Abstract
Dictyostelium discoideum is an amoebozoa that exists in both a free-living unicellular and a multicellular form. It is situated in a deep branch in the evolutionary tree and is particularly noteworthy in having a very A/T-rich genome. Dictyostelium provides an ideal system to examine the extreme to which nucleotide bias may be employed in organizing promoters, genes, and nucleosomes across a genome. We find that Dictyostelium genes are demarcated precisely at their 5′ ends by poly-T tracts and precisely at their 3′ ends by poly-A tracts. These tracts are also associated with nucleosome-free regions and are embedded with precisely positioned TATA boxes. Homo- and heteropolymeric tracts of A and T demarcate nucleosome border regions. Together, these findings reveal the presence of a variety of functionally distinct polymeric A/T elements. Strikingly, Dictyostelium chromatin may be organized in di-nucleosome units but is otherwise organized as in animals. This includes a +1 nucleosome in a position that predicts the presence of a paused RNA polymerase II. Indeed, we find a strong phylogenetic relationship between the presence of the NELF pausing factor and positioning of the +1 nucleosome. Pausing and +1 nucleosome positioning may have coevolved in animals.
Among the social amoebae, Dictyostelium discoideum is the most studied species. All of these organisms live in the forest soil as solitary cells, feed on bacteria, and proliferate by binary fission (Chisholm and Firtel 2004). Upon starvation, free-living cells aggregate to form multicellular structures that undergo morphogenesis and cell type differentiation. The major transition of gene expression during development occurs 4–6 h after onset of starvation (Van Driessche et al. 2002). This transition not only comprises the turning on of developmental genes but also a shutdown of basic cellular functions. At this stage, the cells are committed to development and are no longer able to revert to the growth phase (Katoh et al. 2007). Development ends with the formation of a basal disc and stalk, which supports a fruiting body. Fruiting bodies contain spores that can survive starvation conditions.
Dictyostelium is located deep in the phylogenetic tree of life, diverging just after plants but before the divergence of fungi and animals (Eichinger and Noegel 2003; Williams et al. 2005). Since fungi and animals organize their chromatin differently, Dictyostelium should provide some insight into this major evolutionary shift.
The 34-Mb and ∼12,500-gene genome of Dictyostelium is unusual in that it is highly A/T-rich (78%), surpassed only by the 81% A/T-richness of Plasmodium falciparum (Eichinger et al. 2005). The evolutionary forces that drove these organisms to such an A/T-rich extreme are unclear. Analysis of the Dictyostelium genome structure may, therefore, provide insight into how highly preferred usage of adenosine and thymine bases leads to the organization of chromatin, genes, and regulatory elements.
Like most eukaryotes, Dictyostelium utilizes nucleosomes to package its nuclear genome into chromatin. A nucleosome is composed of a histone protein core with ∼147 bp of DNA wrapped around its exterior. Typically, nucleosomes are spaced by linker DNA that ranges from 10 to ∼50 bp in length. Nucleosomes may regulate gene expression by allowing or blocking access of transcription factors to regulatory DNA elements such as TATA boxes (Fascher et al. 1990; Moreira and Holmberg 1998). Across eukaryotic species, promoter regions and other transcriptional regulatory sites are generally nucleosome-free (termed NFR), whereas most genic regions are occupied with arrays of well-phased nucleosomes, especially just downstream of the transcription start site (TSS) (Lee et al. 2004; Yuan et al. 2005; Albert et al. 2007; Mavrich et al. 2008b; Schones et al. 2008; Lantermann et al. 2010; Valouev et al. 2011).
As demonstrated in yeast (Ioshikhes et al. 2006; Lee et al. 2007; Kaplan et al. 2009; Tillo and Hughes 2009; Zhang et al. 2009, 2011b), nucleosome occupancy levels (but not positioning) are controlled in large part by the underlying DNA sequence. AA/TT dinucleotide periodicities and GC-richness have been proposed to contribute to nucleosome occupancy. An intriguing question, then, is whether nucleosome occupancy in the highly A/T-rich Dictyostelium genome utilizes similar principles or, instead, has evolved different mechanisms.
Given the depth of the Dictyostelium branch on the tree of life and that it spends part of its life cycle in an independent unicellular state and part in a multicellular state, it is unclear as to whether Dictyostelium organizes its chromatin as a unicellular or a multicellular organism. The unicellular pattern has been defined by Saccharomyces cerevisiae and Schizosaccharomyces pombe, which are evolutionarily divergent by nearly a billion years. These fungi place the upstream border of the first genic nucleosome over the TSS (Albert et al. 2007), where it would be positioned to block access of the transcription machinery. This placement of nucleosomes over the TSS would suggest that loss of histone-DNA contacts around the TSS might precede transcription initiation.
In contrast, multicellular eukaryotes, defined largely by Drosophila and vertebrates, place their first nucleosome further downstream and away from the TSS (Mavrich et al. 2008b; Schones et al. 2008). As a result, the TSS resides in the NFR, which would appear to give unobstructed access of RNA polymerase II (Pol II) to the TSS. Possibly as a consequence of this, Pol II initiates transcription, elongates, then pauses just upstream of the +1 nucleosome, which is also ∼20–50 bp downstream from the TSS (Gilmour and Lis 1986; Rougvie and Lis 1988; Lee et al. 2008). Recent genome-wide studies showed that this promoter-proximal pausing is widespread in animals (Muse et al. 2007; Zeitlinger et al. 2007; Gilchrist et al. 2010) and that it may contribute to cell-to-cell coordination/timing of developmental programs in animals (Levine 2011). Coordinated timing is critical for multicellular development and is also a key element in the Dictyostelium life cycle.
Here, we present the first high-resolution genome-wide nucleosome maps of an amoeba, whose genome is highly A/T-rich and representative of a deep branch in the phylogenic tree. First, we examined the nucleotide composition of Dictyostelium genes, promoter regions, and nucleosome positions. Second, we determined nucleosome organization around genes during its unicellular and multicellular life stages. Third, we addressed the evolutionary relationship between nucleosome organization and potential pausing of the transcription elongation complex.
Results
Dictyostelium start their genes with poly-T and end with poly-A
As a prelude to nucleosome mapping, we first sought to characterize the extreme A/T-richness of the Dictyostelium genome. In particular, we found tracts of poly-T or poly-A to be more prevalent across the Dictyostelium genome than expected by chance (Supplemental Fig. S1A). The same was seen for the highly A/T-rich genome of Plasmodium falciparum and much less so in other genomes (Supplemental Fig. S1B). We, therefore, suspect that A/T-rich genomes impart function to long poly-T/poly-A tracts.
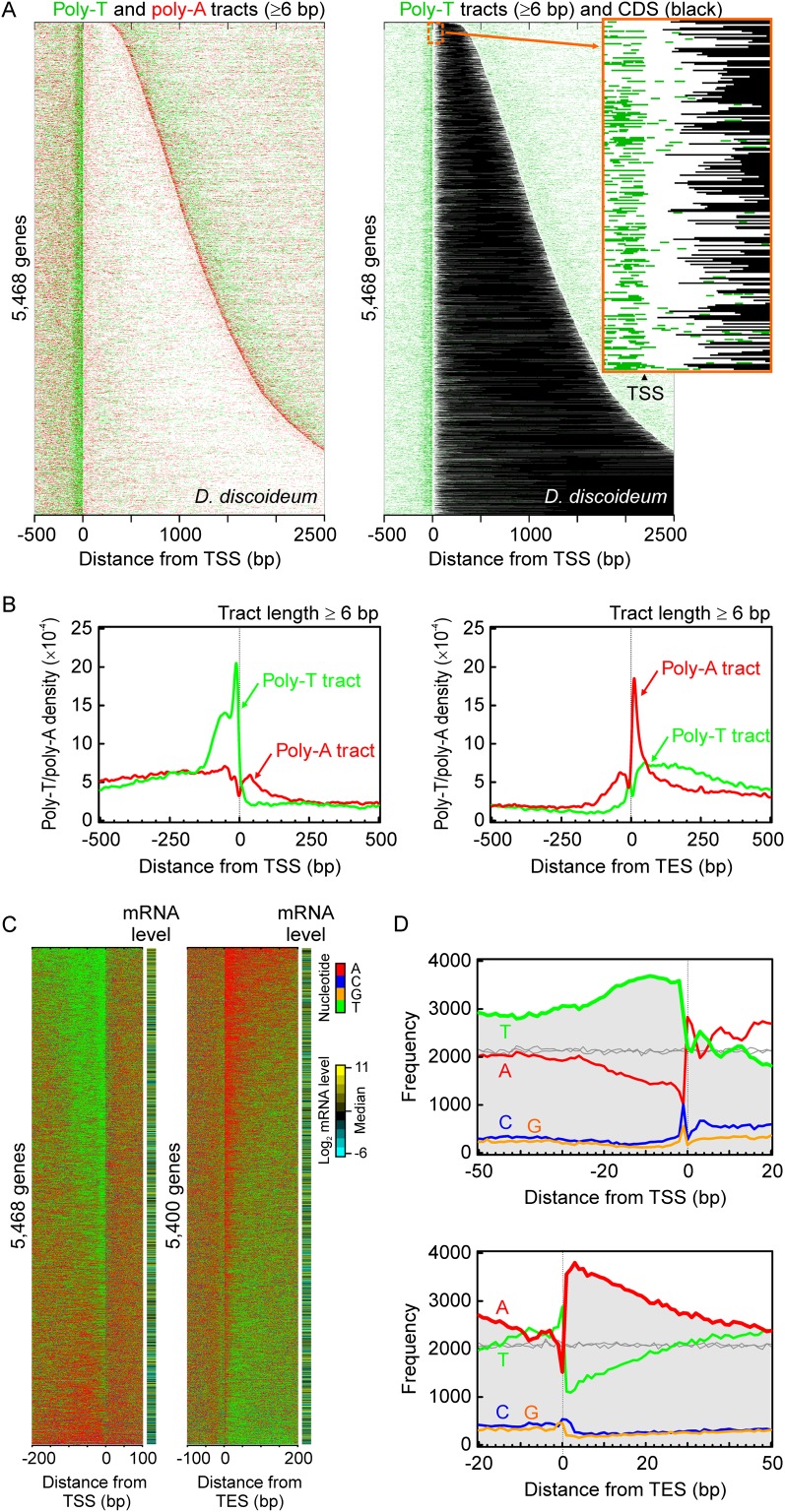
Next, we examined the distribution of poly-T or poly-A tracts around transcription start (TSS) and end (TES) sites. This necessitated the production of a high-resolution transcriptome map of Dictyostelium, which was accomplished by RNA-seq. Four observations regarding the distribution of T and A nucleotides surprised us. First, poly-T tracts were highly enriched on the sense (or nontemplate) strand near the start of genes, immediately upstream of the TSS (Fig. 1), as seen previously on a small number of Dictyostelium genes (Kimmel and Firtel 1983; Hori and Firtel 1994). Second, the 3′ ends of these poly-T tracts were tightly distributed ∼2 bp upstream of the TSS. Third and fourth, poly-A tracts were highly enriched on the sense strand at the end of genes and started ∼1 bp downstream from the TES. Remarkably, poly-T and poly-A on the sense strand precisely demarcated the start and end of transcribed regions, respectively. The precision of poly-T and poly-A tracts with respect to the TSS and TES highlights the accuracy of the transcriptome map.
Figure 1.
Dictyostelium gene organization with poly-T/poly-A enrichment. (A) Distribution of poly-T/poly-A tracts (≥6 bp) in 5468 genes with the annotated TSS. Each track represents the DNA sequence of a gene, fetched from −500 to 2500 relative to the TSS. Genes were aligned by their TSS, and their DNA sequence in the sense strand was oriented in the 5′ to 3′ direction. If any poly-T/poly-A tract of length six or more was found in the DNA sequences, the poly-T sequence was colored in green and the poly-A sequence in red, and all the others were drawn as white in the sequence. The genes were sorted in ascending order of length. The right panel shows poly-T tracts as green and coding sequences as black. A zoom-in screenshot demonstrates the positional details in poly-T enrichment at TSS and translation start sites. (B) Composite distribution of the location of poly-T/poly-A tracts around the 5′ and 3′ end of Dictyostelium genes. The density distribution of poly-T/poly-A tracts (≥6 bp) was displayed as a function of the distance (bp) on the sense strand between the midpoint of each nonoverlapping tract and the given TSS (or TES). The density of the occurrence of poly-T (green trace) and poly-A tracts (red trace) is shown in the y-axis, which was estimated by Gaussian kernel and a smoothing bandwidth of 5. The density curve was calculated within the range from −1500 to 1500 relative to the TSS (or TES) and only the −500 to 500 region shown in the figure. (C) Transcription start (TSS) and end (TES) site are linked to high T and A enrichment, respectively, in the adjacent intergenic sequence. Each track represents the sense strand of a gene, fetched from 200 bp intergenic to 100 genic relative to the TSS (or TES) of 5468 (or 5400) protein-coding genes. Genes are aligned by the TSS (left) or TES (right), in the 5′ to 3′ direction from left to right. Transcript abundance is shown for each gene in an adjacent column. Genes were ordered by descending T (left) or A (right) density of their extracted sequence (301 bp). Color codes for the four nucleotides are indicated. This trend was not observed with randomization simulations (not shown). (D) Frequency distribution of A, C, G, and T relative to the TSS and TES. Shown is a summation of columns from panel C, over the indicated distance, color-coded as in panel C. The same simulation was applied to a set of TSSs or TESs randomly positioned across the Dictyostelium genome (gray traces, shown for A and T).
The lengths of these poly-T and poly-A tracts did not correlate with transcript abundance or with each other (Supplemental Figs. S2, S3), the latter indicating that these complementary nucleotides were not quantitatively linked between the 5′ and 3′ ends of genes. Other organisms lacked an equivalent organization (Supplemental Fig. S4), although other patterns were evident, in accord with the literature (Wu and Li 2010). Plasmodium lacked a strand-specific enrichment of poly-T/poly-A (Supplemental Fig. S5), and thus A/T-richness alone cannot explain the striking poly-T/poly-A demarcation of genes in D. discoideum. The A/T-rich genome of Dictyostelium might be the product of a distinct evolutionary path toward A/T-richness, which Plasmodium did not encounter (Szafranski et al. 2005).
Poly-T tracts in promoters are interrupted by TATA boxes
The TATA box promoter element consists of an 8-bp consensus in Saccharomyces: TATA(A/T)A(A/T)(A/G) (Basehoar et al. 2004). In Dictyostelium, the A/T-richness of the genome raises the question as to whether TATA boxes are sufficiently unique to specify the location of the transcription initiation complex. If so, we would expect bona fide TATA boxes to be precisely positioned upstream of the TSS, as they are in other organisms (∼30 bp in metazoans and ∼60 bp in Saccharomyces). As in metazoans, we found a tight distribution of TATAAA(A/T)(A/T) elements centered ∼29 bp upstream of Dictyostelium TSSs (Fig. 2A). The sequence directionality of these elements was strongly biased toward the sense strand. A highly focused enrichment at the canonical metazoan location, with a strong strand bias, provides three lines of evidence that they are functional. As a fourth line of evidence, no strand-specific enrichment was found downstream from the TES at the 3′ ends of genes (Fig. 2B). In contrast, fortuitous TATA elements may be present on the genic side of the TSS-proximal poly-T (e.g., 5′-TTTTTATA-3′ is the reverse complement of a TATA box) and TES-proximal poly-A tracts (e.g., 5′-TATAAAAA-3′).
Figure 2.
Frequency distribution of TATA elements as TATAAA(A/T)(A/T) midpoint locations around the 5′ and 3′ ends of Dictyostelium genes. TATA elements were counted on the sense (black trace) and template (red trace) strand. Their frequency was binned at the 5-bp interval and plotted as a function of the distance from the TSS (A) and TES (B). The frequency distribution of poly-T/poly-A tract locations (≥12 bp) was produced in the same way and is shown in gray.
Dictyostelium nucleosomes have polymeric-A/T borders
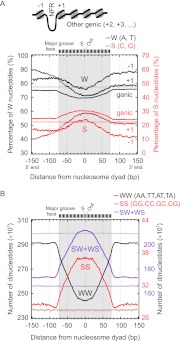
Nucleosomes were mapped across the Dictyostelium genome using MNase-seq (Supplemental Fig. S6), before and after the major transition in gene expression during the two distinct phases of its life cycle: as a free-living unicellular amoeba and as a multicellular aggregate. Dyad-enriched G/C and the presence of 10-bp repeats of WW-type (W = A/T) dinucleotides are important determinants of nucleosome formation. Indeed, Dictyostelium nucleosomes were relatively enriched with G/C near their midpoints (Fig. 3; Supplemental Fig. S7). However, 10-bp periodic WW dinucleotides were not evident (Fig. 3B), despite these dinucleotides being among the most abundant in the genome (Supplemental Table S1). Similar observations were made for human nucleosomes (Tolstorukov et al. 2009), and auto correlation analysis of dinucleotides within nucleosomal sequences also indicated a general lack of such periodicities (data not shown).
Figure 3.
Nucleosomal DNA properties of Dictyostelium nucleosomes. (A) 73,396 nucleosome dyad locations were grouped as −1 (N = 3230), +1 (N = 7285), and all other genic nucleosomes (N = 63,643) (see diagram). For the three assigned nucleosome groups, the W (= A/T in black) and S (= G/C in red) nucleotide percentage at each position was calculated on the sense strand (5′ → 3′ from left to right), so as to maintain directionality of the frequency patterns with respect to the TSS. Plots were smoothed via a 3-bp moving average. (Gray and light red) Randomized distributions. The schematic bar in the upper part represents the rotational orientation of the major groove against the histone octamer surface. (B) Frequency distribution of the WW, SS, and SW+WS dinucleotides relative to nucleosome dyads, as described in Albert et al. (2007) and with the same randomization.
Toward the nucleosome borders and linker region, we observed a high incidence of homo- and heteropolymeric-A/T (meaning any continuous combination of A or T along a strand) (Supplemental Fig. S8). Such nucleosome placement was observed in all classes of nucleosomes including the −1 and +1 nucleosomes that flank promoter regions (data not shown). Thus, Dictyostelium may utilize a combination of both homo- and heteropolymeric-A/T near nucleosome peripheries and relative G/C-richness (albeit sparse on an absolute level) toward the dyad to promote nucleosome formation. This trend is similar to but perhaps a more extreme version of what is evident in fungi and metazoans.
Dictyostelium organizes its nucleosomes on genes similar to animals
We next examined the organization of Dictyostelium nucleosomes around their transcription start and end sites and addressed the following questions: (1) Does Dictyostelium package the 5′ ends of its genes basically in the same way, by having canonical nucleosome positions relative to the TSS? (2) Are NFRs present at the 5′ and 3′ end of genes, as seen in other organisms? (3) Is the +1 nucleosome placed over the TSS, as in single-celled fungi, or downstream, as in multicellular animals and plants? (4) Is nucleosome spacing uniform within genic nucleosomal arrays, as seen in other organisms? (5) Does nucleosome organization change between the unicellular and multicellular state?
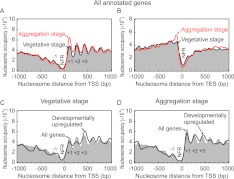
Figure 4A displays a composite distribution of nucleosomes in vegetative amoebae and multicellular aggregates aligned by the transcriptional start site of 5468 protein-coding genes. No canonical organization was seen upstream of the TSS, where the −1 nucleosome typically resides, although the region appeared rather depleted of nucleosomes compared to genic regions. A 5′ NFR, centered 54 bp upstream of the TSS, was evident. This region is where the TATA box and poly-T/poly-A tracts reside and is the likely site of transcription complex assembly. At the ends of genes, a 3′ NFR was found as seen in other organisms (Mavrich et al. 2008a; Shivaswamy et al. 2008; Yadon et al. 2010). This region might be important for transcription termination. The depletion of nucleosomes upstream of the TSS is unlikely to be due to an inability to extract those portions of the genome, because whole genome 454 Life Sciences (Roche) sequencing of Dictyostelium does not reveal such bias (Supplemental Fig. S9). Within genic regions, nucleosomes were, indeed, organized into a canonical arrangement relative to the TSS. The first genic nucleosome (+1) was centered at approximately +115 relative to the TSS.
Figure 4.
Nucleosome organization around the 5′ and 3′ end of the Dictyostelium genes. (A,B) Composite distribution of in vivo nucleosome positions relative to the TSS and TES. The midpoints of all nucleosomal sequence reads were distributed around the TSS (or TES) of 5468 (or 5400) protein-coding genes, plotted as described in Figure 1B and smoothed with a bandwidth of 15 bp. The free-living (vegetative) and multicellular (aggregation) stages are indicated by black and red traces, respectively. (C,D) Nucleosome organization around the TSS of 325 developmentally up-regulated genes (black trace) in the vegetative and aggregation stages, compared to all nucleosomes (gray fill). Traces were smoothed with a 30-bp bandwidth.
Vegetative and aggregating cells showed a similar nucleosome organization in the genome, which indicates that the same overall chromatin structure predominates at Dictyostelium genes and is maintained regardless of its uni- vs. multicellular status (Fig. 4C,D). However, cells that developed for 6 h (aggregation stage) appeared to have a higher nucleosome occupancy at the beginning (+1 position) and end of genes compared to the unicellular state. The +1 nucleosome position is among the most highly dynamic, at least in yeast (Rando and Ahmad 2007; Rufiange et al. 2007). During cellular differentiation imposed by starvation, most genes become quiescent (Loomis and Shaulsky 2011). Nucleosome acquisition at this position might therefore play a role in this process.
We further examined genes whose expression is up-regulated during the aggregation phase (measured by RNA-seq) and found that they displayed the same basic nucleosome organization as the average gene in the aggregation state. Thus, changes in gene expression are not generally associated with major or global changes in chromatin organization, although transient changes are not excluded (and are, indeed, likely). The main conclusion is that, like other organisms, Dictyostelium has a canonical nucleosome organization that is largely invariant, regardless of the activity of relevant genes and through at least two distinct life cycle states.
An evolutionarily conserved change in genic nucleosome spacing upon starvation
Despite similarities in overall organization during the unicellular and aggregated states, we observed a modal shift in the spacing between genic nucleosomes in the two states (Fig. 5). Canonical spacing increased on average from 162 bp in the unicellular state to 169 in the multicellular state (Supplemental Table S2B). This increase in spacing is reminiscent of a similar increase in spacing seen in yeast during a starvation response (Zhang et al. 2011a) and the increase seen in human at cells quiescent/repressed regions of the genome (Vaillant et al. 2010; Valouev et al. 2011). In Dictyostelium, a starvation response is essentially the basis for development into a multicellular organism and a shutdown in gene expression. Thus, an increase in genic inter-nucleosome spacing may be an evolutionarily conserved response to starvation, which may be associated with transcriptional quiescence and the production of refractory chromatin.
Figure 5.
Comparison of inter-nucleosomal distances between five species in their nucleosome arrangement within the genic region. The canonical inter-nucleosomal distance (bp) between two neighboring nucleosomes (for example, +1 and +2) was calculated as the peak-to-peak distance, which was measured in the composite distribution of in vivo nucleosome locations. The nucleosome distribution of each species was produced as described in Methods. The nucleosomal repeat length (as an averaged distance between neighboring nucleosomes) is reported in Supplemental Table S2B.
Dictyostelium organizes genic nucleosomes into pairs
We examined nucleosome spacing more quantitatively in Figure 5, by plotting the canonical inter-nucleosomal distance between the +1 and +2 positions, between +2 and +3, and so on. We also did the same in other organisms whose nucleosomes had been mapped at high resolution. Strikingly, Dictyostelium nucleosomes appeared to be arranged into dinucleosome units where +1 and +2 are closely spaced, whereas +2 and +3 were more separated. Similarly, +3 and +4 were closely spaced compared to their flanking nucleosomes. These differences were not due to a position-specific alteration in the size or sequence composition of the nucleosomal DNA (Supplemental Fig. S9). Such dinucleosome patterns were not evident in yeast and other organisms, suggesting that Dictyostelium undergoes an unusual organizational pattern involving pairs of closely spaced nucleosomes (150 to ∼155 bp), which are separated from adjacent pairs by a somewhat greater distance (170 to ∼180 bp).
Evolutionary linkage between RNA polymerase II pausing and a downstream placement of the +1 nucleosome
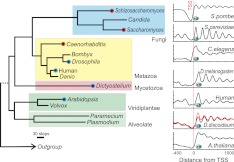
One of our driving questions concerning D. discoideum, was whether the placement of the first genic nucleosome (or +1) resembled that of multicellular eukaryotes or that defined in unicellular fungi. The placement of the first nucleosome has implications with respect to unicellular versus multicellular gene regulatory strategies. A downstream placement has been linked to the existence of an adjacent upstream paused RNA polymerase (Mavrich et al. 2008b; Valouev et al. 2011). In Figure 6, we compare the canonical genic nucleosome organization across the tree of life, with particular emphasis on the +1 nucleosome position. Seven species were chosen in this study, which come from four major phylogenetic branches of eukaryotes (Baldauf et al. 2000) and which range widely across the evolutionary spectrum—S. cerevisiae, S. pombe, human, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, and also D. discoideum. For D. discoideum, genic nucleosomal arrays started at approximately the same distance from the TSS as seen in other multicellular organisms, with the apparent exception of C. elegans. The canonical pattern in C. elegans was rather weak. Most of the TSSs used for analyses in C. elegans are actually ORF starts, since C. elegans undergoes trans-splicing, which obfuscates the location of the TSS (Blumenthal 1995; von Mering and Bork 2002; Valouev et al. 2008). Our results suggest that chromatin organization in Dictyostelium is more like that of multicellular organisms rather than unicellular fungi.
Figure 6.
Chromatin structure around the 5′ end of genes evolutionarily conserved across major eukaryotes. The data of genome-wide nucleosome positions in vivo were curated from the literature and the composite distributions of nucleosome locations were shown for each species in the right panel, aligned by the TSS. The life tree was adapted from the kingdom-level phylogenetic tree of eukaryotes (Baldauf et al. 2000) and simplified in the figure. (Red circle) Those having a substantial portion of the nucleosome over the TSS; (blue circle) those where the TSS is located in the NFR. See Supplemental Table S2A for consensus positions.
We, therefore, wondered whether placement of a canonical +1 nucleosome in the “downstream” position is associated with the presence of a paused RNA polymerase II. To address this possibility, we searched for genes encoding the NELF pausing factor. NELF is a complex of proteins that is responsible for pausing RNA polymerase II at a fixed distance immediately downstream from the TSS (Wu et al. 2003; Gilchrist et al. 2008; Lee et al. 2008). As reported (Narita et al. 2003), this type of pausing has been documented only in metazoans and not in fungi, although a retention of RNA polymerase II in the promoter region has been observed (Venters and Pugh 2009).
As expected of a role of NELF in pausing, NELF homologs were found in metazoans (Supplemental Fig. S11; summarized in Fig. 6). However, NELF homologs were not found in fungi (Saccharomyces or Schizosaccharomyces), and also, surprisingly, not detected in C. elegans. Importantly, NELF homologs were found in Dictyostelium (Supplemental Fig. S11), which suggests that Dictyostelium is likely to undergo RNA polymerase II pausing, and is consistent with the downstream placement of the +1 nucleosome. Thus, there appears to be a strong linkage between having NELF (presumed to reflect the existence of pausing) and having a +1 nucleosome positioned downstream, next to where polymerase is expected to pause. Remarkably, this relationship did not appear to have a phylogenetic continuum, suggesting that the positioning of the +1 nucleosome and its linkage to RNA polymerase pausing may have shifted multiple times during evolution.
NELF was also not found in plants, despite plants seemingly having a downstream placement of the +1 nucleosome. However, plants reside on a more distant branch of the evolutionary tree compared with all other multicellular eukaryotes tested, and thus a NELF homolog may be present but sufficiently diverged to be undetectable by our homology search algorithm. Taken together, these findings provide evidence for a deep evolutionary linkage between placement of the +1 nucleosome in a downstream position and the presence of an adjacent upstream paused polymerase that perhaps arose as part of a genomic gene regulatory mechanism that is imposed by the requirements of multicellularity. This is consistent with the notion that RNA polymerase pausing is a means of coordinating the developmental timing between adjacent cells in multicellular eukaryotes (Levine 2011).
Discussion
Is Dictyostelium moving toward a binary genomic code?
The unusual A/T-richness of the Dictyostelium genome makes it well-suited to evaluate the constraints placed on the utilization of G/C vs A/T nucleotides. Most organisms have a more-or-less balanced usage of the four nucleotides to encode not only genetic information but also for the regulation of the maintenance and expression of that information. For Dictyostelium, we see some very biased and unusual usages of A/T to the point where it raises the question as to whether this organism is evolving toward a binary code. Inasmuch as G or C is critical for certain codons, this seems unlikely to ultimately occur. Nonetheless, the extent to which different combinations of A and T can be used for different purposes is remarkable.
The diverse usage of A/T nucleotides is illustrated in the following examples. First, we find that the 3′ ends of poly-T tracts, located on the sense strand upstream of ORF starts, demarcate transcriptional start sites. Second, the 5′ ends of poly-A tracts, when located on the sense strand downstream from ORF ends, demarcate transcript end sites. Third, embedded within these poly-T tracts resides the TATA box, which most typically has the sequence TATAAA(A/T)(A/T). In general, the TATA box is located 25–30 bp upstream of the transcription start site and, together with the general transcription factors, helps determine TSS selection (Fairley et al. 2002). Fourth, further upstream of the TATA box reside poly-T and poly-A tracts which disfavor nucleosome formation and may help maintain nucleosome-free promoter regions. Fifth, as in most other organisms, AATAAA near the 3′ ends of genes signifies the site of transcript cleavage, and sixth, to some extent in some organisms, periodic AA/TT dinucleotides may promote nucleosome formation.
We do not know to what extent poly-T tracts define promoter regions and why poly-T rather than poly-A is preferred on the sense strand. Certainly, we would expect such tracts to exclude nucleosomes around the TATA box. This suggests that nucleosomes may not be as influential in controlling TATA box accessibility and pre-initiation complex formation in Dictyostelium as might occur in more GC-rich genomes. One model of transcription initiation is that RNA polymerase II moves away from the pre-initiation complex in search of a start site (Giardina and Lis 1993; Li et al. 1994; Pardee et al. 1998; Kuehner and Brow 2006). Such movement might be accompanied by abortive transcription, where short RNA transcripts are released until a bona fide start site is found. After the start site, the nascent transcript may be more stable in an RNA/DNA hybrid to allow productive transcript elongation. Thus, it is conceivable that abortive transcripts consisting of poly-rU are particularly dissociable and thus preferred just upstream of the TSS. On the other end of the transcript, higher RNA/DNA stability imparted by a tract of poly-rA might slow down RNA polymerase, making it more susceptible to termination mechanisms. While these ideas are speculative, studies on RNA/DNA hybrid stability support these ideas. In particular, poly-rU/poly-dA hybrids (akin to those occurring just upstream of the TSS) are the least stable, and poly-rA/poly-dT hybrids (akin to the downstream arrangement) are the most stable of all RNA/DNA hybrid tracts consisting of only A or T (or U) (Sugimoto et al. 1995; Kraeva et al. 2007).
While G/C-richness near the nucleosome dyad or AA/TT dinucleotide periodicities may be contributory toward nucleosome assembly, we only observed G/C-enrichment near the dyad of Dictyostelium nucleosomes, with little or no AA/TT periodicity. The latter is surprising given the extreme A/T-richness of the genome, but such periodicities also appear to be weak or nonexistent in other organisms (Mavrich et al. 2008a; Tolstorukov et al. 2009; Valouev et al. 2011). Dyad-enriched G/C was also weak on an absolute scale. However, homo- and heteropolymeric A/T tracts were enriched near nucleosome borders, which raises the question as to what extent these sequence preferences contribute to nucleosome formation.
Dictyostelium organizes genic nucleosomes as in animals
Nucleosome organization in the Dictyostelium genome is most similar to that of metazoans, where the first nucleosome downstream from the TSS places the TSS in the NFR. As such, the transcription machinery would be generally free to initiate transcription without direct influence of the +1 nucleosome. This contrasts with fungi where one edge of the +1 nucleosome resides over the TSS, and thus nucleosome displacement is expected to be a prerequisite to initiation.
A downstream placement of the +1 nucleosome raises the question as to whether RNA polymerase II pauses as it approaches that nucleosome. More generally, the question arises as to whether RNA polymerase II pausing and downstream placement of the +1 nucleosomes have coevolved, perhaps originating from constraints imposed by multicellularity occurring at some stage of the life cycle. We examined this by comparing whether downstream placement of the +1 nucleosome is evolutionarily linked to the presence of NELF-encoding genes. NELF is a key determinant of pausing. Consistent with the link between pausing and nucleosome placement, fungi (S. cerevisiae and S. pombe, representing deep phylogenetic branches in fungi) lacked both, whereas metazoans and Dictyostelium contained both. This linkage ties in and agrees well with the recent notion that RNA polymerase II pausing functions to coordinate the timing of multicellular development (Levine 2011), and this would include Dictyostelium. One rationale is that many events are needed for assembly of the transcription machinery at promoters, and the combination of these events may have some stochastic elements. Thus, response to cell–cell signaling in development may be less coordinated among cells if left solely to the regulation of pre-initiation complex assembly. In contrast, a paused polymerase may only need a release factor to promote the expression of the gene, and in principle, this would be a more effective mechanism to ensure coordination among cells.
Methods
Nucleosome mapping
Strain AX2-214 (Harloff et al. 1989; Bloomfield et al. 2008) was grown axenically in liquid nutrient medium. Aggregated cells were grown in the same way, then harvested and incubated in Soerensen phosphate buffer at 22°C for 6 h. Cells were then subjected to formaldehyde crosslinking. Cells were harvested, washed, and lysed in 1% Triton-×-100.
Mononucleosomes were prepared essentially as described (Albert et al. 2007; Mavrich et al. 2008b). Briefly, washed chromatin pellets were treated with MNase to produce mainly mononucleosomes, which were then gel-purified and subjected to deep sequencing using the 454 Life Sciences (Roche) GS20/FLX sequencer. This sequencer was utilized to take advantage of its long-read capability, which may improve mappability for potentially low complexity (high A/T-rich) genomes. Sequencing reads essentially covered the entire nucleosome, thereby reporting on the location of both of its ends. We took the read midpoint to reflect the nucleosome dyad.
Transcriptome sequencing and analysis
RNA was isolated from wild-type strain NC4, which is the parent of AX4 and AX2 derivatives. Cells were grown on bacteria, then plated onto phosphate agar plates (0 timepoint). RNA was extracted at 0, 4, 8, 12, 16, 20, and 24 h of development using the RNeasy Mini Kit from Qiagen, then converted to cDNA using the Evrogen Mint Kit. cDNA libraries were sequenced using the Roche GS20/FLX system.
To obtain an unbiased location of transcripts, all seven RNA-seq data of 0, 4, 8, 12, 16, 20, and 24 h were compiled together, and the 5′- and 3′-most base of the matching region of a gene were defined as the TSS and TES, respectively (Supplemental Table S3), and were compared to TSS locations that pre-exist in the literature (Supplemental Table S4). Read counts for each gene at the indicated time point of development were taken as a measure of expression levels. Expression data were normalized to total read counts and to gene length. Differential expression was assumed if the read numbers between the start point of development and 4 and/or 8 h into the cycle were at least fivefold different. Among the 5468 genes with the annotated TSS, 325 genes were finally grouped as developmentally up-regulated genes, which were highly expressed during the first 8 h of starvation (Supplemental Table S3).
Data analysis
To identify overrepresented TATA sequence motifs in core promoters, we initially used the D. discoideum consensus TATA box TATAAA[T/A]A, which was reported in 15 genes (Kimmel and Firtel 1983). We located TATA boxes with this consensus as a preliminary screen and aligned their locations by the TSS. We extracted the DNA sequences in the core promoter region from −37 to −21 bp upstream of the TSS, where bona fide TATA boxes are highly likely to be located. We considered the TATA sequences only in the sense (or nontemplate) strand (Basehoar et al. 2004). We looked for overrepresented motifs with those sequences using the MEME software (Bailey and Elkan 1994).
In accordance with the systematic nomenclature of nucleosome positions in the study of Jiang and Pugh (2009), we demarcated the border of the −1, NFR, and +1 zones around transcription start sites. The TES of some genes was not available, and the ORF end site was used instead.
In conducting the NPS (nucleosome-positioning sequence) analysis with nucleosomal core particle DNA sequences, the frequency of 10 dinucleotide sets (Satchwell et al. 1986) was compiled as a function of the distance from the nucleosome dyad. Each dinucleotide was defined by reading their sequence in the 5′- to 3′-end direction (Albert et al. 2007; Mavrich et al. 2008a,b).
Data access
The raw nucleosomal sequencing reads have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA044979. The raw RNA sequencing reads have been submitted to the NCBI Sequence Read Archive (SRA) under accession number SRA045580.
Acknowledgments
This work was supported by NIH grant HG004160 to B.F.P., by DFG grant Gl235-1 to G.G., and by CECAD to A.A.N.
Author contributions: G.S.C. conducted most of the analyses and generated the graphs; A.A.N. provided material, evaluated the data, and contributed to manuscript writing; T.N.M., E.W., and R.M. prepared nucleosomes, L.T. sequenced nucleosomal DNA; M.F. and C.J. conducted data analysis; L.E. contributed to data analysis and manuscript writing; G.G. conducted data analysis, provided transcript data, and contributed to manuscript writing; S.C.S. supervised nucleosome sequencing; A.A.N., S.C.S., and B.F.P. designed the research, B.F.P. supervised the project and data analysis and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131649.111.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977 [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bloomfield G, Tanaka Y, Skelton J, Ivens A, Kay RR 2008. Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum. Genome Biol 9: R75 doi: 10.1186/gb-2008-9-4-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet 11: 132–136 [DOI] [PubMed] [Google Scholar]
- Chisholm RL, Firtel RA 2004. Insights into morphogenesis from a simple developmental system. Nat Rev Mol Cell Biol 5: 531–541 [DOI] [PubMed] [Google Scholar]
- Eichinger L, Noegel AA 2003. Crawling into a new era—the Dictyostelium genome project. EMBO J 22: 1941–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairley JA, Evans R, Hawkes NA, Roberts SG 2002. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol Cell Biol 22: 6697–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher KD, Schmitz J, Horz W 1990. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J 9: 2523–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C, Lis JT 1993. DNA melting on yeast RNA polymerase II promoters. Science 261: 759–762 [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 22: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol 6: 3984–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harloff C, Gerisch G, Noegel AA 1989. Selective elimination of the contact site A protein of Dictyostelium discoideum by gene disruption. Genes Dev 3: 2011–2019 [DOI] [PubMed] [Google Scholar]
- Hori R, Firtel RA 1994. Identification and characterization of multiple A/T-rich cis-acting elements that control expression from Dictyostelium actin promoters: The Dictyostelium actin upstream activating sequence confers growth-phase expression and has enhancer-like properties. Nucleic Acids Res 22: 5099–5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF 2006. Nucleosome positions predicted through comparative genomics. Nat Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109 doi: 10.1186/gb-2009-10-10-r109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Chen G, Roberge E, Shaulsky G, Kuspa A 2007. Developmental commitment in Dictyostelium discoideum. Eukaryot Cell 6: 2038–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Firtel RA 1983. Sequence organization in Dictyostelium: Unique structure at the 5′-ends of protein coding genes. Nucleic Acids Res 11: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraeva RI, Krastev DB, Roguev A, Ivanova A, Nedelcheva-Veleva MN, Stoynov SS 2007. Stability of mRNA/DNA and DNA/DNA duplexes affects mRNA transcription. PLoS ONE 2: e290 doi: 10.1371/journal.pone.0000290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA 2006. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J Biol Chem 281: 14119–14128 [DOI] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS 2008. NELF and GAGA factor are linked to promoter proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Flanagan PM, Tschochner H, Kornberg RD 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263: 805–807 [DOI] [PubMed] [Google Scholar]
- Loomis WF, Shaulsky G 2011. Developmental changes in transcriptional profiles. Dev Growth Differ 53: 567–575 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008a. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008b. Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JM, Holmberg S 1998. Nucleosome structure of the yeast CHA1 promoter: Analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J 17: 6028–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al. 2003. Human transcription elongation factor NELF: Identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 23: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee TS, Bangur CS, Ponticelli AS 1998. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem 273: 17859–17864 [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K 2007. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol 19: 250–256 [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA 1986. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191: 659–675 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR 2008. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65 doi: 10.1371/journal.pbio.0060065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211–11216 [DOI] [PubMed] [Google Scholar]
- Szafranski K, Lehmann R, Parra G, Guigo R, Glockner G 2005. Gene organization features in A/T-rich organisms. J Mol Evol 60: 90–98 [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442 doi: 10.1186/1471-2105-10-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ 2009. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res 19: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant C, Palmeira L, Chevereau G, Audit B, d'Aubenton-Carafa Y, Thermes C, Arneodo A 2010. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res 20: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. 2008. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche N, Shaw C, Katoh M, Morio T, Sucgang R, Ibarra M, Kuwayama H, Saito T, Urushihara H, Maeda M, et al. 2002. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 129: 1543–1552 [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Bork P 2002. Teamed up for transcription. Nature 417: 797–798 [DOI] [PubMed] [Google Scholar]
- Williams JG, Noegel AA, Eichinger L 2005. Manifestations of multicellularity: Dictyostelium reports in. Trends Genet 21: 392–398 [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Li H 2010. Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters. Genome Res 20: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadon AN, Van de Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T 2010. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol 30: 5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ma H, Pugh BF 2011a. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res 21: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF 2011b. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]