Abstract
BACKGROUND
In the RV144 trial, the estimated efficacy of a vaccine regimen against human immunodeficiency virus type 1 (HIV-1) was 31.2%. We performed a case–control analysis to identify antibody and cellular immune correlates of infection risk.
METHODS
In pilot studies conducted with RV144 blood samples, 17 antibody or cellular assays met prespecified criteria, of which 6 were chosen for primary analysis to determine the roles of T-cell, IgG antibody, and IgA antibody responses in the modulation of infection risk. Assays were performed on samples from 41 vaccinees who became infected and 205 uninfected vaccinees, obtained 2 weeks after final immunization, to evaluate whether immune-response variables predicted HIV-1 infection through 42 months of follow-up.
RESULTS
Of six primary variables, two correlated significantly with infection risk: the binding of IgG antibodies to variable regions 1 and 2 (V1V2) of HIV-1 envelope proteins (Env) correlated inversely with the rate of HIV-1 infection (estimated odds ratio, 0.57 per 1-SD increase; P = 0.02; q = 0.08), and the binding of plasma IgA antibodies to Env correlated directly with the rate of infection (estimated odds ratio, 1.54 per 1-SD increase; P = 0.03; q = 0.08). Neither low levels of V1V2 antibodies nor high levels of Env-specific IgA antibodies were associated with higher rates of infection than were found in the placebo group. Secondary analyses suggested that Env-specific IgA antibodies may mitigate the effects of potentially protective antibodies.
CONCLUSIONS
This immune-correlates study generated the hypotheses that V1V2 antibodies may have contributed to protection against HIV-1 infection, whereas high levels of Env-specific IgA antibodies may have mitigated the effects of protective antibodies. Vaccines that are designed to induce higher levels of V1V2 antibodies and lower levels of Env-specific IgA antibodies than are induced by the RV144 vaccine may have improved efficacy against HIV-1 infection.
In clinical trials that show the efficacy of a vaccine, the identification of immune responses that are predictive of trial outcomes generates hypotheses about which of those responses are responsible for protection.1–3 The RV144 phase 3 trial in Thailand (ClinicalTrials.gov number, NCT00223080) was an opportunity to perform such a hypothesis-generating analysis for a human immunodeficiency virus type 1 (HIV-1) vaccine.4 Studies involving patients with HIV-1 infection in whom the disease did not progress in the long term have shown that cellular responses control the disease,5 and passive infusion of neutralizing antibodies prevents infection with chimeric simian–human immunodeficiency virus (SHIV).6,7 Antibodies as well as T-cell responses to HIV-1 have been shown to protect vaccinated nonhuman primates from infection with simian immunodeficiency virus (SIV) or SHIV.8–15 An analysis of a phase 3 trial of a glycoprotein 120 (gp120) B/B vaccine (AIDSVAX B/B), which did not show efficacy against HIV-1, showed that vaccine-specific neutralizing antibody, antibody inhibition of CD4 molecule binding to HIV-1 envelope proteins (Env), and antibody-dependent, cell-mediated viral inhibition were associated with reduced infection rates among vaccine recipients.16,17
The RV144 trial of the canarypox vector vaccine (ALVAC-HIV [vCP1521]) plus the gp120 AIDSVAX B/E vaccine showed an estimated vaccine efficacy of 31.2% for the prevention of HIV-1 infection over a period of 42 months after the first of four planned vaccinations.4 This result enabled a systematic search for immune correlates of infection risk that may be relevant for protection. Building on prior work,18,19 our consortium conducted a two-stage evaluation of vaccine-evoked antibody responses, innate immune responses, and cellular immune responses.20 First, 17 assay types were selected from 32 pilot assay types on the basis of reproducibility, ability to detect postvaccine responses, and uniqueness of responses detected, from which 6 primary assay variables were selected. Second, the selected assays in primary analyses (6 assays) and secondary analyses (152 assays) were performed on cryopreserved blood samples from vaccinees who became infected (case patients) and on a frequency-matched set of samples from uninfected vaccinees (controls) to determine the association of immune-response variables with HIV-1 infection risk.
METHODS
STUDY PROCEDURES
Case–Control Sampling Design
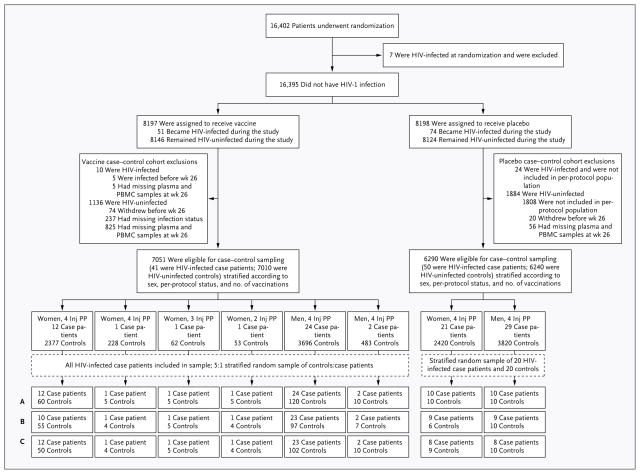
Patients enrolled in the RV144 trial were vaccinated at weeks 0, 4, 12, and 24, and immune responses at week 26 were evaluated as immune correlates of infection risk4 (Fig. 1). We assessed vaccine-induced immune responses at peak immunogenicity (week 26 [2 weeks after the final immunization]) in vaccinees who acquired HIV-1 infection after week 26 (41 vaccinated case patients) as compared with vaccinees who did not acquire infection over a follow-up period of 42 months (205 vaccinated controls). Vaccinated case patients were documented to be free of HIV-1 infection at week 24 and to have later received a diagnosis of infection.4 The control vaccinees were selected from a stratified random sample of vaccine recipients who were documented to be free of HIV-1 infection at 42 months. Patients were stratified according to sex, the number of vaccinations received (of four planned), and per-protocol status, as previously defined.4 For each of the eight strata, the number of vaccinated case patients was noted, and samples from five times as many vaccinated controls were obtained. The assays were also performed on random samples from 20 infected placebo recipients and 20 uninfected placebo-recipient controls (Fig. 1).
Figure 1. Sample Selection for the Case–Control Study.
Patients enrolled in the RV144 study were vaccinated at weeks 0, 4, 12, and 24, and immune responses at week 26 were evaluated as immune correlates of infection risk. The vaccinated case patients were documented as not having HIV-1 infection at week 24 and as having later received a diagnosis of infection. The vaccine recipients who served as controls were selected from a stratified random sample of vaccine recipients who were documented as not having HIV-1 infection at the last study visit, at 42 months. Of the 7010 HIV-uninfected vaccinated controls eligible for the case–control sample, only those for whom plasma and peripheral-blood mononu-clear cell (PBMC) specimens were available at all later time points and who were not part of previous immunogenicity-testing cohorts (6899 patients) were included. For vaccine recipients who were included in the sample, 6 strata with 1 or more case patients are shown; the remaining strata had 0 case patients and 111 controls. All humoral assays were performed in plasma samples from all case patients and all controls (row A). Data on intra-cellular cytokine staining of PBMCs (row B) were missing for 15% of patients (owing to assay quality-control issues, including an aberrant batch of samples in 24 patients and high values for the assay negative control in 18). Data on multiplex bead assay (Luminex) of PBMCs (row C) were missing for 13% of patients (owing to high values for the assay negative control in 36 patients). Inj denotes injection with vaccine or placebo, and PP per-protocol cohort (i.e., patients who received all four injections as previously described4).
Immune-Response Variables and Tiered Structure of the Correlates Analysis
The correlates study was preceded by pilot studies from November 2009 through July 201120 (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Pilot assays were performed on samples taken at baseline and week 26 from 50 to 100 uninfected RV144 participants (80% of whom were vaccine recipients and 20% of whom were placebo recipients) and scored according to four statistical criteria: a low false positive rate on the basis of samples from placebo and vaccine recipients at baseline, a large dynamic range of vaccine-induced immune responses, nonredundancy of responses (low correlations), and high reproducibility.
Of the 32 types of antibody, T-cell, and innate immunity assays evaluated in pilot studies, 17 met these criteria, from which 6 primary variables were chosen for assessment as correlates of infection risk. The purpose was to restrict the primary analysis to a limited number of variables in order to optimize the statistical power for showing a correlation of risk between vaccinated persons who acquired versus those who did not acquire HIV-1. The primary variables included 5 Env-specific antibody responses and 1 cellular response: the binding of plasma IgA antibodies to Env, the avidity of IgG antibodies for Env, antibody-dependent cellular cytotoxicity, HIV-1 neutralizing antibodies, the binding of IgG antibodies to variable regions 1 and 2 (V1V2) of the gp120 Env, and the level of Env-specific CD4+ T cells (for details, see the Supplementary Appendix). All 17 types of immune assays and their 152 component variables were also included in the secondary correlates analyses (Tables S1 and S2 in the Supplementary Appendix).
Secondary variables were drawn from the remaining 152 assays selected from pilot assay studies; they were evaluated to help interpret the results of the primary analysis and to generate additional hypotheses (Table S1 in the Supplementary Appendix). For the sensitivity analysis, immune-response variables that were closely related to the six primary variables (within the same assay type) were substituted for each of the primary variables into the multivariable model (eight variables, with three individual variables paired to the primary variable of neutralizing antibodies) (Table S2 in the Supplementary Appendix). All assays were performed by personnel who were unaware of treatment assignments and case–control status.
STATISTICAL ANALYSIS
The statistical analysis plan was finalized before data analysis, and the primary results were confirmed by an independent statistical group (EMMES). This statistical analysis plan prescribed the statistical methods and the definitions of the immune-response variables (for details, see the Supplementary Appendix). In the primary analysis, logistic-regression and Cox proportional-hazards models that accounted for the sampling design were used.21,22 The analyses controlled for sex and baseline self-reported behavioral risk factors, as defined previously.4
The six primary variables were evaluated in multivariate and univariate models. The immune-response variables were modeled quantitatively and with the use of categories based on thirds of response (low, medium, and high) in the vaccine group. The q value is the minimal false discovery rate at which a statistical test result may be called significant. The q values were used for multiplicity correction, with a significance threshold of less than 0.20, indicating that any detected correlate can have up to a 20% chance of false positivity. This approach was designed to optimize the discovery of correlates at the expense of an acceptable risk of false positive results.
Because of the small number of infected vaccinees, this study had statistical power to detect only strong correlates of infection risk, with 80% power to detect a 50% reduction in the infection rate per 1-SD increment in normally distributed immune responses. The 152 secondary variables were assessed with the same univariate regression analyses as the primary variables were, to generate exploratory hypotheses for further study (Table S1 in the Supplementary Appendix).
RESULTS
PRIMARY VARIABLES IN CASE–CONTROL ANALYSES
Vaccine-induced immune responses were detected with all primary assay variables, with sufficient dynamic ranges to support regression analyses (Fig. 2). Figure S2 in the Supplementary Appendix shows that the six primary variables were only weakly correlated with each other, verifying that the process for selecting the primary variables yielded nonredundant primary immune-response variables.
Figure 2. Distribution of the Six Primary Immune-Response Variables in Infected and Uninfected Vaccine and Placebo Recipients in the Case–Control Study.
Panel A includes the two identified immune correlates of risk. Panel B includes the remaining four primary immune-response variables. Box plots show the 25th percentile (lower edge of the box), 50th percentile (horizontal line in the box), and 75th percentile (upper edge of the box) for the six primary variables, with patients stratified according to HIV-1 infection status and treatment assignment. Additional characteristics of these patients, including sex and immune-response categories, are indicated by the color and shape of the points. Low, medium, and high immune responses at week 26 were used to divide the vaccine group into thirds; medium response is indicated by the gray horizontal bar. Optical density was measured by means of an enzyme-linked immunosorbent assay at a wavelength of 405 nm. Log MFI is the natural log transformation of the median fluorescence intensity (MFI). The avidity score is [response units × (1 ÷ dissociation rate in seconds)] × 10−5. The partial area between the curves is the sum of the differences over the first four dilutions between the readouts at week 0 and week 26, measured in log10-transformed relative light units (RLUs). The area under the magnitude–breadth curve (AUC-MB) is the average log10-transformed 50% inhibitory concentration in response to a panel of six pseudoviruses. The net percentage of cytokine-expressing CD4+ T cells is the percentage of live CD3+ CD4+ T cells expressing CD154, interleukin-2, interferon-γ, or tumor necrosis factor α minus the negative control value. The I bars indicate the most extreme data points, which are no more than 1.5 times the interquartile range from the box. The distribution plots of the six primary variables and sensitivity variables are shown in Figures S5 through S11 in the Supplementary Appendix.
First, when we analyzed the six quantitative variables together in multivariate logistic-regression models, there was a trend toward the prediction of infection risk by the variables (P = 0.08 for all six variables together). In this model, IgG avidity, antibody-dependent cellular cytotoxicity, neutralizing antibodies, and level of Env-specific CD4+ T cells did not significantly predict the HIV-1 infection rate (q>0.20). However, IgG binding to a scaffolded V1V2 antigen was inversely correlated with infection (estimated odds ratio, 0.57 per 1-SD increase; P = 0.02; q = 0.08), and composite IgA antibody binding to an Env panel was directly correlated with infection (estimated odds ratio, 1.54 per 1-SD increase; P = 0.03; q = 0.08) (Table 1). The univariate analyses of V1V2 and IgA responses yielded odds-ratio estimates of 0.70 and 1.39, respectively, with slightly reduced significance (P = 0.06, q=0.19, and P=0.05, q= 0.19, respectively) (Table 1).
Table 1.
Odds Ratios for HIV-1 Infection in Univariate and Multivariate Analyses of the Six Primary Variables.*
| Variable | Multivariate Logistic Regression | Univariate Logistic Regression | ||||
|---|---|---|---|---|---|---|
| Odds Ratio (95% CI) | P Value | q Value | Odds Ratio (95% CI) | P Value | q Value | |
| Quantitative variables | ||||||
| IgA antibodies binding to Env | 1.54 (1.05–2.25) | 0.03 | 0.08 | 1.39 (1.00–1.93) | 0.05 | 0.19 |
| Avidity of IgG antibodies for Env | 0.81 (0.50–1.30) | 0.37 | 0.56 | 0.93 (0.65–1.34) | 0.70 | 0.81 |
| ADCC | 0.92 (0.62–1.37) | 0.68 | 0.68 | 0.96 (0.68–1.35) | 0.81 | 0.81 |
| Neutralizing antibodies | 1.37 (0.82–2.27) | 0.23 | 0.45 | 1.08 (0.76–1.54) | 0.67 | 0.81 |
| gp70-V1V2 binding | 0.57 (0.36–0.90) | 0.02 | 0.08 | 0.70 (0.49–1.02) | 0.06 | 0.19 |
| CD4+ T cells | 1.09 (0.79–1.49) | 0.61 | 0.68 | 1.08 (0.79–1.47) | 0.64 | 0.81 |
| Categorical variables | ||||||
| IgA antibodies binding to Env | ||||||
| Medium vs. low response | 0.68 (0.25–1.83) | 0.45 | 0.71 (0.28–1.77) | 0.46 | ||
| High vs. low response | 1.89 (0.75–4.74) | 0.17 | 1.55 (0.69–3.48) | 0.29 | ||
| Overall response† | 0.07 | 0.23 | 0.18 | 0.54 | ||
| Avidity of IgG antibodies for Env | ||||||
| Medium vs. low response | 0.67 (0.26–1.75) | 0.42 | 0.79 (0.34–1.84) | 0.59 | ||
| High vs. low response | 0.83 (0.30–2.30) | 0.72 | 0.87 (0.38–2.01) | 0.75 | ||
| Overall response† | 0.71 | 0.84 | 0.86 | 0.86 | ||
| ADCC | ||||||
| Medium vs. low response | 0.93 (0.39–2.19) | 0.86 | 0.96 (0.43–2.14) | 0.92 | ||
| High vs. low response | 0.59 (0.22–1.59) | 0.30 | 0.60 (0.24–1.48) | 0.27 | ||
| Overall response† | 0.57 | 0.84 | 0.53 | 0.71 | ||
| Neutralizing antibodies | ||||||
| Medium vs. low response | 2.08 (0.78–5.57) | 0.14 | 1.58 (0.66–3.76) | 0.31 | ||
| High vs. low response | 2.43 (0.77–7.67) | 0.13 | 1.29 (0.53–3.14) | 0.57 | ||
| Overall response† | 0.26 | 0.52 | 0.59 | 0.71 | ||
| gp70-V1V2 binding | ||||||
| Medium vs. low response | 0.68 (0.28–1.62) | 0.38 | 0.84 (0.38–1.84) | 0.67 | ||
| High vs. low response | 0.29 (0.10–0.86) | 0.02 | 0.43 (0.18–1.05) | 0.06 | ||
| Overall response† | 0.08 | 0.23 | 0.16 | 0.54 | ||
| CD4+ T cells | ||||||
| Medium vs. low response | 0.84 (0.35–2.02) | 0.69 | 0.62 (0.27–1.45) | 0.27 | ||
| High vs. low response | 0.77 (0.31–1.91) | 0.57 | 0.51 (0.21–1.23) | 0.13 | ||
| Overall response† | 0.84 | 0.84 | 0.28 | 0.57 | ||
For each variable, the odds ratio is reported per 1-SD increase and for the comparison of medium and high responses with low responses. ADCC denotes antibody-dependent cellular cytotoxicity, and gp70-V1V2 the binding level of IgG antibodies to HIV-1 Env V1V2.
To test the overall response, a two-sided P value from a Wald test of the null hypothesis of an equal infection rate in the low, medium, and high response subgroups was calculated.
Parallel multivariate analyses with the Cox model yielded similar results, with an overall multivariate P value of 0.06 and multivariate hazard-ratio estimates of 0.57 for the V1V2 response (P = 0.01, q = 0.06) and 1.58 for the IgA response (P = 0.02, q = 0.06) (Table S3 in the Supplementary Appendix). When the multivariate analysis was repeated with only the V1V2 and IgA immune-response variables, the overall P value was 0.01 for both the logistic-regression and Cox regression models.
The logistic-regression analyses of the six primary variables categorized into low, medium, and high levels of response yielded odds-ratio estimates that were consistent with those in the quantitative variable analysis. There was no evidence that IgG avidity, antibody-dependent cellular cytotoxicity, neutralizing antibodies, or Env-specific CD4+ T cells were associated with infection risk (q>0.20). Comparison of high and low levels showed an inverse correlation between V1V2 antibody levels and the risk of infection (estimated odds ratio, 0.29; P = 0.02) and a trend toward a direct correlation of Env-specific IgA antibody level with infection risk (estimated odds ratio, 1.89; P=0.17) (Table 1). However, these categorical-model results had reduced significance levels for testing an equal infection rate across low, medium, and high responses (V1V2 antibodies, q = 0.23; Env-specific IgA antibodies, q = 0.23), which may be related to the division of responses into thirds, which can reduce statistical power.
Figure 3 shows curves for the cumulative incidence of HIV-1 infection with each primary variable among vaccine recipients according to the level of response (low, medium, or high) and for all placebo recipients who were negative for HIV-1 infection at week 24. These curves underscore the increased rate of infection among vaccine recipients with high levels of Env-specific IgA antibodies, as compared with other vaccine recipients, and the decreased rate of infection among vaccine recipients with high levels of V1V2 antibodies.
Figure 3. Estimated Cumulative HIV-1 Incidence Curves for the Six Primary Immune-Response Variables.
Because the overall infection rate in the RV144 trial was low, at 0.234 cases per 100 person-years for the vaccine and placebo groups combined, expression of the cumulative HIV-1 incidence curves for the six primary immune-response variables on a scale of infection probabilities from 0.0 to 1.0 does not allow for an analysis of relative cumulative incidences (indicated by the flat cumulative-incidence curves at the bottom of each graph in Panels A and B). The inset for each graph, which shows an expanded lower range of the infection probability scale, reveals patterns of cumulative risk across the participant groups. Panel A includes the two identified immune correlates of risk.
Panel B includes the remaining four primary immune-response variables. For each primary variable, 41 vaccinated case patients were stratified into subgroups divided into thirds according to the immune response (low, medium, and high) at week 26 in the vaccine group in the case–control study. The estimated cumulative incidence of HIV-1 infection over time since the measurement of immune response at week 26 is shown for the three vaccine subgroups and for placebo recipients who were negative for HIV-1 infection at week 24.
RISK OF INFECTION WITH VACCINE, ACCORDING TO V1V2 OR ENV-SPECIFIC IGA ANTIBODY LEVEL, AS COMPARED WITH PLACEBO
Env-specific IgA responses were directly associated with infection risk in the vaccine group, raising the possibility that a vaccine-elicited plasma Env-specific IgA response increased the risk of infection in the RV144 trial. To evaluate this possibility, we used logistic and Cox regression to estimate vaccine efficacy as 1 minus the odds (hazard) ratio for infection among vaccinees with low, medium, and high Env-specific IgA responses, as compared with all placebo recipients who were HIV-1-negative at week 24 (Fig. S3 in the Supplementary Appendix). We found that neither low levels of V1V2 antibodies nor high levels of Env-specific IgA antibodies in vaccinees were associated with higher rates of infection than were found among placebo recipients (Fig. S3 in the Supplementary Appendix). These data suggest that vaccine-induced IgA levels did not confer an added risk of infection, as compared with placebo, and therefore were not infection-enhancing antibodies.
Interaction analyses were performed with logistic-regression and Cox regression models to test for interactions of Env-specific IgA antibodies and of V1V2 antibodies with the other five primary variables. The analysis showed no interaction of any primary variables with V1V2 antibodies but did show significant interactions of Env-specific IgA antibodies with IgG avidity, antibody-dependent cellular cytotoxicity, neutralizing antibodies, and CD4+ T cells (q<0.20). Thus, in the presence of high levels of Env-specific IgA antibodies, none of these four variables correlated with the risk of infection, whereas with low levels of Env-specific IgA antibodies, all four variables had inverse correlations with the risk of infection that were of borderline significance (Table S4 in the Supplementary Appendix).
SECONDARY AND EXPLORATORY ANALYSES
In the sensitivity analysis that substituted each of the secondary analysis variables with the primary variable, the significance levels tended to be similar or lower, with the exceptions that neutralization of TH023.6, neutralization of clade AE viruses in the A3R5 assay, and magnitude of induced cytokines measured in peripheral-blood mononuclear cells had q values of less than 0.20 (although at P>0.05) (Tables S2 and S5 in the Supplementary Appendix).
Of the 152 secondary variables analyzed, only 2 had q values of less than 0.20. These 2 variables were IgA antibody binding to group A consensus Env gp140 (odds ratio for positive vs. negative responses, 3.71; P = 0.001; q = 0.10) and IgA antibody binding to a gp120 Env first constant (C1) region peptide (MQEDVISLWDQSLKP-CVKLTPLCV) (odds ratio for positive vs. negative responses, 3.15; P = 0.003; q = 0.13) (Table S1 in the Supplementary Appendix).
DISCUSSION
We report the results of an immune-correlates analysis of the RV144 HIV-1 vaccine efficacy trial. This correlates study was designed to be hypothesis-generating and sensitive for discovering strong correlates of infection risk.23 An identified correlate of infection risk could be a cause of vaccine-induced protection against HIV-1 infection, a surrogate for other unidentified immune responses that are actually responsible for protection, or a marker of HIV-1 exposure or susceptibility to infection.1–3 To determine whether a correlate of infection risk is a cause of vaccine protection, it must be tested in additional clinical vaccine efficacy trials or tested in animal models.1–3 Extensive pilot immunogenicity studies revealed 17 T-cell, antibody, and innate immunity assays that were prioritized into prespecified primary and secondary analyses in order to maximize statistical power in the primary analysis to detect correlates of infection risk.
Of the six assay variables chosen for the primary analysis, two showed significant correlations with infection among vaccine recipients: IgG antibody binding to scaffolded V1V2 Env correlated inversely with infection, and IgA antibody binding to Env correlated directly with infection. These two correlates of risk, taken together, were highly correlated with the infection rate and may generate important hypotheses about immune responses required for protection from HIV-1,1–3 improve the selection of primary end points in subsequent HIV-1 vaccine trials,24 and lead to improved vaccines. If protection conferred by V1V2 IgG antibodies can be confirmed, then the design of vaccines to induce high levels of V1V2 antibodies and low levels of Env-specific IgA antibodies might augment vaccine efficacy.
Several lines of evidence suggest that vaccine-induced antibodies recognize conformational epitopes in the scaffolded V1V2 reagent, which has been shown to detect conformational V1V2 antibodies.25,26 The results of an analysis of breakthrough viruses from patients in the RV144 trial were consistent with immune pressure focused on amino acid patterns in and flanking the V1V2 region of HIV-1 Env.27 This region serves critical functions, such as participating in CD4-receptor and chemokine-receptor binding, binding to α4β7 integrin,28 and serving as the binding site of neutralizing antibodies.29–32
In the Step HIV-1 vaccine trial (ClinicalTrials.gov number, NCT00095576), which was designed to induce HIV-1 T-cell responses, the hazard ratio for HIV-1 infection in the vaccine group as compared with the placebo group was higher in selected subgroups of vaccine recipients.33 Although the notion of antibody-mediated enhancement of intrauterine infection has been raised in a clinical trial of treatment of HIV-1–infected pregnant women with infusion of immune globlulin,34 no vaccine-associated increase in the risk of infection was seen in the RV144 trial, and in analyses that compared infection rates in vaccine-recipient subgroups with the rate in the placebo group, no increase was seen with high levels of vaccine-induced Env-specific plasma IgA antibodies (Fig. S3 in the Supplementary Appendix). However, a limitation of the analyses that compared infection risk among vaccine and placebo recipients is that the comparator groups could not be randomized, and there may have been residual confounding because the analysis controlled only for sex and baseline behavioral risk factors.
The significant interactions of Env-specific IgA antibodies with other primary variables further support the importance of IgA-binding antibodies in predicting the risk of infection (Table S4 in the Supplementary Appendix). In vaccinees with low levels of Env-specific IgA antibodies, four of the other five primary variables — IgG avidity, antibody-dependent cellular cytotoxicity, neutralizing antibodies, and Env-specific CD4+ T cells — were inversely correlated with infection, whereas in vaccinees with high levels of Env-specific IgA antibodies, there was no correlation between these variables and infection (Table S4 in the Supplementary Appendix). The observed interactions generated the hypothesis that plasma IgA antibody levels interfere with protective IgG effector functions, a phenomenon that has been observed with other pathogens,35,36 in the regulation of autoantibody function,37 and in immune responses to cancer.38
We found that vaccinees with IgA antibodies to the first conserved region (C1) of gp120 had a higher risk of infection than vaccinees without these antibodies (odds ratio, 3.15; P = 0.003; q = 0.13). The gp120 C1 region contains an epi-tope that can be a target on the surface of virus-infected cells for antibodies that mediate antibody-dependent cellular cytotoxicity.39 Another possible scenario is that high levels of Env-specific IgA antibodies is a surrogate marker for HIV-1 exposure that was not fully accounted for by adjustment for baseline self-reported behavioral risk factors in the regression models. The primary variable of Env-specific IgA antibodies was not significantly associated with baseline behavioral risk factors (P = 0.28), nor did IgA antibodies to the individual Env proteins included in the primary IgA variable correlate with baseline behavioral risk factors (Table S6 in the Supplementary Appendix). Plasma IgA is primarily monomeric IgA, whereas mucosal IgA is primarily dimeric.40 Any protective role of mucosal dimeric IgA in the context of HIV-1 vaccination could not be evaluated in the RV144 trial, because mucosal samples were not collected.
The relevance of these findings to different HIV-1 risk populations receiving ALVAC-HIV, AIDSVAX B/E, or other HIV-1 vaccine regimens cannot be inferred and must be prospectively determined. Moreover, further studies are required to determine causality — whether V1V2 antibodies mediate vaccine-induced protection from infection or whether Env-specific IgA antibodies interfere with protection. Nonetheless, the identification of immune correlates of the risk of HIV-1 infection in the RV144 trial provides plausible biologic hypotheses for the original clinical observation of vaccine efficacy.4 Elucidation of the potential roles of V1V2 and Env-specific IgA antibodies in the modulation of HIV-1 infection risk may accelerate the clinical development of vaccine candidates that can improve on the results of the RV144 clinical trial.
Supplementary Material
Acknowledgments
Supported in part by grants from the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (to Drs. Haynes, Koup, and Montefiori); an interagency agreement between the U.S. Army Medical Research and Materiel Command and the NIAID (Y1-AI-2642-12); a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the Department of Defense (W81XWH- 07-2-0067); and grants from the NIAID to the Center for HIV/AIDS Vaccine Immunology (U01-AI-067854) and the HIV Vaccine Trials Network Laboratory Program (UM1-AI-068618).
We thank Judith T. Lucas, Vicki C. Ashley, R. Glenn Overman, Claire Beard, Michele Donathan, Doris Murray, Kelly Seaton, Yong Lin, Robert Parks, Amanda Martelli, Corrine Dary, Wenhong Feng, Xing Yuan, Kara John, Dora de Fonseca, and Robert O’Connell for project management or assay performance; Cristine Cooper-Trenbeath, Cheryl DeBoer, Cindy Molitor, Mark Bollenbeck, Linda Harris, and Craig Magaret for assay data processing; Daryl Morris, Alicia Sato, Ying Huang, Xuesong Yu, Raphael Gottardo, Barbara Metch, Nidhi Kochar, Hasan Ahmed, Zoe Moodie, and Maggie Wang for statistical analysis; Drienna Holman, Laura Saganic, Huguette Redinger, Kelli Greene, Hongmei Gao, Raul Louzao, Whitney Binz, Marcella Sarzotti-Kelsoe, and Caroline Cockrell for project management or project quality control or both; Corey Nathe, Shane Coultas, Scott Langley, and Sravani Cheeti for Atlas portal design; John Mascola for Env proteins; Dennis Burton and Ann Hessell for IgA 1b12 control antibody; Rebecca Putnam, Dave Friedrich, Don Carter, Olivier Defawe, Julie Williams, Jason Stuckey, Terry Stewart, Nathaniel Chartrand, Paul Newling, Jane Vasilyeva, Paula Walsh, Rebecca Smith, Jonathan Karademos, Carol Marty, David Chambliss, Leo French, and Bryce Manso for the management and conduct of the cellular assays; Ryan Wiley and Anne Mullin for discussions; Donald Francis, Carter Lee, Keith Higgins, and Faruk Sinangil for Env gp120s; James Tartaglia, Sanjay Gurunathan, and Sanjay Phogat for advice and discussions; the RV144 Humoral/Innate and Cellular RV144 Working Groups for helpful discussions and pilot analyses; and the RV144 Steering Committee for discussions and oversight (see the Supplementary Appendix for full lists of the Working Groups and Steering Committee).
APPENDIX
The authors’ affiliations are as follows: the Duke University Human Vaccine Institute and the Center for HIV/AIDS Vaccine Immunology, Duke University School of Medicine, Durham, NC (B.F.H., G.D.T., S.M.A., D.C.M., H.-X.L., G.F., K.A.S., N.L.Y., X.S.); the Statistical Center for HIV/AIDS Research and Prevention (P.B.G., Y.F., H.J., A.D., Y.H.) and the Clinical Research Division, Vaccine and Infectious Disease Division (M.J.M., N.F., S.C.D.R.), Fred Hutchinson Cancer Research Center, Seattle; the Veterans Affairs New York Harbor Health-care System and the Department of Pathology, New York University School of Medicine — both in New York (S.Z.-P., C.W.); the Department of Microbiology and Immunobiology, Harvard Medical School, New England Regional Primate Center, Southborough, MA (D.T.E., M.D.A.); the U.S. Military Research Program, Walter Reed Army Institute of Research, Silver Spring (M.R., E.B., M.L.R., R.P., C.A., N.L.M., J.H.K.), and the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda (R.T.B., R.A.K.) — both in Maryland; the U.S. Army Component (C.K., N.K., V.N., M.S.S.) and the Royal Thai Army (S.N.), Armed Forces Research Institute of Medical Sciences, Bangkok, the Department of Microbiology, National HIV Repository and Bioinformatic Center, Siriraj Hospital, Bangkok (R.S.), and the Faculty of Tropical Medicine, Mahidol University, Bangkok (P.P., J.K.), and the Department of Disease Control, Ministry of Public Health, Nonthaburi (S.R.-N.) — all in Thailand; the Department of Biomolecular Engineering, University of California, Santa Cruz, Santa Cruz (P.W.B.); the Institute of Human Virology, University of Maryland School of Medicine, Baltimore (A.L.D., G.K.L.); and the University of Medicine and Dentistry of New Jersey, New Jersey Medical School, Newark (A.P.).
Footnotes
The authors’ affiliations are listed in the Appendix.
The views expressed in this article are those of the authors and should not be construed as official or as representing the views of the Department of Health and Human Services, the National Institute of Allergy and Infectious Diseases (NIAID), the Department of Defense, the Department of the Army, or the Department of Veterans Affairs.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–9. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 2.Idem. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–65. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin L, Gilbert PB, Corey L, McElrath MJ, Self SG. A framework for assessing immunological correlates of protection in vaccine trials. J Infect Dis. 2007;196:1304–12. doi: 10.1086/522428. [DOI] [PubMed] [Google Scholar]
- 4.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 5.Walker BD. Elite control of HIV Infection: implications for vaccines and treatment. Top HIV Med. 2007;15:134–6. [PubMed] [Google Scholar]
- 6.Mascola JR. Defining the protective antibody response for HIV-1. Curr Mol Med. 2003;3:209–16. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- 7.Hessell AJ, Rakasz EG, Tehrani DM, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84:1302–13. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnett SW, Burke B, Sun Y, et al. Antibody-mediated protection against mucosal simian-human immunodeficiency virus challenge of macaques immunized with alphavirus replicon particles and boosted with trimeric envelope glycoprotein in MF59 adjuvant. J Virol. 2010;84:5975–85. doi: 10.1128/JVI.02533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnett SW, Srivastava IK, Kan E, et al. Protection of macaques against vaginal SHIV challenge by systemic or mucosal and systemic vaccinations with HIV-envelope. AIDS. 2008;22:339–48. doi: 10.1097/QAD.0b013e3282f3ca57. [DOI] [PubMed] [Google Scholar]
- 10.Letvin NL, Mascola JR, Sun Y, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–3. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letvin NL, Rao SS, Montefiori DC, et al. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci Transl Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SG, Ford JC, Lewis MS, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–7. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picker LJ, Hansen SG, Lifson JD. New paradigms for HIV/AIDS vaccine development. Annu Rev Med. 2012;63:95–111. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole KS, Rowles JL, Jagerski BA, et al. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J Virol. 1997;71:5069–79. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barouch DH, Liu J, Li H, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forthal DN, Gilbert PB, Landucci G, Phan T. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of HIV-1 in the presence of Fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J Immunol. 2007;178:6596–603. doi: 10.4049/jimmunol.178.10.6596. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert PB, Peterson ML, Follmann D, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–77. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 18.Karnasuta C, Paris RM, Cox JH, et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV-1 vaccine trial in Thailand. Vaccine. 2005;23:2522–9. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Nitayaphan S, Pitisuttithum P, Karnasuta C, et al. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190:702–6. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 20.Rolland M, Gilbert P. Evaluating immune correlates in HIV type 1 vaccine efficacy trials: what RV144 may provide. AIDS Res Hum Retroviruses. 2011 Sep 27; doi: 10.1089/aid.2011.0240. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breslow NE, Holubkov R. Maximum likelihood estimation of logistic regression parameters under two-phase, outcome-dependent sampling. J R Stat Soc B. 1997;59:447–61. [Google Scholar]
- 22.Borgan O, Langholz B, Samuelsen SO, Goldstein L, Pogoda J. Exposure stratified case-cohort designs. Lifetime Data Anal. 2000;6:39–58. doi: 10.1023/a:1009661900674. [DOI] [PubMed] [Google Scholar]
- 23.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
- 24.Gilbert P, Hudgens MG, Wolfson J. Commentary on “Principal stratification — a goal or a tool?” by Judea Pearl. Int J Biostat. 2011;7:36. doi: 10.2202/1557-4679.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinter A, Honnen WJ, Kayman SC, Trochev O, Wu Z. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine. 1998;16:1803–11. doi: 10.1016/s0264-410x(98)00182-0. [DOI] [PubMed] [Google Scholar]
- 26.Zolla-Pazner S, Cardozo T, DeCamp A, et al. V2-Reactive antibodies in RV144 vaccinees’ plasma. AIDS Res Hum Retro-viruses. 2011;27:A-21. abstract. [Google Scholar]
- 27.Edlefsen P, Hertz T, Magaret C, DeCamp A, Gilbert P. Sieve analysis of RV144. Presented at AIDS Vaccine; Bangkok, Thailand. September 12–15, 2011; 2011. abstract. [Google Scholar]
- 28.Newaz F, Cicala C, Van Ryk D, et al. The genotype of early-transmitting HIV gp120s promotes α (4) β(7)-reactivity, revealing α (4) β(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pat-hog. 2011;7(2):e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny MK, Stamatatos L, Volsky B, et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J Virol. 2005;79:5232–7. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Changela A, Wu X, Yang Y, et al. Crystal structure of human antibody 2909 reveals conserved features of quaternary structure-specific antibodies that potently neutralize HIV-1. J Virol. 2011;85:2524–35. doi: 10.1128/JVI.02335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLellan JS, Pancera M, Carrico C, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onyango-Makumbi C, Omer SB, Mubiru M, et al. Safety and efficacy of HIV hyperimmune globulin for prevention of mother-to-child HIV transmission in HIV-1-infected pregnant women and their infants in Kampala, Uganda (HIVIGLOB/NVP STUDY) J Acquir Immune Defic Syndr. 2011;58:399–407. doi: 10.1097/QAI.0b013e31822f8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiss JM, Goroff DK. IgA blocks IgM and IgG-initiated immune lysis by separate molecular mechanisms. J Immunol. 1983;130:2882–5. [PubMed] [Google Scholar]
- 36.Jarvis GA, Griffiss JM. Human IgA1 blockade of IgG-initiated lysis of Neisseria meningitidis is a function of antigen-binding fragment binding to the polysaccharide capsule. J Immunol. 1991;147:1962–7. [PubMed] [Google Scholar]
- 37.Quan CP, Watanabe S, Forestier F, Bouvet JP. Human amniotic IgA inhibits natural IgG autoantibodies of maternal or unrelated origin. Eur J Immunol. 1998;28:4001–9. doi: 10.1002/(SICI)1521-4141(199812)28:12<4001::AID-IMMU4001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 38.Mathew GD, Qualtiere LF, Neel HB, III, Pearson GR. IgA antibody, antibody-dependent cellular cytotoxicity and prognosis in patients with nasopharyngeal carcinoma. Int J Cancer. 1981;27:175–80. doi: 10.1002/ijc.2910270208. [DOI] [PubMed] [Google Scholar]
- 39.Ferrari G, Pollara J, Kozink D, et al. An HIV-1 gp120 envelope human mono-clonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol. 2011;85:7029–36. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mestecky J, Moro I, Kerr MA, Woof JM. Mucosal immunoglobulins. In: Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, Mayer L, editors. Mucosal immunology. 3. Burlington, MA: Elsevier Academic Press; 2005. pp. 153–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.