Abstract
MicroRNAs (miRNAs) and transcription factors control eukaryotic cell proliferation, differentiation, and metabolism through their specific gene regulatory networks. However, differently from transcription factors, our understanding of the processes regulated by miRNAs is currently limited. Here, we introduce gene network analysis as a new means for gaining insight into miRNA biology. A systematic analysis of all human miRNAs based on Co-expression Meta-analysis of miRNA Targets (CoMeTa) assigns high-resolution biological functions to miRNAs and provides a comprehensive, genome-scale analysis of human miRNA regulatory networks. Moreover, gene cotargeting analyses show that miRNAs synergistically regulate cohorts of genes that participate in similar processes. We experimentally validate the CoMeTa procedure through focusing on three poorly characterized miRNAs, miR-519d/190/340, which CoMeTa predicts to be associated with the TGFβ pathway. Using lung adenocarcinoma A549 cells as a model system, we show that miR-519d and miR-190 inhibit, while miR-340 enhances TGFβ signaling and its effects on cell proliferation, morphology, and scattering. Based on these findings, we formalize and propose co-expression analysis as a general paradigm for second-generation procedures to recognize bona fide targets and infer biological roles and network communities of miRNAs.
MicroRNAs (miRNAs) are small noncoding RNAs that have basic roles in the control of gene expression (Bushati and Cohen 2007). They carry out their functions in animal cells by binding, with imperfect base pairing, to complementary sequences in the 3′-untranslated regions (3′UTRs) of their target mRNAs. This results in down-regulation of target expression, at either the transcript or the translational level (Baek et al. 2008; Selbach et al. 2008; Guo et al. 2010).
Over the last decade, miRNAs have emerged as important and evolutionarily conserved regulators of various physiopathological processes, from development to cancer (Meola et al. 2009; Visone and Croce 2009). As in the case of transcription factors, target identification is key to an understanding of the functions of miRNAs. The analogies between these two classes of regulatory molecules include the specificity of the sequences they target and a certain degree of flexibility in the composition of these sequences. However, decades of molecular studies on transcription factors have revealed that their actions are largely combinatorial, i.e., their specific effect—activation or repression of gene expression—is strictly dependent on the local chromatin microenvironment, which in turn is an expression of the combination of multiple factors, such as cell type and a plethora of internal and external stimuli. In this regard, one of the most notable features is that the same transcription factor can activate or repress gene expression and even change binding specificities according to its dynamic interactions with other transcription factors and coactivators (Chen et al. 2011). Combinatorial effects multiply the complexity of transcription-factor gene regulatory networks, as well as the efforts needed for their dissection. In contrast, miRNAs appear to have less flexible specificities and effects: They basically repress gene expression through binding to a few subtypes of target sequences, the compositions of which are dictated by their “seed” sequence (Bartel 2009). Moreover, they do not appear to have the same combinatorial logic as transcription factors, but rather more plain synergic or additive effects when multiple miRNAs target the same mRNA (Tsang et al. 2010).
These simpler features give much more appeal to the dissection of the miRNA regulatory networks through the computational identification of their targets. Indeed, the first tools for miRNA target identification were developed shortly after the emergence of miRNAs as regulatory factors of cellular metabolic processes and animal development. These “first-generation” tools have taken into account sequence-based features, like miRNA–mRNA complementarity at the seed region (Rehmsmeier et al. 2004; Krek et al. 2005; Miranda et al. 2006; Betel et al. 2008; Maragkakis et al. 2009; Thomas et al. 2010), the evolutionary conservation of target sequences (Friedman et al. 2009), their numbers (John et al. 2004), and their accessibility, as predicted by analysis of secondary structures (Kertesz et al. 2007). However, current computational methods have intrinsic limitations due to the imprecise complementarity between mRNA/miRNA sequences in animal systems and an overall low specificity that results in a large number of false targets among the predictions (Didiano and Hobert 2006). Moreover, a substantial lack of overlap between the various algorithms has been reported (Saito and Saetrom 2010), which suggests that additional parameters should be considered for the development of more comprehensive prediction algorithms. Recent methods for reducing the number of false positives include expression analysis to detect inverse correlations between miRNA and mRNA transcriptional behaviors, and they have required the use of specific microarray platforms that contain probes for miRNAs (Huang et al. 2007; Hausser et al. 2009; Ulitsky et al. 2010). Alternative methods included the use of miRNA host genes as proxy for measuring the expression of the embedded miRNAs (Gennarino et al. 2009).
Here, we perform a comprehensive analysis of human miRNA regulatory networks by focusing on the expression relationships among miRNA targets. We have developed a strategy based on Co-expression Meta-analysis of miRNA Target genes (CoMeTa) to integrate expression data from hundreds of cellular systems and multiple tissues. CoMeTa analysis of 675 human miRNAs was used to effectively select bona fide miRNA target genes by ranking them according to their degree of co-expression. Subsequent analyses of clusters of miRNA targets have led to the association of specific miRNAs with biological function(s) at high resolution. Furthermore, network analysis has resulted in a comprehensive map of miRNA–miRNA functional interactions based on the overlap among their target cohorts of genes. We validated the CoMeTa procedure by experimental assays focused on the control on the TGFβ pathways exerted by three previously uncharacterized miRNAs.
Results
The CoMeTa procedure
We hypothesized that the targets of a given miRNA are co-expressed with each other, at least in certain tissues/conditions, i.e., they belong to the same gene regulatory network. Based on this assumption, we devised a strategy for the general inference of miRNA downstream regulatory networks through analysis of the expression correlations of their putative targets (CoMeTa).
A scheme for the rationale of CoMeTa is shown in Figure 1A. For each miRNA, the procedure was seeded by using the predicted targets from three sequence-based prediction tools, miRanda (Betel et al. 2008), PicTar (Krek et al. 2005), and TargetScan (Friedman et al. 2009). The overlap among these algorithms is often limited, and thus together they should ensure consistent coverage for target prediction. The expression relationships between predicted targets were calculated by analyzing thousands of publicly available expression microarray experiments that are representative of multiple tissues and conditions. For each miRNA target, a co-expression list was calculated, where any other gene was ranked according to its positive expression correlation with the given target. We recently used a similar approach to identify a gene network that regulates lysosomal biogenesis and function (Sardiello et al. 2009).
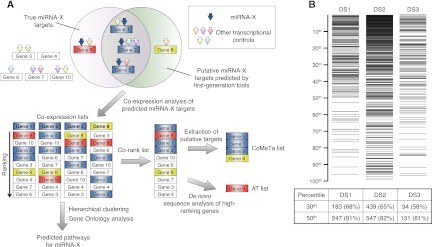
Figure 1.
The CoMeTa procedure. (A) For simplicity, the strategy is described on a subgroup of 10 genes. Multiple transcriptional controls (arrows) for these genes are shown, including a specific miRNA (miRNA-X, blue arrows). In the example, Gene 8 (yellow box) and Gene 9 (red box) are, respectively, a false positive and a false negative result of sequence-based software predictions, used to seed the analysis. In co-expression lists, the ranking of the genes is an index of their expression correlations with the probe gene. A co-rank list is obtained by averaging the co-expression lists. The false-positive target ranks low in the co-rank list, whereas the false-negative target ranks high and can be identified by subsequent de novo sequence analyses (additional targets [AT] list). The CoMeTa output consists of the list of predicted targets ranked by expression analysis. Co-expression lists are subsequently used for further analysis (see text). (B) Distribution of known miRNA targets (horizontal lines in the top panel) within CoMeTa lists, for three independent data sets of miRNA targets (DS1, DS2, DS3). Counts within the first 30th and 50th percentiles are provided for each data set in the lower table. The average number of targets for each miRNA present in the first 50th percentile of CoMeTa lists is 750 (DS1), 850 (DS2), and 900 (DS3). P-values: <10−39 (DS1), <10−29 (DS2), and <10−23 (DS3).
Using this ranking system, a gene under the control of a given miRNA is expected to generate a co-expression list that is enriched with other target genes of the same miRNA at the top positions because of the positive correlations among their expression. However, most genes are subject to multiple transcriptional controls. This implies that a gene under the control of N factors is expected to generate a co-expression list that is enriched at the top positions with genes under the control of the same N factors, including the targets of the given miRNA (Fig. 1A). The co-expression lists associated with a collection of putative miRNA targets will therefore be enriched for genes controlled by multiple factors, specific to each target. However, they will be collectively enriched for the targets of the given miRNA (see Fig. 1A). Based on this hypothesis, for each collection of putative targets we generated a “co-rank” list by taking into account the average target ranking in their respective lists. This procedure was expected to produce two notable outcomes: First, the true targets, including genes missed by sequence-based prediction tools (the false negatives), would rank higher than nontarget genes in the co-rank list because of their higher average ranking in the single co-expression lists (see Fig. 1A). Second, based on the same principle, genes that are not targets of the given miRNA would rank low, including false positives from sequence-based prediction tools.
We applied this procedure to all of the known human miRNAs (n = 675; miRBase, release 13.0) and we obtained the corresponding lists of the coregulated targets ranked according to their expression concordance (Co-rank and CoMeTa lists) (Fig. 1A).
CoMeTa is effective in the recognition of true positive miRNA targets
To test the efficacy of the procedure, we built three independent data sets of previously validated miRNA targets (DS1, DS2, DS3) that were derived from the analysis of available data. DS1 was built using high-confidence miRNA–target pairs that had been validated experimentally and includes 270 target genes coupled with 84 miRNAs (Supplemental Table 1) (Papadopoulos et al. 2009); DS2 includes 671 target genes coupled with eight miRNAs, identified through pSilac experiments (Baek et al. 2008; Selbach et al. 2008); and DS3 includes 162 target genes coupled with three miRNAs, identified by transcriptome analysis (Lim et al. 2005).
The analysis of the CoMeTa lists showed that >90% of the validated targets from DS1 and >80% of the targets identified by high-throughput analyses (DS2, DS3) fall within the first 50th percentile of their respective ranked lists (P < 10−20 for all of the data sets) (Fig. 1B), thus demonstrating the validity of the procedure. A comparison with the scoring systems of TargetScan, PicTar, and miRanda showed that CoMeTa's ranking system improves miRNA target prediction efficiency in all three data sets analyzed (DS1, DS2, and DS3; Supplemental Fig. S1).
We also evaluated the performance of the CoMeTa procedure for the identification of validated targets that escaped recognition by the sequence-based prediction algorithms used to seed the procedure (n = 25 in DS1; Supplemental Table S1). Interestingly, we found that most of these targets have high rankings in their respective miRNA co-rank lists (21 out of 25 above the 50th percentile; P < 10−3). Therefore, to identify putative, additional targets for each miRNA, we carried out a de novo analysis of the 3′UTRs of all genes to search for canonical miRNA seeds (7-mer-A1, 7-mer-m8, or 8-mer sites) (Bartel 2009). The lists of these additional targets (AT lists) (Fig. 1A) are available through the CoMeTa website (http://cometa.tigem.it/site/index.php), along with their ranking positions.
Inference of miRNA gene networks and association with biological functions
We hypothesized that co-expression analyses can drive the prediction of the functional pathways controlled by miRNAs. To test this hypothesis, for each miRNA we clustered the targets that showed the highest extent of co-expression, a procedure hereafter referred to as CO-Operational Level (COOL) analysis. We systematically carried out COOL analyses for all of the human miRNAs and found that predicted targets tended to aggregate in discrete co-expression clusters, compared with random groups of miRNA target genes of similar size (Fig. 2A; Supplemental Fig. S2). We then selected the co-expression clusters that showed greater significance over the control clusters (R-squared [R2] ≤0.91; see Methods for details). A total of 508 co-expression clusters (for 508 miRNAs) were retained as the most statistically significant. Of note, these clusters were significantly enriched for the validated miRNA targets in DS1 (77% of the total; P < 10−5) (Fig. 2A), which indicates that the miRNA targets indeed tend to aggregate in co-expression clusters. As an additional control, we mapped on the COOL clusters the genes that we previously found to be down-regulated upon transient overexpression of miR-26b and miR-98 in HeLa cells (Gennarino et al. 2009). Gene-set enrichment analysis of these genes showed that down-regulated genes (i.e., the most likely direct targets) were significantly enriched in clusters with R2 <0.91 (Fig. 2B,C).
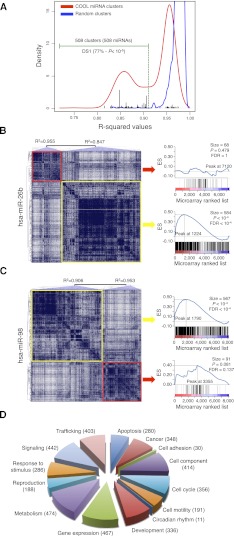
Figure 2.
COOL analysis of miRNA-predicted target transcriptional networks. (A) Kernel density estimation of R2 values for the normal probability plot analysis of miRNA clusters (red line) and size-matched random clusters (blue line). An R2 value of 0.912 represents the lowest value for the random clusters, and this was used as the threshold (green dotted line) to select the significant miRNA clusters. (Black bars) R2 values of DS1-associated clusters. (B,C) COOL heat-maps (left) and their enrichment plots generated by gene-set enrichment analysis (right) for the data set of probes differentially expressed after miR-26b transfection (B) and miR-98 transfection (C). (Blue lines in the plots) Enrichment scores; (vertical black bars below the plots) positions of the probes from the analyzed COOL clusters according to the ranking of their expression levels, i.e., from the most down-regulated (left) to the most up-regulated. (ES) enrichment score; (FDR) false discovery rate. (D) Frequency distribution of functions assigned to COOL clusters as grouped in macrocategories. The number of miRNAs associated with each macrocategory is shown in parenthesis.
Next, to assign biological functions to human miRNAs, we performed gene ontology (GO) and KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway analyses of the significant COOL clusters. These analyses associated hundreds of nonredundant developmental or metabolic functions with specific miRNAs. The diagram in Figure 2D shows that miRNAs were associated with virtually every known functional macro category, from cell housekeeping (cell components, trafficking, and metabolism) and regulatory pathways (cell signaling, gene expression, response to stimulus, apoptosis) to development, reproduction, and cancer. In each macrocategory, the resolution of the analysis for specific pathways was remarkable. For example, miRNAs associated with intracellular trafficking could be mapped to several distinct pathways, including endocytosis (miR-1/103/106a/106b/107), ER-to-Golgi (miR-1/1323/19a/23a/23b), and Golgi (let-7c/7e and miR-1182/1183/1202) vesicle transport, phagocytosis (miR-1257/182/524-5p), and axon cargo (miR-103/107/143/16/195). Similarly, miRNAs associated with gene expression were differentially assigned to pathways regulating epigenetic control (miR-1/1202/1253/1266), basal transcription-factor activity (miR-1294/181b/181c/26a/26b) and RNA processing (miR-105/107/1179/1183), silencing (let-7b/7c and miR-1205/184/298), and translation (let-7 family and miR-1183/1205/1236). Functions were assigned with high confidence to all 508 miRNAs with significant co-expression clusters, and the results showed high concordance with the miRNA functions that had been determined experimentally (135 such cases are given in Supplemental Table 2). Examples include: miR-155, which is involved in hematopoiesis (Kluiver et al. 2006) and immunity (Fig. 3A; Rodriguez et al. 2007); miR-1, which has a role in heart development (Fig. 3B; Sayed et al. 2007); miR-130a, which has been identified as a proangiogenic miRNA (Fig. 3C; Chen and Gorski 2008); miR-106b and miR-93, which are known potent inhibitors of transforming growth factor (TGF)β signaling (Petrocca et al. 2008a); and the miR-29 family, whose members miR-29a and miR29c, but not miR-29b, have been associated with regulation of the Wnt pathway (Kapinas et al. 2009). Notably, COOL analyses also correctly distinguished between closely related members of the same family, i.e., miRNAs that share the same seed sequence, as in the case of the miR-29 family. Dozens of critical biological processes were associated for the first time with miRNA regulation, e.g., neuronal migration (hsa-miR-20b), muscle-tissue development (hsa-miR-655), the BMP signaling pathway (hsa-miR-1252), and many others (see Supplemental Table 2).
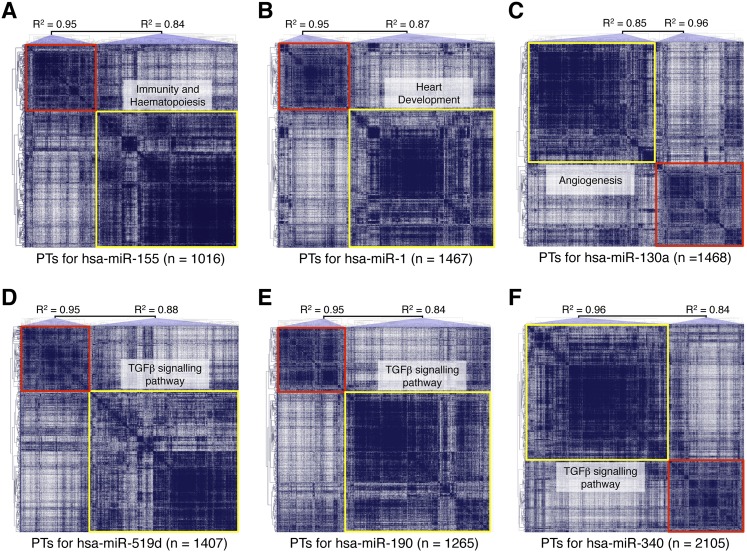
Figure 3.
Overview of COOL clusters with known and predicted functions. Analyses of miR-155 (A), miR-1 (B), miR-130a (C), miR-519d (D), miR-190 (E), and miR-340 (F). The graphs represent the COOL heat-maps of putative targets generated according to their reciprocal expression relationships. (Red and yellow boxes) Clusters with an R2 value higher or lower than the set threshold, respectively. (PTs) Predicted targets. Some of the functional categories enriched in the significant miRNA clusters are indicated.
In addition to providing putative functions to most human miRNAs, these results also strengthen the hypothesis that miRNAs act as global regulators of specific pathways, a function that has classically been attributed to transcription factors.
miR-519d, miR-190, and miR-340 are involved in regulation of the TGFβ signaling pathway
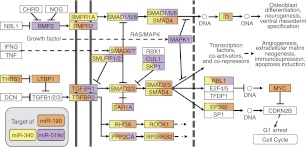
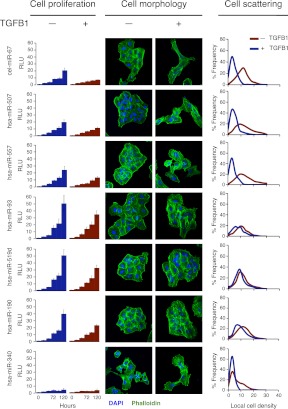
The high concordance between CoMeTa COOL analysis and literature data prompted us to test some of the novel associations with miRNA functions generated by our procedure. Among the miRNAs associated by COOL analysis to TGFβ signaling, miR-519d, miR-190, and miR-340 showed the most significant enrichment (see CoMeTa database) (Fig. 3D–F). A detailed analysis of the CoMeTa associations showed, indeed, that these miRNAs are predicted to target most genes participating in the TGFβ pathway (Fig. 4), which regulates a wide range of biological responses including cell proliferation and differentiation and tumorigenesis. Interestingly, the predicted targets of the three miRNAs include both positive (for example SMAD2/3) and negative (for example SMAD6/7) regulators of the pathway. The A549 non-small cell lung carcinoma cell line is highly sensitive to TGFB1 (TGF-β1) administration, which triggers growth arrest along with cell scattering and invasion (Kasai et al. 2005). To test the activity of the selected miRNAs, we transiently transfected A549 cells with the synthetic RNA duplexes of the mature forms of human miR-519d, miR-190, and miR-340; as a positive control, we used miR-93, a known inhibitor of TGFβ-induced cell cycle arrest (Petrocca et al. 2008b). An unrelated Caenorhabditis elegans miRNA (cel-miR-67) was used as a reference, while miR-507 and miR-557, which were not associated with TGFβ signaling by COOL analysis, were used as negative controls. TGFB1 addition to cells transfected with cel-miR-67, miR-507, and miR-557 resulted in the loss of intercellular adhesion and cell scattering, while transfection of miR-93, miR-519d, and miR-190 resulted in complete inhibition of TGFB1-induced cell scattering. Strikingly, miR-340 did not inhibit these effects of TGFB1, but rather triggered cell scattering, even in the absence of TGFB1 stimulation. These data were confirmed by quantitative analysis of cell scattering, which showed that miR-93, miR-519d, and miR-190 fully antagonized the effects of TGFB1, while miR-340 mimicked the actions of TGFB1 stimulation (Fig. 5). Analysis of cell proliferation revealed that all three of these tested miRNAs significantly affected cell growth. In particular, miR-519d and miR-190 significantly counteracted cell-growth inhibition mediated by TGFB1, similar to what was observed for the positive control, miR-93. In contrast, miR-340 strongly inhibited cell proliferation to a level that could not be further decreased by additional TGFB1 treatment (Fig. 5). In summary, miR-519d, miR-190, and miR-340 were associated with the TGFβ pathway by COOL analysis, and indeed modulated two major biological responses elicited by TGFβ activation, i.e., cell scattering and cell cycle arrest in lung tumor cells.
Figure 4.
miR-519d, miR-190, and miR-340 in the TGFβ signaling pathway. Schematic of the network of interactions between genes and proteins involved in TGFβ signaling. Putative targets of miR-519d (purple), miR-190 (orange), and miR-340 (yellow) are indicated. (Double vertical lines) The cell membrane; (dotted vertical line) the nuclear membrane; (dotted horizontal lines) links to other cellular pathways.
Figure 5.
miR-519d, miR-190, and miR-340 modulate TGFβ signaling. Analysis of cell proliferation, cell morphology, and cell scattering following miRNA transfection in A549 cells, with or without TGFB1 addition. All data and confocal microscope images are representative of at least three experiments for each miRNA. Cell proliferation is expressed in relative luminescence units (RLU), determined by luminescence-based cell viability assays. Confocal microscopy was performed after staining with FITC-phalloidin (green) and DAPI (blue). Cell scattering was quantified by digital image analysis of local cell density.
Co-expression analysis identifies communities of miRNAs associated with common functions
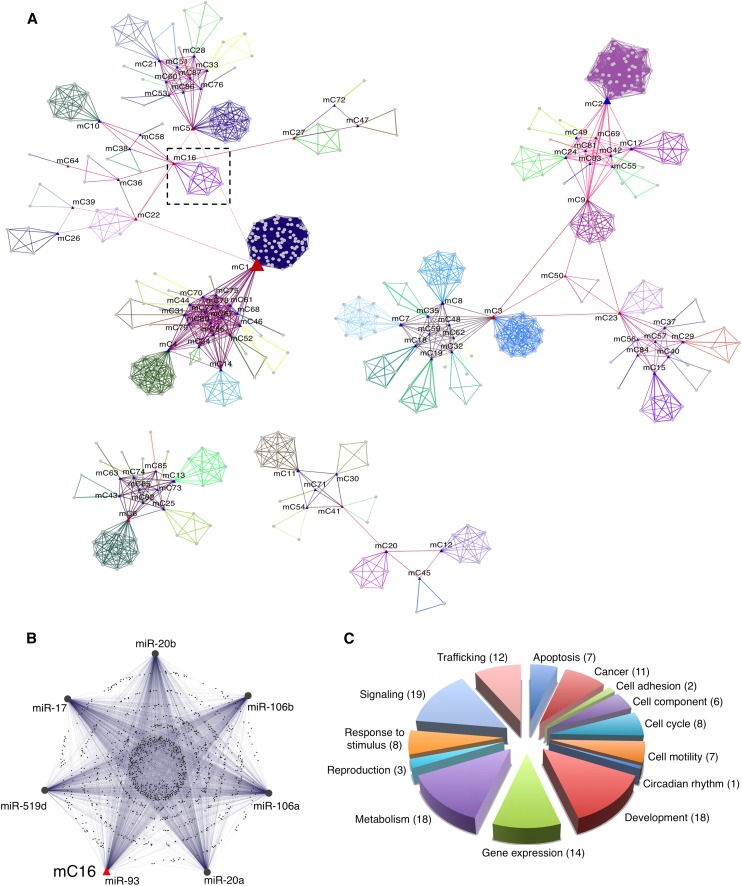
Recent work by Tsang et al. (2010) showed that the members of a same family of miRNAs tend to target common transcripts due to similarities among their seed sequences. miRNA “co-targeting” helped to define the putative function of miRNA families by investigating the functional categories enriched among their targets. To further develop this topic, we defined the concept of “miRNA communities” (miRCos) as groups of miRNA sharing a significant proportion of target genes as revealed by co-expression analysis. To this aim, we measured the proportion of genes shared by all possible pairwise combinations of COOL clusters. This procedure resulted in the identification of 87 miRNA communities (miRCo1-87) (Fig. 6A). Most of these miRCos are composed of only a few members: eight communities include >10 miRNAs (Supplemental Fig. 3), with two of them (miRCo1, miRCo2) containing more than 20 miRNAs. Remarkably, miR-519d and miR-93, which behaved similarly in our experimental analysis, mapped to the same community, miRCo16 (Fig. 6B). This community also includes miR-17, miR-20, and miR-106, which were previously described as being involved in the regulation of TGFβ signaling (Petrocca et al. 2008b).
Figure 6.
miRNA community networks. (A) Graphical representation of the community organization of human miRNA downstream transcriptional networks. (Gray circles) miRNAs. miRNAs that belong to the same community are linked with edges of the same color. For each community an exemplar (center of the miRNA community) was chosen and is indicated with a blue triangle. (Red triangle) miRNAs that link together different communities. (B) Enlarged section from A, showing the composition of miRCo16 and the mutual relationships of its miRNAs and shared target genes. (C) Frequency distribution of functions assigned to miRCOs as grouped in macrocategories. The number of miRCOs associated with each macrocategory is shown in parenthesis.
We inferred the biological functions associated with each miRCo by calculating their hypergeometric enrichment of GO and KEGG terms compared with the whole set of significant co-expression clusters (Supplemental Table 3). Emerging functional macrocategories included the same wide spectrum of biological functions as single miRNAs, and the relative proportions were also similar (Fig. 6C). Exceptions were categories associated with development and signaling, which were relatively more represented in miRCos (∼70% and ∼40% more frequent than in single miRNA analysis, respectively). This could be interpreted as a lower tendency of these categories to group into communities with shared targets, which copes with their regulatory role, i.e., differentiated by definition. Literature analysis confirmed the reliability of the functional categories associated with miRCos (Supplemental Table 4). miRNA communities are likely to be involved in the synergistic modulation of cohorts of genes that regulate similar processes, which adds a new layer of complexity to the regulatory functions of human miRNAs.
In summary, our results demonstrate that co-expression meta-analysis performed by using widespread, non-miRNA-specific microarray platforms is a powerful tool to define miRNAs' downstream gene networks, biological roles, and functional communities.
CoMeTa website
To enable researchers to retrieve associations between miRNAs, genes, and biological functions of interest, we have organized all of the information generated by CoMeTa into an interactive on-line database that is publicly available at http://cometa.tigem.it/site/index.php. The website includes the CoMeTa corank lists and additional targets for all of the human miRNAs, their associated pathways resulting from COOL analysis, and miRNA communities with their corresponding enriched functional categories. The CoMeTa website is searchable by miRNA, target gene, or biological function of interest, and represents a unique resource to gain insight into miRNA-controlled gene networks and functions.
Discussion
It was previously suggested that different miRNAs might contribute to the regulation of the same functions by cotargeting similar sets of genes (Tsang et al. 2010; Ulitsky et al. 2010; Sass et al. 2011; Su et al. 2011). Here, we have introduced co-expression-based gene network analysis as a means for inferring genes and functions associated with the transcriptional control of specific miRNAs. Network analyses were performed by elaborating the information associated with hundreds of different cellular and tissue conditions, an ensemble that is capable of capturing an impressive number of relationships between gene regulatory dynamics. Previous computational methods for the identification of miRNA targets have solely relied on sequence analysis of miRNA–mRNA target sites (Bartel 2009). More recently, a number of tools introduced the use of high-throughput expression analysis to improve predictions of miRNA targets (Huang et al. 2007; Ulitsky et al. 2010) and the identification of gene networks controlled by miRNAs (Friard et al. 2011; Huang et al. 2011; Jayaswal et al. 2011; Le Bechec et al. 2011; Liu et al. 2011; Xu et al. 2011). All of the above procedures are based on the comparison of paired data sets of miRNA and mRNA expression data generated from specific microarray platforms. CoMeTa is the first tool to integrate computational and expression analysis by relying exclusively on the extraordinary resource of mRNA transcriptome data sets available in public databases. Our tool does not require expression data from miRNA-specific probes and is effective in the recognition of miRNA targets, including those missed by sequence-based prediction tools (Fig. 1B; Supplemental Table 1). Recent observations reported that target mRNA abundance may dilute miRNA activity based on concentration/competition effects (Arvey et al. 2010), which could limit the inference of miRNA-target mRNA relationship based on the observation of expression variations. However, CoMeTa is based on the use of Pearson correlation scores among variations in mRNA expression, which depend on relative and not absolute variations. Therefore, CoMeTa's performance is unaffected by possible concentration/competition effects, as long as the effect of a miRNA on its target genes is measurable by microarray experiments.
Interestingly, while testing the CoMeTa procedure, we did not observe any significant differences in the behaviors of targets previously ascertained at the translational level (DS2) versus targets reportedly controlled at the transcript level (DS3). Since the CoMeTa procedure is based on the analysis of transcript levels, our results indicate that the effects of miRNAs on the expression levels of their mRNA targets is a widespread phenomenon that is not limited to a restricted subset of targets, thus strengthening the emerging view that miRNA-mediated regulation acts predominantly at the transcript level (Lim et al. 2005; Guo et al. 2010).
The central hypothesis of our study was that genes targeted by the same miRNA are co-expressed with each other under multiple conditions. We demonstrated this hypothesis by showing that miRNAs identify clusters of co-expressed genes, which were subsequently used to infer miRNA functions. We assigned specific biological roles to more than 500 human miRNAs that identified significant co-expression clusters. The high overlap with miRNA functions supported by published experiments (135 cases, see Supplemental Table 2) demonstrates that our procedure is both reliable and general, and endorses the predictions associated with miRNAs for which a biological role has not experimentally been established yet. However, one cannot exclude that in some instances a significant degree of co-expression between miRNA predicted targets may reflect the presence of alternative sources of transcriptional controls such as transcription factors. Therefore, to further demonstrate the causal relationship between gene co-expression clusters and miRNA functions, we decided to experimentally investigate three miRNAs (miR-519d/miR-190/miR-340), which our procedure associated with modulation of the TGFβ pathway. We indeed showed that miR-519d and miR-190 inhibit, whereas miR-340 mimics the effects of TGFB1 on cell proliferation, morphology, and scattering. Interestingly, miR-519d is part of a miRNA community with several miRNAs (miR-17/20a/106b/93) known to be involved in TGFβ regulation (Fig. 6B; Petrocca et al. 2008a), which supports the concept that these communities underlie a common regulatory function. While no information is available on the role of miR-190 in cell proliferation and tumorigenesis, a recent study showed that miR-340 has tumor-suppressive roles in the aggressive variants of breast cancer, in which miR-340 expression inversely correlates with tumor progression and metastasis (Wu et al. 2011). In summary, we have characterized miR-519d, miR-190, and miR-340 as novel regulators of the TGFβ pathway, thus providing potential therapeutic targets for the treatment of invasive tumors.
The analysis of other miRNA communities illustrated the multiplicity of functions associated with synergic miRNA control. We identified 87 such communities based on the overlap among miRNA gene networks: analysis of the available literature showed an impressive concordance with the functions associated by cluster enrichment analysis (Supplemental Table 4). This analysis revealed that all of the miRNAs examined (with only one exception) are associated in communities with other miRNAs, indicating that the sharing of downstream regulatory networks is a general tendency of human miRNAs. Our analysis also established connections between different miRNA communities, which resulted in a general assessment of the network of interactions of the entire human miRNome.
As previously stated, CoMeTa relies on the analysis of a vast data set of publicly available transcriptome data generated from hundreds of different cellular and tissue conditions, which ensures an appropriate coverage of the diverse biological roles controlled by miRNAs. This particular aspect further distinguishes CoMeTa from a number of previous efforts, which utilized gene expression analysis to infer putative miRNA functions as the latter focused on the evaluation of more restricted, and often tissue-specific, expression data sets (Ulitsky et al. 2010; Jayaswal et al. 2011; Liu et al. 2011; Su et al. 2011). While, on one hand, ensuring the identification of a broader variety of miRNA-controlled biological functions, the use of such a massive expression data set could lead, on the other hand, to a slightly less-efficient performance of CoMeTa in dissecting miRNA-controlled pathways that are specific for human tissues (e.g., the retina) that are, as yet, poorly represented in the expression data set used to seed the procedure. In those instances, it will be necessary to generate a more comprehensive starting data set of transcriptome data. However, it is expected that, thanks to the expected increase of high-resolution transcriptome data facilitated by the advances and cost reduction of next-generation sequencing approaches (Ozsolak and Milos 2011), the current gap in adequate transcriptome coverage of some human tissues will be filled, and therefore this possible caveat for the efficacy of the CoMeTa procedure will soon be overcome.
In summary, we have used gene network analyses to assign hundreds of functions to human miRNAs, of which only a small fraction had been previously reported. Our data indicate that miRNAs control an important portion of cellular metabolism and accurately describe known and novel functions of specific miRNAs and miRNA communities. The resulting biological hypotheses and novel functional associations, along with the development of an innovative research paradigm, represent valuable resources for future investigations aimed at dissecting out miRNA functions.
Methods
The CoMeTa procedure
The full set of human miRNAs was retrieved from miRBase (release 13.0) (Griffiths-Jones et al. 2008). For co-expression data analysis, 250 microarray data sets were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/), based on the Affymetrix HG-U133A GeneChip array (GPL96, Feb 19, 2002). Each data set was preprocessed and normalized independently, and the data matrices with less than three arrays, or more than 200 missing values, were removed. This resulted in a final list of 217 data sets. For each human miRNA, we collected the full list of predicted targets from miRanda (Betel et al. 2008) (September 2008 release), and the targets conserved in mammals from TargetScan (Friedman et al. 2009) (release 5.1; April 2009) and PicTar (Krek et al. 2005) (March 2007 release). All targets were pooled together in a single list of predicted targets. To evaluate co-expression, we first associated each gene with a “co-expression” list consisting of all other genes of the Affymetrix platform, ranked by their Pearson correlation score relative to the expression behavior in each single experiment. Then, for each gene on the list we generated a co-expression score that was set equal to its number of occurrences in the top third percentile of each ranked list, across all expression data sets analyzed (Gennarino et al. 2009; Sardiello et al. 2009; Palmieri et al. 2011). Thus, each pair of genes had a unique co-expression score as a result of this procedure. Subsequently, to generate the co-rank list associated to each single miRNA, we collected the co-expression lists of all pooled putative targets, averaged the co-expression scores of all ranked genes, and extracted only the data associated with the specific putative targets.
Additional miRNA target genes
Human 3′UTR sequences were retrieved from the Primate UTRef collection of the UTRdb (Grillo et al. 2010). The 3UTRef.Pri.dat flatfile database downloaded from http://utrdb.ba.itb.cnr.it contains 3′UTRs from the RefSeq database (http://www.ncbi.nlm.nih.gov/RefSeq/). To associate each human 3′UTR with its corresponding probe on the HG-U133A GeneChip, we used the Affymetrix annotation file HG-U133A.na30.annot.csv.zip, downloaded from the Affymetrix website (http://www.affymetrix.com). Collected human 3′UTRs were searched for canonical, 7–8-nt seed-matched sites (Bartel 2009). Custom Perl scripts were used to perform the analyses, making extensive use of the Bioperl toolkit.
COOL analysis
For each miRNA, the collected predicted targets were used to generate a co-expression matrix according to the co-expression scores obtained with the CoMeTa procedure. This matrix was then processed with MultiExperiment Viewer, to obtain the hierarchical clustering (Saeed et al. 2006). The first node was selected to obtain two clusters of predicted targets, for which two respective co-rank lists were calculated. We reasoned that co-expressed genes would generate a co-rank list with nonrandom enrichment of co-expressed entities in the top positions, and that their associated values would deviate from normality because of this enrichment. To test this hypothesis, we compared the distributions of co-expression values from each cluster with a randomly generated gene list of the same size using Quartile-Quartile (Q-Q) plot analysis (Chambers et al. 1983) against a hypothetical standardized normal distribution. To evaluate the deviation from the normality hypothesis, a normal probability plot was drawn for each COOL cluster (see Supplemental Fig. 2) and regression analysis was performed. The corresponding R2 value was used as the index for measuring the deviation of the co-rank list from normality: The lower the R2 value, the greater the deviation from normality. Using this procedure, we observed that the distribution of co-expression values associated with random lists were close to normality, with R2 values ranging from 0.91 to 0.98. However, R2 values associated with COOL clusters were distributed in a biphasic fashion, typically with only one of the two clusters for each miRNA associated with an R2 value lower than 0.91 (and therefore far from randomness). We only considered clusters with R2 ≤0.91 for the subsequent analyses. Overall, for the COOL analysis we started from a total number of 410 million possible pairs of miRNA target gene co-expression interactions. Following all of the above described filtering steps, only 30% of these interactions were left for further analysis.
Gene-set enrichment analysis
Gene-set enrichment analysis was performed as previously described (Gennarino et al. 2011). The cumulative distribution function was constructed by performing 1000 random gene-set membership assignments. A nominal P-value <0.01 and a false discovery rate (FDR) <0.25 were used to assess the significance of the enrichment scores. The microarray expression data used in this study have GEO accession numbers GSE12091 and GSE12092 (Gennarino et al. 2009).
miRCos procedure
To identify the communities of miRNAs, we used the Affinity Propagation clustering algorithm, APcluster (Frey and Dueck 2007), which groups items into communities of items. APcluster considers a similarity (or distance) between the items and iteratively groups them via a message passing paradigm, minimizing a scoring function until convergence is reached. APcluster does not require the number of communities to be specified by the user, which is the main advantage compared with other clustering algorithms. The algorithm generated a list of 87 communities and automatically assigned an “exemplar” node to each community. The network of miRNAs (Shannon et al. 2003). In the graphical representation, only the interactions among miRNAs from the same community were kept.
Gene Ontology (GO) analysis
GO analysis was performed using the “database for annotation, visualization, and integrated discover” (DAVID) web tool and default parameters (http://david.abcc.ncifcrf.gov/). We used Biological Process FAT (BP_FAT) and KEGG pathway analysis to infer enriched terms. Only BP_FAT categories with FDR ≤5 and KEGG pathways with FDR ≤20 were retained. For miRCos, enrichments were calculated by considering significant clusters from COOL analysis as the background.
Cell transfection assay
Non-small cell lung cancer A549 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 ng/mL) at 37°C in an atmosphere of 5% CO2. SiRNA transfection of these A549 cells was performed using Interferin (Polyplus transfection), according to the manufacturer's protocol. The cells were transfected with miRIDIAN Dharmacon microRNA Mimics at a final concentration of 20 nM.
Cell proliferation assays
Transfected cells were seeded in triplicate in opaque-walled 96-well plates (Corning). The medium was changed the following day and supplemented with 5 ng/mL of TGF-β1 (TGFB1) (Sigma-Aldrich) where indicated. Viable cells were counted using the CellTiter-Glo Luminescent Cell Viability Assay (Promega Corporation).
Immunofluorescence and cell-scattering analysis
Cells were transfected with miRNA Mimics and seeded on coverslips in 24-well plates (Corning). After 24 h, the medium was changed and supplemented with 5 ng/mL TGF-β1 (TGFB1) where required. After 72 h, the cells were fixed with 4% paraformaldehyde and stained with FITC-phalloidin (Sigma-Aldrich) and DAPI. Imaging was performed using a 10× objective on a Zeiss LSM710 confocal microscope. Local cell density was evaluated as the number of cell nuclei within a square area (10,000 px2 ≅ 18,000 μm2) centered on each cell nucleus detected in pictures using CellProfiler 2.0 software. An average of ∼1000 cells was analyzed for each condition.
Acknowledgments
We thank Graciana Diez-Roux, Alberto Luini, Huda Y. Zoghbi, Christian P. Schaaf, Adriano Flora, Ivone Bruno, and Justin Bottsford-Miller for critical reading of the manuscript. We are grateful to Giampiero Lago for the database website design. This work was supported by the Italian Telethon Foundation (grants TSBP41TELC and TSBSB2TELD to S.B.) and by AIRC (grant MFAG 10585 to G.D.A. and grant 10489 to P.V.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.130435.111.
References
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS 2010. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 6: 363 doi: 10.1038/msb.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C 2008. The microRNA.org resource: Targets and expression. Nucleic Acids Res 36: D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM 2007. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Chambers J, Cleveland W, Kleiner B, Tukey P 1983. Graphical methods for data analysis. Wadsworth International Group, Belmont, CA [Google Scholar]
- Chen Y, Gorski DH 2008. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 111: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen ST, Fuh CS, Juan HF, Huang HC 2011. Coregulation of transcription factors and microRNAs in human transcriptional regulatory network. BMC Bioinformatics 12: S41 doi: 10.1186/1471-2105-12-S1-S41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Hobert O 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 13: 849–851 [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D 2007. Clustering by passing messages between data points. Science 315: 972–976 [DOI] [PubMed] [Google Scholar]
- Friard O, Re A, Taverna D, De Bortoli M, Cora D 2011. CircuitsDB: A database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinformatics 11: 435 doi: 10.1186/1471-2105-11-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S 2009. MicroRNA target prediction by expression analysis of host genes. Genome Res 19: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Sardiello M, Mutarelli M, Dharmalingam G, Maselli V, Lago G, Banfi S 2011. HOCTAR database: A unique resource for microRNA target prediction. Gene 480: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. 2010. UTRdb and UTRsite (RELEASE 2010): A collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38: D75–D80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M 2009. MirZ: An integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res 37: W266–W272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD 2007. Using expression profiling data to identify human microRNA targets. Nat Methods 4: 1045–1049 [DOI] [PubMed] [Google Scholar]
- Huang GT, Athanassiou C, Benos PV 2011. mirConnX: Condition-specific mRNA-microRNA network integrator. Nucleic Acids Res 39: W416–W423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaswal V, Lutherborrow M, Ma DD, Yang YH 2011. Identification of microRNA-mRNA modules using microarray data. BMC Genomics 12: 138 doi: 10.1186/1471-2164-12-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human MicroRNA targets. PLoS Biol 2: e363 doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinas K, Kessler CB, Delany AM 2009. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J Cell Biochem 108: 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Allen JT, Mason RM, Kamimura T, Zhang Z 2005. TGF-β1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir Res 6: 56 doi: 10.1186/1465-9921-6-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kluiver J, Kroesen BJ, Poppema S, van den Berg A 2006. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia 20: 1931–1936 [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Le Bechec A, Portales-Casamar E, Vetter G, Moes M, Zindy PJ, Saumet A, Arenillas D, Theillet C, Wasserman WW, Lecellier CH, et al. 2011. MIR@NT@N: A framework integrating transcription factors, microRNAs and their targets to identify sub-network motifs in a meta-regulation network model. BMC Bioinformatics 12: 67 doi: 10.1186/1471-2105-12-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Liu B, Liu L, Tsykin A, Goodall GJ, Green JE, Zhu M, Kim CH, Li J 2011. Identifying functional miRNA-mRNA regulatory modules with correspondence latent dirichlet allocation. Bioinformatics 26: 3105–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. 2009. DIANA-microT web server: Elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Gennarino VA, Banfi S 2009. microRNAs and genetic diseases. Pathogenetics 2: 7 doi: 10.1186/1755-8417-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I 2006. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM 2011. RNA sequencing: Advances, challenges and opportunities. Nat Rev Genet 12: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20: 3852–3866 [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG 2009. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res 37: D155–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM 2008a. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor β signaling. Cancer Res 68: 8191–8194 [DOI] [PubMed] [Google Scholar]
- Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, et al. 2008b. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286 [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J 2006. TM4 microarray software suite. Methods Enzymol 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Saito T, Saetrom P 2010. MicroRNAs–targeting and target prediction. New Biotechnol 27: 243–249 [DOI] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. 2009. A gene network regulating lysosomal biogenesis and function. Science 325: 473–477 [DOI] [PubMed] [Google Scholar]
- Sass S, Dietmann S, Burk UC, Brabletz S, Lutter D, Kowarsch A, Mayer KF, Brabletz T, Ruepp A, Theis FJ, et al. 2011. MicroRNAs coordinately regulate protein complexes. BMC Syst Biol 5: 136 doi: 10.1186/1752-0509-5-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M 2007. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100: 416–424 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WL, Kleinhanz RR, Schadt EE 2011. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol 7: 490 doi: 10.1038/msb.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Lieberman J, Lal A 2010. Desperately seeking microRNA targets. Nat Struct Mol Biol 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Tsang JS, Ebert MS, van Oudenaarden A 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell 38: 140–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Laurent LC, Shamir R 2010. Towards computational prediction of microRNA function and activity. Nucleic Acids Res 38: E160 doi: 10.1093/nar/gkg570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Croce CM 2009. MiRNAs and cancer. Am J Pathol 174: 1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J, Zhao JJ, Mao SS, Zhang GH, Xu XC, Zhang N 2011. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer doi: 10.1002/cncr.25860 [DOI] [PubMed] [Google Scholar]
- Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT, Xu LD, Wang YY, Du L, Zhang YP, et al. 2011. MiRNA-miRNA synergistic network: Construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res 39: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]