Abstract
Eukaryotic RNA polymerase II and Escherichia coli RNA polymerase possess an intrinsic ribonuclease activity that is stimulated by the polymerase-binding proteins SII and GreB, respectively. This factor-activated hydrolysis of nascent RNA has been postulated to be involved in transcription elongation as well as removal of incorrect bases misincorporated into RNA. Little is known about the frequency of misincorporation by RNA polymerases in vivo or about the mechanisms involved in improving RNA polymerase accuracy. Here we have developed a luciferase reporter system in an effort to assay for base misincorporation in living Saccharomyces cerevisiae. The assay employs a luciferase open reading frame that contains a premature stop codon. The inactive truncated enzyme would become active if misincorporation by RNA polymerase II took place at the stop triplet. Yeast lacking SII did not display a significant change in reporter activity when compared with wild-type cells. We estimate that under our assay conditions, mRNAs with a misincorporation at the test site could not exceed 1 transcript per 500 cells. The reporter assay was very effective in detecting the previously described process of nonsense suppression (translational read-through) by ribosomes, making it difficult to determine an absolute level of basal (SII-independent) misincorporation by RNA polymerase II. Although these data cannot exclude the possibility that SII is involved in proofreading, they make it unlikely that such a contribution is physiologically significant, especially relative to the high frequency of translational errors.
Many DNA polymerases contain a nuclease activity that allows them to excise, from newly replicated DNA, bases that are misincorporated with respect to Watson-Crick base pair rules. With the recognition that many, if not all, multisubunit DNA-dependent RNA polymerases contain a nuclease activity that operates on nascent RNA came the suggestion that RNA polymerases proofread RNA misincorporation events using this activity (1–3). For bacterial RNA polymerase, this nuclease activity is stimulated by small RNA polymerase-binding proteins called GreA and GreB (4). A similar activity in eukaryotic RNA polymerases is stimulated by a small RNA polymerase II-binding protein called SII (also known as TFIIS) that has been found in all eukaryotes so far investigated. The greA and greB genes are not essential for Escherichia coli; nor is the SII-encoding gene essential for Saccharomyces cerevisiae (5, 6). In vitro, these proteins can reactivate RNA polymerase enzymes that lapse into an elongation-incompetent form (7). They do so by activating cleavage of the nascent RNA chain by RNA polymerase. The vast majority of nascent RNA is assembled according to strict Watson-Crick base pairing rules. Hence, these proteins have been considered transcription elongation factors. In vivo evidence consistent with this role has been described (8–14).
In vitro, misincorporation of nucleotides into RNA by RNA polymerase can be detected by experimental manipulations such as providing a high level of a nucleotide other than that called for by the DNA template. For example, GTP can be incorporated into RNA at a low frequency when poly(dA-dT) · poly(dA-dT) is used as a template for bacterial RNA polymerase (15, 16). In vitro, RNA polymerase II poised at a specific template thymine has been shown to incorporate a G residue in lieu of A when ATP is absent (2). Similarly, E. coli RNA polymerase will incorporate C instead of U when UTP is absent (17). Both polymerases have been shown to misincorporate a U instead of C on a synthetic template (18). GreA, GreB, and SII stimulate the cleavage of nascent RNA containing misincorporated bases (2, 17, 18). Thus, it has been suggested that these factors could assist in the fidelity of transcription by activating RNA polymerase to excise these misincorporated bases (2, 18). However, there is no direct evidence that RNA polymerases employ this factor-activated nuclease activity in vivo. The misincorporation rate of RNA polymerase in vivo has not been accurately measured in eukaryotic cells. A frequency of ~10−5 has been estimated for misincorporation in E. coli; however, the rate varies as a function of the nucleotide substituted, the identity of the template base, the divalent cation, and presumably, sequence context around the substitution (15, 16, 19). Neither the biological sequelae of misincorporation nor the extent to which nuclease-stimulating factors participate in proofreading are well understood, particularly in eukaryotes.
Errors in protein synthesis have been described and estimated previously in bacteria and yeast using reporter systems (19–22). Parameters that influence translational fidelity include genetic background, antibiotics, and epigenetic states. Typically, the fidelity of translation has been considered less stringent than that of transcription (19, 23–25).
In an effort to examine the contribution of transcriptional and translational errors and SII’s potential role in proofreading, we have designed a reporter system to detect misincorporation events mediated by RNA polymerase II at an artificial stop codon engineered into a plasmid introduced into yeast. Using yeast strains with a deletion or a disruption of the SII-encoding gene (DST1), we were unable to find evidence that this cleavage-activating factor participates in proofreading. On the other hand, translational read-through of stop codons was readily detected using pharmacological and genetic approaches and could be readily measured. Rates of SII-independent misincorporation by RNA polymerase appear to be small relative to the higher rates of translational errors such as stop codon bypass.
MATERIALS AND METHODS
Plasmids and Strains
The plasmid pGAL-LUC has been described previously (26) and was obtained from A. Caplan (Mt. Sinai School of Medicine, New York). The sequence (… TACAAAGGA …), encoding Lys445 and the flanking amino acids (Tyr upstream and Gly downstream), was changed to (… TCCTAGGGA …) by site-directed mutagenesis (Gene Dynamics, LLC) to generate the plasmid pLuc-Stop. The mutagenesis generates a novel AvrII restriction site (underlined) that also changes the Tyr codon to Ser and the Lys codon to a stop codon. The stop codon in pLuc-Stop was then changed a base at a time to AAG, CAG, GAG, TCG, TGG, TTG, and TAA to generate the plasmids pGAL-LUC-AAG, pCAG-LUC, pGAG-LUC, pTCG-LUC, pTGG-LUC, pTTG-LUC, and pTAA-LUC, respectively. The pLUC-Δ plasmid was made by religating pLuc-Stop that had been digested with AvrII and SacI after treating the linear DNA with T4 DNA polymerase. The plasmid p2X-Stop was generated by site-directed mutagenesis of the sequence around codon 445 in pLuc-Stop from … TCC TAG GGA … to … TCC TAG TGA …
The strains used in this study are described in Table I. Cells were transformed with plasmids by the lithium acetate/polyethylene glycol method (27). DY978, DY2010, DY2014, DY771, DY773, DY775, DY777, DY779, and DY979 were generated from Z96 (10) by transformation with pGAL-LUC-AAG, pGAL-LUC, pLuc-Stop, pCAG-LUC, pGAG-LUC, pTCG-LUC, pTGG-LUC, pTTG-LUC, and pTAA-LUC, respectively. Strains DY969 and DY975 were generated by transforming pLuc-Δ and p2X-Stop, respectively, into BY4741. Strains DY766 and DY770 were generated from BY4741 and BY4741-4411 (Research Genetics, Huntsville, AL), respectively, by transformation with pLuc-Stop. Strain DY2016 was generated from DY100 (12) by transformation with pLuc-Stop. DY971 and DY973 were derived by transforming pLuc-Stop into strains GT81-1C and GT174 (obtained from Dr. Y. Chernoff, Georgia Institute of Technology), respectively.
Table I.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| DY766 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pLuc-Stop (CEN URA3)] |
| DY770 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 dst1::kanMX4 [pLuc-Stop (CEN URA3)] |
| DY771 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pCAG-LUC (CEN URA3)] |
| DY773 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pGAG-LUC (CEN URA3)] |
| DY775 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pTCG-LUC (CEN URA3)] |
| DY777 | MATα ura3–52 leu2–3,112 his3Δ200 rbp2Δb297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pTGG-LUC (CEN URA3)] |
| DY779 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pTTG-LUC (CEN URA3)] |
| DY969 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pLuc-Δ (CEN URA3)] |
| DY971 | MATa ade1–14 leu2–3,112 lys2 trp1Δ ura3–52 his3-Δ200 ψ+ [pLuc-Stop (CEN URA3)] |
| DY973 | MATa ade1–14 leu2–3,112 lys2 trp1Δ ura3–52 his3-Δ200 ψ−pin− [pLuc-Stop (CEN URA3)] |
| DY975 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [p2X-Stop (CEN URA3)] |
| DY978 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pGAL-LUC-AAG (CEN URA3)] |
| DY979 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pTAA-LUC (CEN URA3)] |
| DY2010 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pGAL-LUC (CEN URA3)] |
| DY2014 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2 CEN)] [pLuc-Stop (CEN URA3)] |
| DY2016 | MATα ura3–52 leu2–3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP214 (LEU2 RPB2 CEN)] [pLuc-Stop (CEN URA3)] |
Paromomycin sulfate was obtained from Sigma and dissolved in water.
Luciferase Assay
Cells from a saturated culture were diluted to an A600 of 0.05–0.1 and grown to an A600 of 0.5 at 30 °C with aeration. The culture was treated with galactose (2% (w/v) final concentration), and triplicate samples of 0.5 OD unit of cells were collected at the indicated times and frozen at −80 °C. Cell pellets were thawed at 4 °C for assay and lysed by vortexing (30 s followed by 30 s on ice) 10 times with 40–50 μl of acid-washed glass beads in 60 μl of luciferase lysis buffer (Promega Corp.). Samples were spun for 30 s at 15,000 × g in a microcentrifuge. A standard assay used 40 μl of cell extract in 100 μl of assay substrate (Promega), but samples were diluted in reaction buffer as necessary to keep signals in the linear range of the luminometer. Samples were mixed, and luminescence was read immediately at 22 °C in a Optocomp I Luminometer (GEM Biomedical, Pineville, NC). Recombinant luciferase was purchased from Promega.
Northern Analysis
Total RNA was isolated from thawed cell pellets by the hot phenol extraction method and quantitated by measuring absorbance at 260 nm (28). Total RNA (15 μg) was resolved on a 1% (w/v) agarose, 7% (v/v) formaldehyde gel and blotted onto Zeta-probe GT nylon membrane (Bio-Rad). Filters were baked at 80 °C for 2 h and prehybridized for a minimum of 3 h at 42 °C in 5 × SSC (1 × SSC: 0.15 m NaCl, 0.015 sodium citrate), 5 × Denhardt’s solution, 50% (v/v) form-amide, 1% (w/v) SDS, and 100 μg/ml salmon sperm DNA. Filters were hybridized under the same conditions with ~108 cpm of 32P-labeled DNA probe for 15–18 h. The filters were washed twice at 22 °C in 2 × SSC, 0.1% SDS for 5 min each and twice in 0.2 × SSC, 0.1% SDS for 5 min each, followed by two 0.2 × SSC, 0.1% SDS washes at 42 °C for 20 min each. The washed filters were exposed to X-Omat film and quantitated with a Fuji BAS1000 phosphorimaging system. The luciferase probe was a polymerase chain reaction product generated from pGAL-LUC using the oligonucleotides 5′-TTCCATCTTCCAGGGATACG-3′ and 5′-TCGCGGTTGTTACTTGCATG-3′. The probe was labeled to a specific activity of ~107 to 108 cpm/μg with Klenow DNA polymerase (Promega, Madison, WI), random hexamer primers (Invitrogen), and [α-32P]dATP (Amersham Biosciences).
RESULTS
Rationale
We sought to establish an in vivo assay that would provide a positive readout of a transcriptional misincorporation event. S. cerevisiae was selected because of a relatively well developed understanding of RNA polymerase II elongation from genetic, biochemical, and molecular biological analyses. Using transformation, we introduced a reporter plasmid into wild-type cells and cells deleted or disrupted for DST1, the gene that encodes transcription elongation factor SII. Yeast is also advantageous, since a large number of cells can be analyzed, and under inducing conditions the GAL1 promoter can be used to generate numerous reporter transcripts per cell, thereby optimizing the ability to detect a rare event. Firefly luciferase was chosen as a reporter, since the activity assay is simple to perform and has a high signal/noise ratio. The lacZ reporter, which has been used for similar purposes in bacteria, may be atypical with respect to transcription and particularly elongation in yeast cells (29–32).
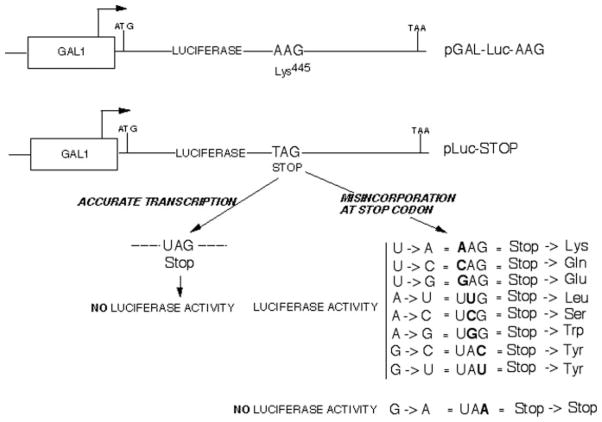
We constructed a selectable centromeric plasmid (pLuc-Stop) that contained the inducible GAL1 promoter driving transcription of the 550-amino acid luciferase open reading frame with a premature stop codon replacing lysine 445 (Fig. 1). Introduction of this stop codon has been shown previously to destroy the protein’s activity, and it has been used as a reporter in E. coli to assess transcription on mismatch-containing DNA (33, 34). Nine different RNA polymerase II misincorporation events are possible at the introduced UAG stop codon triplet: A, G, or C instead of U at position 1; U, C, or G instead of A in position 2; and A, C, or U instead of G at position 3 (Fig. 1). One of these would yield another stop codon (UAG → UAA). The others would result in transcripts that would lead to the substitution of Gln, Glu, Leu, Ser, Trp, or Tyr (two events) for Lys (Fig. 1). One of the changes would restore a codon for lysine (AAG), the natural residue. If misincorporation did not result in abortion of full-length transcript synthesis, and the aforementioned amino acid substitutions led to active luciferase (see below), misincorporation should be detectable as luciferase activity after galactose induction in cells harboring the stop codon-containing plasmid. If the SII protein encoded by the DST1 gene contributes to proofreading, then a deletion of DST1 should result in an increase in luciferase activity, since misincorporation levels would be higher in these cells. Stop codon read-through by the translation machinery would be an alternative way to yield full-length protein and luciferase activity.
Fig. 1. Experimental design of the luciferase reporter assay.
The potential products of misincorporation and the respective codons that would result are shown at the lower right.
Luciferase Activity in Cells Harboring Reporter Plasmids
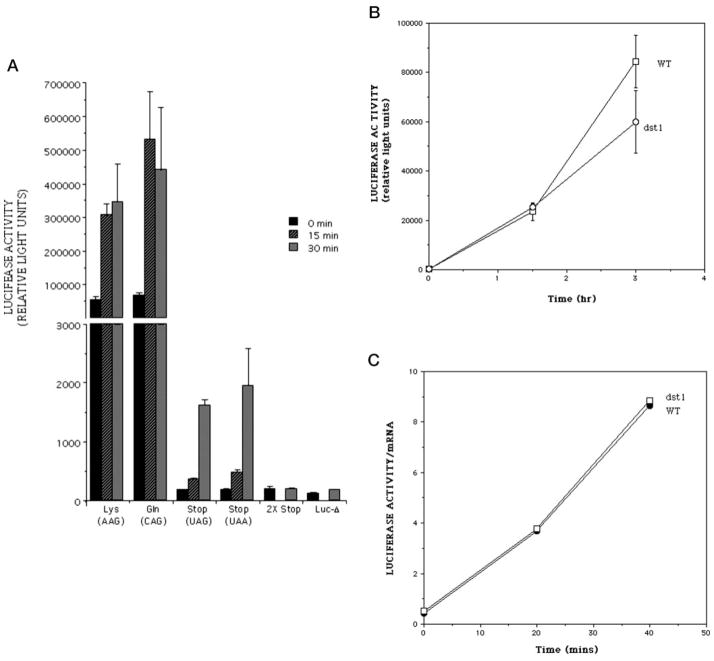
Yeast cells harboring a reporter with an intact luciferase reading frame were able to generate luciferase activity that was inducible upon exposure to galactose (Lys, Fig. 2A) (26). A strain bearing the plasmid encoding a UAG stop codon at position 445 was severely compromised in its ability to generate luciferase, producing a level of activity ~300-fold lower than Lys445 after 30 min of galactose induction (Fig. 2A, compare Lys with Stop; note broken y axis scale). Longer induction times resulted in the additional accumulation of luciferase exhibiting ~105 relative light units in a standard assay (data not shown). Similar activity levels were measured in cells with a plasmid containing the UAA stop codon (Fig. 2, UAA). Activity from the pLuc-Stop plasmid could result from a residual level of enzyme activity of the truncated luciferase polypeptide. Alternatively, “leaky” read-through of the stop codon-containing mRNA by the translation machinery and/or a basal level of misincorporation by RNA polymerase II could yield full-length active protein from the mutation-containing plasmid.
Fig. 2. Induction of luciferase activity with galactose in DST1+ (A) and dst1− (B and C) cells.
A, yeast strains containing the plasmids encoding Lys445 luciferase (DY978), the Lys → Gln substitution (DY771), luciferase with a premature UAG stop codon (DY2014), luciferase with a premature UAA stop codon (DY979), luciferase with tandem stop codons 445 and 446 (DY975, 2X-Stop), and luciferase sequence extending only up to residue 445 (DY969, Luc-Δ), were exposed to galactose. Cell extracts were prepared at the indicated times and assayed for luciferase activity. Experiments were performed in triplicate, and the mean and S.D. (error bars) were calculated and plotted. Only 0- and 30-min points were taken for the 2X-Stop and Luc-Δ samples. B, strains DY766 (WT) and DY770 (dst1) were grown in raffinose-containing medium. Galactose was added at time 0, and extract was prepared for luciferase assay at the indicated times. Triplicate samples were averaged, and S.D. are shown as error bars. C, normalization of the luciferase enzyme activity to luciferase mRNA levels. The enzyme activity values shown in B were divided by the relative levels of luciferase mRNA determined by Northern blotting and phosphorimaging.
To test whether the truncated protein had residual enzyme activity, two control plasmids were created. In one, the luciferase coding sequence downstream of the stop codon at position 445 was deleted (pLUC-Δ). This plasmid could only yield truncated protein regardless of the efficiency of transcriptional or translational read-through. The second control plasmid (p2X-Stop) had a second stop codon engineered in tandem with the first. Thus, two sequential stop codon read-through events or misincorporation events would be required on the same transcript to obtain functional full-length luciferase, an event with a very low probability of occurrence. Cells containing either of these plasmids expressed only background activity (Fig. 2A, Luc-Δ and 2 × Stop). Western blotting showed that the amount of each of the truncated proteins generated in cells carrying pLUC-Δ or p2X-Stop was similar to that produced by cells with full-length luciferase (data not shown). This suggests that the majority of the enzyme activity generated by the pLuc-Stop plasmid comes from translational read-through of the stop codon or transcriptional misincorporation and not from residual activity of the truncated proteins.
To test if active luciferase can be produced by the possible amino acid substitutions expected from transcriptional misincorporation (Fig. 1), we generated a family of plasmids with the cognate single base mutations that change the stop codon to Leu, Trp, Gln, Glu, or Ser. These were compared with luciferase encoding the natural lysine residue at position 445. Plasmids were introduced individually into wild-type yeast and the level of inducible luciferase activity was measured after galactose induction. The luciferase enzymes produced by these substitutions were as active as that containing lysine445 (shown for the Gln substitution in Fig. 2A; data not shown for the other substitutions). We conclude that active luciferase enzyme can be produced by eight of the nine misincorporation events at the DNA-encoded stop codon, and we should be able to score them using this luciferase reporter system. (The ninth possible change, which yields a UAA stop codon, would not be informative.)
Yeasts Deleted for the SII Gene Do Not Show Enhanced Luciferase Activity from the pLuc-Stop Reporter
To test whether SII plays a role in misincorporation, we transformed the stop codoncontaining luciferase reporter plasmid into a strain of yeast deleted for the DST1 open reading frame. If SII enhanced the fidelity of transcription, cells lacking it would show an increased level of transcripts containing misincorporated bases that should code for active luciferase. However, after a 3-h galactose induction, these cells generated luciferase activity comparable with, or slightly less than, that seen for cells expressing wild-type SII (Fig. 2B).
Cells lacking SII are defective in transcriptional induction of a number of genes (10, 12, 35). To ensure that the luciferase activity determinations were not biased by a difference in luciferase mRNA levels between the DST1 deletant and cells wild-type for DST1, we measured the amount of transcript by Northern blotting (data not shown). When luciferase activity was normalized to the abundance of luciferase mRNA in each sample, the values were indistinguishable for SII-containing and SII-lacking strains (Fig. 2C).
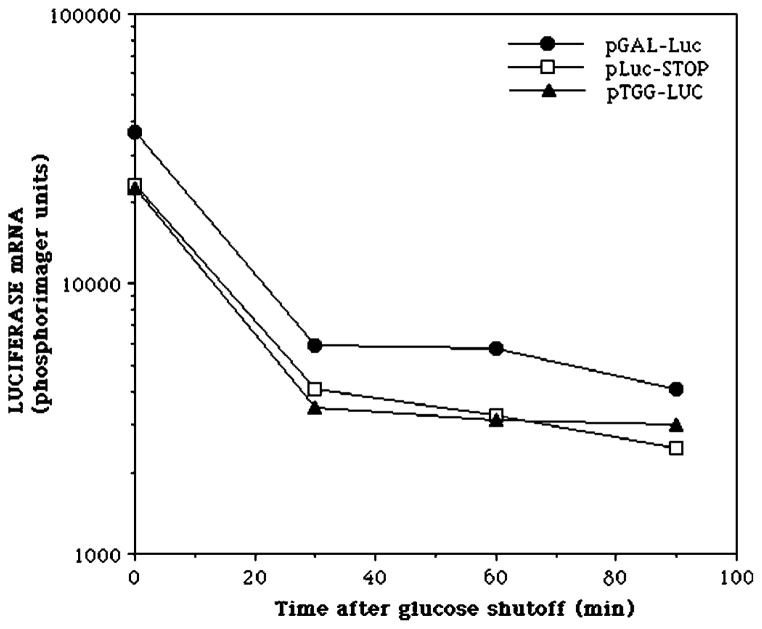
Messenger RNAs containing a premature stop codon can be substrates for the nonsense-mediated mRNA decay pathway (36). To compare the mRNA degradation rates of the luciferase mRNAs bearing the stop codon, lysine codon, or tryptophan codon at position 445, we measured mRNA half-life by inhibiting transcription by the addition of glucose and by quantifying the mRNA decay rate. All of these mRNAs showed similar half-lives (25–30 min), indicating that differential mRNA turnover cannot account for differences in the yield of luciferase activity for mRNAs encoding wild-type, missense-containing, or prematurely truncated luciferases (Fig. 3). From these results, we conclude that there is no evidence that SII affects misincorporation by RNA polymerase II, at least as assessed in this assay system.
Fig. 3. Decay rate of the luciferase mRNA derivatives.
Strains DY2010 (pGAL-LUC), DY2014 (pLuc-Stop), and DY777 (pTGG-LUC) were grown in the presence of galactose for 2 h. RNA was harvested at the indicated times after the addition of glucose (2% w/v) and analyzed by Northern blotting using a probe complementary to the last 400 bp of the luciferase open reading frame. The signals were quantified using a phosphorimager and plotted.
Detection of Active Luciferase Enzyme by Induction of Translational Misreading
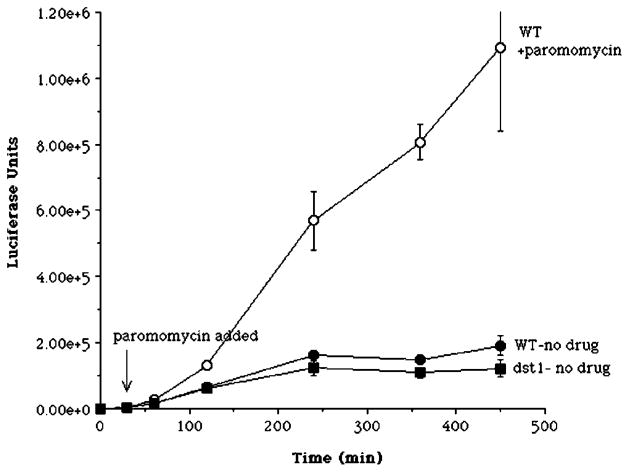
A crucial question is whether this assay would detect transcriptional misincorporation if it took place, especially if translational read-through of the stop codon takes place at high frequency. There are no known genetic changes that lead to an increase or decrease in misincorporation by RNA polymerase II that could serve as a positive control for an error-prone polymerase. Nor are there experimental perturbations proven to alter misincorporation by RNA polymerases in vivo. As an alternative test of the assay’s sensitivity, we resorted to the use of a drug that enhances the frequency of translational read-through of a stop codon. The antibiotic paromomycin has been shown to induce stop codon read-through in yeast as well as in other organisms (see Ref. 20; reviewed in Ref. 37). Wild-type cells harboring pLuc-Stop were induced with galactose to express luciferase and then treated with the antibiotic paromomycin or left untreated. A strong, time-dependent increase in luciferase activity was observed after treatment of wild-type cells with paromomycin compared with untreated cells (Fig. 4, ○). After 7.5 h, the paromomycin-treated cells showed an ~6-fold increase in luciferase activity. Note as well that after this long induction period, untreated yeast with a disruption of DST1 again displayed luciferase activity comparable with wild-type cells with the pLuc-Stop plasmid (Fig. 4, ● versus ■). This latter result provides further confirmation of the data shown in Fig. 2B. Here, however, a different laboratory strain of yeast was used in which the DST1 gene was disrupted by a hisG cassette (10). The paromomycin experiment shows that the assay was able to detect the induction of full-length active luciferase from transcripts containing a stop codon at position 445.
Fig. 4. Effect of paromomycin and DST1 deletion on luciferase induction in a strain containing the stop codon reporter construct.
Strains DY2014 (WT) and DY2016 (dst1) in the logarithmic phase of growth were induced with galactose. After 30 min, an aliquot of the DY2014 culture received paromomycin (200 μm). At the indicated times, extracts were prepared for luciferase assays in triplicate. Values were averaged and plotted, with error bars indicating S.D. Where not visible, error bars are smaller than the symbol.
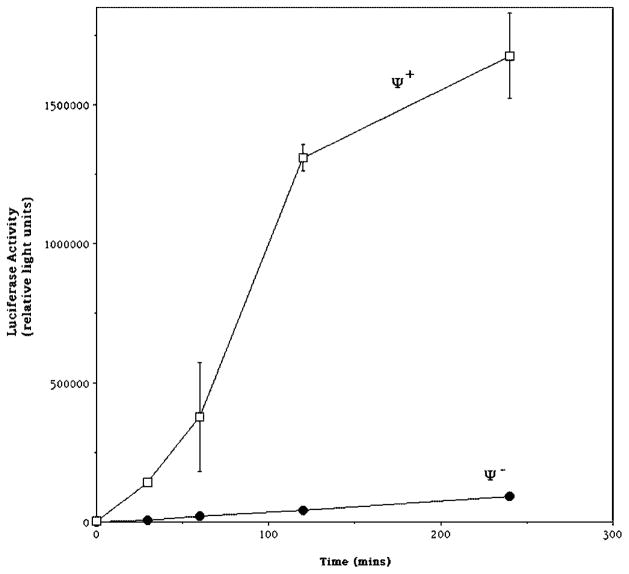
Certain strains designated Ψ+ show a prion phenotype in which the translational termination factor Sup35p becomes aggregated (reviewed in Refs. 38 and 39). Such strains demonstrate nonsense suppression in the absence of known suppressor mutations that is probably due to loss of translation terminating activity. Since our assay appears to be measuring translational read-through, we were interested in testing the effect of the Ψ+ phenotype upon translational read-through of the stop codon-containing luciferase reporter. pLuc-Stop was introduced into a known Ψ+ strain and a cognate Ψ− strain, the cells were induced with galactose, and cells were assayed for luciferase activity. The Ψ+ strain expressed 8–32-fold more enzyme activity than the Ψ− strain depending upon the time of galactose induction (Fig. 5). These results are an independent genetic confirmation of the pharmacological data of Fig. 4, showing that this assay is a sensitive measure of stop codon read-through. The magnitude of luciferase expression in the known Ψ− strain was comparable with that found in DY2014, the strain used above to measure pLuc-Stop expression (Fig. 2A), suggesting that our analysis thus far has taken place in Ψ− strains, although their Ψ phenotype has not been otherwise tested. The plasmid encoding truncated luciferase (pLuc-Δ), which cannot yield full-length luciferase by either translational or transcriptional errors, was equivalently inactive in the Ψ+ and Ψ− strains (data not shown).
Fig. 5. Effect of ψ phenotype upon expression of luciferase activity from stop codon-containing luciferase reporter.
Strains DY971 (Ψ+) and DY973 (Ψ−) were grown in raffinose and exposed to galactose for the indicated period of time before they were lysed and assayed for luciferase activity. Data are the mean of triplicate samples, and error bars represent the S.D. Some error bars were too small to be seen at this scale.
Sensitivity of the Luciferase Assay
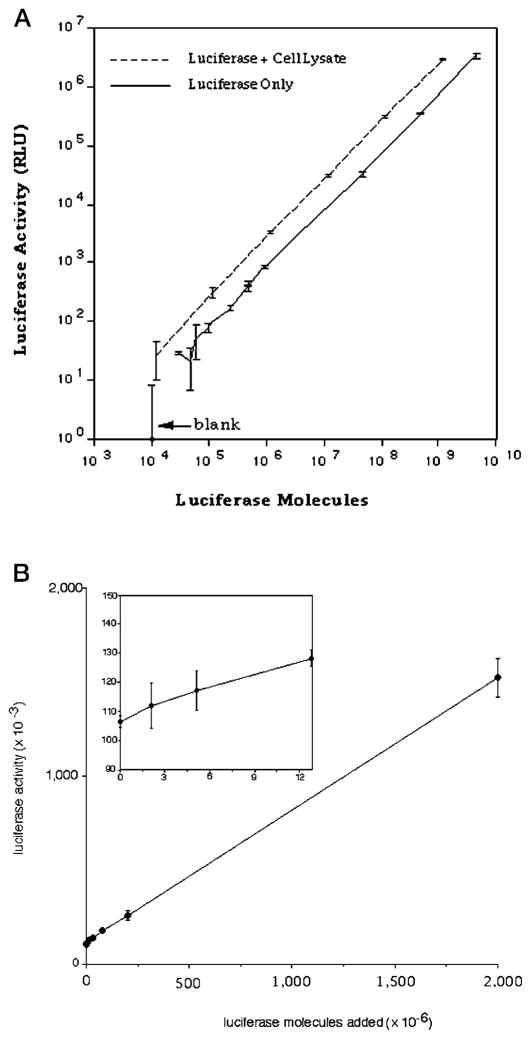
To assess the sensitivity of the luciferase assay under the conditions employed here, we set up a titration curve using wild-type luciferase purified to near homogeneity. The assay was linear over 6 orders of magnitude of enzyme concentration (Fig. 6A). We could reliably measure the signal from 4 fg (39,000 molecules) of luciferase, which was greater than 2 S.D. values above background (Fig. 6A, solid line). To ensure that the activity of purified luciferase was not compromised after cell lysis by inhibitors in the extract, we performed a mixing experiment in which varying amounts of purified luciferase was added to a standard amount of cell extract made from yeast devoid of a luciferase gene (Fig. 6A, dashed line). Signal from the extract containing recombinant purified luciferase was comparable with, and even slightly higher on a per molecule basis than, purified protein alone. This indicates that the enzyme is stable in a yeast cell extract. From this standard curve, we determined that the relative light unit output per molecule of wild type luciferase is 8 × 10−4. The basis of the use of this assay to score for SII-dependent misincorporation requires the detection of an increase in luciferase reporter activity over that detected in cells bearing the pLuc-Stop plasmid. To determine the sensitivity with which we could observe full-length luciferase against the pLuc-Stop signal, we repeated the mixing experiment, this time adding increasing amounts of purified recombinant luciferase to an extract from cells with pLuc-Stop plasmid that had been induced with galactose (Fig. 6B). From this graph, we conservatively (>11 S.D. values) estimate that we can detect 12 × 106 molecules of wild-type luciferase over the basal luciferase activity of this extract (Fig. 6B, inset).
Fig. 6. Activity of purified recombinant luciferase in yeast cell extracts.
A, the indicated amounts of purified recombinant luciferase were added to buffer (solid line) or extract from a strain that lacks any luciferase-encoding plasmid (BY4741; dotted line) and assayed for luciferase. Triplicate measurements were made, and the mean and S.D. (error bars) were calculated. B, the indicated amounts of purified recombinant luciferase (Promega) were added to an extract from strain DY2014 (pLuc-Stop) and assayed for luciferase activity. Triplicate measurements at each point were used to calculate the means and S.D. (error bars). Inset, the first four points on the line are plotted on an expanded scale. Where error bars are not visible, they were smaller than the symbol for that point.
DISCUSSION
Our initial efforts at designing an in vivo transcriptional misincorporation assay in yeast provided no evidence for a role of SII in RNA polymerase II proofreading. Since this is negative evidence, we cannot exclude the possibility that SII and the nuclease activity it activates are involved in transcriptional fidelity in vivo. The results, however, suggest that the SII-activated nuclease activity and misincorporation itself are not biologically robust processes in yeast and that they may be masked by a higher rate of translational errors.
A more direct measurement of misincorporation, such as nucleotide sequencing of cDNAs or RT-PCR products derived from cells containing or lacking SII, would be technically forbidding, since the best estimates of in vivo RNA polymerase misincorporation frequencies suggest that it is low (~10−5) (19, 23, 24, 40). Furthermore, base changes due to misincorporation by RNA polymerase II would have to be detectable against a comparable background of misincorporation by reverse transcriptase and DNA polymerase during reverse transcription and PCR. As a positive indicator of the efficacy of the assay presented here, we induced translational misreading of a reporter transcript in yeast using an antibiotic known to alter stop codon read-through. In prior work, paromomycin increased translation errors by ~20-fold at 200 μm in yeast (20). Our ability to detect an effect of paromomycin in this reporter assay indicates that we can observe rate changes of this size. Similarly, experiments on yeast with a known nonsense suppression phenotype (Ψ+) support the idea that the assay is robust in its ability to detect nonsense codon read-through by the translation machinery.
We can use the data presented herein to make two estimates. The first is the maximal level of RNA polymerase misincorporation that would be required if the activity from pLuc-Stop in a DST1+ cell is assumed to be entirely due to misincorporation by RNA polymerase II. The second is the maximal number of misincorporation-containing transcripts that would be detectable in a standard reaction.
Since luciferase truncated at codon 445 is inactive (Fig. 2A), we can estimate a maximal frequency of transcriptional misincorporation that would be needed to account for all of the enzyme activity derived from pLuc-Stop plasmid. Although it is possible, and perhaps likely, that little or none of this activity is due to misincorporation, this would provide an upper limit. Using our standard curve for purified authentic wild-type luciferase activity (Fig. 6A, solid line), we calculate that a molecule of Lys445 expressed from pGAL-LUC-AAG corresponds to 8 × 10−3 relative light units. (This value has been corrected for the reduced activity, relative to authentic luciferase, of the Lys445 luciferase expressed from the plasmid, which also contains a Tyr444 → Ser substitution due to creation of a restriction site during cloning (data not shown)). This allows us to estimate that the pLuc-Stop extract contains at most 1.1 × 107 molecules (90,000 relative light units in a 4-h induction) of luciferase. This is in comparison with 4 × 1010 luciferase molecules measured in yeast expressing luciferase from pGAL-LUC-AAG with the natural lysine 445. Thus, a maximal frequency of misincorporation by RNA polymerase II of 2.8 × 10−4 can be estimated by dividing the maximal number of active luciferase molecules derived from pLuc-Stop, 1.1 × 107, by the number of total luciferase enzymes present in the pGAL-LUC-AAG extract, 4 × 1010. The actual frequency would be lower if activity from the pLuc-Stop plasmid was due to other causes, one of which would be stop codon read-through.
The deletion or disruption of SII did not change the yield of luciferase reporter activity in two independent dst1− strains assayed here. It is important to know what level of misincorporation the assay can detect had there been an effect. Since we can readily detect 1.2 × 108 molecules of Lys445 luciferase above background in this extract (Fig. 6B, corrected for the substitution at 444) and abundant transcripts are translated 4000 times each on average (41), less than 30,000 transcripts in the 1.7 × 107 cells extracted for a standard assay contain base misincorporations attributable to the loss of SII. This equates to fewer than 0.002 transcripts/cell that would contain a misincorporation at the stop codon tested here. We infer that SII’s contribution to preventing or reversing misincorporation is small as judged by this assay in yeast, at least from a biological perspective.
It should be emphasized that measurements of luciferase activity do not distinguish between translational misreading of the stop codon and transcriptional misincorporation during synthesis of the stop codon. Based upon gross measurements in yeast and E. coli, it seems likely that the majority of the activity derived from pLuc-Stop is due to translational stop codon read-through, which is thought to be ~10-fold higher than transcriptional misincorporation (19, 23–25, 40), although nonsense suppression is variable and nucleotide sequence-dependent (22). In certain cases, translation through stop codons can attain levels as high as 16% in yeast (22). Presumably, in the case presented here, translational read-through takes place by insertion of glutamine at the UAA and UAG stop codons by tRNAGln isoacceptors with the near cognate UUG and CUG anticodons, respectively (21, 42), a substitution that results in active luciferase (Fig. 2A). If the majority of the activity from pLuc-Stop is due to stop codon read-through in our system, then RNA polymerase II misincorporation is very small. Alternatively, if all or most of the read-through of the stop codon, as measured by activity from the pLuc-Stop plasmid, is due to misincorporation by RNA polymerase, SII does not seem to play a major role in changing its frequency.
We offer three plausible explanations for the apparently negligible contribution of SII to luciferase activity in this reporter assay despite in vitro evidence indicating that transcript cleavage factors increase transcriptional fidelity (2, 17, 18). First, in vivo misincorporation rates may be too low for SII-mediated proofreading activity to be recognized, even if the cleavage factor is efficient at reversing misincorporation. Second, SII-activated proofreading may be induced or active only under a specific set of conditions. SII-independent proofreading might operate either through RNA polymerase II’s cleavage activity or via a distinct proofreading system, thereby making SII’s contribution to proofreading appear undetectable. Thus far, only SII has been shown to activate nascent RNA cleavage by RNA polymerase II. In vitro, RNA polymerase shows a low level of transcript cleavage activity in the absence of cleavage-activating factors (5, 43). It is possible that this low level of intrinsic nuclease activity is sufficient to remove misincorporated bases in the absence of SII, or it may be activated by an alternative pathway to a level that may compensate for the loss of SII. Finally, SII and transcript cleavage may simply not participate in proofreading in vivo.
Acknowledgments
We thank Drs. Judy Fridovich-Keil, Charlie Moran, Yury Chernoff, Dean Jones, and the Biochemistry Department Chalk Talk for helpful discussions and Dr. Avrom Caplan for plasmid DNA. We also thank John Mote for technical assistance.
Footnotes
This work was supported by National Institutes of Health Grant GM46331.
References
- 1.Reines D. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas MJ, Platas AA, Hawley DK. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 3.Izban MG, Luse DS. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 4.Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 5.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Proc Natl Acad Sci U S A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 7.Wind M, Reines D. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu LM, Vo NV, Chamberlin MJ. Proc Natl Acad Sci U S A. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archambault J, Lacroute F, Ruet A, Friesen JD. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennon JC, Wind M, Saunders L, Hock MB, Reines D. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartzog GA, Wada T, Handa H, Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw RJ, Reines D. Mol Cell Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davie JK, Kane CM. Mol Cell Biol. 2000;20:5960–5973. doi: 10.1128/mcb.20.16.5960-5973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulish D, Struhl K. Mol Cell Biol. 2001;21:4162–4168. doi: 10.1128/MCB.21.13.4162-4168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozoline ON, Oganesjan MG, Kamzolova SG. FEBS Lett. 1980;110:123–125. doi: 10.1016/0014-5793(80)80038-x. [DOI] [PubMed] [Google Scholar]
- 16.Springgate CF, Loeb LA. J Mol Biol. 1975;97:577–591. doi: 10.1016/s0022-2836(75)80060-x. [DOI] [PubMed] [Google Scholar]
- 17.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 18.Jeon C, Agarwal K. Proc Natl Acad Sci U S A. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberger RF, Hilton J. Mol Gen Genet. 1983;191:207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- 20.Stansfield I, Jones KM, Herbert P, Lewendon A, Shaw WV, Tuite MF. J Mol Biol. 1998;282:13–24. doi: 10.1006/jmbi.1998.1976. [DOI] [PubMed] [Google Scholar]
- 21.Firoozan M, Grant CM, Duarte JA, Tuite MF. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 22.Bonetti B, Fu L, Moon J, Bedwell DM. J Mol Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 23.Ninio J. Biochimie (Paris) 1991;73:1517–1523. doi: 10.1016/0300-9084(91)90186-5. [DOI] [PubMed] [Google Scholar]
- 24.Ninio J. Genetics. 1991;129:957–962. doi: 10.1093/genetics/129.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Libby RT, Gallant JA. Mol Microbiol. 1991;5:999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 26.Brodsky JL, Lawrence JG, Caplan AJ. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- 27.Gietz RD, Woods RA. In: Molecular Genetics of Yeast: Practical Approaches. Johnston JA, editor. Oxford University Press; New York, NY: 1994. pp. 121–134. [Google Scholar]
- 28.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Chapter 4. Greene Publishing Associates/Wiley-Interscience; New York: 1988. [Google Scholar]
- 29.Tabtiang RK, Herskowitz I. Mol Cell Biol. 1998;18:4707–4718. doi: 10.1128/mcb.18.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West RW, Jr, Kruger B, Thomas S, Ma J, Milgrom E. Gene (Amst) 2000;243:195–205. doi: 10.1016/s0378-1119(99)00510-7. [DOI] [PubMed] [Google Scholar]
- 31.Miller LL, Lopes JM. Curr Genet. 2001;39:77–82. doi: 10.1007/s002940100186. [DOI] [PubMed] [Google Scholar]
- 32.Chavez S, Garcia-Rubio M, Prado F, Aguilera A. Mol Cell Biol. 2001;21:7054–7064. doi: 10.1128/MCB.21.20.7054-7064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sala-Newby GB, Campbell AK. Biochim Biophys Acta. 1994;1206:155–160. doi: 10.1016/0167-4838(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 34.Viswanathan A, You H, Doetsch PW. Science. 1999;284:159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 35.Shimoaraiso M, Nakanishi T, Kubo T, Natori S. J Biol Chem. 2000;275:29623–29627. doi: 10.1074/jbc.M910371199. [DOI] [PubMed] [Google Scholar]
- 36.Lykke-Andersen J. Curr Biol. 2001;11:R88–R91. doi: 10.1016/s0960-9822(01)00036-7. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder R, Waldsich C, Wank H. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mugnier P, Tuite MF. Biochemistry (Mosc) 1999;64:1360–1366. [PubMed] [Google Scholar]
- 39.Serio TR, Lindquist SL. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- 40.Blank A, Gallant JA, Burgess RR, Loeb LA. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 41.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beier H, Grimm M. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudd MD, Izban MG, Luse DS. Proc Natl Acad Sci U S A. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]