Abstract
IMP dehydrogenase is a rate-limiting enzyme involved in the synthesis of GTP. In mammalian cells it is regulated with respect to growth rate and is the target of numerous therapeutic agents. Mutations in the RNA polymerase II elongation machinery render yeast sensitive to inhibitors of IMP dehydrogenase and defective in inducing transcription of one of the IMP dehydrogenase-encoding genes, IMD2. Here we show that loss of IMD2, but not IMD1, IMD3, or IMD4, conferred upon yeast the same drug sensitivity found in elongation mutants. We tested whether the drug sensitivity of elongation mutants is due to their inability to induce IMD2 by providing them with exogenous copies of the gene. In some elongation mutants, overexpression reversed drug sensitivity and a transcriptional defect. Overexpression in mutants with a more severe phenotype partially suppressed drug sensitivity but was inconsequential in reversing a defect in transcription. These findings suggest that the drug sensitivity of elongation mutants is largely but not solely attributable to defects in the ability to induce IMD2, because transcription is compromised even when IMD2 mRNA levels are adequate. We describe two DNA sequence elements in the promoter of the gene that regulate it. We also found that IMD2 mRNA abundance is coupled to cell growth rate. These findings show that yeast possess a conserved system that gauges nucleotide pools and cell growth rate and responds through a uniquely regulated member of the IMD gene family.
IMP dehydrogenase is a rate-limiting enzyme involved in de novo GTP biosynthesis. The abundance of the enzymatic activity is correlated with the rate of growth of mammalian cells (1–3). Antagonists of this family of enzymes, one of which is mycophenolic acid, serve as medically important antitumor, antimicrobial, and immunosuppressive agents (4). Characterization of two IMP dehydrogenase genes in mammalian cells has included an analysis of their promoters and the solution of a crystal structure of rodent IMP dehydrogenase in a complex with mycophenolic acid (5–7). Mammalian IMP dehydrogenase (IMPDH)1 II is inducible in a post-transcriptional manner, in response to depressed guanine nucleotide pools (3, 8). Saccharomyces cerevisiae has a family of four closely related genes, IMD1, IMD2 (formerly known as PUR5), IMD3, and IMD4, that encode potential IMP dehydrogenases. They share from 58 – 62% amino acid identity with mammalian IMPDH I and II. The four yeast proteins are themselves highly related with pairwise alignments showing amino acid identities ranging from 80 to 96%. Our understanding of the expression and regulation of this gene family is rudimentary. The proportion of cellular enzyme activity derived from each gene is unknown, and it has not yet been shown that these putative IMPDHs possess enzyme activity.
When cells are treated with drugs that inhibit IMP dehydrogenase, such as 6-azauracil (6AU) or mycophenolic acid, cellular GTP is depleted, and transcription of IMD2 is induced by an as yet unknown mechanism (9 –11). Conversely, providing yeast with extracellular guanine down-regulates constitutive levels of IMD2 and prevents induction by IMP dehydrogenase antagonists (10, 11).
In vitro, ribonucleotide concentration is a direct determinant of transcript elongation rate and the propensity of RNA polymerase II to become arrested during RNA chain elongation (12). Elongation factor SII (also known as TFIIS) is a conserved and well studied protein that allows recovery of RNA polymerase II from arrest (reviewed in Ref. 13). Sensitivity to 6AU and mycophenolic acid is a phenotype characteristic of yeast with mutations in the transcription elongation machinery including a deletion of DST1 (the gene encoding yeast SII, also known as PPR2) (9, 14). Mutations in other elongation factors and RNA polymerase II subunits also confer this phenotype upon yeast (15–24). SII enables RNA polymerase II to complete full-length transcript synthesis in vitro and is implicated in mRNA biosynthesis in living yeast (10, 13, 17, 20, 25). One model suggests that this drug-sensitive phenotype results from the difficulty RNA polymerase II has in elongating nascent transcripts when it is starved for nucleotide substrates in vivo (9, 20). Under such conditions, cell growth becomes dependent upon an optimally functioning elongation machinery. Recently, it has been suggested that the SII-dependent expression of downstream genes may play a role in the effect of the drug upon mRNA synthesis and cell physiology (10, 25). For example, the drug-induced transcription of IMD2 observed in wild-type cells is defective in a number of elongation mutants but not in 6AU-sensitive mutants not known to be involved in elongation (10). Whether defects in transcription observed in elongation mutants are due to direct effects of the mutations upon gene transcription or are secondary to the inability to induce IMP dehydrogenase activity and obtain sufficient GTP levels to support transcription is an open question. It is possible that both mechanisms operate to reduce transcription in the presence of IMPDH inhibitors.
If the drug-sensitive phenotype of elongation mutants is due to their inability to induce IMD2, we would predict that inactivation of IMD2 would lead to a drug-sensitive phenotype. Also, overexpression of IMD2 in elongationally compromised RNA polymerase II and SII mutants might rescue the drug-sensitive phenotypes. Here we test these predictions. Deletion of IMD2 but not the other IMD genes phenocopies the drug sensitivity of elongation mutants. Overexpression of IMD2 in elongation mutants suggests that their drug sensitivity is largely, although not completely, due to their failure to induce IMD2 in the presence of IMPDH inhibitors. We have identified an upstream element responsible for 6AU sensitivity and a region of the IMD2 promoter that is repressive for basal transcription. This and the fact that down-regulation of IMD2 by guanine was not due to altered mRNA turnover indicate that in yeast, unlike in mammalian cells, regulation is at the level of transcription. On the other hand, IMD2 transcription was sensitive to the rate of growth, as has been observed for mammalian cells. These results reveal details of the role of IMD2 in drug sensitivity and as a sensor of guanine pools with the ability to alter rates of mRNA synthesis and perhaps the cell cycle.
MATERIALS AND METHODS
Yeast Strains and Plasmids
pIMD2-S288C was made by inserting a PCR product amplified from S288C genomic DNA (Research Genetics, Huntsville, AL) into the NotI and XhoI sites of the pRS426 vector. The fragment inserted into the vector was generated using the primers 5′-GACTAGTCTCGAGTCGTAAACATAACACCCCATCA-3′ and 5′-GACTAGTGCGGCCGCATCGGTTGAGCGCGATATTA-3′, followed by digestion with XhoI and NotI. The NotI restriction site is located ≈500 bp upstream from the start codon of IMD2, and the XhoI restriction site is located ≈150 bp downstream the stop codon of IMD2. pIMD2-C335A was generated by site-directed mutagenesis (Gene Dynamics, LLC) of TGT to GCT at codon 335 in pIMD2-S288C. The plasmid pUC119-GAL was made by inserting a 187-bp EcoRI-HindIII restriction fragment from a GAL1 PCR product into the EcoRI and HindIII sites of pUC119.
Reporter plasmid constructs were made by PCR amplification, restriction digestion, and insertion into an appropriate yeast shuttle vector. Inserts were confirmed by DNA sequencing. The IMD2 promoter deletion plasmids, pPur5P800Luc, pIMD2-PL2, pIMD2-PL3, and pIMD2-PL4 were created by amplifying from pIMD2-S288C using 5′-CTGATCAGGATCCGGCCATTGCTTTTGCTACTT-3′ as the reverse primer and either 5′-GGGGTACCAAGCTTTGGAACAACAAACACAGTCCA-3′, 5′-GGGGTACCAAGCTTATCGGTTGAGCGCGATATTA-3′, 5′-GGGGTACCAAGCTTTATTGGTTTTCGTAACCGCC-3′, or 5′-GGGGTACCAAGCTTTGGTAAAAATTTCGGCTGGA-3′ as the forward primer, respectively. The products were cut with BamHI and HindIII and ligated into pGAL-Luc (26) digested with the same enzymes. The plasmids pIMD2-PL5, pIMD2-PL6, pIMD2-PL7, and pIMD2-PL8 were created by amplifying from pIMD2 using 5′-CACTAGTGGATCCGGAATAGAATAGAATACGG-3′ as the reverse primer and either 5′-GGGGTACCAAGCTTTGGAACAACAAACACAGTCCA-3′, 5′-GGGGTACCAAGCTTATCGGTTGAGCGCGATATTA-3′, 5′-GGGGTACCAAGCTTTATTGGTTTTCGTAACCGCC-3′, or 5′-GGGGTACCAAGCTTTGGTAAAAATTTCGGCTGGA-3′ as the forward primer, respectively. pIMD2-PGL1 and pIMD2-PGL2 were created by amplifying from pIMD2-S288C using 5′-CGGGATCCTTCCGTATTCTATTCTATTCCTTGC-3′ and 5′-CTGATCAGGATCCGCCATTGCTTTTGCTACTT-3′. The product was cut with BamHI and ligated into pGAL-Luc digested with BamHI. The former plasmid contains the insert in the forward orientation and the latter in the inverse orientation. Similarly, pIMD2-PGL5 and pIMD2-PGL4 were made by amplifying from pIMD2-S288C with 5′-CGGGATCCTTCCGTATTCTATTCTATTCCTTGC-3′ and 5′-CTGATCAGGATCCAACAAAATGCGTTTATGACAGTT-3′ and inserting the product in the. forward and reverse orientation, respectively. pIMD2-PGL3 was created by amplifying from pIMD2-S288C with 5′-GGGGTACCAAGCTTTTCCGTATTCTATTCTATTCCTTGC-3′ and 5′-CTGATCAGGATCCGCCATTGCTTTTGCTACTT-3′. The product was blunted with Klenow DNA polymerase, cut with KpnI, and ligated into pGAL-Luc, which was itself cut with HindIII, blunted with Klenow DNA polymerase, and cut with KpnI.
The yeast strains used in these studies are listed in Table I. DNA transformation was performed as described previously (27). Transformation of ABG-G11 (D. Kaback; University of Medicine and Dentistry of New Jersey) with pIMD2-S288C or pIMD2-C335A generated strains DY835 and DY836, respectively. Strains DY787, DY788, DY789 and DY790 were derived by transformation of Z96, Z106, DY100 (17), and DY102 (17) with pRS426 (28), respectively. Strains DY2065, DY2066, DY2067, and DY2068 were generated by transformation of Z96, DY100, Z106, and DY102 with pIMD2-S288C, respectively. Transformation of BY4741 and BY4437 (Research Genetics), and FY120 and FY1638 (15) with pRS426 yielded strains DY818, DY809, DY812, and DY815, respectively. Strains DY823, DY826, DY828, and DY829 were derived from BY4741, BY4436, FY120, and FY1638 by transformation with pIMD2-S288C, respectively. DY750 was generated by transformation of BY6035 (Research Genetics) with pRS316 (29).
Table I.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| ABGG10a | MATα ade1:Δ2 LEU2Δ4 HIS3 his5 leu2− |
| ABGG11a | MATα Δ4HIS3 trp1−ura3–1 his3–11,15 leu2–3,112 |
| ABGG12a | MATα Δ3LEU2 ade1−ura3–1 his3–11,15 leu2–3,112 |
| BY4741b | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4741–515b | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δimd4::kanMX4 |
| DY103c | MATα ura3–52 leu2–3, 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2 CEN)][pRS316 (URA3)] |
| DY700 | MATα ura3–52 leu2–3, 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2 CEN)][pPur5P800luc (URA3)] |
| DY704 | MATα ura3–52 leu2–3, 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PL4 (URA3)] |
| DY713 | MATα ura3–52 leu2–3, 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PL3 (URA3)] |
| DY714 | MATα ura3–52 leu2–3 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PL7 (URA3)] |
| DY715 | MATα ura3–52 leu2–3, 112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PL8 (URA3)] |
| DY731 | MATα Δ4HIS3 trp1−ura3–1 his3–11,15 leu2–3,112 [pRS316 (URA3)] |
| DY732 | MATα Δ3LEU2 ade1−ura3–1 his3–11,15 leu2–3,112 [pRS316 (URA3)] |
| DY741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pRS316 (URA3)] |
| DY743 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0Δimd4::kanMX4 [pRS316 (URA3)] |
| DY750 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0Δimd3::kanMX4 [pRS316] |
| DY787 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (RPB2 LEU2 CEN)][pRS426 (URA3 2μ)] |
| DY788 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP2–10L (rpb2–10 LEU2 CEN)][pRS426 (URA3 2μ)] |
| DY789 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP214 (RPB2 LEU2 CEN)][pRS426 (URA3 2μ)] |
| DY790 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)][pRS426 (URA3 2μ)] |
| DY809 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δrpb9::kanMX4 [pRS426 (URA3 2μ)] |
| DY812 | MATa his4–912δ, lys2–128δ, leu2Δ1, ura3–52 [pRS426 (URA3 2μ)] |
| DY815 | MATa his4–912δ, lys2–128δ, leu2Δ1, ura3–52, rpb1–221 [pRS426 (URA3 2μ)] |
| DY818 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pRS426 (URA3 2μ)] |
| DY823 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pIMD2-S288C (IMD2 URA3 2μ)] |
| DY826 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δrpb9::kanMX4 [pIMD2-S288C (IMD2 URA3 2μ)] |
| DY828 | MATa his4–912δ, lys2–128δ, leu2Δ1, ura3–52 [pIMD2-S288C (IMD2 URA3 2μ)] |
| DY829 | MATa his4–912δ, lys2–128δ, leu2Δ1, ura3–52, rpb1–221 [pIMD2-S288C (IMD2 URA3 2μ)] |
| DY835 | MATα Δ4HIS3 trp1-ura3–1 his3–11,15 leu2–3,112 [pIMD2-S288C (IMD2 URA3 2μ)] |
| DY836 | MATα Δ4HIS3 trp1-ura3–1 his3–11,15 leu2–3,112 [pIMD2-C335A (URA3 2μ)] |
| DY900 | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PGL1 (URA3)] |
| DY901 | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PGL2 (URA3)] |
| DY902 | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PGL3 (URA3)] |
| DY904 | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PGL5 (URA3)] |
| DY905 | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)][pIMD2-PGL4 (URA3)] |
| DY2010 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (RPB2 LEU2 CEN)][pGAL-LUC (CEN URA3 GAL1 promoter luciferase)] |
| DY2020 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (RPB2 LEU2 CEN)][pMT-Luc (CEN URA3 luciferase)] |
| DY2065 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (RPB2 LEU2 CEN)][pIMD2-S288C (IMD2 URA3 2μ)] |
| DY2066 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP214 (RPB2 LEU2 CEN)][pIMD2-S288C (IMD2 URA3 2μ)] |
| DY2067 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP2–10L (rpb2–10 LEU2 CEN)][pIMD2-S288C (IMD2 URA3 2μ)] |
| DY2068 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP2–10L (rpb2–10 LEU2 CEN)][pIMD2-S288C (IMD2 URA3 2μ)] |
| Z460d | MATa ura3–52 leu2–3,112 his3Δ200 his4–912 lys2–128 rpb1D187::HIS3 [pRP1–1U (CEN URA3 rpb1–1)] |
| Z96d | MATα ura3–52 leu2–3,112his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] |
Growth Assays
For solid medium growth assays, cells picked from a single colony were grown to saturation in 5 ml of synthetic complete medium lacking uracil (SCura−; Ref. 30). The cells were diluted to an initial A600 of 0.01 followed by five consecutive 1:10 dilutions in a 96-well plate. Ten μl of each dilution were spotted onto SCura− plates containing no drug or mycophenolic acid at the indicated concentrations. The plates were incubated at 30 °C for 3– 4 days. To measure GAL1 induction, 5-ml cultures were started from single colonies and grown to saturation at 30 °C in SCura− with raffinose (2% w/v). The cells were diluted to an A600 of 0.1 into fresh medium and grown to an A600 of 0.4 – 0.6. Mycophenolic acid (15 μg/ml) was added to the cultures where indicated. The cultures were grown for 30 min with or without drug at 30 °C, followed by the addition of galactose (2% w/v) to the medium. To test IMD2 mRNA decay, 5-ml cultures of Z460 were started from single colonies in SCura− and grown to saturation at 25 °C. The cells were diluted, grown to an A600 of 0.4 – 0.6, and induced with 6-azauracil (75 μg/ml) for 2 h at 25 °C. The cells were shifted to 37 °C for 5 min, and guanine (1 mM) was added to the medium where indicated.
Northern Analysis
Total RNA was isolated from thawed cell pellets by the hot phenol extraction method and quantitated by measuring absorbance at 260 nm (31). Total RNA (15 μg) was resolved on a 1% (w/v) agarose, 7% formaldehyde gel and blotted onto Zeta-probe GT nylon membrane (Bio-Rad). Filters were baked at 80 °C for 2 h and then prehybridized for a minimum of 3 h at 42 °C in 5× SSC (1× SSC = 0.15 M NaCl, 0.015 sodium citrate), 5× Denhardt’s solution (31), 50% (v/v) formamide, 1% (w/v) SDS, and 100 μg/ml salmon sperm DNA. Filters were hybridized under the same conditions with ~108 cpm of 32P-labeled DNA probe for 15–18 h. The filters were washed twice at 22 °C in 2× SSC/0.1% SDS, for 5 min each and twice in 0.2% SSC, 0.1% SDS for 5 min each, followed by two 0.2%SSC, 0.1% SDS washes at 42 °C for 20 min each. The washed filters were exposed to X-Omat film and quantitated with a Fuji BAS1000 imaging system. The IMD2 probe was prepared using PCR with a wild-type yeast genomic DNA template (S288C; Research Genetics) and the oligonucleotides 5′-GTGGTATGTTGGCCGGTACTACCG-3′ and 5′-TCAGTTATGTAAACGCTTTTCGTA-3′. The GAL1 probe was a BamHI-HindIII fragment of GAL1 digested from pUC119-GAL1. ACT1 probes were made by PCR using S288C genomic DNA as template and primers 5′-GAACTTCCAGATGGTCAAGTC-3′ and 5′-TGGAAGATGGAGCCAAAGCG-3′. The luciferase probe was a PCR product generated from pGAL-Luc using 5′-TTCCATCTTCCAGGGATACG-3′ and 5′-TCGCGGTTGTTACTTGCATG-3′. Probes were labeled to a specific activity of ~107–108 cpm/μg with Klenow DNA polymerase (Promega, Madison, WI), random hexamer primers (Life Technologies, Inc.), and [α-32P]dATP (Amersham Pharmacia Biotech).
Primer Extension Analysis
Approximately 1.5 pmol of 5′-32P-labeled oligonucleotide (≈106 cpm/pmol; 5′-TTGGTAAAGTCTAGTGCGGTCTT-3′ for Fig. 7 and 5′-GCCTTATGCAGTTGCTCTCC-3′ for Fig. 9) were annealed to 10 μg of total RNA for 1 h at 50 °C in 2 mM Tris, pH 8, 0.2 mM EDTA, and 250 mM KCl. The primer was extended with 100 units of Superscript II™ RNase H− reverse transcriptase (Life Technologies, Inc.) at 47 °C for 1 h in 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl, 1 mM dithiothreitol, and 50 μM of each dNTP. The samples were ethanol-precipitated, resuspended in 20 μl of 90 mM Tris borate, pH 8, 2.5 mM EDTA, 80% (v/v) formamide, heated to 95 °C, and resolved on an 8% polyacrylamide gel.
Fig. 7. Primer extension mapping of the transcription start site of chromosomal IMD2.
DY103 was treated with 6-azauracil (75 μg/ml) for the indicated times, and RNA was isolated for primer extension analysis (left panel). The position of labeled 200- and 100-base DNA reference markers are indicated. Total RNA was isolated from the IMD deletion strains ABG-G10, DY731, DY732, and DY743, and two amounts of RNA (10 or 25 μg) were added to a primer extension assay as indicated by the triangles above the lanes. The lane marked tRNA is a negative control in which total yeast RNA was omitted.
Fig. 9. Demonstration of two functional promoter elements in IMD2.
A, strains DY704, DY713, DY714, and DY715 containing the indicated reporter constructs were treated with 75 μg/ml 6-azauracil for 0, 1, 2, or 3 h. RNA was analyzed by Northern blotting with a probe complementary to luciferase sequences. B, the Northern blot signals in A were quantified using a PhosphorImager and plotted. C, the 3-h time point RNAs analyzed by Northern blotting in A were used for primer extension analysis. The sample marked − was from a strain with the plasmid pMT-Luc that lacks any IMD2 sequence upstream of luciferase. The top arrow indicates the extension product representing the normal +1 site. The bottom arrow indicates the major extension product seen when the repressive element is removed. As a negative control, tRNA was used in lieu of yeast total RNA.
RESULTS
The IMD Family of Genes and Drug Sensitivity
Individual deletions of each of the four Saccharomyces cerevisiae IMD genes reveals that none are essential for viability (32, 33). Recently, it has been shown that treatment of yeast with IMP dehydrogenase inhibitors including 6-azauracil and mycophenolic acid results in the induction of IMD2 (10, 11). An earlier report indicated that overexpression of an IMP dehydrogenase gene resulted in mycophenolic acid resistance in Candida albicans (34). This led to the expectation that the deletion of IMD2 or one or more other IMD genes in S. cerevisiae would result in the sensitivity of yeast growth to IMP dehydrogenase antagonists. Indeed, yeast lacking IMD2, but not strains lacking IMD1, IMD3, or IMD4, were unable to grow in the presence of mycophenolic acid (Fig. 1A). Transforming the IMD2-deleted strain with a copy of the gene restored growth in the presence of the drug (Fig. 1B). Catalysis by IMP dehydrogenase involves the formation of a covalent adduct between the substrate and an active site cysteine (35) that is completely conserved among over 30 IMP dehydrogenase enzymes across phyla and is represented by cysteine 335 in S. cerevisiae IMD2. Removal of this thiol by mutagenesis inactivates the activity of the Tritrichomonas foetus enzyme (36). We created a cysteine to alanine substitution at position 335 of IMD2 to test the prediction that the enzymatic activity of Imd2p was important for conferring mycophenolic acid resistance upon yeast. Indeed, IMD2 with the C335A substitution was unable to complement the drug-sensitive phenotype of an IMD2 deletion strain (Fig. 1B).
Fig. 1. Growth of yeast deleted for IMD genes in the presence of mycophenolic acid.
A, yeast strains DY741 (WT), DY732 (ΔIMD1), DY731 (ΔIMD2), DY750 (ΔIMD3), DY743 (ΔIMD4), and ABG-G10 (ΔIMD1 ΔIMD2) were streaked onto SCura− medium lacking or containing 7.5 μg/ml mycophenolic acid and incubated at 30 °C for 3 days. B, yeast strains DY731 (−), DY835 (+pIMD2), and DY836 (+pC335A) were grown to saturation and diluted to an A600 of 0.01. Ten μl of this and serial 10-fold dilutions thereof were plated onto SCura− medium lacking or containing 30 μg/ml mycophenolic acid and grown at 30 °C for 4 days.
Expression of IMD2 in Elongation Mutants of Yeast
Mutations in elements of the transcription elongation machinery result in drug sensitivity and a failure to induce IMD2 (10). To test whether this was the cause of drug sensitivity, we examined whether introducing into cells a plasmid containing the intact IMD2 gene driven by its native regulatory sequences could suppress the drug-sensitive phenotype. A 2μ plasmid containing 500 bp upstream and 150 bp downstream of the IMD2 open reading frame was transformed into each of four previously characterized strains bearing mutations in the elongation machinery. The cognate isogenic wild-type strains were similarly transformed. The four mutations included: a disruption of DST1 (SII), a deletion of RPB9 (the gene encoding the ninth subunit of RNA polymerase II), a point mutation in the second largest subunit of RNA polymerase II called rpb2-10, and, a point mutation in the largest subunit of RNA polymerase II, called rpb1-221. Strains harboring each of these mutations are sensitive to both 6AU and mycophenolic acid (Refs. 14, 15, 18, 20, and 22 and see below). There is biochemical evidence implicating each in the process of elongation by RNA polymerase II. Elongation by yeast RNA polymerase II in vitro is stimulated by the yeast SII protein (37). Δrpb9 results in an RNA polymerase II enzyme that has altered arrest properties and altered responsiveness to SII (22, 38). The rpb2-10 mutation leads to an arrest-prone RNA polymerase II enzyme with a slowed average elongation rate (18). The rpb1-221 mutation shows a genetic interaction with spt5, whose gene product has both a positive and negative effect upon elongation in vitro (15, 39). All four of these mutant strains are defective in the induction of IMD2 in response to drug challenge (Ref. 10 and see below). In each case, the growth sensitivity of the mutants in the presence of mycophenolic acid was suppressed when the cells harbored a 2μ plasmid containing the IMD2 gene (Fig. 2). Growth of the cognate wild-type strain was also modestly improved on drug-containing medium.
Fig. 2. Growth of elongation mutants containing or lacking IMD2 on a 2μ plasmid in the absence and the presence of mycophenolic acid.
Yeast strains containing vector (pRS426) or pIMD2-S288C were grown to saturation and diluted to an A600 of 0.01. Ten μl of this and 10-fold serial dilutions thereof were spotted onto SCura− medium lacking or containing 15 μg/ml (rpb9 and rpb1-221 plates) or 30 μg/ml mycophenolic acid.
Yeast strains with the dst1, Δrpb9, rpb2-10, or rpb1-221 mutations have a reduced ability to induce transcription of GAL1 (Ref. 40 and see below). We asked whether providing IMD2 on a 2μ plasmid could suppress this transcriptional defect. The strains disrupted for DST1 (the SII encoding gene) or containing the rpb2-10 (“slow” polymerase) mutation were relatively mildly affected for GAL1 induction in either the presence or the absence of drug (Fig. 3A, pRS426; charted in Fig. 3B). In these strains, high copy IMD2 improved the efficacy of GAL1 induction in both the presence and the absence of drug (Fig. 3, compare pRS426 with pIMD2 in the NO DRUG or MYCOPHENOLIC ACID panels). For the more severely affected strains, including the rpb1-221 mutation (Fig. 4), Δrpb9 (Fig. 5), and the dst1, rpb2-10 double mutant (Fig. 3), providing IMD2 in high copy was only slightly effective or unable to rescue the GAL1 response in either the presence or the absence of drug. Curiously, we detected a reproducible stimulation of GAL1 induction for wild-type strains when they received IMD2 on this high copy plasmid (Figs. 3–5).
Fig. 3. Induction of GAL1 by elongation mutants (rpb2-10, dst1, and rpb2-10 dst1) and control strains containing or lacking an IMD2–2μ plasmid.
A, yeast strains with vector (pRS426) or pIMD2-S288C were treated with mycophenolic acid (15 μg/ml) or mock treated and challenged with galactose for 0, 0.5, or 2 h. RNA was isolated and analyzed by Northern blotting using a probe for GAL1 mRNA. B, the Northern blots in A were quantified using a PhosphorImager and charted. wt, wild type.
Fig. 4. Induction of GAL1 in rpb1-221 and control strains containing or lacking an IMD2–2μ plasmid.
A, yeast strains containing vector (pRS426) or pIMD2-S288C were treated with mycophenolic acid (15 μg/ml) or mock treated and challenged with galactose for the indicated times. RNA was isolated and analyzed by Northern blotting using a probe for GAL1 mRNA. B, the Northern blots in A were quantified using a PhosphorImager and charted.
Fig. 5. Induction of GAL1 in Δrpb9 and control strains containing or lacking an IMD2–2μ plasmid.
A, yeast strains containing vector (pRS426) or pIMD2-S288C were treated with mycophenolic acid (15 μg/ml) or mock treated and challenged with galactose for the indicated times. RNA was isolated and analyzed by Northern blotting using a probe for GAL1 mRNA. B, the Northern blots in A were quantified using a PhosphorImager and charted.
Because these mutants have transcription defects, it was important to gauge the extent to which IMD2 expression itself had increased as a function of the 2μ-IMD2 plasmid. In Fig. 6, we probed the identical RNA samples employed in Figs. 3–5 for IMD2 mRNA (Fig. 6). Quantitation of the blots revealed that although the mutants generally produced less IMD2 mRNA than wild-type cells, the presence of the 2μ-IMD2 plasmid still resulted in IMD2 mRNA levels in the mutant strains that were at least 10-fold higher at any time point than the natural level of IMD2 mRNA in wild-type cells in the absence of drug (Fig. 6). On these brief autoradiographic exposures, the endogenous level of uninduced mRNA is not apparent. (e.g. left portion of Fig. 6A). The IMD2-containing plasmid generates a doublet of IMD2 transcripts, including the native mRNA (lower band), and a larger transcript that probably results from the use of a secondary plasmid-derived polyadenylation site downstream of IMD2 sequences. Quantitation of these blots was confined to the lower band corresponding to the size of the natural transcript and hence represents a minimal estimate of overexpression. In the presence of drug, the dst1 rpb2-10 mutant with the 2μ-IMD2 plasmid contained 6-, 3-, and 1.2-fold more IMD2 mRNA than the natural level found in wild-type cells at the 0-, 0.5-, and 2-h time points, respectively (Fig. 6A). Similarly, the untreated rpb1-221 mutant with the high copy plasmid generated IMD2 transcript levels that were comparable with those found in wild-type cells with the plasmid and that were 4 –7-fold higher than wild-type cells lacking the plasmid (Fig. 6B). Depending upon the time point examined, the drug-treated rpb1-221 strain had at least as much, and up to 3-fold more, IMD2 transcript than treated wild-type cells lacking the plasmid (Fig. 6B). With or without drug exposure, the Δrpb9 strain with the 2μ-IMD2 plasmid accumulated 2–3-fold more transcript than its cognate wild-type control strain lacking the plasmid (Fig. 6B). In summary, although the mutant strains showed the expected reduced capacity to transcribe IMD2 when they harbored the 2μ plasmid, they consistently possessed at least as much, and usually more, than the natural level of transcript found in wild-type cells because of the additional copies of the gene. Any inability of the elongation mutants to induce GAL1 cannot be due to their incapacity to generate wild-type levels of IMP dehydrogenase mRNA from the additional copies of the gene provided by the plasmid.
Fig. 6. IMD2 mRNA levels in mutant and control strains containing or lacking an IMD2–2μ plasmid and grown in the presence or the absence of mycophenolic acid.
A, 15 μg of RNA obtained from the cells analyzed in Fig. 3 were analyzed in independent Northern blots using a probe complementary to IMD2. B, same as in A but with RNA from cells analyzed in Figs. 4 and 5. wt, wild type.
Regulation of IMD2 Expression
Of the four IMP dehydrogenase genes in S. cerevisiae, only IMD2 is strongly induced when GTP pools are compromised by drug treatment (10, 11). As an initial step toward characterizing the regulation of this gene, we mapped the transcription start site using primer extension analysis (Fig. 7). To ensure that IMD2 transcripts and not other family members were being examined, we isolated RNA from wild-type cells or cells deleted for different IMD genes and from cells grown in the presence or the absence of 6AU to examine the inducibility of any primer extension products we detected. Indeed, a 6AU-inducible extension product was detected in cells containing, but not lacking, the IMD2 gene (Fig. 7). The size of this product placed the major transcription start site (+1) at an adenine 104 bp upstream of the start codon of the open reading frame.
Yeast DNA upstream of the IMD2 open reading frame and extending to the next open reading frame on chromosome VIII (PHO12) drives 6AU-inducible expression of a luciferase reporter (10). To identify promoter elements involved in the drug-inducible transcription of IMD2, we generated reporter plasmids containing ≈700, 400, 300, or 170 bp of DNA upstream of +1 linked to firefly luciferase (Fig. 8). Drug-inducible luciferase activity was detected for the constructs containing 700, 400, and 300 bp of upstream sequence but was lost when only 170 bp of upstream DNA were used (data not shown). Northern blot analysis with a luciferase probe confirmed that −300 to +104 IMD2 DNA could drive inducible transcription but that the −170 to +104 region could not (Fig. 9A; plotted with squares in Fig. 9B). Thus, the cellular guanine nucleotide-sensing machinery ultimately operates through a response element in the promoter.
Fig. 8. Schematic depiction of the IMD2 promoter.
The open boxes depict IMD2 sequences annotated using the transcription start site as +1 (bent arrow). The shaded boxes indicate the luciferase open reading frames. The large and small cross-hatched boxes illustrate the positions of the drug-responsive and the repressive elements described in the text, respectively. The strain names and the plasmids they contains are to the left of the diagram, and the IMD2 sequences they contain are indicated in parentheses.
Deletions into the IMD2 promoter from its 3′ side led to the unexpected finding of a repressive element in the proximal promoter region of IMD2. Removing IMD2 sequences from −32 to +104 (Fig. 8) resulted in a large increase in the uninduced level of luciferase activity (data not shown) and transcript (Fig. 9A, compare zero time point for −300 to −104 with that for −300 to −32; Fig. 9B, plotted with solid symbols). Nevertheless, the construct remained 6AU-inducible (Fig. 9, −300 to −32). Indeed, the maximal extent of induction was significantly higher in the construct lacking the repressive sequences as long as the response element was also present (Fig. 9, −300 to −32 versus −300 to −104). Because the repressive region between −32 and +104 contains the transcription start site, we tested whether it was involved in positioning the transcription start site by primer extension analysis. Removal of this ≈140-bp region resulted in a new major start site (Fig. 9C, lower arrow) that migrated into luciferase DNA by a distance commensurate with the deletion size, suggesting that the TATA box or other determinants of start site selection were not within the deleted region. There was some loss of positioning activity as a number of new weaker start sites also became detectable (Fig. 9C, −300 to −32 lane). We conclude that TATA box function has been spared in this deletion derivative based upon the presence of a preferred start site and the ability of the remaining sequences to support high levels of uninduced (constitutive) and activated transcription.
To determine whether the sequences between −31 and +104 were sufficient for the repressive function and to test whether the element functioned autonomously, we placed it both upstream and downstream of the inducible GAL1 promoter in a luciferase reporter plasmid (Fig. 10). Luciferase enzyme activity (data not shown) and Northern blot analysis (Fig. 10) showed that induction of the GAL1 promoter was strongly compromised when −31 to +104 of IMD2 was placed downstream of the GAL1 promoter (Fig. 10, E versus F). The repressive element was inactive when situated upstream of the GAL1 promoter (Fig. 10, A versus F). A smaller sequence representing −31 to +35 of IMD2 was as active as −31 to +104 (Fig. 10, C versus E), from which we conclude that the repressive function is contained within a 66-bp region centered around the transcription start site. Repression was strictly orientation-dependent for both fragments (Fig. 10, B versus C and D versus E).
Fig. 10. The repressive element from IMD2 functions in a heterologus context.
The indicated IMD2 sequences, indexed relative to +1, were inserted into the indicated GAL1-luciferase reporter plasmid and introduced into yeast strains DY902 (A), DY904 (B), DY905 (C), DY901 (D), DY900 (E), and DY2010 (F). The cells were grown in raffinose medium and challenged with galactose (2% w/v) for the indicated times. RNA was harvested and analyzed by Northern blotting with a probe that detected luciferase sequences.
Guanine and Growth Regulation of IMD2
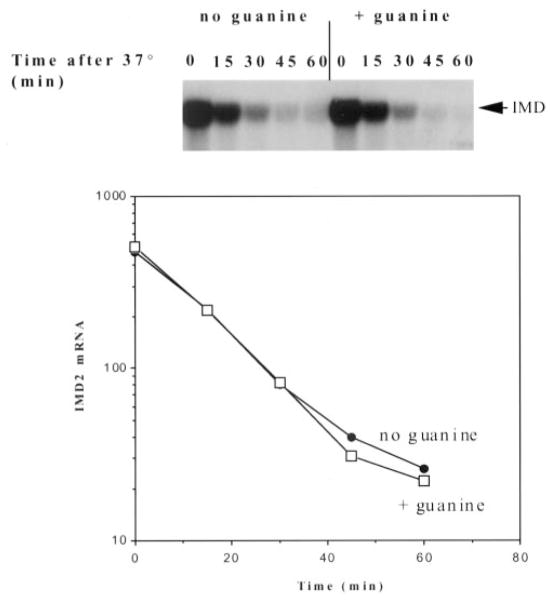
Extracellular guanine can be imported and used by a salvage pathway to synthesize guanine nucleotides. Hence, the presence of guanine in the growth medium obviates IMD2 induction in the presence of IMP dehydrogenase inhibitors (10, 11, 41). Because repression of mammalian IMD2 transcription has been reported to operate at a post-transcriptional level, we tested whether guanine altered the half-life of IMD2 mRNA (8). We employed a temperature-sensitive strain of yeast with a mutation (rpb1-1) in the largest subunit of RNA polymerase II that, upon shift to the nonpermissive temperature, rapidly ceases transcription (42). To facilitate the analysis, cells were induced with 6AU at the permissive temperature to obtain high levels of IMD2 mRNA (Fig. 11, zero time points). Transcript decay in the presence and the absence of guanine was monitored after transcription was shut off by shifting the cells to the nonpermissive temperature (Fig. 11). The presence of guanine did not influence the decay rate of the IMD2 transcript, showing that guanine does not down-regulate IMD2 at the post-transcriptional level in yeast.
Fig. 11. IMD2 mRNA decay in the presence and absence of guanine.
The yeast strain Z460 containing the rpb1-1 mutation was grown at 25 °C to an A600 of 0.5 in SCura−, challenged with 75 μg/ml 6-azauracil for 2 h, and shifted to 37 °C for the indicated times. RNA was harvested and analyzed by Northern blotting using a probe complementary to IMD2. The signals were quantified using a PhosphorImager and plotted.
In a number of mammalian cell types, particularly tumor cells and T lymphocytes, IMP dehydrogenase activity is positively correlated with cell growth rate (2, 43– 46). To test whether this was the case for S. cerevisiae, we grew cultures of wild-type cells that either were permitted to approach the diauxic shift (Fig. 12A, UNDILUTED) or were reseeded into fresh medium such that they maintained an optical density (600 nm) of ≈0.5 and a logarithmic growth rate (Fig. 12A, DILUTED). In contrast to the reseeded cells, IMD2 mRNA is lost from cells approaching diauxie with a half-life of ≈1.5 h (Fig. 12, B and D). This is in contrast to ACT1 mRNA, which maintains a fairly constant level under both growth conditions (Fig. 12, B and C). Hence, the IMD2 transcript level is sensitive to the physiological state of the yeast cell.
Fig. 12. IMD2 mRNA abundance is regulated as a function of growth.
A, strain DY103 was grown to saturation in SCura−, diluted to an A600 of ≈0.5, and split into two equal cultures that were incubated at 30 °C. One was diluted every hour into fresh medium (diluted). The other was allowed to grow to diauxie (undiluted). A600 was measured and plotted. B, RNA was prepared from aliquots of cells removed at the indicated times and analyzed by Northern blotting with probes against IMD2 and ACT1. The ACT1 and IMD2 signals were quantified using a PhosphorImager and plotted in C and D, respectively.
DISCUSSION
To understand the basis of the drug sensitivity conferred by mutations in the transcription elongation machinery, we have explored the IMD family of genes in yeast that encode the class of enzyme targeted by these drugs. Loss of IMD2 is sufficient to manifest mycophenolic acid sensitivity. Overexpression of IMD2 suppressed to different extents two phenotypes of S. cerevisiae elongation mutants: sensitivity of growth to inhibitors of IMP dehydrogenase and transcription of an inducible gene. This analysis suggests that these mutations have additional biologically significant consequences apart from compromising the IMD2 transcriptional response, although loss of the ability to up-regulate IMPDH is an important part of the phenotype. Transcriptional control was attributable to two separate sequence elements in the promoter. The guanine-mediated repression of IMD2 observed previously (10, 11) is not due to a change in mRNA half-life. Hence, regulation is not likely to operate at a post-transcriptional level as seen in mammalian cells (8). These data emphasize that IMD2 is a privileged member of a four member gene family in yeast with a major role in sensing and responding to nucleotide levels and growth rate.
Deletion of IMD2, one of the four genes encoding very similar IMP dehydrogenases, rendered cells sensitive to mycophenolic acid, proving that loss of the expression of this gene is sufficient to obtain drug sensitivity. Only loss of IMD2 conferred this phenotype, indicating that it contributes to growth in the presence of drug to a larger extent than the other three IMD genes and that it is uniquely regulated among the family members. This is consistent with the finding that IMD2 is the most responsive to drug treatment and guanine repression (10, 11). Presumably, loss of all four IMD genes would result in guanine auxotrophy; however, the quadruple IMD deletant (or a triple deletant) has not yet been described.
It has been suggested that yeast with mutations in the elongation machinery display sensitivity to drugs that inhibit IMP dehydrogenase because of the pharmacological reduction of GTP, a critical substrate for RNA polymerase II (9, 20). Reduced GTP levels would be expected to reduce elongation rates, increase the potential for RNA polymerase II arrest, and precipitate a stronger requirement for elongation factors (9, 20). With the identification of the inductive response of IMD2, it became important to examine whether the drug sensitivity was due solely to the inability of the elongation mutants to respond to drug treatment via the transcriptional up-regulation of IMD2. Alternatively, transcriptional defects in the presence of drug could result from an indirect and general effect upon RNA synthesis of limiting GTP levels. The data presented here argue that both are the case.
Our approach has been to bypass the inability of elongation mutants to induce IMD2 by providing the gene on a high copy plasmid. Strains with mild drug sensitivity and reduced transcription, as monitored by GAL1 induction, showed improved growth on mycophenolic acid when transformed with a high copy plasmid containing IMD2, suggesting that a boost in GTP levels might resolve transcriptional problems. On the other hand, for more severely affected mutants, high copy expression of IMD2 incompletely suppressed drug sensitivity and reversed GAL1 induction little, if at all. This suggests that intrinsic elongation defects persist even when levels of IMD2 mRNA, and presumably GTP, are present at levels sufficient for normal growth. Thus, this class of mutations would appear to result in a defect that is independent of the IMD2 induction deficit. Presumably this involves transcription elongation on genes other than IMD2.
While our work was ongoing, the mapping of a guanine response element (GRE) in the IMD2 promoter was reported (11). Our findings are in good general agreement with those of Escobar-Henriques and Daignan-Fornier (11) with one exception. These investigators found that removal or mutation of the GRE led to a loss of induction. This resulted from an increase in IMD2 basal transcription leading to mRNA levels comparable to that derived from the native promoter following mycophenolic acid induction. We found that removal of this region resulted in the loss of drug inducibility but did not elevate basal transcription. Instead, removal of a separate sequence downstream from the GRE derepressed the basal (uninduced or constitutive) level of transcription of IMD2. It is possible that derepression by guanine nucleotide depletion operates through this element. Interestingly, the derepressed promoter remained inducible in a GRE-dependent manner, indicating that an additional mechanism can activate transcription. Thus, in our assays, the GRE played a positive role and a second element near the transcription start site played a negative one. Differences in the reporter assays used by Escobar-Henriques and Daignan-Fornier (11) and us may account for this disparity. The function of the repressive sequence was portable to a heterologus inducible promoter and showed strict orientation dependence. Although the repressive sequence contains a good match to a consensus TATA box (TATAA), the TATAA sequence is immediately adjacent to the transcription start site, a spacing that makes it an unlikely candidate for TBP binding and traditional TATA box function. The region does not function as a TATA box because its removal leads to elevated basal activity. Furthermore, inducible promoter function remained intact in its absence. Also, substantial start site positioning activity can be attributed to the remaining promoter sequences after deletion of the repressive element. It is unclear how either promoter element functions or what proteins might be their cognate ligands. Bas1p and Bas2p, transcription factors that regulate adenine metabolism, are not involved in IMD2 induction, nor are the transcription factors Swi5p, Hap3p, or Gcn4p, for which consensus binding sites are present in the promoter (11).2 The −31 to +35 region is highly conserved between IMD1 and IMD2 with only four nucleotide differences. IMD1 is thought to be poorly expressed because of its proximity to the telomere of chromosome I (32). The repressive element is less well conserved between IMD2 and IMD3 (63% identity) and is essentially unrecognizable in the IMD4 promoter.
The extent to which the IMD1-IMD4 gene products contribute to cellular IMPDH activity is unknown. IMD2 is the dominant IMD mRNA species under induced and uninduced conditions based upon experiments in IMD deletion strains (10, 32). This suggests that it is the major contributor to enzyme abundance and activity in vivo. However, determinations of enzyme levels or a direct measurement of IMPDH activity has not been made in such strains.
Our analysis of GAL1 induction led to the unexpected observation that wild-type cells showed an increased ability to induce GAL1 when they harbored additional copies of IMD2. This suggests that IMP dehydrogenase activity and GTP levels might be rate-limiting for RNA synthesis under normal vegetative growth conditions and is interesting in light of the correlation between IMP dehydrogenase activity and mammalian cell proliferation including tumor cell growth. Unlike logarithmically growing cells, cultures allowed to approach saturation showed a precipitous loss of the IMD2 mRNA. Thus, IMD2 transcription appears to be sensitive to the growth state of yeast. The mechanism by which this occurs is unclear but is observed in a culture long before the general decline in transcription and mRNA levels seen in post-diauxic and stationary phase yeast cultures (47). We considered the possibility that IMD2 overexpression could accelerate the rate of cell growth. However, the presence of high copy plasmid-borne IMD2 was not sufficient to increase the growth rate of wild-type yeast.3 Further analysis will be required to determine the mechanism by which cellular growth state is related to IMD2 transcription.
Acknowledgments
We thank Nancy Hannett, Dr. Richard Young, and Dr. Scott Devine for yeast strains and Dr. Avrom Caplan for plasmid DNA. We are also grateful to Drs. R. Kahn, A. Corbett, and J. Boss for comments and a critical reading of this manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM46331.
The abbreviations used are: IMPDH, IMP dehydrogenase; 6AU, 6-azauracil; PCR, polymerase chain reaction; bp, base pair(s); GRE, guanine response element.
R. Shaw, unpublished results.
J. Wilson, unpublished results.
References
- 1.Collart F, Huberman E. Blood. 1990;75:570–576. [PubMed] [Google Scholar]
- 2.Huberman E, Glesne D, Collart F. Purine and Pyrimidine Metabolism in Man VIII. In: Sahota A, Taylor M, editors. Plenum Press; New York: 1995. pp. 741–746. [Google Scholar]
- 3.Zimmermann AG, Gu JJ, Laliberte J, Mitchell BS. Prog Nucleic Acids Res Mol Biol. 1998;61:181–209. doi: 10.1016/s0079-6603(08)60827-2. [DOI] [PubMed] [Google Scholar]
- 4.Franchetti P, Grifantini M. Curr Med Chem. 1999;6:599–614. [PubMed] [Google Scholar]
- 5.Sintchak MD, Fleming MA, Futer O, Raybuck SA, Chambers SP, Caron PR, Murcko MA, Wilson KP. Cell. 1996;85:921–930. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann AG, Spychala J, Mitchell BS. J Biol Chem. 1995;270:6808–6814. doi: 10.1074/jbc.270.12.6808. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann AG, Wright KL, Ting JP, Mitchell BS. J Biol Chem. 1997;272:22913–22923. doi: 10.1074/jbc.272.36.22913. [DOI] [PubMed] [Google Scholar]
- 8.Glesne DA, Collart FR, Huberman E. Mol Cell Biol. 1991;11:5417–5425. doi: 10.1128/mcb.11.11.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exinger F, Lacroute F. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 10.Shaw RJ, Reines D. Mol Cell Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Escobar-Henriques M, Daignan-Fornier B. J Biol Chem. 2001;276:1523–1530. doi: 10.1074/jbc.M007926200. [DOI] [PubMed] [Google Scholar]
- 12.Uptain SM, Kane CM, Chamberlin MJ. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 13.Wind M, Reines D. BioEssays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 15.Hartzog GA, Wada T, Handa H, Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 17.Lennon JC, III, Wind M, Saunders L, Hock MB, Reines D. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell W, Reines D. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Awrey DE, Edwards AM, Archambault J, Friesen JD. Proc Natl Acad Sci U S A. 1996;93:1152–1157. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archambault J, Lacroute F, Ruet A, Friesen JD. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costa PJ, Arndt KM. Genetics. 2000;156:535–547. doi: 10.1093/genetics/156.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemming SA, Jansma DB, Macgregor PF, Goryachev A, Friesen JD, Edwards AM. J Biol Chem. 2000;275:35506–355011. doi: 10.1074/jbc.M004721200. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. Mol Cell Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davie JK, Kane CM. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimoraiso M, Nakanishi T, Kubo T, Natori S. J Biol Chem. 2000;275:29623–29627. doi: 10.1074/jbc.M910371199. [DOI] [PubMed] [Google Scholar]
- 26.Brodsky JL, Lawrence JG, Caplan AJ. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- 27.Gietz D, St Jean A, Woods RA, Schiestl RH. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Gene (Amst ) 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 29.Sikorski R, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology, Appendix 2. Greene Publishing Associates/Wiley-Interscience; New York: 1988. [Google Scholar]
- 32.Barton A, Bussey H, Storms R, Kaback D. Yeast. 1997;13:1251–1263. doi: 10.1002/(sici)1097-0061(199710)13:13<1251::aid-yea174>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 34.Kohler GA, White TC, Agabian N. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huete-Perez JA, Wu JC, Whitby FG, Wang CC. Biochemistry. 1995;34:13889–94. doi: 10.1021/bi00042a021. [DOI] [PubMed] [Google Scholar]
- 36.Digits JA, Hedstrom L. Biochemistry. 1999;38:2295–2306. doi: 10.1021/bi982305k. [DOI] [PubMed] [Google Scholar]
- 37.Christie KR, Awrey DE, Edwards AM, Kane CM. J Biol Chem. 1994;269:936–943. [PubMed] [Google Scholar]
- 38.Awrey DE, Weilbaecher RG, Hemming SA, Orlicky SM, Kane CM, Edwards AM. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 39.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wind-Rotolo M, Reines D. J Biol Chem. 2001;276:11531–11538. doi: 10.1074/jbc.M011322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lecoq K, Konrad M, Daignan-Fornier B. Genetics. 2000;156:953–961. doi: 10.1093/genetics/156.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson RC, Weber G, Morris HP. Nature. 1975;256:331–333. doi: 10.1038/256331a0. [DOI] [PubMed] [Google Scholar]
- 44.Dayton JS, Lindsten T, Thompson CB, Mitchell BS. J Immunol. 1994;152:984–991. [PubMed] [Google Scholar]
- 45.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. J Biol Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 46.Liu Y, Bohn SA, Sherley JL. Mol Biol Cell. 1998;9:15–28. doi: 10.1091/mbc.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werner-Washburne M, Braun E, Johnston GC, Singer RA. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]