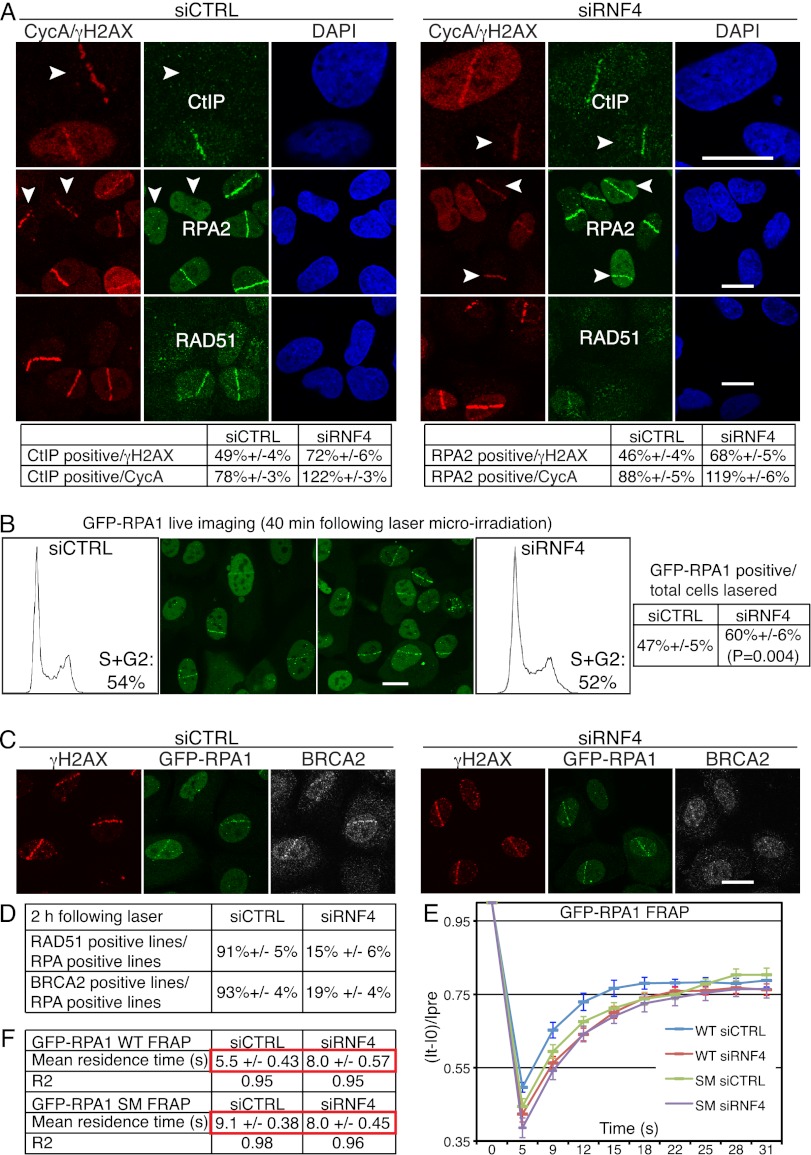
RNF4 regulates RPA turnover and BRCA2-mediated RAD51 loading onto damaged DNA. (A) RNF4 is required for normal accumulation of CtIP and RPA2 and for efficient accumulation of RAD51 at DNA damage sites. U2OS cells transfected with siRNAs were laser-micro-irradiated, fixed after 2 h, and then analyzed by immunofluorescence as indicated; arrows mark cyclinA (CycA)-negative cells. For additional images of RPA staining and cyclinA immunoblotting in control and RNF4-depleted cells, see Supplemental Figure S5A. Quantification numbers represent proportions of cells showing CtIP or RPA2 accumulation out of γH2AX- or cyclinA-positive cells ±SED (n > 100). (Note that the staining of RPA2 and CtIP in cyclinA-negative cells is unlikely to be left over from the previous G2, as these cells would have to have been lasered/damaged in S/G2 and would then have had to progress through mitosis with high amounts of damage even though they possess an intact G2/M checkpoint [see Fig. 3F], all within the 2-h incubation step of this experiment.) (B) U2OS cells stably expressing GFP-RPA1 (wild-type [WT]) were transfected with siRNAs and laser-micro-irradiated 48 h later (to allow subsequent quantification, the numbers of cells lasered were documented). GFP-RPA1 lines were live-imaged and counted 40 min post-irradiation. Proportions of cells displaying GFP-RPA1 lines out of total cells irradiated are presented ±SED with the corresponding flow cytometry profiles (n > 150 cells per siRNA, accumulated over three independent experiments). (C) RNF4 is required of efficient accumulation of BRCA2 at DNA-damaged sites. U2OS cells stably expressing GFP-RPA1 were treated as in A and stained with the indicated antibodies. For additional images of RAD51 and BRCA2 accumulation in GFP-RPA1 control and RNF4-depleted cells, see Supplemental Figure S5, B and C. (D) Quantifications of RAD51- and BRCA2-positive cells in GFP-RPA1-positive control and RNF4-deleted cells (n ≥ 100 cells per siRNA, ±SED). (E,F) U2OS cells stably expressing GFP-RPA1 wild-type (WT) or GFP-RPA1K449R,K577R (SM) were transfected with siRNAs and, 48 h later, laser-micro-irradiated and subjected to FRAP analysis 40 min post-irradiation (n = 24 independent measurements; error bars, ±SED). For equations and calculations, see the Materials and Methods. For siRNA depletions, see Supplemental Figure S11A.
Strikingly, while flow cytometry revealed that RNF4-depleted cells effectively mediated a G2–M cell cycle checkpoint arrest after IR treatment (Fig. 3F), they reproducibly displayed a pronounced delay in checkpoint recovery, as evidenced by hindered resumption of cell cycle progression (cf. the red-boxed profiles in Fig. 3F). These data, combined with the fact that RNF4 depletion caused persistent MDC1 and γH2AX accumulation at DSB sites, suggested that defective DSB repair in RNF4-depleted cells causes persistent DNA damage checkpoint activation. In accord with the impact of RNF4 on NHEJ and HR, clonogenic cell survival assays established that RNF4 depletion caused hypersensitivity to IR, which kills cells primarily through generating DSBs (Fig. 3G), and to chronic hydroxyurea (HU) treatment, which causes replication fork stalling and also yields some DSBs that are repaired by HR (Fig. 3H) (we note that the hypersensitivity of RNF4-depleted cells to HU could also partially reflect roles for RNF4 in promoting responses to replication fork stalling).
RNF4 promotes RPA turnover and RAD51 loading on ssDNA
As RNF4 was required for effective HR, we investigated whether RNF4 depletion influenced the accumulation of HR components at DNA damage sites. To do this, we employed laser micro-irradiation conditions in which DNA damage-induced accumulation of HR-promoting factors such as CtIP, RPA, BRCA2, and RAD51 only occurred in cyclinA-positive cells (Fig. 4A, note that CtIP, RPA2, and RAD51 accumulate at DNA damage sites in the S/G2 cell exhibiting pan-nuclear cyclinA [CycA] staining but not in the cell lacking detectable cyclinA, marked with an arrow). While RNF4 depletion did not appreciably affect CtIP recruitment to DNA damage sites in S/G2 cells (Fig. 4A, top right row), unlike the situation in control cells, CtIP was also recruited to DNA damage sites in some RNF4-depleted cells that displayed weak or no cyclinA staining (Fig. 4A, top right row). Similarly, accumulation of the second-largest subunit of RPA (RPA2) at sites of laser damage, reflecting the resection of DSBs to produce ssDNA, took place effectively in RNF4-depleted cells, but unlike in control cells, this occurred not only in cyclinA-positive cells, but also in a subset of cyclinA-negative cells (Fig. 4A, middle rows; Supplemental Fig. S5A). Importantly, only some (∼20%) cyclinA-negative, RNF4-depleted cells exhibited RPA2 or CtIP recruitment (see quantification tables in Fig. 4A; also note that Fig. 4A and Supplemental Fig. S5A only display examples of cyclinA-negative cells that show CtIP or RPA2 recruitment). These data therefore implied that RNF4 restrains DSB resection in cells that are in late G1 and/or early S phase. In agreement with higher proportions of RNF4-depleted cells showing DNA damage accumulation of CtIP and RPA compared with control cells, a higher proportion of these cells also displayed ssDNA at DNA damage sites, as measured by 5-bromo-2′-deoxyuridine (BrdU) staining under nondenaturing conditions, a technique that detects resected but not nonresected DNA (see Supplemental Fig. S7 for representative images and quantifications; Bekker-Jensen et al. 2006). In accord with these data, when we analyzed a cell line stably expressing green fluorescent protein (GFP) fused to RPA1, whereas 47% of control cells exhibited RPA recruitment to sites of laser-induced DNA damage, this figure was increased to 60% in RNF4-depleted cells (Fig. 4B, note that the cell cycle distributions of control and RNF4-depleted cells were essentially equivalent).
In striking contrast to the situation with CtIP and RPA, RNF4-depleted cells were markedly defective in the accrual of RAD51 at ssDNA sites (Fig. 4A, bottom rows; see Supplemental Fig. S5A,B for additional representative images and for immunoblotting analysis showing that IR and/or RNF4 depletion did not detectably affect cyclinA levels). Indeed, while ∼91% of cells displaying RPA recruitment in control cell populations also exhibited RAD51 recruitment, this figure was reduced to 15% upon RNF4 depletion (Fig. 4D). Furthermore, interfering with RNF4 function by stably expressing E3 ligase-defective RNF4 (but not wild-type RNF4) also caused accumulation of RPA2 in some cyclinA-negative cells and impaired RAD51 accumulation at DNA damage sites (data not shown). In line with the connection between RAD51 and BRCA2 loading onto resected ssDNA (Jensen et al. 2010; Liu et al. 2010; Thorslund et al. 2010), we also observed significant reductions in BRCA2 accumulation at DNA damage sites in RNF4-depleted cells (Fig. 4C; see Fig. 4D for quantification of these effects; see Supplemental Fig. S5C for additional representative images). Because cells with impaired RNF4 function did not efficiently accumulate RAD51 or BRCA2 on resected DNA but nevertheless displayed effective CtIP and RPA1/2 recruitment plus effective ssDNA generation, as measured by BrdU staining, these data collectively indicated that RNF4-depleted cells were able to resect DSBs but were unable to effectively replace RPA with RAD51 and BRCA2 at ssDNA sites.
In light of the above results, we tested whether RNF4 affected the dynamics of RPA at DNA damage sites by carrying out fluorescence recovery after photo-bleaching (FRAP) analyses on a cell line stably expressing RPA1 fused to GFP. This revealed that the residence time of GFP-tagged wild-type RPA1 at DNA damage sites was significantly higher in RNF4-depleted cells than in control cells (Fig. 4E,F) (that is, fluorescence intensity in the damaged region took longer to recover in RNF4-depleted cells than in control cells). This suggested that RNF4 promotes RPA turnover at DNA damage sites, thereby facilitating RAD51 loading. Of potential relevance given that RNF4 is a SUMO-dependent ubiquitin ligase, it has been reported that a small proportion of RPA1 is SUMO2/3-modified on Lys 449 and Lys 577 and that these sites promote RAD51 binding, induction of RAD51 ATP hydrolysis in cells, and DSB repair by HR (Dou et al. 2010). To explore whether the observed effects of RNF4 on RPA might be mediated via these sites, we established a cell line stably expressing GFP-RPA1 mutated in the sumoylation motifs (generating a nonsumoylatable mutant [SM]). Analysis of these cells revealed that the residence time of SM RPA1 at DNA damage sites was higher than that of the wild-type protein and was similar to that of wild-type RPA1 upon RNF4 depletion (Fig. 4E,F). Furthermore, unlike the situation for wild-type RPA1, the residence time of nonsumoylatable RPA1 was not increased upon RNF4 depletion (Fig. 4E,F). Additionally, when we depleted endogenous RPA1 in cells stably expressing GFP-RPA1 wild-type or SM, we found that the SUMO sites (Lys 449 and Lys 577) were important for efficient accumulation of RAD51 (as previously suggested by Dou et al. 2010) as well as BRCA2 at DNA damage sites induced by laser micro-irradiation (Supplemental Fig. S6). Because the effects of the RPA1 SUMO site mutations on RAD51 and BRCA2 accumulation were not as strong as those of RNF4 depletion, this suggested that RNF4 likely affects additional factors playing roles in replacing RPA with RAD51.
Collectively, the above data supported a model in which SUMO-modified RPA is targeted by RNF4 in a manner that enhances RPA turnover at DNA damage sites, thereby promoting BRCA2-mediated RAD51 loading and ensuing HR events. In accord with this, when we treated cells with camptothecin, which is thought to yield DSBs only in S phase, the mean intensity of RPA accumulation in γH2AX-positive cells was higher in RNF4-depleted cells than in control cells (Fig. 5A). Because RNF4 can target proteins for degradation, we assessed whether RPA1 levels were affected in response to DNA damage and, if so, whether this was RNF4-dependent. Indeed, while RNF4 depletion did not appreciably affect RPA1 levels under control conditions, in cells that had been IR-treated, RPA1 levels were reproducibly higher in RNF4-depleted cells than in control cells (Fig. 5B, note that the cells were cultured in the presence of cycloheximide to prevent de novo protein synthesis). Further highlighting the connection between RNF4 and RPA, the two proteins coimmunoprecipitated in a manner that was substantially impaired when the SIM region of RNF4 was deleted (Fig. 5C).
To explore the mechanism by which RNF4 depletion leads to excessive DNA end resection in late G1 and/or early S phase (cyclinA-negative cells) (see Fig. 4A,B; Supplemental Figs. S5, S7), we assessed the effect of RNF4 depletion on 53BP1 accumulation at DNA damage sites. The rationale for this was that 53BP1 hinders resection in BRCA1-deficient cells (Bunting et al. 2010) and that we found that RNF4 was required for effective accumulation of ubiquitin conjugates at DSB sites, which are in turn needed for efficient 53BP1 accrual (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Doil et al. 2009; Stewart et al. 2009). Notably, 53BP1 accumulation at regions of DNA damage was lower in RNF4-depleted cells than in control cells, as shown by measuring either 53BP1 intensity at damage sites or the proportion of cells displaying 53BP1 IRIF (Fig. 5D,E). To see whether the reduced 53BP1 accrual upon RNF4 depletion might be linked to excessive resection under these conditions, we established cell lines stably expressing GFP-53BP1 or GFP-53BP1-MB, the latter of which comprises 53BP1 with its BRCT domains replaced with the BRCT region of MDC1, a region that is necessary and sufficient for MDC1 recruitment to IRIF (Stucki et al. 2005). Strikingly, while RNF4 depletion with two different siRNAs enhanced markers of resection in control cells (Figs. 4A,B, 5F; Supplemental Figs. S5, S7) and in cells expressing GFP-53BP1 (Fig. 5F; Supplemental Fig. S8), this had little or no effect on resection in cells expressing GFP-53BP1-MB (Fig. 5F; Supplemental Fig. S9). These data therefore indicated that forcing 53BP1 recruitment to DNA damage sites via the MDC1 BRCT region overcomes the effect of RNF4 on resection, thereby supporting a model in which the effect of RNF4 depletion on resection is mediated at least in part by reduced 53BP1 accumulation at DNA damage sites.
RNF4 regulates MDC1 turnover at DNA damage sites
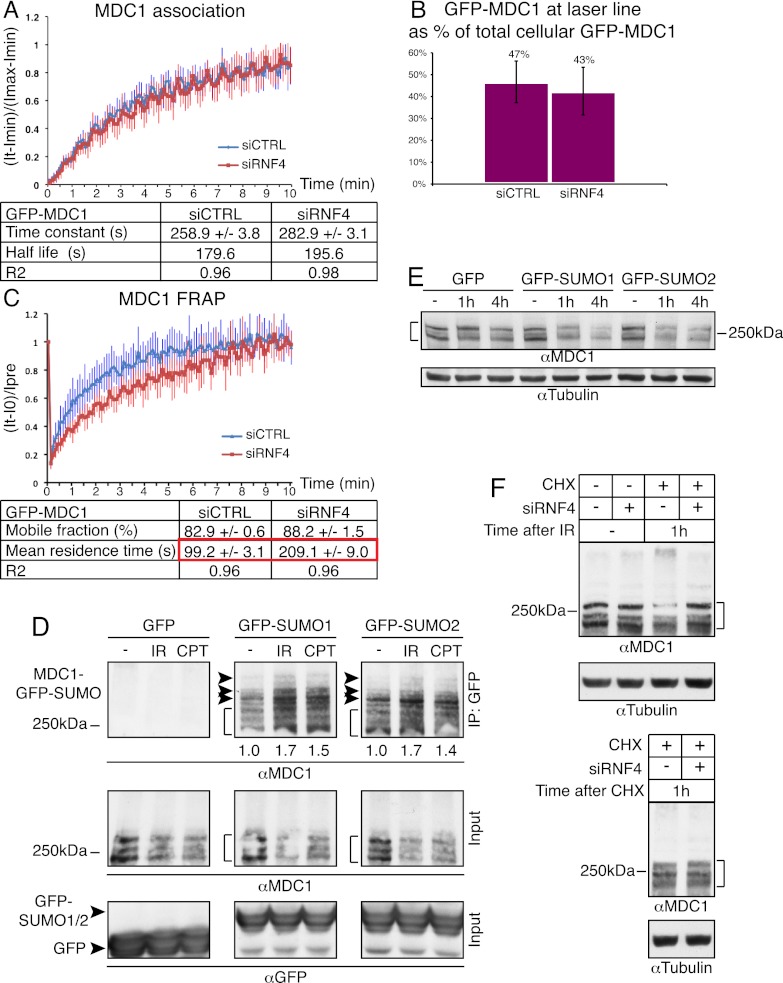
Although the effects of RNF4 on RPA, BRCA2, and RAD51 helped to explain the HR defects of RNF4-depleted cells, they could not easily explain the NHEJ defects of such cells. We therefore used micro-irradiation and FRAP experiments to see whether RNF4 affected the DSB recruitment and/or dynamics of factors known to influence NHEJ, including MDC1. By using a cell line stably expressing GFP-tagged MDC1 in live-cell imaging studies coupled with laser micro-irradiation, we observed that the initial association kinetics of MDC1 and the proportion of MDC1 accumulated at DNA damage sites at early time points were not significantly affected by RNF4 depletion (Fig. 6A,B). In contrast, FRAP assays revealed that the mean residence time of GFP-MDC1 at damaged sites was considerably higher in RNF4-depleted cells than in control cells (209 vs. 99 sec, respectively) (Fig. 6C). These data indicated that, as we had observed for RPA, RNF4 promotes MDC1 turnover and/or removal from DNA damage sites. Furthermore, because RNF4 recognizes sumoylated substrates (Sun et al. 2007; Lallemand-Breitenbach et al. 2008; Tatham et al. 2008), these findings suggested that MDC1 might be sumoylated. Indeed, by using a GFP-SUMO immunoprecipitation strategy (employing high stringency conditions) (see the Materials and Methods), we found that MDC1 was modified by both SUMO1 and SUMO2 in a manner that was enhanced when cells were treated with IR or camptothecin (Fig. 6D, note that although RNF4 preferably binds poly-SUMO2/3 chains, it might recognize multiple adjacent SUMO1 sites or mixed chains containing SUMO1 as a chain terminator). Taken together with our other data, these findings suggested that RNF4 likely impacts on HR and NHEJ by regulating the functions and turnover of multiple DDR components, including RPA and MDC1.
Figure 6.
RNF4 regulates MDC1 turnover at DNA damage sites. (A,B) Cells stably expressing GFP-MDC1 were transfected with siRNAs. Forty-eight hours later, cells were laser-micro-irradiated, and then MDC1 association kinetics (A) and its accumulated amounts (B) were calculated (n = 24 independent measurements; error bars, ±SED). MDC1 accumulation reached a steady-state level after 10 min. For equations and calculations, see the Materials and Methods. (C) As in B, but cells were subjected to FRAP analysis (n = 10 independent measurements; error bars, ±SED). For equations and calculations, see the Materials and Methods. (D) HEK293 cells were transfected with GFP-SUMO1, GFP-SUMO2, or GFP. Extracts were prepared 1 h after mock treatment or treatment with 10 Gy or IR or 1 μM camptothecin (CPT). GFP-SUMO immunoprecipitations were with GFP-Trap-A beads; samples were analyzed by 4%–15% gradient SDS-PAGE and immunoblotting as indicated. Arrows mark SUMO-modified MDC1, while the brackets mark the triplet of MDC1 species. Quantifications of immunoblot signals of SUMO-modified MDC1 forms were with ImageJ software. (E) U2OS cells stably expressing GFP, GFP-SUMO1, or GFP-SUMO2 were mock-treated (−) or treated with 10 Gy of IR. Whole-cell extracts were prepared after the indicated times and samples analyzed as in D, but with 4%–20% gradient SDS-PAGE. (F) U2OS cells were transfected with siRNAs. Forty-eight hours later, they were mock-treated (−) or treated with 100 μg/mL cycloheximide (CHX) and 10 Gy of IR (top panel) or with cycloheximide alone (CHX; bottom panel). Extracts were prepared after the indicated times, and samples were analyzed as in D. For siRNA depletions, see Supplemental Figure S11A.
Because RNF4 can target proteins for degradation (Lallemand-Breitenbach et al. 2008; Tatham et al. 2008), we tested whether MDC1 protein levels were affected in response to DNA damage. Indeed, MDC1 levels were slightly but reproducibly reduced in IR-treated cells, with this reduction being more evident when SUMO1 or SUMO2 was stably overexpressed in cells (Fig. 6E). Moreover, through combining cycloheximide with IR treatments in RNF4-depleted or control cells, we found that levels of full-length MDC1 were reduced in response to IR and that this was accompanied by an increase in higher-molecular-weight forms of the protein (Fig. 6F). Furthermore, and consistent with a model in which RNF4 targets MDC1, these IR-induced changes in MDC1 were RNF4-dependent (Fig. 6F). Although we were able to detect coimmunoprecipitation of MDC1 with RNF4 in a SIM-dependent manner (data not shown), we could not reproducibly observe a clear increase in this interaction following DNA damage, possibly because RNF4 binding leads to rapid MDC1 degradation.
Links between RNF4 and proteasome functions at DNA damage sites
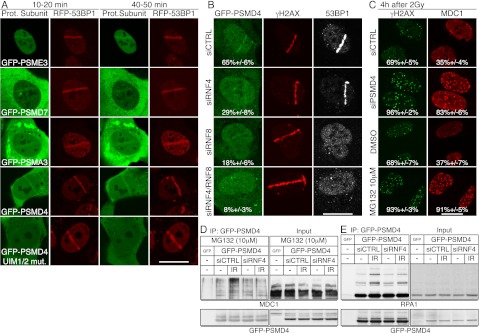
Given that RNF4 can target proteins for proteasome-mediated degradation (Lallemand-Breitenbach et al. 2008; Tatham et al. 2008), we speculated that the effects of RNF4 on DSB repair might be associated with proteasome recruitment to DNA damage sites. In line with this idea and published observations (Blickwedehl et al. 2007, 2008; Ben-Aroya et al. 2010; Levy-Barda et al. 2011), we observed that various GFP-tagged proteasome subunits were recruited to laser-induced DNA damage (Fig. 7A). These included the proteasome activator PSME3, implicated in MDM2-mediated p53 degradation (Zhang and Zhang 2008) and recently in the DDR (Levy-Barda et al. 2011); PSMD7, a core component of the 19S proteasome regulator (PA700); the peptidase PSMA3, a core component of the proteolytic 20S proteasome; and PSMD4, another component of the 19S proteasome regulator (Fig. 7A) (PSME1, a component of the 11S immunoproteasome activator [Stadtmueller and Hill 2011], was not detectably recruited to DNA damage regions [data not shown]).
Figure 7.
Accumulation of the proteasome component PSMD4 at DNA damage sites requires its UIM domains, RNF4 and RNF8. (A) U2OS cells stably expressing RFP-53BP1 were transfected with GFP-PSME3, GFP-PSMD7, GFP-PSMA3, GFP-PSMD4, or GFP-PSMD4L218A,L220A,Y289A,M291A (UIM1/2 mut.). Forty-eight hours later, cells were laser-micro-irradiated, and live cells were imaged at the indicated times. (B) U2OS cells stably expressing GFP-PSMD4 transfected with siRNAs were laser-micro-irradiated, fixed after 2 h, and then analyzed by immunofluorescence. Quantifications: Numbers represent proportion of cells showing GFP-PSMD4 accumulation out of γH2AX-positive, ±SED (n > 100). (C) PSMD4 depletion or proteasome inhibition causes persistence of MDC1 and γH2AX IRIF. U2OS cells were transfected with siRNAs or treated with MG132 or DMSO immediately after irradiation (2 Gy of IR), fixed after 4 h, and analyzed by immunofluorescence as indicated. Quantifications: Numbers represent proportion of cells showing γH2AX or MDC1 foci, ±SED (n > 100). For time course and quantifications, see Supplemental Figure S10 (MDC1 detection was as in Fig. 2C). (D,E) PSMD4 interaction with MDC1 and RPA1 following IR is RNF4-dependent. (D) U2OS cells stably expressing GFP-PSMD4 were transfected with siRNAs and HA-tagged ubiquitin. Forty-eight hours later, cells were exposed to 10 Gy of IR and MG132. Extracts were prepared 4 h later and used for GFP-PSMD4 or GFP immunoprecipitations by GFP-Trap-A beads. Samples were analyzed by 4%–12% SDS-PAGE and immunoblotted as indicated. (E) As in D, but cells were not transfected with HA-Ub or treated with MG132. For siRNA depletions, see Supplemental Figure S11, A and C.
To study proteasome recruitment to DNA damage sites in more detail, we focused on PSMD4 because it is the main ubiquitin-binding component of the 19S proteasome regulator and is phosphorylated by the DDR protein kinases ATM and/or ATR following DNA damage induction (Matsuoka et al. 2007). Notably, we found that point mutations in the ubiquitin-interacting motifs (UIMs) of PSMD4 that abrogate their ability to bind ubiquitin (Young et al. 1998) significantly reduced PSMD4 accumulation at DNA damage sites (Fig. 7A, bottom panels). Furthermore, by using a cell line stably expressing GFP-tagged PSMD4, we found that effective PSMD4 accumulation at DNA damage sites required RNF4 and RNF8 (Fig. 7B) (RNF8 deletion had the strongest effect on PSMD4 accrual, probably because it promotes RNF4 recruitment to DNA damage [see Figure 1C; Supplemental S1A] and also mediates DNA damage-induced recruitment of the RNF168 and BRCA1 ubiquitin E3 ligases [Huen et al. 2007; Stewart et al. 2009]). Consistent with there being functional connections between RNF4, PSMD4, and the proteasome, as was the case for RNF4 depletion (Fig. 2A), PSMD4 depletion or proteasome inhibition with the compound MG132 caused persistent MDC1 and γH2AX foci following IR (Fig. 7C; Supplemental Fig. S10). Moreover, in accord with MDC1 and RPA1 being sumoylated and targeted by RNF4, we found that GFP-PSMD4 interacted with MDC1 and RPA1 in a manner that was enhanced after IR and was RNF4-dependent (Fig. 7D,E, note that GFP-PSMD4 mainly bound the slower-migrating forms of MDC1). Unlike GFP-RPA1 binding, however, the GFP-PSMD4–MDC1 interaction was not clearly and reproducibly observed without proteasome inhibition, possibly reflecting faster proteasomal turnover of modified MDC1 than of modified RPA1. Taken together with our other findings, these data supported a model in which RNF4 accrual at DNA damage sites leads to PSMD4-targeted, proteasome-mediated MDC1 and RPA1 turnover.
Discussion
We showed that the STUbL RNF4 is rapidly recruited to DSB sites and promotes DSB repair. Consistent with its known biochemical functions, RNF4 recruitment to DNA damage requires its tandem SIM region, DDR mediator proteins, and the SUMO E3 ligases PIAS1 and PIAS4. Furthermore, our data indicate that RNF4 and specifically its ubiquitin E3 ligase activity and SIMs are needed for effective DSB repair. Additionally, we showed that RNF4 and its ubiquitin E3 ligase activity are also needed for effective ubiquitin adduct formation at DNA damage regions. These findings therefore suggest a model in which RNF4 promotes DSB repair by mediating RNF4-SIM-targeted ubiquitylation of sumoylated DDR components at sites of DNA damage.
Crucially, we established that RNF4 depletion impairs both NHEJ and HR and is associated with defective RAD51 and BRCA2 loading on ssDNA, with MDC1 and γH2AX persistence at DNA damage sites, and defective recovery from IR-induced cell cycle arrest. Although RNF4 very likely operates at multiple levels in the DDR, our data suggest that one of its key functions is to affect the dynamics of DDR–protein complexes. Thus, we found that it enhances RPA turnover at DNA damage sites, with this effect being abrogated when RPA1 is mutated on Lys 449 and Lys 557 so that it can no longer be effectively sumoylated. Notably, previous work has shown that cells expressing this nonsumoylatable RPA1 derivative are defective in RAD51 loading and HR (Dou et al. 2010), and in our experiments, we found that this mutated RPA1 derivative was defective in supporting efficient RAD51 and BRCA2 accrual at DNA damage sites. Because RNF4 depletion also causes such defects, this suggests that RNF4-mediated RPA turnover promotes the exchange of RPA with RAD51 and BRCA2 on ssDNA so that HR can proceed effectively. We also established that RNF4 enhances the rate of turnover of MDC1 at DNA damage sites. By analogy to its effects on RPA, RAD51, and BRCA2, we speculate that RNF4-mediated MDC1 turnover facilitates the access of other DDR factors to damaged DNA. This could allow coordinated and potentially temporally controlled transitions in DDR protein complexes, thus promoting effective DSB repair—including NHEJ, where MDC1 has its most pronounced effects (Stucki et al. 2005; Xie et al. 2007)—and perhaps also the subsequent re-establishment of normal chromatin structure. Our data also indicate that RNF4 suppresses DNA end resection in late G1 and/or early S phases and that this at least in part reflects RNF4 facilitating 53BP1 accrual at DNA damage sites. Clearly, RNF4 could also target other DDR proteins, many of which are known to be sumoylated and/or ubiquitylated (for reviews, see Bergink and Jentsch 2009; Al-Hakim et al. 2010; Ciccia and Elledge 2010; Polo and Jackson 2011). Indeed, we recently obtained evidence that SUMO-modified BLM is also targeted by RNF4. It will be of interest to determine whether this influences BLM function and whether RNF4 also regulates the activities of additional DDR factors. It will also be interesting to determine whether RNF4 counterparts in yeast and other organisms promote DDR events by mechanisms analogous to those mediated by human RNF4.
Our findings suggest that RNF4 acts at least in part in conjunction with proteasomal components to enhance DSB repair. Proteasome function has been linked to the DDR previously by studies investigating the effects of proteasome inhibitors on IRIF and DNA repair by HR (Jacquemont and Taniguchi 2007; Murakawa et al. 2007). Furthermore, a previous study reported enhanced ubiquitylation of MDC1 on chromatin following IR, linking this to proteasome-mediated disassembly of MDC1 and γH2AX foci, although the ubiquitin E3 ligases remained unknown (Shi et al. 2008). In addition, cellular fractionation approaches have shown that proteasomes can accumulate on the chromatin of damaged cells (Blickwedehl et al. 2007, 2008), although the kinetics of proteasome component accumulation at DNA damage sites and the factors required have not hitherto been investigated. We established that multiple proteasome subunits, including the proteasome activator PSME3 that has been recently linked to the DDR while this work was ongoing (Levy-Barda et al. 2011), are detectable at DNA damage sites as early as 10 min following DSB induction. Moreover, we showed that the main ubiquitin-binding subunit of the 19S proteasome regulator complex PSMD4 accumulates at DNA damage sites in a manner that requires its intact UIMs together with RNF4 and RNF8. Consistent with this and our other data, we found that PSMD4 binds to MDC1 and RPA1 and that these interactions are enhanced following IR in an RNF4-dependent manner. Collectively, these results suggest that ubiquitin conjugates on DDR proteins are recognized by certain types of proteasome (such as the 19S–20S–PSME3 complex). Furthermore, because we found that PSMD4 depletion, like RNF4 depletion, leads to MDC1 and γH2AX persistence at DSB sites, this supports a model in which proteasome accrual mediated by the actions of RNF4 and other ubiquitin ligases is important for effective DSB repair. While it seems likely that such events are associated with enhanced protein turnover at DNA damage sites and involve proteasome-mediated degradation of DDR factors, it is also possible that proteasome recruitment to DNA damage sites also promotes DSB repair by nonproteolytic mechanisms.
While much is known about the mechanisms governing the assembly of DDR proteins at DSB sites, we know considerably less about events affecting the dynamics and/or disassembly of DDR–protein complexes and IRIF. Our findings identify RNF4 as a human DDR factor that specifically functions by modulating the dynamics of DDR foci. Accordingly, our findings suggest that the dynamic properties of IRIF are not mere consequences of their composition but are of functional importance. Indeed, it is tempting to speculate that RNF4-dependent, proteasome-mediated remodeling of DDR protein complexes—through catalyzing key protein handovers such as those that we have characterized—facilitates molecular transitions that are necessary for effective progression of NHEJ and HR through multiple, discrete stages. Such molecular handover mechanisms triggered by ubiquitylation and/or sumoylation could help explain the apparently paradoxical observation that these modifications often have pervasive effects on the function of a protein, even though only a small proportion of that protein is modified at a given time (Hay 2005). Finally, we believe that it will be interesting to establish whether RNF4 dysfunction is linked to cancer, whether this affects the ability of cancer cells to respond to DNA-damaging and/or DDR-targeting drugs, and whether RNF4 could itself represent a target for developing novel anti-cancer agents.
Materials and methods
Cell culture
HEK293 and U2OS cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; BioSera), 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma-Aldrich). U2OS cells stably expressing GFP, GFP-MDC1 (Kolas et al. 2007), RFP-53BP1, RFP-SUMO1, RFP-SUMO2, GFP-PSMD4, GFP-RPA1 wild-type and SM, GFP-53BP1 (Galanty et al. 2009), and GFP-53BP1 in which its BRCTs have been replaced with MDC1 BRCTs (GFP-53BP1-MB) were grown in standard U2OS medium supplemented with 0.5 mg/mL G418 (GIBCO; Invitrogen). U2OS cells stably expressing human or rat YFP-RNF4 (siRNA-resistant or not) wild-type, LD, or SIM-deleted (ΔSIM) were grown in standard U2OS medium supplemented with 2 μg/mL blasticidin (InvivoGen).
siRNA transfections and sequences
siRNA duplexes were from MWG Biotech or Qiagen (Supplemental Table S1). Two consecutive rounds of siRNA transfections were carried out with Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol unless otherwise specified. siRNA transfected cells were assayed 48 or 72 h after transfection for MDC1, 53BP1, BRCA1, ATM, and RPA1 depletions. For cotransfection with siRNA and expression constructs, cells were first transfected with siRNA, followed by plasmid transfection 8 h later by using FuGENE6 (Roche) according to manufacturer's protocol; 24 h after the plasmid transfection, a second round of siRNA transfection was made. Cells were assayed 48 h after plasmid transfections.
Plasmids and cloning
SUMO1 and SUMO2 cloning was as previously described (Galanty et al. 2009). R.T. Hay (Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dundee, UK) provided the mammalian expression clones of human YFP-RNF4 and rat YFP-RNF4 wild-type, LD, or ΔSIM. Rat YFP-RNF4 wild-type, LD, and ΔSIM siRNA-resistant clones were obtained by inserting the four nucleotide mismatches underlined (GAAUGGACGUCUCAUCGUU) into rat RNF4 cDNA by using QuikChange site-directed mutagenesis kit (Stratagene). PSME1, PSME3, PSMA3, and PSMD4 were PCR-amplified from a human fetal brain cDNA library and cloned into pEGFP-N1 and pEGFP-C3. The PSMD4 L218A,L220A,Y289A,M291A bacterial expression construct has been described previously (Young et al. 1998) and was provided by Patrick Young (Stockholm University, Sweden); this construct was used as a template for PCR amplification and cloning into pEGFP-C1. Sylvie Urbé (Cellular and Molecular Physiology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK) provided the GFP-PSMD7 mammalian expression plasmid. GFP-RPA1 wild type was PCR-amplified from a human fetal brain cDNA library and cloned into pEGFP-C2, GFP-RPA1K449R,K577R (SM) as previously described (Dou et al. 2010), and was generated using QuikChange site-directed mutagenesis kit (Stratagene). Construction of pEGFP-53BP1 with BRCT domains replaced by the BRCT domains of MDC1; the C-terminal half of 53BP1 was PCR-amplified and then inserted in BamHI/XhoI-digested pEGFP-53BP1 plasmid. This deleted BRCT domains of 53BP1 and introduced the XhoI site following nucleotide 5160 (amino acid 1720) of the 53BP1 ORF. The BRCT domains of MDC1 were PCR-amplified, digested with SalI and XhoI, and inserted at the XhoI site of pEGFP-53BP1 ΔBRCT. Primers were obtained from Sigma-Aldrich (Supplemental Table S2).
Laser micro-irradiation and imaging of live and fixed cells
For generation of localized damage in cellular DNA by exposure to a UV-A laser beam (Limoli and Ward 1993; Bekker-Jensen et al. 2006), cells were plated on glass-bottomed dishes (Willco-Wells) and presensitized with 10 μM BrdU (Sigma-Aldrich) in phenol red-free medium (Invitrogen) for 24 h (48 h for all BrdU staining-based assays) at 37°C. Laser micro-irradiation was performed by using a FluoView 1000 confocal microscope (Olympus) equipped with a 37°C heating stage (Ibidi) and a 405-nm laser diode (6 mW) focused through a 60× UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 mm. The time of cell exposure to the laser beam was ∼250 msec (fast scanning mode). Laser settings (0.4-mW output, 50 scans) were chosen to generate a detectable damage response restricted to the laser path in a presensitization-dependent manner without noticeable cytotoxicity. For all experiments involving RPA, RAD51, BRCA2, and BrdU, a laser power of 0.20- to 0.25-mW output and 50 scans were used. Imaging of live and fixed cells was done on the same microscope by using the objective and software as described above.
Immunofluorescence
Cells were washed three times with PBS/0.1% Tween-20 followed by pre-extraction for 10 min (pre-extraction buffer: 25 mM HEPES at pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100). Cells were fixed with 2% paraformaldehyde (w/v) in PBS for 20 min. Following three washes with PBS 0.1% Tween-20, cells were blocked for 1 h with 5% BSA in PBS/0.1% Tween-20, costained with the indicated antibodies (Supplemental Table S3) in blocking solution overnight, and then coimmunostained with the appropriate secondary antibodies (Supplemental Table S3) in blocking solution. The pre-extraction step was omitted and permeabilization (0.5% Triton X-100 in PBS) was performed after fixation for imaging of cyclinA, RAD51, BRCA2, RNF4, YFP-RNF4, GFP-PSMD4, and BrdU. Mean fluorescence intensity was calculated with Volocity 6.0 or Olympus FV-10 software. Bars are 10 μm throughout. All quantifications include ±SED. The P-value was calculated using Student's t-test.
FRAP and association kinetics
FRAP analyses were performed on the microscope used for laser micro-irradiation when the accumulation of the GFP-tagged protein on the laser track reached its maximal steady-state level. After a series of three prebleach images, a rectangular region placed over half of the laser-damaged line was subjected to a bleach pulse (five scans with 488-nm argon laser focused through a 60× UPlanSApo/1.35 oil objective, main scanner, 100% AOTF [acousto–optical tunable filter], slow scanning mode), followed by image acquisition at 6-sec intervals for GFP-MDC1 and fastest speed for GFP-RPA1. Average fluorescent intensities in the bleached region were normalized against intensities in an undamaged nucleus in the same field after background subtraction to correct for overall bleaching of the GFP signal due to repetitive imaging. For mathematical modeling of GFP-tagged protein mobility, (It − I0)/Ipre values were plotted as a function of time, where I0 is the fluorescence intensity immediately after bleaching, and Ipre is the average of the three prebleach measurements. Estimation of mobile protein fraction (A) and residence time (τ) were performed using Prism 4 software, assuming the existence of one protein population using the following equation: y(t) = A[1 − exp(−t/τ)]. The association kinetics of GFP-MDC1 at sites of laser micro-irradiation were monitored on the same microscope by measuring GFP fluorescence over time in the damaged region using the 488-nm argon laser. To correct for overall bleaching of the signal due to repetitive imaging, fluorescence intensities were normalized against intensities measured in an undamaged nucleus in the same field after background subtraction. Variations in fluorescence intensity (I) were plotted as a function of time (t) using Microsoft Excel software. Plotted data are averaged values of a minimum of 24 cells accumulated over three independent experiments. To compare between different experimental conditions, data were normalized against the fluorescence intensity at the time of micro-irradiation. For mathematical modeling of protein mobility, the association kinetics were determined from (It − Imin)/(Imax − Imin) plotted as a function of time, where Imax and Imin are the maximum and minimum values measured, respectively. Curve fitting and determination of the time constant (τ) for the association kinetics were carried out with Prism4 software, based on a first-order exponential model using the following equation: f(t) = 1 − exp(−t/τ). Calculation of the total GFP-MDC1 and GFP-53BP1 accumulated at sites of laser micro-irradiation was determined using (Imax line − background line)/(Imax cell − background cell), where background line and background cell are intensities of the image at the time that line intensities are at their maximum steady-state levels. Images were acquired at 6-sec intervals for GFP-MDC1 and 20-sec intervals for GFP-53BP1. The P-value was calculated using Student's t-test.
Treatment with DNA-damaging agents
Camptothecin and HU were obtained from Sigma-Aldrich. IR was performed with a Faxitron X-ray machine (Faxitron X-ray Corporation).
Immunoprecipitation and immunoblotting
Cell extracts were prepared on plates using lysis buffer containing 2% SDS, 50 mM Tris-HCl (pH 7.5), 10 mM N-ethylmaleimide (Sigma-Aldrich), and protease inhibitor cocktail (Roche), followed by sonication or passage of extracts through a 19-gauage needle-mounted syringe. For monitoring MDC1, cell extracts were prepared by using lysis buffer containing 20 mM HEPES (pH 7.4), 500 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Tween20, 10% glycerol, serine/threonine phosphatase inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail (Roche), and 10 mM N-ethylmaleimide (Sigma-Aldrich). Subsequently, extracts were sonicated and diluted 1:2 with the same buffer lacking NaCl. For GFP and YFP-RNF4 immunoprecipitation using GFP-Trap-A beads (ChromoTek GmbH), cell extracts were prepared using lysis buffer containing 20 mM HEPES (pH 7.4), 500 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Tween20, 10% glycerol, serine/threonine phosphatase inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail (Roche), and 10 mM N-ethylmaleimide (Sigma-Aldrich). Extracts were then sonicated and diluted 1:2 with the same buffer lacking NaCl. For all immunoprecipitations, extracts were supplemented with 200 μg/mL ethidium bromide to prevent nonspecific binding via DNA and cleared using centrifugation at 16,000g for 60 min at 4°C. Overnight incubation/binding with GFP-Trap-A beads at 4°C was followed by five washes, alternating three washes with immunoprecipitation buffer (250 mM NaCl) and two washes with lysis buffer (500 mM NaCl), and 5 min boiling in 1.5× SDS sample buffer. Proteins were resolved by 4%–18% SDS-PAGE (unless otherwise specified) and transferred to PVDF membrane (GE Healthcare). Immunoblotting was performed with the indicated antibodies (Supplemental Table S3). Immunoblotting for BRCA1 was done using a 1:1 mix of the rabbit antibodies in Supplemental Table S3. For coimmunoprecipitation of YFP-RNF4 with RPA as well as for GFP-PSMD4 with MDC1 and RPA1, cells were lysed on plates in Benzonase nuclease buffer: 20 mM Tris-HCl, 40 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.5% NP-40, and EDTA free protease inhibitor cocktail (Roche) supplemented with 100 U/mL Benzonase nuclease (Novagen) and 10 mM N-ethylmaleimide. Extracts were then collected, and the NaCl concentration was increased to 450 mM followed by 20 min of incubation on ice. The extracts were then cleared using centrifugation at 16,000g for 60 min at 4°C. The NaCl concentration was reduced to 225 mM and supplemented with 10 mM N-ethylmaleimide (Sigma-Aldrich) and serine/threonine phosphatase inhibitor cocktail (Sigma-Aldrich). The resulting extracts were subjected to an overnight incubation with GFP-Trap-A beads at 4°C followed by five washes with Benzonase nuclease buffer with 225 mM NaCl. Samples were subsequently analyzed by SDS-PAGE as described above. Quantifications of immunoblotting signals for SUMO-modified MDC1 and RPA1 levels were normalized to tubulin signals and were acquired by ImageJ software.
Flow cytometry and S-phase index measurements
To determine cell cycle distribution, cells were fixed with 70% ethanol, incubated for 30 min with RNase A (250 μg/mL) and propidium iodide (10 μg/mL) at 37°C, and analyzed by flow cytometry. Data were analyzed using FlowJo software to reveal the percentage of cells in each cell cycle phase. The S-phase index was determined using Click-iT EdU Alexa Fluor 647 flow cytometry assay kit (Invitrogen, A10202) according to the manufacturer's protocol.
Random plasmid integration assay
Assays were performed as previously described (Galanty et al. 2009). Briefly, 1 d after transfection with siRNA, U2OS cells were transfected with BamHI–XhoI-linearized pEGFP-C1 (Clontech). The following day, cells were collected, counted, and plated on three plates, one of which contained 0.5 mg/mL G418. One day after plating, cells on the plate lacking G418 were fixed to assess transfection efficiency, and the other two plates were incubated for 10–14 d at 37°C to allow colony formation. Colonies were stained with 0.5% crystal violet/20% ethanol and counted. Random plasmid integration events were normalized to transfection and plating efficiencies. The P-value was calculated using Student's t-test.
HR assay
A U2OS clone with the integrated HR reporter DR-GFP was generated as described previously (Pierce et al. 2001; Sartori et al. 2007). One day after transfection with siRNA, U2OS-DR-GFP cells were cotransfected with an I-SceI expression vector (pCBA-I-SceI) and a vector expressing monomeric RFP (pCS2-mRFP). The latter plasmid was added in a 1:10 ratio to mark the I-SceI-positive cells. Cells were harvested 1 d after I-SceI transfection and subjected to flow cytometric analysis to examine recombination induced by I-SceI digestion. Only RFP-positive cells were analyzed for HR efficiency to circumvent possible differences in transfection efficiencies. FACS data were analyzed using Summit version 4.3 software to reveal the percentage of GFP-positive cells relative to the number of transfected cells (RFP-positive). The data were normalized to a control siRNA treatment in each individual experiment. The cut-off between GFP (HR)-positive and -negative cells was set to 0.5% background level of GFP-positive cells in the internal control (RFP-positive, not transfected with I-SceI). This gate was then applied to the RFP/I-SceI-positive samples to determine HR efficiency. Results are presented as a percentage of control siRNA. The P-value was calculated using Student's t-test.
Comet assay
U2OS, U2OS stably expressing vector only, siRNA-resistant YFP-RNF4 wild-type, LD, or ΔSIM cells were transfected with siRNA, exposed to 10 Gy IR, and harvested at the indicated time points. For the 0 time point, cells were irradiated on ice to slow down repair and processed immediately following treatment. Neutral comet assays were performed using the comet assay kit from Trevigen (catalog no. 4250-050-K). Gelbond film (0.22 mm thick, 85 × 100 mm, LONZA catalog no. 53734, agarose gel support medium) was used instead of the standard glass slides. Images were collected using a 10× UPlanFLN objective mounted on an Olympus IX71 fluorescence microscope equipped with a CCD camera; comets were scored using Comet score software, and data were analyzed using Microsoft Excel.
Cell survival assay
U2OS cells were transfected with siRNAs and exposed to IR or HU at the indicated doses. Cells were incubated for 10–14 d at 37°C to allow colony formation. Colonies were stained with 0.5% crystal violet/20% ethanol and counted. Results were normalized to plating efficiencies.
Acknowledgments
We thank S.P.J. laboratory members for support, in particular J. Harrigan, J. Forment, and A. Kaidi, for helpful discussions; J. Forment for determining the S-phase index of control and RNF4-depleted cells; S. Polo for setting up the laser system and helping in the analysis of MDC1 FRAP and association kinetics experiments; and K. Dry for help with the manuscript. We also thank R.T Hay for YFP-tagged RNF4 constructs and siRNA sequences targeting RNF4, P. Young for PSMD4 UIM1/2 point mutant, J. Palvimo for RNF4 antibody, T. Halazonetis for RNF8 antibody, R. Baer for CtIP antibody, Y. Shiloh for ATM antibody, and R. Walker for help with FACS and cell sorting. Research in the S.P.J. laboratory is supported by grants from Cancer Research UK (C6/A11226), the European Research Council, the European Community's Seventh Framework Program (GENICA and DDResponse), and core infrastructure funding from Cancer Research UK and the Wellcome Trust. S.P.J. receives his salary from the University of Cambridge, supplemented by Cancer Research UK. R.B. cloned the RNF4 siRNA-resistant, proteasome component cDNAs, GFP-RPA1 (wild type and SM), and GFP-53BP1-MB; generated RFP-53BP1 and GFP-53BP1-MB stable cell lines; participated in testing siRNA efficiencies; provided help with the processing of laser experiments and in the establishment and testing of stable-cell-lines; and engaged in helpful discussions. J.C. extensively helped with cell survival and HR and NHEJ experiments and provided support with tissue culture maintenance and stable cell line generation. Y.G. initiated and led the project and performed all other experiments. Y.G. and S.P.J. conceived the study and wrote the paper. All authors discussed and commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.188284.112.
References
- Al-Hakim A, Escribano-Diaz C, Landry MC, O'Donnell L, Panier S, Szilard RK, Durocher D 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 9: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N 2009. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86 [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P 2010. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet 6: e1000852 doi: 10.1371/journal.pgen.1000852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N 2007. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res 167: 663–674 [DOI] [PubMed] [Google Scholar]
- Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N 2008. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci 105: 16165–16170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X 2007. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27: 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Dou H, Huang C, Singh M, Carpenter PB, Yeh ET 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 39: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A 2007. Chromatin dynamics and the preservation of genetic information. Nature 447: 951–958 [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakli M, Lorick KL, Weissman AM, Janne OA, Palvimo JJ 2004. Transcriptional coregulator SNURF (RNF4) possesses ubiquitin E3 ligase activity. FEBS Lett 560: 56–62 [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Scully R 2009. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J 423: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT 2005. SUMO: A history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Holloman WK 2011. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P 2010. DNA resection in eukaryotes: Deciding how to fix the break. Nat Struct Mol Biol 17: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont C, Taniguchi T 2007. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res 67: 7395–7405 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Dikic I 2007. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol 19: 199–205 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy A, Calonge TM, Outwin EA, O'Connell MJ 2007. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem 282: 20388–20394 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H 2008. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10: 547–555 [DOI] [PubMed] [Google Scholar]
- Levy-Barda A, Lerenthal Y, Davis AJ, Chung YM, Essers J, Shao Z, van Vliet N, Chen DJ, Hu MC, Kanaar R, et al. 2011. Involvement of the nuclear proteasome activator PA28γ in the cellular response to DNA double-strand breaks. Cell Cycle 10: 4300–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Ward JF 1993. A new method for introducing double-strand breaks into cellular DNA. Radiat Res 134:160–169 [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Morris JR 2010a. More modifiers move on DNA damage. Cancer Res 70: 3861–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR 2010b. SUMO in the mammalian response to DNA damage. Biochem Soc Trans 38: 92–97 [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Motegi A, Murakawa Y, Takeda S 2009. The vital link between the ubiquitin-proteasome pathway and DNA repair: Impact on cancer therapy. Cancer Lett 283: 1–9 [DOI] [PubMed] [Google Scholar]
- Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, et al. 2007. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res 67: 8536–8543 [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Gomez-Gonzalez B, Aguilera A 2009. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15: 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, Powell S 2008. Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. J Biol Chem 283: 31608–31616 [DOI] [PubMed] [Google Scholar]
- Stadtmueller BM, Hill CP 2011. Proteasome activators. Mol Cell 41: 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T 2007. Conserved function of RNF4 family proteins in eukaryotes: Targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17: 1263–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V, Hoffman L, Pratt G, Rechsteiner M 2002. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J 21: 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S 2009. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res 37: 2176–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar SR, Keusekotten K, Krause A, Horst C, Springer HM, Gottsche K, Dohmen RJ, Praefcke GJ 2008. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett 582: 3174–3178 [DOI] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan GL, D'Andrea AD 2010. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J Biol Chem 285: 19085–19091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M 1998. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem 273: 5461–5467 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang R 2008. Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 27: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell 25: 663–675 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]