Abstract
Background
Using the Breast Cancer Association Consortium, the authors previously reported that the single nucleotide polymorphism 7q21-rs6964587 (AKAP9-M463I) is associated with breast cancer risk. The authors have now assessed this association more comprehensively using 16 independent case–control studies.
Methods
The authors genotyped 14 843 invasive case patients and 19 852 control subjects with white European ancestry and 2595 invasive case patients and 2192 control subjects with Asian ancestry. ORs were estimated by logistic regression, adjusted for study. Heterogeneity in ORs was assessed by fitting interaction terms or by subclassifying case patients and applying polytomous logistic regression.
Results
For white European women, the minor T allele of 7q21-rs6964587 was associated with breast cancer risk under a recessive model (OR 1.07, 95% CI 1.00 to 1.13, p = 0.04). Results were inconclusive for Asian women. From a combined analysis of 24 154 case patients and 33 376 control subjects of white European ancestry from the present and previous series, the best-fitting model was recessive, with an estimated OR of 1.08 (95% CI 1.03 to 1.13, p = 0.001). The OR was greater at younger ages (p trend = 0.01).
Conclusion
This may be the first common susceptibility allele for breast cancer to be identified with a recessive mode of inheritance.
INTRODUCTION
In a previous publication, we reported that the M463I variant in the A-kinase anchoring protein 9 gene (AKAP9) on chromosome 7 (7q21-rs6964587) was associated with breast cancer risk, based on a study of 9523 patients with breast cancer and 13 770 control subjects from seven independent European and Australian studies.1 The estimated OR for rare TT homozygotes compared to GG homozygotes was 1.17 (95% CI 1.08 to 1.27, p = 0.0003). We aimed to assess this association more comprehensively by extending the study of this single nucleotide polymorphism to an additional 17 438 female patients with invasive breast cancer and 22 044 female control subjects from 16 independent studies participating in the Breast Cancer Association Consortium, by testing additional genetic models and by considering different breast cancer subtypes defined by immunohistochemical tumour markers.
MATERIALS AND METHODS
Eleven studies were conducted in Europe, two in the USA, one in Australia and two in Asia (table 1). All studies provided information on disease status and ethnic group (white European, Asian, other), as well as age at diagnosis and family history of breast cancer for case patients; all except one (Karolinska Breast Cancer Study) provided age at interview for control subjects. Patients with ‘genetically enriched’ breast cancer were defined as those aged younger than 40 years at diagnosis, with bilateral breast cancer and/or with at least one first-degree relative affected with breast cancer, corresponding to ‘familial’ cases in the original publication.1 Oestrogen receptor (ER) status and progesterone receptor (PR) status were provided for a subset of 12 385 (77% positive) and 11 347 (63% positive) white European case patients, respectively, while human epidermal growth factor receptor 2 (HER2) status was provided for 5322 (15% positive) white European case patients (table 1). These variables were also obtained for 6228 (77% positive), 5400 (67% positive) and 3614 (18% positive) white European case patients, respectively, from studies that contributed data to the previously published analysis.1 This histopathology information was generally abstracted from medical reports. Subjects who reported having ethnicity other than white European were excluded, with the exception of those from the two Asian studies (Seoul Breast Cancer Study and Taiwanese Breast Cancer Study), for which only subjects of Asian origin were included. All subjects gave written informed consent, and each study was approved by relevant local institutional review boards.
Table 1.
Participating studies, number of subjects and genotyping methods used
| Study acronym | Study name (reference) | Country | Controls (n) | Cases (n) | ER | PR | HER2 | Genotyping method* |
|---|---|---|---|---|---|---|---|---|
| Women of European origin (previously published series)1 | ||||||||
| ABCFS | Australian Breast Cancer Family Study | Australia | 368 | 540 | 473 (67) | 474 (70) | 0 | iPLEX |
| BBCS | British Breast Cancer Study | UK | 2635 | 580 | 0 | 0 | 0 | Sentrix |
| GENICA | Gene Environment Interaction and Breast Cancer in Germany | Germany | 995 | 983 | 934 (78) | 932 (70) | 604 (27) | TaqMan |
| GFBCS | German Familial Breast Cancer Study | Germany | 1115 | 1083 | 0 | 0 | 0 | TaqMan |
| kConFab/AOCS | Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer/Australian Ovarian Cancer Study | Australia | 714 | 271 | 119 (71) | 100 (73) | 0 | iPLEX |
| MARIE | Mammary Carcinoma Risk Factor Investigation | Germany | 3187 | 1608 | 1646 (76) | 1644 (64) | 1472 (20) | TaqMan |
| SEARCH | Study of Epidemiology and Risk Factors in Cancer Heredity | UK | 4510 | 4246 | 3056 (79) | 2250 (67) | 1538 (11) | TaqMan |
| Women of European origin (replication series) | ||||||||
| BBCC | Bavarian Breast Cancer Cases and Controls2 | Germany | 965 | 1251 | 995 (73) | 992 (65) | 880 (16) | TaqMan |
| CGPS | Copenhagen General Population Study3 | Denmark | 6541 | 1931 | 1711 (82) | 1149 (58) | 0 | TaqMan |
| CNIO-BCS | Spanish National Cancer Centre Breast Cancer Study4 | Spain | 807 | 696 | 240 (75) | 254 (57) | 145 (26) | TaqMan |
| GESBC | Genetic Epidemiology Study of Breast Cancer by Age 505 | Germany | 560 | 511 | 431 (60) | 423 (57) | 0 | TaqMan |
| HEBCS | Helsinki Breast Cancer Study6 | Finland | 1273 | 2233 | 2216 (81) | 2215 (65) | 1304 (16) | iPLEX |
| KARBAC | Karolinska Breast Cancer Study7 | Sweden | 855 | 796 | 433 (83) | 365 (76) | 0 | TaqMan |
| KBCP | Kuopio Breast Cancer Project8 | Finland | 404 | 467 | 440 (76) | 438 (62) | 398 (13) | TaqMan |
| MCBCS | Mayo Clinic Breast Cancer Study9 | USA | 1152 | 1045 | 1079 (83) | 1074 (73) | 735 (20) | TaqMan |
| MCCS | Melbourne Collaborative Cohort Study10 | Australia | 760 | 663 | 605 (73) | 606 (57) | 396 (12) | TaqMan |
| ORIGO | Leiden University Medical Centre Breast Cancer Study11 | Netherlands | 1419 | 552 | 403 (76) | 355 (59) | 0 | TaqMan |
| PBCS | NCI Polish Breast Cancer Study12 | Poland | 2323 | 1941 | 1808 (65) | 1802 (52) | 1268 (11) | TaqMan |
| SASBAC | Singapore and Sweden Breast Cancer Study13 | Sweden | 1458 | 1217 | 833 (82) | 812 (74) | 0 | iPLEX |
| SBCS | Sheffield Breast Cancer Study14 | UK | 822 | 727 | 505 (78) | 185 (57) | 196 (9) | TaqMan |
| UCIBCS | UCI Breast Cancer Study15 | USA | 513 | 813 | 686 (80) | 677 (70) | 0 | TaqMan |
| Total (white Europeans) | 33 376 | 24 154 | 18 613 (77) | 16 747 (64) | 8936 (16) | |||
| Studies of Asian women (replication series) | ||||||||
| SEBCS | Seoul Breast Cancer Study16 | South Korea | 1114 | 1689 | 0 | 0 | 0 | TaqMan |
| TWBCS | Taiwanese Breast Cancer Study17 | Taiwan | 1078 | 906 | 779 (63) | 779 (56) | 347 (33) | TaqMan |
| Total (Asians) | 2192 | 2595 | 779 (63) | 779 (56) | 347 (33) | |||
TaqMan, nuclease assay (TaqMan®), with reagents designed by Applied Biosystems (http://www.appliedbiosystems.com/) as Assays-by-DesignSM and genotyping performed using the ABI PRISM 7900HT, 7700 or 7500 Sequence Detection Systems according to manufacturer’s instructions; Sentrix, customised Illumina Sentrix Bead Arrays (Illumina, San Diego, California, USA); iPLEX, matrix-assisted laser desorption/ionization time of flight mass spectrometry for the determination of allele-specific primer extension products using Sequenom’s MassARRAY system and iPLEX technology (Sequenom, San Diego, California, USA), with oligonucleotide design carried out according to the guidelines of Sequenom and performed using MassARRAY Assay Design software (V.3.1).
ER, oestrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
ER, number of cases with known ER status (percentage positive in parentheses); PR, number of cases with known PR status (percentage positive in parentheses); HER2, number of cases with known HER2 status (percentage positive in parentheses).
The method used by each study to genotype 7q21-rs6964587 is provided in table 1. All studies complied with the Breast Cancer Association Consortium genotyping quality control standards by including at least 2% of samples in duplicate and a common set of 93 CEPH (Centre d’Etude du Polymorphisme Humain) DNAs used by the HapMap Consortium (HAPMAPPT01; Coriell Institute for Medical Research, Camden, New Jersey, USA).
The association of 7q21-rs6964587 with breast cancer risk was assessed by estimating genotype-specific and per-allele ORs using multivariate logistic regression, with study as a categorical covariate. Dominant and recessive models were also considered. Additional adjustment for age made no substantial difference to the results. The best-fitting genetic model was identified using Akaike’s Information Criterion (AIC), which is defined as AIC = −2*(ln(likelihood))+2*(number of parameters).18 Between-study heterogeneity in ORs was assessed using a likelihood ratio test (LRT) comparing the model with interaction terms for the per-allele, dominant or recessive (df = 1), or genotype-specific (df = 2), log-OR by study to the model with no interaction terms. Differences in ORs by ethnicity and age were evaluated using a similar LRT. Differences in ORs between case patient groups defined by ER, PR and HER2 status were tested for white Europeans by an LRT comparing polytomous logistic regression models with and without the per-allele, dominant or recessive (all df = 1) or genotype-specific (df = 2) log-OR constrained to be equal for the two corresponding case patient groups. This LRTwas also used to test the enrichment of the putative risk genotype(s) in AKAP9-rs6964587 in selected case patients, even though the OR estimate for genetically enriched case patients cannot be interpreted as a RR.19 All statistical tests were two-sided. The term ‘statistically significant’ implies p<0.05. All analyses were carried out using Stata: Release 10 (StataCorp).
RESULTS
A minimum genotype concordance of 98% for duplicated samples and 95% for the CEPH samples was observed in all 16 studies, as were minimum genotype calls of 95% for case patients and control subjects. Based on Pearson’s χ2 test applied to control subjects, statistical evidence of departure from Hardy–Weinberg equilibrium was observed for two studies (Genetic Epidemiology Study of Breast Cancer by Age 50 (GESBCS) and Mayo Clinic Breast Cancer Study; p = 0.03 and 0.02, respectively); for both studies, cluster plots were double-checked visually and determined to be of high quality, and all their genotype data were therefore included in the final analysis.
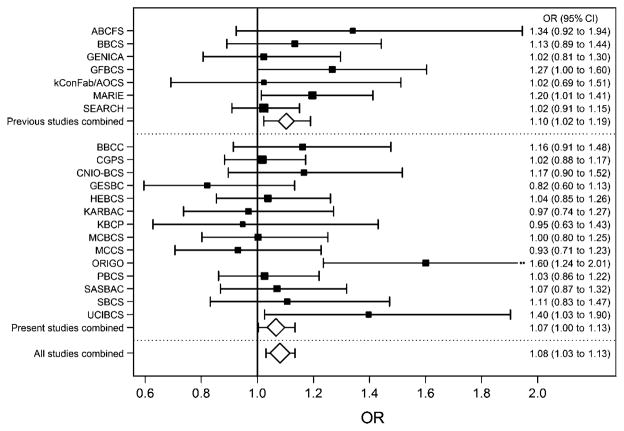
Initial analyses were based on 14 843 case patients and 19 852 control subjects with white European ancestry and 2595 case patients and 2192 control subjects with Asian ancestry. The estimated frequency of the minor (T) allele at 7q21-rs6964587 in control subjects was 0.39 for white Europeans (range among studies 0.37 to 0.42) but lower for Asians (0.17 in both studies) (supplementary table 1). The OR estimate for white Europeans was 1.01 (95% CI 0.98 to 1.04, p = 0.5) per T allele, 0.97 (95% CI 0.92 to 1.02, p = 0.2) for genotype GT versus GG and 1.05 (95% CI 0.98 to 1.12, p = 0.2) for TT versus GG. The corresponding estimates for Asians were 1.07 (95% CI 0.96 to 1.19, p = 0.2), 1.06 (95% CI 0.93 to 1.20, p = 0.4) and 1.16 (95% CI 0.83 to 1.62, p = 0.4), although ORs did not differ by ethnicity (p>0.4). In contrast to the previous analysis,1 there was no evidence of association for white Europeans under a dominant model (OR 0.99, 95% CI 0.94 to 1.03, p = 0.6). However, there was evidence of increased risk under a recessive model (OR 1.07, 95% CI 1.00 to 1.13, p = 0.04). Study-specific OR estimates are provided in figure 1 and in supplementary figure 1. As observed in the previous publication1 for the models tested, the recessive OR estimate was greater when case patients with genetically enriched breast cancer were compared to controls (OR 1.13, 95% CI 1.03 to 1.24, p = 0.01). We reanalysed the data from Frank et al1 based on 9311 case patients and 13 524 control subjects of white European ancestry and obtained consistent estimates under the recessive model (OR 1.10, 95% CI 1.02 to 1.19, p = 0.01).
Figure 1.
OR estimates and their associated 95% CIs under a recessive model, by study. The area of the box/diamond is inversely proportional to the standard error of the log-OR estimate.
When we combined the data from the 21 studies of white Europeans in the present replication series and those in Frank et al1 (24 154 case patients and 33 376 control subjects in total), we obtained OR estimates of 1.01 (95% CI 0.97 to 1.05, p = 0.6) for genotype GT versus GG and 1.09 (95% CI 1.03 to 1.15, p = 0.002) for TT versus GG. While a log-additive model could not be rejected (per-allele OR 1.04, 95% CI 1.01 to 1.06, p = 0.006, AIC = 74 083.4), the best-fitting model was recessive (AIC = 74 080.6), giving an estimated OR of 1.08 (95% CI 1.03 to 1.13, p = 0.001). The combined recessive OR estimate was higher (OR 1.14, 95% CI 1.06 to 1.22, p = 0.0005), but not statistically significantly so (p = 0.09), when case patients with genetically enriched breast cancer were compared to controls. However, the recessively increased risk was stronger for younger women, with estimated ORs of 1.22 (95% CI 1.02 to 1.45), 1.11 (95% CI 1.03 to 1.19) and 1.01 (95% CI 0.93 to 1.09) for women aged <40, 40–59 and 60 years or older, respectively (p trend = 0.01). There was no evidence of heterogeneity in ORs under any model by study (p≥0.2), and results were consistent across analyses excluding each study, one by one (supplementary table 2), suggesting that no single study was driving them. There was also no evidence of heterogeneity in ORs by tumour ER, PR or HER2 status (p≥0.2) (supplementary table 3).
DISCUSSION
The present study has found independent evidence of an association between 7q21-rs6964587 and breast cancer risk for white women of European origin. This combined analysis of more than 57 000 white European women suggests that homozygotes for the T allele have an average 8% increased risk compared to G allele homozygotes, with no evidence of an increased risk for heterozygotes, and this increased risk was greater for younger women. The results were inconclusive for Asian women, which was not surprising given the smaller sample size and that the Tallele is less frequent; the estimated power to detect a recessive OR of 1.08 at 5% statistical significance was 6%. Given that the replication study was 50% larger than the previous study,1 that no evidence of log-additive or dominant association was found (p≥0.5) and that the results of the previous study were consistent with the association being recessive, it seems reasonable to assume that the previously reported increased risk for genotype GT versus GG was due to chance.
This may be the first common variant found to be associated with breast cancer risk under a recessive mode of inheritance. However, because T allele homozygotes are relatively uncommon, further large studies will be needed to estimate the associated RR reliably. The 7q21-rs6964587 variant is a potentially deleterious20 non-synonymous coding single nucleotide polymorphism in AKAP9. It is in strong linkage disequilibrium (r2 = 0.971) with 7q21-rs6960867 (AKAP9-N2792S), which has also been suggested to be potentially deleterious.20 However, it is not clear that either variant is causal. They are located in a region of high linkage disequilibrium that spans beyond AKAP9, and so the association, if real, may be due to a causal relationship with a variant in another gene nearby. Again, further studies will be required to identify a causal variant.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. In particular, we thank Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez, Anna González-Neira, Jonathan Morrison, Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner, Eija Myöhänen, Helena Kemiläinen, Kirsimari Aatonen, Hanna Jäntti, Irja Erkkilä, the Finnish Cancer Registry, Irene Masunaka, PEA Huijts, E Krol-Warmerdam, J Blom, Ursula Eilber, Tanya Koehler, Simon Cross, Helen Cramp, Dan Connley, Heather Thorne, Eveline Niedermayr, the Australian Ovarian Cancer Study Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green and P Webb), the Australian Cancer Study Management Group (A Green, P Parsons, N Hayward, P Webb and D Whiteman), Eileen Williams, Elaine Ryder-Mills, Kara Sargus, Rita K Schmutzler, Claus R Bartram, Tracy Slanger, Elke Mutschelknauss, Ramona Salazar, Sabine Behrens, Renate Birr, Belinda Kaspereit, Nicole Knese, Maggie Angelakos, Judi Maskiell, Gillian Dite, the SEARCH (Study of Epidemiology and Risk Factors in Cancer Heredity) and European Prospective Investigation into Cancer and Nutrition (EPIC) teams, Christina Justenhoven, Hiltrud Brauch, Volker Harth, Sylvia Rabstein, Thomas Brüning and Hans-Peter Fischer.
Footnotes
Competing interests None.
Ethics approval Ethics approval was provided by the relevant local institutional review boards for each of the 23 studies that contributed data.
Provenance and peer review Not commissioned; externally peer reviewed.
Additional materials and the funding statement are published online only. To view these files please visit the journal online (http://jmg.bmj.com).
References
- 1.Frank B, Wiestler M, Kropp S, Hemminki K, Spurdle AB, Sutter C, Wappenschmidt B, Chen X, Beesley J, Hopper JL, Meindl A, Kiechle M, Slanger T, Bugert P, Schmutzler RK, Bartram CR, Flesch-Janys D, Mutschelknauss E, Ashton K, Salazar R, Webb E, Hamann U, Brauch H, Justenhoven C, Ko YD, Bruning T, Silva Idos S, Johnson N, Pharoah PP, Dunning AM, Pooley KA, Chang-Claude J, Easton DF, Peto J, Chenevix-Trench G, Fletcher O, Burwinkel B Australian Breast Cancer Family Study Investigators; Houlston RGene Environment Interaction and Breast Cancer in Germany Group, Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer Investigators, Australian Ovarian Cancer Study Management Group. Association of a common AKAP9 variant with breast cancer risk: a collaborative analysis. J Natl Cancer Inst. 2008;100:437–42. doi: 10.1093/jnci/djn037. [DOI] [PubMed] [Google Scholar]
- 2.Schrauder M, Frank S, Strissel PL, Lux MP, Bani MR, Rauh C, Sieber CC, Heusinger K, Hartmann A, Schulz-Wendtland R, Strick R, Beckmann MW, Fasching PA. Single nucleotide polymorphism D1853N of the ATM gene may alter the risk for breast cancer. J Cancer Res Clin Oncol. 2008;134:873–82. doi: 10.1007/s00432-008-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weischer M, Bojesen SE, Tybjaerg-Hansen A, Axelsson CK, Nordestgaard BG. Increased risk of breast cancer associated with CHEK2*1100delC. J Clin Oncol. 2007;25:57–63. doi: 10.1200/JCO.2005.05.5160. [DOI] [PubMed] [Google Scholar]
- 4.Milne RL, Ribas G, Gonzalez-Neira A, Fagerholm R, Salas A, Gonzalez E, Dopazo J, Nevanlinna H, Robledo M, Benitez J. ERCC4 associated with breast cancer risk: a two-stage case–control study using high-throughput genotyping. Cancer Res. 2006;66:9420–7. doi: 10.1158/0008-5472.CAN-06-1418. [DOI] [PubMed] [Google Scholar]
- 5.Chang-Claude J, Eby N, Kiechle M, Bastert G, Becher H. Breastfeeding and breast cancer risk by age 50 among women in Germany. Cancer Causes Control. 2000;11:687–95. doi: 10.1023/a:1008907901087. [DOI] [PubMed] [Google Scholar]
- 6.Heikkinen T, Karkkainen H, Aaltonen K, Milne RL, Heikkila P, Aittomaki K, Blomqvist C, Nevanlinna H. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15:3214–22. doi: 10.1158/1078-0432.CCR-08-3128. [DOI] [PubMed] [Google Scholar]
- 7.Margolin S, Werelius B, Fornander T, Lindblom A. BRCA1 mutations in a population-based study of breast cancer in Stockholm County. Genet Test. 2004;8:127–32. doi: 10.1089/gte.2004.8.127. [DOI] [PubMed] [Google Scholar]
- 8.Hartikainen JM, Tuhkanen H, Kataja V, Dunning AM, Antoniou A, Smith P, Arffman A, Pirskanen M, Easton DF, Eskelinen M, Uusitupa M, Kosma VM, Mannermaa A. An autosome-wide scan for linkage disequilibrium-based association in sporadic breast cancer cases in eastern Finland: three candidate regions found. Cancer Epidemiol Biomarkers Prev. 2005;14:75–80. [PubMed] [Google Scholar]
- 9.Olson JE, Ma CX, Pelleymounter LL, Schaid DJ, Pankratz VS, Vierkant RA, Fredericksen ZS, Ingle JN, Wu Y, Couch F, Sellers TA, Weinshilboum RM, Vachon CM. A comprehensive examination of CYP19 variation and breast density. Cancer Epidemiol Biomarkers Prev. 2007;16:623–5. doi: 10.1158/1055-9965.EPI-06-0781. [DOI] [PubMed] [Google Scholar]
- 10.Giles GG, English DR. The Melbourne Collaborative Cohort Study. IARC Sci Publ. 2002;156:69–70. [PubMed] [Google Scholar]
- 11.Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM, Blom JC, Pooley KA, Klijn JG, Tollenaar RA, Devilee P, van Asperen CJ. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Closas M, Brinton LA, Lissowska J, Chatterjee N, Peplonska B, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Blair A, Kalaylioglu Z, Rymkiewicz G, Mazepa-Sikora D, Kordek R, Lukaszek S, Sherman ME. Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer. 2006;95:123–9. doi: 10.1038/sj.bjc.6603207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–49. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP, Cox A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–9. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- 15.Ziogas A, Gildea M, Cohen P, Bringman D, Taylor TH, Seminara D, Barker D, Casey G, Haile R, Liao SY, Thomas D, Noble B, Kurosaki T, Anton-Culver H. Cancer risk estimates for family members of a population-based family registry for breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:103–11. [PubMed] [Google Scholar]
- 16.Han S, Lee KM, Choi JY, Park SK, Lee JY, Lee JE, Noh DY, Ahn SH, Han W, Kim DH, Hong YC, Ha E, Yoo KY, Kang D. CASP8 polymorphisms, oestrogen and progesterone receptor status, and breast cancer risk. Breast Cancer Res Treat. 2008;110:387–93. doi: 10.1007/s10549-007-9730-5. [DOI] [PubMed] [Google Scholar]
- 17.Ding SL, Yu JC, Chen ST, Hsu GC, Kuo SJ, Lin YH, Wu PE, Shen CY. Genetic variants of BLM interact with RAD51 to increase breast cancer susceptibility. Carcinogenesis. 2009;30:43–9. doi: 10.1093/carcin/bgn233. [DOI] [PubMed] [Google Scholar]
- 18.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–23. [Google Scholar]
- 19.Byrnes GB, Southey MC, Hopper JL. Are the so-called low penetrance breast cancer genes, ATM, BRIP1, PALB2 and CHEK2, high risk for women with strong family histories? Breast Cancer Res. 2008;10:208. doi: 10.1186/bcr2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.