Abstract
Experiments were performed to examine the feasibility of MS depth profiling of animal tissue by ~75 fs, 800 nm laser pulses to expose underlying layers of tissue for subsequent MS analysis. Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) was used to analyze phospholipids and proteins from both intact bovine eye lens tissue and tissue ablated by ultrashort laser pulses. Laser desorption postionization (LDPI-MS) with 10.5 eV single photon ionization was also used to analyze cholesterol and other small molecules in the tissue before and after laser ablation. Scanning electron microscopy was applied to examine the ablation patterns in the tissue and estimate the depth of the ablation craters. Ultrashort pulse laser ablation was found able to remove a layer of several tens of micrometers from the surface of eye lens tissue while leaving the underlying tissue relatively undamaged for subsequent MS analysis. MS analysis of cholesterol, phospholipids, peptides, and various unidentified species did not reveal any chemical damage caused by ultrashort pulse laser ablation for analytes smaller than ~6 kDa. However, a drop in intensity of larger protein ions was detected by MALDI-MS following laser ablation. An additional advantage was that ablated tissue displayed up to an order of magnitude higher signal intensities than intact tissue when subsequently analyzed by MS. These results support the use of ultrashort pulse laser ablation in combination with MS analysis to permit depth profiling of animal tissue.
I. INTRODUCTION
Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) is a very popular MS technique for molecular profiling and imaging of intact biological tissue samples.1,2 Depth profiling of biological tissue samples can be performed by tissue slicing and separate MALDI-MS analysis of individual, 5 – 20 µm thick slices. However, this process is both time consuming and laborious. Furthermore, it can be difficult to correlate 2D images obtained from different slices, especially if interlayer mixing occurs during microtoming.
Ultrashort (sub-picosecond) laser pulses are commonly employed for laser surgery because they allow superior control of incisions and induce less thermal and mechanical damage3,4 compared to nanosecond (ns) laser pulses. The reduced damage produced by ~100 fs laser pulses has been attributed to a non-linear laser-tissue interaction mechanism that allows for ablation from a small volume localized around the beam waist. Ultrashort pulsed laser irradiation can be employed for ablation of both transparent and non-transparent substrates due to multiphoton absorption mechanisms thought to occur at high irradiances. Previous studies by some of the authors and others have evaluated the precision of the ablation pattern and the level of mechanical and thermal damage induced by ultrashort pulse ablation of eye tissue and collagen samples.5–8
This paper evaluates the feasibility of MS depth profiling of biological tissues by ultrashort laser pulses to expose underlying layers of tissue for subsequent MS analysis. MS analysis of ablated tissue was performed with both MALDI-MS and laser desorption postionization MS (LDPI-MS) in separate instruments.9–11 Such studies are particularly important because other workers have reported that ultrashort laser pulses can desorb molecular analytes from biological samples, permitting mass analysis of both direct ions12,13 and desorbed neutrals after postionization.14–17
The use of lasers for depth profiling has been limited by mechanical and thermal degradation of material surrounding the ablation crater. Mechanical and thermal side effects are especially prominent when the pulse duration is longer than the characteristic times for thermal diffusion and stress confinement (on the order of several picoseconds). These times depend on the absorption coefficient and thermal diffusivity of the substrate and are typically exceeded in the case of ns laser pulses. However, thermal and stress confinement may be safely achieved using ultrashort laser pulses above the ablation threshold.3,4,18 A possible unwanted side-effect of using ultrashort pulses is photochemical decomposition within tissue surface layers that may cause chemical damage within buried tissue layers not directly affected by ablation. Such effects may also result from shockwaves generated by very high temperatures and pressures in the ablated outer layer as well as by plasma-induced chemical reactions.3,4,18 A requirement for depth profiling is that ultrashort laser pulses do not induce any chemical damage in the tissue surrounding the ablation crater. However, relatively little prior work has examined the extent of chemical damage by ultrashort pulse ablation.19 By contrast, the extent of chemical damage during cluster ion depth profiling in secondary ion mass spectrometry has been extensively explored.20–24
Some of the authors previously showed that when the antibiotic sulfadiazine was introduced into a microbial biofilm, it survived subsequent ablation by ~75 fs, 800 nm Ti-sapphire laser irradiation.25 That finding indicated that ultrashort, near-infrared laser pulses might be used for depth profiling of microbial biofilms and other biological tissues. However, that prior study did not examine the effect of ultrashort laser pulses on larger endogenous biological molecules naturally occurring in the biofilm. Furthermore, that study did not examine the effect of laser ablation on animal tissue, a very popular target of MS imaging.
The present work evaluates whether endogenous phosopholipids (phosphatidylcholines and sphingomyelins) and proteins (crystallins) in bovine eye lens tissue remain unaltered by ultrashort pulsed laser ablation. The choice of sample was based on the composition of phospholipids and proteins in the bovine eye lens, which was previously probed by MALDI-MS 26–29 and which facilitated analysis here. The present study then used MALDI-MS to analyze phospholipids and proteins from the intact tissue and laser ablated tissue. LDPI-MS was also used to analyze small molecules in the tissue before and after laser ablation.9–11 Mass spectra obtained from the intact and laser ablated surfaces were compared to estimate the extent of chemical damage introduced by laser ablation. Scanning electron microscopy was also used to examine the ablation patterns in the tissue and estimate the depth of the ablation craters.
II. EXPERIMENTAL METHODS
A. Eye lens tissue handling and sample preparation
Eye lenses were dissected from bovine eyeballs taken from animals younger than 30 months and stored at −80°C after flash freezing in liquid nitrogen.27 Sample thawing from −80°C to −20°C was done at least 24 hrs in advance and then samples were transferred to a −12°C cryostat chamber (Microm Vacutome/HM500OM, Thermo Scientific, Kalamazoo, MI, USA) at least two hours before sectioning. Eye lenses were mounted on the top of a microtome sample stub by means of optimal cutting temperature media (OCT, Tissue-Tek 4583, Sakura Finetek USA Inc., Torrance, CA, USA). After allowing for the OCT to freeze, the sides of the eye lens were removed with a razorblade, leaving on the stub a cube-shaped central part of the eye lens of approximately 1 cm3 in volume. 300 µm thick sections cut parallel to the equatorial plane from around the lens nuclei were mounted atop cold (−12°C) stainless steel MALDI plates by means of vacuum-compatible double-sided copper adhesive tape.
B. Ultrashort pulsed laser ablation
45 fs, 800 nm pulses generated by a Ti-Sapphire oscillator (Spectra-Physics Tsunami, Newport corporation, Irvine, CA, USA) and amplified by a regenerative amplifier (Spectra-Physics Spitfire) were used to ablate sections of the eye lens tissue (see Supporting Information for schematic diagram).25,30 The laser beam was focused above the surface with a 100 mm focal length lens, so that the spot-diameter on the surface was 200 µm. The pulse duration at the sample surface was found by autocorrelation to be ~75 fs. The focal radius of the beam (radius at the e−2 point of the intensity) was determined by a scanning knife edge to be 55 µm, and the Rayleigh range was 0.8 mm. The laser power was measured by a commercial power meter (Ophir 30A-SH-V1, Ophire-Spiricon, North Logan, UT, USA). A He-Ne laser (1125, JDSU, Milpitas, CA, USA) was used as a guiding beam. Experiments were monitored by a surgical microscope (Universal S3, Carl Zeiss, Oberkochen, Germany) equipped with a CCD camera. The ablation laser was rastered across the tissue surface in a single-shot mode with a step size of 10 µm. Ablation of samples was performed over square areas ranging from 0.2 to 1.5 mm on a side.
C. Scanning electron microscopy (SEM)
Electron micrographs of all tissue sections were obtained by SEM (Hitachi S-3000N, Hitachi High Technologies America, Pleasanton, CA, USA) with a tungsten electron source operating at 15 keV in high vacuum mode after spin coating with an 8 nm thick layer of Pt/Pd (4:1). All samples were air-dried, without fixation or dehydration. Images of ablation patterns created with different laser fluences were taken from sections that were not treated by a MALDI matrix. SEM images of ablation patterns for MALDI-MS analysis of proteins were taken from control sections that were laser ablated and treated by a MALDI matrix in the identical way as sections analyzed by MALDI-MS. While most SEM were recorded in imaging mode at 0° tilt, some were also recorded in topography mode at 45° tilt to estimate the depth of ablation and the roughness of the ablated tissue.
D. MALDI-MS
For analysis of phospholipids, tissue sections were manually sprayed with several cycles of a mixture of acetonitrile and water (1:1) that served to adhere tissue sections to the substrate.27 Sections were then sprayed by several cycles of 20 mg/mL solution of matrix p-nitroaniline in ethanol applied by an airbrush (Testors Corp., Rockford, IL, USA) and allowed to dry.26 For protein analysis, the spraying procedure gave poor results (data not shown). Tissue sections were instead spotted with 5 droplets of 0.2 µl of 15 mg/ml solution of sinapinic acid in a mixture of acetonitrile, water, and formic acid (50:40:10), as reported in earlier MALDI-MS work.28 Care was taken so that droplets were delivered only within the region of the laser ablated-pattern. All MALDI mass spectra were collected by a commercial instrument (Applied Biosystems 4700, AB Sciex, Foster City, CA, USA) equipped with a 355 nm desorption laser operating at 200 Hz, focused down to a spot size of ~150 µm. See Supporting Information for more details of the MALDI-MS analysis.
E. LDPI-MS
Laser desorption vacuum ultraviolet postionization MS data were collected using a home-built instrument10 equipped with a new 10.5 eV vacuum ultraviolet source for single photon ionization (see Supporting Information for more details).9 A 349 nm Nd:YLF pulsed laser (Spectra-Physics Explorer, Newport corporation, Irvine, CA, USA) with a pulse energy ranging between 1 and 10 µJ/pulse and a laser spot diameter of ~15 µm was used for desorption. The desorbed neutrals were ionized using the 118 nm ninth harmonic of a Nd:YAG laser (Tempest 10 Hz 355 nm, New Wave Research, Fremont, CA, USA), which was generated by focusing the 355 nm third harmonic into a cell filled with ~6.5 Torr of Xe.9,31,32
III. RESULTS
A. Optimization of Ablation Using Scanning Electron Microscopy (SEM)
The threshold for ablation of bovine eye lens tissue by ~75 fs, 800 nm laser pulses was evaluated microscopically by SEM. Samples were also examined using a surgical microscope equipped with a CCD camera (data not shown). Changes in electron and optical micrographs of the tissue were assessed after rastering the 200 µm diameter laser beam across the surface in a single shot mode with a step size of 10 µm. Such small step sizes were employed because at lower fluences only the center of the beam induced observable ablation (see below).
Microscopically observable changes in the tissue occurred at a threshold fluence of 0.14 J/cm2, corresponding to an irradiance of 1.9 × 1012 W/cm2. All fluences reported here are given by Φ = 2E/ π r2,25 where E is the laser pulse energy measured with the power meter and r is the beam radius as measured with a scanning knife edge (see Supporting Information). Losses in the optics are also accounted for in the quoted fluences. These results are consistent with those reported for plasma-mediated ablation of soft biological tissues.3,4,18
Panel A of Figure 1 shows the SEM image of an ablation pattern created by rastering a 200 µm diameter, 0.15 J/cm2 fluence laser beam across a 200 × 200 µm region with a step size of 10 µm. Individual craters had diameters of only ~10 µm, indicating that only the center of the 200 µm diameter laser beam lead to ablation at this fluence (see Supporting Information). Further support for the ~10 µm ablation spot diameter was displayed in the similar width of the horizontal rows formed in the tissue at this fluence.
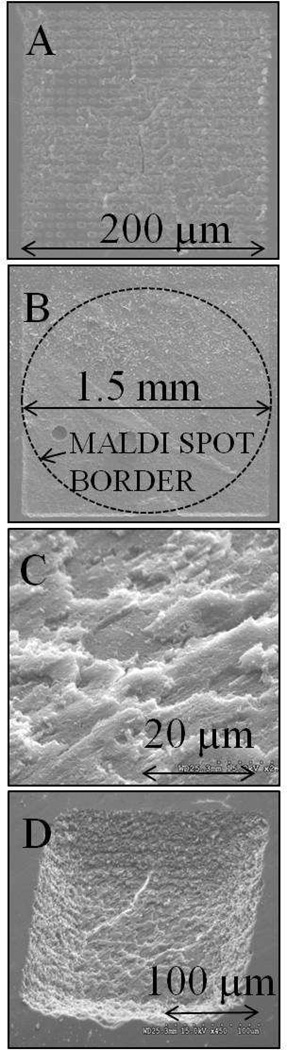
Figure 1.
Scanning electron micrographs (SEM) of bovine eye lens tissue showing ablation patterns created by rastering a laser focused to a beam diameter of 200 µm with 10 µm step in a single shot mode. A: After ablation by laser fluence of 0.15 J/cm2 (×250 magnification). B: Tissue immediately after laser ablation by 0.30 J/cm2, with dashed line showing area where matrix would be applied for subsequent MALDI analysis (×40). C: Topographic mode (45° tilt) of tissue after ablation by fluence of 0.30 J/cm2 (×2000). D: Topographic SEM of 200 × 200 µm ablation pattern created by 0.30 J/cm2 fluence laser pulses.
Fluences higher than 0.15 J/cm2 formed individual ablation craters larger than 10 µm in diameter, which, when combined with a 10 µm spacing between laser shots, overlapped to form a relatively smooth surface at the bottom of the ablated region. Panel B of Figure 1 is an SEM image of a 1.5 × 1.5 mm ablation pattern created by rastering the laser at a fluence of 0.30 J/cm2 across the surface of the tissue section, where the dashed line delineates the area that was subsequently used for matrix application and analysis by MALDI-MS. An SEM image of the tissue surface ablated with a fluence of 0.30 J/cm2 is shown in panel C of Figure 1. This image was acquired by SEM in topography mode under 45° tilt. The roughness of ablated surface measured by the size of the laser-generated, flaky features at the flat bottom of the laser ablated region appears to be on the order of a few µm (<5 µm).
The depth of the ablation patterns was estimated by varying the laser fluence and by SEM imaging in topography mode. Panel D of Figure 1 shows a topographic SEM image of a 200 × 200 µm wide ablation pattern created by laser pulses having a fluence of 0.30 J/cm2 and spaced by 10 µm. Geometric considerations indicate a crater depth of ~40 µm at 0.30 J/cm2. Similar experiments found crater depths ranged from ~20 µm at 0.17 J/cm2 to ~55 µm at 0.60 J/cm2.
B. MALDI-MS analysis of phospholipids
The fluence used for ablation prior to all MS analyses was 0.30 J/cm2. When combined with the 200 µm beam diameter and the 10 µm spacing between laser shots, it is clear that each point at the surface of the ablated tissue was exposed to multiple laser shots, although the intensity over most of the laser spot area was insufficient to cause ablation. MALDI-MS requires that a matrix solution be applied to a sample prior to analysis: p-nitroaniline crystals was applied for phospholipid analysis and sinapinic acid was applied for protein analysis.
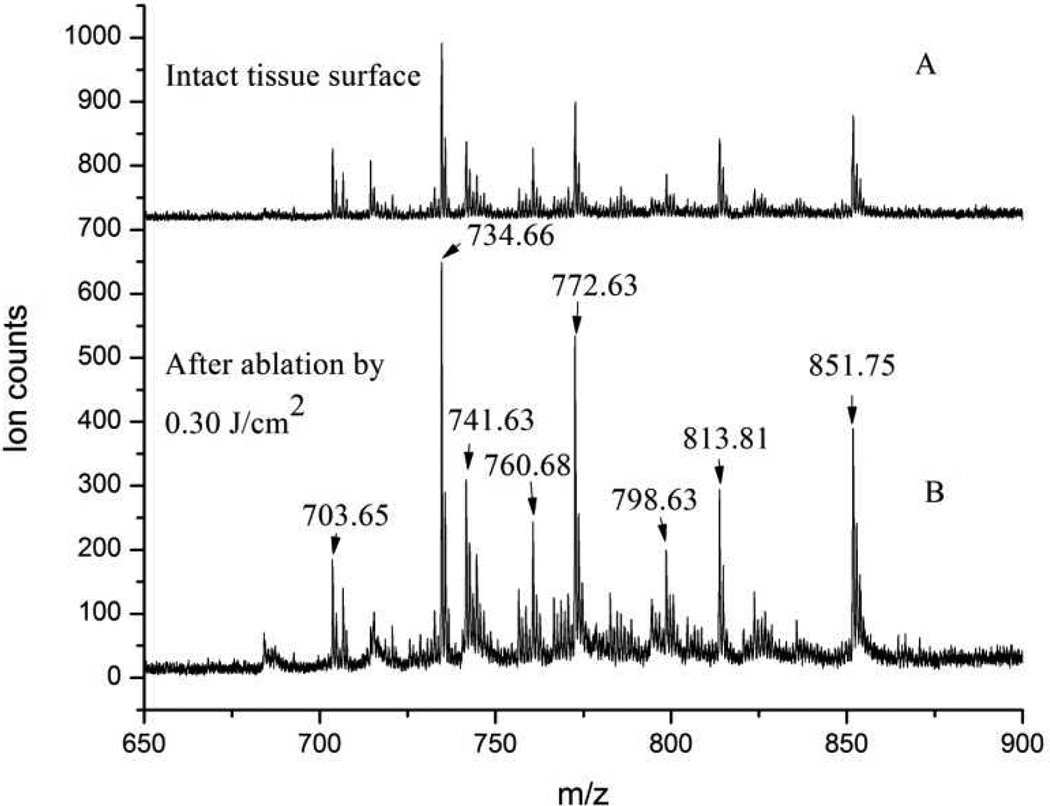
Figure 2 displays MALDI mass spectra of intact and laser-ablated eye lens tissue, showing similar phosphatidylcholines and sphingomyelins distributions. Spectrum A of Figure 2 was obtained from the intact tissue prior to ablation, whereas spectrum B was obtained from the laser-ablated tissue. Many of the peaks in both spectra correspond to those previously observed and assigned as phosopholipids.26 Table 1 summarizes the m/z values corresponding to the detected series of sphingomyelins (SM) and phosphatidylcholines (PC). Since prior work found that the phosopholipid composition does not vary across a few mm in eye lens tissue,26 it was expected that their composition would also not vary between adjacent MALDI spots.
Figure 2.
MALDI-MS of phospholipids obtained from eye lens tissue. A: intact tissue; B: after laser ablation by 0.30 J/cm2.
Table 1.
Predicted and observed m/z values in phospholipid MALDI-MS.26
| Theoretical m/z | Observed m/z | Assignment | Ion type |
|---|---|---|---|
| 703.58 | 703.65 | SM (16:0) | [MH]+ |
| 734.57 | 734.66 | PC (32:0) | [MH]+ |
| 741.53 | 741.63 | SM (16:0) | [MK]+ |
| 760.59 | 760.68 | PC (34:1) | [MH]+ |
| 772.53 | 772.63 | PC (32:0) | [MK]+ |
| 798.54 | 798.63 | PC (34:1) | [MK]+ |
| 813.68 | 813.81 | SM (24:1) | [MH]+ |
| 851.64 | 851.75 | SM (24:1) | [MK]+ |
Peak intensity ratios between individual peaks from intact tissue (spectrum A of Figure 2) are similar to those from laser-ablated tissue (spectrum B). For example, the ratio of ion intensities at m/z 734.7 to 813.8 is ~2.2 in both spectra, the ratio of m/z 772.6 to 798.6 is ~2.6, and the ratio of m/z 741.6 to m/z 760.7 is ~1.2. Slight differences in peak intensity ratios between spectra obtained from similarly treated tissue occurred randomly between experimental runs and were attributed to run-to-run fluctuations rather than to any laser ablation effects.
The overall ion intensity was 2–3 times higher when mass spectra were recorded from the laser-ablated tissue compared with intact tissue, as shown in Figure 2.
C. MALDI-MS analysis of proteins
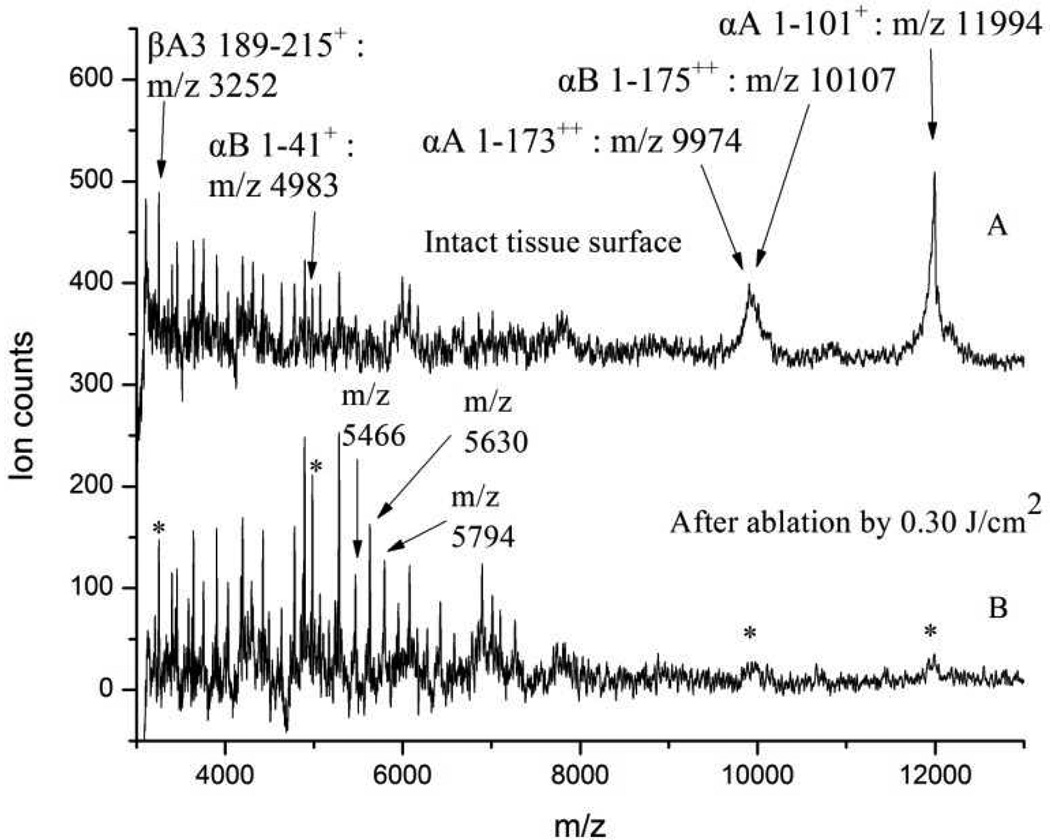
The most abundant lens tissue proteins, crystallins and their truncation products, were also observed to survive ultrafast laser ablation at a fluence of 0.30 J/cm2, albeit in a somewhat mass-dependent fashion. Figure 3 displays MALDI-MS of proteins obtained from bovine eye lens tissue in the mass range between m/z 3,000 and 13,000. Analysis of all collected MALDI-MS data showed that the intensities of peaks falling below approximately m/z 6,000 were preserved, while those above that mass displayed decreased intensity from the laser-ablated tissue. The most intense peaks appearing from the intact tissue surface (in spectrum A of Figure 3) were m/z 9,974; 10,107; and 11,994. These peaks were assigned to α-crystallin (αA 1–1732+), β-crystallin (αB 1–1752+), and the most abundant truncation product of α-crystallin (αA 1–101+), respectively.28 These three peaks almost completely disappeared from the laser-ablated tissue (spectrum B of Figure 3). Some known crystallin truncation products appearing at m/z 7,012 and 7,741, corresponding to (αA 1–58+) and (αA 1–65+),27 appeared consistently in MALDI-MS of both intact and ablated tissue surfaces. However, these two intermediate mass peaks appeared at rather low intensities and are therefore not discussed further. Table 2 shows the peak assignments of some of the prominent species seen by MALDI-MS analysis of eye lens tissue proteins.
Figure 3.
MALDI-MS spectra of proteins obtained from eye lens tissue. A: intact tissue; B: after laser ablation by 0.30 J/cm2.
Table 2.
| Theoretical m/z | Measured m/z | Assignement | Ion type |
|---|---|---|---|
| 3252.7 | 3252 | C terminal βA3 (189–215)peptide | [MH]+ |
| 4982 | 4983 | αB (1–41) peptide | [MH]+ |
| 7012 | 7012 | αA (1–58) peptide | [MH]+ |
| 7741 | 7739 | αA (1–65) peptide | [MH]+ |
| 9588 | 9588 | αA (1–80) peptide | [MH]+ |
| 9978 | 9974 | αA–crystallin | [MH2]+ |
| 10102 | 10107 | αB–crystallin | [MH2]+ |
| 11995 | 11994 | N terminal αA (1–101) peptide | [MH]+ |
The region of the protein MALDI-MS between m/z 3,000 and 6,000 displays a characteristic fingerprint of the protein content in the eye lens tissue (see Figure 3 and Supporting Information). Only a few of the species in this m/z region were identified, namely the m/z 4983 and m/z 3252 peaks attributed to (αB 1–41+) and (βA3 189–215+), respectively.28,33 However, the same overall pattern of peaks with similar relative intensities was obtained from all intact and ablated tissue analyzed.
Peak intensities below approximately m/z 6,000 increased when probed from the ablated region (Figure 3, spectra B), similar to the peak intensity increase found in the phospholipid spectra after ablation. This intensity increase was especially prominent for the three peaks labeled as m/z 5,466; 5,630; and 5,794.
D. LDPI-MS
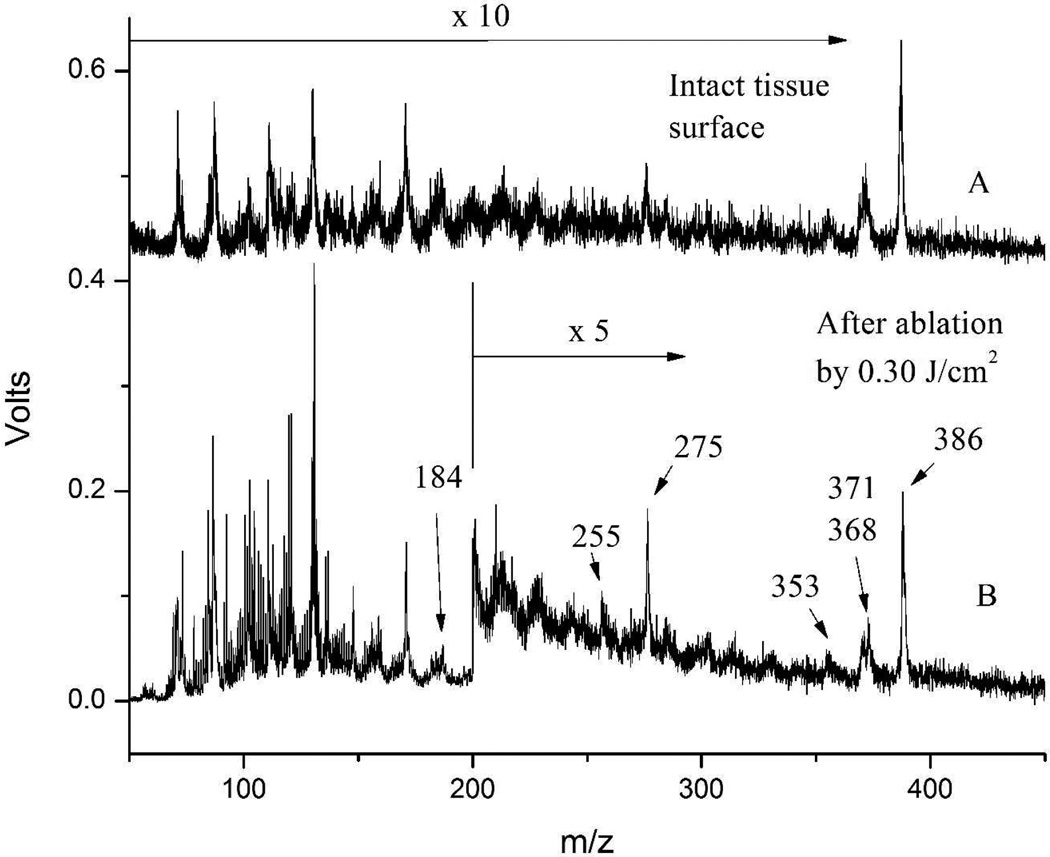
Figure 4 shows LDPI-MS analysis of intact tissue and after ablation by ultrashort laser pulses at a fluence of 0.30 J/cm2. There are no missing peaks, and no new peaks emerge from the laser ablated tissue. Rather, spectra from the intact and the laser-ablated tissue resemble each other to a high level of detail. The labeled peaks are thought to correspond to cholesterol and some of its known fragments, namely m/z 386 corresponding to M+, m/z 371 to [M-CH3]+, m/z 368 to [M-H2O]+, m/z 353 to [M-CH3-H2O]+, m/z 275 to [M-C7H11O]+ and m/z 255 to [M-C8H17-H2O]+.34 Table 3 summarizes the calculated and observed m/z values for cholesterol and its fragmentation products. Phosphocholine, a polar head group of phosphatidylcholines and sphingomyelins, was assigned to the peak at m/z 184.35
Figure 4.
LDPI-MS of eye lens tissue. A: intact tissue, peak intensities were multiplied by factor of 10; B: after laser ablation by 0.30 J/cm2, peak intensities above m/z 200 were multiplied by factor of 5.
Table 3.
Predicted and observed m/z values for cholesterol and its fragmentation products from LDPI-MS.34
| Theoretical m/z | Observed m/z | Assignment |
|---|---|---|
| 386.65 | 386 | M+ |
| 371.62 | 371 | [M-CH3]+ |
| 368.63 | 368 | [M-H2O]+ |
| 353.60 | 353 | [M-CH3-H2O]+ |
| 275.49 | 275 | [M-C7H11O]+ |
| 255.42 | 255 | [M-C8H17-H2O]+ |
Comparison of spectra obtained from the intact and laser-ablated tissue found that the overall peak intensity for the latter increased by an order of magnitude in the region below m/z 200 (enlarged spectrum shown in Supporting Information) and by a factor of two for the region between m/z 200 and 450.
IV. DISCUSSION
The results presented here show that ultrashort pulse laser ablation can remove several tens of micrometers of eye lens tissue while leaving the underlying tissue with relatively little chemical damage, as determined by subsequent MS analyses. The surface of the tissue remaining at the bottom of the laser-ablated area had a slightly rougher texture than the intact tissue. However, chemical analysis of cholesterol, phospholipids, and proteins did not reveal any chemical damage caused by ablation for analytes smaller than ~6 kDa. The observation that the mass spectra from tissue at the bottom of the ablation crater remained relatively unaltered in this mass range supports the use of ultrashort pulsed laser ablation for depth profiling. Little prior experimental data has directly probed the extent of chemical damage in tissue following such ablation (see below). The results presented here are in agreement with studies of ultrafast laser vaporization of blood and eggs followed by electrospray ionization, which found molecules as large as lysozyme (MW ~14,500 Da) can be vaporized intact.15,17 A notable decrease in sensitivity to proteins larger than ~6 kDa found here might be attributed to causes other than laser ablation-induced degradation. For example, lower mass analytes may be more easily desorbed from tissue surface previously treated by ultrashort laser ablation, resulting in higher relative peak intensity.
The increased intensity of mass spectra when sampling laser ablated tissue (two to threefold for MALDI-MS and up to an order of magnitude for LDPI-MS) may be attributed to lysing of the tissue by the ultrashort laser pulses. The lysed tissue would facilitate the extraction of analytes into the crystallized matrix during the sample preparation for MALDI-MS analysis. Additionally, in the case of LDPI-MS, these lysed species are more readily desorbed than when still bound within the intact tissue. The apparent increase in ion intensity in spectra obtained from laser- ablated samples suggests that ultrashort pulse laser ablation might serve as a tool to prepare biological tissue prior to MS imaging analysis.
Both MALDI-MS and LDPI-MS analyses were conducted after ablation at fluences twice that of the ablation threshold, using laser beam diameters ~20 times greater than the spacing between individual shots. These experiments show that although each point at the sample surface was actually exposed to multiple laser shots, little change was observed in the subsequent MS other than an increase in ion intensity. Clearly, lower laser fluence per pulse would cause even less damage to the tissue.
Several prior studies argued for the potential use of ultrashort pulse laser ablation for non-destructive eye surgery in the plasma-mediated regime.5–8 Prior work argued that the damage inflicted by <100 fs, 800 nm laser pulses in plant tissue was superficial, suggesting the possibility of in-vivo analysis.13 Others have argued that the advantages of ultrashort pulse laser ablation of polymeric coatings and biological tissues include both precision and a reduced zone of damage.3–5,8,13,18,19,36 However, these prior studies generally did not employ direct chemical analysis to demonstrate the absence of chemical damage. Furthermore, a previous study by some of the authors showed that a small organic molecule introduced into a bacterial biofilm remained intact after ultrashort pulse laser ablation, as analyzed by LDPI-MS.25
The depth of the laser-ablated volume at a fluence of 0.30 J/cm2, used prior to all MS analyses, was estimated to be ~40 µm, and the ablated area displayed a roughness of ~5 µm. Lower fluences yielded shallower ablation depths, with a minimum depth of ~20 µm at 0.17 J/cm2 fluence, which is slightly above the ablation threshold of 0.14 J/cm2. It follows that ultrashort pulse laser ablation should display a maximum depth resolution on the order of 20 ± 5 µm under the conditions employed here. Use of MALDI-MS for subsequent analysis will degrade this resolution further, as the matrix can extract compounds from up to 40 µm within tissue.37
These penetration depth estimates do not reflect the full potential for depth profiling. Earlier studies using resonant 2.94 µm ns laser pulses reported a depth resolution of ~30 – 50 µm.38 A rough comparison of the number of pulses needed to penetrate a plant leaf with infrared ns pulses38 and near infrared fs pulses16 indicated that the etch depth per pulse may be almost an order of magnitude smaller for fs ablation.
Panel D of Figure 1 shows an ablation pattern ~40 µm deep, created by rastering a laser beam at a fluence of 0.30 J/cm2 with a step of 10 µm. By dividing the total ablated volume (200 × 200 × 40 µm) by the total number of pulses (20 × 20), the volume ablated per pulse was estimated to be ~4 × 103 µm3.
In the present experiments, the laser beam was focused to a diameter of 200 µm at the surface, whereas only a ~10 µm diameter crater formed at the threshold fluence for ablation. This indicated that only a small portion of the Gaussian-shaped beam close to the laser axis was effective in ablating the tissue (see panel A of Figure 1). A similar result was reported previously for the ablation of microbial biofilms.25 Collectively, these results suggest that crater diameters and etch depths even smaller than 10 µm could be obtained if the laser beam was focused to a smaller diameter at the surface and the fluence was more carefully controlled.39–41
Finally, the irradiance threshold for ablation may be estimated from the relation41
Taking ω0 = 55 µm, ωz = 100 µm, an ablation radius rab = 5 µm, and peak irradiance of I0 = 2 × 1012 W/cm2 leads to a threshold irradiance of 6 × 1011 W/cm2.
V. CONCLUSIONS
Molecular depth profiling of layers thinner than 1 µm with a depth resolution of few tens of nm are so far achievable only by cluster ion sputtering, which still suffers from damage accumulation problems and cannot be used for analysis of layers thicker than a few µm.20–24 It is shown here that ultrashort pulse laser ablation should be feasible for depth profiling of tissue with a few tens of microns depth resolution when used in combination with an MS analysis method. Furthermore, prior work with ultrashort pulse micromachining indicates a potential for sub-µm depth resolution.39–41
Ultrashort laser pulses can desorb molecular analytes from biological samples, permitting mass analysis of both direct ions created in the ablation event12,13 and desorbed neutrals after postionization.14–17 Combining depth profiling by ultrashort pulse laser ablation with analysis of either direct ions or ablated neutrals postionized by vacuum ultraviolet postionization9–11 or electrospray ionization14–17 would allow rapid molecular tomography of biological tissues and other types of samples.
It is acknowledged that the use of separate instruments for ultrashort pulse ablation and MS imaging analysis is unwieldy. Furthermore, the need to add matrix to the sample would limit the use of MALDI-MS for such a depth profiling analysis. A distinct and more attractive strategy would be to directly analyze species desorbed during ultrashort pulse ablation, which does not require the addition of matrix.12–17 Depth profiling would then consist of rapid 2D imaging of a biological sample followed by cumulative analysis of the newly exposed surface until a complete 3D tomographic map of the sample volume is obtained. Such an experiment should be possible using a single instrument, which is currently under construction.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Biomedical Imaging and Bioengineering via grant EB006532. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health. The authors acknowledge the support of the mass spectrometry, electron microscopy, and histology core facilities of the Research Resources Center at UIC for assistance in tissue microtoming as well as collection of the SEM and MALDI-MS data.
Footnotes
SUPPORTING INFORMATION
The following figures and adjacent text can be found in Supporting Information.
Figure S1. The optical delivery system for ultrafast pulsed laser ablation.
Figure S2. Scanning electron micrograph of bovine eye lens tissue showing ablation patterns and crystals of sinapinic acid matrix.
Figure S3. M/z 3000 – 6000 region of MALDI-MS of proteins obtained from eye lens tissue.
Figure S4. M/z 60 – 195 region of the LDPI-MS of tissue.
Figure S5. Example of knife edge measurement of laser beam profile.
Figure S6. Beam profile from knife edge measurement.
Figure S7. SEM image showing the size of individual ablation craters in eye tissue.
REFERENCES
- 1.Seeley EH, Caprioli RM. Proceedings of the National Academy of Sciences, U.S.A. 2008;105:18126. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chughtai K, Heeren RMA. Chemical Reviews. 2010;110:3237. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel A, Venugopalan V. Chemical Reviews. 2003;103:577. doi: 10.1021/cr010379n. [DOI] [PubMed] [Google Scholar]
- 4.Vogel A, Noack J, Huttman G, Paltauf G. Applied Physics B. 2005;81:1015. [Google Scholar]
- 5.Toyran S, Liu Y, Singha S, Shan S, Cho MR, Gordon RJ, Edwards DP. Experimental Eye Research. 2005;81:298. doi: 10.1016/j.exer.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Sun S, Singha S, Cho MR, Gordon RJ. Biomaterials. 2005;26:4597. doi: 10.1016/j.biomaterials.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 7.Stachs O, Schumacher S, Hovakimyan M, Fromm M, Heisterkamp A, Lubatschowski H, Guthoff R. Journal of Cataract & Refractive Surgery. 2009;35:1979. doi: 10.1016/j.jcrs.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Kessel LE, van der Poel M, Larsen M. PLoS ONE. 2009:5. doi: 10.1371/journal.pone.0009711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanley L, Zimmermann R. Analytical Chemistry. 2009;81:4174. doi: 10.1021/ac8013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akhmetov A, Moore JF, Gasper GL, Koin PJ, Hanley L. Journal of Mass Spectrometry. 2010;45:137. doi: 10.1002/jms.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasper GL, Takahashi LK, Zhou J, Ahmed M, Moore JF, Hanley L. Analytical Chemistry. 2010;82:7472. doi: 10.1021/ac101667q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry JI, Sun S, Dou Y, Wucher A, Winograd N. Analytical Chemistry. 2003;75:5146. doi: 10.1021/ac034375p. [DOI] [PubMed] [Google Scholar]
- 13.Coello Y, Jones AD, Gunaratne TC, Dantus M. Analytical Chemistry. 2010;82:2753. doi: 10.1021/ac9026466. [DOI] [PubMed] [Google Scholar]
- 14.Brady JJ, Judge EJ, Levis RJ. Rapid Communications in Mass Spectrometry. 2009;23:3151. doi: 10.1002/rcm.4226. [DOI] [PubMed] [Google Scholar]
- 15.Judge EJ, Brady JJ, Levis RJ. Analytical Chemistry. 2010;82:10203. doi: 10.1021/ac102409k. [DOI] [PubMed] [Google Scholar]
- 16.Judge EJ, Brady JJ, Barbano PE, Levis RJ. Analytical Chemistry. 2011;83:2145. doi: 10.1021/ac102978f. [DOI] [PubMed] [Google Scholar]
- 17.Brady JJ, Judge EJ, Levis RJ. Proceedings of the National Academy of Sciences. 2011;108:12217. doi: 10.1073/pnas.1105673108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franjic K, Cowan ML, Kraemer D, Miller RJD. Opt. Express. 2009;17:22937. doi: 10.1364/OE.17.022937. [DOI] [PubMed] [Google Scholar]
- 19.Lim YC, Johnson J, Fei Z, Wu Y, Farson DF, Lannutti JJ, Choi HW, Lee LJ. Biotechnology and Bioengineering. 2011;108:116. doi: 10.1002/bit.22914. [DOI] [PubMed] [Google Scholar]
- 20.Bolotin IL, Tetzler SH, Hanley L. Journal of Physical Chemistry C. 2007;111:9953. [Google Scholar]
- 21.Fletcher JS. Analyst. 2009;134:2204. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 22.Wucher A, Cheng J, Zheng L, Winograd N. Analytical and Bioanalytical Chemistry. 2009;393:1835. doi: 10.1007/s00216-008-2596-5. [DOI] [PubMed] [Google Scholar]
- 23.Mao D, Wucher A, Winograd N. Analytical Chemistry. 2010;82:57. doi: 10.1021/ac902313q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickerman JC. Analyst. 2011;136:2199. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- 25.Milasinovic S, Liu Y, Gasper GL, Zhao Y, Johnston JL, Gordon RJ, Hanley L. Journal of Vacuum Science Technology A. 2010;28:647. doi: 10.1116/1.3397736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rujoi M, Estrada R, Yappert MC. Analytical Chemistry. 2004;76:1657. doi: 10.1021/ac0349680. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Schey KL. IOVS. 2006;47:2990. doi: 10.1167/iovs.05-1529. [DOI] [PubMed] [Google Scholar]
- 28.Grey AC, Schey KL. IOVS. 2009;50:4319. doi: 10.1167/iovs.09-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma KK, Santoshkumar P. Biochim. Biophys. Acta. 2009;1790:1095. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura H, Liu Y, Witt TE, Edward DP, Gordon RJ. Investig. Ophth. Vis. Sci. 2009:50. doi: 10.1167/iovs.07-1536. [DOI] [PubMed] [Google Scholar]
- 31.Van Bramer SE, Johnston MVJ. Am. Soc. Mass Spectrom. 1990;1:419. doi: 10.1016/1044-0305(90)85024-G. [DOI] [PubMed] [Google Scholar]
- 32.Zhou M, Wu C, Edirisinghe PD, Drummond JL, Hanley L. Journal of Biomedical Materials Research. 2006;77A:1. doi: 10.1002/jbm.a.30591. [DOI] [PubMed] [Google Scholar]
- 33.Su SP, McArthur JD, Aquilina JA. Experimental Eye Research. 2010;91:97. doi: 10.1016/j.exer.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Partridge LG, Djerassi C. Journal of Organic Chemistry. 1977;42:2799. doi: 10.1021/jo00437a001. [DOI] [PubMed] [Google Scholar]
- 35.Vaezian B, Anderton CR, Kraft ML. Analytical Chemistry. 2010;82:10006. doi: 10.1021/ac101640c. [DOI] [PubMed] [Google Scholar]
- 36.Keene LT, Fiero T, Clayton CR, Halada GP, Cardoza D, Weinacht T. Polymer Degradation and Stability. 2005;89:393. [Google Scholar]
- 37.Crossman L, McHugh NA, Hseih Y, Korfmacher WA, Chen J. Rapid Communications in Mass Spectrometry. 2006;20:284. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 38.Nemes P, Barton AA, Vertes A. Analytical Chemistry. 2009;81:6668. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 39.Kuper S, Stuke M. Applied Physics Letters. 1989;54:4. [Google Scholar]
- 40.Konig K, Riemann I, Fritzsche W. Optics Letters. 2001;26:819. doi: 10.1364/ol.26.000819. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell S, Mazur E. Medical Laser Application. 2005;20:193. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.