Abstract
High-throughput genotyping and sequencing techniques are rapidly and inexpensively providing large amounts of human genetic variation data. Single Nucleotide Polymorphisms (SNPs) are an important source of human genome variability and have been implicated in several human diseases, including cancer. Amino acid mutations resulting from non-synonymous SNPs in coding regions may generate protein functional changes that affect cell proliferation. In this study, we developed a machine learning approach to predict cancer-causing missense variants. We present a Support Vector Machine (SVM) classifier trained on a set of 3163 cancer-causing variants and an equal number of neutral polymorphisms. The method achieve 93% overall accuracy, a correlation coefficient of 0.86, and area under ROC curve of 0.98. When compared with other previously developed algorithms such as SIFT and CHASM our method results in higher prediction accuracy and correlation coefficient in identifying cancer-causing variants.
Keywords: Single Nucleotide Polymorphisms, Cancer-causing variants, Gene Ontology, Machine-learning, Support Vector Machine
1. Introduction
Single Nucleotide Polymorphisms (SNPs) are a specific class of genomic variation responsible for about 90% of human variability [1]. In particular the SNPs occurring in coding regions may have higher impact affecting the function of the transcribed proteins [2]. More efficient sequencing and genotyping techniques are detecting a large amount of human genetic variation data [3]. Different international consortiums are collecting information about variations in human genome. The HapMap consortium [4] is characterizing common variation and linkage disequilibrium patterns that can be related to common diseases [5,6]. The Human Variation Project [5] has been funded to collect, curate, and make accessible information on genetic variations affecting human health. International institutions are collaborating in the 1000 Genomes Project (http://www.1000genomes.org/) to produce the most complete catalog of genetic variations in human population [7]. In 2005, the Wellcome Trust Case Control Consortium (WTCCC) has been established to understand the relationship between human genome sequence variation and disease. Using high-throughput technologies, WTCCC collaborators have genotyped about 14,000 patients for seven common diseases performing one of the largest Genome-Wide Association Study (GWAS) [8]. This effort results in an increasing number of SNPs data stored in databases available online. Currently, the dbSNP database at the NCBI [9] collects about 20 million of validated human SNPs. The manually curated SwissVar database [10] reports the possible pathologic effect of about 61,000 missense SNPs and the public version of the HGMD database [11] includes more than 74,000 mutations causing or associated with human inherited disease, plus disease-associated/functional polymorphisms. It is evident that there is a need of computational methods to analyze and identify functionally important variants and describe their molecular effects. During the last decade several bioinformatics methods has been developed to predict the effect of a particular class of SNPs resulting in Single Amino acids Polymorphisms (SAPs) [12–14]. In general, computational methods for the prediction of the impact of SAPs use empirical rules [15,16], Hidden Markov Models [17], Neural Networks[18,19], Decision Tree [20,21], Random Forest [22–26] and Support Vector Machines [27–31], algorithms relying on amino acid sequence, structure and evolutionary information. The amino acid sequence provides information about the physico-chemical properties of the mutated residues such as hydrophobicity, charge, polarity, bulkiness etc. Structural information describes the structural environment of the mutation and has been successfully used to predict the protein stability change upon mutation [32,33]. The most important source of information for the characterization of the effect of SAPs is the evolutionary information. The main hypothesis presumes that important amino acids will be conserved in the protein family, and so changes at well-conserved positions tend to be predicted as deleterious. Recently, a second generation of algorithms that includes also knowledge-based information [24,25,28] has shown better performances with respect to older predictors. The first developed methods SIFT [15] and PolyPhen [16] use different representation of evolutionary information. For each mutated site, SIFT scores the normalized probabilities for all possible substitutions using a multiple sequence alignment between homolog proteins and PolyPhen evaluates the impact of SAPs calculating different sequence-based features and a Position Specific Independent Counts (PSIC) matrix from a multiple sequence alignment. Protein family HMM models implemented in PANTHER [34] have been used to predict deleterious mutations and recently, protein three-dimensional structure features have been shown to improve the performance of SAPs prediction algorithms [22,27,31]. Machine learning-based methods such as PhD-SNP [30] and SNAP [18] have shown better results with respect to traditional methods. The new class of predictors relying on knowledge-based information results in overall accuracy higher than 80%. SNPs&GO [28] includes a new function annotation score calculated using GO terms and MutPred [25] evaluates the impact of a given variant considering the output of several machine learning approaches. A selected list of web available tools for the detection of deleterious missense variants is reported in Supplementary Table 1.
Although available methods are producing valuable results in the detection of disease-related mutations they do not provide any information about the associated pathology. Only MutPred [25] is the first attempt of algorithm able to provide information about the disease mechanism.
To address this problem, we propose a new class of disease-specific predictors trained on a subset of SAPs related to specific disease classes. One of the highest causes of mortality and morbidity in the developed countries is cancer. Although several advances have been made in cancer therapy [35,36], the disease mechanism is still largely unclear. Unlike Mendelian disease where the pathology is principally related to one gene, cancer is a complex disease that often involves several genes. Although it is difficult to dissect the contribution of each gene, individual variants could be indicators of disease risk [37]. To address this problem, two machine learning-based methods have been proposed to predict cancer-causing mutation [23,24]. CanPredict [38] combines SIFT output, a PFAM [39] and a functional-based scores [38] to predict cancer-causing mutations and CHASM [23] takes in input several sequence and profile features to discriminate between passenger and driver variants. These methods are addressing two different aspects of the problem: CanPredict discriminates deleterious mutations occurring in cancer genes from neutral variants from dbSNP database and CHASM detects driver SAPs in cancer-related proteins. To reduce possible over-estimation of the performances [40], we tested our method considering all the driver cancer variants of the same protein either in training or testing set. Our disease-specific machine learning-based predictor, which has been extensively tested on a large set of manually annotated from different sources, results in good level of accuracy when compared with previously implemented methods.
2. Material and methods
2.1. Terminology
In this work we use as synonymous the words single amino acid polymorphism (SAP), missense variant and SNP although the term variant is more general and includes also missense SNP with allele frequency lower than 0.01. We distinguished three classes of variants: cancer-causing, neutral polymorphisms and other disease-related SAPs. We refer to cancer-causing SAPs as the driver variants identified to play a functional role in oncogenic cell transformation and used to test and train CHASM algorithm [23]. The missense SNPs without any evidences of association to disease in SwissVar and recently selected as negative cases [41] are indicated as neutral variants or polymorphisms. We also used Synthetic passengers SAPs generated by CHASM as neutral polymorphisms. A set of variants associated to pathologies not related to the MeSH term “neoplasms” are referred as other disease-related variants. In the binary classification problem addressed in this paper, all the variants are classified in Disease and Neutral. The driver cancer variants belong to the class Disease (D). Passenger, neutral and other disease-related variants, that are not associated with the insurgence of cancer, are classified as Neutral (N).
2.2. Datasets
The selection of a representative set of variants for the training and testing of SAPs prediction methods is a key issue. The performances of the algorithms are strongly dependent on the selected set of neutral and disease-related polymorphisms [42]. For this study, we collected SAPs data from different sources. Cancer-causing variants are selected from breast, colorectal, pancreatic tumor resequencing studies [43–45] and COSMIC database [46] that are provided with CHASM package. Neutral variants are from Swiss-Prot database [47] or generated by CHASM. Other disease-related variants are non “neoplasms” disease-related variants annotated in SwissVar database [10].
In particular the neutral polymorphisms and other disease-related variants from SwissVar have been selected according to a recently described procedure [41]. We built three main datasets to train and test the ability of our method to detect cancer-causing variants. The CNO dataset (Cancer and Neutral missense variants only) with a total number of 6326 variants is composed by 3163 cancer-causing variants and an equal number of neutral polymorphisms. The 3163 cancer-causing mutations from 74 proteins in CNO dataset have been selected from the set of driver cancer mutation used to train CHASM algorithm [23]. The 3163 neutral polymorphisms included in the CNO dataset have been randomly selected from the subset of neutral SAPs in SwissVar database with allele frequency higher than 0.01 and chromosome sample count higher than 49 from the dbSNP database [9] build 131. The performance of our method has been evaluated on the subsets of the CNO dataset with primary histology annotated in the COSMIC database as Carcinoma, Hematopoietic Neoplasm, Lymphoid Neoplasm, Glioma and Malignant Melanoma. The Carcinoma, Hematopoietic, Lymphoid, Glioma and Melanoma subsets are composed respectively by 1899, 461, 441, 384 and 257 driver cancer variants and an equal number of neutral polymorphisms. To test the performance of our predictor in the discrimination between cancer and other disease-causing variants, we build the CND dataset (Cancer, Neutral and other Disease-related missense variants) substituting 50% of neutral polymorphisms with same number of randomly selected from disease-related variants in SwissVar not associated to the MeSH term “neoplasms”.
We have also tested our method in the discrimination between driver and passenger cancer variants building the Synthetic dataset composed by the 3163 driver mutations included in the previous two datasets and an equal number of passenger variants generated by CHASM algorithm. The composition of the three datasets and subsets used in this work is summarized in Table 1.
Table 1.
Datasets composition.
| Dataset | Drivers | Passengers | Neutral | Other disease | Total |
|---|---|---|---|---|---|
| CNO | 3163 | – | 3163 | – | 6326 |
| Carcinoma | 1899 | – | 1899 | – | 3798 |
| Hematopoietic | 461 | – | 461 | – | 922 |
| Lymphoid | 441 | – | 441 | – | 882 |
| Glioma | 384 | – | 384 | – | 768 |
| Melanoma | 257 | – | 257 | – | 514 |
| CND | 3163 | – | 1581 | 1582 | 6326 |
| Synthetic | 3163 | 3163 | – | – | 6326 |
The CNO, CND and Synthetic datasets are composed by the same set of driver cancer variants and respectively only neutral polymorphisms (CNO), neutral and other disease-related variants (CND) and passenger cancer variants generated by CHASM algorithm (Synthetic). Carcinoma, Hematopoietic, Lymphoid, Glioma and Melanoma are subsets of CNO composed by driver cancer variant with primary histology description annotated in COSMIC database.
2.3. Implemented SVM-based predictors
The proposed task is to predict whether a given missense variant is a neutral or involved in the insurgence of cancer. The task is treated as a binary classification problem for the protein variants. The Support Vector Machine (SVM) classifies SAPs in cancer-causing (desired output set to 0) and neutral polymorphism (desired output set to 1). The SVM output is a number between 0 and 1 and the decision threshold has been set to 0.5. The input features of our algorithm (SPF-Cancer) include: the amino acid mutation, its local sequence environment, sequence-profile derived features, the output of PANTHER algorithm [34] and a cancer-specific functional-based log-odd score calculated considering the GO slim ontology. The final input vector consists of 51 values:
40 components encoding for the mutation and the local sequence environment (Seq).
5 inputs features derived from sequence profile (Prof)
4 elements vector from the PANTHER output
2 elements encoding for the number of GO slim terms associated to the protein sequence and the GO slim log-odd score (LGO).
Two other predictors have been developed considering subset of features: mutation site specific method (SeqProf) with input features composed by the 45 elements vector corresponding to Seq and Prof data and protein specific method (F-Cancer) with 2 elements vector features encoding for the cancer-specific functional score (LGO). A third predictor (SPF-All) has been developed calculating a generic functional log-odd score on the whole set of SwissVar SAPs including all type of diseases.
2.4. Encoding sequence information
The input vector portion relative to sequence information consists of 40 values: the first 20 (the 20 residue types) explicitly define the mutation by setting to –1 the element corresponding to the wild type residue and to 1 the newly introduced residue (all the remaining elements are kept equal to 0). The last 20 input values encode for the mutation sequence environment (again the 20 elements represent the 20 residue types). Each input is provided as the number of the encoded residue type, to be found inside a window centered at the residue that undergoes mutation and that symmetrically spans the sequence to the left (N-terminus) and to the right (C-terminus) with a length of 19 residues [30].
2.5. Encoding profile information
We derive for each mutation: the frequency of the wild type, the frequency of the mutated residue, the number of totally and locally aligned sequences and a Conservation Index (CI) [48] for the position at hand: the more a residue is functionally important the more is conserved over evolution. The Conservation Index is calculated as:
| (1) |
where fa(i) is the relative frequency of residue a at mutated position i and fa is the overall frequency of the same residue in all the alignmed positions. The sequence profile is computed from the output of the BLAST program [49], running on the uniref90 database (release 13.3 April 2008) (E-value threshold = 10−9, number of runs = 1).
2.6. PANTHER features
The 4 elements vector from PANTHER [32] output is composed by the probability of deleterious mutation, the frequencies of the wild-type and new residues in the PANTHER family alignment and the number of independent counts. In case that PANTHER does not return any output the probability of deleterious mutation is set to 0.5 and the remaining value has been set to 0.
2.7. Computing the LGO score
The Gene Ontology log-odds score (LGO) is computed to derive information related to the correlation among a given SAPs effect (cancer-causing and neutral) and the protein function. The annotation data are relative to the Gene Ontology [50] Database version 1.37 and are retrieved at the web resource hosted at the European Bioinformatics Institute (EBI). The version of gene ontology classification we used (Dec 2009) contains 30,304 Gene Ontology (GO) terms. To reduce the number of terms and have more general functional terms we consider the GO slim annotation. The GO slim is a simplified version of the GO ontology containing a subset of the terms in the whole GO. They give a broad overview of the ontology content without the detail of the specific terms. In this work we used the generic GO slim ontology (release Sep. 2009) that consists of 132 different GO terms. The generic GO slim file has been downloaded from the Gene Ontology web site (http://www.geneontology.org/GO_slims/goslim_generic.obo). To calculate the LGO, first we derived the GO terms (from all the three branches: molecular function, biological process and cellular components, when available) for all our proteins in the dataset (CNO). For each annotated term the appropriate ontology tree was used to retrieve all the parent terms with the GO-TermFinder-0.8 tool (http://search.cpan.org/dist/GO-TermFinder/) [51] and counting a GO term only once. When all the GO terms for each protein have been collected, we mapped them on the generic GO slim terms using the map2slim.pl script downloaded from the Gene Ontology web site. The LGO is finally calculated as the log-odds score associated to each protein:
| (2) |
where fGO is the frequency of occurrence of a given GO slim term for the cancer-causing (D) and neutral mutations (N) adding one pseudocount to each class. The LGO scores are evaluated considering fGO values computed over the training sets without including in the GO slim term counts of the corresponding test set. This strategy avoids overfitting in the cross-validation procedure.
2.8. Support Vector Machine software
The LIBSVM package (http://www.csie.ntu.edu.tw/~cjlin/libsvm/) has been used for the SVM implementation [52]. The selected SVM kernel is a Radial Basis Function (RBF) kernel K(xi,xj) = exp(−γ∥xi − xj∥2) and γ and C parameters are optimized performing a grid like search. After input rescaling the values of the best parameters are C = 8 and γ = 0.03125.
2.9. Scoring the performance
The results obtained with our SVM methods are evaluated using a cross-validation procedure on the CNO dataset. The reported data for the classification task performed by the SVM methods are obtained adopting a 20-fold cross-validation procedure in such a way that the ratio of the disease-related to the neutral polymorphism mutations is similar to the original distribution of the whole set. Furthermore, all the proteins in the CNO datasets are clustered according to their sequence similarity with the blastclust program in the BLAST suite [49]by adopting the default value of length coverage equal to 0.9 and the percentage similarity threshold equal to 30%. We kept the mutations detected on the same protein cluster s in the same training set to prevent an overestimation of the results. In the comparison with CHASM and SIFT, the methods are tested using a similar strategy used in the CHASM paper [23]. The whole Synthetic dataset is divided in two similar subsets composed same number of drivers and passenger cancer variants. The accuracy measures are calculated using a 2-fold cross validation procedure. In this paper, the efficiency of the predictors is scored using the following statistical indexes.
The overall accuracy is:
| (3) |
where CP is the total number of correctly predicted mutations and T is the total number of mutations.
The Matthews correlation coefficient C is defined as:
| (4) |
where D is the normalization factor:
| (5) |
for each class s (D and N, stand for cancer-causing and neutral polymorphism, respectively); p(s) and n(s) are the total number of correct predictions and correctly rejected assignments, respectively, and u(s) and o(s) are the numbers of false negative and false positive for the class s.
The coverage S (sensitivity) for each discriminated class s is evaluated as:
| (6) |
where p(s) and u(s) are the same as in Eq. (5).
The probability of correct predictions P (or positive predictive values) is computed as:
| (7) |
where p(s) and o(s) are the same as in Eq. (5) (ranging from 0 to 1).
Finally, it is very important to assign a reliability score to each prediction. For each output O(D), this is obtained by computing:
| (8) |
Other standard scoring measures, such as the area under the ROC curve (AUC) and the true positive rate (TPR = Q(s)) at 10% of False Positive Rate (FPR = 1-P(s)) are also computed [53].
3. Results
3.1. Method accuracy
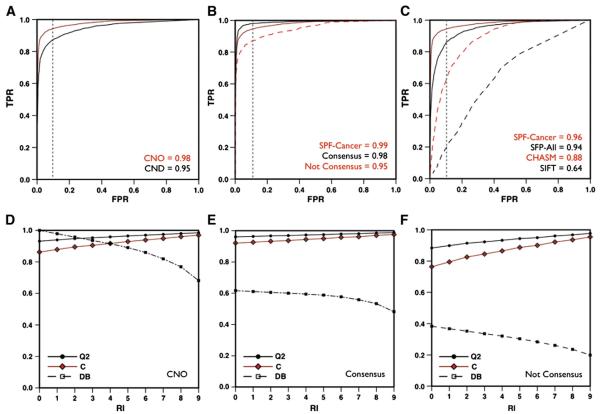
We evaluated our method for predicting cancer-causing missense variants (SPF-Cancer) using a 20-fold cross-validation procedure on the CNO dataset. The SPF-Cancer predictor reaches 93% of overall accuracy, 0.86 correlation coefficient and area under the ROC curve 0.98 (see Table 2). When 10% of false positive are accepted the true positive rate is 0.94 (see Fig. 1 panel A). If predictions with reliability index (RI) higher than 4 are selected, the method results in ~96% accuracy and 0.92 correlation coefficient on 91% of the datasets (see Fig. 1 panel D). We also evaluated the accuracy of our algorithm on the subsets of variants associated to different histology description in COSMIC database. In comparison with the results on CNO dataset, our predictor shows similar performances on the Carcinoma, Lymphoid and Glioma subsets. Contrarily, SPF-Cancer results in 2% higher accuracy and 0.04 higher correlation coefficient on the Melanoma subset and 3% lower accuracy and 0.06 lower correlation coefficient on the Hematopoietic subset with respect to CNO dataset (see Table 2).
Table 2.
Performances of the method.
| Dataset | Q2 | P[D] | S[D] | P[N] | S[N] | C | AUC |
|---|---|---|---|---|---|---|---|
| CNO | 0.93 | 0.93 | 0.93 | 0.93 | 0.93 | 0.86 | 0.98 |
| Carcinoma | 0.93 | 0.93 | 0.94 | 0.94 | 0.93 | 0.87 | 0.98 |
| Hematopoietic | 0.90 | 0.93 | 0.87 | 0.88 | 0.93 | 0.80 | 0.96 |
| Lymphoid | 0.93 | 0.93 | 0.92 | 0.92 | 0.93 | 0.85 | 0.98 |
| Glioma | 0.94 | 0.93 | 0.96 | 0.96 | 0.93 | 0.89 | 0.99 |
| Melanoma | 0.95 | 0.93 | 0.98 | 0.98 | 0.93 | 0.90 | 0.99 |
| CND | 0.90 | 0.87 | 0.93 | 0.92 | 0.86 | 0.79 | 0.95 |
Overall accuracy (Q2), positive predictive value (P) Sensitivity, Correlation coefficient (C) and area under the ROC curve (AUC) are defined in methods section. D (Disease) and N (Neutral) are respectively cancer-causing and neutral variants in CNO dataset. In CND dataset N (Neutral) variants are both neutral and other disease-related.
Fig. 1.
Performance of SPF-Cancer method. ROC curve of SPF-Cancer method on CNO and CND (panel A) on CNO dataset and Consensus and Not Consensus subsets (panel B).In panels C, ROC curves of SIFT, CHASM, SPF-All and SPF-Cancer on the Synthetic dataset. Plot of the accuracy (Q2), correlation coefficient (C) and percentage of the dataset (DB) as a function of the reliability index (RI) for SPF-Cancer method on CNO dataset (panel D) and Consensus (panel E) and Not Consensus subsets (panel F).
The ability of SPF-Cancer in the classification of cancer-causing missense variants, has been tested using the CND dataset that includes 25% of variants from other diseases. In Table 2, we show that the accuracy and the AUC of SPF-Cancer on CND dataset are only 3% lower with respect to those on the CND dataset.
3.2. Using of filters to improve the performance
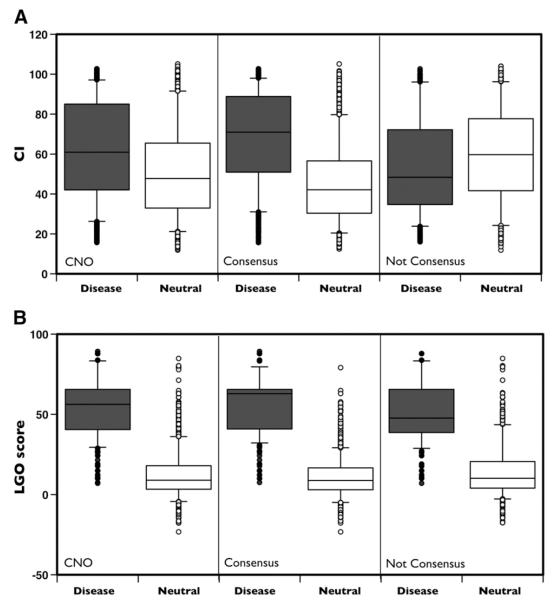
To score the improvement of accuracy resulting from the combination of protein sequence, evolutionary and functional information, the SPF-Cancer method has been compared with simpler SVM-based approaches including either protein sequence and profile information (SeqProf) or only functional information (F-Cancer). On CNO dataset SeqProf and F-Cancer methods result in 64% and 92% overall accuracies and 0.28 and 0.85 correlation coefficients respectively (see Table 3). Thus, SFP-Cancer that includes all the input features results in 1% more accurate predictions and 0.02 higher correlation coefficient with respect to F-Cancer. More interestingly, the SeqProf and F-Cancer results can be used as a filter to select high reliable predictions. In ~62% of the variants in CNO dataset, for which the predictions of SeqProf and F-Cancer methods agree (Consensus), the overall accuracy of SPF-Cancer reaches 96% of accuracy, 0.92 correlation coefficient and 0.99 AUC (see Fig. 1 panel E). On the remaining subset of variants (~38%) where the predictors disagree (notConsensus), SPF-Cancer results only in 88% overall accuracy and 0.76 correlation coefficient (see Fig. 1 panel F). To explain the different level of accuracy between Consensus and notConsensus subset we plot the distributions of the CI values for cancer-causing and neutral variant (see Fig. 2 panel A) and calculated distances (d) between the cumulative distribution for the Kolmogorov–Smirnov (KS) test. The resulting distances are 0.21, 0.44 and −0.22 for the CNO, Consensus and notConsensus datasets respectively. We observed similar trend plotting the distributions of the LGO-scores (see Fig. 2 panel B). In this case, the distances associated to the KS test are 0.87, 0.92 and 0.78 respectively. In Table 4 we reported the summary of the comparison between the CI and LGO distributions.
Table 3.
Selecting more accurate predictions.
| Method | Q2 | P[D] | S[D] | P[N] | S[N] | C | AUC | PM |
|---|---|---|---|---|---|---|---|---|
| SeqProf | 0.64 | 0.66 | 0.58 | 0.63 | 0.70 | 0.28 | 0.70 | 100 |
| F-Cancer | 0.92 | 0.92 | 0.93 | 0.93 | 0.92 | 0.85 | 0.97 | 100 |
| SPF-Cancer | 0.93 | 0.93 | 0.93 | 0.93 | 0.93 | 0.86 | 0.98 | 100 |
| Consensus | 0.96 | 0.96 | 0.95 | 0.96 | 0.97 | 0.92 | 0.99 | 62 |
| NotConsensus | 0.88 | 0.90 | 0.90 | 0.87 | 0.87 | 0.76 | 0.95 | 38 |
Overall accuracy (Q2), positive predictive value (P) Sensitivity, Correlation coefficient (C) and area under the ROC curve (AUC) are defined in Methods section. D (Disease) and N (Neutral) are referred to cancer-causing and neutral variants. PM is the percentage predicted variants of CNO dataset.
Fig. 2.
Distributions of the Conservation Index and LGO on CNO dataset. Boxplot of the distributions for the Conservation Index (Panel A) and LGO scores (panel B) on CNO and Consensus and NotConsensus subset respectively for cancer-causing (Disease) and neutral variants (Neutral).
Table 4.
Comparison of the distribution of Conservation Index and LGO score.
| Dataset | Conservation Index (CI) |
LGO score |
||||||
|---|---|---|---|---|---|---|---|---|
| M[D] | M[N] | d | p-value | M[D] | M[N] | d | p-value | |
| CNO | 61.0 | 47.8 | 0.21 | 2.5×10−61 | 56.2 | 9.0 | 0.87 | 0 |
| Consensus | 71.0 | 42.1 | 0.44 | 0 | 62.9 | 8.8 | 0.92 | 0 |
| Not Consensus | 48.8 | 63.1 | −0.22 | 9.4×10−26 | 47.6 | 10.1 | 0.78 | 0 |
M[D] and M[N] are the median values for cancer-causing (D) and neutral (N) variants respectively. d is the distance between the cumulative distributions of cancer-causing and neutral variants. P-value is associated to Kolmogorov–Smirnov test.
3.3. Comparison with other predictors
We compared the performance of SPF-Cancer with those obtained by SIFT, CHASM and a similar SVM-based predictor with generic GO slim-based score (SPF-All) calculated using whole set of disease-related variants (see Table 5). On the Synthetic dataset, SIFT and CHASM result in 68% and 80% overall accuracies and 0.22 and 0.60 correlation coefficients respectively. Thus, SPF-Cancer shows more than 10% higher accuracy and correlation coefficient with respect to CHASM. SPF-Cancer also results in 2% higher overall accuracy and 0.06 higher correlation coefficient when compared with SPF-All. To estimate the significance of the differences between the four predictors, we calculated the χ2 obtained comparing the confusion matrix SPF-Cancer with those of SPF-All, CHASM and SIFT. The associated probabilities to observe this differences by chance are 3.4×10−5, 8.6×10−82 and 0 respectively for SPF-All, CHASM and SIFT.
Table 5.
Comparison with other methods.
| Dataset | Q2 | P[D] | S[D] | P[N] | S[N] | C | AUC | PM |
|---|---|---|---|---|---|---|---|---|
| SIFT | 0.61 | 0.62 | 0.66 | 0.60 | 0.56 | 0.22 | 0.64 | 95 |
| CHASM | 0.80 | 0.85 | 0.73 | 0.76 | 0.87 | 0.60 | 0.88 | 100 |
| SPF-All | 0.88 | 0.88 | 0.87 | 0.87 | 0.88 | 0.75 | 0.94 | 100 |
| SPF-Cancer | 0.90 | 0.91 | 0.90 | 0.90 | 0.91 | 0.81 | 0.96 | 100 |
Overall accuracy (Q2), positive predictive value (P) Sensitivity, Correlation coefficient (C) and area under the ROC curve (AUC) are defined in Methods section. D (Disease) and N (Neutral) are respectively driver and passenger cancer variants. The latter have been generated by CHASM. PM is the percentage predicted variants for the Synthetic dataset. The confidence interval for Q2, C and AUC calculated on the two subsets is ≤0.01.
3.4. GO score analysis
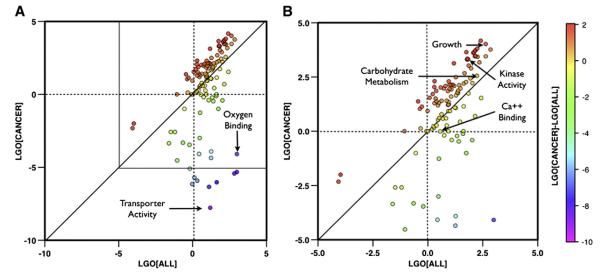
The GO score used in this work, has been calculated using GO slim terms. To better understand the ability of the method to correctly classify cancer-causing mutations score, we compare the values of cancer-specific and generic LGO scores. In particular the comparison between the LGO values calculated on the dataset driver cancer variants and on the dataset including all disease-related variants has been used to detect GO terms associated to cancer. Although the LGO scores are dependent on the training set, their relative values obtained in comparison with generic LGO scores provide an estimation of the GO terms’ occurrences. Thus, a positive difference between the cancer-specific and generic LGO scores indicates an enrichment of the relative GO terms in the cancer specific dataset while negative difference corresponds to GO terms more abundant in the dataset including all disease-related variants. In Fig. 3 the scatter plot of the generic LGO score versus the cancer-specific LGO score for each GO slim term. The interesting GO functions are those corresponding to the points far from the diagonal. The points with negative generic LGOs and positive cancer-specific LGOs are those with GO slim functions related to cancer. The points with cancer-specific LGOs close to zero and higher generic LGOs are those with GO slim functions generally associated to the all the pathologies in SwissVar dataset. For example, in our study we observed that Growth (GO:0040007) and Kinase Activity (GO:0016301) GO slim terms have stronger association to cancer showing respectively cancer-specific LGOs 4.02 and 3.30 and generic LGOs 2.63 and 1.78. Other interesting GO slim terms associated to all the diseases are the Transporter Activity (GO:0005215) and Oxygen Binding (GO:0019825) which have respectively cancer LGOs −7.77 and −4.09 and generic LGOs 1.20 and 2.99. There are also GO slim terms that have similar values for cancer and generic diseases LGO scores. Two examples are the Carbohydrate Metabolic Process (GO:0005975) that has similarly related cancer and all the diseases in our dataset resulting in LGO scores respectively 2.55 and 2.23, and the Calcium Ion Binding (GO:0005509) that is not related to cancer and slightly associated to all the diseases showing LGO scores −0.01 and 0.56 respectively.
Fig. 3.
General and cancer-specific LGO scores. Scatter plot of the generic vs the cancer-specific LGO scores (LGO[All] and LGO[Cancer]) for each GO slim term (panel A). Color scale is related to the value of LGO[Cancer]-LGO[ALL]. In panel B, zoom of the plot in the region of LGO scores between −5 and 5.
4. Discussion
In general cancer-specific prediction methods have been trained either to discriminate between passenger and driver cancer-causing SAPs in a known cancer-related protein or to detect cancer-causing using a negative set of neutral SAPs in proteins with different functions. SPF-Cancer method has been tested on both tasks. We built the CNO dataset selecting all the cancer-causing variants used to train and test CHASM method and an equal number of randomly selected neutral polymorphism from a curated set of variants recently used to test the performances of predictive algorithms [41]. The results obtained on this dataset should be considered as upper bound performances since we selected only neutral variants with allele frequencies higher than 0.01 for which their annotation is expected to be more accurate. To compare our methods against previously developed algorithms we use the Synthetic dataset for which neutral missense variants are generated by CHASM algorithm.
The SPF-Cancer predictor tested in cross-validation on CNO dataset, resulting in 93% overall accuracy and 0.86 correlation coefficient. With respect to the whole CNO dataset, our algorithm shows better performance in the detection of variants annotated as Malignant Melanoma and lower performances on Hematopoietic Neoplasm variants. When compared against CHASM on the Synthetic dataset, SPF-Cancer shows about 10% better accuracy and 0.2 better correlation coefficient. The development of cancer-specific predictor is justified by the improvement of 2% in overall accuracy and 0.06 in correlation coefficient resulting from the cancer-specific LGO scores. Differences between cancer-specific and generic predictors are higher when other disease-related variants are included in the dataset (data not shown). Although SPF-Cancer shows 3% lower accuracy on the Synthetic dataset with respect to CNO dataset, this difference can be due to the unknown annotation of the passenger variants generated by CHASM.
SPF-Cancer is also able to discriminate between cancer-causing variant and other disease-related mutations, while reaching 90% of accuracy on the CND dataset where 50% of the neutral polymorphisms are replaced with variants related to non “neoplasm” diseases. The improvement of the performances resulting from the combination of site-dependent sequence and profile features and functional information can be quantified in 1% higher accuracy and 0.02 higher correlation coefficient with respect to the GO score-based method. In addition, using two different methods it is possible to select a subset of highly accurate predictions. In 62% of the mutations where the sequence and profile-based (SeqProf) and GO score-based (F-Cancer) predictions agree, SPF-Cancer results in 3% better accuracy and 0.06 better correlation with respect to the performance on the whole CNO dataset. On the subset of variants where predictions are in disagreement (NotConsensus) the low performances are justified by the reverse trend in the distributions of the Conservation Index for cancer-causing and neutral variants (d = −0.22). Finally, the comparison between cancer-specific and generic LGO score values allows the estimation of the functional enrichment in cancer-related proteins. For example we observed enrichment of GO terms Growth and Kinase Activity in cancer-related proteins and Transporter Activity and Oxygen Binding in the whole set of disease-related proteins.
In conclusion, we present a new machine learning-based algorithm (SPF-Cancer) to predict cancer-causing variants. The SPF-Cancer method that has been extensively tested on a large set of variants is a valid alternative to previously developed algorithms. Considering that cancer is a complex disease that can involve multiple genes, SPF-Cancer reaches a good level of accuracy also when compared with previously developed algorithms such as SIFT and CHASM. The comparison between SPF-Cancer and SPF-All method indicates that cancer-specific LGO term score improves the prediction accuracy. The calculation of cancer-specific LGO values allows to rank with higher scores those proteins annotated with GO term functions involved in the development of cancer. This suggests new strategies for the development of the next generation of disease-specific algorithms able to discriminate between the genetic variants related to a specific disease and other class of pathologies. Finally, scoring the deleterious effect of missense variants using sequence profile-based and functional-based methods allows to select higher confident predictions where both methods predictions agree. For this subset of high quality predictions (62%), the SPF-Cancer method results in 96% overall accuracy and 0.92 correlation coefficient.
Supplementary Material
Acknowledgments
The authors thank anonymous reviewers for their suggestions that allowed to improve the quality of this work. EC acknowledges support from the Marie Curie International Outgoing Fellowship program (PIOF-GA-2009-237225). RBA would like to acknowledge the following funding sources: NIHLM05652 and GM61374.
Footnotes
Appendix A. Supplementary data Supplementary data to this article can be found online at doi:10.1016/j.ygeno.2011.06.010.
References
- [1].Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- [2].Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- [3].Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cotton RG, Auerbach AD, Axton M, Barash CI, Berkovic SF, Brookes AJ, Burn J, Cutting G, den Dunnen JT, Flicek P, Freimer N, Greenblatt MS, Howard HJ, Katz M, Macrae FA, Maglott D, Moslein G, Povey S, Ramesar RS, Richards CS, Capriotti DE, Altman / Seminara RB, Smith TD, Sobrido MJ, Solbakk JH, Tanzi RE, Tavtigian SV, Taylor GR, Utsunomiya J, Watson M. GENETICS. The Human Variome Project. Science. 2008;322:861–862. doi: 10.1126/science.1167363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, Kruglyak L, Stein L, Hsie L, Topaloglou T, Hubbell E, Robinson E, Mittmann M, Morris MS, Shen N, Kilburn D, Rioux J, Nusbaum C, Rozen S, Hudson TJ, Lipshutz R, Chee M, Lander ES. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- [7].1000 genomes project consortium, a map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].W.T.C.C. Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yip YL, Famiglietti M, Gos A, Duek PD, David FP, Gateau A, Bairoch A. Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase. Hum. Mutat. 2008;29:361–366. doi: 10.1002/humu.20671. [DOI] [PubMed] [Google Scholar]
- [11].Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Karchin R. Next generation tools for the annotation of human SNPs. Brief. Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief. Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- [14].Tavtigian SV, Greenblatt MS, Lesueur F, Byrnes GB. In silico analysis of missense substitutions using sequence-alignment based methods. Hum. Mutat. 2008;29:1327–1336. doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- [20].Dobson RJ, Munroe PB, Caulfield MJ, Saqi MA. Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC Bioinformatics. 2006;7:217. doi: 10.1186/1471-2105-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Krishnan VG, Westhead DR. A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics. 2003;19:2199–2209. doi: 10.1093/bioinformatics/btg297. [DOI] [PubMed] [Google Scholar]
- [22].Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21:2185–2190. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- [23].Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kaminker JS, Zhang Y, Watanabe C, Zhang Z. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007;35:W595–W598. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wainreb G, Ashkenazy H, Bromberg Y, Starovolsky-Shitrit A, Haliloglu T, Ruppin E, Avraham KB, Rost B, Ben-Tal N. MuD: an interactive web server for the prediction of non-neutral substitutions using protein structural data. Nucleic Acids Res. 2010;38(Suppl):W523–W528. doi: 10.1093/nar/gkq528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yue P, Moult J. Identification and analysis of deleterious human SNPs. J. Mol. Biol. 2006;356:1263–1274. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- [28].Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- [29].Capriotti E, Arbiza L, Casadio R, Dopazo J, Dopazo H, Marti-Renom MA. Use of estimated evolutionary strength at the codon level improves the prediction of disease-related protein mutations in humans. Hum. Mutat. 2008;29:198–204. doi: 10.1002/humu.20628. [DOI] [PubMed] [Google Scholar]
- [30].Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- [31].Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- [32].Capriotti E, Fariselli P, Casadio R. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20(Suppl 1):I63–I68. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- [33].Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Maris JM. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Petrelli NJ, Winer EP, Brahmer J, Dubey S, Smith S, Thomas C, Vahdat LT, Obel J, Vogelzang N, Markman M, Sweetenham JW, Pfister D, Kris MG, Schuchter LM, Sawaya R, Raghavan D, Ganz PA, Kramer B. Clinical Cancer Advances 2009: major research advances in cancer treatment, prevention, and screening—a report from the American Society of Clinical Oncology. J. Clin. Oncol. 2009;27:6052–6069. doi: 10.1200/JCO.2009.26.6171. [DOI] [PubMed] [Google Scholar]
- [37].Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat. Rev. Cancer. 2011;10:353–361. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- [38].Kaminker JS, Zhang Y, Waugh A, Haverty PM, Peters B, Sebisanovic D, Stinson J, Forrest WF, Bazan JF, Seshagiri S, Zhang Z. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- [39].Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Boulesteix AL. Over-optimism in bioinformatics research. Bioinformatics. 2010;26:437–439. doi: 10.1093/bioinformatics/btp648. [DOI] [PubMed] [Google Scholar]
- [41].Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- [42].Care MA, Needham CJ, Bulpitt AJ, Westhead DR. Deleterious SNP prediction: be mindful of your training data! Bioinformatics. 2007;23:664–672. doi: 10.1093/bioinformatics/btl649. [DOI] [PubMed] [Google Scholar]
- [43].Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- [45].Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- [46].Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- [49].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO:: TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chang CC, Lin CJ. Training nu-support vector classifiers: theory and algorithms. Neural Comput. 2001;13:2119–2147. doi: 10.1162/089976601750399335. [DOI] [PubMed] [Google Scholar]
- [53].Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.