Abstract
Symbiosis involves responses that maintain the plant host and symbiotic partner’s genetic program; yet these cues are far from elucidated. Here we describe the effects of lumichrome, a flavin identified from Rhizobium spp., applied to lotus (Lotus japonicus) and tomato (Solanum lycopersicum). Combined transcriptional and metabolite analyses suggest that both species shared common pathways that were altered in response to this application under replete, sterile conditions. These included genes involved in symbiosis, as well as transcriptional and metabolic responses related to enhanced starch accumulation and altered ethylene metabolism. Lumichrome priming also resulted in altered colonization with either Mesorhizobium loti (for lotus) or Glomus intraradices/G. mossea (for tomato). It enhanced nodule number but not nodule formation in lotus; while leading to enhanced hyphae initiation and delayed arbuscule maturation in tomato.
Keywords: ethylene, flavin, lumichrome, mycorrhiza, nodulation, starch, symbiosis
Introduction
As photosynthetic organisms, plants are a major conduit of reduced or fixed carbon to the soil. Soil microbiota are recipients of the majority of this carbon that enters the roots and is responsible for releasing factors that liberate the fixed carbon from the roots. One of the ways to achieve this may be by manipulating the expression of sugar efflux transporters (Chen et al., 2010); while other mechanisms may also involve interference in endogenous hormone metabolism leading to enhanced root proliferation (De Salamone et al., 2001; Bottini et al., 2004; Uppalapati et al., 2005; Remans et al., 2008) or microbial mineral solubilization (reviewed in Bloemberg and Lugtenberg, 2001). These result in better plant growth, increased photosynthesis, and more fixed carbon being translocated to the roots and the rhizosphere. Numerous plant growth promoting rhizobacteria (PGPR) and other microbes have been isolated from the rhizosphere and some of the mechanisms by which they manipulate plants are known (reviewed in Lugtenberg and Kamilova, 2009). However, there is much to learn in this field of research not only about the range of microorganisms that manipulate plants, but also about the precise mechanisms they employ.
Rhizobia are soil bacteria that are best known for their ability to form a nitrogen-fixing endosymbiosis with legumes. However, they can be found in the rhizosphere of many species, including non-legumes. The rhizosphere is also laden with fungal partners that form symbiotic relationships with plant roots and aid in phosphate uptake through highly effective mechanisms (Bucher, 2007). In fungal-plant interactions, plant roots constitutively exude strigolactones leading to enhanced hyphal branching and stimulation of fungal mitochondrial metabolism (Besserer et al., 2006, 2008). Strigolactone production is further stimulated upon phosphate deficiency leading to events facilitating arbuscular mycorrhiza (AM) colonization (Akiyama et al., 2005; López-Ráez et al., 2008; Balzergue et al., 2011). In an analogous manner, flavonoids exuded by the host plants are perceived by rhizobacteria that produce lipochitin-oligosaccharide (Nod) factors as recognition signals during symbiotic nitrogen fixation. These Nod signals are perceived via the Nod-factor receptors (the LysM-type serine/threonine receptor kinases, NRF1 and NRF5) in the root hair epidermis and trigger downstream responses including Ca2+ fluxes and spiking, leading to the initiation of nodulation (Madsen et al., 2010, and references therein). While the molecular mechanism(s) leading to colonization in both symbiotic systems is being unraveled, the complete picture remains largely fragmentary. It is, however, clear that after initial rhizobacterial (by Nod factors) or mycorrhizal (by Myc factors; Maillet et al., 2011) perception, both nodulation and mycorrhization share a common symbiotic pathway (CSP; Parniske, 2008) which includes genetic components such as leucine-rich repeat receptor kinases (e.g., SYMRK that perceives both mycorrhizal and rhizobacterial signals; Endre et al., 2002; Stracke et al., 2002), plastidial cation channels (Castor and Pollux; Ané et al., 2004; Imaizumi-Anraku et al., 2005; Charpentier et al., 2008), nucleoporins (Nup85 and Nup133; Kanamori et al., 2006; Saito et al., 2007), calcium calmodulin-dependent protein kinases (CCaMK; Levy et al., 2004; Tirichine et al., 2006) and the nuclear localized coiled-coil domain protein product (CYCLOPS; Yano et al., 2008) that facilitate a transcriptional response in the host nucleus (Kosuta et al., 2008; Parniske, 2008). While genetic interference in the CSP components are indispensable for infected nodulation (Madsen et al., 2010), mutants deficient in symbiont perception (nrf1 and nrf5) are still able to form nodules (although with a lower frequency) due to an epidermal mode of transfection (Madsen et al., 2010), suggesting that the CSP operates autonomously or through multiple independent pathways. In addition, the extent and interaction between the components of the CSP remains unknown.
While several transcriptional regulators, including transcription factors (Smit et al., 2005; Heckmann et al., 2006) and miRNA molecules (Devers et al., 2011, and references therein) have been identified to play a role during the post-perception phase, post-transcriptional regulation (apart from Nod and Myc factor signal perception) has not been addressed. Several chemical signals have been suggested to play a role during symbiosis; yet their identity and specific roles have not been identified to date. Furthermore, plant metabolic perturbations that are evident during this phase of symbiosis include altered primary carbohydrate metabolism (including enhanced photosynthesis, respiration, and starch accumulation) and structural re-organization (of especially cell wall components; reviewed in Reinhardt, 2007), yet the identity of regulators of these processes during symbiosis are lacking. The Nod factor [Nod Bj V (C(18:1)), MeFuc] (LCO) has been previously shown to interact with a chemical called lumichrome (LC; Khan et al., 2008). Lumichrome (7,8-dimethylalloxazine) is presumed to be a light-sensitive breakdown product of riboflavin; however, enzymatic conversion has been measured in Pseudomonas (Yanagita and Foster, 1956), M. loti and Sinorhizobium meliloti (van der Merwe and Lloyd, unpublished data). While the corresponding genes and proteins responsible for LC biosynthesis still remain to be identified, the loss of symbiotic effectiveness in auxotrophic riboflavin mutants in Rhizobium trifolii has been observed (Pankhurst et al., 1974). Furthermore, while both riboflavin and lumichrome exudation are significantly influenced by salinity, pH, and temperature, concentrations measured from bacterial isolate typically range between 0.1 and 15 nM (Kanu and Dakora, 2009). Interestingly, the simultaneous application of LCO and LC lead to enhanced photosynthetic rates in corn and soybean plants (Khan et al., 2008). LC application also stimulates root respiration in alfalfa plants and it has been suggested that LC enhances PEP carboxylase activity and CO2 evolution from roots to increase plant growth (Phillips et al., 1999). The lack of a clear physiological response in the range of crop species investigated thus far (Phillips et al., 1999); however, implies either that LC function is not conserved across all genera or that these effects might rather be related to secondary effects associated with LC. Assessing the mobilization and accumulation properties of LC in the xylem sap of cowpea plants further revealed that the translocation of LC only occurs when inoculated in combination with Bradyrhizobium (Matiru and Dakora, 2005), suggesting that LC could participate as a ligand during symbiosis.
In order to examine this further, we investigated the physiological, molecular, and biochemical responses of the model legume Lotus japonicus and the non-legume model Solanum lycopersicum upon LC treatment. We show that LC treatment alone was able to elicit shared overlapping transcriptional and post-transcriptional responses related to enhanced starch and ethylene metabolism in both species. Furthermore, genetic factors involved in symbiosis were also altered; pertaining not only to elements belonging to the CSP but also individual components related to nodulation or AM colonization, although the experiments were conducted upon nutrient sufficient and sterile (aseptic) conditions. Lastly, in order to more directly assess the role of LC in symbiosis, lotus and tomato plants were subjected to nitrogen and phosphate starvation conditions, respectively, and the degree and dynamics of colonization by rhizobacteria and fungi assessed. LC pre-treatment could increase nodule number, as well as accelerate the rate of hyphal infection thread formation. Taken together, the data suggests that LC plays a more direct role in symbiotic relationships than previously thought, presumably acting downstream of Nod or Myc factor signal perception and leading to altered symbiotic organogenesis.
Materials and Methods
Plant material and growth conditions
Lotus japonicus B-129 (cv. Gifu) and Solanum lycopersicum (cv. Moneymaker) seeds were surface-sterilized and grown in sterilized vermiculite in a 16/8 h day/night regime at 25°C and fertilized every second day with half-strength Hoagland solution (Sigma Aldrich Co., St Louis, MO, USA; supplemented with 1 mM NH4NO3 for replete conditions) with or without 5 nM LC (CAS 1086-80-2)1; applied either through direct root drenching (Phillips et al., 1999) and/or mist foliar (Khan et al., 2008) applications (final amount of 250 pmol per application). For the ethylene inhibitor studies, 10 μM AgNO3 was applied weekly in parallel to the LC treatments described above. Plant material was harvested after 5 weeks and either flash frozen, homogenized in liquid nitrogen and stored at −80°C for biochemical analyses or dried at 65°C for 2 days for dry matter determination. For nodulation and mycorrhization experiments, 5-week-old lotus and tomato plants were subjected to nitrogen- or phosphate-limited conditions, respectively. For this purpose, lotus plants were inoculated with M. loti (strain MAFF 303099) and watered occasionally with B&D medium (Broughton and Dilworth, 1971). For fungal colonization, phosphate-starved plants were supplemented with Biocult G granules (consisting of a clay pellet containing G. intraradices and G. mossea spores)2, and watered with modified Hoagland solution containing 1 μM KH2PO4. Roots were assessed weekly for nodule organogenesis and mycorrhiza colonization, respectively.
Photosynthetic parameters
Gas exchange parameters were measured on 5-week-old plants using a LI-6400XT Portable Photosynthesis System (LiCor Inc, Lincoln, NE, USA). The whole shoot (in a specialized LiCor 6400-05 Conifer Chamber) and third mature leaf (average of individual leaflets) were measured for lotus and tomato, respectively. Data were recorded after the leaf acclimated to the desired conditions of light intensity (1400 μmol m–2 s–1), temperature (25°C) and CO2 concentration (850 μmol mol−1), and measurements made midday under ambient humidity.
Radiolabel activity measurements
Fresh root pieces (500 mg) from 5-week-old plants were washed in 10 mM MES-KOH buffer (pH 6.5) and incubated for 3 h in 10 mM MES-KOH buffer (pH 6.5) containing 1 μCi of unilabeled [14C] glucose (specific activity of 8.11 MBq mmol–1). The 14CO2 liberated and label fractionations were quantified as previously described (van der Merwe et al., 2010).
Bacterial colony forming units and fungal colonization monitoring
Bacterial cultures were monitored as described in Weisskopf et al. (2005). For the fungal growth measurements, rhizospheric and mulched roots were grown for 14 days in minimal M media (Bécard and Fortin, 1988), and hyphae growth monitored.
For microscopic analysis of mycorrhization in tomato roots, samples were cleared in 10% (w/v) KOH, acidified in 2% (v/v) HCl and stained with 0.05% (w/v) acid fuchsin in a 1:1:1 (v/v/v) glycerine:water:lactic acid solution, and assessed via Trouvelot et al. (1986). Root tips from control and lumichrome-treated roots were also stained with Lugol solution for visualization of starch granules. All images were observed with a motorized light microscope (Leica DM 1000) and images captured by LAS EZ (v1.5.0) software.
RNA isolation and transcript analysis
Total RNA was isolated according to the CTAB method of White et al. (2008). RNA cleanup and cDNA synthesis was conducted using SuperScript III Reverse Transcriptase (Invitrogen Co., Carlsbad, CA, USA) according to the manufacturer.
Probe labeling, chip hybridization and scanning for lotus roots were performed using the Affymetrix GeneChip® Lotus Genome Array (Affymetrix, Santa Clara, CA, USA) as previously described (Benedito et al., 2008). Microarray analysis for tomato roots was conducted as described by van der Merwe et al. (2009) on the TOM1 array (Alba et al., 2004). Three biological replicates were hybridized independently in each treatment. All array data has been deposited in ArrayExpress.3
Semi-quantitative RT PCR on EIN3 expression was performed on AgNO3 treated root cDNA using standard protocols with primer combinations of 5′-CCCAAATTGGTTTGCCTAAA-3′ (Lj_AK339680.1_FW)and 5′-CTGGGCTGAGAAAGGTTTTG-3′ (Lj_AK339680.1_REV), and 5′-CACCTTTGTCCTCAGGTGGT-3′ (Sl_BT013783.1_FW) and 5′-TCTTGCTTCGTCACGTCATC-3′ (Sl_BT013783.1_REV) for EIN3 targets in lotus and tomato, respectively, and 5′-GTGTTTTGCTTCCGCCGTT-3′ (PP2A_FW) and 5′-CCAAATCTTGCTCCCTCATCTG-3′ (PP2A_REV), 5′-CGGAGAGGGAGCCTGAGAA-3′ (Sl18S rRNA_FW) and 5′-CCCGTGTTAGGATTGGGTAATTT-3′ (Sl18S rRNA_REV) encoding for serine-threonine protein phosphatase 2A and 18S ribosomal RNA subunit for house-keeping genes of lotus and tomato, respectively.
Statistical methods and array processing
For comparative analyses microarray data were normalized via robust multichip averaging (RMA; Irizarry et al., 2003) and significant differences were corrected for false discovery rate (FDR) according to Benjamini and Hochberg (1995) in R (Gentleman et al., 2004). In order to test the validity of comparative analyses, the EST sequences spotted on the TOM1 microarray of the significantly expressed tomato transcripts were evaluated via the blast algorithm (Altschul et al., 1990) to those of the lotus genome. For all the significantly expressed transcripts from tomato at least one of the probe sequences corresponded to a significant hit to the lotus genome. However, due to fragmentary gene ontology information, both lotus and tomato arrays were re-assigned to match Arabidopsis putative annotations (Kanz et al., 2005; Quevillon et al., 2005), based on sequence similarity to the Arabidopsis genome. For MAPMAN analysis, mapping files adapted for lotus (Sanchez et al., 2008) and tomato (Urbanczyk-Wochniak et al., 2006) were visualized with a P filter <0.05 after duplicate array IDs were manually removed from the lotus results file obtained from R. Lastly, lotus and tomato mapping were utilized for PAGEMAN visualization of over-representation of functional gene categories using the Fisher’s exact test (P < 0.05; Usadel et al., 2006).
Ethylene biosynthetic enzyme activities
Proteins for ethylene biosynthetic enzymes were extracted according to Nakatsuka et al. (1997). ACC synthase and ACC oxidase activity was measured as described by Nakatsuka et al. (1997), malonyl-ACC transferase as outlined by Su et al. (1985) with modifications described by Guo et al. (1993), with the addition of 10 μg protein for all reactions (see ACC and ethylene determinations for further details on ethylene measurements).
Crude and plastidial protein preparations for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity measurements
Non-phosphorylating NAD-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein extraction and activity determination was done according to Kelly and Gibbs (1973).
Crude and plastidial-enriched protein extractions for NAD(P)-dependent GAPDH enzyme activity measurements were conducted exactly as described by Muñoz-Bertomeu et al. (2009), with the exception that the assay reaction was scaled down to a volume of 200 μl and the reaction initiated with 10 μg total protein for each enzyme activity determination. All reactions were determined with and without 20 mM sodium arsenate.
All protein concentrations were measured according to Bradford (1976), using bovine serum albumin (BSA) as a standard.
Phytohormone profiling
ACC was extracted, chemically converted to ethylene and measured as described by Lizada and Yang (1979), with modifications specified in Tucker et al. (2010). Ethylene levels were measured exactly as described in Tucker et al. (2010).
Auxin, cytokinin, jasmonic acid, salicylic acid, and abscisic acid levels were extracted and profiled according to Björklund et al. (2007) and Schmelz et al. (2003), respectively. Quantification was done by means of linear calibration curves of authentic standards.
Soluble sugar and starch measurements
Starch and soluble sugars were extracted according to Müller-Röber et al. (1992), soluble sugars quantified as specified by Müller-Röber et al. (1992), and starch as described by Smith and Zeeman (2006).
Primary metabolite extraction and profiling
Primary metabolites were extracted and analyzed as described by Roessner et al. (2000) optimized for lotus and tomato according to Desbrosses et al. (2005) and Roessner-Tunali et al. (2003), respectively. Gas chromatography coupled to electron impact ionization/time-of-flight mass spectrometry (GC EI TOF-MS) analysis was performed using an Agilent 6890N24 gas chromatograph splitless injection connected to a Pegasus III time-of-flight mass spectrometer (LECO), ran, and evaluated according to Erban et al. (2007) using TagFinder 4.0 software (Luedemann et al., 2008) and the Golm metabolome database (Kopka et al., 2005; Schauer et al., 2005).
Statistical analysis
Unless otherwise specified, statistical analyses were performed using Student’s t-test embedded in the Microsoft Excel software (Microsoft, Seattle). Only the return of a P value <0.05 was designated significant. Analysis of variance (ANOVA) followed by Fisher’s least significant difference test was conducted in R 2.10.0 software (R Development Core Team, 2009), and the return of a P value <0.05 was designated significant.
Results
Biomass accumulation and physiological assessment in lumichrome-treated lotus and tomato plants
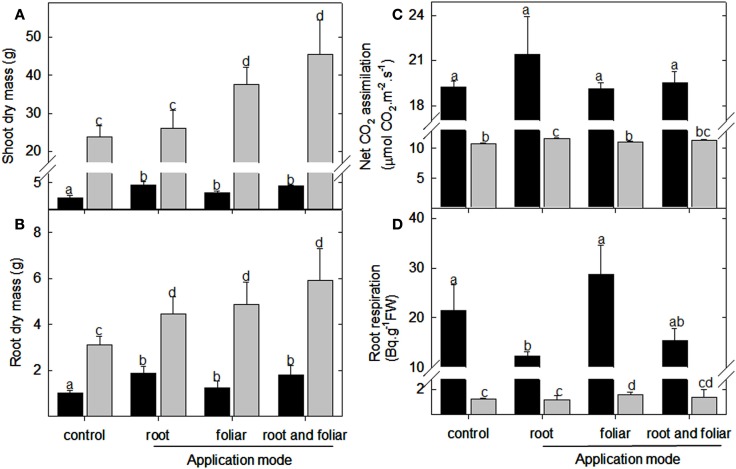
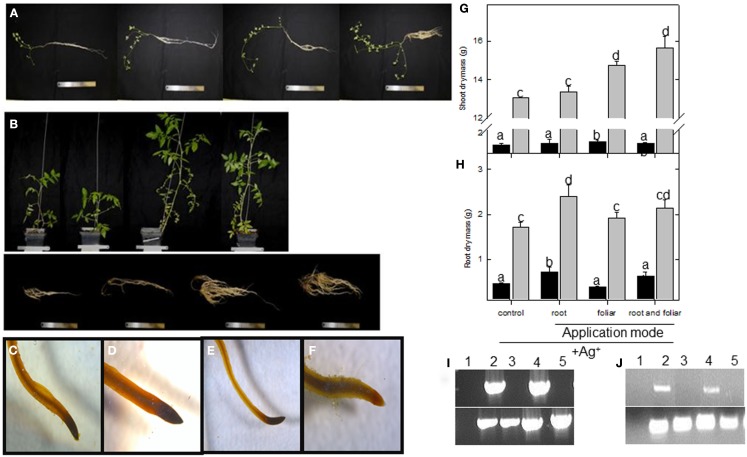
In order to investigate the role of LC on plant performance, the compound was applied every second day to lotus and tomato plants (final concentration 5 nM) grown in vermiculite under full nutrient supplementation. Analyses of 5-week-old plant material (for representative photographs see Figures A1A,B in Appendix) showed that the shoot dry mass of lotus plants increased upon root drenching, foliar, and a combination of foliar and root drenching application (Figure 1A). In contrast, in tomato, only the shoot biomass was increased upon the two types of foliar application (Figure 1A). Assessing the root biomass, it was evident that upon all three application modes (root drenching, foliar misting, and the combination of these treatments) a significant increase was observed in both lotus and tomato (Figure 1B). Concerning root architectural differences, it was further observed that the foliar (and the combination) applications led to enhanced lateral root proliferation which was not observed in the root drenching application (Figures A1A,B in Appendix).
Figure 1.
Physiological assessment lumichrome treatment on lotus and tomato plants. Biomass accumulation of (A) shoot and (B) root organs of 5-week-old lotus (black bar) and tomato (gray bar), as well as (C) net CO2 assimilation rate and (D) root respiration as radiolabeled carbon dioxide evolution were also measured in plants grown in a 16-h photoperiod. The values are the mean ± SE (n = 5). The data was analyzed using one way ANOVA followed by Fisher’s least significant difference, bars that do not share letters are significantly different (P < 0.05) from each other. The results were confirmed in at least four independent experimental trials.
Next, we evaluated photosynthetic performance and respiratory efficiency. This indicated that root drenching with LC resulted in no significant difference in the photosynthetic performance of lotus but led to enhanced CO2 assimilation in the tomato leaves (Figure 1C). Furthermore, the foliar applications of this compound did not alter the photosynthetic rates of the respective plant species (Figure 1C). A significant decrease in respiration rate was observed for the lotus roots upon root drenching (but not with the foliar applications) whilst this was not observed in the tomato roots (Figure 1D). In contrast, upon foliar application of LC to the tomato foliage an increase in root respiration was observed (Figure 1D).
Evaluation of gene expression profiles of lotus and tomato roots upon lumichrome treatments
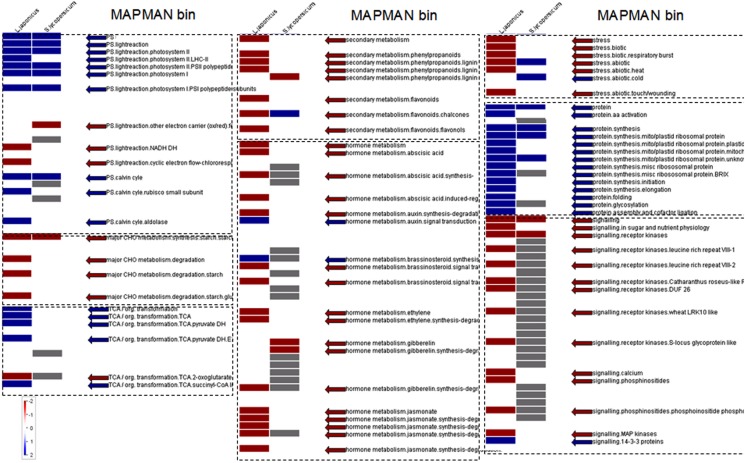
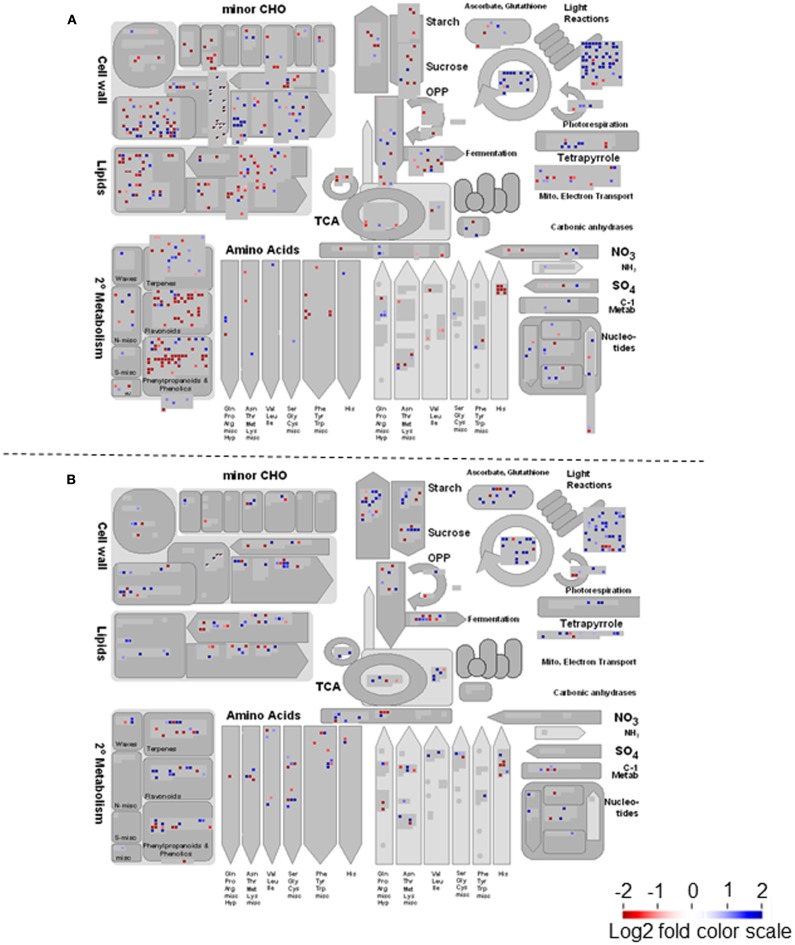
To further investigate these responses, microarray analyses were conducted on lumichrome-treated and control roots. Statistical analyses showed that 656 and 1643 genes were significantly up- and down-regulated, respectively, between control and treated lotus roots. In contrast, 229 and 136 genes were up- and down-regulated respectively, in tomato. In a next step Fisher’s exact test for gene functional over-representation (Usadel et al., 2006) was performed on the data sets, and indicated significant shifts in gene classes associated with primary carbon metabolism including starch and organic acids (only in lotus roots; Figure 2). Furthermore, flavonoid metabolism also seems to be significantly affected in the lotus roots while numerous genes involved in the biosynthesis of various phytohormones were also functionally overrepresented (Figure 2). This was not observed in the tomato metabolism. Lastly, genes involved in mitochondrial or plastidial protein translation were also positively affected in both lotus and tomato, while receptor kinase signaling categories were generally down-regulated within both species (Figure 2).
Figure 2.
Comparison of significant overrepresented functional gene categories (and sub-categories) of lumichrome-treated lotus and tomato roots determined by PAGEMAN. The values are representative of the log2 transformed values of three individual hybridizations, statistically analyzed via robust multichip averaging (RMA) in R, and classed according to the MAPMAN classifications (Thimm et al., 2004; Usadel et al., 2005) and over-representation classification assessed via Wilcoxon rank sum test (P < 0.05; Usadel et al., 2006). Red indicates a decrease whereas blue indicates an increase (see color scale).
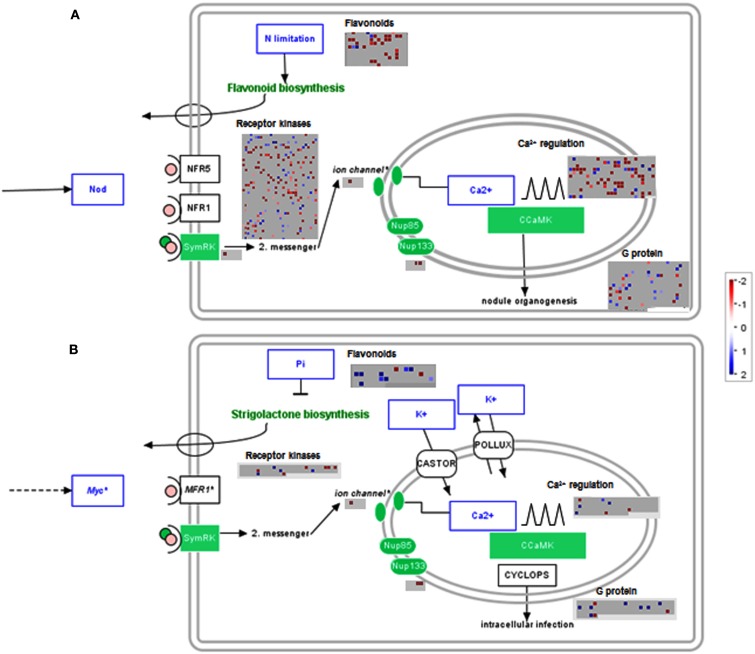
The data was visualized next on a metabolic overview pathway and genes showing no significant change were filtered out. MAPMAN bins for Calvin cycle genes were significantly increased in both lotus and tomato, while major carbohydrate metabolism appears to be largely reduced in lotus and enhanced in tomato roots (Figure A2 in Appendix). In addition, lotus roots had significant decreases in gene classes belonging to cell wall, lipid, and secondary metabolism, including flavonoid and phenylpropanoid metabolism (Figure A2A in Appendix). In order to visualize this in the context of genes associated with the symbiotic process, significantly altered genes related to either nodulation (for lotus) or mycorrhiza colonization (for tomato) were further illustrated in a graphical overview of symbiosis (Figure 3). This reiterated the number of significant transcript changes associated with either nodulation (Figure 3A) or mycorrhiza colonization (Figure 3B) upon LC treatment compared with the untreated control.
Figure 3.
Filtered gene responses (P < 0.05) of a MAPMAN overview of genes associated with symbiotic processes of lumichrome-treated (A) lotus and (B) tomato roots. The values are representative of the log2 transformed values of three individual hybridizations, statistically analyzed via robust multichip averaging (RMA) in R, and classed according to the MAPMAN classifications (Thimm et al., 2004; Usadel et al., 2005) adapted for lotus (Sanchez et al., 2008) and tomato (Urbanczyk-Wochniak et al., 2006), respectively. Red indicates a decrease whereas blue indicates an increase (see color scale).
On the individual gene expression level several other similarities were further apparent (Table 1). In order to apply a more stringent criteria for random noise that may be present in the data set, as well as gain more confidence in the identities of the probe set annotations, both lotus and tomato IDs were compared to Arabidopsis homolog candidates, and only those significant genes (adj. P < 0.05) filtered out that were: (i) concurrently altered in both plant species, as well as (ii) shared a significant similarity to an Arabidopsis homolog (Table 1; for full list of significantly altered genes corresponding to probe IDs, Table S1 in Supplementary Material).Both lotus and tomato roots had reduced transcripts levels involved in ethylene metabolism (ethylene response factor/elements, ACC oxidase and a putative two-component response regulator; Table 1). Similarly, several transcription factors associated with ethylene signal transduction were down-regulated, including those that share similarity with Arabidopsis orthologs belonging to proteins predicted to contain an ethylene-responsive element, including an AP2/EREBP transcription factor (At4g34410) and a C2H2 zinc finger protein (At2g37430; Table 1). Furthermore, genes involved in primary carbon metabolism, namely glyceraldehyde 3-phosphate dehydrogenase (GAPDH; two plastidial isoforms, namely At1g12900 and At1g42970), glucosyltransferases (At3g15350 and At3g22250) and nuclear-encoded subunits of plastidial gene products (At5g38430, At1g67090, At5g38430, At2g30570, and At1g67090) were also affected upon treatment (Table 1).
Table 1.
Orthologous gene comparisons between 5-week-old lumichrome-treated lotus (L. japonicus) and tomato (S. lycopersicum) roots.
|
L. japonicus |
S. lycopersicum |
AGI code | A. thaliana functional annotation | ||||
|---|---|---|---|---|---|---|---|
| Array ID | logFC | Adj. P-value | Array ID | logFC | Adj. P-value | ||
| PHYTOHORMONE METABOLISM | |||||||
| chr5.CM0052.19_at | −1.89 | 0.031 | 4.1.18.1 | −2.64 | 0.011 | At5g64750 | ABA repressor 1 |
| chr6.CM0114.20_at | −3.44 | 0.016 | At1g28360 | Ethylene response factor12 (ERF12) | |||
| chr5.CM0909.73_at | −6.22 | 0.007 | At5g61590 | Ethylene-responsive element binding factor-like | |||
| chr4.CM0288.8_at | −2.38 | 0.064 | 6.1.16.2 | −3.72 | 0.02 | At1g05010 | 1-Aminocyclopropane-1-carboxylate oxidase |
| chr3.TM1367.15_at | 2.64 | 0.019 | 5.1.3.12 | 1.92 | 0.04 | At2g41310 | Putative two-component response regulator 3 protein |
| chr5.CM0335.12_at | −4.31 | 0.017 | 8.4.19.1 | −3.11 | 0.04 | At1g20510 | 4-Coumarate:CoA ligase (OPCL1) |
| TRANSCRIPTION FACTOR (-LIKE) | |||||||
| chr2.CM0132.7_at | 4.85 | 0.021 | 2.3.10.17 | 5.81 | 0.007 | At4g37730 | bZip transcription factor 7(AtbZip7) |
| chr4.CM0227.6_at | −9.95 | 0.028 | 2.1.19.2 | −3.11 | 0.03 | At4g34410 | AP2 domain-containing transcription factor |
| chr5.CM0311.11_at | 2.26 | 0.013 | 4.4.14.11 | 4.55 | 0.04 | At1g31050 | AtbHLH111 |
| Ljwgs_064987.2_at | −6.02 | 0.001 | 7.3.19.10 | −2.41 | 0.04 | At4g27280 | Ca2 + binding EF hand family protein |
| chr3.TM1463.16_at | 1.77 | 0.018 | 5.4.2.3 | 1.87 | 0.03 | At5g54630 | Zinc finger related protein |
| Ljwgs_020980.2_at | −8.48 | 0.03 | 8.4.20.16 | −8.97 | 0.04 | At2g37430 | Putative C2H2-type zinc finger protein |
| CARBOHYDRATE METABOLISM | |||||||
| chr1.CM0222.6_at | 3.83 | 0.008 | 8.4.10.12 | 4.56 | 0.04 | At1g12900 | Glyceraldehyde 3-phosphate dehydrogenase A (GAPA-2) |
| chr1.CM0113.81_at | 3.26 | 0.011 | At1g42970 | Glyceraldehyde 3-phosphate dehydrogenase (GAPB) | |||
| Ljwgs_054325.1_at | −3.77 | 0.0004 | 8.2.19.12 | −1.7 | 0.05 | At3g15350 | Glycosyltransferase family 14 |
| chr2.CM0102.62_at | −5.62 | 0.007 | 5.4.19.4 | −2.7 | 0.02 | At3g22250 | UDP-glucoronosyl/-glucosyl transferase |
| Ljwgs_031056.1_at | 1.96 | 0.021 | 2.2.7.6 | 4.94 | 0.009 | At5g38430 | RuBisCO small subunit 1b |
| chr2.TM1655.9_at | 4.98 | 0.001 | 8.3.12.20 | 2.17 | 0.04 | At1g67090 | RuBisCO like protein, small subunit |
| Ljwgs_031056.1_at | 1.96 | 0.021 | At5g38430 | RuBisCO small chain 1b precursor | |||
| chr5.TM1574.11_at | 3.51 | 0.002 | 8.1.9.9 | 9.65 | 0.02 | At2g30570 | Photosystem II reaction center 6.1KD protein |
| chr2.TM1655.9_at | 4.98 | 0.001 | 5.4.7.9 | 7.08 | 0.05 | At1g67090 | RuBisCO like protein, small subunit |
| Ljwgs_057808.1_at | 1.71 | 0.012 | 4.2.10.10 | 7.18 | 0.04 | At5g54190 | NADPH:protochlorophyllide oxidoreductase A (PORA) |
| FATTY ACID METABOLISM | |||||||
| chr3.CM0135.11_at | 1.81 | 0.029 | 2.4.15.7 | 2.76 | 0.01 | At2g05990 | Enoyl-ACP reductase (ENR1) |
| MISCELLANEOUS | |||||||
| TM0802.5_at | 3.84 | 0.015 | 6.2.1.16 | 3.98 | 0.008 | At5g65360 | Histone H3 |
| chr5.TM0431.19_at | −2.13 | 0.014 | 8.2.17.12 | −2.56 | 0.05 | At1g10390 | Nucleoporin family |
| chr5.TM0431.19_at | −2.13 | 0.014 | 7.1.19.6 | −3.27 | 0.02 | At1g10390 | Nucleoporin family |
| Ljwgs_042055.1_at | −11.2 | 0.027 | 8.4.17.18 | −2.87 | 0.04 | At1g43710 | Histidine decarboxylase, embryo defective 1075 |
| chr1.TM0811.11_at | −3.71 | 0.056 | 6.1.19.5 | −5.64 | 0.02 | At4g02380 | Senescence associated gene 12 (SAG12) |
| chr1.CM0104.34_at | −5.99 | 0.007 | 8.1.16.1 | −4.43 | 0.03 | At5g18270 | NAM (no apical meristem)-like protein |
| chr1.CM0349.17_at | −11.4 | 0.023 | 5.1.13.1 | −4.01 | 0.01 | At1g14560 | Mitochondrial carrier like protein |
| Ljwgs_018600.1_at | −2.09 | 0.049 | At1g20860 | Putative inorganic phosphate transporter protein | |||
| Ljwgs_020324.1_at | −3.21 | 0.03 | 7.2.19.14 | −2.81 | 0.03 | At5g53130 | Cyclic nucleotide-regulated ion channel |
| chr1.TM1573.13.1_at | −2.98 | 0.021 | 8.3.19.13 | −4.03 | 0.02 | At3g24550 | ATP binding protein kinase, AtPERK1 |
| UNKNOWN | |||||||
| chr3.CM0142.58_at | −3.49 | 0.049 | 8.1.20.1 | −6.95 | 0.003 | At3g15810 | Unknown protein |
| chr3.CM0142.58_at | −3.49 | 0.049 | 8.2.17.16 | −3.55 | 0.004 | At3g15810 | Unknown protein |
| chr1.CM0181.3_s_at | 1.85 | 0.005 | 5.1.4.16 | 4.97 | 0.02 | At1g21680 | Unknown protein |
| chr2.CM0608.80_at | −2.6 | 0.038 | 1.2.17.11 | −5.29 | 0.03 | At1g27100 | Unknown protein |
| Ljwgs_054723.1_at | −2.44 | 0.032 | 7.1.19.9 | −2.05 | 0.04 | At3g26580 | Unknown protein |
Values are represented as log2 fold changes (logFC) of gene expressions normalized to control roots of three independent hybridizations and the P value adjusted (adj. P value) to correct for the probability of false positives using the FDR correction (Benjamini and Hochberg, 1995; P value <0.05 was deemed significant). Gene annotations was based on probe oligosequence similarity to the Arabidopsis genome, and represented by the Arabidopsis genome initiative (AGI) code.
Interestingly, this comparative analysis also allowed for the simultaneous identification of a nucleoporin (similar to At1g10390), plastidial cyclic ion channel (similar to At5g53130) and phosphate transporter (similar to At1g20860, AtPht1;8) in response to lumichrome in lotus and tomato roots (Table 1). The expression of all of these genes were reduced compared to the untreated controls (Table 1).
Evaluation of ethylene metabolism and other classical phytohormones in relation to lumichrome promoted growth stimulation
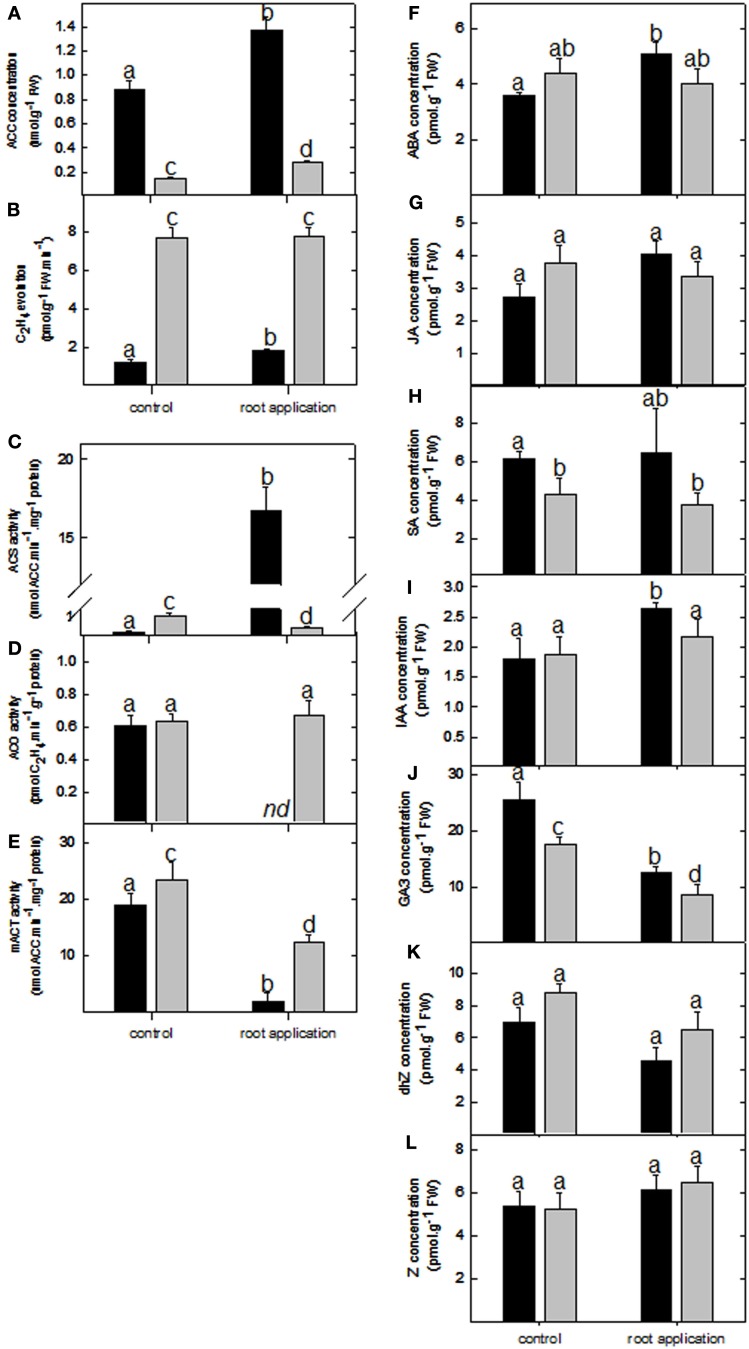
Due to the number of ethylene-associated genes affected by LC treatment, the biosynthesis and signal transduction of this gaseous hormone were further evaluated. Analysis of the ACC levels were significantly enhanced in both lotus and tomato roots upon LC treatment (Figure 4A); however, C2H4 levels were only enhanced in the lotus treated roots (Figure 4B). This is surprising since it is assumed that ACC synthase (responsible for ACC biosynthesis) is one of the key regulatory enzymes in ethylene synthesis. In light of this, the corresponding biosynthetic enzymatic activities were also determined (Figures 4C–F). This further revealed enhanced ACC synthase activity in the lotus plants (which exhibited enhanced C2H4 levels; Figure 4B) but a significant decrease in the catalytic activity in tomato roots under the conditions measured (Figure 4C). Furthermore, ACC oxidase activity was unaltered in tomato roots, whereas the activity was undetectable in LC-treated lotus roots (Figure 4D). As an alternative, steady state levels of ACC can also be maintained via conjugation reactions. In this regard, measurement of malonyl-ACC transferase indicated a significant reduction in its activity in both lotus and tomato roots (Figure 4E). In order to evaluate whether C2H4 levels could partially modulate the increased root biomass previously observed (Figure 1B), ethylene signal transduction was further inhibited by application of silver ions (by monitoring the expression of the EIN3 target downstream of the ethylene receptor CTR1; Figures A1I,J in Appendix; Rodriguez et al., 1999). This still resulted in enhanced biomass accumulation in the respective organs affected in Figures 1A,B following lumichrome treatment (see Figures A1G,H in Appendix).
Figure 4.
Ethylene biosynthesis and phytohormone levels upon lumichrome treatment in lotus and tomato. (A) 1-Aminocyclopropane-1-carboxylic acid (ACC) and (B) ethylene (C2H4) concentrations in roots of 5-week-old lotus (black bar) and tomato (gray bar). Correspondingly, (C) ACC synthase, (D) ACC oxidase, (E) and malonyl-ACC transferase maximal catalytic activity upon root drenching. (F–L) Absolute levels of abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), indole 3-acetic acid (IAA), gibberellic acid A3 (GA3), dihydrozeatin (dhZ) and zeatin (Z). The values are the mean ± SE (n = 6). The data was analyzed using one way ANOVA followed by Fisher’s least significant difference, bars that do not share letters are significantly different (P < 0.05) from each other. nd, not detected.
In addition to the evaluation of C2H4, other phytohormone levels were also evaluated in the control and treated roots of both lotus and tomato. This revealed little alteration in absolute levels of the stress hormone ABA, SA, and JA, with only the former enhanced in the lotus root drenching treatment (Figures 4F–H). Furthermore, indole-3-acetic acid (IAA) levels were also enhanced in the lotus roots (Figure 4I). In contrast, gibberellin A3 (GA3) levels were consistently decreased in both lotus and tomato roots (Figure 4J). Cytokinin levels (dihydrozeatin and trans-zeatin) remained invariable in both the lotus and tomato roots (Figures 4K,L).
Investigating key aspects in primary metabolism following lumichrome treatment in lotus and tomato roots
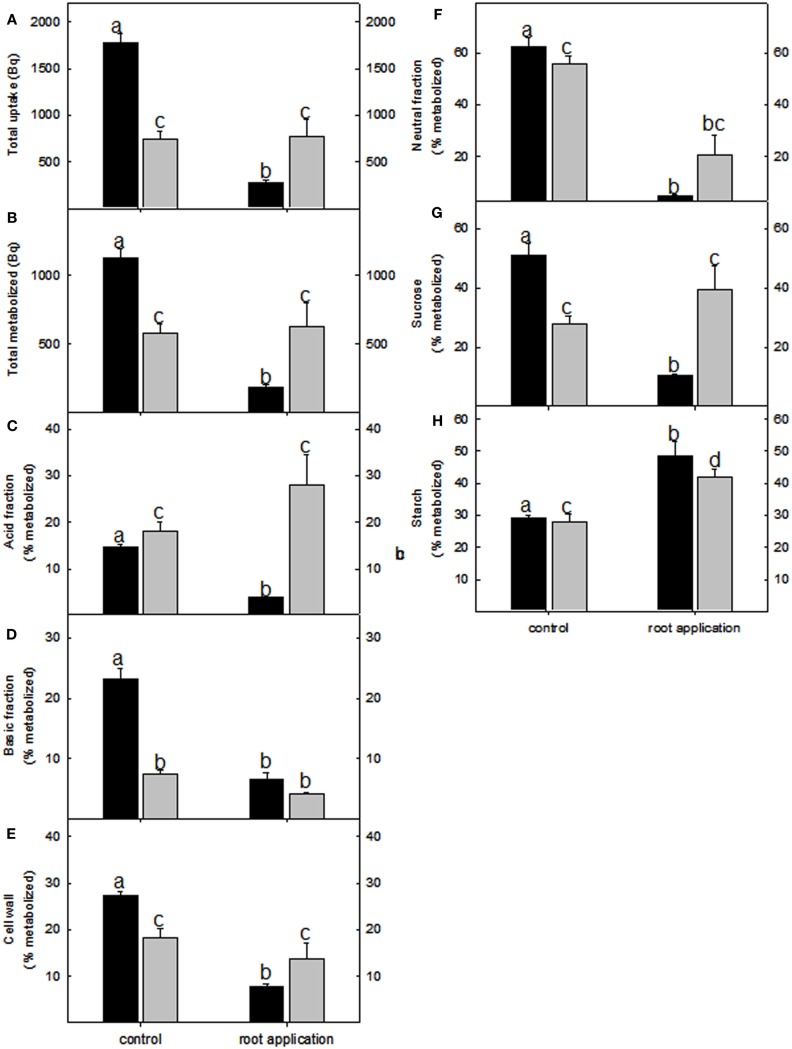
Due to the fact that there were few overlapping changes between lotus and tomato roots in ethylene and other phytohormones that could explain the observed biomass, our attention turned to investigating primary carbon metabolism. Incubating root pieces with [U-14C] glucose indicated that, while lotus roots had significantly reduced label uptake and incorporation in most fractions analyzed (Figures 5A–G); both lotus and tomato roots had a significant increase in radiolabel partitioned to the starch fraction (Figure 5H). This was also further reflected in the amounts of starch and soluble sugars measured. Starch levels were increased between 25 and 30% in lotus- and tomato-treated roots (Table 2). The tomato roots were also characterized by increased glucose levels following LC treatment (Table 2). Lugol staining of the root tips of lotus and tomato further indicated that LC did not lead to a change in the distribution of starch within the lotus; however, perhaps surprisingly, less starch was observed in the tomato root tip, suggesting that, LC further led to a change from indeterminate to determinate root growth in tomato roots (Figures A1C–F in Appendix).
Figure 5.
Redistribution of radiolabel in lotus (black bar) and tomato (gray bar) root metabolic fractions following lumichrome treatment and incubation with 2 mM [U-14C] glucose. (A) Total label uptake, (B) total label metabolized and label distribution to (C) the acid fraction, (D) basic fraction, (E) cell wall fraction, as well as (F) neutral fraction was determined after a 3-h incubation period. Label distribution toward the (G) sucrose and (H) starch pools. The values are expressed as a percentage of the total radiolabel metabolized and are the mean ± SE (n = 6). The data was analyzed using one way ANOVA followed by Fisher’s least significant difference, bars that do not share letters are significantly different (P < 0.05) from each other.
Table 2.
Effect of 5 nM lumichrome root application on soluble sugar and starch content of 5-week-old lotus and tomato roots.
|
L. japonicus |
S. lycopersicum |
|||
|---|---|---|---|---|
| Control | Root application | Control | Root application | |
| Glucose | 322.46 ± 20.02 | 284.39 ± 20.40 | 279.78 ± 10.76 | 411.97 ± 16.96 |
| Fructose | 49.44 ± 8.17 | 54.48 ± 4.08 | 71.59 ± 3.62 | 83.38 ± 4.59 |
| Sucrose | 18.03 ± 0.83 | 17.92 ± 3.86 | 76.99 ± 16.96 | 75.41 ± 9.12 |
| ΣSoluble sugars (hexose equivalents) | 407.97 ± 38.58 | 370.03 ± 26.45 | 505.36 ± 24.28 | 646.17 ± 30.16 |
| Starch | 17.76 ± 6.63 | 25.11 ± 0.81 | 26.47 ± 0.76 | 35.19 ± 0.67 |
Data is representative of the mean ± SE on six individual plants per line. Values in bold type were determined by Student’s t-test to be significantly different (P < 0.05) from the untreated control. Values are presented as μmol g−1 FW, with the exception of starch in milligram glucose equivalents per gram FW.
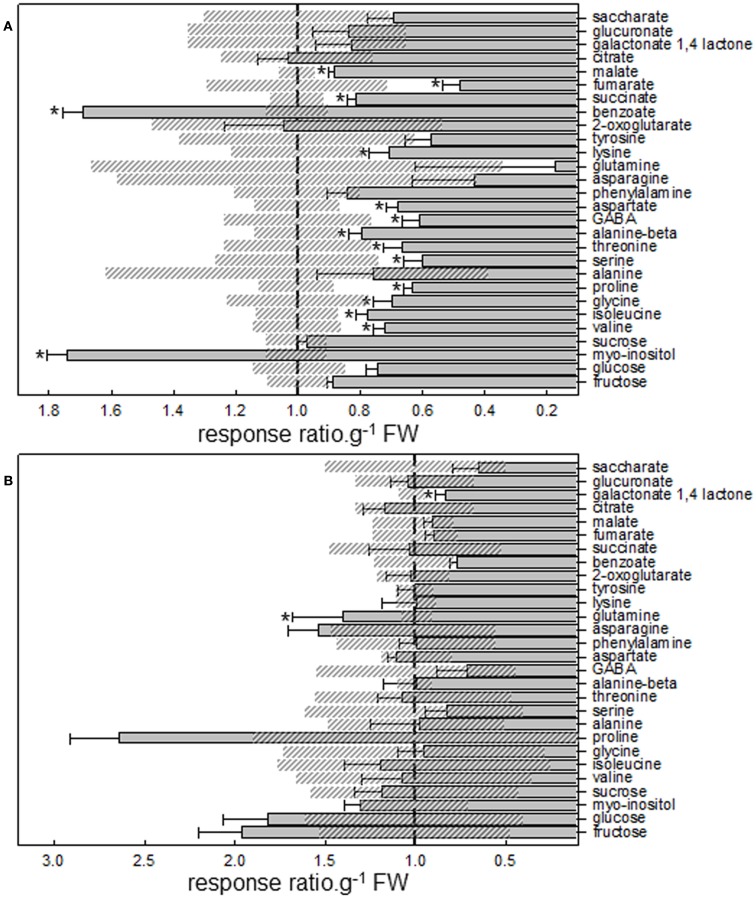
GC MS based metabolite profiling further reflected the radiolabel data; it indicated that lotus treated roots had major reductions in organic acids (malate, fumarate, and succinate) and amino acids (Lys, Asp, GABA, β-Ala, Thr, Ser, Pro, Gly, Iso, and Val; Figure A3A in Appendix). In addition, benzoate and myo-inositol levels were significantly increased upon LC treatment in the lotus roots (Figure A3A in Appendix). In contrast, the primary metabolites from lumichrome-treated tomato roots remained largely unaltered from the untreated controls, with the exception of a significant increase in Gln and decrease in galactonate-1,4-lactone levels (Figure A3B in Appendix).
Due to the apparent switch in metabolism differentiating between major carbohydrate and downstream glycolytic pathways, as well as the gene expression changes of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) upon LC treatment (Table 1), activities of this protein was also measured (Table 3). This indicated firstly that the major cytosolic non-phosphorylating NAD-dependent activity was significantly decreased upon LC treatment in lotus roots but not those of tomato (Table 3). However, plastidial enrichments and determination of phosphorylating NADP-dependent GAPDH activity was significantly increased in the crude and plastidial-enriched fractions of tomato roots, which was not reflected in the corresponding lotus roots (Table 3).
Table 3.
Specific GAPDH activity in lotus and tomato roots upon lumichrome treatment assayed under non-phosphorylating and phosphorylating conditions.
| Fraction |
L. japonicus |
S. lycopersicum |
|||
|---|---|---|---|---|---|
| Control | Root application | Control | Root application | ||
| Non-phosphorylating GAPDH | 19.72 ± 1.20 | 14.24 ± 0.71 | 4.26 ± 0.29 | 3.67 ± 0.14 | |
| Phosphorylating NADP-GAPDH | Crude | 1.39 ± 0.33 | 0.59 ± 0.18 | 0.85 ± 0.07 | 1.43 ± 0.35 |
| Plastid | 0.16 ± 0.07 | 0.42 ± 0.17 | 0.21 ± 0.03 | 0.86 ± 0.25 | |
| Supernatant | 1.52 ± 0.08 | 1.08 ± 0.23 | 0.96 ± 0.24 | 1.89 ± 0.42 | |
| Phosphorylating NAD-GAPDH | Crude | 9.62 ± 1.79 | 10.35 ± 1.23 | 24.87 ± 1.69 | 42.39 ± 4.03 |
| Plastid | 0.97 ± 0.01 | 2.23 ± 0.73 | 1.56 ± 0.39 | 4.32 ± 0.57 | |
| Supernatant | 11.63 ± 1.08 | 9.25 ± 1.67 | 15.83 ± 1.97 | 29.44 ± 8.09 | |
Under phosphorylating conditions, enzyme activity of crude extracts, plastid fractions and resultant supernatant fractions were assayed from root material. The values are the mean ± SE (n = 6) and values in bold type were determined by Student’s t-test to be significantly different (P < 0.05) from the respective untreated control. Non-phosphorylating and phosphorylating NADP-dependent enzyme activity is presented in nmol NAD(H/P) min−1 mg−1 protein, while phosphorylating NAD-dependent GAPDH values are expressed in μmol NAD min−1 mg−1 protein.
Colonization in lumichrome-primed lotus and tomato roots
In order to ensure that these effects were truly related to changes in aseptic endogenous plant metabolism, serial dilutions of root-associated and bacterial colony forming units (CFU) from the sterilized growth media were determined and this indicated that the CFU count in LC treatments were invariant, and negligible, from the control (data not shown). In addition, clearing and staining of lotus and tomato roots did not indicate any obvious signs of the presence of microorganisms.
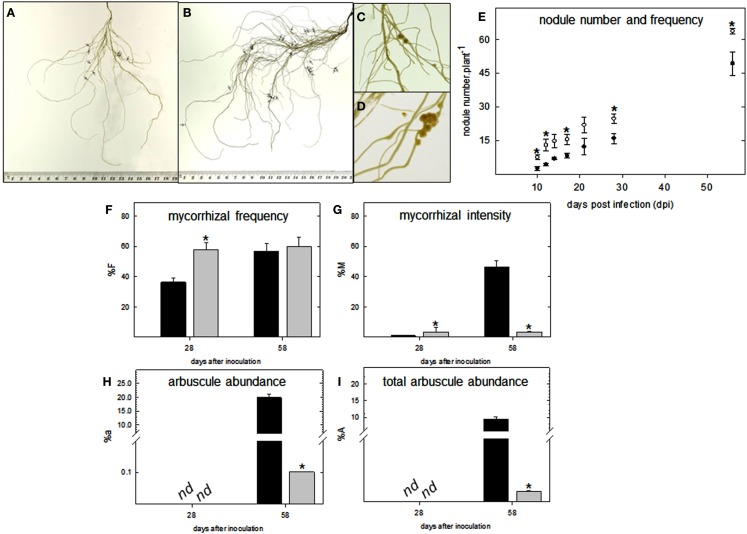
Having determined this, plants were further subjected to nitrogen and phosphate starvation conditions, respectively, and inoculated with M. loti and a mixture of G. intraradices and G. mossea to determine whether lumichrome pre-treatment had a noticeable effect on symbiotic efficiency. This indicated an increase in nodule number but no noticeable difference in the timing of nodule formation during early colonization in lotus (Figures 6A–E). In contrast, in tomato roots an acceleration in infection thread formation caused an increase in myorrhizal frequency and intensity at 28 days after inoculation (dai; Figures 6F,G). However, the LC pre-treated roots formed less arbuscules per root fragment (%a ∼ 0.1%) and total root system (%A ∼ 0.003%) compared to the control at 58 dai (Figures 6H,I).
Figure 6.
Colonization parameters of pre-treated lumichrome roots of lotus and tomato. (A) Untreated control and (B) lumichrome pre-treated lotus roots following 18 days post-infection (dpi) with M. loti. Little arrow heads indicate the presence of nodules. Close-up of (C) control and (D) pre-treated nodules from lotus 18 dpi. (E) Time-course (58 days) of nodule development in lotus following pre-treatment with lumichrome (white circle) or without (black circle). (F–I) Mycorrhization parameters of (F) frequency of mycorrhiza In the root system (%F), (G) intensity of mycorrhizal colonization in the root system (%M), (H) arbuscule abundance in mycorrhizal parts of root fragments (%a) and (I) arbuscule abundance in the root system (%A) of tomato root controls (black bar) and pre-treated with lumichrome (gray bar) at 28 and 58 days after inoculation with Glomus spp. The values are the average ± SE for four independent replicates [of 15 root fragments each, (F–I)] and an asterisk indicates a value that was determined by Student’s t-test (P < 0.05) to be significantly different from the control. nd, not detected.
Discussion
Modulation of growth and development through chemical stimulation has aided in understanding fundamental processes in plants (Kaschani and van der Hoorn, 2007). In this study the application of LC has been assessed in order to assess a role during symbiosis. We demonstrated that an application of LC at a concentration of 5 nM led to a significant enhancement of biomass in both lotus and tomato plants (Figure 1). Previous studies have similarly shown that LC promotes seedling growth at low concentrations whereas the application of higher doses (range between 10 and 50 nM) elicits less pronounced results (Phillips et al., 1999; Matiru and Dakora, 2005). Furthermore, based on the over-representation analysis of functional gene classes (Figure 2); as well as assessing carbon partitioning and steady state metabolite levels, it was apparent that LC led to a significant increase in starch levels and associated label distribution in both species (Table 2; Figure 5H; Figure A3 in Appendix). Lotus japonicus TILLING mutants deficient in starch biosynthetic and degradation enzymes show no observable differences in nodulation efficiency or development (Vriet et al., 2010), suggesting that starch are not required for organogenesis. Rather, as has been observed for tomato roots treated with fungal spore exudates, a transient increase in starch levels could be acting as a carbon overflow mechanism resulting from sugar efflux in the absence of a fungal partner (Gutjahr et al., 2009); thus acting as a marker in sensing the sugar status of the symbiotic host. In agreement, roots that have been pre-treated with LC showed altered colonization properties with either rhizobacteria or mycorrhizae (Figure 6). The observation that the application of LC, in the absence of microorganisms, led to starch accumulation in lotus and tomato roots, strongly suggests that LC act as a modulator of carbon fluxes within the symbiotic plant cell. In agreement with this, in lotus roots LC application led to a significant reduction in glycolytic and respiratory flux, as observed for a reduction in root respiration (Figure 1) and overall metabolic performance (Figure 5; Figure A3 in Appendix). Furthermore, LC also led to a significant reduction in the activity of the non-phosphorylating cytosolic isoform of GAPDH (Table 3). Higher plants contain four distinct isoforms of GAPDH; two cytosolic and two plastidial, and all four are expressed in varying (but low) levels in roots. The cytosolic, phosphorylating, NAD-dependent GAPDH (GAPC; constituting the majority of GAPDH activity) catalyzes the interconversion of glyceraldehyde 3-phosphate (G3P) to 1,3 bisphosphoglycerate, utilizing NAD+ and inorganic P (Pi). The cytosolic non-phosphorylating NADP-dependent isoform, on the other hand, catalyzes the oxidation of G3P to 3-phosphoglycerate (NP-GAPDH), bypassing an ATP-generation step in glycolysis. NP-GAPDH has also been postulated to supply NAPDH during gluconeogenesis (Kelly and Gibbs, 1973). In heterotrophic metabolism, perturbations in total GAPC activity does not lead to major phenotypical alterations although sucrose levels and other glycolytic metabolites are significantly affected (Hajirezaei et al., 2006). More recently GAPC has been shown to be involved in novel regulatory roles, essential for ATP supply and fertility in Arabidopsis (Rius et al., 2008). During lotus LC treatment, altered NP-GAPDH activity led to a significant shift in carbohydrate and organic acid metabolism. Similarly, it has been shown that Arabidopsis double mutants in the plastidial NAD-dependent GAPDH isoforms, gapcp1/gapcp2 drastically affects sugar and amino acid balances leading to enhanced starch accumulation (Muñoz-Bertomeu et al., 2009). Upon LC treatment, tomato roots had increased plastidial GAPDH transcript and enhanced plastidial NAD-dependent enzyme activity (Tables 1 and 3); however, with the exception of increases in starch and soluble sugars only minor changes in amino acid metabolism were observed (Figure 5), suggesting that a less severe perturbation was experienced. One plausible explanation for this is that enhanced photosynthesis (Figure 1C) and enhanced glucose levels (Table 2) in the tomato root drenching treatment leads to altered sink-source partitioning and thereby improving root metabolism.
Apart from altered primary carbon metabolism, LC treatments also led to alterations in ethylene (C2H4) responses (Table 1; Figure 4). C2H4 is a potent phytohormone that regulates diverse developmental processes, including fruit ripening, senescence, biotic and abiotic stress responses, root hair development, and abscission (for review, Kendrick and Chang, 2008; Zhu and Guo, 2008). Although the mechanism is not completely understood, enhanced ethylene and ACC levels inhibit nodulation in certain legumes (including L. japonicus; Penmetsa and Cook, 1997; Nukui et al., 2000) whilst having no effect on others (Lee and LaRue, 1992; Schmidt et al., 1999). Furthermore, lotus plants expressing the etr1-1 mutant allele from Arabidopsis, one of the dominant ethylene receptors, exhibita hyper-nodulation phenotype (Lohar et al., 2009). During fungal colonization, ethylene has also been implicated in modulating arbuscule development, although via alterations in ABA metabolism (Martín-Rodríguez et al., 2011). However, under nutrient replete LC additions, only lotus roots increased ethylene evolution rates, while that of tomato remained invariable from the control (Figure 3). In addition, in both species ACC levels increased significantly (Figure 3). One plausible reason for this difference is that the two species in question differ in the post-transcriptional regulation of ethylene biosynthetic enzymes, which was confirmed by the maximal catalytic activities of the respective biosynthetic proteins (Figure 3). In both lotus and tomato roots the ACC-conjugating enzyme activity was also down-regulated which could explain the increase in ACC levels (Figure 3). While the reason for this remains unclear, we speculate that the interaction between LC and C2H4 metabolism was a result of a transient redox mimicry (as observed during symbiosis), and independent from the biomass accumulation observed (Figure 1). In agreement with this, LC-treated roots treated with silver ions showed that the dry weight of the lotus and tomato roots was still significantly increased compared with the control; however, this treatment resulted in a significant decrease in biomass in general, suggesting that C2H4 metabolism is not the primary cause for the increase in biomass (Figure A1 in Appendix). In further support, LC applied to the foliar organs also enhanced biomass (Figure 1), suggesting that the effects of LC are pertinent throughout carbon partitioning in both autotrophic and heterotrophic metabolism. C2H4 is known to interact with several other phytohormones, including auxin (Stepanova et al., 2005, 2007), brassinosteroids (De Grauwe et al., 2005) and gibberellins (Achard et al., 2003; Vandenbussche et al., 2007). Absolute phytohormone levels showed little consistent responses in lotus and tomato roots; the exception to this was the reciprocal relationship between ACC and GA3 levels in both species (Figure 3). During symbiosis, exogenous gibberellin applications to the hyper-nodulated calcium calmodulin-dependent protein kinase mutant (ccamk) reduce nodulation significantly (Maekawa et al., 2009), while the overexpression of SLEEPY1, a positive regulator of GA signaling, also results in a reduction in nodule number in lotus (Maekawa et al., 2009). Our data further suggest that GA might play a role during mycorrhization; the molecular targets of which still remain to be identified. In addition, it is also likely that alterations in C2H4 metabolism could lead to an alteration in auxin distribution (which would not be reflected in the absolute levels) and could explain some of the variance. It would be useful to study the spatial accumulation pattern of several phytohormones during symbiosis and compare these with LC treatment in future. Furthermore, it has been recently proposed that ACC, the precursor of C2H4, might act as a unique signaling molecule with roles independent from C2H4 (Tsuchisaka et al., 2009; Tsang et al., 2011). While molecular evidence regarding this are still lacking, it suggests that LC and ACC could interact in a similar phytohormone cascade, and this pathway and its significance remains to be elucidated in future.
Lastly, upon LC application several transcripts associated with early phase symbiosis were repressed in both species (Table 1). Furthermore, an enhancement in nodule number or the timing of hyphal initiation, respectively (Figure 6), as well as an alteration in primary carbon, ethylene and gibberellin metabolism in the absence of colonization partners was observed in the lotus- and tomato-treated roots. These modulations involve both symbiotic dependent processes, as well as general increases in primary carbon metabolism. However, the exact molecular partner(s) LC interacts with to initiate this cascade of events still needs to be elucidated. In addition, it is not clear whether LC is essential for symbiosis or rather just alters the plant’s response toward symbiosis. It is also apparent that other downstream regulators of the symbiosis pathway need to be activated during AM colonization, making the use of LC an extremely useful tool in order to study these in future. In the context of the defined symbiosis, future elucidation and characterization of LC biosynthetic mutants and transgenic lines would greatly aid in addressing these questions.
In conclusion, lumichrome-associated responses include a reconfiguration in primary carbon and phythormone metabolism suggesting that it participates in aspects of symbiosis and general plant growth. Lotus japonicus showed a significant improvement in biomass and nodule number pre-treated with this compound. In contrast, while the timing of hyphae initiation during early post-infection phases was increased upon application in S. lycopersicum, this also led to a reduction in arbuscule maturation. While the mechanism for this discrepancy remains unclear at present, future work elucidating the similarities and differences in genetic factors involved in nodulation and mycorrhization events, respectively, will shed light on the protein targets affected by lumichrome, and its downstream effects, during symbiosis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/plant_physiology/10.3389/fpls.2012.00120/abstract
List of all expressed genes (based on array ID identifier), log2 fold and P value in lotus and tomato following lumichrome root drenching.
Acknowledgments
This work was supported by the National Research Foundation of South Africa (Liezel M. Gouws, Eileen Botes, Anna J. Wiese, Paul N. Hills, James R. Lloyd, Jens Kossmann, Margaretha J. van der Merwe), Harry Crossley Foundation (Liezel M. Gouws), Deutscher Akademischer Austauschdienst (Margaretha J. van der Merwe), Max Planck Institute (Björn Usadel, Sandra Trenkamp, Alisdair R. Fernie, Margaretha J. van der Merwe), and Noble Foundation (Ivone Torres-Jerez, Yuhong Tang). The Central Analytical Facility (Stellenbosch University) and Renate Smit (Horticultural Science, Stellenbosch University) are acknowledged for the use of chromatography equipment. We are grateful to Michael Udvardi (Noble Foundation, Ardmore, USA) for helpful comments, Jens Stougaard (Department of Molecular Biology, Aarhuis, Denmark) for lotus (Gifu B-129) seeds, Christian Sohlenkamp (Center for Genomics Sciences, Cuernavaca, Mexico) for M. loti (MAFF 303099) strain, and Alexander Erban (Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany) for technical assistance with metabolite data.
Appendix
Figure A1.
Physiological assessment of lumichrome treatment on lotus and tomato plants. Phenotypes of control, 5 nM lumichrome root-drenched, foliar, and a combination of root and foliar applications (from left to right) of (A) lotus plants and (B) tomato shoots (top) and roots (bottom) grown under a 16-h photoperiod. Lugol staining of mature lateral root tips of (C) control and (D) lumichrome-treated lotus roots, and (E) control and (F) lumichrome-treated tomato roots. (G) Shoot and (H) root dry mass was recorded upon a weekly 10 μM AgNO3 treatment in parallel with untreated controls (see Figure 1) for lotus (black bar) and tomato (gray bar). Semi-quantitative RT PCR of EIN3 targets of (I) lotus and (J) tomato roots after silver ion inhibition for water control (1) untreated control (2), Ag+ untreated control (3), LC-treated (4), and Ag+ LC-treated (5) roots (from left to right). Bottom panel represents PP2A and 18S rRNA expression for lotus and tomato, respectively. The values are the mean ± SE (n = 6). The data was analyzed using one way ANOVA followed by Fisher’s least significant difference and bars that do not share letters are significantly different (P < 0.05) from each other.
Figure A2.
Filtered gene responses (P < 0.05) of a MAPMAN overview of heterotrophic metabolism of lumichrome-treated (A) lotus and (B) tomato roots. The values are representative of the log2 transformed values of three individual hybridizations, statistically analyzed via robust multichip averaging (RMA) in R, and classed according to the MAPMAN classifications (Thimm et al., 2004; Usadel et al., 2005) adapted for lotus (Sanchez et al., 2008) and tomato (Urbanczyk-Wochniak et al., 2006), respectively. Red indicates a decrease whereas blue indicates an increase [see color scale in (B)].
Figure A3.
Relative primary metabolite levels of (A) lotus and (B) tomato roots treated with 5 nM lumichrome. Data are normalized with respect to mean response calculated for the respective untreated control (to allow statistical assessment in the same way; indicated by vertical striped line). The values are the average ± SE (SE for control indicated with diagonal striped boxes) reported for five independent replicates and an asterisk indicates a value that was determined by Student’s t-test (P < 0.05) to be significantly different from the respective untreated control.
Footnotes
References
- Achard P., Vriezen W. H., van der Straeten D., Harberd N. P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825 10.1105/tpc.015685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Alba R., Fei Z., Payton P., Liu Y., Moore S. L., Debbie P., Cohn J., D’Ascenzo M., Gordon J. S., Rose J. K., Martin G., Tanksley S. D., Bouzayen M., Jahn M. M., Giovannoni J. (2004). ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J. 39, 697–714 10.1111/j.1365-313X.2004.02178.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Ané J. M., Kiss G. B., Riely B. K., Penmetsa R. V., Oldroyd G. E. D., Ayax C., Lévy J., Debellé F., Baek J-M., Kalo P., Rosenberg C., Roe B. A., Long S. R., Dénarié J., Cook D. R. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367 10.1126/science.1092986 [DOI] [PubMed] [Google Scholar]
- Balzergue C., Puech-Pagès V., Bécard G., Rochange S. F. (2011). The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 62, 1049–1060 10.1093/jxb/erq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G., Fortin J. A. (1988). Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108, 211–218 10.1111/j.1469-8137.1988.tb03698.x [DOI] [PubMed] [Google Scholar]
- Benedito V. A., Torres-Jerez I., Murray J. D., Andriankaja A., Allen S., Kakar K., Wandrey M., Verdier J., Zuber H., Ott T., Moreau S., Niebel A., Frickey T., Weiller G., He J., Dai X., Zhao P. X., Tang Y., Udvardi M. K. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513 10.1111/j.1365-313X.2008.03519.x [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- Besserer A., Bécard G., Jauneau A., Roux C., Séjalon-Delmas N. (2008). GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148, 402–413 10.1104/pp.108.121400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A., Puech-Pages V., Kiefer P., Gomez-Roldan V., Jauneau A., Roy S., Portais J. C., Roux C., Bécard G., Séjalon-Delmas N. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4, e226. 10.1371/journal.pbio.0040226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund S., Antti H., Uddestrand I., Moritz T., Sundberg B. (2007). Cross-talkbetween gibberellin and auxin in development of Populus wood: gibberellinstimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 52, 499–511 10.1111/j.1365-313X.2007.03250.x [DOI] [PubMed] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- Bottini R., Cassán F., Piccoli P. (2004). Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 65, 497–503 10.1007/s00253-004-1696-1 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye-binding. Anal. Biochem. 72, 248–252 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Dilworth M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173, 11–26 10.1111/j.1469-8137.2006.01935.x [DOI] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479 10.1105/tpc.108.063255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., Guo W. J., Kim J. G., Underwood W., Chaudhuri B., Chermak D., Antony G., White F. F., Somerville S. C., Mudgett M. B., Frommer W. B. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grauwe L., Vandenbussche F., Tietz O., Palme K., van der Straeten D. (2005). Auxin, ethylene and brassinosteroids: tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46, 827–836 10.1093/pcp/pci111 [DOI] [PubMed] [Google Scholar]
- De Salamone I. E. G., Hynes R. K., Nelson L. M. (2001). Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can. J. Microbiol. 47, 404–411 10.1139/w01-029 [DOI] [PubMed] [Google Scholar]
- Desbrosses G. G., Kopka J., Udvardi M. K. (2005). Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for thestudy of plant-microbe interactions. Plant Physiol. 137, 1302–1318 10.1104/pp.104.054957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers E. A., Branscheid A., May P., Krajinski F. (2011). Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156, 1990–2010 10.1104/pp.111.172627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G. B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966 10.1038/nature00842 [DOI] [PubMed] [Google Scholar]
- Erban A., Schauer N., Fernie A. R., Kopka J. (2007). “Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles,” in Metabolomics: Methods and Protocols, ed. Weckwerth W. (Totowa: Humana Press Inc.), 19–38 [DOI] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004). Bioconductor: open softwaredevelopment for computational biology and bioinformatics. Genome Biol. 5, R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Phillips A. T., Arteca R. N. (1993). Amino acids N-malonyltransferasesfrom mung beans: action of 1-aminocyclopropane-1-carboxylic acid and D-phenylalanine. J. Biol. Chem. 268, 25389–25394 [PubMed] [Google Scholar]
- Gutjahr C., Novero M., Guether M., Montanari O., Udvardi M., Bonfante P. (2009). Presymbiotic factors released by the arbuscular mycorrhizal fungus Gigaspora margarita induce starch accumulation in Lotus japonicus roots. New Phytol. 183, 53–61 10.1111/j.1469-8137.2009.02871.x [DOI] [PubMed] [Google Scholar]
- Hajirezaei M. R., Biemelt S., Peisker M., Lytovchenko A., Fernie A. R., Sonnewald U. (2006). The influence of cytosolic phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPC) on potato tuber metabolism. J. Exp. Bot. 57, 2363–2377 10.1093/jxb/erj207 [DOI] [PubMed] [Google Scholar]
- Heckmann A. B., Lombardo F., Miwa H., Perry J. A., Bunnewell S., Parniske M., Wang T. L., Downie J. A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750 10.1104/pp.106.089508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., Takeda N., Charpentier M., Perry J., Miwa H., Umehara Y., Kouchi H., Murakami Y., Mulder L., Vickers K., Pike J., Downie J. A., Wang T., Sato S., Asamizu E., Tabata S., Yoshikawa M., Murooka Y., Wu G-J., Kawaguchi M., Kawasaki S., Parniske M., Hayashi M. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433, 527–531 10.1038/nature03237 [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Kanamori N., Madsen L. H., Radutoiu S., Frantescu M., Quistgaard E. M. H., Miwa H., Downie J. A., James E. K., Felle H. H., Lindegaard L. H., Jensen T. H., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 359–364 10.1073/pnas.0508883103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanu S., Dakora F. D. (2009). Thin-layer chromatographic analysis of lumichrome, riboflavin and indole acetic acid in cell-free culture filtrate of Psoralea nodule bacteria grown at different pH, salinity and temperature regimes. Symbiosis 48, 173–181 10.1007/BF03179996 [DOI] [Google Scholar]
- Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro M., Cochrane G., Duggan K., Eberhardt R., Faruque N., Gamble J., Diez F. G., Harte N., Kulikova T., Lin Q., Lombard V., Lopez R., Mancuso R., McHale M., Nardone F., Silventoinen V., Sobhany S., Stoehr P., Tuli M., Tzouvara K., Vaughan R., Wu D., Zhu W., Apweiler R. (2005). The EMBL nucleotide sequence database. Nucleic Acids Res. 33, D29–D33 10.1093/nar/gki397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F., van der Hoorn R. (2007). Small molecule approaches in plants. Curr. Opin. Chem. Biol. 11, 88–98 10.1016/j.cbpa.2006.11.038 [DOI] [PubMed] [Google Scholar]
- Kelly G. J., Gibbs M. (1973). Nonreversible D-glyceraldehyde 3-phosphate dehydrogenase of plant tissues. Plant Physiol. 52, 111–118 10.1104/pp.52.6.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick M. D., Chang C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485 10.1016/j.pbi.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan W., Prithiviraj B., Smith D. L. (2008). Nod factor [Nod Bj (C18:1, MeFuc)] and lumichrome enhance photosynthesis and growth of corn and soybean. J. Plant Physiol. 165, 1342–1351 10.1016/j.jplph.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmuller E., Dormann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A. R., Steinhauser D. (2005). GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 10.1093/bioinformatics/bti236 [DOI] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R. J., Downie J. A., Oldroyd G. E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105, 9823–9828 10.1073/pnas.0803499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., LaRue T. A. (1992). Exogenous ethylene inhibits nodulation of Pisum sativum L. cv Sparkle. Plant Physiol. 100, 1759–1763 10.1104/pp.100.3.1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., Journet E-P., Ané J-M., Lauber E., Bisseling T., Dénarié J., Rosenberg C., Debellé F. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364 10.1126/science.1093038 [DOI] [PubMed] [Google Scholar]
- Lizada M. C. C., Yang S. F. (1979). A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal. Biochem. 100, 140–145 10.1016/0003-2697(79)90123-4 [DOI] [PubMed] [Google Scholar]
- Lohar D., Stiller J., Kam J., Stacey G., Gresshoff P. M. (2009). Ethylene insensitivity conferred by a mutated Arabidopsis ethylene receptor gene alters nodulation in transgenic Lotus japonicus. Ann. Bot. 104, 277–285 10.1093/aob/mcp132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez J. A., Charnikhova T., Gómez-Roldán V., Matusova R., Kohlen W., De Vos R., Verstappen F., Puech-Pages V., Bécard G., Mulder P., Bouwmeester H. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178, 863–874 10.1111/j.1469-8137.2008.02406.x [DOI] [PubMed] [Google Scholar]
- Luedemann A., Strassburg K., Erban A., Kopka J. (2008). TagFinder for the quantitative analysis of gas chromatography - mass spectrometry (GC-MS) based metabolite profiling experiments. Bioinformatics 24, 732–737 10.1093/bioinformatics/btn023 [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Madsen L. H., Tirichine L., Jurkiewicz A., Sullivan J. T., Heckmann A. B., Bek A. S., Ronson C. W., James E. K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1, 10. 10.1038/ncomms1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194 10.1111/j.1365-313X.2008.03774.x [DOI] [PubMed] [Google Scholar]
- Maillet F., Poinsot V., Andre O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., Martinez E. A., Driguez H., Bécard G., Dénarié J. (2011). Fungal lipochitooligosaccharide symbiotic signals inarbuscular mycorrhiza. Nature 469, 58–63 10.1038/nature09622 [DOI] [PubMed] [Google Scholar]
- Martín-Rodríguez J. A., León-Morcillo R., Vierheilig H., Ocampo J. A., Ludwig-Müller J., García-Garrido J. M. (2011). Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol. 190, 193–205 10.1111/j.1469-8137.2010.03610.x [DOI] [PubMed] [Google Scholar]
- Matiru V. N., Dakora F. D. (2005). Xylem transport and shoot accumulation of lumichrome, a newly recognized rhizobial signal, alters root respiration, stomatal conductance, leaf transpiration, and photosynthetic rates in legumes and cereals. New Phytol. 165, 847–855 10.1111/j.1469-8137.2004.01254.x [DOI] [PubMed] [Google Scholar]
- Müller-Röber B., Sonnewald U., Willmitzer L. (1992). Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J. 11, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Mulet J. M., Baroja-Fernández E., Pozueta-Romero J., Kuhn J. M., Segura J., Ros R. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151, 541–558 10.1104/pp.109.143701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka A., Shiomi S., Kubo Y., Inaba A. (1997). Expression and internal feedback regulation of ACC synthase and ACC oxidase genes in ripening tomato fruit. Plant Cell Physiol. 38, 1103–1110 10.1093/oxfordjournals.pcp.a029094 [DOI] [PubMed] [Google Scholar]
- Nukui N., Ezura H., Yuhashi K-I., Yasuta T., Minamisawa K. (2000). Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 41, 893–897 10.1093/pcp/pcd011 [DOI] [PubMed] [Google Scholar]
- Pankhurst C. E., Schwinghamer E. A., Thorne S. W., Bergersen F. J. (1974). The flavin content of clovers relative to symbiosis with a riboflavin-requiring mutant of Rhizobium trifolii. Plant Physiol. 53, 198–205 10.1104/pp.53.2.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775 10.1038/nrmicro1987 [DOI] [PubMed] [Google Scholar]
- Penmetsa R. V., Cook D. R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530 10.1126/science.275.5299.527 [DOI] [PubMed] [Google Scholar]
- Phillips D. A., Joseph C. M., Yang G-P., Martínez-Romero E., Sanborn J. R., Volpin H. (1999). Identification of lumichrome as a Sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl. Acad. Sci. U.S.A. 96, 12275–12280 10.1073/pnas.96.18.9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing [Google Scholar]
- Reinhardt D. (2007). Programming good relations - development of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 10, 98–105 10.1016/j.pbi.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Remans R., Beebe S., Blair M., Manrique G., Tovar E., Rao I., Croonenborghs A., Torres-Gutierrez R., El-Howeity M., Michiels J., Vanderleyden J. (2008). Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil 302, 149–161 10.1007/s11104-007-9462-7 [DOI] [Google Scholar]
- Rius S. P., Casati P., Iglesias A. A., Gomez-Casati D. F. (2008). Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 148, 1655–1667 10.1104/pp.108.128769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F. I., Esch J. J., Hall A. E., Binder B. M., Schaller G. E., Bleecker A. B. (1999). A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283, 996–998 10.1126/science.283.5404.996 [DOI] [PubMed] [Google Scholar]
- Roessner U., Wagner C., Kopka J., Trethewey R. N., Willmitzer L. (2000). Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23, 131–142 10.1046/j.1365-313x.2000.00774.x [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A. R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133, 84–99 10.1104/pp.103.023572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Yoshikawa M., Yano K., Miwa H., Uchida H., Asamizu E., Sato S., Tabata S., Imaizumi-Anraku H., Umehara Y., Kouchi H., Murooka Y., Szczyglowski K., Downie J. A., Parniske M., Hayashi M., Kawaguchi M. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19, 610–624 10.1105/tpc.106.046938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D. H., Lippold F., Redestig H., Hannah M., Erban A., Krämer U., Kopka J., Udvardi M. K. (2008). Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 53, 973–987 10.1111/j.1365-313X.2007.03381.x [DOI] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner Tunali U., Forbes M. G., Willmitzer L., Fernie A. R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 10.1016/j.febslet.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Schmelz E. A., Engelberth J., Alborn H. T., O’Donnell P., Sammons M., Toshima H., Tumlinson J. H., III. (2003). Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 10552–10557 10.1073/pnas.1633615100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. S., Harper J. E., Hoffman T. K., Bent A. F. (1999). Regulation of soybean nodulation independent of ethylene signaling. Plant Physiol. 119, 951–959 10.1104/pp.119.3.951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791 10.1126/science.1111025 [DOI] [PubMed] [Google Scholar]
- Smith A. M., Zeeman S. C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1, 1342–1345 10.1038/nprot.2006.232 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Hoyt J. M., Hamilton A. A., Alonso J. M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242 10.1105/tpc.105.033365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Yun J., Likhacheva A. V., Alonso J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185 10.1105/tpc.107.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962 10.1038/nature00841 [DOI] [PubMed] [Google Scholar]
- Su L. Y., Liu Y., Yang S. F. (1985). Relationship between 1-aminocyclopropane carboxylate malonyltransferase and D amino acid malonyltransferase. Phytochemistry 24, 1141–1145 10.1016/S0031-9422(00)81090-0 [DOI] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer A., Kruger S., Selbig J., Muller L. A., Rhee S. Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L. H., Miwa H., Nakagawa T., Sandal N., Albrektsen A. S., Kawaguchi M., Downie A., Sato S., Tabata S., Kouchi H., Parniske M., Kawasaki S., Stougaard J. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156 10.1038/nature04862 [DOI] [PubMed] [Google Scholar]
- Trouvelot A., Kough J. L., Gianinazzi-Pearson V. (1986). “Estimation of VA mycorrhizal infection levels. Research for methods having a functional significance,” in Physiological and Genetical Aspects of Mycorrhizae, eds Gianinazzi-Pearson V., Gianinazzi S. (Paris: INRA Press; ), 217–221 [Google Scholar]
- Tsang D. L., Edmond C., Harrington J. L., Nühse T. S. (2011). Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 156, 596–604 10.1104/pp.111.175372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Yu G., Jin H., Alonso J. M., Ecker J. R., Zhang X., Gao S., Theologis A. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003 10.1534/genetics.109.107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. L., Xue P., Yang R. (2010). 1-Aminocyclopropane-1-carboxylic acid (ACC) concentration and ACC synthase expression in soybean roots, root tips, and soybean cyst nematode (Heterodera glycines)-infected roots. J. Exp. Bot. 61, 463–472 10.1093/jxb/erp317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S. R., Ayoubi P., Weng H., Palmer D. A., Mitchell R. E., Jones W., Bender C. L. (2005). The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 42, 201–217 10.1111/j.1365-313X.2005.02366.x [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E., Usadel B., Thimm O., Nunes-Nesi A., Carrari F., Davy M., Bläsing O., Kowalczyk M., Weicht D., Polinceusz A., Meyer S., Stitt M., Fernie A. R. (2006). Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol. Biol. 60, 773–792 10.1007/s11103-005-5772-4 [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Blasing O. E., Redestig H., Sreenivasula N., Krall L., Hannah M. A., Poree F., Fernie A. R., Stitt M. (2006). PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinform. 7, 535. 10.1186/1471-2105-7-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., Selbig J., Hannemann J., Piques M. C., Steinhauser D., Scheible W-R., Gibon Y., Morcuende R., Weicht D., Meyer S., Stitt M. (2005). Extension of the visualizationtool MapMan to allow statistical analysis of arrays display of corresponding genesand comparison with known responses. Plant Physiol. 138, 1195–1204 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M. J., Osorio S., Araujo W. L., Balbo I., Nunes-Nesi A., Maximova E., Carrari F., Bunik V. I., Persson S., Fernie A. R. (2010). TCA cycle activity regulates tomato root growth via effects on secondary cell wall production. Plant Physiol. 153, 611–621 10.1104/pp.109.149047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M. J., Osorio S., Moritz T., Nunes-Nesi A., Fernie A. R. (2009). Decreased mitochondrial activities of malate dehydrogenase and fumarase inSolanum lycopersicum lead to altered root growth and architecture via diversemechanisms. Plant Physiol. 149, 653–669 10.1104/pp.108.130518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Vancompernolle B., Rieu I., Ahmad M., Phillips A., Moritz T., Hedden P., Van Der Straeten D. (2007). Ethylene-induced Arabidopsis hypocotyls elongation is dependent on but not mediated by gibberellins. J. Exp. Bot. 58, 4269–4281 10.1093/jxb/erm288 [DOI] [PubMed] [Google Scholar]
- Vriet C., Welham T., Brachmann A., Pike M., Pike J., Perry J., Parniske M., Sato S., Tabata S., Smith A. M., Wang T. L. (2010). A suite of Lotus japonicus starch mutants reveals both conserved and novel features of starch metabolism. Plant Physiol. 154, 643–655 10.1104/pp.110.161844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf L., Fromin N., Tomasi N., Aragno M., Martinoia E. (2005). Secretionactivity of white lupin’s cluster roots influences bacterial abundance, functionand community structure. Plant Soil 268, 181–194 10.1007/s11104-004-0264-x [DOI] [Google Scholar]
- White E. J., Venter M., Hiten N. F., Burger J. T. (2008). Modified cetyltrimethlammonium bromide method improves robustness and versatility: the benchmark for plant RNA extraction. Biotech. J. 3, 1424–1428 10.1002/biot.200800207 [DOI] [PubMed] [Google Scholar]
- Yanagita T., Foster J. W. (1956). A bacterial riboflavin hydrolase. J. Biol. Chem. 221, 593–607 [PubMed] [Google Scholar]
- Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., Markmann K., White C., Schuller B., Sato S., Asamizu E., Tabata S., Murooka Y., Perry J., Wang T. L., Kawaguchi M., Imaizumi-Anraku H., Hayashi M., Parniske M. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105, 20540–20545 10.1073/pnas.0806858105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Guo H. (2008). Genetic basis of ethylene perception and signal transduction in Arabidopsis. J. Integr. Plant Biol. 50, 808–815 10.1111/j.1744-7909.2008.00710.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all expressed genes (based on array ID identifier), log2 fold and P value in lotus and tomato following lumichrome root drenching.