Abstract
Over the past few years, biochemical and genetic studies have shed considerable light on the structure and function of the RNA polymerase II (pol II) elongation complex and the transcription factors that control it. Novel elongation factors have been identified and their mechanisms of action characterized in increasing detail; new insights into the biological roles of elongation factors have been gained from genetic studies of the regulation of mRNA synthesis in yeast; and intriguing links between the pol II elongation machinery and the pathways of DNA repair and recombination have emerged.
Introduction
Eukaryotic mRNA synthesis is catalyzed by the multisubunit RNA polymerase II (pol II) and proceeds through multiple stages designated pre-initiation, initiation, and elongation. Although substantial evidence argues that expression of most genes is controlled, at least in part, at the level of transcription elongation [1,2,3•,4,5], efforts to elucidate the mechanisms by which specific elongation factors regulate elongation by pol II in cells have been hampered by lack of suitable assays. Many transcriptional regulatory proteins, such as sequence-specific DNA binding transactivators and the general initiation factors, function through interactions with DNA sequences in the promoter-regulatory regions of genes. In contrast, most of the known elongation factors appear to expedite transcription either by promoting efficient elongation by pol II through chromatin (for example, FACT [facilitates chromatin transcription] and SWI/SNF family members [6•,7•]) or by targeting pol II directly and suppressing transient pausing (for example, TFIIF, Elongin, ELL, ELL2, CSB [Cockayne syndrome B] [8•], and Tat stimulatory factor 1 [Tat-SF-1] [9•]) or arrest (for example, SII, P-TEFb [positive transcription elongation factor b]) [4,5,6•–9•]. Because specific DNA sequences that mediate the activity of the known elongation factors have not been identified, development of DNA-sequence-based reporter assays for elongation factor activity in cells has not been possible. Thus, little information about how the known elongation factors contribute to regulation of transcription of specific genes is available. Nevertheless, ongoing biochemical studies are leading to the discovery of a growing list of potential elongation factors and genetic studies are beginning to provide substantive insights into the roles of these proteins in control of gene expression in cells. In addition, recent studies suggest that the pol II elongation complex interacts functionally with proteins involved in DNA repair and recombination and serves as a recruitment platform for factors involved in the capping, splicing, and polyadenylation of nascent transcripts (see reviews by Bentley, pp 347–351 and Minivielle-Sebastia and Keller pp 352–357, this volume).
This short article is a commentary on recent developments in biochemical and genetic studies on the mechanism and regulation of transcriptional elongation by pol II. Pol II elongation factors and mechanisms have recently been reviewed in detail [4,5]. Here, we do not attempt a comprehensive review of the literature but, instead, describe recent findings that are shaping our current view of the elongation stage of eukaryotic mRNA synthesis.
Emerging evidence for functions of specific elongation factors in cells
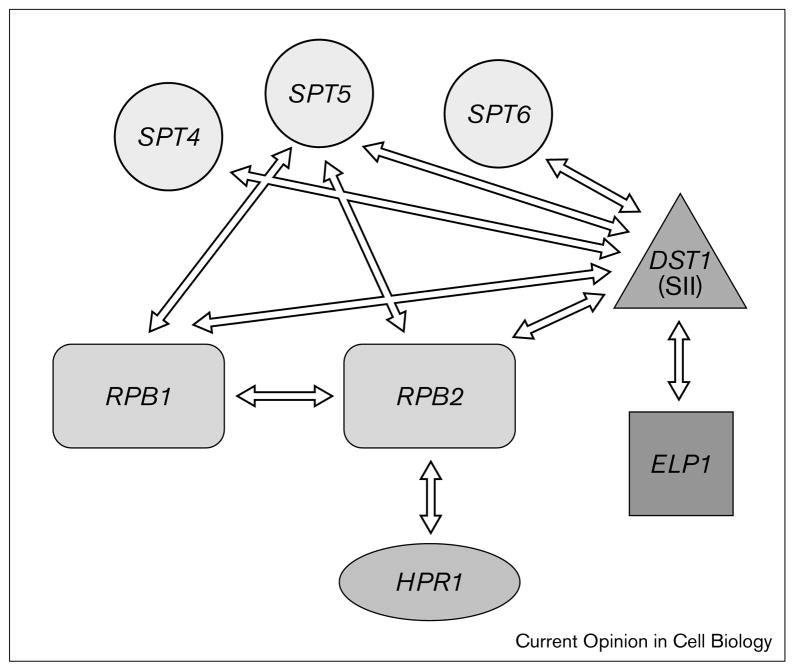
Over the past few years, studies on the roles of elongation factors in cells have taken advantage of the experimentally tractable simple eukaryote Saccharomyces cerevisiae and the HIV-1 viral model of Tat-dependent elongation. Tat is a sequence-specific RNA-binding protein encoded by HIV. Tat binds to the transactivation response (TAR) element at the 5′-end of HIV transcripts and functions together with cellular proteins to promote efficient elongation of HIV transcripts by pol II. These studies have brought to light genetic interactions among genes encoding subunits of pol II and elongation factors SII, elongator, and DRB sensitivity inducing factor (DSIF) (Spt4–Spt5) and have provided new insights into the roles of these and other elongation factors in gene expression (Figure 1).
Figure 1.
Genetic interactions between pol II and elongation factors in yeast. Genetic interaction (arrows) between pol II subunits (rectangles), elongation factors (square, triangle) and chromatin-related transcription factors (circle) that have been recently shown in S. cerevisiae are depicted. A factor involved in elongation and DNA recombination (HPR1, oval) has also been shown to interact with RNA pol II in yeast.
SII
SII was initially identified biochemically by its ability to promote synthesis of long transcripts by purified pol II [10]. SII enables pol II to transcribe through a variety of transcriptional impediments, including intrinsic arrest sites in DNA and nucleoprotein complexes. Mechanistic studies have shown that SII accomplishes this by interacting with arrested pol II and activating cleavage of the nascent transcript by a latent endoribonuclease intrinsic to the polymerase [11]. Following cleavage, pol II can re-extend the nascent transcript, allowing repeated attempts at elongation through arrest sites and, eventually, clearance of the impediment. In light of evidence that pol II arrests when the 3′-OH of the nascent transcript loses contact with the polymerase catalytic site, it has been proposed that SII-induced nascent transcript cleavage reactivates arrested pol II by realigning polymerase catalytic site residues with the 3′-OH of the nascent transcript [11].
The S. cerevisiae gene encoding SII (designated DST1 or PPR2) is not essential for viability; however, growth of yeast strains lacking SII is weakly sensitive to drugs such as 6-azauracil (6-AU) and mycophenolic acid, which are known to reduce intracellular NTP pools [12,13] and thus are expected to decrease overall rates of elongation by pol II in cells and to increase the likelihood that pol II will suffer arrest. Genetic evidence for a functional interaction between pol II and SII has come from several observations. Overexpression of SII rescues the drug sensitivity of a yeast strain harboring a pol II mutant that exhibits a decreased binding affinity for SII in vitro [12,14]. In addition, Archambault and coworkers [15] identified an allele of RPB1, encoding the largest pol II subunit, that confers synthetic lethality in yeast lacking SII. Finally, Lennon and coworkers [16•] recently found that combination of two mutations (deletion of DST1 and the pol II mutant rpb2-10), each of which confers weak 6-AU and mycophenolic acid sensitivity upon yeast, leads to a dramatic decrease in growth rate in the presence of drug. The rpb2-10 allele contains a point mutation that changes a conserved proline residue near the pol II catalytic pocket to serine. Pol II with the rpb2-10 mutation elongates transcripts more slowly in vitro and is more prone to arrest than wild-type pol II [17], suggesting that the phenotype of the double mutant may be due to an inability of the mutant pol II enzyme to overcome arrest induced by nucleosomes, by other DNA-bound proteins, or by intrinsic arrest sites in the DNA.
Evidence for a broad role for SII in regulation of transcription in vivo is provided by the observation that the levels of total poly A+ RNA — as well as of a number of specific transcripts — have been shown to decrease dramatically following treatment of the rpb2-10, dst1 deletion strain with 6-AU [16•]. It is not yet clear whether SII directly regulates expression of many genes or whether its primary targets are a few important transcriptional regulators. It is noteworthy, however, that the kinetics of reduction of steady state mRNA levels upon addition of drug to the rpb2-10, dst1 strain are very similar to those observed when yeast bearing temperature-sensitive alleles of genes encoding basal transcription factors or subunits of the pol II holoenzyme are shifted to the non-permissive temperature. The development of SII-deficient strains with severe phenotypes and the identification of potential target genes should provide a foundation for future studies on the role of SII in gene regulation in cells.
Elongator
Elongator is a three-subunit complex composed of polypeptides of ~150, ~90, and ~60 kDa. Elongator was purified from yeast chromatin preparations containing the elongating form of pol II. Evidence suggests that the interaction of elongator with pol II requires phosphorylation of the pol II carboxy-terminal domain (CTD) [18•,19•]. Like the gene encoding SII, the ELP1 gene, encoding the largest subunit of elongator, is not essential for yeast viability. Yeast lacking ELP1 are weakly sensitive to 6-AU but yeast lacking both DST1 and ELP1 are hypersensitive to the drug, suggesting a functional interaction between the two proteins. A variety of genes, including GAL1-10, PHO5, and INO2 — but not HIS3 and CHA1 — exhibit delayed activation in cells lacking the ELP1 gene [19•]. On the basis of these observations, together with previous work from several laboratories demonstrating that DNA binding transactivators can control the efficiency of elongation [1,2,3•], it has been proposed that Elp1 might function as a co-activator in post-initiation events [19•].
DSIF, P-TEFb and regulation of elongation by HIV Tat
The nucleotide analog DRB potently inhibits synthesis of cellular heterogeneous nuclear (hn)RNA in vivo and of long transcripts in crude and partially fractionated transcription systems [20,21]. In addition, expression of the Tat-activated HIV-1 polyprotein gene is exquisitely sensitive to DRB [22,23]. DRB inhibits RNA synthesis in these systems by inducing elongating pol II to arrest shortly after initiating transcription. Recent studies on the mechanism of DRB-sensitive elongation have led to the identification of a negatively-acting DRB-sensitivity inducing factor, DSIF [24•], and a positively-acting factor, P-TEFb, which counteracts DSIF activity.
Mammalian DSIF is composed of ~14 and ~160 kDa polypeptides, which are homologs of the S. cerevisiae Spt4 and Spt5 proteins, respectively [24•]. Early genetic experiments implicated Spt4 and Spt5, as well as a genetically related gene product, Spt6, in transcription and modification of chromatin in yeast [25]. Consistent with the idea that DSIF (Spt4–Spt5) functions as a negative elongation factor in cells, some conditional spt5 mutations are suppressed when combined with the elongation-defective pol II rpb2-10 mutant or when cells are grown in 6-AU [26•]. In addition, mutant alleles of SPT4, SPT5 and SPT6 confer 6-AU sensitivity on yeast, and combinations of mutations in SPT5 and RPB1, SPT5 and RPB2, SPT4 and DST1, SPT5 and DST1, and SPT6 and DST1 have been shown to produce synthetic phenotypes in yeast [26•] (Figure 1). Together with results implicating Spt4, Spt5, and Spt6 in chromatin modification, these results raise the possibility that DSIF and SII may help to enable pol II to transcribe through nucleosomes [26•].
The inhibitory effect of DSIF on productive elongation by pol II in vitro is antagonized by pTEFb [27•], a DRB-sensitive pol II CTD kinase composed of cdk9 (also known as PITALRE) and cyclin T [28•,29–32]. Phosphorylation of pol II CTD by P-TEFb appears to block functional interactions between DSIF and pol II, on the basis of findings indicating that a mutant P-TEFb lacking kinase activity is transcriptionally inactive, that the pol II CTD is essential for P-TEFb activity [27•,31,32], and that both DSIF transcription inhibitory activity and the binding of DSIF to pol II are blocked upon pol II CTD phosphorylation by P-TEFb [27•].
A possible explanation for the extreme DRB sensitivity of Tat-dependent elongation of the HIV-1 polyprotein transcript has come from studies implicating the pol II CTD kinase P-TEFb in this process. Tat-dependent transcription depends on the presence of an intact CTD [33–35], and immunodepletion experiments demonstrate that P-TEFb is required for Tat-dependent transcription in vitro [28•,36]. In addition, a high throughput screen for drugs that block Tat-dependent HIV-1 transcription led to the identification of compounds that specifically inhibit P-TEFb kinase activity [37•].
Genetic support for the idea that P-TEFb plays a critical role in Tat function comes from the observation that expression of human cyclin T1 rescues Tat function in rodent cells, which are normally refractory to HIV-1 infection and to Tat-dependent transcriptional activation [38•]. Human but not murine cyclin T1 is capable of forming a ternary complex with Tat and the transactivation response (TAR) element [38•,39•], suggesting that Tat may subvert normal cellular elongation control mechanisms by interacting with P-TEFb and recruiting it to the TAR element, thereby specifically activating elongation of the HIV-1 transcript.
Links between elongation and DNA repair and recombination
Substantial evidence links the process of transcription elongation to a variety of DNA transactions, including transcription-coupled DNA repair, somatic hypermutation of immunoglobulin genes, homologous DNA recombination, and maintenance of genome stability [40–44,45•]. Although the mechanisms linking these processes are poorly understood, proteins with roles in both transcription elongation and either DNA repair or recombination have been identified.
The gene encoding the mammalian CSB protein was initially identified by its requirement in transcription-coupled nucleotide excision repair of damaged DNA [46]. Recently, the CSB protein was found to function in vitro as an elongation factor that interacts directly with transcribing pol II [47] and stimulates the overall elongation rate [6•].
Mutations in the S. cerevisiae HPR1 and THO2 genes induce a hyper-recombination phenotype [48,49,50•]. Evidence that the Hpr1 and Tho2 proteins have roles in transcription by pol II has come from findings indicating that the hyper-recombination phenotype of HPR1 and THO2 mutant yeast strains can be suppressed by mutations in genes encoding pol II subunits, the mediator component SRB2, and initiation factor TFIIB [51] and further that the Hpr1 protein can be purified in association with a pol II-containing complex [52]. Evidence that the hyper-recombination phenotype is due to a defect in transcription elongation has come from several findings. These indicate, first, that the hyper-recombination phenotype of HPR1 and THO2 mutant yeast strains can also be suppressed by insertion of transcription termination signals upstream of recombinogenic regions of transcribed DNA [53•]. Second, that on the basis of the results of nuclear run-on experiments transcription elongation by pol II is impaired in yeast strains lacking HPR1 or THO2 [50•,53•], and third, that the hyper-recombination phenotype of HPR1 and THO2 mutant yeast strains is enhanced when cells are grown in 6-AU [45•]. On the basis of these observations, it has been proposed that the hyper-recombination phenotype of HPR1 or THO2 mutant strains results from recruitment of the recombination machinery to the vicinity of inappropriately paused elongation complexes [50•]. Whether paused pol II elongation complexes play a similar role in normal recombination events remains unclear; however, it is intriguing that somatic hypermutation of an artificial immunoglobulin gene is enhanced upon insertion of a DNA sequence predicted to induce transcriptional pausing [41].
Conclusions and prospects for the future
Over the past few years, our understanding of the mechanism and regulation of elongation by pol II has improved substantially. During this time, novel elongation factors have been identified, their mechanisms of action in vitro characterized in increasing detail and, in some cases, their cellular roles defined. In the future, we expect that investigations of transcription elongation will focus more and more on understanding the mechanisms by which pol II transcribes its natural template, chromatin. These studies are now well underway with the development of purified, reconstituted chromatin transcription systems and the discoveries of important roles for the SWI/SNF chromatin remodeling complex and for novel elongation factors such as FACT in transcription of nucleosomal templates by pol II in vitro [7•,8•,54–56]. It is likely that these new avenues of research will continue to gain momentum and will bring us closer to a mechanistic understanding of the elongation stage of eukaryotic mRNA synthesis.
Abbreviations
- 6-AU
6-azauracil
- CSB
Cockayne syndrome B
- CTD
carboxy-terminal domain
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole
- DSIF
DRB sensitivity inducing factor
- ELL
11–19 lysine-rich in leukemia
- FACT
facilitates chromatin transcription
- pol II
RNA polymerase II
- P-TEFb
positive transcription elongation factor b
- TAR
transactivation response
- Tat-SF1
Tat-stimulatory factor 1
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 2.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 3•.Brown SA, Weirich CS, Newton EM, Kingston RE. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. This paper presents evidence that some transcriptional activation domains have distinct subdomains responsible for their effects on transcription initiation and elongation. These findings provide important insight into the nature of DNA binding transactivators and how they function in the regulation of elongation by pol II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A, Conaway JW, Conaway RC. Mechanism and regulation of transcriptional elongation and termination by RNA polymerase II. Curr Opin Genet Dev. 1997;7:199–204. doi: 10.1016/s0959-437x(97)80129-3. [DOI] [PubMed] [Google Scholar]
- 6•.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. This paper reports the discovery that the CSB protein is a pol II elongation factor. The authors present evidence that CSB stimulates the overall rate of elongation by suppressing transient pausing by pol II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Orphanides G, Leroy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. See annotation [8•] [DOI] [PubMed] [Google Scholar]
- 8•.Leroy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. These papers by Orphanides et al. [7•] and Leroy et al. [8•] report reconstitution with purified proteins of promoter-dependent transcription by pol II on nucleosomal templates. In addition, the authors report discovery of a novel elongation factor, FACT, required for this process. [DOI] [PubMed] [Google Scholar]
- 9•.Li XY, Green MR. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 1998;12:2992–2996. doi: 10.1101/gad.12.19.2992. This paper reports the discovery that Tat-SF1, which was originally identified as a Tat binding protein, is a pol II elongation factor. The authors present evidence that Tat-SF1 stimulates the overall rate of elongation by suppressing transient pausing by pol II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekimizu K, Kobayashi N, Mizuno D, Natori S. Purification of a factor from Ehrlich Ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976;15:5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- 11.Reines D. Nascent RNA cleavage by transcription elongation complexes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and Regulation. New York: Raven Press; 1994. pp. 263–278. [Google Scholar]
- 12.Archambault J, LaCroute F, Ruet A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Exinger G, LaCroute F. 6-azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;142:749–759. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Awrey DE, Edwards AM, Archambault J, Friesen JD. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:11552–11557. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archambault J, Jansma DB, Kawasoe JH, Arndt KT, Greenblatt J, Friesen JD. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J Bacteriol. 1998;180:2590–2598. doi: 10.1128/jb.180.10.2590-2598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Lennon JC, Wind M, Saunders L, Hock MB, Reines D. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. This paper reports identification of specific genes whose transcription depends upon the elongation factor SII. The authors’ findings set the stage for development of gene-specific assays for SII-dependent elongation in vivo and in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. See annotation [19•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Otero G, Fellows J, Li Y, De Bizemont T, Dirac A, Gustafsson C, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80179-3. These papers by Svejstrup et al. [18•] and Otero et al. [19•] report discovery and purification of elongator from yeast. Interaction of elongator with pol II is shown to be regulated by phosphorylation of the pol II CTD. In addition, the authors present genetic evidence that elongator is involved in transcription elongation and activation in vivo. [DOI] [PubMed] [Google Scholar]
- 20.Fraser NW, Sehgal PB, Darnell JE. DRB-induced premature termination of late adenovirus transcription. Nature. 1978;272:590–593. doi: 10.1038/272590a0. [DOI] [PubMed] [Google Scholar]
- 21.Chodosh LA, Fire A, Samuels M, Sharp PA. 5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem. 1989;264:2250–2257. [PubMed] [Google Scholar]
- 22.Braddock M, Thorburn AM, Kingsman AJ, Kingsman SM. Blocking of Tat-dependent HIV-1 RNA modification by an inhibitor of RNA polymerase II processivity. Nature. 1991;350:439–441. doi: 10.1038/350439a0. [DOI] [PubMed] [Google Scholar]
- 23.Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. See annotation [27•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson MS, Winston F. SPT4, SPT5, and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. See annotation [27•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. Together, the papers by Wada et al. [24•,27•] and Hartzog et al. [26•] provide genetic and biochemical evidence that DSIF (Spt4–Spt5), the DRB-sensitivity inducing factor, is a negative regulator of elongation by pol II. In addition, Wada et al. [27•] demonstrate that the positive elongation factor P-TEFb functions at least in part by counteracting the activity of DSIF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall NF, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. See annotation [37•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng J, Zhu Y, Milto JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 31.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun RF, Jeang KT. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 34.Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Herrman CH, Rice AP. The human immunodeficiency virus tat proteins specifically associate with TAK in vivo and require the carboxy-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gold MO, Yang X, Herrmann CH, Rice AP. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Mancebo HSY, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Price DH, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. These papers by Zhu et al. [28•] and Mancebo et al. [37•] elegantly demonstrate that the pol II CTD kinase P-TEFb plays an essential role in Tat-dependent activation of the HIV-1 mRNA synthesis. This work provides the first direct evidence that P-TEFb regulates expression of specific genes in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Wei P, Garber ME, Fang S, Fischer WH, Jones KA. A novel CDK9 associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. See annotation [39•] [DOI] [PubMed] [Google Scholar]
- 39•.Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. These papers by Wei et al. [38•] and Fujinaga et al. [39•] demonstrate that human, but not mouse, cyclin T interacts with the HIV-1 Tat protein and mediates its binding to the TAR element in the HIV-1 polyprotein transcript. This finding provides a mechanistic explanation for the long-standing observation that the HIV-1 is capable of infecting human but not rodent cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedberg EC. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 41.Storb U, Peters A, Klotz E, Kim N, Shen HM, Hackett J, Rogerson B, O’Brien R, Martin TE. Immunoglobulin transgenes as targets for somatic mutation. Int J Dev Biol. 1998;42:977–982. [PubMed] [Google Scholar]
- 42.Wilson PC, Capra JD. The insertion/deletion phenotype in somatic hypermutation and a new model for somatic hypermutation. The Immunologist. 1998;6:48–53. [Google Scholar]
- 43.Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 44.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 45•.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. See annotation [53•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Gool AJ, Verhage R, Swagemakers SMA, Van De Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers JHJ. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene. ERCC6 EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilera A, Klein HL. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos-Rosa H, Aguilera A. Increase in incidence of chromosome instability and non-conservative recombination between repeats in Saccharomyces cerevisiae hpr1 delta strains. Mol Gen Genet. 1994;245:224–236. doi: 10.1007/BF00283271. [DOI] [PubMed] [Google Scholar]
- 50•.Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. See annotation [53•] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan HY, Cheng KK, Klein KL. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang M, French-Cornay D, Fan H, Klein H, Denis C, Jaehning JA. A complex containing RNA polymerase II, paf1p, cdc73p, hpr1p, and ccr4p plays a role in protein kinase C signalling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Prado F, Piruat JI, Aguilera A. Recombination between DNA repeats in yeast hpr1 δ cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. Together, these papers by Chavez and Aguilera [45•], Piruat and Aguilera [50•], and Prado et al. [53•] make a strong case for a mechanistic connection between elongation by pol II, recombination, and maintenance of genome stability. In addition, the authors findings implicate the HPR1 and THO2 gene products in control of transcription elongation in cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 55.Brown SA, Kingston RE. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]