Related to the study by Galanty et al. (this issue), this work by Hay and colleagues investigates RNF4's role in mammalian DNA damage response. Sumoylated MDC1 recruits RNF4 to damage foci, where it then mediates ubiquitin signaling. The work elucidates RNF4's crucial role in integrating sumoylation and ubiquitin signaling during DNA repair.
Keywords: RNF4, SUMO, ubiqutin, DNA damage
Abstract
Here we demonstrate that RNF4, a highly conserved small ubiquitin-like modifier (SUMO)-targeted ubiquitin E3 ligase, plays a critical role in the response of mammalian cells to DNA damage. Human cells in which RNF4 expression was ablated by siRNA or chicken DT40 cells with a homozygous deletion of the RNF4 gene displayed increased sensitivity to DNA-damaging agents. Recruitment of RNF4 to double-strand breaks required its RING and SUMO interaction motif (SIM) domains and DNA damage factors such as NBS1, mediator of DNA damage checkpoint 1 (MDC1), RNF8, 53BP1, and BRCA1. In the absence of RNF4, these factors were still recruited to sites of DNA damage, but 53BP1, RNF8, and RNF168 displayed delayed clearance from such foci. SILAC-based proteomics of SUMO substrates revealed that MDC1 was SUMO-modified in response to ionizing radiation. As a consequence of SUMO modification, MDC1 recruited RNF4, which mediated ubiquitylation at the DNA damage site. Failure to recruit RNF4 resulted in defective loading of replication protein A (RPA) and Rad51 onto ssDNA. This appeared to be a consequence of reduced recruitment of the CtIP nuclease, resulting in inefficient end resection. Thus, RNF4 is a novel DNA damage-responsive protein that plays a role in homologous recombination and integrates SUMO modification and ubiquitin signaling in the cellular response to genotoxic stress.
The integrity of the genome is under constant threat as intrinsic and extrinsic agents modify and break the DNA. If unrepaired, these toxic lesions can lead to cell death or induce chromosomal translocations and mutations that can ultimately lead to cancer. A double-strand break (DSB) is one of the most dangerous lesions that the genome can sustain, and such breaks can be generated by ionizing radiation (IR), as a consequence of damage during DNA replication, during recombinational rearrangement of the immunoglobulin genes, and in meiosis. Given the toxicity of DSBs, cells have evolved highly conserved and overlapping mechanisms to repair the damaged DNA. Homologous recombination (HR) and nonhomologous end-joining (NHEJ) are two distinct pathways that are differentially used during distinct phases of the cell cycle to repair DSBs. While repair of DNA damage via HR is highly accurate, NHEJ is regarded as operating at a lower fidelity. Although NHEJ is the predominant form of DSB repair in G1, it can function at any stage of the cell cycle, whereas HR functions predominantly in S/G2 and has a role in the repair of damaged replication forks. DSBs to be repaired by NHEJ are recognized and bound by the heterodimeric Ku protein, which allows recruitment and activation of the DNA-dependent protein kinase (DNA-PK). Tethering of active DNA-PK at the DNA ends allows assembly of a complex containing DNA ligase IV, XRCC4, and XLF/Cernunnos protein, which ligates the DNA ends (Hiom 2010). As cells pass from S to G2 phase, the sister chromatid resulting from DNA replication in S phase can function as a template for accurate repair of the damaged DNA using HR. In this scenario, the damaged DNA is recognized by the Mre11–Rad50–Nbs1 (MRN) complex, which maintains the broken ends in close proximity (Williams et al. 2009) and allows recruitment of the ATM kinase, which initiates a signaling cascade that results in the phosphorylation of many substrates, including H2AX (Uziel et al. 2003). This process also activates several ubiquitin E3 ligases that ubiquitylate chromatin components. These signaling events appear to be coordinated by scaffold proteins such as mediator of DNA damage checkpoint 1 (MDC1) (Goldberg et al. 2003; Lou et al. 2003; Stewart et al. 2003). Together, the phosphorylation and ubiquitylation signaling cascades allow the assembly of a supramolecular complex in the vicinity of the DNA break that facilitates DNA end resection and generates a long 3′ overhanging region of ssDNA that is the key substrate for HR. Although there is significant redundancy in the mechanisms that lead to end resection, it is clear that the nuclease and helicase functions of the MRN complex and the nuclease activity of the BRCA1-associated protein CtIP play important roles in the initial processing of the DNA ends (Sartori et al. 2007). More extensive resection of the DNA ends (Zierhut and Diffley 2008) appears to be carried out by the processive single-strand exonuclease Exo1 (Sugiyama and Kowalczykowski 2002; Mimitou and Symington 2008). As the ssDNA is generated, it is bound by the single-strand-specific DNA-binding protein replication protein A (RPA), which is then replaced with Rad51 in a concerted reaction involving Rad52 and BRCA2 (Chen et al. 1998; Sugiyama and Kowalczykowski 2002). Once formed, the Rad51 nucleoprotein filament searches for homology in the sister chromatid and facilitates the exchange of homologous DNA strands to generate a Holliday junction, which provides a primer and template to initiate DNA synthesis. This can then be resolved by either synthesis-dependent strand annealing (SDSA) or HR. In SDSA, the de novo synthesized DNA switches back to its initial partner, where it can be used as a template to complete repair. Repair of the DNA by HR involves branch migration, DNA polymerase-mediated extension of 3′ ends, and resolution of Holliday junctions (Hiom 2010).
As indicated above, DNA damage signaling uses a variety of post-translational modifiers as molecular switches to regulate the signaling network (Zhou and Elledge 2000). Ubiquitin and, more recently, small ubiquitin-like modifier (SUMO) have been shown to be important mediators of this response (Haglund and Dikic 2005; Golebiowski et al. 2009), although molecular targets and mechanism of action remain to be determined. SUMO modification, mediated by the SUMO E3 ligases PIAS1 and PIAS4, is required for an effective response to DNA damage in mammalian cells, and all three SUMO paralogs (1, 2, and 3) accumulate at sites of DNA damage (Galanty et al. 2009; Morris et al. 2009). However, the downstream effectors that recognize and mediate the functional consequences of SUMO modification remain to be identified. The conserved SUMO-targeted ubiquitin E3 ligases (STUbLs) include mammalian RNF4, budding yeast Slx5–Slx8, and fission yeast Rfp1/Rfp2–Slx8. These proteins target polySUMO-modified proteins for ubiquitin-mediated proteolysis (Tatham et al. 2008; Geoffroy and Hay 2009; Heideker et al. 2009), and in yeast, the STUbLs play an important role in the maintenance of genome stability (Prudden et al. 2007; Sun et al. 2007; Uzunova et al. 2007; Xie et al. 2007). Here we show that RNF4 plays a role in the response of mammalian cells to DNA damage. Cells in which RNF4 expression has been eliminated are hypersensitive to certain types of DNA damage. Recruitment of RNF4 to DNA damage sites is dependent on its SUMO interaction motifs (SIMs) and RING domain and the upstream molecules NBS1, MDC1, RNF8, 53BP1, and BRCA1, which are involved in sensing and generating a DNA damage signal. Depletion of RNF4 did not compromise recruitment of these factors to sites of DNA damage, although their clearance from DNA damage-induced foci was delayed. Proteomic analysis of SUMO substrates indicated that DNA damage induced SUMO modification of MDC1, which in turn recruited RNF4 to mediate ubiquitin deposition at DNA damage sites. Failure to recruit RNF4 resulted in defective loading of RPA and Rad51 onto ssDNA. This appeared to be a consequence of reduced recruitment of the CtIP nuclease, resulting in inefficient end resection and indicating that RNF4 plays a role in HR.
Results
Depletion of RNF4 sensitizes cells to genotoxic stress
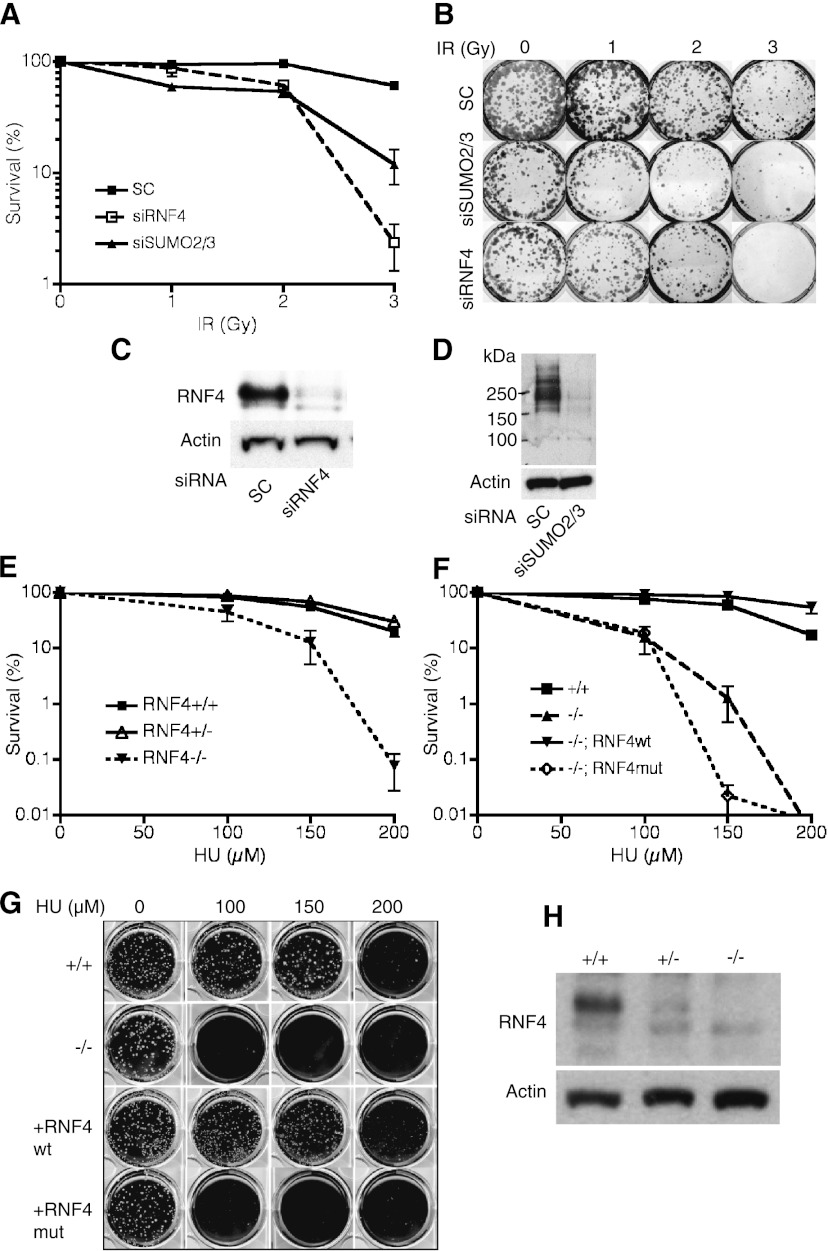
Given the involvement of ubiquitylation and SUMO modification in the DNA damage response and the role of the yeast STUbLs in genome stability (Heideker et al. 2009), we tested the possibility that RNF4 might be the factor linking these two important modifications. To examine the role of RNF4 in the mammalian DNA damage response, the expression of RNF4 and its SUMO-2/3 substrate was ablated with siRNA in U2OS cells, and the cells were exposed to IR. Clonogenic survival assays revealed that ablation of RNF4 and SUMO-2/3 expression sensitized cells to IR (Fig. 1A–D), supporting a role for RNF4 in the maintenance of genomic integrity. To obtain a complete knockout of RNF4 expression and to rule out potential off-target effects of the siRNA, we used HR to eliminate both alleles of RNF4 in chicken DT40 cells (Supplemental Fig. S1). Clonogenic survival assays revealed that while the RNF4−/− DT40 cells displayed a modest sensitivity to cisplatin (data not shown), they were highly sensitive to hydroxyurea (HU) (Fig. 1E–H). Reintroduction of wild-type RNF4 into the RNF4−/− cells rescued the HU sensitivity, while reintroduction of an RNF4 variant (M140A or R181A) that was unable to engage the E2 ubiquitin-conjugating enzyme and thus lacked ubiquitin E3 ligase activity (Plechanovova et al. 2011) failed to rescue the HU sensitivity (Fig. 1F,G). As expected, RNF4 expression was not detectable in RNF4−/− DT40 cells (Fig. 1H).
Figure 1.
Depletion of RNF4 sensitizes cells to genotoxic stress. (A,B) RNF4-depleted U2OS cells are sensitive to IR. U2OS cells were transfected with siRNA against SUMO-2/3 (siSUMO2/3), RNF4 (siRNF4), or a nontarget siRNA (SC) for 3 d and then exposed to IR at the indicated dose. Survival rates were determined by colony formation. The data represent the mean + SD (n = 3). (C,D) The efficiency of RNF4 or SUMO-2/3 depletion by siRNA in A and B was determined by Western blotting. (E–H) Chicken DT40 RNF4−/− cells display an increased sensitivity to genotoxic stress. Clonogenic survival assays were performed, and the number of surviving colonies was counted after chronic HU exposure. The assay was repeated with stably transfected clones expressing wild-type RNF4 (RNF4wt) or an E2-binding mutant of the rat ortholog of RNF4 (RNF4mut). The data represent the mean of three independent experiments, and the error bars indicate the SD. (H) Depletion of RNF4 in DT40 was confirmed by Western blotting (see also Supplemental Fig. S1).
RNF4 is recruited to sites of DNA damage
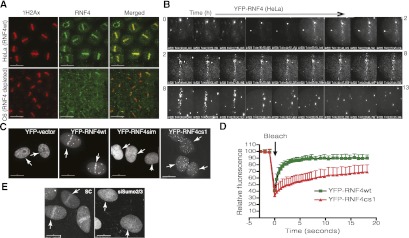
To examine the recruitment of RNF4 to sites of DNA damage, cells were sensitized with BrdU and subjected to laser micro-irradiation (Lukas et al. 2003). After laser micro-irradiation, endogenous RNF4 accumulated at the DNA damage sites, colocalizing with phosphorylated H2AX (γH2AX). RNF4 staining at the damage site was eliminated when RNF4 expression was ablated stably by shRNA, confirming the specificity of the antibody (Fig. 2A). To observe the dynamic recruitment of RNF4 to sites of DNA damage, HeLa cells expressing close to endogenous levels of YFP-RNF4 were laser-micro-irradiated, and RNF4 recruitment was continuously monitored by live-cell imaging. RNF4 recruitment is detectable by 15 min post-irradiation and is maintained for 9–11 h, whereafter it declines as the damage is repaired (Fig. 2B; Supplemental Movies 1, 2). Mutation of either the SIMs (Song et al. 2004) or the RING domain of RNF4 (Tatham et al. 2008) resulted in a failure of RNF4 to be recruited to sites of DNA damage. While RNF4 mutants unable to interact with SUMO displayed a uniform distribution throughout the nucleus, RING mutants displayed a punctate localization, suggesting that they were already bound to SUMO but could not be released (Fig. 2C). This hypothesis was tested by fluorescence recovery after photobleaching (FRAP) on cells expressing either wild-type YFP-RNF4 or YFP-RNF4 containing a mutation in the RING domain. In both cases, punctate localizations of RNF4 were photobleached, and the time taken for fluorescence to recover was determined. Wild-type YFP-RNF4 was fairly mobile and recovered quickly (half-time of recovery [t1/2], 0.63 sec), but the catalytically inactive form of YFP-RNF4 was less mobile and recovered fluorescence slowly (t1/2, 6.9 sec) (Fig. 2D; Supplemental Fig. S2). SUMO-2/3 recruitment to sites of damage preceded that of RNF4 (Supplemental Movie 3), and SUMO-2/3 expression was required for RNF4 recruitment (Fig. 2E).
Figure 2.
RNF4 is recruited to sites of DNA damage. (A) Endogenous RNF4 colocalizes with γH2AX after DNA damage. HeLa cells or HeLa with RNF4 stably depleted (C6) were subjected to laser microirradiation. Immunofluorescence staining was performed using anti-RNF4 and anti-γH2AX antibodies. Specificity of the RNF4 antibody was confirmed by Western blotting after siRNA depletion. (B) Time-lapse experment showing the dynamic recruitment of RNF4. HeLa cells expressing YFP-RNF4 were laser-micro-irradiated, and redistribution of YFP-RNF4 was recorded for 13 h at 1-min intervals (Materials and Methods; see also Supplemental Movies S1, S2). (C) RNF4 recruitment to sites of micro-irradiation-induced DNA damage depends on its SIM and RING domains. HeLa cells were transfected with wild-type RNF4 (YFP-RNF4wt), SIM domain mutant RNF4 (YFP-RNF4sim), or a RING domain mutant (YFP-RNF4cs1) for 24 h and then laser-micro-irradiated. Time-lapse microscopy monitored recruitment of YFP-RNF4 for 2 h. The end point image is shown. Arrows indicate laser movement during micro-irradiation. (D) Quantitative FRAP analysis of YFP-RNFwt and YFP-RNF4cs1. The FRAP curves were derived from 20 cells for each condition. Normalized florescence intensity was compared. Error bars, SD. Data were analyzed by Student's t-test, and P-values were calculated (Materials and Methods; see also Supplemental Fig. S2). (E) RNF4 retention at sites of DNA damage requires SUMO modification. U2OS cells stably expressing YFP-RNF4 were transfected with SUMO-2/3-targeting siRNA or a nontarget siRNA control for 3 d, laser-micro-irradiated, and monitored for the recruitment of YFP-RNF4 along the laser track (arrows) using time-lapse microscopy. The image was derived from the 1-h time point. Bars: A, 30 μm; C,E, 15 μm.
RNF4 mediates repair and ubiquitin conjugation of DSBs
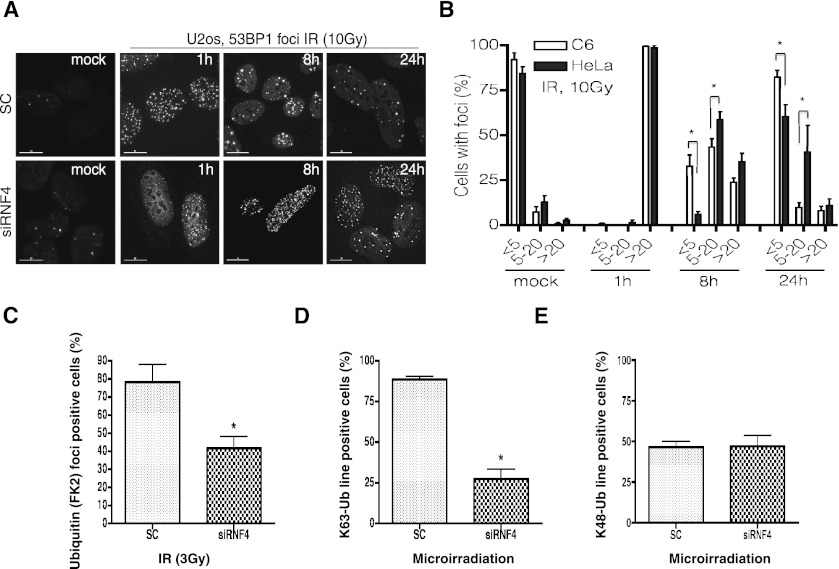
To establish which components of the DNA damage response are dependent on RNF4 for recruitment to sites of DNA damage, RNF4 expression was ablated with siRNA, and recruitment was visualized by immunofluorescence after either IR or laser micro-irradiation. RNF4 depletion did not dramatically influence the initial recruitment of γH2AX, NBS1, MRE11, MDC1, RNF8, RNF168, BRCA1, or RAP80 (Paull et al. 2000; Stewart et al. 2003, 2009; Lukas et al. 2004; Stucki and Jackson 2004; Boulton 2006; Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Doil et al. 2009), although at later times after DNA damage substantially increased, staining for RNF8 and RNF168 was apparent in the absence of RNF4 (Supplemental Fig. S3). In contrast, it was noted that depletion of RNF4 had a significant impact on the kinetics of 53BP1 accumulation at sites of DNA damage. In the absence of RNF4, 53BP1 recruitment was delayed and persisted for substantially longer than in the presence of RNF4 (Fig. 3A,B; Supplemental Fig. S4). This was also evident in a stable cell line expressing a fluorescent protein-tagged version of 53BP1 (Supplemental Fig. S4a). It was also apparent that RNF4 ablation blocked the accumulation of ubiquitin conjugates at sites of DNA damage induced by IR (Fig. 3C). Analysis of ubiquitin deposition at sites of laser micro-irradiation using antibodies specific for K63- or K48-linked ubiquitin chains indicated that RNF4 depletion resulted in a reduction of K63-linked ubiquitin at sites of damage (Fig. 3D), whereas K48-linked ubiquitin chains were unaffected (Fig. 3E).
Figure 3.
RNF4 mediates repair and ubiquitin conjugation at sites of DNA damage. (A) RNF4 depletion delays DNA repair. U2OS cells were transfected with siRNA to RNF4 or a nontargeting siRNA (SC) for 3 d, exposed to 10 Gy of IR either prior to irradiation (mock) or 1, 8, or 24 h after irradiation, and immunostained with antibody against 53BP1 (see also Supplemental Fig. S4). (B) Quantification of data presented in Supplemental Figure S4c. At least 100 cells for each time point were scored. Cells were grouped by 53BP1 foci number: less than five, five to 20, and >20. Error bar, SD. (*) P < 0.05 (see also Supplemental Fig. S4c). (C) HeLa cells transfected with siRNA to RNF4 or a nontargeting siRNA (SC) for 72 h were exposed to 3 Gy of IR and allowed to recover for 1 h, and damage foci were visualized by staining with antibodies against γH2AX or conjugated ubiquitin (FK2). The percentage of γH2AX-positive cells that were also FK2-positive was determined; >100 cells were scored per condition. Data represent mean + SE from three independent experiments. (*) P < 0.05. (D) RNF4 mediates K63 ubiquitin chain formation after damage. Cells were treated with siRNA as in C and micro-irradiated before immunostaining using K63 ubiquitin-specific antibody and γH2AX antibody. The percentage of γH2AX-staining cells that were also stained for K63 ubiquitin along the laser track was calculated. More than 100 cells were scored per condition. Data represent mean + SE from two independent experiments. (*) P < 0.05. (E) Cells were treated with siRNA and micro-irridiation as in D. Immunostaining was performed using K48 ubiquitin-specific antibody. The percentage of cells with K48-Ub-positive laser tracks was determined as in D. More than 100 cells were scored per condition. Data represent mean + SE. (*) P < 0.05.
To determine which components of the DNA damage response were required for the recruitment of RNF4, expression of individual components was ablated by siRNA, and recruitment of RNF4 to sites of laser micro-irradiation-induced DNA damage was determined by immunofluorescence. Depletion of NBS1, RNF8, 53BP1, and BRCA1 all abrogated RNF4 accumulation at sites of DNA damage (Supplemental Fig. S5).
RNF4 binds to SUMO-modified MDC1 after DNA damage
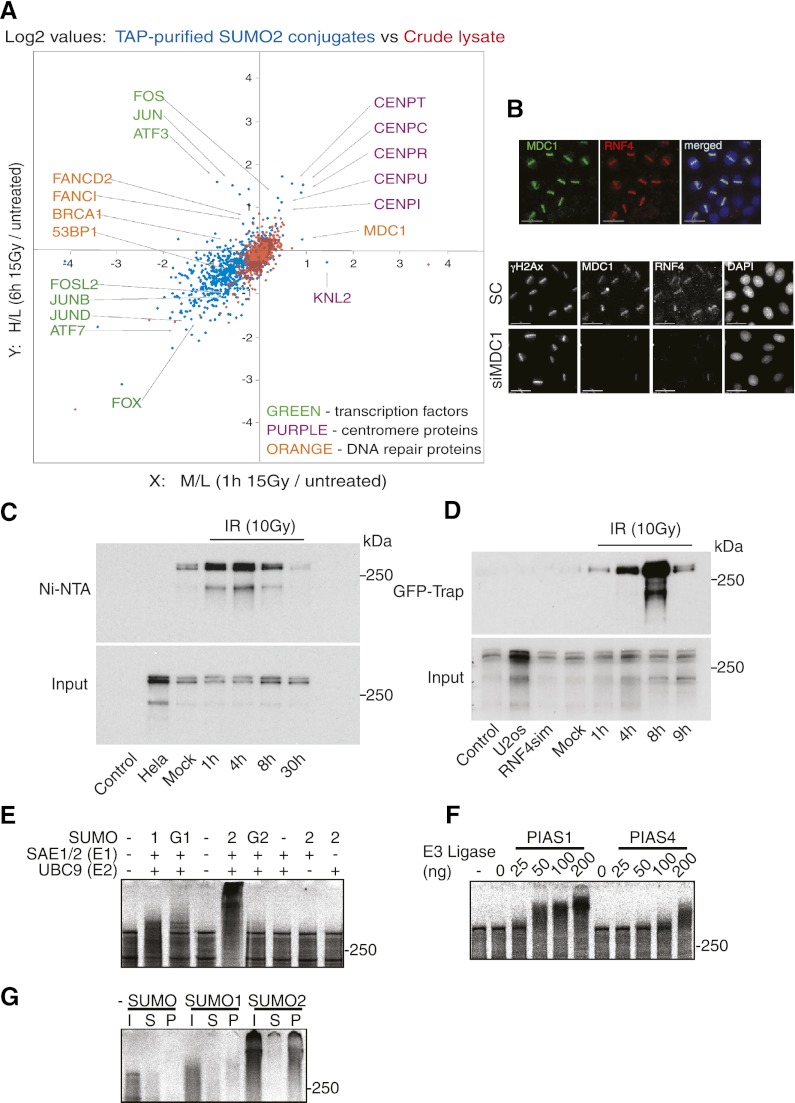
In an attempt to identify potential SUMO-modified substrates of RNF4, a SILAC-based quantitative proteomic analysis of SUMO-modified substrates was carried out comparing the SUMO modification status (Golebiowski et al. 2009) of untreated cells with cells examined 1 h or 6 h after exposure to 15 Gy of IR. While many SUMO-modified substrates were detected, MDC1 showed a twofold increase in SUMO modification 1 h post-irradiation (Fig. 4A). Ablation of MDC1 expression, as expected, blocked RNF4 accumulation at sites of DNA damage (Fig. 4B). To confirm the IR-induced SUMO modification of MDC1, HeLa cells stably expressing 6His-SUMO-2 (Tatham et al. 2009) were exposed to IR, and SUMO-modified proteins were isolated by binding to Ni-NTA beads under denaturing conditions. Western blotting for endogenous MDC1 revealed that the protein was inducibly modified by SUMO-2 in response to IR (Fig. 4C). To determine whether SUMO-2-modified MDC1 is a substrate for RNF4, U2OS cells stably expressing YFP-RNF4 were exposed to IR, proteins associated with RNF4 were collected by immunoprecipitation with an affinity reagent for YFP, and endogenous MDC1 was detected by Western blotting. IR induced a strong association between MDC1 and RNF4 (Fig. 4D). To determine whether the interaction between RNF4 and MDC1 was SUMO-dependent and direct, we established an in vitro system for the SUMO modification of in vitro translated MDC1 with purified components (Tatham et al. 2005). MDC1 was modified by SUMO-1 and more efficiently with SUMO-2 (Fig. 4E). Given the previously established role of PIAS1 and PIAS4 in SUMO modification in response to DNA damage, we purified recombinant PIAS1 and PIAS4 and titrated them into an in vitro modification reaction containing limiting amounts of the E2 SUMO-conjugating enzyme Ubc9. Under these conditions, both PIAS1 and PIAS4 dramatically increased the efficiency of MDC1 SUMO modification (Fig. 4F). To test whether SUMO-modified MDC1 was a substrate for RNF4, the modified material was incubated with beads covalently linked to the SIM containing the N-terminal region of RNF4 (Bruderer et al. 2011). MDC1 modified with SUMO-2 was bound by the SIM containing the fragment of RNF4, whereas SUMO-1-modified MDC1 was not bound by RNF4, suggesting that, like promyelocytic leukemia (PML) (Tatham et al. 2008), it is polySUMO-2-modified MDC1 that is bound by RNF4.
Figure 4.
RNF4 binds to SUMO-modified MDC1 after DNA damage. (A) SILAC-based quantitative proteomic analysis (see the Materials and Methods) of SUMO modification after IR. TAP-SUMO-2 HeLa cells were grown in either normal medium, medium containing Lys4 and Arg6, or medium containing Lys8 and Arg10. Cells in normal medium were untreated, cells in Lys4 and Arg6 were exposed to 15 Gy of IR and harvested after 1 h, and cells in Lys8 and Arg10 were exposed to 15 Gy of IR and harvested after 6 h. Samples were mixed, and SUMO-modified proteins were analyzed by mass spectrometry. Crude lysates were also analyzed by mass spectrometry. (B) MDC1 is required for RNF4 recruitment to damage sites. The top panel shows that endogenous MDC1 and RNF4 colocalize after DNA damage. Laser-micro-irradiated HeLa cells were costained for endogenous MDC1 and RNF4. The bottom panel shows that RNF4 retention at DNA damage depends on MDC1. HeLa cells were treated with siRNA against MDC1 or a nontarget siRNA (SC) for 3 d, laser-micro-irradiated, and, after 1 h, immunostained with the indicated antibodies. Depletion of MDC1 by siRNA is shown in Supplemental Figures S6. (Bottom) Bars, 15 μm. (C) MDC1 is SUMO-modified after DNA damage in vivo. HeLa cells stably expressing His-SUMO2 were treated with or without 10 Gy of IR, collected at the indicated time point, and analyzed by Ni-NTA affinity chromatogaphy under denaturing condition, followed by Western blotting with MDC1-specific antibody. (D) RNF4 binds to MDC1 after DNA damage in vivo. U2OS stably expressing YFP-RNF4wt or its SIMs mutant (YFP-RNF4sim) were exposed to IR (10 Gy). After recovery at the indicated time point, nuclear fractions were isolated, and the YFP-containing complexes were purified on GFP-Trap-agarose and analyzed by immunoblotting with antibodies against MDC1. (E) MDC1 is SUMO-modified in vitro. In vitro translated and radiolabeled 35S-methionine full-length Homo sapiens MDC1 migrated on acrylamide gel as a species above 250 kDa. Modified forms of MDC1 were seen only in the presence of SUMO1 (lane 1), GST-SUMO-1 (lane G1), SUMO-2 (lane 2), and GST-SUMO-2 (lane G2) in the presence of Ubc9 and Sae1/2 (see the Materials and Methods). (F) PIAS1 and PIAS4 enhance SUMO modification of MDC1 in vitro. SUMOylation reactions were performed similar to as in E but with limiting levels of Ubc9 and the indicated amounts of PIAS1 and PIAS4. (G) A SIM-containing fragment of RNF4 binds to SUMO-modified MDC1. 35S-labeled unmodified MDC1 or MDC1 modified by either SUMO-1 or SUMO-2 as in E was incubated with the N-terminal region of H. sapiens. RNF4 containing the four SIMs cross-linked to Sepharose beads. Input (I), supernatant (S), and beads (P) were analyzed by SDS-PAGE and PhosphorImaging.
RNF4 is required for the recruitment of RPA70 and Rad51 to ssDNA
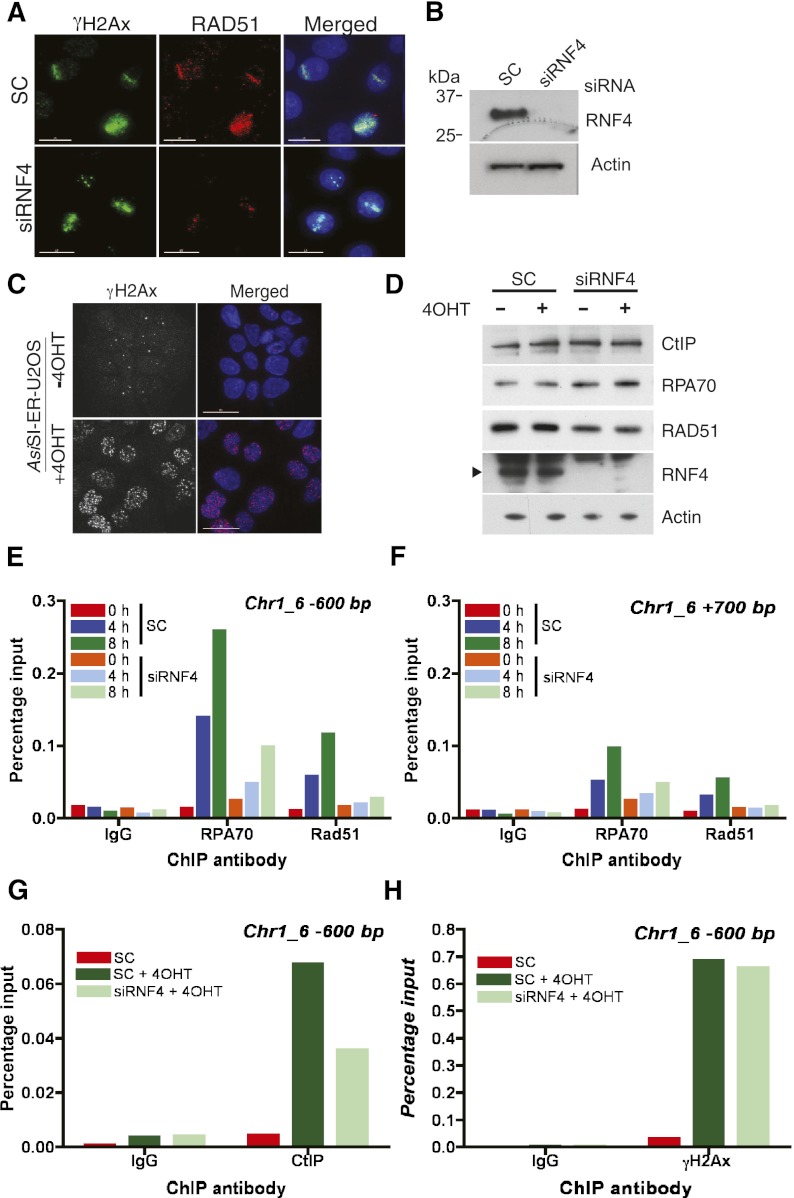
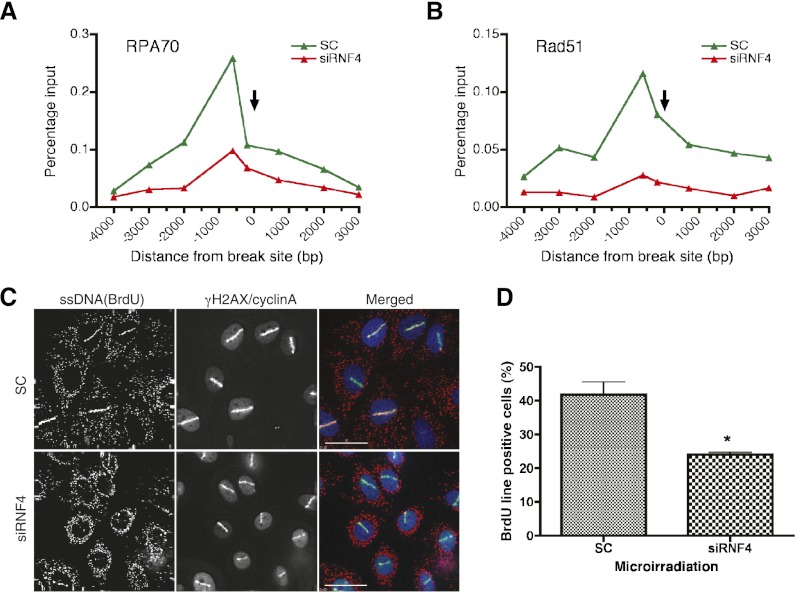
Analysis of the factors recruited to damaged DNA in the absence of RNF4 indicated that the initial recruitment of DNA damage sensors and signaling components was unaffected by RNF4 depletion. As foci of 53BP1 displayed delayed clearance after DNA damage in the absence of RNF4, and RNF4 was not rapidly recruited to DNA damage foci, we suspected that RNF4 might have a more direct role in the repair process, particularly HR. To test this, we therefore asked whether RNF4 was required for the recruitment of the Rad51 protein that polymerizes on the resected ssDNA and for strand invasion during HR. HeLa cells treated with either a nontargeting siRNA or a siRNA directed against RNF4 were therefore laser-micro-irradiated, and recruitment of Rad51 to sites of DNA damage was determined by immunofluorescence. In cells depleted of RNF4, Rad51 recruitment to sites of DNA damage was severely impaired, although the same cells displayed normal recruitment of γH2AX (Fig. 5A). Western blotting indicated that the RNF4 depletion was effective (Fig. 5B). While immunofluorescence indicates that Rad51 is recruited into the vicinity of damaged DNA, it is not direct proof that it is bound to damaged DNA. To address this question, we made use of a U2OS cell line that stably expresses the 8-base-pair (bp) cutter restriction enzyme AsiSI fused to a modified estrogen receptor (ER) hormone-binding domain (Iacovoni et al. 2010). Upon treatment of the AsiSI-ER-U2OS cells with 4-hydroxy tamoxifen (4OHT), the AsiSI-ER translocates to the nucleus and induces DSBs in DNA at defined sites (Fig. 5C). As the AsiSI-ER induced sites of cleavage are known, it is possible to use chromatin immunoprecipitation (ChIP) to directly monitor recruitment of repair factors onto the damaged chromatin. As this approach has been used successfully to define the genomic landscape of γH2AX after DNA damage (Iacovoni et al. 2010), we therefore adapted it to investigate the DNA damage-induced recruitment of factors dependent on the activity of RNF4. To validate this approach for RNF4 substrates, we induced DSBs in the AsiSI-ER-U2OS cells and used antibodies to SUMO-1 and SUMO-2 to carry out ChIP. Quantitative PCR (qPCR) revealed strong accumulation of both SUMO-1 and SUMO-2 to regions close (100–200 bp) to break sites on chromosome 1 (Supplemental Fig. S7a) and chromosome 6 (Supplemental Fig. S7b) but not to regions distant (2 Mb) from the break sites (Supplemental Fig. S7c). The robust accumulation of SUMO-1 and SUMO-2 to sites of DNA damage was eliminated by siRNA-mediated ablation of Ubc9, the single SUMO E2 (Supplemental Fig. S7d,e), but was unaffected by depletion of RNF4 (data not shown). To establish whether the failure to recruit Rad51 to sites of laser micro-irradiation induced DNA damage in RNF4-depleted cells was due to the instability of Rad51, cell extracts were analyzed by Western blotting for Rad51 and the RPA70 component of the single-strand-specific DNA-binding protein RPA. Levels of RPA70 and Rad51 were not substantially altered when RNF4 expression was ablated (Fig. 5D). ChIP analysis of Rad51 after AsiSI induced double-strand cleavage indicated that Rad51 was robustly recruited to both sides of the DNA breakage and that this recruitment was ablated in the absence of RNF4 (Fig. 5E,F). As Rad51 loading is dependent on the concerted deposition and removal of RPA on the resected ssDNA, RNF4 could be required for the initial recruitment of RPA or for its removal. This was therefore tested by ChIP analysis after induction of DSBs in DNA by AsiSI-ER. Robust RPA loading at the same sites used to analyze Rad51 binding was detected on both sides of the break. siRNA-mediated ablation of RNF4 expression resulted in a substantial reduction of RPA loading on both sides of the broken DNA (Fig. 5E,F). Based on these results, it appears that Rad51 loading fails in RNF4-depleted cells due to a failure to load RPA. This is most likely to be due to a failure to resect the DNA ends and generate the ssDNA to which RPA binds. A key player in end resection is the nuclease CtIP (Sartori et al. 2007), and we therefore determined whether its recruitment to damaged DNA was influenced by RNF4. Robust CtIP loading was detected by ChIP analysis after induction of DSBs in DNA by AsiSI-ER. siRNA-mediated ablation of RNF4 expression resulted in reduced recruitment of CtIP onto the broken DNA (Fig. 5G). The reduced loading of RPA, Rad51, and CtIP observed after RNF4 depletion was not a consequence of reduced activity of the AsiSI-ER restriction enzyme, as γH2AX recruitment to damaged DNA was unaffected by RNF4 depletion (Fig. 5H). During DNA repair, the 5′ end of the broken DNA is resected for some considerable distance (Zierhut and Diffley 2008), and the remaining ssDNA is bound by RPA. To determine whether RNF4 depletion affects the extent of DNA resection/RPA loading, we carried out ChIP analysis with RPA and Rad51 using primers targeting regions between 200 bp and 4000 bp from the AsiSI-induced break site. Recruitment of both RPA and Rad51 appears to be asymmetric on either side of the break, but recruitment of both RPA and Rad51 is maximal at 600 bp from the break and decreases with increasing distance from the break. As previously reported (Zierhut and Diffley 2008), there appeared to be some loss of genetic material at the DNA end as a consequence of nucleolytic digestion of the 3′ end of the DNA. This was manifest in reduced recruitment of RPA and Rad51 to DNA 100 bp from the break site compared with DNA 600 bp from the break site (Fig. 6A,B). While RPA and Rad51 could be clearly detected at 3000 bp from the break site, they were not detected 4000 bp from the break site (Fig. 6A,B). In the absence of RNF4, it appeared that RPA and Rad51 loading was only detected, albeit at reduced levels, within 700 bp on either side of the break point (Fig. 6A,B). It therefore seems likely that in the absence of RNF4, RPA loading is inefficient and is restricted to the ends of the DNA, probably as a consequence of a failure to resect the damaged DNA. To test this directly, micro-irradiated U2-OS cells previously incubated in the presence of BrdU and treated with siRNA to RNF4 or a nontargeting siRNA were stained with an antibody to BrdU. Under native conditions, only extensive stretches of ssDNA are detected by this antibody. The cells were also stained with an antibody to γH2AX to mark the damage track and an antibody to cyclin A to mark cells in the S/G2 phase of the cell cycle. In cells treated with nontargeting siRNA, tracks of BrdU staining are clearly detected, but only in cells that are in S/G2 as judged by cyclin A staining (Fig. 6C). This is consistent with micro-irradiation generating DSBs that are repaired by homology-directed mechanisms involving extensive end resection in S/G2. Ablation of RNF4 expression reduced BrdU staining (Fig. 6C,D), indicating that in the absence of RNF4, there is a defect in end resection.
Figure 5.
RNF4 is required for loading of RPA70, Rad51, and CtIP onto damaged DNA. (A) RAD51 loading to sites of laser micro-irradiation-induced DNA damage requires RNF4. HeLa cells were treated with siRNA to RNF4 or nontargeting siRNA (SC) for 3 d, micro-irradiated, and stained with the indicated antibodies. Nuclei were counterstained with DAPI. Bar, 15 μm. (B) Depletion of RNF4 in A was confirmed by Western blotting with β-actin as a loading control. (C) Generation of dsDNA breaks by AsiSI. AsiSI-ER-U2OS cells were treated with 4OHT for 4 h, and DSBs were visualized by γH2AX staining. Nuclei were stained with DAPI. Bar, 30 μm. (D) AsiSI-ER-U2OS cells were transfected with siRNA to RNF4 or nontargeting siRNA for 3 d and treated with 4OHT for 4 h, and protein levels were determined by Western blotting with the indicated antibodies. (E–H) RNF4 mediates the loading of CtIP, RPA, and RAD51 to DSBs. AsiSI-ER-U2OS cells were transfected with nontargeting siRNA (SC) or siRNA to RNF4 for 72 h prior to treatment. ChIP analysis was performed before (0 h) and after 4OHT treatment (300 nM) for 4 or 8 h using antibodies specific for RPA70, Rad51, CtIP, or γH2AX. In each case, a matched nonimmune IgG was used as a control. Enrichment of RPA70, Rad51, and CtIP was determined by RT-qPCR using primers located 600 bp to the left or 700 bp to the right of the AsiSI site on chromosome 1 at position 89231183 (see also Supplemental Fig. S7f,g).
Figure 6.
Distribution of RPA70 and Rad51 along a DSB region and ssDNA formation after damage. (A,B) AsiSI-ER-U2OS cells were treated with nontargeting siRNA and siRNA to RNF4 and treated with 4OHT for 8 h. ChIP analysis was performed, and enrichment of RPA70 (A) and Rad51 (B) was assessed by RT-qPCR using primers at indicated distances from the AsiSI site on chromosome 1 at position 89231183. The arrow indicates the location of the AsiSI site. (C,D) RNF4 depletion impairs the generation of ssDNA at DSBs. HeLa cells were treated with siRNA to RNF4 or a nontargeting siRNA for 2 d and subjected to laser micro-irradiation. (C) ssDNA was detected by staining with an antibody to BrdU under nondenaturing conditions (see the Materials and Methods; see the text). Cells were costained with antibodies to γH2AX to visualize the sites of DNA damage and with antibodies to cyclin A to mark cells in the S/G2 phase of the cell cycle. (D) The percentage of cells that were positive for both γH2AX and BrdU was calculated. For each condition, >100 cells were scored. Data represent the mean + SE from two independent experiments. (*) P < 0.05. (C) Bar, 30 μM.
Discussion
Our findings suggest that the STUbL RNF4 is a regulator of the DNA damage response in human cells. Both SUMO and ubiquitin modifications have been shown to contribute to DNA damage repair programs in yeast (Bergink and Jentsch 2009), in part through the action of the STUbLs Rfp1, Rfp2, and Slx5/Slx8 (Heideker et al. 2009). Our data demonstrate that MDC1, a key regulator of the DNA damage response, is modified by SUMO-2/3 and, as a consequence, recruits RNF4 to sites of DNA damage. The most striking defect in RNF4-deficient cells is a failure to load RPA and Rad51 onto DNA at the break site. As both RPA and Rad51 bind to ssDNA, it seemed likely that the defect in RNF4-depleted cells is a failure to efficiently carry out the 5′-to-3′ exonuclease-mediated resection from the DSB. Consistent with this, depletion of RNF4 also led to a reduction in the recruitment of CtIP to the damaged DNA. CtIP has been shown to possess nuclease activity and has a central role in DNA end resection (Sartori et al. 2007). Failure to recruit CtIP would thus provide an explanation for the defect in RPA and Rad51 recruitment, as the ssDNA to which they bind would not be efficiently generated. Evidence that RNF4 depletion led to a defect in the generation of ssDNA came from direct detection of ssDNA with antibodies to BrdU. Staining of micro-irradiation-induced DNA damage tracks with BrdU was evident in cells in the S/G2 phases of the cell cycle, and this was reduced when RNF4 was depleted. The precise mechanism by which the DNA is resected from the break point remains to be established, but MDC1 is centrally placed to influence this process, as it directly interacts with the MRN complex, which has intrinsic nuclease activity and interacts via Nbs1 with CtIP (Lloyd et al. 2009; Williams et al. 2009). The ability of MDC1 to interact with the MRN complex could influence the activity of the BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN, DNA end resection machineries that appear to function in human cells (Nimonkar et al. 2011). SUMO modification of both BLM (Ouyang et al. 2009) and RPA70 (Dou et al. 2010) has been shown to influence their role in the DNA damage response, but it is likely that these functions are downstream from the RNF4 requirement described here.
While microhomology-mediated end-joining (MMEJ) and NHEJ can take place throughout the cell cycle, they are error-prone mechanisms of DNA repair, and there are clear advantages for the cell to switch to template-directed HR in S/G2 once the DNA has been replicated. Although CtIP participates both in MMEJ in G1 and HR during S/G2 (Sartori et al. 2007; Huertas and Jackson 2009; Yun and Hiom 2009), its levels are low during G1, where there is less of a requirement for extensive end resection, but its levels are increased in S phase, where extensive end resection is required for HR. Not only do CtIP levels increase in S phase, but it is subject to phosphorylation that appears to increase the activity of CtIP (Huertas and Jackson 2009; Yun and Hiom 2009). Thus, CtIP plays a central role in the decision-making process that triggers the switch from error-prone to error-free repair mechanisms. The role of RNF4 in recruiting CtIP to sites of DNA damage could also allow it to influence such key decisions.
RNF4 mediates ubiquitin conjugation at sites of DNA damage, and previous in vitro studies have indicated that RNF4 can catalyze formation of K11-, K48-, and K63-linked ubiquitin in the presence of UbcH5 in vitro (Tatham et al. 2008). However, depletion of RNF4 did not influence the deposition of K48-linked ubiquitin chains at sites of DNA damage, but did lead to a significant decrease in the K63-linked ubiquitin chains that were deposited at sites of DNA damage. Further studies will be required to understand the complex interplay between ubiquitin chain formation and SUMO modification during the process of DNA repair.
Materials and methods
Plasmids
Rattus norvegicus RNF4 and mutant (RNF4cs1 and RNF4sim) cDNAs were generated as described before (Tatham et al. 2008). Homo sapiens RNF4 cDNAs were amplified by RT–PCR from the total RNA of HeLa cells and then were cloned into a pBOSYFP expression vector. The primer pairs were used as ATCCGTCGACATGAGTACAAGAAAGCGTC and CGGTGGATCCCCTATATAAATGGGGTGGTAC.
The Gallus gallus RNF4 sequence was obtained from the NCBI database (gene ID: NC_006091.2). To prepare the RNF4 targeting construct, a 2.4-kb SalI/BamHI fragment from the RNF4 upstream genomic region to part of exon 1 and a 2.3-kb BamHI/NotI fragment from part of exon 7 to downstream from RNF4 were cloned into the SalI and NotI sites of the pBluescript vector. Puromycin or blasticidin antibiotic resistance cassettes were then inserted into the BamHI site between the two genomic fragments. For the rescue experiments, a Flag-tagged RNF4 from R. norvegicus was cloned into pcDNA3.1-Zeo (Sigma). The E2-binding mutant contains two mutated amino acid residues (M140A and R181A) and was generated by site-directed mutagenesis.
Full-length H. sapiens MDC1 was amplified by PCR (Expand Long Template PCR system; Roche) from HeLa Kyoto cDNA (gift from S.c. Moser) using forward and reverse primers R64 (GGGCGGCCGCATGGAGGACACCCAGGCTATTGACTGG) and R63 (GGGCGGCCGCTCAGGTGGATGACATCTCCAAAGGG), respectively. The PCR fragment was cloned into pSC-B vector (Strataclone Blunt PCR cloning kit; Stratagene). The plasmid pA338 encodes full-length MDC1 cDNA (NP_055456.2) under T7 polymerase promoter containing six previously reported amino acid change polymorphisms, respectively [Glu371(GAG>AAG)Lys, Pro386(CCA>CTA)Leu, Ser1180(TCT>CCT)Pro, Tyr1266(TAT>TCT)Ser, Met1316(ATG>ACG)Thr, and Ser1540(TCC>CCC)Pro], and three previously reported silent polymorphisms, respectively [Thr1548(ACC>ACT), Thr1157(ACC>ACG), and Phe2046(TTC>TTT)], plus a new silent polymorphism (or silent mutation) [Ile2051(ATT>ATC)].
Cell culture and stable cell line generation
HeLa and U2OS were grown in normal conditions as described previously (Tatham et al. 2008). DT40 cells were grown in RPMI-1640 (GIBCO) medium under normal conditions with 10% fetal bovine serum, 1% heat-inactivated chicken serum, 2 mM L-glutamine (GIBCO), and 10 μM β-mercaptoethanol. Culture and treatment of AsiSI-ER-U2OS were carried out as described previously (Iacovoni et al. 2010).
For the stable DT40 cell line, cells were transfected with 30 μg of targeting construct that was linearized with NotI and electroporated at 550 V and 25 μF using the Bio-Rad Gene Pulser Xcell electroporation system. Targeted clones were selected with 0.5 μg/mL puromycin or 50 μg/mL blasticidin, while nontargeted rescue clones were selected with 0.5 mg/mL zeocin.
For the YFP-RNF4 stable cell line, U2OS cells were transfected with an expression plasmid of human or rat wild-type YFP-RNF4 or YFP-RNF4 mutants using FuGENE HD transfection reagent (Roche) following the protocol of the manufacturer. Twenty-four hours after transfection, stable transfectants were selected using 5 μg/mL blasticidin.
siRNA methods
siRNA was from Dharmocon. We used established protocols for Lipofectamine RNAiMAX (Invitrogen). For Ubc9 knockdown in ChIP experiments, a sequential transfection was performed on the first and fourth days, respectively, and then harvested on the sixth day.
Antibodies
The antibodies used in the study are shown in Supplemental Table 1.
Treatment with DNA-damaging agents
IR treatment was performed using a self-shielded γ irradiator containing 65 TBq of cesium 137 (IBL437C, CIS Biointernational). HU was from Sigma. For DT40, cells were treated with HU chronically.
Clonogenic survival assay
U2OS cells were transfected with RNF4- or SUMO-targeting siRNAs for 3 d and treated with DNA-damaging agents or IR. Cells were left for 10–14 d at 37°C to allow colony formation. DT40 cells treated with DNA-damaging agents were plated on methylcellulose, and viable colonies were counted after 14 d. Colonies were stained with Giemsa and counted. Survival rates are normalized relative to an untreated control.
Immunofluorescence staining
Cells were fixed with 2% PFA for 20 min or in cold methanol for <10 min. Cells were then permeabilized with 0.5% Triton X-100 PBS for 10 min and blocked in 3% BSA and 0.1% Tween 20 in PBS for 1 h. Primary and secondary antibodies were incubated for 1 h at room temperature, respectively.
Time-lapse imaging
Cells were seeded onto a Lab-TekII cover glass chamber (Nalge Nunc international) and cultured in normal conditions as above. The next day, cells were transfected with 0.2 μg of plasmid cDNA encoding RNF4 wild type or RNF4 mutants for 24 h. Before imaging, cells were transferred to CO2-independent medium (Invitrogen) with 10% feta calf serum (FCS) throughout image acquisition. Images were acquired on a restoration microscope (DeltaVision Spectris; Applied Precision) with a 60× 1.40 NA objective and a cooled charge-coupled device camera (CoolSNAP HQ; Roper Scientific). Data sets of 512 × 512 pixels with 2 × 2 binning (or 1 × 1), and 30 Z sections spaced by 0.2 μm were acquired every 1 or 5 min on a microscope fitted with a 37°C environmental chamber (Solent). Image analysis was performed by SoftWorx software (Applied Precision). Time courses were presented as maximum intensity projections of deconvolved three-dimensional data sets. Images were displayed using Photoshop (Adobe) or QuickTime Player.
Laser micro-irradiation
For laser micro-irradiation, cells were placed in μ-Dish35mm, high Grid-500 (ibidi) and sensitized using 10 μM BrdU for 24 h. DNA damage was created in a defined subnuclear region using a 405-nm diode laser beam through a 60× 1.4 NA Plan-Apochromat lens (Olympus). Typically, an average of 60 cells was micro-irradiated within 15 min, and each cell was exposed to the laser beam for the average of 10 sec. After recovery at the times indicated in the figure legends, cells were fixed. DNA damage was visualized by indirect immunofluorescence staining. Fluorescence images were captured using the same lens and microscope as the micro-irradiation. Obtained data were analyzed using the image analysis tools included in SoftWorx. For the ssDNA test (BrdU staining) and K63 and K48 ubiquitin chain detection, micro-irradiation was performed using a Zeiss PALM Microbeam system. Energy output was <25%. For each condition, >100 cells were micro-irradiated within 5 min.
FRAP
FRAP analysis was performed on the microscope described above. A circle region was placed over foci with comparable intensity between YFP-RNF4 and YFP-RNF4cs1. After a series of three prebleach images, these foci were subject to a bleach pulse with a 488-nm argon laser focused through the same lens described above, followed by image acquisition in ∼0.5-sec intervals. Average fluorescent intensities in the bleached region were normalized against intensities in an unbleached area. Estimations of the mobile protein fraction (Fm) and half-time of recovery (t1/2) were performed using SoftWorx, Excel, and Graphpad Prism software. The values were expressed as mean + SD.
Quantitative proteomics
The quantitative proteomic experiment was performed with the SILAC technique as described (Golebiowski et al. 2009). Briefly, cells were grown in Dulbecco's modified Eagle's medium lacking all amino acids except L-lysine and L-arginine, which were replaced with stable isotope (SILAC) forms (Cambridge Isotope Laboratories) depending on the treatment (see below). The medium was supplemented with 10% dialyzed FCS. The experiment was performed on tandem affinity purification (TAP)-SUMO-2-containing HeLa cells in the following three SILAC conditions: untreated (Lys0 and Arg0), 15 Gy of IR with 1 h (Lys4 and Arg6), and 6 h (Lys8 and Arg10) of recovery. Cells were grown in 50 150-mm diameter dishes per SILAC condition. The TAP procedure as well as mass spectrometry and quantitative data analyses were performed as previously described (Golebiowski et al. 2009).
GFP pull-down
Cell stably expressing YFP-targeted RNF4 or its SIM mutant were fractionated in buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 200 mM iodoacetamide, and protease inhibitor cocktail (EDTA-free; Roche). The lysate was passed through a 25- to 26-gauge needle and spun down. The nuclear pellet was lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 50 mM sodium floride, 5 mM sodium pyrophosphate, 1 mM sodium ortho vanadate, 1 mM benzamidine, 0.2 mM phenylmethanesulfonylfluoride (PMSF), 0.1% β-mercaptoethanol, 0.27 M sucrose, protease inhibitor cocktail, and 200 mM iodoacetamide; sonicated; and diluted 1:2 with the same buffer. After preclearing with agarose beads, the nuclear lysates were incubated with GFP-TrapA beads overnight at 4°C followed by three washes with lysis buffer and 5 min of boiling in 2× SDS sample buffer. Proteins were resolved by 4%–12% NuPAGE Novex Bis-Tris gels (Invitrogen) and transferred to PVDF membrane (GE Healthcare). Immunoblotting was performed with the appropriate antibodies as indicated.
Ni-NTA bead pull-down
HeLa cells stably expressing 6x-his-tagged SUMO2 were lysed as described before (Tatham et al. 2009). In brief, the lysate was sonicated and incubated with 50 μL of Ni-NTA agarose bead suspension (Qiagen, Inc.) overnight at 4°C. After washing, the beads were incubated with elution buffer containing 200 mM imidazole, 5% SDS, 150 mM Tris-HCl (pH 6.7), 30% glycerol, 720 mM β-mercaptoethanol, and 0.0025% Bromophenol blue for 20 min at room temperature. Immunoblot analysis was performed using antibody against MDC1.
SUMOylation assay and pull-down in vitro
Full-length MDC1 was in vitro translated using the Flexi Rabbit Reticulocyte lysate system (Promega) and radiolabeled using 35S-L-methionine (Perkin Elmer). SUMOylation assays were performed by mixing 2 μL of radiolabeled MDC1 with recombinant proteins SUMO1/2 (2 μg), UBC9 (E2, 1.2 μg), and SAE1/2 (E1, 0.12 μg) as described previously (Tatham et al. 2005). Gels were dried, and the radioactivity of in vitro translated protein was measured by a PhosphorImager (Fuji). To determine SUMO E3 ligase activity, SUMOylation reactions were performed with 0.1 μg of Ubc9 per assay where no residual SUMOylation of MDC1 could be observed without ligase addition. Recombinant PIAS1/4 were then added to the reaction at 25, 50, 100, and 200 ng.
For RNF4 pull-down, in vitro translated and radiolabeled full-length MDC1 samples were SUMOylated with SUMO-1/2 as described above. SUMOylated reactions were then mixed with 10 μL of agarose beads containing the four SIM domains of RNF4. Suspension were incubated for 20 min at 4°C (Bruderer et al. 2011). After one wash, beads were resuspended in sample buffer and boiled. Equal amounts of input, bead supernatant, and beads were loaded on a 4%–12% NuPAGE gel, and radioactivity was measured by PhosphorImager (see above).
ChIP and RT-qPCR
ChIP assays were carried out according to the Upstate protocol (Millipore) with the following modifications: Approximately 2 × 106 to 3 × 106 cells were cross-linked with 1% formaldehyde for 10 min, followed by incubation with 0.125 M glycine for 5 min. Cells were scraped into PBS and lysed in 300 μL of buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris (pH 8.1), and protease inhibitors. Chromatin was sheared by sonication for 14 cycles at the high setting using a Bioruptor (Diagenode). Lysates were then diluted 1/10 with dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris at pH 8.1). One-third of the lysate was incubated overnight with 1 μg of control IgG (Santa Cruz Biotechnology) or antibody specific for SUMO-1, SUMO-2 (Invitrogen), RPA70 (Cell Signaling), Rad51 (Santa Cruz Biotechnology), or CtIP (Bethyl Laboratories). Immunocomplexes were captured by incubation with protein G Sepharose (Sigma) for 1 h. Reversal of cross-links was carried out by incubation in the presence of 200 mM sodium chloride overnight at 65°C and subsequent incubation with 0.25 μg/μL proteinase K (Roche) for 1 h at 55°C. DNA was purified using a PCR purification kit (Qiagen).
Enrichment of chromatin-binding factors was assessed by RT-qPCR using specific primers and Perfecta SYBR Green Fastmix (Quanta Biosciences) on an ABI7500 real-time PCR machine.
End processing assay
To detect ssDNA by microscopy, HeLa cells were treated with siRNA to RNF4 or a nontargeting siRNA for 2 d. Cells were cultivated for 24 h in the presence of 10 μM BrdU before micro-irradiation. After 2 h of recovery, cells were fixed and immunostained with anti-BrdU, anti-γH2AX, and anti-cyclinA antibodies without any preceding DNA denaturation or nuclease treatment. The energy output of micro-irradiaton was controlled at the minimum that causes DSBs. This was further tested by Cyclin A staining showing that BrdU lines were only detected in the S/G2 phase of the cell cycle, indicating the DSB resection.
Statistical analysis
Two groups of mean comparisons were performed using Student's t-test. A P-value <0.05 was considered as significant.
Acknowledgments
We thank John Rouse for invaluable provision of cells and antibodies and many helpful discussions. Sam Swift's help in setting up laser micro-irradiation and Arno Alpi's help with DT40 work are greatly appreciated. We thank Penny Jeggo and Atsushi Shibata for advice on the detection of ssDNA. We thank Ellis Jaffray for provision of recombinant, purified SUMO-1, SUMO-2, SAE1/2, Ubc9, PIAS1, and PIAS4; and Marie-Claude Geoffroy for provision of RNF4-targeting constructs. This work was supported by Cancer Research UK, Medical Research Council, and a European Regional Development Fund grant for an Innovation Pipeline for Translational Science (LUPS/ERDF/2008/2/1/0429). J.S.C. was supported by the A-STAR PhD student program of Singapore. A.S. set up and executed the ChIP analysis. J.S.C. generated the RNF4−/− DT40 cells and analyzed their DNA damage phenotype. J.-F.M. generated MDC1 constructs, carried out in vitro SUMO modification reactions, and performed RNF4 pull-down experiments. F.G. carried out the quantitative proteomic analysis of SUMO substrates in response to DNA damage. The project was led by Y.Y., who carried out all other experiments. R.T.H. conceived the study. Y.Y. and R.T.H. wrote the paper with help from A.S., J.S.C., J.-F.M., and F.G. All authors discussed the data and contributed to the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.189274.112.
References
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Boulton SJ 2006. Cellular functions of the BRCA tumour-suppressor proteins. Biochem Soc Trans 34: 633–643 [DOI] [PubMed] [Google Scholar]
- Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT 2011. Purification and identification of endogenous polySUMO conjugates. EMBO Rep 12: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci 95: 5287–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Dou H, Huang C, Singh M, Carpenter PB, Yeh ET 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 39: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10: 564–568 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT 2009. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24 doi: 10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I 2005. Ubiquitylation and cell signaling. EMBO J 24: 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideker J, Perry JJ, Boddy MN 2009. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst) 8: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K 2010. Coping with DNA double strand breaks. DNA Repair (Amst) 9: 1256–1263 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Wu X, Chen J 2003. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421: 957–961 [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J 23: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA 2009. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 7: e1000252 doi: 10.1371/journal.pbio.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, Hay RT 2011. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol 18: 1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP 2004. MDC1/NFBD1: A key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst) 3: 953–957 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem 277: 31663–31672 [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T 2007. Conserved function of RNF4 family proteins in eukaryotes: Targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Kim S, Jaffray E, Song J, Chen Y, Hay RT 2005. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat Struct Mol Biol 12: 67–74 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT 2009. Detection of protein SUMOylation in vivo. Nat Protoc 4: 1363–1371 [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, et al. 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11–Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M 2007. The yeast Hex3·Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184 [DOI] [PubMed] [Google Scholar]
- Yun MH, Hiom K 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459: 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
- Zierhut C, Diffley JF 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J 27: 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]