Growth factor-mediated proliferation and self-renewal maintain tissue-specific stem cells and are frequently dysregulated in cancers. Platelet-derived growth factor (PDGF) ligands and receptors (PDGFRs) are commonly overexpressed in gliomas and can initiate tumors in genetically engineered models. In this study by Rich and colleagues, the authors investigate the role of PDGFRs in intratumoral glioblastoma heterogeneity. They demonstrate that while PDGFRα is expressed only in a subset of gliomas, PDGFRβ is more commonly expressed by self-renewing tumorigenic glioma stem cells (GSCs), which are known to drive tumor growth and promote therapeutic resistance. In addition, inhibiting PDGFRβ activity repressed GSC self-renewal in vitro and tumor growth in vivo. Thus, this study provides novel insight into the mechanisms controlling GSC function.
Keywords: PDGFRβ, Stat3, cancer stem cell, glioblastoma, invasion
Abstract
Growth factor-mediated proliferation and self-renewal maintain tissue-specific stem cells and are frequently dysregulated in cancers. Platelet-derived growth factor (PDGF) ligands and receptors (PDGFRs) are commonly overexpressed in gliomas and initiate tumors, as proven in genetically engineered models. While PDGFRα alterations inform intertumoral heterogeneity toward a proneural glioblastoma (GBM) subtype, we interrogated the role of PDGFRs in intratumoral GBM heterogeneity. We found that PDGFRα is expressed only in a subset of GBMs, while PDGFRβ is more commonly expressed in tumors but is preferentially expressed by self-renewing tumorigenic GBM stem cells (GSCs). Genetic or pharmacological targeting of PDGFRβ (but not PDGFRα) attenuated GSC self-renewal, survival, tumor growth, and invasion. PDGFRβ inhibition decreased activation of the cancer stem cell signaling node STAT3, while constitutively active STAT3 rescued the loss of GSC self-renewal caused by PDGFRβ targeting. In silico survival analysis demonstrated that PDGFRB informed poor prognosis, while PDGFRA was a positive prognostic factor. Our results may explain mixed clinical responses of anti-PDGFR-based approaches and suggest the need for integration of models of cancer as an organ system into development of cancer therapies.
Aberrant growth factor receptor signaling promotes multiple hallmarks of cancer (Hanahan and Weinberg 2011), but anti-growth factor therapies often display therapeutic efficacy limited to rare patient subgroups associated with receptor expression or mutation (Lynch et al. 2004; Paez et al. 2004; Mellinghoff et al. 2005; Gerber and Minna 2010). The additional intricacies of growth factor receptor functions in cancer are derived from the complexity of tumors that are not simply neoplastic cells, but rather multicellular tissues (Reya et al. 2001; Hanahan and Weinberg 2011). Growth factors provide instructive cues in normal development and organ homeostasis that become destructive when dysregulated; e.g., expression of growth factors in the brain stimulates proliferation of neural and glial progenitors to induce the formation of glioma-like growths (Doetsch et al. 2002; Jackson et al. 2006; Assanah et al. 2009). Indeed, many genetically engineered brain tumor mouse models have demonstrated that forced expression of growth factors or their receptors can initiate tumors (Furnari et al. 2007). While epidermal growth factor receptor (EGFR) has been the focus of many brain tumor studies, it is notable that expression of wild-type or constitutively active mutant EGFR is rarely oncogenic as a single lesion (Holland et al. 1998; Weiss et al. 2003; Wei et al. 2006), whereas expression of platelet-derived growth factor (PDGF) ligands can induce tumors as a single driving event (Uhrbom et al. 1998; Dai et al. 2001; Shih et al. 2004). Expression of PDGFs and PDGF receptors (PDGFRs) is found even in low-grade gliomas (Nister et al. 1982; Pantazis et al. 1985; Harsh et al. 1990; Maxwell et al. 1990; Hermanson et al. 1992; Plate et al. 1992; Guha et al. 1995), suggesting that this pathway is possibly an early oncogenic event, in contrast to EGFR, which is much more commonly found in high-grade gliomas (Furnari et al. 2007).
Gliomas are an attractive model to study the role of growth factors in tumor cell heterogeneity, as these tumors are frequently lethal, have been characterized in their genetics, display intratumoral heterogeneity, and commonly have aberrant growth factor pathways. Indeed, a recent study has shown that a mutant form of EGFR (EGFRvIII) maintains tumor heterogeneity through induction of interleukin-6 (Inda et al. 2010), which we demonstrated promotes glioblastoma (GBM) stem cell (GSC) maintenance (Wang et al. 2009). Systematic gene expression and sequencing GBM (World Health Organization grade IV gliomas) studies have informed a greater granularity of this disease with at least two very strong tumor subgroups (proneural and mesenchymal) with two other possible groups (classical/proliferative and neural), according to the work of Heidi Phillips and The Cancer Genome Atlas (TCGA) (Phillips et al. 2006; The Cancer Genome Atlas Research Network 2008; Verhaak et al. 2010). These subgroups are associated with specific alterations in growth factor receptors: The strongest association has been made between overexpression, amplification, and mutation of PDGFRα and the proneural subtype, with a more modest association between EGFR and the classical/proliferative tumor group (Verhaak et al. 2010). Based on this background, we hypothesized that PDGF and PDGFR signaling may also serve a role in intratumoral heterogeneity.
There are four PDGF ligands (PDGF-A, PDGF-B, PDGF-C, and PDGF-D) that dimerize and bind to PDGF receptors (PDGFRα and PDGFRβ) (Fredriksson et al. 2004). PDGF-A, PDGF-B, and PDGF-C bind to PDGFRα, while PDGF-B and PDGF-D bind to PDGFRβ. Ligand binding induces autophosphorylation of the PDGFR and propagation of intracellular signals, resulting in changes in cellular behaviors, including proliferation, survival, and migration. Within the CNS, PDGF maintains neural stem cells (NSCs) with differential receptor expression based on developmental stage. Ishii et al. (2008) reported that NSCs located in the subventricular zone (SVZ) of an early postnatal mouse brain express PDGFRβ and that PDGFRβ-mediated signaling is not essential for ex vivo NSC proliferation, but rather their survival, migration, and neural differentiation (Ishii et al. 2008). However, this same group (Ishii et al. 2006) demonstrated that a brain-specific disruption of PDGFRβ using a nestin-Cre model displayed grossly normal development, but with cognitive and socio-emotional deficits (Nguyen et al. 2011) and hippocampal neuronal dendrite alterations (Shioda et al. 2011). In contrast, Jackson et al. (2006) reported that PDGFRα was the only PDGFR isoform expressed in SVZ NSCs located in the adult mouse brain, and Smits et al. (1991) reported that PDGFRβ-expressing neuronal cells are the cortical neurons but not in the SVZ. In GBM, PDGF ligands and PDGFRα are overexpressed in human cell lines and patient specimens, whereas PDGFRβ is detected in adjacent vascular cells (Nister et al. 1982; Harsh et al. 1990; Hermanson et al. 1992; Plate et al. 1992). Overexpression of PDGF-B in mouse neural progenitors induces glioma formation associated with proliferation of PDGFRαhigh cells (Jackson et al. 2006). PDGFRα expression is also associated with poor survival in patients with low-grade gliomas (Varela et al. 2004), while PDGFRβ and activated PDGFRα were associated with malignant histology in pediatric gliomas (Thorarinsdottir et al. 2008). While these data suggest the importance of the PDGF/PDGFR axis in tumor initiation, the role of PDGFRs in glioma intratumoral variation is not defined.
Discovery of differences between GBMs is complemented by identification of highly tumorigenic subpopulations of glioma cells within an individual tumor (Ignatova et al. 2002; Hemmati et al. 2003; Galli et al. 2004; Singh et al. 2004). Functionally defined self-renewing and tumorigenic GSCs may be clinically important, as several studies have shown an inverse relationship between the frequency of GSCs and patient survival and resistance to therapy (Murat et al. 2008; Pallini et al. 2008; Laks et al. 2009; Kappadakunnel et al. 2010; Metellus et al. 2011; Svendsen et al. 2011), although this is not uniform (Kim et al. 2011). While the cancer stem cell hypothesis and, by extension, GSCs have been controversial due to universally informative enrichment markers and cell-of-origin and optimized assays for functional identification (Rahman et al. 2011), GBMs have proven a largely reliable model of a hierarchical model of intratumoral heterogeneity. GSCs are potentially additionally important in clinical paradigms, as they have a greater angiogenic and invasive potential than nonstem glioma cells (Bao et al. 2006; Folkins et al. 2007; Cheng et al. 2011). Thus, identification of GSC-dependent pathways may provide new opportunities for targeting important intratumoral subpopulations that may have been underappreciated in prior studies (e.g., targeting of subpopulations of cells that express inducible nitric oxide synthase) (Eyler et al. 2011). We investigated the role of PDGFRs in GSCs and determined that PDGFRβ specifically correlated with intratumoral heterogeneity. Our data demonstrate that PDGFR signals differ within glioma subpopulations, suggesting that not all PDGF signals are equivalent within the tumor. Furthermore, PDGFRβ is likely to be a viable target for anti-glioma therapies, even in GBM subgroups that do not express high levels of PDGFRα. These results demonstrate that growth factor receptors may function on different levels of the complex systems in cancer.
Results
PDGFRβ is preferentially expressed in glioma stem cells
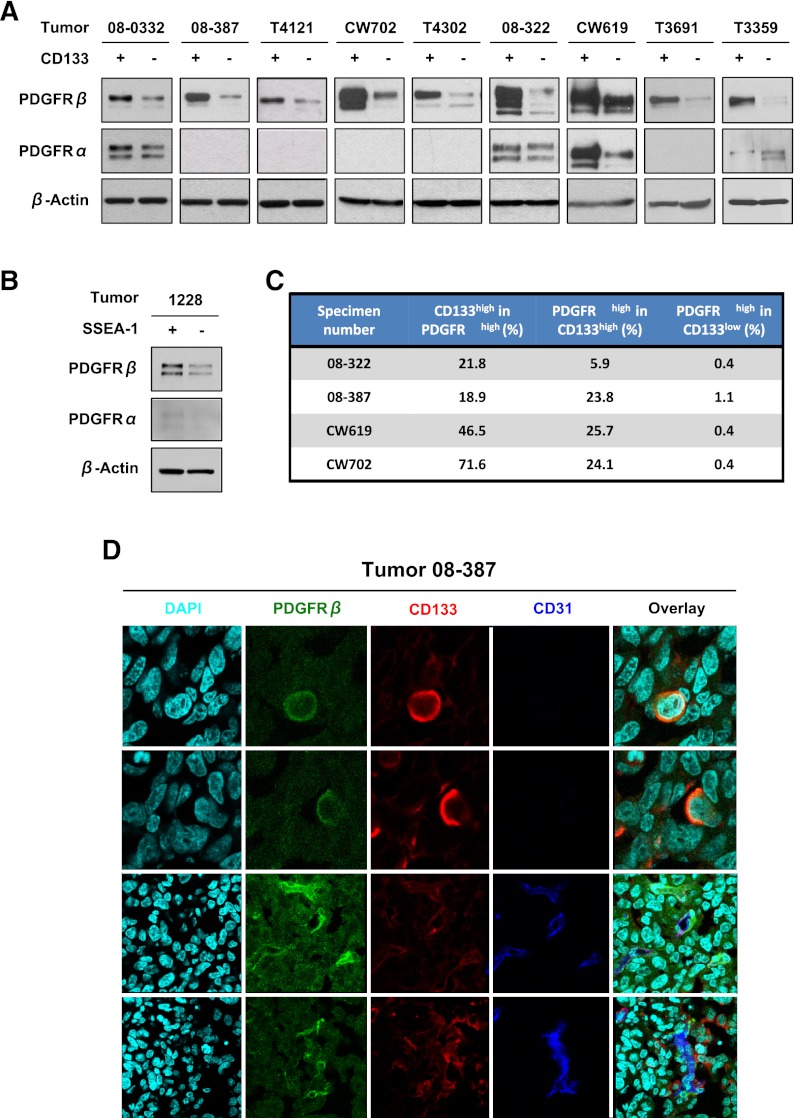
To determine the expression of PDGFRs in the complex neoplastic compartment, we measured levels of PDGFRα and PDGFRβ via immunoblotting in cells briefly cultured (less than five passages) and previously functionally validated as GSCs (self-renewing, expressing stem cell markers, and tumorigenic) or non-GSCs isolated from the same tumor. PDGFRα was expressed only in a subset of tumors, with modest variation between GSCs and non-GSCs (Fig. 1A,B). In contrast, PDGFRβ was detected in all specimens evaluated regardless of PDGFRα expression, with some variance of basal PDGFRβ levels (Fig. 1A,B). GSCs consistently displayed a strong elevation of PDGFRβ expression in comparison with matched non-GSCs regardless of the enrichment method (Fig. 1A,B), suggesting that differences in intratumoral PDGFRβ expression patterns reach beyond a single marker. Together, these data suggest that PDGFRβ, but not PDGFRα, correlates with intratumoral subpopulations of GBM cells.
Figure 1.
PDGFRβ is elevated in GSCs. Immunoblotting assay comparing PDGFRα and PDGFRβ expression in GSCs and GBM nonstem cells sorted using CD133 (A) and CD15 (B) antibodies reveals increased PDGFRβ in the GSC fraction. (C) Summary of fluorescence-activated cell sorting (FACS) analysis demonstrating coexpression of PDGFRβ and CD133. (D) Immunofluorescence staining of PDGFRβ and CD133 antibodies in glioma specimens demonstrates coexpression. PDGFRβ-positive cells are green, CD133 cells are red, and CD31 cells are blue.
To confirm that PDGFRβ is highly expressed on GSCs, we performed double labeling with PDGFRβ and a putative GSC marker followed by flow cytometry (Fig. 1C). Analysis of bulk tumor cells from five different GBMs showed that 18.9%–71.6% of PDGFRβhigh cells were CD133high. Enrichment for coexpression was also determined; 5.9%–25.7% of CD133high cells were PDGFRβhigh, whereas only 0.4%–1.1% of CD133low cells were PDGFRβhigh (Fig. 1C). To rule out cell culture effects on PDGFRβ expression, tumor sections were stained with antibodies against PDGFRβ and a putative GSC marker. PDGFRβ and CD133 frequently marked cells in the perivascular niche, a region enriched for GSCs (Calabrese et al. 2007), as well as pericytes. However, a subset of PDGFRβ;CD133 double-positive cells were found without adjacent vasculature in the tumor sections (Fig. 1D). Collectively, these data suggest that GSCs express PDGFRβ.
PDGFRβ regulates expression of glioma stem cell markers
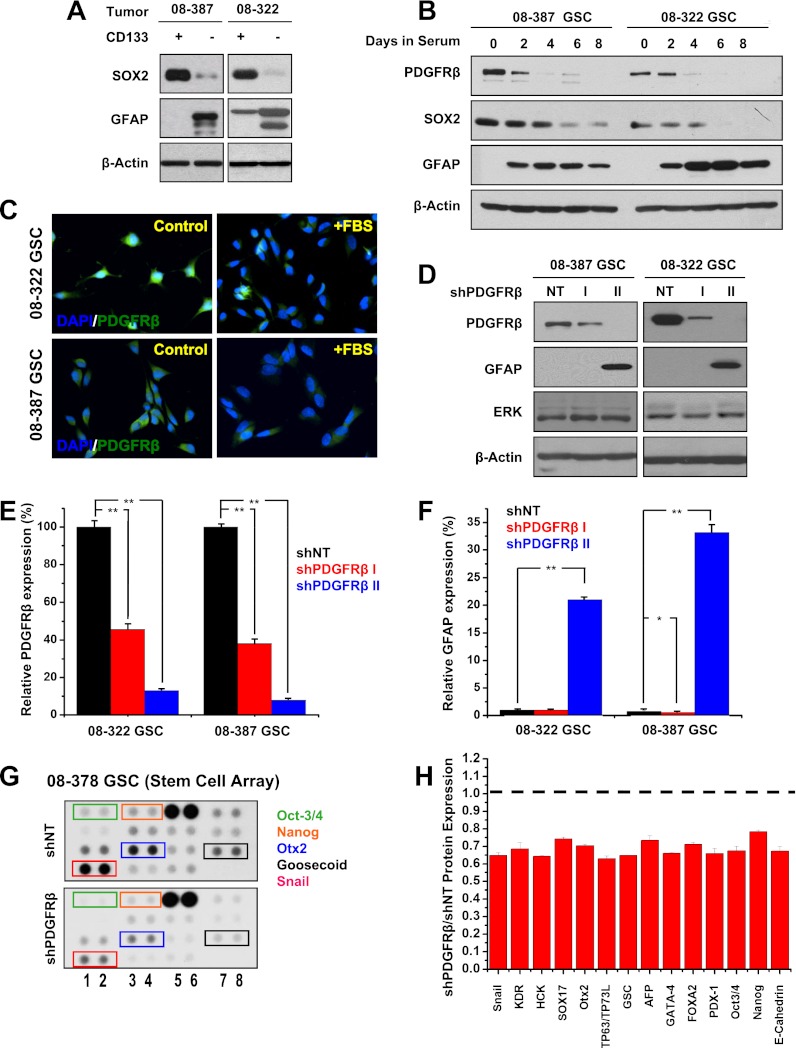
Cancer stem cells often share developmental programs with normal stem cells, both embryonic and adult, with regulation by core stem cell machinery that has also been linked to induced pluripotency. Independent of PDGFRα levels, PDGFRβhigh GSCs strongly expressed SOX2 (SRY [sex determining region Y]-box 2), with reduced levels of the astrocytic lineage marker GFAP (glial fibrillary acidic protein) compared with GSC-depleted fractions (Fig. 2A,B). Upon the induction of differentiation using serum or retinoic acid, GSCs lost expression of PDGFRβ and SOX2 while gaining GFAP expression within 4 d, as determined by immunoblotting and immunofluorescence (Figs. 2B,C; Supplemental Fig. S1a).
Figure 2.
PDGFRβ and GSC marker expression correlate. (A) Sox2 and GFAP protein expression in GSCs and GBM nonstem cells determined via Western confirmed differences in the expression of these stem and differentiation markers. (B) PDGFRβ, Sox2, and GFAP expression monitored using Western after FBS addition demonstrated PDGFRβ and Sox2 decreased while GFAP increased with differentiation. (C) Immunofluorescence demonstrated that PDGFRβ expression decreased after differentiation. (D) PDGFRβ knockdown was confirmed via Western after introduction of two different PDGFRβ shRNAs (shPDGFRβ I and shPDGFRβ II) in comparison with nontargeting (NT) control. PDGFRβ knockdown associated with increased GFAP expression. (E) Efficiency of PDGFRβ knockdown was quantitatively measured by real-time PCR after exposure to lentivirus expressing shPDGFRβ I, shPDGFRβ II, or a nontargeting control shRNA (shNT). (F) Real-time PCR demonstrated increased expression of the astrocyte differentiation marker GFAP in cells with shPDGFRβ II. (G) Representative image of stem cell arrays after exposure to lysate from GSCs expressing nontargeting shRNA (shNT) or shPDGFRβ demonstrating decreased levels of many stem factors after PDGFRβ knockdown. (H) Quantification of the relative expression of stem cell factors in shPDGFRβ versus nontargeting shRNA (shNT).
To determine whether modulating PDGFRβ expression could influence GSC marker levels, we used two nonoverlapping shRNAs against PDGFRβ (designated shPDGFRβ I and shPDGFRβ II) that reduced PDGFRβ expression at both the protein and mRNA levels compared with the nontargeting control shRNA sequence (shNT), with variation in efficacy permitting dose response studies (Fig. 2D,E). shPDGFRβ II was more efficient, with >80% knockdown, whereas shPDGFRβ I reduced PDGFRβ expression by >50% (Fig. 2D,E). The potent knockdown produced by shPDGFRβ II caused an increase in GFAP protein (Fig. 2D) and mRNA (Fig. 2F) expression.
To evaluate the dependence of GSC pathways on PDGFRβ beyond SOX2, we measured the expression of stem cell regulators in GSCs targeted by shPDGFRβ using a stem cell array (Fig. 2G,H). Knockdown of PDGFRβ reduced the expression of several transcription factors known to regulate stem cell biology, including Oct-3/4 and Nanog, which form a transcriptional complex in embryonic stem cells with SOX2 (Fig. 2G,H). The broad reduction of stem cell regulatory pathways in GSCs upon the loss of PDGFRβ supports a functional role for PDGFRβ in maintaining a stem-like state in cancer.
PDGFRβ critically regulates glioma stem cell growth and survival
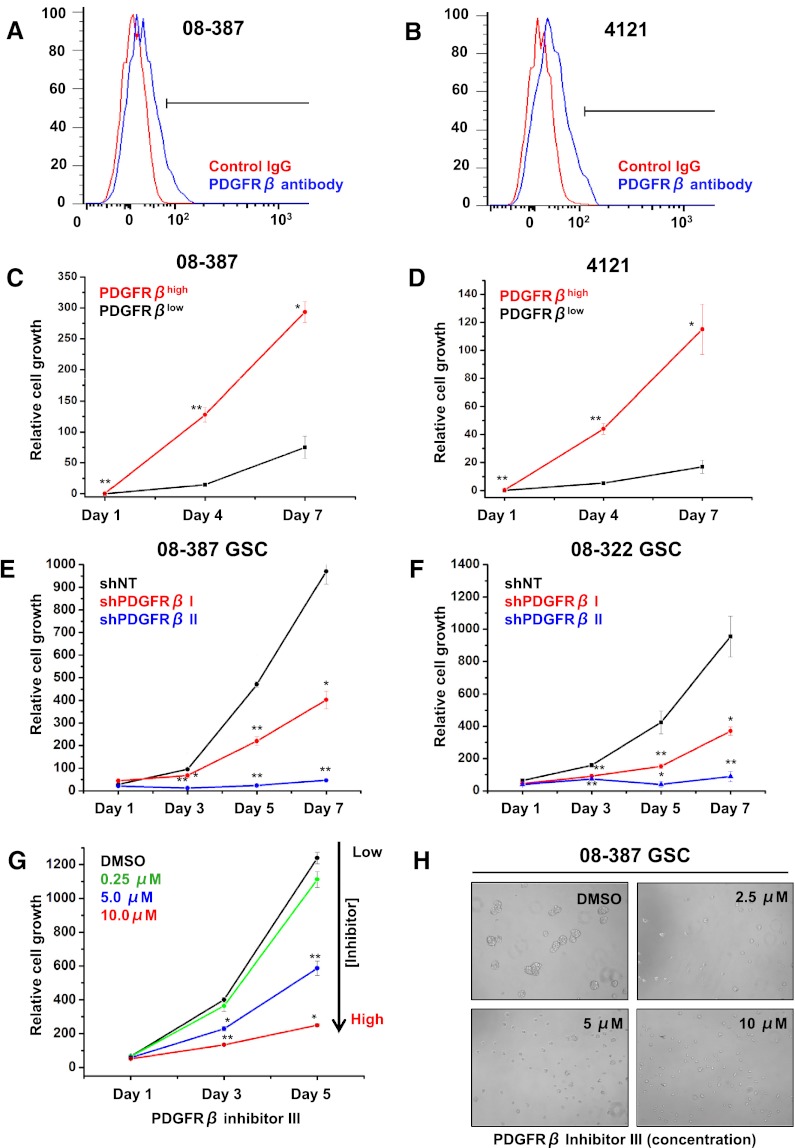
Receptor tyrosine kinases, including PDGFRβ, commonly promote cell proliferation and survival, so we determined the dependence of GSC growth on PDGFRβ signaling. PDGFRβhigh GBM cells enriched by fluorescence-activated cell sorting (FACS) were more proliferative than PDGFRβlow cells (Fig. 3A–D). Targeting PDGFRβ expression in GSCs by shRNA decreased cell growth in a dose-dependent manner in comparison with a nontargeting control (Fig. 3E,F). We validated these results in GSCs treated with PDGFRβ-specific inhibitors with a concentration-dependent effect (Fig. 3G,H).
Figure 3.
PDGFRβ promotes GSC growth. Representative FACS plots of 08-387 (A) and 4121 (B) cells demonstrating isolation of PDGFRβhigh cells. Growth of PDGFRβhigh and PDGFRβlow cells isolated via FACS from 08-387 (C) or 4121 (D) cells over time was measured using adenosine triphosphate (ATP) content in accordance with the cell titer assay. PDGFRβhigh cells grow faster than PDGFRβlow cells. Growth of 08-387 (E) or 08-322 (F) GSCs expressing two different shRNAs directed against PDGFRβ (shPDGFRβ I and shPDGFRβ II) was lower than GSCs expressing nontargeting shRNA (shNT) as measured over time using the cell titer assay. (G) Growth of GSCs exposed to increasing concentrations of PDGFRβ inhibitor III was decreased in the cell titer assay. (H) Representative images of GSCs exposed to increasing concentrations of PDGFRβ inhibitor III.
We further investigated the role of PDGFRβ in regulating cell cycle progression and survival. Using EdU labeling, we found that targeting PDGFRβ expression reduced the proportion of cells in the S phase of the cell cycle (Figs. 4A–C; Supplemental Fig. S1b). This decrease in the fraction of cycling cells was associated with increases in cells arrested in the G1 phase and present in the sub-G0 fraction (Fig. 4A,B). As the potent increase in the sub-G0 fraction with the most efficient shPDGFRβ (4%–49%) suggested an apoptotic component to the changes in cell growth, terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assays were used to quantify the percentage of apoptotic cells with PDGFRβ targeting. Both shPDGFRβs increased the apoptotic fraction of GSCs, with the most efficient knockdown of PDGFRβ resulting in a sevenfold to 10-fold increase in cell death (Fig. 4D–F). These results were consistent with the reduction of GSC growth that occurred with targeting of PDGFRβ and demonstrate that PDGFRβ signaling is critical for regulating GSC survival.
Figure 4.
PDGFRβ regulates GSC survival. Cell cycle analysis of EdU-labeled 08-387 (A) and 08-322 (B) GSCs expressing nontargeting shRNA (shNT) or two different shRNAs directed against shPDGFRβ (shPDGFRβ I and shPDGFRβ II) shows that the percentage of S-phase cells is decreased and the percentage of sub-G1 cells increased with shPDGFRβ. (C) Representative images of EdU-positive cells (red) with a DAPI costain (blue). The percentage of apoptotic cells in 08-387 (D) and 08-322 (E) GSCs was increased with shPDGFRβ in the TUNEL assay. (F) Representative images of TUNEL-positive cells (red) with a DAPI costain (blue).
PDGFRβ promotes glioma stem cell self-renewal
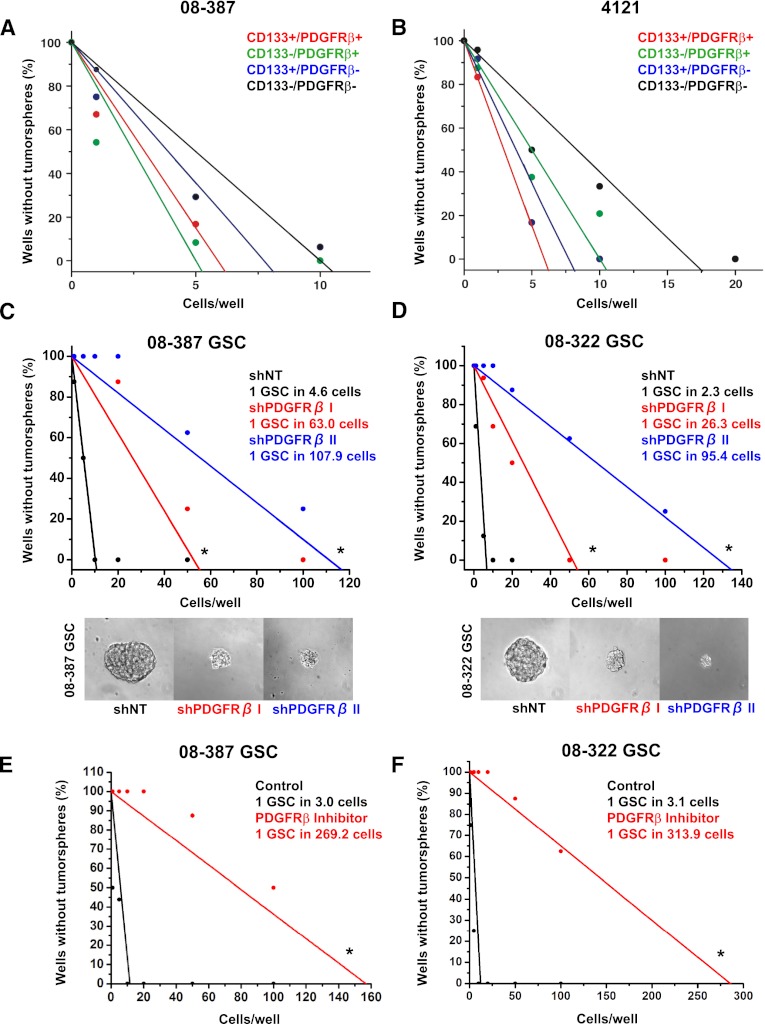
Self-renewal is a defining characteristic of cancer stem cells (Reya et al. 2001). Although the tumorsphere formation assay must be interpreted with caution (Pastrana et al. 2011), sphere formation is associated with poor clinical outcome and tumor propagation (Laks et al. 2009). We therefore determined the effect of targeting PDGFRβ on tumorsphere formation using an in vitro limiting dilution assay. CD133 (a putative GSC marker) and PDGFRβ antibodies were used to isolate four different populations of cells using FACS sorting, such as CD133high/PDGFRβhigh, CD133high/PDGFRβlow, CD133low/ PDGFRβhigh, and CD133low/PDGFRβlow cells (Fig. 5A,B). Both 4121 and 08-387 showed that CD133high/PDGFRβhigh cells possessed a higher capacity of tumorsphere formation than CD133high/PDGFRβlow cells. This result was consistent with SSEA-1 (CD15), known as another GSC marker (Supplemental Fig. S2). SSEA-1high/PDGFRβhigh cells were more likely to form tumorspheres than SSEA-1high/PDGFRβlow cells. Consistent with a functional role of PDGFRβ in self-renewal, knockdown of PDGFRβ caused a >10-fold decrease in sphere-forming efficiency in all GSC cultures tested (Fig. 5C,D), and we again noted that the reduction in sphere formation correlated with the efficiency of the shRNA. These results were further validated in pharmacological studies of a PDGFRβ inhibitor with potent reduction in the ability of GSCs to form tumorspheres by >80-fold (Fig. 5E,F). In contrast to the dependence on PDGFRβ, targeting PDGFRα expression minimally reduced sphere formation in PDGFRα-expressing GSCs and was dispensable for GSCs without PDGFRα (Supplemental Fig. S3). These data support a role for PDGFRβ in tumorsphere formation and implicate PDGFRβ in GSC self-renewal.
Figure 5.
Genetic or pharmacological targeting of PDGFRβ decreases tumorsphere formation. In vitro limiting dilution assay with 08-387 (A) and 4121 (B) demonstrated that higher PDGFRβ expression led to increasing tumorsphere formation when CD133 was used as a GSC marker. In vitro limiting dilution assays with 08-387 (C) and 08-322 (D) GSCs expressing nontargeting shRNA (shNT) or two different shRNAs directed against shPDGFRβ (shPDGFRβ I and shPDGFRβ II) demonstrated that tumorsphere formation decreases with shPDGFRβ. The tumorsphere formation capacity of 08-387 (E) and 08-322 (F) GSCs is decreased with PDGFRβ Inhibitor III treatment in the in vitro limiting dilution assay. Limiting dilution analyses were performed using Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda). (*) P < 0.0001.
PDGFRβ maintains glioma stem cells through STAT3 activation
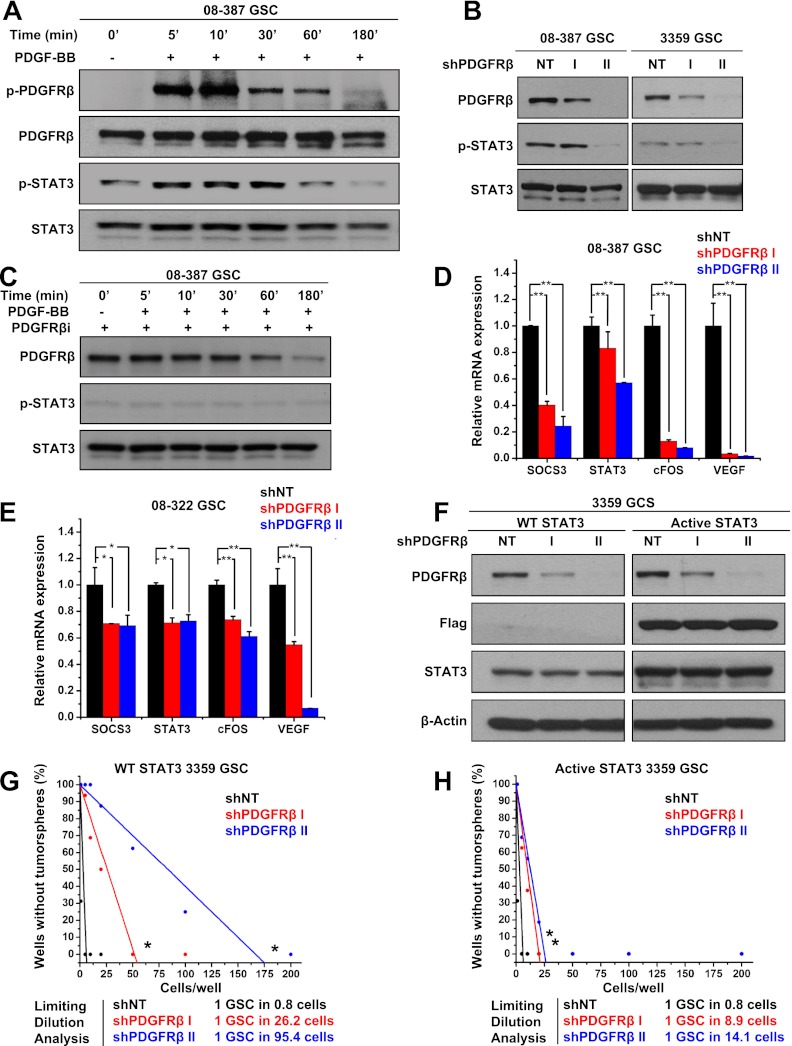
Activated PDGFRβ transduces intracellular signals to modify cellular phenotypes through several mediators that may contribute to GSC maintenance. We therefore screened potential candidates downstream from PDGFRβ through a phosphoprotein array screen comparing GSCs transduced with shPDGFRβ and the control nontargeting shRNA sequence. Several targets displayed modest phosphorylation changes with PDGFRβ knockdown, but phosphorylation of Src and signal transducer and activator of transcription 3 (STAT3) were each reduced by >50% (Supplemental Fig. S4a). As Src may serve as an intermediary between PDGFRβ and STAT3, these results suggested the potential importance of this pathway in mediating PDGFRβ effects. Furthermore, STAT3 has been suggested as a critical signaling node in cancer stem cells in general and GSCs in particular in the maintenance of a stem-like state (Sherry et al. 2009; Wang et al. 2009; Cao et al. 2010; Marotta et al. 2011). GSCs treated with PDGF-BB to specifically activate PDGFRβ displayed an induction of activating STAT3 phosphorylation (Fig. 6A). Immunoprecipitation confirmed that PDGF-BB induced the formation of a PDGFRβ/STAT3 complex in GSCs (Supplemental Fig. S4b). RNAi (Fig. 6B) or pharmacological (Figs. 6C; Supplemental Fig. S4c) inhibition of PDGFRβ reduced the activation of STAT3 in GSCs, as determined by immunoblotting. We extended these results to mRNA analysis of STAT3 and its target genes (Fig. 6D,E). STAT3 transcriptional activity after transduction with shPDGFRβ was reduced, as demonstrated through the reduced expression of STAT3 targets, including suppressor of cytokine signaling 3 (SOCS3), cFOS, and vascular endothelial growth factor (VEGF) (Fig. 6D,E). These data support STAT3 as a downstream effector of PDGF-B/PDGFRβ signaling in GSCs.
Figure 6.
PDGFRβ activates STAT3 to promote GSC tumorsphere formation capacity. (A) PDGF-BB induced activation of PDGFRβ and STAT3 in GSCs, as demonstrated with phopho-specific antibodies via Western. (B) Immunoblotting showed decreased phospho-STAT3 in cells expressing shRNA directed against PDGFRβ (shPDGFRβ) in comparison with a nontargeting control shRNA (NT). (C) Western analysis demonstrated that PDGFRβ inhibitor prevented PDGF-BB-induced phosphorylation of STAT3. mRNA expression of STAT3 target genes was decreased in 08-387 (D) or 08-322 (E) GSCs expressing shPDGFRβ in comparison with nontargeting control shRNA (shNT). (F) Immunoblotting showed successful knockdown of shPDGFRβ in comparison with nontargeting control shRNA (shNT) in GSCs expressing GFP (Control) or a constitutively active Flag-tagged STAT3 (Active STAT3). The in vitro limiting dilution assay with control GSCs (G) or GSCs expressing constitutively active STAT3 (H) demonstrated that activated STAT3 could compensate for the knockdown of PDGFRβ by restoring the ability of GSCs to form tumorspheres. Limiting dilution analyses were performed using Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda). (*) P < 0.0001.
To interrogate the role of STAT3 in PDGFRβ regulation of GSCs, we determined whether constitutively active STAT3 would functionally rescue the effects of PDGFRβ knockdown. GSCs engineered to express either GFP control or a predimerized constitutively active, Flag-tagged STAT3 were transduced with nontargeting or PDGFRβ-directed shRNAs (Fig. 6F). Introduction of shPDGFRβ caused a loss of tumorsphere formation capacity as above in parental cells (Fig. 6G). In contrast, constitutively active STAT3 rescued the effects of PDGFRβ knockdown in GSCs (Fig. 6G,H). Thus, we conclude that PDGFRβ signals through STAT3 in GSCs and that STAT3 is a transcription factor important for PDGFRβ-mediated regulation of the GSC stem-like behavior.
PDGFRβ promotes glioma stem cell invasion
Gliomas display a striking propensity to invade into a normal brain, preventing curative resection and providing a pool of tumor cells resistant to conventional therapies due to relative quiescence. Several studies suggest that GSCs display a greater invasive potential than their nonstem counterparts (Wakimoto et al. 2009; Cheng et al. 2011). As PDGF can stimulate migration in glioma cells (Shih and Holland 2006), we explored the possibility that PDGFRβ promotes GSC invasion.
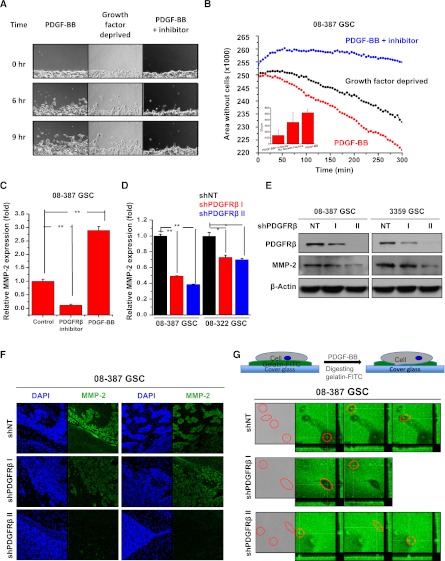
To first confirm that PDGF-BB could regulate GSC migration in vitro, we performed a wound healing assay (Fig. 7A,B). Growth factor-deprived GSCs attached on extracellular matrix displayed an increased ability to migrate when treated with PDGF-BB, as determined via light microscopy (Fig. 7A) and quantification of the open space remaining in the scratched area over time (Fig. 7B). Migration potency was reduced by removing PDGF-BB. Addition of a PDGFRβ inhibitor to the cells in the scratch assay completely blocked GSC migration (Fig. 7A,B), indicating a requirement for PDGFRβ signaling.
Figure 7.
GSC migration and invasion is dependent on PDGFRβ. (A) Representative images of GSCs in the scratch assay. (B) Calculation of the area remaining without cells in the scratch assay demonstrated that GSCs migrated in response to PDGF-BB treatment and that this movement was prevented by PDGFRβ inhibitor. (C) Quantitative real-time PCR demonstrated that MMP-2 mRNA was increased by PDGF-BB and reduced by PDGFRβ inhibitor in GSCs. (D) Quantitative real-time PCR demonstrated that MMP-2 mRNA was decreased in GSCs expressing shPDGFRβ in comparison with a nontargeting control shRNA (shNT). (E) Immunoblotting showed decreased MMP-2 expression in cells expressing shRNA directed against PDGFRβ (shPDGFRβ) in comparison with a nontargeting control shRNA (NT). (F) Representative immunofluorescent images of sections of glioma xenografts showed that GSCs treated with shPDGFRβ had reduced levels of MMP-2 and were unable to form invasive islets in vivo. (G) Activity of MMPs as determined by loss of fluorescence from FITC-gelatin was decreased in GSCs expressing shPDGFRβ in comparison with nontargeting control shRNA (shNT).
To define molecular mediators of the migratory effects of PDGFRβ in GSCs, we next evaluated the expression of a potential transcriptional target, matrix metalloproteinase-2 (MMP-2), which is known to mediate receptor tyrosine kinase regulation of invasion and metastasis. Analysis of MMP-2 mRNA (Fig. 7C,D) and protein (Fig. 7E) demonstrated that pharmacological (Fig. 7C) or genetic (Fig. 7D,E) inhibition of PDGFRβ led to reduction of MMP-2 expression. In contrast, activation of PDGFRβ by its ligand significantly increased MMP-2 (Fig. 7C). Immunofluorescence further confirmed that MMP-2 was decreased with transduction of shPDGFRβ in GSC-derived tumors (Fig. 7F), suggesting that MMP-2 is an important regulator of shPDGFRβ-mediated invasion.
To verify that decreases in MMP-2 expression with shPDGFRβ translated into reduced MMP-2 activity, we visualized gelatin digestion by MMPs upon PDGFRβ knockdown (Fig. 7G). In this assay, we measured MMP activity of GSCs by FITC-gelatin digestion, resulting in a local decrease of fluorescent signal, as determined with confocal microscopy. GSCs treated with nontargeting control shRNA produced localized reductions in fluorescence caused by gelatin digestion, but these signals were diminished by shPDGFRβ treatment such that GSCs transduced with shPDGFRβ did not show any MMP activity (Fig. 7G). Together, these experiments demonstrate a role for PDGFRβ-induced MMP-2 activity in GSC migration.
PDGFRβ knockdown impairs glioma stem cell tumor propagation
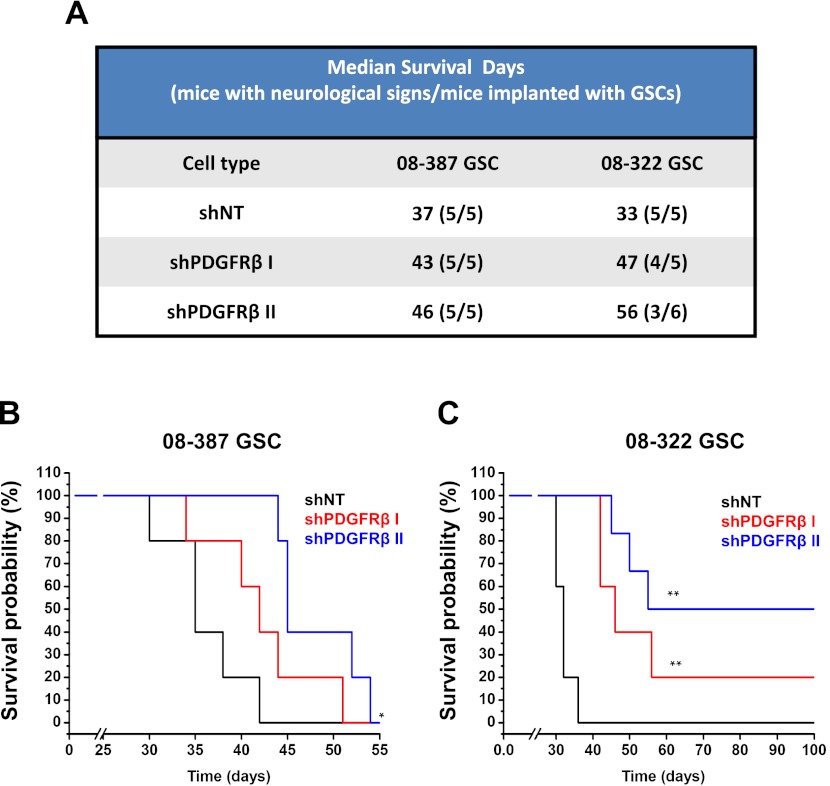
Our in vitro studies demonstrated that down-regulation of PDGFRβ expression or activity decreased GSC maintenance. To verify that these effects were sufficient to produce changes in GSC tumor propagation in vivo, we compared the ability of GSCs to initiate tumors in immunocompromised mice after transduction with shPDGFRβ or a nontargeting control sequence shRNA (Fig. 8). After shRNA incorporation, identical numbers of viable GSCs were intracranially implanted into mouse brains, and animals were monitored over time for evidence of neurological signs. Using two different xenograft models, survival of mice was prolonged with PDGFRβ targeting in comparison with nontargeting controls (Fig. 8). Median survival was increased for mice bearing either 08-387 (Fig. 8A,B) or 08-322 (Fig. 8A,C) xenografts derived from GSCs expressing shPDGFRβ. The number of tumors formed was also decreased when 08-322 shPDGFRβ II-expressing cells were implanted (Fig. 8A,C). The extension of animal survival and reduction in tumor propagation with shPDGFRβ demonstrate that PDGFRβ regulates the tumorigenic potential of GSCs. Analysis of the Repository of Molecular Brain Neoplasia Data (REMBRANDT) also demonstrates that elevation of PDGFRβ (Supplemental Fig. S5) but not PDGFRα (Supplemental Fig. S6) in GBM patient specimens is associated with poor survival, but any effects of aberrant PDGFR protein expression or activation on outcome cannot be reflected by these mRNA expression data. When taken together with our cell culture data, our experimental results suggest that PDGFRβ plays a more critical role in glioma biology than previously understood through the regulation of the GSC phenotype.
Figure 8.
PDGFRβ promotes GSC in vivo tumor propagation. (A) The median survival and number of tumors formed are shown for 08-387 and 08-322 GSCs expressing nontargeting shRNA (shNT) or shRNA directed against PDGFRβ (shPDGFRβ I and shPDGFRβ II). Kaplan-Meier survival curves for 08-387 (B) and 08-322 (C) GSCs expressing nontargeting shRNA (shNT) or shPDGFRβ demonstrate delayed tumor growth with shPDGFRβ.
Discussion
Functional contribution of PDGFRs in GBM intratumoral and intertumoral heterogeneity
Comprehensive understanding of key oncogenic signaling pathways has been advanced with the discovery of genetic changes unique to GBM subtypes (Phillips et al. 2006; Wang et al. 2009; Verhaak et al. 2010) and molecular mechanisms enhanced in GSC subpopulations (Bao et al. 2006; Folkins et al. 2007; Cheng et al. 2011; Eyler et al. 2011). Our studies build on these advances by demonstrating that different PDGFRs distinguish not only between GBMs (intertumoral heterogeneity), but also among the tumor cells within a tumor (intratumoral heterogeneity). PDGFRα expression was highly variable among glioma samples, whereas expression of PDGFRβ was more closely associated with differences in cellular subsets within a tumor. This is important because prior studies had demonstrated autocrine activation for PDGFRα in glioma, while the role of PDGFRβ was less clear. As PDGFRα and PDGFRβ can stimulate distinct pathways when activated by PDGFs, our results indicate that PDGF signals through these closely related molecules are unlikely to produce similar effects in all GBMs. It will therefore be important to continue to examine the effects of PDGF on glioma cellular biology and signaling in the context of the different PDGFR isoforms and with the recently described PDGFRα fusion (Ozawa et al. 2010).
Expression of PDGFRβ and other growth factor receptor kinases in GSCs
Our data demonstrate that PDGFRβ expression is relatively higher within GSCs, while data from the literature suggest that enrichment for GSCs may be achieved with EGFR (Mazzoleni et al. 2010) or c-Met (Li et al. 2011). Very recently, two independent groups demonstrated that GBMs display mosaic amplification of EGFR and PDGFRA (Snuderl et al. 2011; Szerlip et al. 2012). These results are highly complementary to our findings and those of others, as they suggest that growth factor pathways may have both genetic and nongenetic causes of intratumoral heterogeneity (models presented as Supplemental Fig. S7). As there may be different pools of cancer stem cells within a single tumor, we interrogated the expression of other growth factor receptors in CD133highPDGFRβ−/low GBM cells and found that c-met and several other receptors were not differentially expressed, but EGFR was highly expressed (Supplemental Fig. S8). These results suggest that activation of EGFR in CD133highPDGFRβlow/− cells is an alternative pathway through which cancer stem cell-driven phenotypes and/or biologies can be mediated. Therefore, combinations of receptor antagonists or a common regulatory node will be required for optimal efficacy against cancer stem cells.
The molecular mechanisms regulating the levels of these growth factor receptors in GSCs have not been determined, but it is possible that a change in a common pathway contributes. For example, activated receptors are typically internalized via endocytosis and targeted for degradation by the lysosome. Circumvention of these pathways, as through receptor mutation, is known to prolong cell signaling and contribute to oncogenesis. It would therefore not be surprising if GSCs had a perturbation of one or more components of the receptor degradation process that allowed for sustained cell surface expression. However, previously identified changes in the transcription factor profiles of GSCs are also likely to lead to increased mRNA expression of some receptors. For example, the NSC transcription factor SOX2 can increase EGFR expression (Hu et al. 2010), although the transcriptional regulation of PDGFRβ is less clear and should be further evaluated.
PDGFRβ as a regulator of cellular plasticity
Recent evidence suggests that GSCs may represent a highly plastic cellular subset that is capable of differentiating toward an endothelial cell lineage. GSCs expressed vascular markers in vitro and were capable of becoming incorporated into the vasculature in xenograft models in vivo (Ricci-Vitiani et al. 2010; Soda et al. 2011). Recent evidence also suggests that PDGFRβ is an important regular of mural cell plasticity, as mice with PDGFRβ-activating mutations show changes in the differentiation of pericytes and aortic vascular smooth muscle cells (Olson and Soriano 2011). It therefore is interesting to speculate that PDGFRβ could contribute to the regulation of GSC plasticity to promote tumor growth. If PDGFRβ signaling promotes vascular smooth muscle cell-like behaviors, this GSC phenotype would be expected to impact patient outcome, as changes in vascular smooth muscle size and density correlate with tumor grade (Sato et al. 2011).
PDGFRβ signaling: STAT3 and its target genes in GSCs
Activation of STAT3 has been shown to be elevated in GSCs (Sherry et al. 2009; Wang et al. 2009; Cao et al. 2010), and STAT3 mediates the effects of cytokines, including erythropoietin (Cao et al. 2010), interleukin-6 (Wang et al. 2009), and now PDGFs on GSCs. We found that PDGF-BB stimulated phosphorylation of STAT3 in a PDGFRβ-dependent manner in GSCs and knockdown of PDGFRβ decreased the expression of STAT3 target genes. Constitutively active STAT3 also prevented the reduction in tumorsphere formation capacity produced by PDGFRβ knockdown, suggesting that inhibition of STAT3 and PDGFRβ would provide benefits for patients. While the option to target STAT3 is being explored for clinical treatments, combinatorial therapies targeting mediators downstream from STAT3 may also be reasonable. For example, the STAT3 target gene MMP-2 (Xie et al. 2004) is well known to regulate metastasis, and our data demonstrate an important role for MMPs in PDGFRβ-regulated invasion. mRNA levels of the STAT3 target and critical angiogenesis regulator VEGF (Niu et al. 2002) were also reduced in GSCs by PDGFRβ knockdown. Targeting of VEGF is already approved for GBM therapy with the anti-VEGF antibody bevacizumab (Avastin), suggesting this is one signal downstream from PDGFRβ and STAT3 that can already be targeted in the clinic. PDGFRβ inhibition also decreased levels of SOCS3, but the significance of SOCS3 in glioma is still being determined. While some evidence demonstrates that targeting SOCS3 may sensitize glioma cells to radiotherapy (Zhou et al. 2007), other reports suggest that SOCS3 inactivation may promote glioma cell invasion (Lindemann et al. 2011). Further research will therefore be needed to determine the importance of STAT3 transcriptional targets regulated by PDGFRβ for GSC therapeutic resistance.
Targeting GBM heterogeneity by PDGFRs
Our data demonstrate that targeting PDGFRβ in GSCs reduces the ability of these cells to propagate tumors in vivo and suggests the potential of anti-PDGFRβ-based therapies. While broad tyrosine kinase inhibitors such as imatinib mesylate (Gleevec) have not demonstrated strong efficacy against GBM (Wen et al. 2006), newly developed drugs specifically inhibiting PDGFRα and PDGFRβ such as crenolanib (CP-868,596) are being evaluated in clinical trials for glioma. The identification of glioma subtypes with amplification of PDGFRA suggests that tumor genetic profiles may predict patients particularly sensitive to anti-PDGFRα-based approaches (e.g., ramucirumab). However, our data suggest that inhibition of PDGFRβ may still provide benefit against tumors in which PDGFRA is not amplified. Furthermore, the PDGFRs are likely to be differentially expressed with respect to developmental stage and cell type in the NSC compartment. PDGFRα is expressed throughout development, and PDGFRβ expression may be elevated in the postnatal brain, with expression decreasing in adulthood. We compared the expression of PDGFRs in normal brains and found more cell type-specific expression of PDGFRα in adult SVZ NSCs, but human fetal neuroprogenitors expressed both PDGFRs at levels similar to GSCs (Supplemental Figs. S9, S10). Of note, the function of PDGFRβ appears to differ between NSCs and GSCs, as tumors display proliferation dependence in contrast to normal brains. Collectively, these results suggest that PDGFRβ may be targetable with limited toxicity.
While no single therapy is likely to eliminate a GBM or tumor recurrence, treatment with PDGFRβ inhibitors in combination with established regimes of surgery, chemotherapy, and radiotherapy may prove to be more broadly effective in targeting GSCs and improve patient outcomes. We therefore believe that continued development of anti-PDGFR-based strategies with a focus on PDGFRβ holds value.
Materials and methods
Isolation and culture of cells
GBM cells were derived from specimens of neurosurgical resection directly from patients in accordance with a Cleveland Clinic Institutional Review Board-approved protocol. GSCs and nonstem glioma cells were separated from GBM surgical specimens or xenografts as previously described (Bao et al. 2006). The cancer stem cell phenotype of these cells was confirmed by functional assays of self-renewal (serial tumorsphere passage), stem cell marker expression (CD133, OLIG2, SOX2, and Musashi1), and tumor propagation (in vivo limiting dilution assay) (Bao et al. 2006). The CD133-depleted cells did not share these properties and were used in matched assays as nonstem tumor cells.
Immunoblotting analysis and coimmunoprecipitation
Immunoblotting analysis or homogenized tissues were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors (Roche) and were analyzed via Western as previously described (Snuderl et al. 2011). For coimmunoprecipitation experiments, GSCs growth factor-deprived overnight were treated with PDGF-BB (R&D Systems), washed in ice-cold PBS, and lysed in NET buffer (150 mM NaCl, 50 mM Tris-HCl at pH 7.4, 5 mM EDTA, 1% NP-40) supplemented with protease and phosphatase inhibitors. One milligram of precleared protein lysates was mixed with 5 μg of anti-PDGFRβ antibodies (Cell Signaling) or normal rabbit IgG (Santa Cruz Biotechnology) and incubated overnight at 4°C with gentle rocking. Immunocomplexes were captured with Protein A/G Plus agarose beads (Santa Cruz Biotechnology) and eluted using Laemmli sample buffer subjected to immunoblotting analysis as described above. Antibodies against PDGFRβ, phospho-PDGFRβ (pTyr751), STAT3, and phospho-STAT3 (pY705) were from Cell Signaling. Antibodies against SOX2 (R&D Systems), α-tubulin (Santa Cruz Biotechnology), GFAP, and Flag (M2) (Sigma-Aldrich) were also used for immunoblotting analysis.
Immunofluorescent staining
For immunostaining analysis at the single-cell level, cells were plated onto Geltrex-coated glass coverslips, allowed to attach overnight, and then fixed with 4% formaldehyde for 15 min; they were then post-fixed/permeabilized with cold methanol for 20 min. Alternatively, cells were permeabilized in 0.25% Triton X-100 for 15 min at room temperature. Nonspecific binding was blocked by incubation in 5% goat serum for 30 min. Samples were incubated with primary antibodies overnight at 4°C, followed by the appropriate isotype-specific or highly cross-adsorbed secondary fluorescently labeled antibodies (Invitrogen Molecular Probes) for 1 h at room temperature. Nuclei were counterstained with DAPI. For immunostaining analysis of tissue sections, 10-μm frozen sections were fixed in 4% formaldehyde for 15 min at room temperature followed by a cold methanol fixation/permeabilization step for 20 min and were processed as described above. Images were taken using wide-field fluorescence microscope (Leica) or Leica SP-5 confocal microscope.
Differentiation assay
GSCs plated on Geltrex-coated plates or coverslips were induced to differentiate through the addition of 10% serum in stem cell medium and then harvested at indicated time points. Harvested cells were subjected to immunoblotting analysis or fixation and processed as described above.
Vectors and lentiviral transfection
Lentiviral clones expressing PDGFRβ shRNAs and control shRNA (SHC002) were purchased from Sigma-Aldrich. shPDGFRβ 1 sequence: 5′-CCGGGCTCACCATCATCTCCCTTATCTCGAGATAAGGGAGATGATGGTGAGCTTTTT-3′; shPDGFRβ 2 sequence: 5′-CCGGGCTGGAACAGTTGCCGGATTCCTCGAGGAATCCGGCAACTGTTCCAGCTTTTTTG-3′. A lentiviral construct expressing constitutively active STAT3 was generated by subcloning a PCR-amplified fragment into the XbaI and SalI restriction sites of pLCMV-Flag-neo (a kind gift of P. Chumakov) in-frame with the N-terminal Flag sequence. Viral particles were produced in 293T cells with the pPACK set of helper plasmids (System Biosciences) in stem cell medium. Viral stocks were concentrated.
Antibody arrays
Human pluripotent stem cell antibody array (catalog no. ARY010) and human phospho-kinase antibody array (catalog no. ARY003) were purchased from R&D Systems. Assays were performed as per the manufacturer's instructions.
Proliferation assays
The cell proliferation was performed using Cell-Titer Glow (Promega) as per the manufacturer's instructions.
In vivo tumor initiation assay
GSCs were transduced with lentiviral vectors expressing shPDGFRβ targeting or nontargeting control shRNA for knockdown experiment. After puromycin selection, cells were counted, and 1000 viable cells were engrafted intracranially into athymic/nude immunocompromised mice. Animals were maintained until manifestation of neurological signs or for 180 d, when they were sacrificed. Harvested brains were photographed, fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and cryosectioned. All animal procedures conformed to the Cleveland Clinic Institutional Animal Care and Use Committee-approved protocol.
Rescue experiments with constitutively active STAT3
Activation of STAT3 requires phosphorylation of its Y705, followed by the formation of homodimers. A form of STAT3 harboring two cysteine substitutions within the C-terminal loop of the SH2 domain (STAT3-C) allowed for rendering the transcription factor constitutively active (Szerlip et al. 2012). The constitutively active STAT3 retroviral expression construct was generated by subcloning the STAT3-C with a C-terminal Flag tag followed by a TGA stop codon into the HindIII restriction site within the pLEGFP-N1 vector (BD Biosciences). The resulting construct did not express GFP. For rescue experiments, CD133-enriched GSCs were transduced by retroviral particles packaged with STAT3-C-Flag retroviral construct or pLEGFP empty vector and allowed to recover for 48 h. Neomycin-resistant cells were selected by exposure to G418 for 7 d. Stable cell populations expressing STAT3-C-Flag or EGFP were transduced to express either control shRNA or shPDGFRβs. Forty-eight hours post-infection, cells were plated to assess proliferation potential, self-renewal capacity, or expression of stem cell factors or intracranially injected for tumor-initiation studies.
Quantitative RT–PCR
Total cellular RNA was isolated with the RNeasy kit (Qiagen) and reverse-transcribed into cDNA using the SuperScript III Reverse Transcription kit (Invitrogen). Real-time PCR was performed on an Applied Biosystems 7900HT cycler using SYBR Green Master mix (SA Biosciences) and intron-spanning, gene-specific primers as follows: β-actin forward (5′-AGAAAATCTGGCACCACACC-3′) and reverse (5′-AGAGGCGTACAGGGATAGCA-3′), SOCS3 forward (5′-AGACTTCGATTCGGGACCAGCCCC-3′) and reverse (5′-GAGCCAGCGTGGATCTGCGC-3′), STAT3 forward (5′-GGGTGGAGAAGGACATCAGCGGTAA-3′) and reverse (5′-GCCGACAATACTTTCCGAATGC-3′), c-Fos forward (5′-GAGGGGCAAGGTGGAACAGTTATCT-3′) and reverse (5′-TCCTCCGGTTGCGGCATTTGG-3′), PDGFRβ forward (5′-CGTCAAGATGCTTAAATCCACAGC-3′) and reverse (5′-TGATGATATAGATGGGTCCTCCTTTG-3′), MMP-2 forward (5′-GCCCCAGACAGGTGATCTTG-3′) and reverse (5′-GCTTGCGAGGGAAGAAGTTGT-3′), VEGF-A forward (5′-TTTGCTTGCCATTCCCCACT-3′) and reverse (5′-GGGGCGGTGTCTGTCTGTCT-3′), and GFAP forward (5′-TGTGTGAGTAAGAAGGGACCGCAA-3′) and reverse (5′-GCAGGGCATGACTTGTCCCATTT-3′).
Statistical analysis
All grouped data are presented as mean ± standard deviation. Difference between groups was assessed by Student's t-test or ANOVA using GraphPad InStat software. Kaplan-Meier curves were generated and log-rank analysis was performed using MedCalc software. (*) P < 0.05; (**) P < 0.005.
Acknowledgments
We appreciate flow cytometry assistance from C. Shemo and S. O'Bryant, and the tissue provided by the Cleveland Clinic Foundation Tissue Procurement Service and S. Staugatis and M. McGraw. We thank the sources of our funding, including the American Brain Tumor Association (to Y.K.), grants from the National Institutes of Health (CA129958, CA116659, and CA154130 for J.N.R.; CA151522 for A.B.H.; K99/R00 Pathway to Independence Award CA157948 for J.D.L.; CA137443, NS063971, CA128269, CA101954, and CA116257 for A.E.S.; and NS073425 for A.N.). We also thank the Goldhirsh Foundation (to J.N.R.), James S. McDonnell Foundation (to J.N.R.), and the Ohio Department of Development Tech 09-071 (to A.E.S.). We thank the Melvin Burkhardt Chair in Neurosurgical Oncology and the Karen Colina Wilson Research Endowment within the Burkhardt Brain Tumor and Neuro-oncology Center at the Cleveland Clinic Foundation for additional support and funding (to R.J.W.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.193565.112.
References
- Assanah MC, Bruce JN, Suzuki SO, Chen A, Goldman JE, Canoll P 2009. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia 57: 1835–1847 [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN 2006. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res 66: 7843–7848 [DOI] [PubMed] [Google Scholar]
- Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, et al. 2007. A perivascular niche for brain tumor stem cells. Cancer Cell 11: 69–82 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Lathia JD, Eyler CE, Wu Q, Li Z, Wang H, McLendon RE, Hjelmeland AB, Rich JN 2010. Erythropoietin receptor signaling through STAT3 is required for glioma stem cell maintenance. Genes Cancer 1: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Wu Q, Guryanova OA, Huang Z, Huang Q, Rich JN, Bao S 2011. Elevated invasive potential of glioblastoma stem cells. Biochem Biophys Res Commun 406: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC 2001. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 15: 1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A 2002. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron 36: 1021–1034 [DOI] [PubMed] [Google Scholar]
- Eyler CE, Wu Q, Yan K, MacSwords JM, Chandler-Militello D, Misuraca KL, Lathia JD, Forrester MT, Lee J, Stamler JS, et al. 2011. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell 146: 53–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkins C, Man S, Xu P, Shaked Y, Hicklin DJ, Kerbel RS 2007. Anticancer therapies combining antiangiogenic and tumor cell cytotoxic effects reduce the tumor stem-like cell fraction in glioma xenograft tumors. Cancer Res 67: 3560–3564 [DOI] [PubMed] [Google Scholar]
- Fredriksson L, Li H, Eriksson U 2004. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor Rev 15: 197–204 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A 2004. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64: 7011–7021 [DOI] [PubMed] [Google Scholar]
- Gerber DE, Minna JD 2010. ALK inhibition for non-small cell lung cancer: From discovery to therapy in record time. Cancer Cell 18: 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Dashner K, Black PM, Wagner JA, Stiles CD 1995. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Int J Cancer 60: 168–173 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Harsh GR, Keating MT, Escobedo JA, Williams LT 1990. Platelet derived growth factor (PDGF) autocrine components in human tumor cell lines. J Neurooncol 8: 1–12 [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI 2003. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci 100: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin CH, Westermark B, Nistér M 1992. Platelet-derived growth factor and its receptors in human glioma tissue: Expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res 52: 3213–3219 [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12: 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Zhang L, Wen J, Wang S, Li M, Feng R, Yang X, Li L 2010. The EGF receptor-sox2-EGF receptor feedback loop positively regulates the self-renewal of neural precursor cells. Stem Cells 28: 279–286 [DOI] [PubMed] [Google Scholar]
- Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA 2002. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39: 193–206 [DOI] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 24: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Oya T, Zheng L, Gao Z, Kawaguchi M, Sabit H, Matsushima T, Tokunaga A, Ishizawa S, Hori E, et al. 2006. Mouse brains deficient in neuronal PDGF receptor-β develop normally but are vulnerable to injury. J Neurochem 98: 588–600 [DOI] [PubMed] [Google Scholar]
- Ishii Y, Matsumoto Y, Watanabe R, Elmi M, Fujimori T, Nissen J, Cao Y, Nabeshima Y, Sasahara M, Funa K 2008. Characterization of neuroprogenitor cells expressing the PDGF β-receptor within the subventricular zone of postnatal mice. Mol Cell Neurosci 37: 507–518 [DOI] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A 2006. PDGFRβ-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 51: 187–199 [DOI] [PubMed] [Google Scholar]
- Kappadakunnel M, Eskin A, Dong J, Nelson SF, Mischel PS, Liau LM, Ngheimphu P, Lai A, Cloughesy TF, Goldin J, et al. 2010. Stem cell associated gene expression in glioblastoma multiforme: Relationship to survival and the subventricular zone. J Neurooncol 96: 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Lee KH, Kim HS, Moon KS, Jung TY, Jung S, Lee MC 2011. The presence of stem cell marker-expressing cells is not prognostically significant in glioblastomas. Neuropathology 31: 494–502 [DOI] [PubMed] [Google Scholar]
- Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, Yong WH, Vinters HV, Liau LM, Lazareff JA, et al. 2009. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells 27: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cázares H, Eberhart CG, et al. 2011. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci 108: 9951–9956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann C, Hackmann O, Delic S, Schmidt N, Reifenberger G, Riemenschneider MJ 2011. SOCS3 promoter methylation is mutually exclusive to EGFR amplification in gliomas and promotes glioma cell invasion through STAT3 and FAK activation. Acta Neuropathol 122: 241–251 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, et al. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J Clin Invest 121: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell M, Naber SP, Wolfe HJ, Galanopoulos T, Hedley-Whyte ET, Black PM, Antoniades HN 1990. Coexpression of platelet-derived growth factor (PDGF) and PDGF-receptor genes by primary human astrocytomas may contribute to their development and maintenance. J Clin Invest 86: 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, Falini A, De Palma M, Bulfone A, Poliani PL, et al. 2010. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res 70: 7500–7513 [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. 2005. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012–2024 [DOI] [PubMed] [Google Scholar]
- Metellus P, Nanni-Metellus I, Delfino C, Colin C, Tchogandjian A, Coulibaly B, Fina F, Loundou A, Barrie M, Chinot O, et al. 2011. Prognostic impact of CD133 mRNA expression in 48 glioblastoma patients treated with concomitant radiochemotherapy: A prospective patient cohort at a single institution. Ann Surg Oncol 18: 2937–2945 [DOI] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. 2008. Stem cell-related ‘self-renewal' signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 26: 3015–3024 [DOI] [PubMed] [Google Scholar]
- Nguyen PT, Nakamura T, Hori E, Urakawa S, Uwano T, Zhao J, Li R, Bac ND, Hamashima T, Ishii Y, et al. 2011. Cognitive and socio-emotional deficits in platelet-derived growth factor receptor-β gene knockout mice. PLoS ONE 6: e18004 doi: 10.1371/journal.pone.0018004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nister M, Heldin CH, Wasteson A, Westermark B 1982. A platelet-derived growth factor analog produced by a human clonal glioma cell line. Ann NY Acad Sci 397: 25–33 [DOI] [PubMed] [Google Scholar]
- Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, et al. 2002. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 21: 2000–2008 [DOI] [PubMed] [Google Scholar]
- Olson LE, Soriano P 2011. PDGFRβ signaling regulates mural cell plasticity and inhibits fat development. Dev Cell 20: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, et al. 2010. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev 24: 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. 2004. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, et al. 2008. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res 14: 8205–8212 [DOI] [PubMed] [Google Scholar]
- Pantazis P, Pelicci PG, Dalla-Favera R, Antoniades HN 1985. Synthesis and secretion of proteins resembling platelet-derived growth factor by human glioblastoma and fibrosarcoma cells in culture. Proc Natl Acad Sci 82: 2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F 2011. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8: 486–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173 [DOI] [PubMed] [Google Scholar]
- Plate KH, Breier G, Farrell CL, Risau W 1992. Platelet-derived growth factor receptor-β is induced during tumor development and upregulated during tumor progression in endothelial cells in human gliomas. Lab Invest 67: 529–534 [PubMed] [Google Scholar]
- Rahman M, Deleyrolle L, Vedam-Mai V, Azari H, Abd-El-Barr M, Reynolds BA. 2011. The cancer stem cell hypothesis: Failures and pitfalls. Neurosurgery68: 531–545. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105–111 [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, et al. 2010. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature 468: 824–828 [DOI] [PubMed] [Google Scholar]
- Sato S, Sato Y, Hatakeyama K, Marutsuka K, Yamashita A, Takeshima H, Asada Y 2011. Quantitative analysis of vessels with smooth muscle layer in astrocytic tumors: Correlation with histological grade and prognostic significance 219. Histol Histopathol 26: 497–504 [DOI] [PubMed] [Google Scholar]
- Sherry MM, Reeves A, Wu JK, Cochran BH 2009. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 27: 2383–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Holland EC 2006. Platelet-derived growth factor (PDGF) and glial tumorigenesis. Cancer Lett 232: 139–147 [DOI] [PubMed] [Google Scholar]
- Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC 2004. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res 64: 4783–4789 [DOI] [PubMed] [Google Scholar]
- Shioda N, Moriguchi S, Oya T, Ishii Y, Shen J, Matsushima T, Nishijo H, Sasahara M, Fukunaga K 2011. Aberrant hippocampal spine morphology and impaired memory formation in neuronal platelet-derived growth factor β-receptor lacking mice. Hippocampus doi: 10.1002/hipo.20973 [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. 2004. Identification of human brain tumour initiating cells. Nature432: 396–401. [DOI] [PubMed] [Google Scholar]
- Smits A, Kato M, Westermark B, Nistér M, Heldin CH, Funa K 1991. Neurotrophic activity of platelet-derived growth factor (PDGF): Rat neuronal cells possess functional PDGF β-type receptors and respond to PDGF. Proc Natl Acad Sci 88: 8159–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. 2011. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20: 810–817 [DOI] [PubMed] [Google Scholar]
- Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, Pastorino S, Yang M, Hoffman RM, Kesari S, et al. 2011. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci 108: 4274–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen A, Verhoeff JJ, Immervoll H, Brøgger JC, Kmiecik J, Poli A, Netland IA, Prestegarden L, Planagumà J, Torsvik A, et al. 2011. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol 122: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. 2012. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci 109: 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorarinsdottir HK, Santi M, McCarter R, Rushing EJ, Cornelison R, Jales A, MacDonald TJ 2008. Protein expression of platelet-derived growth factor receptor correlates with malignant histology and PTEN with survival in childhood gliomas. Clin Cancer Res 14: 3386–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrbom L, Hesselager G, Nister M, Westermark B 1998. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res 58: 5275–5279 [PubMed] [Google Scholar]
- Varela M, Ranuncolo SM, Morand A, Lastiri J, De Kier Joffé EB, Puricelli LI, Pallotta MG 2004. EGF-R and PDGF-R, but not bcl-2, overexpression predict overall survival in patients with low-grade astrocytomas. J Surg Oncol 86: 34–40 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, et al. 2009. Human glioblastoma-derived cancer stem cells: Establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res 69: 3472–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lathia JD, Wu Q, Wang J, Li Z, Heddleston JM, Eyler CE, Elderbroom J, Gallagher J, Schuschu J, et al. 2009. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells 27: 2393–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, Karim Z, Bock NA, Reti R, Swoboda R, et al. 2006. High-grade glioma formation results from postnatal PTEN loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res 66: 7429–7437 [DOI] [PubMed] [Google Scholar]
- Weiss WA, Burns MJ, Hackett C, Aldape K, Hill JR, Kuriyama H, Kuriyama N, Milshteyn N, Roberts T, Wendland MF, et al. 2003. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res 63: 1589–1595 [PubMed] [Google Scholar]
- Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, Abrey LE, Raizer J, Cloughesy TF, Fink K, et al. 2006. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99-08. Clin Cancer Res 12: 4899–4907 [DOI] [PubMed] [Google Scholar]
- Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S 2004. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene 23: 3550–3560 [DOI] [PubMed] [Google Scholar]
- Zhou H, Miki R, Eeva M, Fike FM, Seligson D, Yang L, Yoshimura A, Teitell MA, Jamieson CA, Cacalano NA 2007. Reciprocal regulation of SOCS 1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res 13: 2344–2353 [DOI] [PubMed] [Google Scholar]