Abstract
Objective
Susceptibility to inflammatory arthritis is determined by a complex set of environmental and genetic factors, but only a portion of the genetic effect can be explained. Conventional genome-wide screens of arthritis models using crosses between inbred mice have been hampered by the low resolution of results and by the restricted range of natural genetic variation sampled. We sought to address these limitations by performing a genome-wide screen for determinants of arthritis severity using a genetically heterogeneous cohort of mice.
Methods
Heterogeneous Stock (HS) mice derive from eight founder inbred strains by serial intercrossing (N>60), resulting in fine-grained genetic variation. With a cohort of 570 HS mice, we performed a genome-wide screen for determinants of severity in the K/BxN serum-transfer arthritis model.
Results
We mapped regions on chromosomes 1, 2, 4, 6, 7 and 15 that contain QTLs influencing arthritis severity at a resolution of a few Mb. In several instances, these regions proved to contain 2 QTLs: the region on chromosome 2 includes the C5 fraction of complement known to be required for K/BxN arthritis, but also contained a second adjacent QTL, for which an intriguing candidate is Ptgs1 (Cox-1). Interesting candidates on Chr4 include the Padi gene family, encoding peptidyl-arginine-deiminases responsible for citrulline protein modification; suggestively, Padi2 and Padi4 RNA expression was correlated with arthritis severity in HS mice.
Conclusions
These results provide a broad overview of the genetic variation that controls the severity of K/BxN arthritis and suggest intriguing candidate genes for further study.
The pathogenesis of rheumatoid arthritis (RA) remains poorly understood, and its complex genetic basis has been difficult to dissect. Genome-wide association studies (GWAS) have identified more than thirty genomic regions that contribute to the susceptibility to RA (1). Although the causal polymorphism has been formally identified in a few cases, most of the regions identified in GWAS studies remain ambiguous, and not connected to a functionally-relevant polymorphism. Similarly, genome-wide screens in mouse models of RA have implicated over 50 loci in the pathogenesis of inflammatory arthritis, but only a few genes within these regions have been identified (2;3).
The K/BxN serum-transfer arthritis model is well-suited to genetic analysis. K/BxN mice develop a progressive inflammatory arthritis that shares the major histologic features of RA (4;5). K/BxN T and B cells recognize a ubiquitously expressed protein, glucose-6-phosphate isomerase (GPI), leading to very high production of anti-GPI antibodies. The transfer of serum from arthritic K/BxN mice or of purified anti-GPI antibodies into healthy animals, even lymphocyte-deficient mice, provokes arthritis within 1–4 days after injection, peaks at 10–14 days, and resolves slowly over the next 2 weeks (6). There is considerable variation among inbred strains in the susceptibility to arthritis induced by transfer of K/BxN serum (7–9). The serum-transfer model isolates the inflammatory effector-phase cascade from the early immunologic initiation events, thereby reducing the complexity of the factors under examination. Indeed, we and others have been able to pinpoint a few loci where natural genetic variation conditions the severity of arthritis induced by K/BxN serum: the Hc gene which encodes the C5 fraction of complement (10;11); the Il1b locus, an allele of which is present in wild-derived mice and in a minority of inbred strains, which results in a 5-fold higher expression of Il-1β upon challenge, and to more aggressive arthritis (9).
QTL mapping experiments in crosses of inbred mouse strains are limited to studying the regions and polymorphisms that differ between a pair of strains. In addition, such intercross or backcross studies have poor resolving power because the distances between informative recombinants are great. As a consequence, fine-resolution mapping for complex traits requires either impossibly large numbers of animals in a single cross or many generations of intercrossing to achieve sub-centimorgan resolution (12).
The Northport Heterogeneous Stock (HS) was generated from 8 founder inbred strains by serial intercrossing for more than 50 generations (13). This breeding scheme accumulates recombinants and turns each HS chromosome into a fine-grained mosaic of the founder strain genomes. HS mice allow for quantitative trait locus (QTL) mapping at very high resolution because the distance between recombinants is small (~2 cM), sampling a diverse cross-section of the genetic variability in mice. The potential of genetic mapping in HS mice was exploited in a large-scale QTL mapping, in which 843 QTLs for 97 traits were mapped to an average interval of 2.8 Mb (14). Here, we attempted to exploit these characteristics and performed a whole-genome scan in a cohort of HS mice tested for sensitivity to arthritis by transfer of arthritogenic K/BxN serum, and identified a number of genomic regions that contribute to the effector-phase of inflammatory arthritis
METHODS
Mice
The HS/NPT mice tested were from the 53rd generation of circular breeding from an initial combination of eight inbred strains (A/J, AKR/J, BALB/cJ, C3H/HeJ, C57BL/6J, DBA/2J, CBA/J and Lp/J) (13). Breedings were set up to avoid the production of homozygous C5-deficient mice, and 570 mice were challenged by K/BxN serum injection (150 microliters on days 0+2). The thickness of each ankle was measured and a clinical score for each paw (0–3) was determined on days 0–10. Four phenotypes were calculated: MAT: maximum ankle thickness achieved minus thickness on Day 0, MCI: maximum clinical index (Days 0–10), IAT: integrated ankle thickening from Day 0–10, and ICI: integrated clinical index from Day 0 –10. On Day 14, mice were sacrificed, spleens snap-frozen for RNA isolation, and serum collected for determination of anti-GPI titers by ELISA (7). Genotyping of tail DNA for 1449 SNPs was performed with Illumina mouse medium density linkage panel. Genotyping of additional SNPs for higher density analysis was performed by Sequenom-iplex (15) or by allele-specific fluorogenic-PCR (16).
Genomewide QTL
For linkage disequilibrium (LD) mapping of QTL, the underlying haplotype structure of each mouse was inferred and then tested for association with MAT. Haplotype descent was inferred using the hidden Markov model (HMM) implemented in HAPPY (17), using known founder genotypes and recombination distances to provide a probabilistic estimate of haplotype descent at each interval for each mouse. For mouse i at marker interval m, HAPPY computes a vector gi(m) containing the expected proportion of genetic material descended from each of the 36 possible distinct pairs of founder haplotypes. This vector is then used to characterize variation at the locus and for tests of phenotype association.
Genetic studies of outbred populations such as the HS are prone to confounding from unveven genetic relatedness between individuals, requiring careful treatment to avoid false-positive associations (18) from familial structure (sibship), common environment (cage) and the day of testing (cohort). Following (19), we modeled the effect of a putative QTL at locus m on the MAT of animali as:
| (Eq 1) |
where C is a set of fixed effects covariates that includes sex, xi(c) is the value of covariate c for individual i, εi~N(0,σ2) is the residual, and sibshipk[i], cohorth[i], and cagea[i]are normally distributed random effects. For (unconditional) mapping of single QTL, we obtained the nominal significance of the QTL at each locus m by comparing the fit of a null model, Q = 0, and alternative (QTL) model, Q = βTgi(m), via a likelihood ratio test (LRT; procedures as described in (19)). Variation at the Hc locus affects K/BxN arthritis (9;10). To determine QTL with variation uncorrelated with Hc and independent QTL in the region around Hc, we controlled for the mean effect of the Hc heterozgyote by including C5 genotype status among the covariates in C of Eq 1.
Dissecting QTL regions: fine-mapping of imputed SNPs
To maximize resolution, we imputed ungenotyped SNPs in regions of interest, based on strain distribution patterns (SDP) are available for HS founder strains (http://www.sanger.ac.uk/cgi-bin/modelorgs/mousegenomes/snps.pl), using a variant of the approach of (20): probabilities in gi(m) are combined with the SDP s to give the expected number of high alleles (additive dosage; ai(s)) and the probability of being heterozygote (dominance dosage; di(s)). The association of a SNP with MAT controlling for C5 was tested by comparing the fit of Eq 1 with Q = ai(s) + di(s) vs Q = 0 via an LRT as above.
Dissecting QTL regions: modeling joint action of SNPs by local multilocus analysis
To identify plausible sets of independent signals within local regions we performed a multilocus analysis using the resample model averaging approach of (18). Starting with the mixed model in Eq 1, we incorporated the effects of additional multiple SNPs into Q, choosing SNPs by stepwise selection that minimized the BIC (21). We applied this procedure to 200 random 63%-subsamples, recording for each SNP the proportion of subsamples in which it was selected. This resample model inclusion probability (RMIP) provides a measure of how robust an association is to correlations among other candidate SNPs and to finite sampling of the individuals.
Due to LD, our set of imputed SNPs was highly redundant such that ~100K SNPs might reduce to ~3K unique representatives. Our selection scheme chose between equally best fitting SNPs at random. To identify regions in which SNPs are consistently included but where high correlation meant that no single SNP predominated we used an RMIP sliding-window based on the range probability described by (18): for each SNP position, we report the average number of SNPs included in a 0.5Mb radius.
Expression analysis
Genome-wide expression profiles for whole spleens of each of the 8 HS founder strains were determined on the MuGene ST 1.0 microarray (Affymetrix). Transcripts from the Padi gene family were quantitated by Taq-Man RT-PCR in splenic RNA from inbred mice and 278 of the experimental HS mice. Timed expression profiles from synovial fluid and synovium of C57BL/6J mice after K/BxN serum transfer have been reported (22).
RESULTS
K/BxN arthritis in an HS cohort
The HS mice tested were from the 53rd generation of the pedigree derived from a combination of eight inbred strains (13), which include high as well as low arthritis responders (8). Three of the 8 strains harbor a spontaneous mutation in the Hc gene, which encodes complement factor 5 (C5) previously determined to have an important impact on arthritis in the K/BxN serum-transfer model (10). Therefore, the parents of the HS mice used in this study were genotyped for the Hc null allele, and breedings were set up to avoid the production of homozygous C5-deficient mice. Of the resulting progeny, 570 C5+/+ or C5+/o mice at 4 weeks of age were tested by injection of K/BxN serum. The experiment was performed over a 7 week period, and a strict operating protocol was adopted to minimize noise and drift, with a single operator and set times to minimize confounders from diurnal variation. A single lot of serum was used throughout, at a dose determined to yield robust disease in C57Bl/6 mice.
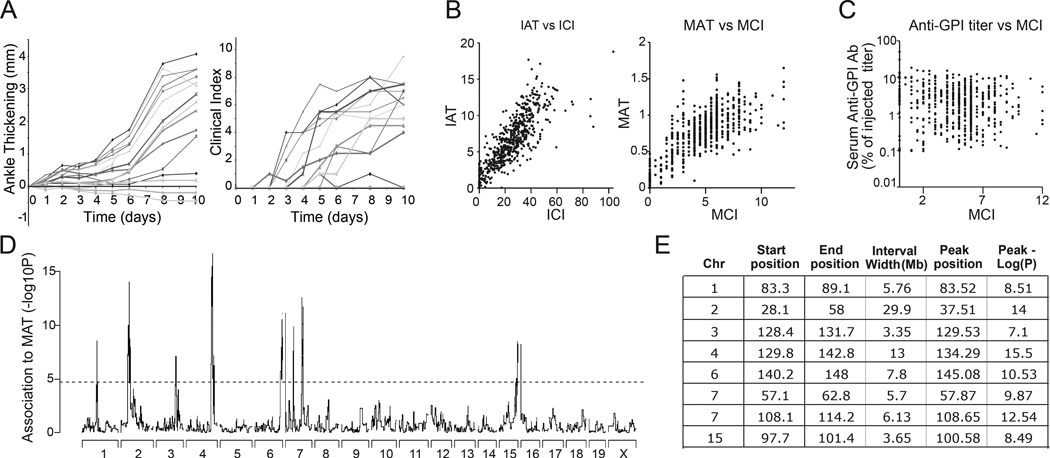
A wide spectrum of arthritis severity was exhibited by the experimental mice, as shown for 16 representative mice in Fig 1A, B. Some exhibited strong disease, with scores as high as in the strongest responders inbred strains. Fifty mice showed no arthritis whatsoever, but this was unlikely to be due to misinjection because residual anti-GPI titers in the serum of these non-responders were not markedly different from those of responder mice (Fig. 1C). Some mice with low residual anti-GPI exhibited robust arthritis, suggesting that residual titers might also be due to accelerated clearance or deposition in some cases. Therefore, all mice were included in the genetic analysis. The different metrics from measured ankle thickening (MAT, IAT) or from operator evaluation (MCI, ICI) were all highly correlated.
Figure 1.
Arthritis phenotype distribution for the HS mice. A. Change in the ankle thickness (left) and clinical index (right) over time in 16 representative mice. B. Correlation between phenotypes. Integrated ankle thickness (IAT) versus integrated clinical index (ICI) and Maximum ankle thickness (MAT) versus maximum clinical index (MCI) C. The anti-GPI titer in the serum of the experimental mice at Day 10, expressed as a percentage of the original GPI-titer of the injected serum, plotted versus the MCI. D. Genome-wide marker-trait association to the MAT phenotype, for the whole set of 570 HS mice tested E. Position of regions exceeding the genome-wide significance threshold (−log10P = 4.67) for MAT. The start and end of the interval are defined as the first and last SNPs to exceed the genome-wide significance threshold. The “Peak position” is the location of the SNP with the highest −log10P within the interval for the full genetic model. Genome-wide QTL Analysis for the phenotype Maximum Ankle Thickening (MAT). Significance is indicated on the y-axis by the −log10P value. Location in Mb is on the x-axis. Threshold for genome-wide associated is indicated by a dashed line (−log10P = 4.67).
Genetic loci associated with arthritis severity in the HS cohort
We identified quantitative trait loci (QTL) for arthritis severity using LD mapping of inferred haplotypes in combination with hierarchical modeling of family and covariate effects (19). This used the HAPPY model (17) of genetic association, in which the phenotype is associated not with individual markers but instead with inferred descent between markers. We collected genotypes in two stages, first performing a mid-density screen to identify major regions of association, then genotyping at higher density around peaks identified in the first stage. For the first stage, we genotyped 1449 single-nucleotide polymorphisms (SNPs) (average spacing of 1.5 Mb) including a SNP that identified the C5 loss-of-function mutation in the Hc gene. The proportion of mice successfully genotyped varied from 90–100 % for the different SNPs.
Given the high degree of correlation among the different arthritis measures, we focused our QTL analysis on MAT. Potential QTL identified in the mid-density scan as exceeding the genome-wide significance threshold were found on chromosomes 1, 2, 3, 4, 6, 7 and 15 and were subjected to higher density genotyping at an average spacing of one marker every 200 kb, yielding the overall profile depicted in Fig 1D–E.
As expected, a highly significant peak on Chr2 at 34.9 Mb, the location of the Hc gene, was detected. This region proved much broader than others, suggesting more complexity. The 83.3 to 89.0 Mb region on Chr1 may coincide with that identified on distal Chr1 in a B6xNOD F2 intercross (8), and in a selected backcross in the BALB/cxSJL combination (9). The other regions are novel with respect to K/BxN arthritis, but may correspond with potential QTLs in the collagen- or pristane-induced arthritis models (Cia3 on Chr6, Cia41 and Cia7 on Chr7, Pgia9/Cia36 on Chr15 (3)).
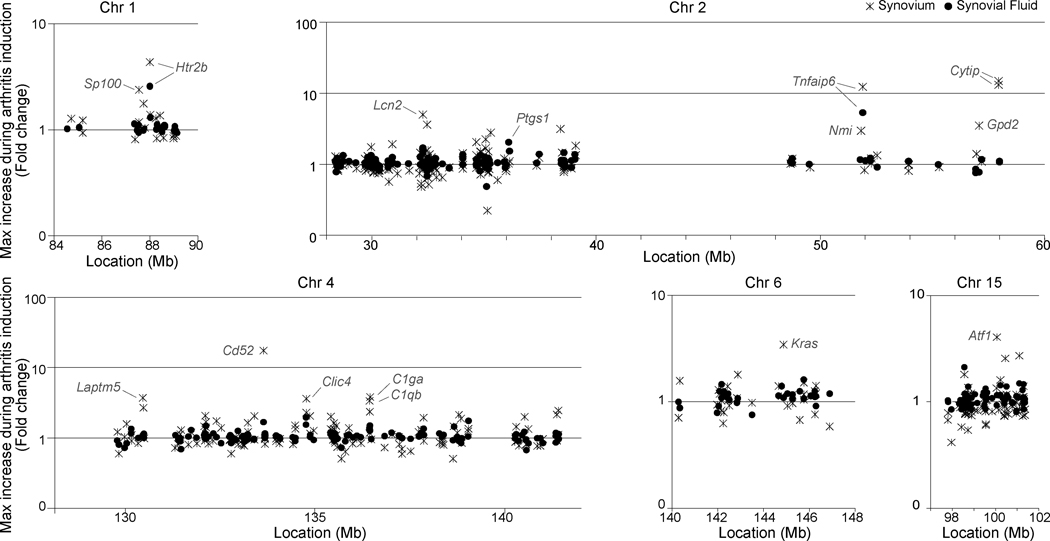
To complement the genetic analysis, we searched previously reported gene expression profiles of joint tissue (synovium and synovial fluid) from C57BL/6J mice at different stages of K/BxN serum transfer arthritis (22), under the assumption that causal variants would have a higher probability of corresponding to transcripts altered by arthritis (Fig. 2). This identified a number of genes whose expression changed robustly in the course of K/BxN arthritis. Some intriguing candidates, mapping to the peak of the relevant intervals include the TNF-induced Tnfaip6 on Chr2 (a protease-inhibitor cofactor that partakes in the protease network associated with inflammation), Cd52 on Chr4 (a cell surface activating protein, against which a depleting monoclonal antibody is currently in clinical trials for RA), or the GTPase oncogene Kras on Chr6.
Figure 2.
Expression of genes within associated loci. C57BL/6J mice were injected with K/BxN serum. Synovium was collected on day 0, 1, 3, 7, 12 and 18 and synovial fluid on day 3, 7, 12, and 18, RNA was isolated and microarray analysis was performed. Each panel represents the genes encoded within one of the disease-associated QTLs. X axis = chromosome location (bp). Y axis = fold change in expression. “x” = the maximum fold change in gene expression in synovium tissue, compared to day 0. “•” = the maximum fold change in synovial fluid compared to synovial fluid day 3.
Focused Analysis of Chromosome 2
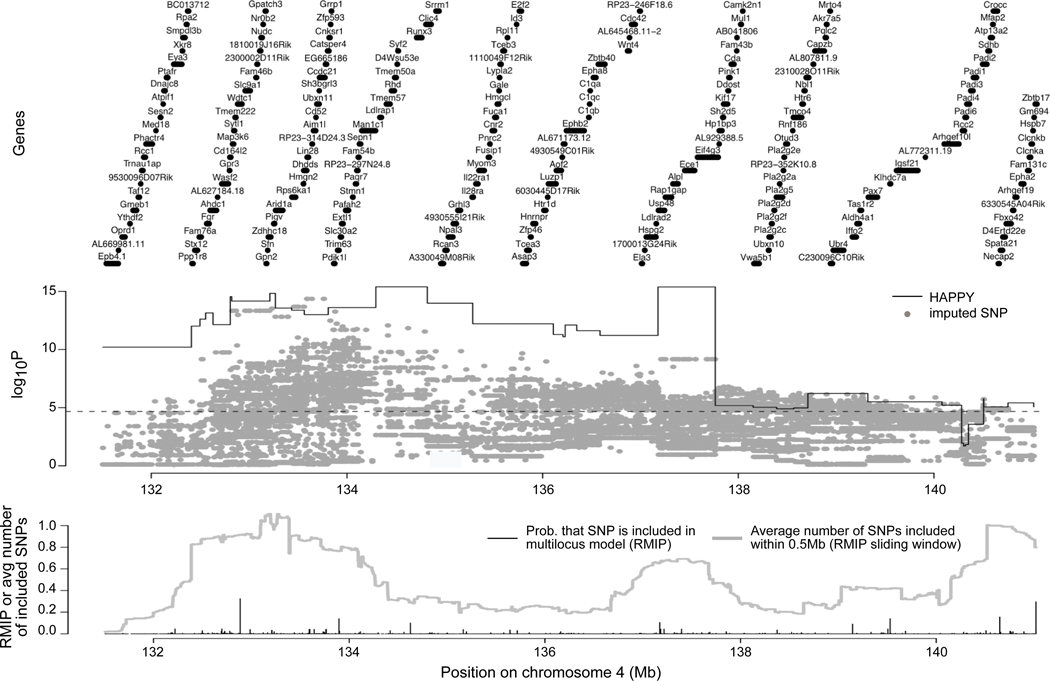
The region of association on Chr2 included Hc, a known determinant of arthritis susceptibility in several mouse models, but the large interval suggested the possibility that it contained more than one QTL. In order to determine whether Hc was the only determinant of arthritis severity in this region, a scan conditioning on the genotype of the informative Hc SNP was performed for the region from 25.95104 to 54.01145Mb (Fig 3, middle panel). Higher density SNP data was generated (97 markers spaced at 200 kb interval on average), and we imputed an additional 54,891 informative SNPs in the region and tested each for association with MAT, again controlling for C5 (Fig 3).
Figure 3.
Fine-mapping of QTL in the region of the C5 gene (Hc) on Chr2 using HAPPY and imputed SNP association. Top panel shows the locations of annotated genes. Middle panel shows QTL mapping of HAPPY haplotype descent probabilities in 79 marker intervals (stepped lines) and 54,891 imputed SNPs (dots). Thin lines show mapping using single interval mapping (continuous), and the corresponding genomewide threshold for significance (dashed; logP=4.67), thick lines show results of single interval mapping conditional on Hc with corresponding genomewide threshold (dashed; logP=4.62), gray dots show mapping of imputed SNPs for both additive and dominance genotypes, and a cross indicates the association of the C5 genotype (i.e., setting Q as Hc in Eq 1; logP=10.4, see Methods). Bottom plot shows multilocus mapping of the imputed SNPs. Black spikes show the proportion of times (RMIP) each SNP was included in a multiple-SNP model predicting the phenotype. Because RMIPs are often split among neighboring SNPs that have been rendered indistinguishable by close linkage, the gray line provides a score (RMIP sliding window) that aggregates inclusions local to each SNP.
With a density of SNPs so much higher than the expected density of recombinations in 53 generations, many of the imputed SNPs were highly or completely correlated, producing a large number of strongly associated but mostly confounded candidate variants. In order to prioritize individual SNPs or small SNP regions, we used multiple QTL (multilocus). This provided, for each SNP, a reliability score (RMIP) corresponding to the probability it would be selected for inclusion in a parsimonious model of joint action. An imputed SNP with a high RMIP represents an association that is robust and poorly explained by other SNPs or consistently more associated with MAT than its correlates. To identify general regions in which a SNP is often chosen but no one SNP stands out, we also report a region-based score (RMIP sliding window), which estimates for a given SNP location the number of SNPs that would be selected within 0.5Mb.
Figure 3 (lower panel) shows the RMIPs for individual SNPs (black spikes) and the RMIP sliding window (gray line). The RMIP sliding window strongly implicates the subregion centered at 36Mb as containing a robust association (~1 inclusion in an average subsample). This subregion includes three SNPs (one in Mrrf; two nongenic) that are selected most often in the 30Mb region, supporting a hypothesis of close linkage with a causal variant independent of Hc. Of note, within this subregion is the gene Ptgs1 (encoding cyclooxygenase 1 Cox-1). K/BxN arthritis is dependent on Cox-1, as mice deficient in this protein are completely resistant (23). Although a SNP within Ptgs1 is not selected, the selected SNPs may be in LD with a variant in Ptgs1, which remains an attractive candidate for an additional genetic determinant in the Chr2 locus.
The RMIP sliding window also highlights the subregion centered on 51.45Mb as being frequently included (0.571 inclusions on average), albeit with no single SNP distinguished by the RMIP due to high within-region correlation. This subregion, which is significantly associated with MAT in the unconditional single marker interval scans (Fig. 3 middle panel, lines), may represent a weak but independent association. It contains the protease inhibitor Tnfaip6 noted above. In contrast, the associations around 30Mb and 47Mb in the single marker interval scans are not well supported by the multilocus analysis, suggesting their association is readily explained by LD with other loci within the cohort’s family structure.
Focused Analysis of Chromosome 4
We also analyzed in more detail the 129–142 Mb interval on Chr4 which might also encompass several QTLs, with Padi4 as an attractive a priori candidate based on its association to RA in some human cohorts (24;25); its product catalyzes arginine to citrulline conversion, and anti-citrulline antibodies are arguably the most specific serum biomarker of RA (25). We therefore sought to refine the analysis and prioritize candidate SNPs within the Chr4 locus using the same fine-mapping procedure used for Chr2. Higher density SNP genotypes were generated approximately every 200 kb, we imputed 102,809 SNPs from 131.506544 to 141.019465Mb, and performed both single SNP and multi-SNP analyses described for the Chr 2 (not conditioning on C5). The Padi genes (Padi 1, 2, 3, 4 and 6) are encoded from 140.283270 to 140.507916Mb, with Padi4 located from 140.301767 to 140.330027Mb. The single locus analysis (Fig 4, middle panel) shows significant association in the region around the Padi genes that is (at least visually) dwarfed by a broad multimodal region of strong association from ~132 to ~138Mb. The multilocus analysis (Fig. 4, bottom) suggests an alternative prioritization: it provides support for three independent sources of association, centered around 133.5, 137.5 and 140.5Mb. The Padi genes are all within a region with a high probability of being associated with K/BxN arthritis in the multi-locus model. The RMIPs for individual SNPs in this region are generally low (the maximum being RMIP=0.15, residing in Spata21), indicating that associations among the imputations in this region are highly confounded. The highest associated single imputed SNPs (gray dots) and the HAPPY model (black line) are within Padi2 (HAPPY logP=5.76, SNP logP=5.04) or immediately adjacent (SNP logP=5.72 at 140.5088Mb). Also, the region containing the Padi genes is more efficiently explained by the imputed SNPs than by a more flexible (but less precise) model allowing for multiallelic effects, as evidenced by the SNP logPs scoring higher than the HAPPY model for the interval.
Figure 4.
A. Fine-mapping of QTL on Chr4 using HAPPY and imputed SNP association. Top panel shows the locations of annotated genes. Middle panel shows QTL mapping of HAPPY haplotype descent probabilities in 38 marker intervals (stepped lines) and 102,809 imputed SNPs (dots). Data series are plotted as for Fig 3 with HAPPY single interval mapping (thin black line) and genomewide threshold (dashed line; logP=4.67) and association of imputed SNPs (gray dots). Bottom panel shows multilocus mapping of imputed SNPs, with inclusion probabilities (RMIP) for individual SNPs (black spikes), and an aggregate score (RMIP sliding window) for neighboring sets of SNPs (gray line; see Fig 3 legend and main text).
The other regions of high RMIP score on Chr4 include, as intriguing candidates, Cd52 (the protease regulator noted above) and Lin28, an RNA binding protein which regulates miRNA levels and forms, together with IL6 and NF-Kb, a strong positive-feedback loop connecting inflammation, cell growth and tumorigenesis (26).
Analysis of Padi gene expression
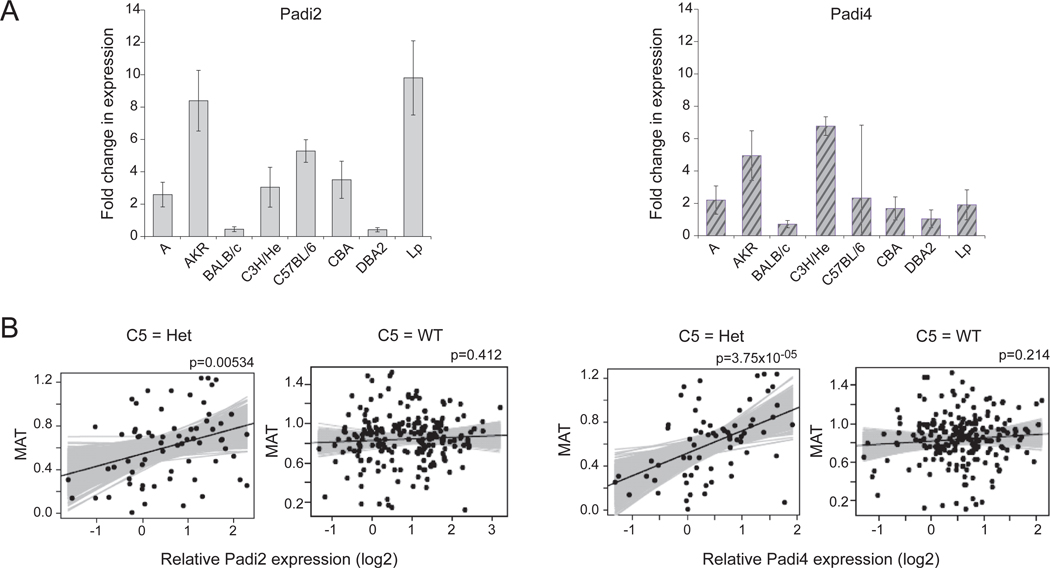
Given this association with the Padi gene region, we evaluated Padi genes as candidates, searching for variation in either protein-coding sequence or expression levels that could underpin the genetic association. Mining sequencing data (http://www.sanger.ac.uk/) revealed 11 non-synonymous SNPs in the Padi genes. One of these (in Padi3) had been genotyped in our original screen. We selected an additional 4 coding SNPs in Padi2 and Padi4, for which the minor allele was present in more than one of the HS parental strains. However, none of these SNPs showed highly significant marker-trait association (data not shown), suggesting that none of these individual SNPs were uniquely responsible for the association. We then asked whether Padi mRNA expression varied in the HS founder mice, analyzing spleen RNA which would not be confounded by variation in arthritis. Microarray gene expression profiles from whole spleen of all 8 founder strains were generated, and the expression of Padi1, 2, 3, 4 and 6 were extracted. Two-fold difference in expression among the strains was detected in Padi2 and Padi4, but not the other genes (data not shown) and these differences were verified by qPCR for these transcripts (Figure 5A). To test for a potential relationship between Padi gene expression and K/BxN arthritis, we assayed Padi2 and Padi4 expression by qPCR in spleen RNA from 278 of the experimental HS mice, stratifying by Hc genotype. High Padi2 or Padi4 expression in the spleen was associated with more severe arthritis in those mice that were heterozygous at the C5 locus, although the trend was not present in mice homozygous for the wild-type C5 allele (Fig. 5B).
Figure 5.
A. Real-time RT-PCR expression of Padi2 and Padi4 in spleen RNA of the 8 HS founder strains. Each bar represents the average of 5 or 6 individual mice, error bars represent standard deviation. B. Real-time RT-PCR expression of Padi2 and Padi4 in the spleens of 278 of the experimental HS mice. The solid black line represents the best fit, with the gray lines representing lines with alternative slopes that are also compatible with the data. The p-value is the probability that the slope of the black line is due to chance and that there is no association (a horizontal line).
DISCUSSION
We exploited the unique genetic characteristics of HS mice to delineate the range of QTLs that contribute to the severity of K/BxN serum-transferred arthritis. The two key advantages of HS mice over traditional F2 intercrosses between inbred strain pairs proved beneficial. First, the broad representation of genetic variants inherited by HS mice from their 8 founder strains allowed us to define novel regions of association not previously observed in K/BxN screens based on inbred pairs. Second, the associated intervals defined in this study, particularly after the use of multiple QTL modeling to reduce confounding signals from SNPs in tight LD, were only a few Mb in length, considerably smaller than the 15–40 Mb intervals that are the norm in F2 analyses. When a longer interval was initially observed (e.g. on Chr2 and Chr4), further analysis showed these to represent combined effects of several likely QTLs.
Table 1 shows a comparison between the regions associated with arthritis in GWAS meta-analyses (1) and in the syntenic regions of HS mice. The concordance is limited, although two corresponding regions are found, around TRAF1/C5 and around PADI4. GWAS only address the effect of SNPs frequently represented in the population, and it may be of interest to analyze more directly, in the next-generation resequencing programs, the impact of candidate loci identified here.
Table 1.
| Human Location |
Likeliest Candidate gene |
Mouse genome Location |
Assoc in HS |
|---|---|---|---|
| 2q11 | AFF3 | Chr 1, 38 | No |
| 2q32 | STAT4 | Chr 1, 52 | No |
| 2q33 | CD28 | Chr 1, 61 | No |
| 2q33 | CTLA4 | Chr 1, 61 | No |
| 5q21 | C5orf30 | Chr 1, 100 | No |
| 1q31 | PTPRC | Chr 1, 140 | No |
| 10p15 | PRKCQ | Chr 2, 11 | No |
| 10p15 | IL2RA | Chr 2, 12 | No |
| 9q33 | TRAF1, C5 | Chr 2, 35 | YES |
| 1p13 | CD2, CD58 | Chr 2, 101; no CD58 | No |
| 11p12 | TRAF6 | Chr 2, 102 | No |
| 20q13 | CD40 | Chr 2, 165 | No |
| 4q27 | IL2, IL21 | Chr 3, 37 | No |
| 1p13 | PTPN22 | Chr 3, 104 | No |
| 9p13 | CCL21 | Chr 4, 42 | No |
| 1p36 | PADI4* | Chr 4, 140 | YES |
| 1p36 | TNFRSF14 | Chr 4, 157 | No |
| 4p15 | RBPJ | Chr 5, 54 | No |
| 7q32 | IRF5 | Chr 6, 29 | No |
| 12q13 | KIF5A, PIP4K2C | Chr 10, 126 | No |
| 6q23 | TNFAIP3 | Chr 10, 19 | No |
| 6q21 | PRDM1 | Chr 10, 44 | No |
| 2p14 | SPRED2 | Chr 11, 19 | No |
| 2p16 | REL | Chr 11, 24 | No |
| 5q11 | ANKRD55, IL6ST | Chr 13, 113 | No |
| 8p23 | BLK | Chr 14, 64 | No |
| 3p14 | PXK | Chr 14, 90 | No |
| 22q12 | IL2RB | Chr 15, 78 | No |
| 6p21 | HLA-DRB1 (*0401) | Chr 17, 34 | No |
| 6q25 | TAGAP | Chr 17, 81 | No |
| 6q27 | CCR6 | Chr 17, 84 | No |
| 1q23 | FCGR2A | no Fcgr2a in mice | NA |
only in some populations
HS populations pose significant analytic challenges compared with F2 crosses or designs that are similarly outbred but with fewer founders such as advanced intercross lines (18). We built upon the analysis methods developed for previous HS studies, deploying haplotype reconstruction (17), imputation of SNPs (20), mixed models to control for uneven relatedness (19), and simultaneous consideration of multiple loci (14;18). In particular, employing a novel analytic development, we explored the pattern of association within QTL regions and identified independent associations by combining imputation of SNPs with modeling of the joint effect of multiple SNPs. This multilocus imputation mapping, based on the cross-validation (18), prioritizes imputed associations that are conditionally independent and robust to resampling, similar in spirit to Bayesian methods that have proven useful in human fine-mapping studies (27). Importantly, it controls for statistical dependencies between associated SNPs that may not be reflected by local LD (e.g., some SNP pairs may be highly confounded only conditional on family and other SNPs). Nonetheless, it relies on and is sensitive to a number of rather simplistic assumptions about joint action of SNP effects, and is applied to incomplete data subject to imputation uncertainty. Thus, although we consider the multilocus analysis a useful way of characterizing a complex pattern of association we use it as prelude to, rather than a substitute for, more focused subsequent experimental investigation.
Our power to detect QTLs may have been limited by the number of HS mice tested; prior studies used ~2000 mice (14) but a screen of this magnitude for arthritis would pose serious logistical challenges. In most cases, the intervals defined still remained too large for a direct definition of candidate genes to follow up. Thus further definition of QTLs on Chrs 1, 6, 7 and 15 will require further analysis, by an independent screen in HS or outbred mice that would increase resolving power and/or provide independent overlapping intervals, or by tapping future resources such as “Collaborative Cross” mice (28). Yet, two intriguing candidates emerge from the Chr2 and Chr4 intervals.
In-depth analysis of the Chr2 region suggests that a gene or genes in addition to Hc are contributing to this peak. One striking candidate is Ptgs1 (Cox-1), which showed a distinct change in expression over the course of arthritis (Fig. 2). Cyclo-oxygenases are the key synthetic enzymes in the prostaglandin/leukotriene pathway and are primary NSAID targets. Chen at al showed that both cyclo-oxygenase isoforms are found in inflamed joints of K/BxN mice, but Cox-1-deficient mice were fully resistant while Cox-2-deficient mice were susceptible (23). The Ptgs1 candidate thus comes “pre-validated, but our data support the notion that allelic polymorphism in Ptgs1/Cox-1 is a modifier inflammatory arthritis, warranting further exploration in human patients.
The Padi genes are encoded within the Chr4 QTL. They produce enzymes that post-translationally deiminate arginine to citrulline. A subset of RA patients form antibodies to citrullinated peptides, a reactivity quite specific for RA. An RA-associated Padi4 haplotype correlating with increased transcript stability and higher anti-citrulline antibodies has been described, the association being most significant in Asian populations (24;25). Mechanistically, citrullination might contribute to RA pathogenesis by improving the affinity of peptides for disease-associated MHC molecules (29), or by generating neo-epitopes not encountered during T cell maturation in the thymus and thus more likely to evade tolerance pathways, or citrullinated proteins of the joint extracellular matrix may provide additional targets for pathogenic autoantibodies. Arthritis in the K/BxN or other serum-transfer models is due to immune complexes depositing on joint surfaces, and thus limited to events downstream from the auto-antigen recognition by T and B cells during autoimmune activation. If one of the Padi genes is indeed the QTL on Chr4, this may suggest that pre-formed anti-citrulline antibodies in the K/BxN serum are binding citrullinated targets, and that the variation in the degree of citrullination might impact arthritis severity. Alternative mechanisms are also possible, since hypercitrullination by Padi4 mediates chromatin decondensation (30) and citrullination of various other proteins can affect their function, as was demonstrated for CXCL8 (31). Interestingly, the correlation between Padi2/4 expression and arthritis severity was only detectable in C5 heterozygous mice within the HS cohort, suggesting a possible interaction between complement activation and citrullination.
In conclusion, these results provide a broad overview of the genetic variation that controls the severity of K/BxN arthritis in mice, and are likely to be relevant to other antibody-dependent models such as CIA, and to human pathology as well. Confirmation of candidate genes will require assessing susceptibility to K/BxN arthritis in existing knock-out or customized RNAi knock-down mice. Genes shown to be important in controlling K/BxN arthritis may suggest novel molecules and pathways in the pathophysiology of human RA.
ACKNOWLEDGEMENTS
The authors would like to thank Christopher Campbell and Angela Wilcox for their help with genotyping and Catherine LaPlace, who helped with the figures.
Grant Support:
-
-
Scholarship to AJ from the Arthritis National Research Foundation
-
-
K08 AI072044-01A1, NIAID/NIH
-
-
R01 AR055271 from NIAMS/NIH to CB/DM, JF and RH
Abbreviations
- RA
rheumatoid arthritis
- CCP
cyclic citrullinated peptides
- SNP
single-nucleotide polymorphism
- PCR
polymerase chain reaction
- LD
linkage disequilibrium
- GWAS
Genome-wide association study
- HS
“Heterogeneous Stock” mice
- GPI
glucose-6-phosphate isomerase
- Cox
cyclo-oxygenase
- Padi
peptidyl-arginine-deiminase
- MAT
maximum ankle thickening
- MCI
maximum clinical index
- ICI
integration over the clinical index
- IAT
integration over the ankle thickening
- QTL
quantitative trait locus
- LRT
likelihood ratio test
- SDP
strain distribution pattern
- RMIP
resample model inclusion probabililty
Footnotes
These authors made equivalent contributions to this work
REFERENCES
- 1.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nandakumar KS, Holmdahl R. Antibody-induced arthritis: disease mechanisms and genes involved at the effector phase of arthritis. Arthritis Res Ther. 2006;8:223. doi: 10.1186/ar2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlqvist E, Hultqvist M, Holmdahl R. The value of animal models in predicting genetic susceptibility to complex diseases such as rheumatoid arthritis. Arthritis Res Ther. 2009;11:226. doi: 10.1186/ar2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 5.Monach PA, Benoist C, Mathis D. The role of antibodies in mouse models of rheumatoid arthritis, and relevance to human disease. Adv Immunol. 2004;82:217–248. doi: 10.1016/S0065-2776(04)82005-4. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto I, Staub A, Benoist C, Mathis D. Arthritis provoked by linked T and B cell recognition of a glycolytic enzyme. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto I, Maccioni M, Lee DM, Maurice M, Simmons B, Brenner M, et al. How antibodies to a ubiquitous cytoplasmic enzyme may provoke joint-specific autoimmune disease. Nat Immunol. 2002;3:360–365. doi: 10.1038/ni772. [DOI] [PubMed] [Google Scholar]
- 8.Ji H, Gauguier D, Ohmura K, Gonzalez A, Duchatelle V, Danoy P, et al. Genetic influences on the end-stage effector phase of arthritis. J Exp Med. 2001;194:321–330. doi: 10.1084/jem.194.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohmura K, Johnsen A, Ortiz-Lopez A, Desany P, Roy M, Besse W, et al. Variation in IL-1β gene expression is a major determinant of genetic differences in arthritis aggressivity in mice. Proc Natl Acad Sci U S A. 2005;102:12489–12494. doi: 10.1073/pnas.0504325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FMA, Boackle SA, et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 11.Wipke BT, Wang Z, Nagengast W, Reichert DE, Allen PM. Staging the initiation of autoantibody-induced arthritis: a critical role for immune complexes. J Immunol. 2004;172:7694–7702. doi: 10.4049/jimmunol.172.12.7694. [DOI] [PubMed] [Google Scholar]
- 12.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 13.Demarest K, Koyner J, McCaughran J, Jr, Cipp L, Hitzemann R. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet. 2001;31:79–91. doi: 10.1023/a:1010261909853. [DOI] [PubMed] [Google Scholar]
- 14.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 16.Zucchelli S, Holler P, Yamagata T, Roy M, Benoist C, Mathis D. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity. 2005;22:385–396. doi: 10.1016/j.immuni.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdar W, Holmes CC, Mott R, Flint J. Mapping in Structured Populations by Resample Model Averaging. Genetics. 2009 doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solberg Woods LC, Holl K, Tschannen M, Valdar W. Fine-mapping a locus for glucose tolerance using heterogeneous stock rats. Physiol Genomics. 2010;41:102–108. doi: 10.1152/physiolgenomics.00178.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yalcin B, Flint J, Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 2005;171:673–681. doi: 10.1534/genetics.104.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz G. Estimating the dimension of a model. The Annals of Statistics. 1978:461–464. [Google Scholar]
- 22.Jacobs JP, Ortiz-Lopez A, Campbell JJ, Gerard CJ, Mathis D, Benoist C. Deficiency of CXCR2, but not other chemokine receptors, attenuates autoantibody-mediated arthritis in a murine model. Arthritis Rheum. 2010;62:1921–1932. doi: 10.1002/art.27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Boilard E, Nigrovic PA, Clark P, Xu D, Fitzgerald GA, et al. Predominance of cyclooxygenase 1 over cyclooxygenase 2 in the generation of proinflammatory prostaglandins in autoantibody-driven K/BxN serum-transfer arthritis. Arthritis Rheum. 2008;58:1354–1365. doi: 10.1002/art.23453. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 25.Huseyin Uysal. Antibodies to citrullinated proteins: molecular interactions and arthritogenicity. Immunological Review. 2010:9–33. doi: 10.1111/j.0105-2896.2009.00853.x. 233 ed. [DOI] [PubMed] [Google Scholar]
- 26.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 28.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 29.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, et al. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J Exp Med. 2008;205:2085–2097. doi: 10.1084/jem.20080305. [DOI] [PMC free article] [PubMed] [Google Scholar]