Abstract
RNA polymerase II arrested at specific template locations can be rescued by elongation factor SII via RNA cleavage. The size of the products removed from the 3′-end of the RNA varies. The release of single nucleotides, dinucleotides, and larger oligonucleotides has been detected by different workers. Dinucleotides tend to originate from SII-independent complexes and 7–14 base products from SII-dependent complexes (Izban, M. G., and Luse, D. S. (1993) J. Biol. Chem. 268, 12874–12885). Different modes of cleavage have also been recognized for bacterial transcription complexes and are thought to represent important structural differences between functionally distinct transcription intermediates.
Using an elongation complex “walking” technique, we have observed factor-independent complexes as they approach and become arrested at an arrest site. Dinucleotides or 7–9-base (large) oligonucleotides were released from SII-independent or dependent complexes, respectively. The abrupt shift between the release of dinucleotide versus large products accompanied the change from factor-dependent to factor-independent elongation, as described by others. However, not all factor-independent complexes showed cleavage in dinucleotide intervals since oligonucleotides 2–6 bases long were also liberated from elongation-competent complexes. These were all 5′-coterminal oligonucleotides indicating that a preferred phosphodiester bond is targeted for cleavage in a series of related complexes. This is consistent with recent models postulating a large product binding site that can hold RNA chains whose size increases as a function of chain polymerization. A specific transitional complex was identified that acquired the ability to cleave in a large increment one base insertion event prior to attaining the arrested configuration.
Elongation by RNA polymerase II (pol II)1 is a complex process that is poorly understood. Accessory elongation factors have been identified that influence the elongation reaction in vitro (Kane, 1994). Elongation factor SII allows transcription to proceed in vitro from template locations that are intrinsic arrest sites. At such sites RNA polymerase II remains potentially active but unable to elongate RNA chains even though a full complement of nucleotide substrates is present. SII interacts with arrested complexes and activates a nascent RNA cleavage activity that is thought to reside in RNA polymerase II itself (reviewed in Reines (1994)). RNAs are shortened from their 3′-end; the 5′-fragment remains in an elongation complex and can be re-extended. RNA cleavage is required for the resumption of chain elongation from specific template sites (Reines et al., 1992; Izban and Luse, 1993b). The requirement for SII can apparently be circumvented in vitro by pyrophosphate-mediated RNA cleavage (Rudd et al., 1994). The probability of arrest upon chain re-extension is not altered by chain cleavage. Instead, repeated rounds of RNA shortening and re-extension provide additional opportunities for RNA polymerase II to bypass an arrest site, most or all of which are only partial blockages to elongation (Gu et al., 1993; Izban and Luse, 1993b; Guo and Price, 1993).
RNA polymerase from Escherichia coli also has an elongation factor-activated nuclease involved in readthrough of transcriptional blockages (Surratt et al., 1991; Borukhov et al., 1992, 1993; Nudler et al., 1994; Lee et al., 1994; Feng et al., 1994). Two elongation factors that associate with E. coli RNA polymerase, GreA and GreB, activate distinct forms of the cleavage reaction. GreA induces cleavage of 2- and 3-base oligonucleotides while GreB activates releases of large oligonucleotides (Borukhov et al., 1993). This reaction appears to fulfill a similar role in chain elongation to that activated by SII.
A full understanding of the RNA polymerase II-associated ribonuclease requires a characterization of the reaction products. One report indicates that mononucleotides are released from isolated elongation complexes arrested within the adenovirus major late transcription unit (Wang and Hawley, 1993). Other workers report that di- and oligonucleotides are the major products of cleavage (Izban and Luse, 1993a, 1993b; Guo and Price, 1993). It is clear, however, that the cleavage increment is variable. It is a function of both the extent to which elongation is SII-dependent and the sequence of the DNA template (Izban and Luse, 1993a, 1993b). For example, extension of a template strand A tract at an arrest site increases the fraction of elongation complexes that become arrested. RNA cleavage releases proportionally more large oligonucleotide than dinucleotide during transcription on such templates (Izban and Luse, 1993b). The variable size of the cleavage product is probably related to the physicochemical differences between arrested and elongation-competent complexes.
Here we have investigated the characteristics of the cleavage reaction carried out by a nested set of elongation complexes on a portion of a human histone gene. We confirm the preference of SII-independent complexes for removing dinucleotides. However, 5′-coterminal oligonucleotides from 3 to 9 bases long were also released from distinct complexes by scission of a preferred phosphodiester bond. Arrested complexes preferentially removed 7–9-base oligonucleotides by cleavage at this position, proving that cleavage of only a single phosphodiester bond was sufficient to rescue these arrested complexes. A hexanucleotide was released from a transitional factor-independent complex located at a long pause site 1-nucleotide incorporation event before an arrest site. Hence, elongation competence of a pol II complex does not always predict the cleavage increment, and acquisition of the ability to cleave in the large increment can precede the arrested phenotype. This transition may presage the change in elongation potential that the complex experiences upon arrest.
MATERIALS AND METHODS
Proteins and Reagents
RNA polymerase II, SII, and basal transcription factors were isolated from rat liver as described (Conaway et al., 1995). Recombinant α was purified from E. coli as described (Tsuboi et al., 1992). Full-length recombinant human SII was expressed from the plasmid pT7–7/Met (provided by Caroline Kane, University of California, Berkeley) and purified as described for rat liver SII (Conaway et al., 1995).
Fast protein liquid chromatography-purified nucleoside triphosphates (NTPs) were purchased from Pharmacia-LKB Biotechnology (Uppsala, Sweden). Formalin-fixed Staphylococcus aureus (immunoprecipitin) was obtained from Life Technologies Inc. Dinucleotides ApC and CpG were purchased from Sigma. Oligonucleotides CpGpUpUpU-pUpU and CpGpUpUpUpUpUpUpU were synthesized by National Biosciences, Inc. (Plymouth, MN). [α-32P]CTP was obtained from Amersham Corp.
3′-End Labeling of RNA in Elongation Complex
Transcription was reconstituted with partially purified initiation factors and RNA polymerase II as described (Reines et al., 1987). A single reaction contained 100 ng of EcoRI and PstI cut pAdTerm-2 (Reines et al., 1989), rat liver RNA polymerase II (0.5 μg), and fraction D (2 μg) in 20 μl of 20 mM HEPES, pH 7.9, 20 mM Tris-HCl, pH 7.9, 2.2% (w/v) polyvinyl alcohol, 0.5 mg/ml acetylated BSA (New England Biolabs), 150 mM KCl, 2 mM dithiothreitol, and 3% (v/v) glycerol. After 30 min at 28 °C, 30 μl of a solution containing fraction B′ (1 μg) and recombinant α in the same buffer without KCl were added, and incubation continued for another 20 min. Ternary complexes containing a 14-nucleotide transcript were synthesized by adding 7 mM MgCl2 and ATP, UTP, and CTP (20 μM each). Transcription was completed by adding heparin to 10 μg/ml and all four NTPs (800 μM each). After 15 min, elongation complexes were immunoprecipitated by anti-RNA D44 IgG and fixed S. aureus (Eilat et al., 1982; Reines, 1991). Elongation complexes were washed three times with reaction buffer (60 mM KCl, 20 mM Tris-HCl, pH 7.9, 2 mM dithiothreitol, 3 mM HEPES-NaOH, pH 7.9, 0.5 mM EDTA, 2.2% (w/v) polyvinyl alcohol, 0.2 mg/ml acetylated BSA, 3% (v/v) glycerol). SII and MgCl2 were added and the reaction was incubated for 1.5 min at 28 °C. After washing twice with an equal volume of reaction buffer, [α-32P]CTP (20 μCi, 3000 Ci/mmol) was added with 7 mM MgCl2 and 400 μM GTP (unless otherwise indicated) to form G200 complexes labeled at C199. After washing three times, elongation complexes bearing Ia-RNA were obtained by adding 400 μM UTP.
Identification of Cleavage Products
End-labeled elongation complexes were washed twice with, and resuspended in, reaction buffer containing 0.02 mg/ml acetylated BSA, 5% glycerol, and no polyvinyl alcohol. Partially purified rat liver SII or recombinant human SII was added to the reaction with 7 mM MgCl2. After incubation at 28 °C (10 min for rat liver SII and 30 min for recombinant human SII), the supernatants containing the cleavage products were collected by centrifugation for 5 min at 15,000 × g. To concentrate the products and remove salt, the supernatants were twice extracted with 6 volumes of 1-butanol (Cathala and Brunel, 1990; Wang and Hawley, 1993), dried, and dissolved in loading buffer (80% (v/v) formamide, 0.05% (w/v) bromphenol, 0.05% (w/v) xylene cyanol in TBE (89 mM boric acid, 89 mM Tris, pH 8.0, 1 mM EDTA)) and then resolved on a 25% polyacrylamide gel (acrylamide:bisacrylamide, 22:3) in TBE and 8.3 M urea. After electrophoresis the gel was exposed to x-ray film (Kodak XAR) with an enhancing screen (DuPont) or to a PhosphorImager screen (Molecular Dynamics) for quantitation.
Preparation of Dinucleotide and Oligonucleotide Standards
CpG, ApC (0.33 mM dinucleotide), or oligonucleotides (0.033 mM oligonucleotide) were labeled at 37 °C for 30 min with T4 polynucleotide kinase (Promega) and [γ-32P]ATP (10 μCi, 6000 Ci/mmol) in one-phor-all (+) buffer (Pharmacia), phenol-extracted, and used as reference markers.
RESULTS
The template used in these reactions contains the core adenovirus major later promoter and a segment of human histone H3.3 gene containing an intrinsic arrest site called Ia. Transcription in the absence of SII results in approximately half of the RNA polymerase II molecules becoming arrested at positions 205, 206, and 207 (Ia) (+1 is the transcription start site (Gu et al., 1993) (Fig. 1, lane 1)). These arrested elongation complexes require elongation factor SII for further RNA synthesis. Detailed studies have revealed that SII activates a nuclease activity that cleaves the nascent RNA near its 3′-end, which is essential for the resumption of polymerization (Reines et al., 1992; Gu et al., 1993). Preliminary mapping experiments showed that no more than 7–9 nucleotides were removed before the relief of arrest and renewed chain elongation (Gu et al., 1993).
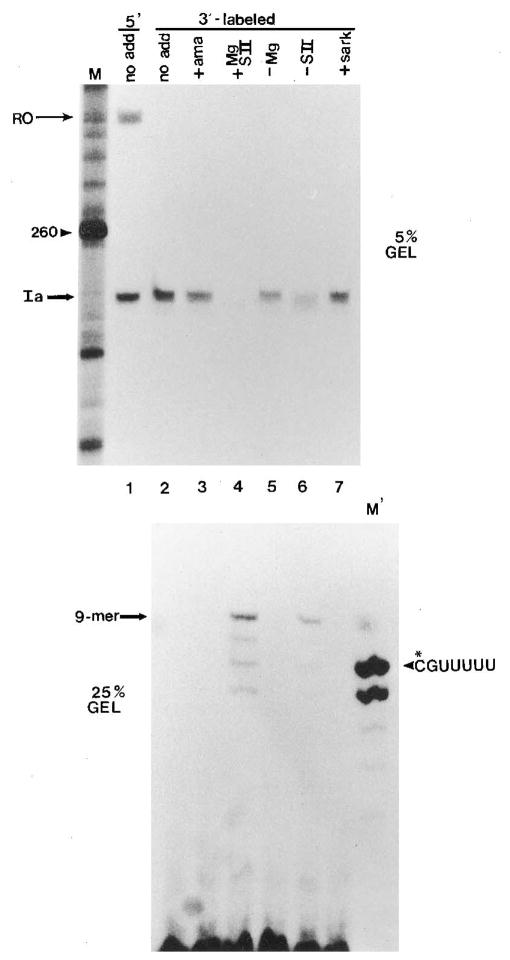
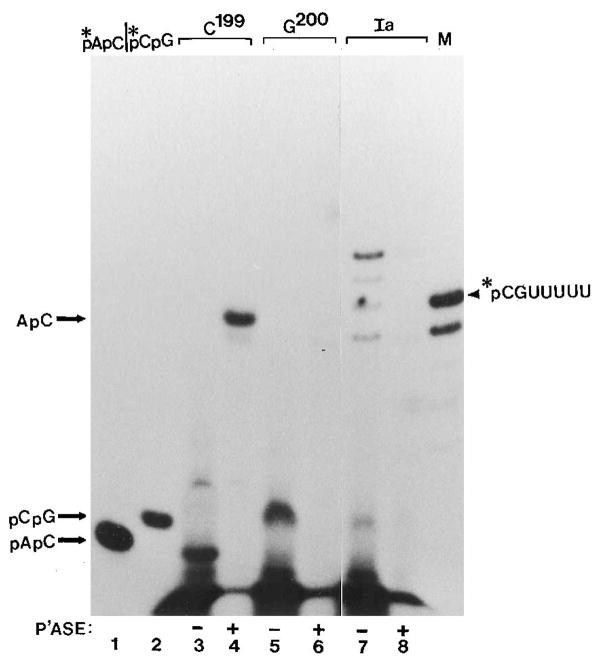
Fig. 1. Identification of cleavage products from arrested (Ia) complexes.
Washed Ia complexes labeled uniquely at position C199 received: no addition (lane 2); SII (0.5 μg), MgCl2 (7 mM) and α-amanitin (1 μ g/ml) (ama, lane 3); SII and MgCl2 (lane 4); SII only (lane 5); MgCl2 only (lane 6); or SII, MgCl2, and 0.25% Sarkosyl (sark, lane 7) for 10 min at 28 °C. Small cleavage products were separated from elongation complexes by centrifugation and resolved on a 25% gel (lower panel). RNA remaining in complexes was isolated and resolved on a 5% gel (upper panel). Lane 1 is 5′-end-labeled RNA showing runoff RNA (RO) and Ia-RNA (Ia). An RNA marker of 260 nucleotides is indicated in lane M. Lane M′ shows 32P-labeled, chemically synthesized pCpGpUpUpUpUpU.
To identify the cleavage products, we have taken advantage of SII-activated nascent RNA cleavage to label elongation complexes (Gu and Reines, 1995). Arrested Ia complexes were assembled on linear templates in the presence of unlabeled NTPs and immunoprecipitated with an anti-RNA antibody (Eilat et al., 1982; Reines, 1991). They were washed free of NTPs, and SII was added to activate nascent RNA cleavage. From previous work, we know that Ia-RNA is shortened to yield a major 5′-product ending in an A residue at position 198 (Gu et al., 1993) (see Fig. 6). In the presence of [α -32P]CTP and GTP, A198 complexes could be extended to G200 with a single 32P atom in the transcript between bases 198 and 199. The G200 complexes could be further extended with UTP to obtain an elongation complex bearing an internally labeled Ia-RNA. Under these conditions, only Ia-RNA was labeled (Gu and Reines, 1995) (Fig. 1, lane 2). These uniquely labeled complexes were the substrates for the analysis of SII-dependent cleavage.
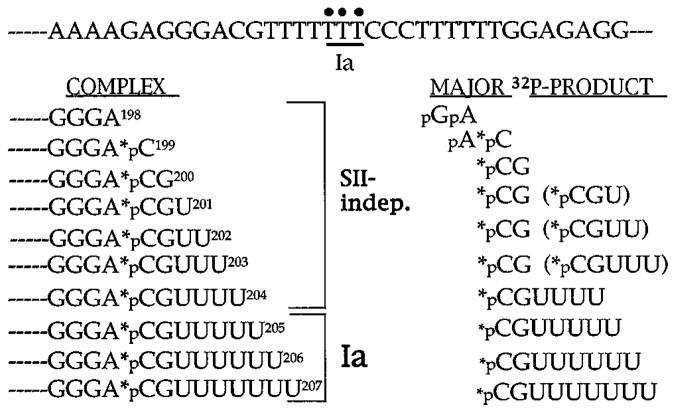
Fig. 6. Summary of the data generated in this study.
The DNA sequence surrounding site Ia is shown at the top. Elongation complexes are shown on the left, and major labeled cleavage products are shown on the right. Minor products are shown in parentheses. The product assignments for complexes U201, U202, and U203 are inferred from the collection of products generated by a mixture of these three complexes. The asterisk indicates the position of the 32P label. Release of pGpA from A198 complexes is indicated from prior mapping experiments (Gu et al., 1993)
After nucleotide depletion and treatment with SII, cleavage products were separated from the elongation complex by precipitation with fixed S. aureus. Precipitated RNA was isolated and resolved on a 5% polyacrylamide gel (Fig. 1, upper panel). Products released from complexes were resolved on 25% polyacrylamide gels (Fig. 1, lower panel). In the presence of both Mg2+ and SII, radioactivity was lost from Ia-RNA and found in 6–9-base oligonucleotides (Fig. 1, lane 4). The formation of these oligonucleotides was strongly dependent upon SII (Fig. 1, lane 6) and inhibited by α -amanitin (Fig. 1, lane 3) and Sarkosyl (Fig. 1, lane 7). The lack of complete dependence upon added SII probably resulted from residual factor used to activate transcript cleavage before labeling. Two of these small products comigrated with authentic, chemically synthesized pCpGpU-pUpUpUpU (Fig. 1, lanes 4 and M′) and pCpGpUpUpUpUpU-pUpU (see Fig. 5A, lanes 7 and M). Therefore, it appeared that 3′-end-labeled Ia complex gave rise to products pCpGpUp-UpUpU, pCpGpUpUpUpUpU, pCpGpUpUpUpUpUpU, and pCpGpUpUpUpUpUpUpU, consistent with the heterogeneous ends of Ia-RNA (Gu et al., 1993). Chemically synthesized oligonucleotide pCpGpUpUpUpUpUpUpU was not degraded when included in a reaction (data not shown) indicating that the 6-, 7-, and 8-base oligonucleotides we observed were not generated by secondary digestion of the nonamer. The same products were liberated by SII-mediated cleavage using rat liver SII (Fig. 2, lanes 1 and 2) or recombinant human SII (Fig. 2, lane 3), both of which were purified to near homogeneity (Fig. 2, right panel and Conaway et al. (1995)). The loss of radioactivity from these 6–9-base oligonucleotides after alkaline phosphatase treatment (Fig. 3, lane 7 versus 8) demonstrated that they were 5′-coterminal resulting from cleavage on the 5′-side of the phosphodiester bond between bases A198 and C199.
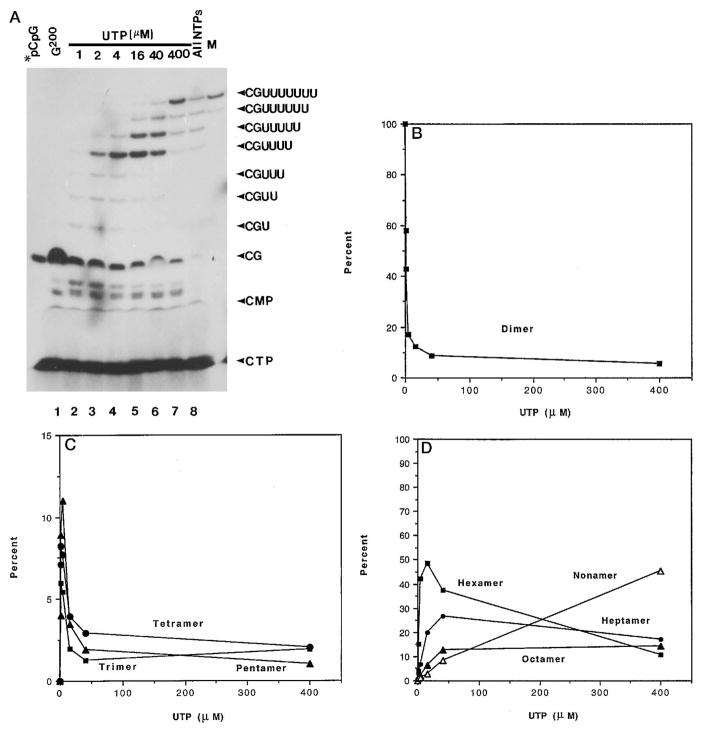
Fig. 5. Identification of cleavage products from a nested set of elongation complexes.
A, labeled G200 complexes were extended with 1 μM (lane 2), 2 μM (lane 3), 4 μM (lane 4), 16 μM (lane 5), 40 μM (lane 6), 400 μM (lane 7) UTP, or 400 μM each of all four nucleotides (lane 8) at 28 °C for 15 min. After washing, these complexes were treated with rat liver SII (phosphocellulose fraction, 0.5 μ g) and 7 mM MgCl2 for 10 min. Cleavage products were isolated and resolved on a 25% polyacrylamide gel. Shown in lane M is [32P]pCpGpUpUpUpUpUpUpU. Label comigrating with CTP and CMP is indicated. The amounts of dinucleotide (B); tri-, tetra-, and pentanucleotide (C); or hexa-, hepta-, octa-, and nonanucleotide (D) were quantitated as a percent of total oligonucleotides released in reactions such as those shown in A and plotted as a function of UTP concentration. Note the expanded y axis of panel C.
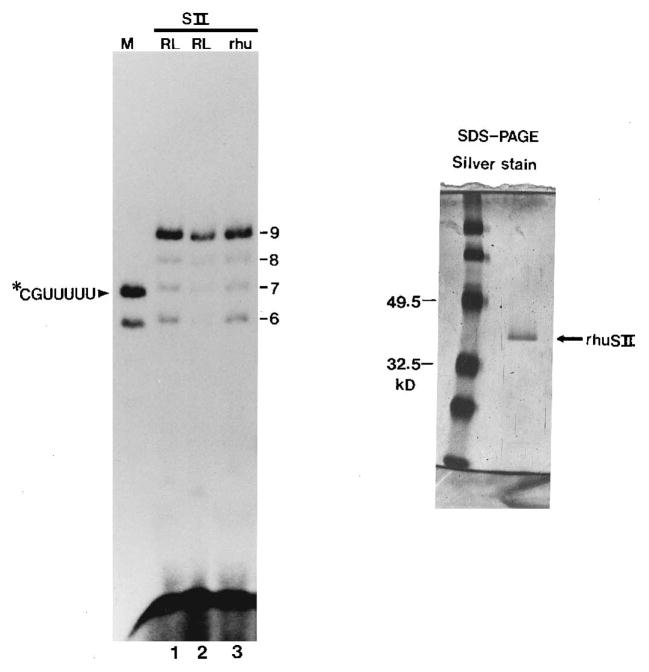
Fig. 2. Cleavage products from Ia complexes treated with native rat and recombinant human SII.
Left, washed Ia complexes labeled at C199 were treated with rat liver (RL) SII (phospho-cellulose fraction, 0.5 μ g, lane 1; Bio-Gel SP-5 PW fraction, approximately 1 ng, lane 2) or recombinant human (rhu) SII (Bio-Gel SP-5 PW fraction, approximately 6 ng, lane 3). Cleavage products were isolated and resolved on a 25% polyacrylamide gel. Lane M shows 32P-labeled, chemically synthesized pCpGpUpUpU-pUpU. Right, human SII was expressed in E. coli, purified to near homogeneity, resolved on a SDS gel, and silver-stained. Two protein standards (Bio-Rad pre-stained SDS-polyacrylamide gel electrophoresis standards, low range) are indicated in kilodaltons.
Fig. 3. Alkaline phosphatase sensitivity of cleavage products.
Complexes C199 (lanes 3 and 4), G200 (lanes 5 and 6), and Ia (lanes 7 and 8) were radiolabeled at position C199 and treated with rat liver SII (phosphocellulose fraction, 0.5 μ g) and 7 mM MgCl2 for 10 min at 28 °C. Cleavage products were separated from the complexes by centrifugation, butanol-extracted, dried, dissolved in 9 μl of 1 mM MgCl2, and treated with (lanes 4, 6, and 8) or without (lanes 3, 5, and 7) 0.05 unit of calf intestinal alkaline phosphatase (Boehringer Mannheim) for 10 min at 37 °C. Samples were dried and resolved on a 25% polyacrylamide gel. Lanes 1, 2, and M are chemically synthesized, 32P-labeled pApC, pCpG, and pCpGpUpUpUpUpU, respectively.
We have previously employed pol II “walking” experiments to assemble a nested set of elongation complexes on this template extending from position 199 to 207 (Gu and Reines, 1995). Labeled C199 and G200 complexes were prepared by incubating unlabeled A198 complexes with [α -32P]CTP (not shown) or [α -32P]CTP and GTP (Fig. 4, lane 3, and Fig. 6), respectively. Elongation by G200 complexes in the presence of increasing amounts of UTP resulted in a series of complexes between U201 and U207 (Fig. 4, lanes 4–7, and Fig. 6). Each set of complexes was washed free of NTPs and incubated with SII and MgCl2. The major product released from G200 complexes comigrated with authentic pCpG (Fig. 5A, lane 1) and was sensitive to alkaline phosphatase (Fig. 3, lane 5 versus 6). Incubation of G200 complexes with 1 μM UTP resulted in a mixture of complexes U201, U202, and U203 (Fig. 4, lane 6). The labeled 3′-products observed from these complexes were predominantly pCpG (72%) with some trimer (pCpGpU) (10%), tetramer (pCpGpUpU) (9%), pentamer (pCpGpUpUpU) (5%), and hexamer (pCpGpUpUpUpUpU) (5%) (Fig. 5A, lane 2). Trimer, tetramer, and pentamer contained 32P at their 5′ terminus since they were sensitive to phosphatase treatment (data not shown). Although these products arose from a mixture of complexes, we infer that the trimer, tetramer, and pentamer came from the U201, U202, and U203 complex, respectively. In any case, they result from the cleavage of the phosphodiester bond between A198 and C199. The size of the released oligonucleotides increased as G200 complexes were extended toward site Ia with increasing amounts of UTP (Fig. 5A, lanes 3–6, 5C, and 5D). In particular, the ratio of hexamer to each of the other oligonucleotides increased commensurate with chain extension (Fig. 5D). When G200 was incubated with 400 μM UTP (and no CTP), 6–9-base oligonucleotides were observed almost exclusively (Fig. 5A, lane 7). Labeled material migrating faster than pCpG was also observed. A portion of this is pApC resulting from unextended C199 complexes (see below and Fig. 3, lanes 1 and 2). Label co-migrating with CTP and CMP was identified as well. Both were present in preparations of complexes prior to SII treatment. CMP in particular could not be removed even after extensive washing and gel filtration (data not shown) as described by others (Rudd et al., 1994; Izban et al., 1995). From this we conclude that CMP can represent at most 10.1 ± 0.6% (mean ± S.D., n = 6) of the total cleavage products identified in the reactions shown in Fig. 5A.
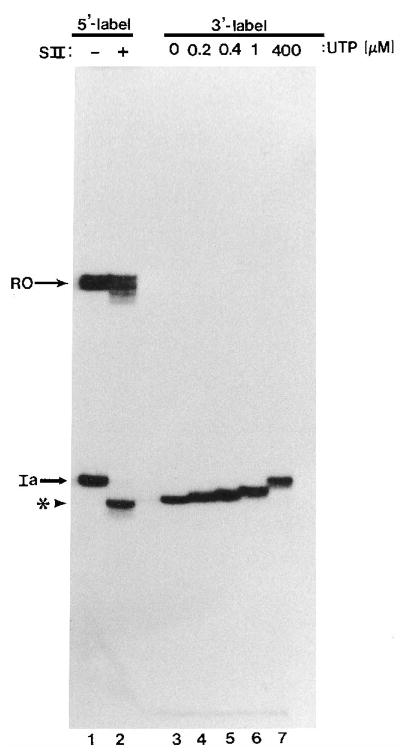
Fig. 4. Assembly and “walking” of elongation complexes.
5′-End-labeled elongation complexes (lane 1) were treated with rat liver SII (phosphocellulose fraction, 0.5 μ g) and 7 mM MgCl2 for 1.5 min at 28 °C to generate A198 complexes (asterisk, lane 2). Radiolabeled G200 complexes (lane 3) were obtained as described under “Materials and Methods” and extended with the indicated amounts of UTP for 15 min at 28 °C (lanes 4–7). RO, runoff RNA.
When this template is transcribed by pol II, about half of the elongation complex becomes arrested nearly equivalently at U205, U206, and U207 while the other half passes through site Ia (Gu et al., 1993). Under conditions where RNA pol II was able to proceed past site Ia, i.e. in the presence of all four NTPs, we observed the expected release of an approximately equivalent amount of 7-, 8-, and 9-base oligonucleotides (Fig. 5A, lane 8). Equivalent amounts of hexanucleotide are also present, probably due to a long pause at site 204 (data not shown). Interestingly, elongation from this complex, although slow, is SII-independent (Gu et al., 1993; Gu and Reines, 1995). Hence, this SII-independent elongation complex released oligonucleotides in a cleavage increment characteristic of arrested elongation complexes 205–207, suggesting that this represents a critical transition between factor-independent and -dependent elongation. In other words, cleavage in the large increment became uncoupled from the acquisition of the arrested state.
The effect of withholding CTP (the nucleotide downstream of site Ia) is to allow the 50% of complexes that would otherwise read through site Ia to become arrested at position 207. Therefore, all the complexes become arrested at positions 205, 206, or 207 when G200 complexes are supplied with only UTP (Gu and Reines, 1995). Indeed, more 9-base oligonucleotide was generated when CTP was absent because those complexes that were SII-independent accumulated at position 207 (Fig. 5A, lane 7 versus 8, and Fig. 6). Hence, there is a good correlation between arrest at position 207 and generation of the 9-base oligonucleotide by SII-dependent cleavage.
These results suggested a preferred cleavage site at the phosphodiester bond between bases 198 and 199 since a nested set of 5′-coterminal oligonucleotides were released as the chain was extended from 200 to 207. We next asked what cleavage products were generated from C199 complexes that contain an RNA chain 1 base longer than this apparent preferred cleavage boundary. Surprisingly, a dinucleotide comigrating with authentic pApC was released after SII treatment (Fig. 3, lane 1 versus 3). Consistent with this assignment, the electrophoretic mobility of the product shifted dramatically after alkaline phosphatase treatment, but its label was not lost since the 32P was located at an internal phosphodiester bond (Fig. 3, lane 3 versus 4, and Fig. 6). This finding emphasizes the extent to which dinucleotide release is favored in SII-independent complexes. The results of this analysis are summarized in Fig. 6.
DISCUSSION
We have identified the products of nascent RNA cleavage from a well characterized arrested elongation complex. The strategy utilized in this study also enabled us to determine how the site of cleavage varies across a set of related elongation complexes. In our experiments, one nucleotide was labeled in RNA chains ranging from 199 to 207 nucleotides long. This approach allowed us to unequivocally identify labeled cleavage products and in particular to define the sequence of the large oligonucleotides.
SII activates RNA cleavage in both SII-independent and SII-dependent complexes although cleavage is more readily accomplished by arrested complexes (Reines et al., 1992). The product of SII-activated RNA cleavage has been identified by others in different transcription systems. Wang and Hawley (1993) reported that exclusively mononucleotides were released by human pol II elongation complexes. They proposed that this activity may serve as a proofreading exonuclease for RNA synthesis in analogy to some DNA polymerases. Luse and co-workers (Izban and Luse, 1993b) identified dinucleotides and oligonucleotides released by this reaction and found that the size of the oligonucleotides varied as a function of the elongation factor dependence of the complexes. A dinucleotide was the predominant product for SII-independent complexes while 7–14-base oligonucleotides were released from SII-dependent complexes (Izban and Luse, 1993a, 1993b). In that work the sequence identity of the large oligonucleotides was not determined. Guo and Price (1993) also showed that SII stimulated the release of dinucleotides from Drosophila elongation complexes.
Our findings confirm the work of Luse and co-workers (Izban and Luse, 1993a, 1993b) in showing that elongation complexes arrested at site Ia released 7–9-base oligonucleotides and elongation-competent complexes such as C199 and G200 cleaved predominantly in a dinucleotide increment. The 7–9-base oligonucleotides were generated from U205, U206, and U207 complexes by cutting the same phosphodiester bond between bases A198 and C199, suggesting that this is a favored cleavage site for arrested complexes. In addition, complexes extending from position 199 to 204 yielded 2-, 3-, 4-, 5-, and 6-base oligonucleotides, all of which have the same 5′-end. Hence, oligonucleotides of many lengths can be released, although dinucleotides are the preferred cleavage product by factor-independent complexes.
Complex U204 is unusual in that it is largely SII-independent, yet cleavage removes a 6-base oligonucleotide almost exclusively. This may represent a transitional complex in that it carries out cleavage as arrested complexes bearing RNAs 1, 2, or 3 bases longer (205, 206, 207) but is not itself an arrested complex. Hence, the increment of cleavage cannot necessarily be predicted for a given elongation complex.
Based on a report that mononucleotides are removed from some pol II complexes (Wang and Hawley, 1993) and that the phosphodiester bond between 198 and 199 was a preferred cleavage position (Fig. 6), we expected that CMP might be released from C199 complexes. Surprisingly, a significant amount of the dinucleotide pApC was observed. Our effort to identify CMP as a product was hampered by a background of [32P]CMP tightly associated with labeled complexes, similar to the finding of others (Izban et al., 1995). Hence, we cannot rule out the possibility that CMP is a cleavage product, albeit a minor one.
Our data are consistent with current models developed for the discontinuous (inchworming) movement of the E. coli elongation complex (Wang et al., 1995; Nudler et al., 1995) and may be related to why RNA pol II becomes arrested at site Ia. At some DNA sequences enzyme translocation becomes uncoupled from chain elongation leading to the development of internal strain in the complex. For a strained E. coli RNA polymerase, transcription terminates when the RNA structure permits (Wang et al., 1995; Nudler et al., 1995). T tracts on the non-template strand appear to be a common signal for the inch-worming cycle (Nudler et al., 1995) and are found at many pol II arrest sites (Hawley and Roeder, 1985; Sato et al., 1986; Kerppola and Kane, 1987; Reines et al., 1987). In our template, the sequence around arrest site Ia is TTTTTTTCCCTTTTTT on the nontemplate strand, which is associated with a bend in this region of the template (Kerppola and Kane, 1990). Previous exonuclease III footprint analysis showed little or no translocation of pol II between the A198 and U207 complexes (Gu et al., 1993). The T tracts could be involved in uncoupling enzyme translocation and chain extension (an inchworming signal). The unusual template structure could contribute to the accumulation of strain until a threshold is encountered whereupon pol II becomes arrested. Strain accumulation may be gradual but appears to rise most dramatically at the U204 to U205 step. As for bacterial RNA polymerase in the strained configuration, the RNA 3′-end is close to the downstream edge of the complex (Gu et al., 1993). Similar uncoupling has also been seen for protein-blocked elongation complexes (Pavco and Steege, 1990; Kuhn et al., 1990; Reines and Mote, 1993; Nudler et al., 1995). The distance between the 3′-end of Ia-RNA and the downstream boundary of pol II is unusually short (10–12 base pairs) compared to that found in elongation-competent complexes (18–20 base pairs) (Gu et al., 1993). The inchworming model describes a product groove on the enzyme in which RNA is polymerized and accessible to elongation factor-activated cleavage (loose binding site (Chamberlin, 1994; Nudler et al., 1995)). Elongation of RNA by our complexes from position 198 to 207 appears to be filling such a groove. We have not detected the upper limit of RNA that can be accommodated in the groove but it is at least 9 bases. The capacity of such a product groove could vary between arrest sites. More experiments will be required to demonstrate if this additional plasticity exists in elongation complexes.
Acknowledgments
We thank J. Mote, Jr. for expert technical assistance and W. Powell for providing Bio-Gel SP-5 PW fraction of rat liver SII. We also thank Dr. C. Kane for the generous gift of human SII expression plasmid pT7–7/Met.
Footnotes
This work was supported by National Institutes of Health Grant GM-46331 and American Cancer Society Grant JFRA-394.
The abbreviations used are: pol II, RNA polymerase II; BSA, bovine serum albumin; Ia, intrinsic arrest site.
References
- Borukhov S, Polyakov A, Nikiforov V, Goldfarb A. Proc Natl Acad Sci U S A. 1992;89:8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Cathala G, Brunel C. Nucleic Acids Res. 1990;18:201. doi: 10.1093/nar/18.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin MJ. Harvey Lect. 1994;88:1–21. [PubMed] [Google Scholar]
- Conaway RC, Reines D, Garrett KP, Powell W, Conaway JW. Methods Enzymol. 1996 doi: 10.1016/s0076-6879(96)73020-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilat D, Hochberg M, Fishel R, Laskov R. Proc Natl Acad Sci U S A. 1982;79:3818–3822. doi: 10.1073/pnas.79.12.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Lee DN, Wang D, Chan CL, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- Gu W, Reines D. J Biol Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Powell W, Mote J, Jr, Reines D. J Biol Chem. 1993;268:25604–25616. [PMC free article] [PubMed] [Google Scholar]
- Guo H, Price DH. J Biol Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- Hawley DK, Roeder RG. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- Kane CM. In: Transcription. Conaway R, Conaway JW, editors. Raven Press; New York: 1994. pp. 279–296. [Google Scholar]
- Kerppola TK, Kane CM. Mol Cell Biol. 1987;8:4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK, Kane CM. Biochemistry. 1990;29:269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Bartsch I, Grummt I. Nature. 1990;344:559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Izban MG, Luse DS. J Biol Chem. 1993a;268:12684–12873. [PubMed] [Google Scholar]
- Izban MG, Luse DS. J Biol Chem. 1993b;268:12874–12885. [PubMed] [Google Scholar]
- Izban MG, Samkurashvili I, Luse DS. J Biol Chem. 1995;270:2290–2297. doi: 10.1074/jbc.270.5.2290. [DOI] [PubMed] [Google Scholar]
- Lee D, Feng G, Landick R. J Biol Chem. 1994;269:22295–22303. [PubMed] [Google Scholar]
- Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Nudler E, Kashlev M, Nikiforov V, Goldfarb A. Cell. 1995;81:351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Pavco PA, Steege DA. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- Reines D. J Biol Chem. 1991;266:10510–10517. [PMC free article] [PubMed] [Google Scholar]
- Reines D. In: Transcription. Conaway R, Conaway JW, editors. Raven Press; New York: 1994. pp. 263–278. [Google Scholar]
- Reines D, Mote J., Jr Proc Natl Acad Sci U S A. 1993;90:1917–1921. doi: 10.1073/pnas.90.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Wells D, Chamberlin MJ, Kane CM. J Mol Biol. 1987;196:299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Reines D, Chamberlin MJ, Kane CM. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- Reines D, Ghanouni P, Li Q-q, Mote J. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- Rudd MD, Izban MG, Luse DS. Proc Natl Acad Sci U S A. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Ito R, Baek KH, Agarwal K. Mol Cell Biol. 1986;47:259–266. doi: 10.1128/mcb.6.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt CK, Milan SC, Chamberlin MJ. Proc Natl Acad Sci U S A. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Conger K, Garrett KP, Conaway RC, Conaway JW, Arai N. Nucleic Acids Res. 1992;20:3250. doi: 10.1093/nar/20.12.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hawley DK. Proc Natl Acad Sci U S A. 1993;90:843–847. doi: 10.1073/pnas.90.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Meier TI, Chan CL, Feng G, Lee DN, Landick R. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]