Abstract
Aberrations of protein-coding genes are a focus of cancer genomics; however, the impact of oncogenes on expression of the ∼50% of transcripts without protein-coding potential, including long noncoding RNAs (lncRNAs), has been largely uncharacterized. Activating mutations in the BRAF oncogene are present in >70% of melanomas, 90% of which produce active mutant BRAFV600E protein. To define the impacts of oncogenic BRAF on the melanocyte transcriptome, massively parallel cDNA sequencing (RNA-seq) was performed on genetically matched normal human melanocytes with and without BRAFV600E expression. To enhance potential disease relevance by verifying expression of altered genes in BRAF-driven cancer tissue, parallel RNA-seq was also undertaken of two BRAFV600E-mutant human melanomas. BRAFV600E regulated expression of 1027 protein-coding transcripts and 39 annotated lncRNAs, as well as 70 unannotated, potentially novel, intergenic transcripts. These transcripts display both tissue-specific and multi-tissue expression profiles and harbor distinctive regulatory chromatin marks and transcription factor binding sites indicative of active transcription. Coding potential analysis of the 70 unannotated transcripts suggested that most may represent newly identified lncRNAs. BRAF-regulated lncRNA 1 (BANCR) was identified as a recurrently overexpressed, previously unannotated 693-bp transcript on chromosome 9 with a potential functional role in melanoma cell migration. BANCR knockdown reduced melanoma cell migration, and this could be rescued by the chemokine CXCL11. Combining RNA-seq of oncogene-expressing normal cells with RNA-seq of their corresponding human cancers may represent a useful approach to discover new oncogene-regulated RNA transcripts of potential clinical relevance in cancer.
The RAS/RAF/MAPK is pathway is hyperactive in ∼30% of human cancers, and activating mutations in key members of this pathway serve as driver mutations in many malignancies (Dankort et al. 2009; Dhomen et al. 2009; Michailidou et al. 2009). Activating mutations in the BRAF oncogene are present in >70% of melanomas, ∼90% of which are BRAFV600E (Davies et al. 2002; Rubinstein et al. 2010). The recent clinical use of selective BRAFV600E inhibitors demonstrates that impairing BRAFV600E signaling evokes tumor regression in patients with BRAF-mutant metastatic melanoma (Bollag et al. 2010; Flaherty et al. 2010). However, patients eventually relapse and succumb to acquired chemoresistance via mechanisms that reactivate MAPK signaling (Johannessen et al. 2010; Nazarian et al. 2010; Villanueva et al. 2010; Wagle et al. 2011). It is therefore clear that a greater understanding of oncogenic BRAF-driven impacts is required to yield alternative and complementary therapeutic targets. Recent DNA sequencing efforts have focused on protein coding exons to provide further insight into genomic mutations in melanoma (Berger et al. 2010; Johannessen et al. 2010; Pleasance et al. 2010; Prickett et al. 2011; Wei et al. 2011); however, areas of the genome that do not encode functional proteins are largely unexplored.
Large-scale transcriptome analyses indicate that ∼50% of transcribed RNAs have no protein-coding potential (Claverie 2005; Kapranov et al. 2007; Mercer et al. 2009); however, the functional importance of untranslated RNA in cancer is not fully understood. Among these transcripts, long noncoding RNAs (lncRNAs) that vary in length from 200 bp to tens of kilobases are a particularly important class of noncoding RNA (ncRNA) that have recently been shown to regulate diverse functions, including X-chromosome silencing (Borsani et al. 1991; Lee et al. 1999), pluripotency (Loewer et al. 2010), epigenetic gene regulation (Rinn et al. 2007; Tsai et al. 2010), TP53 (also known as p53) functions (Huarte et al. 2010), and the DNA damage response (Hung et al. 2011). The regulation and function of lncRNAs in cancer is largely uncharacterized, and whether expression of lncRNAs is regulated by oncogenic BRAFV600E is unknown.
We performed RNA-seq on both normal melanocytes (± ectopic BRAFV600E) and BRAF-mutant human melanomas to define the impact of oncogenic BRAF expression on the melanocyte transcriptome. BRAFV600E regulated expression of 1027 protein-coding transcripts, 39 annotated lncRNAs, and 70 previously unannotated transcripts. These transcripts display both tissue-specific and multi-tissue expression profiles and harbor distinctive regulatory chromatin marks indicative of active transcription. We validated expression of a subset of these transcripts and also confirmed similar expression patterns in another publicly available melanoma RNA-seq data set. One of the novel transcripts most highly induced by oncogenic BRAF, which is also recurrently overexpressed in melanoma, is a putative lncRNA we named BRAF-activated non-coding RNA (BANCR). BANCR regulates a set of genes involved in cell migration, including the chemokine CXCL11, and BANCR is required for full migratory capacity of melanoma cells. CXCL11 rescued defective migration in BANCR-knockdown cells, suggesting that CXCL11 is an important target of BANCR involved in cell migration. These data present a high-resolution profile of an oncogene-remodeled transcriptome and identify oncogene-regulated lncRNAs and novel RNA transcripts of potential interest for future study in human cancer.
Results
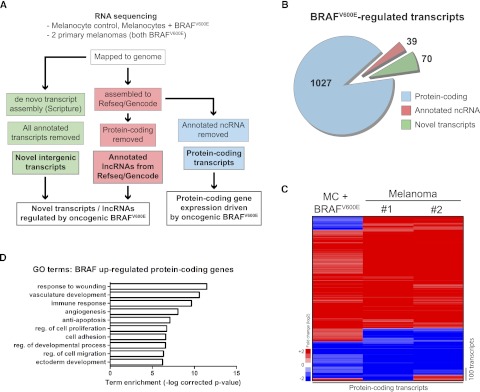
To assess transcriptome-wide changes induced by BRAFV600E, we performed RNA-seq on genetically matched primary human melanocytes after transduction with a lentivirus expressing either mutant oncogenic BRAFV600E or red fluorescent protein (RFP) marker control. As an additional filter for potential clinical relevance, RNA-seq was also performed on two primary human melanomas expressing BRAFV600E (Fig. 1; Supplemental Fig. 1). Transcript assembly of mapped reads was performed using two methods (Fig. 1A): assembly to a reference transcriptome (RefSeq/Gencode) using Cufflinks (Trapnell et al. 2010), or unbiased, de novo assembly using Scripture (Guttman et al. 2010). Assembly to RefSeq yielded expression data for annotated protein-coding transcripts and RefSeq-annotated ncRNAs. Sequencing reads were also mapped to the Gencode reference since it contains additional, curated ncRNA annotations that are absent in RefSeq. Additionally, all annotated transcripts were deducted from Scripture-assembled transcripts to yield a population of novel de novo–assembled transcripts (Fig. 1A). This approach permitted analysis of BRAF-dependent regulation of protein-coding transcripts, annotated lncRNAs, and novel transcripts. BRAFV600E concordantly altered expression twofold or more of 1027 protein-coding transcripts, 39 annotated lncRNAs, and 70 novel transcripts (Fig. 1B). Unsupervised hierarchical clustering in both melanocytes expressing BRAFV600E as well as BRAFV600E-positive human melanoma tissue demonstrated concordant expression changes for the majority of protein coding transcripts (Fig. 1C; Supplemental Table 1). The top 10 unique gene ontology (GO) terms associated with BRAF up-regulated protein-coding transcripts agree with GO terms previously associated with BRAF-mutant melanoma (Fig. 1D; Kannengiesser et al. 2008; Packer et al. 2009), supporting the potential clinical relevance of the RNA-seq data obtained.
Figure 1.
Oncogenic BRAFV600E remodels the transcriptome in melanoma. (A) Schematic of experimental workflow and RNA-seq data analysis. (B) Pie chart showing number and categories of transcripts regulated by BRAFV600E. Numbers of transcripts changed in all three samples in the same direction ± twofold versus pooled normal melanocyte control. (C) Heatmap showing hierarchical clustering of protein-coding transcript expression in primary melanocytes overexpressing BRAFV600E and in primary melanomas (± twofold, changed in all three samples). (D) Top 10 unique gene ontology (GO) terms associated with protein-coding transcripts up-regulated by BRAFV600E.
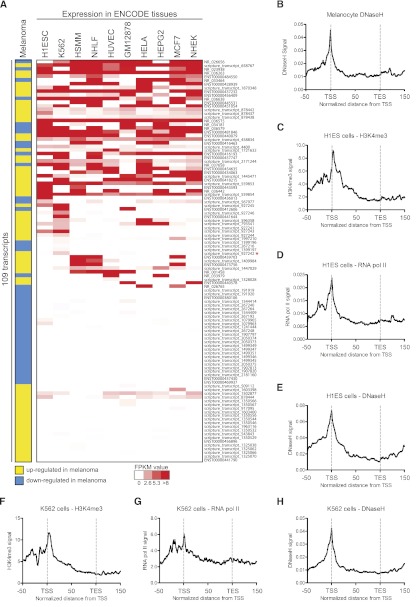
In normal human melanocytes, BRAFV600E significantly altered expression of 39 annotated lncRNAs and 70 novel transcripts that also exhibited twofold or more altered expression in BRAFV600E-expressing primary melanomas (Fig. 1B; Supplemental Tables 2, 3). We validated regulation by BRAF for 12/12 annotated transcripts tested and 13/13 novel transcripts tested (Supplemental Tables 4, 5, respectively). We then assessed expression levels of these 109 transcripts in multiple tissues using publicly available ENCODE RNA-seq data and found that transcripts displayed both tissue-specific and multi-tissue expression profiles (Fig. 2A; Supplemental Table 6). DNase I hypersensitive sites (DNaseH) are indicative of open chromatin and are commonly enriched at transcription start sites (TSSs) and enhancers. We interrogated ENCODE data for DNaseH sites in primary human melanocytes and found enrichment at the TSSs of these transcripts (Fig. 2B). In addition, we also looked for chromatin modifications in other ENCODE cell types where these transcripts are most abundant since no other ENCODE data exist for primary melanocytes. TSSs were enriched for H3K4me3, RNA pol II binding, and DNaseH in H1ES cells (Fig. 2C–E) and in K562 leukemia cells (Fig. 2F–H). Loading of RNA pol II and the presence of chromatin modifications associated with active transcription further validate existence and expression of these annotated lncRNAs and novel transcripts in melanocytes and other tissues. In addition, we also analyzed ChIP-seq data for 77 transcription factors (ENCODE) and found that most transcripts (79/109) bound at least one transcription factor in its promoter region, and positive binding was more highly represented in transcripts with broad tissue expression (Supplemental Fig. 2; Supplemental Table 7). A representative example of an annotated lncRNA regulated by BRAF in melanoma, expressed in other tissues that harbors transcription-associated chromatin marks is shown in Supplemental Figure 3.
Figure 2.
Multi-tissue expression profile of annotated lncRNAs and novel transcripts. (A) Publicly available RNA-seq data (ENCODE project) for multiple cell types were downloaded from the UCSC Genome Browser. The melanoma column expresses an aggregate from our RNA-seq data indicating if each transcript was concordantly up-regulated (yellow) or down-regulated (blue) by BRAF in melanoma. Red/white scaling indicates expression level in other cell types. (B) Melanocyte DNaseH (ENCODE) distribution over the 109 BRAF-regulated transcripts is shown. The following charts (C–H) depict signal distribution (ENCODE) over the 109 BRAF-regulated transcripts. (C) Distribution of H3K4me3 in H1ES cells. (D) Distribution of RNA pol II in H1ES cells. (E) Distribution of DNaseH in H1ES cells. (F) Distribution of H3K4me3 in K562 cells. (G) Distribution of RNA pol II in K562 cells. (H) Distribution of DNaseH in K562 cells. (TSS) Transcription start site; (TES) transcription end site.
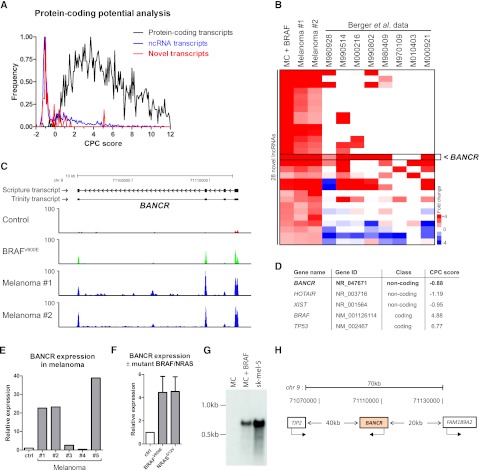
Next, we focused on exploring putative transcripts discovered by unbiased de novo assembly of RNA-seq data using previously validated techniques (Guttman et al. 2010; Trapnell et al. 2010). By use of this approach, 70 unannotated transcripts were identified as concordantly changed twofold or more in both melanocytes overexpressing BRAFV600E and in primary melanoma tissue samples (Fig. 1B; Supplemental Table 3). To assess the protein-coding potential of these transcripts, we used a coding potential calculator (CPC) algorithm previously shown to discriminate coding from noncoding transcripts with high accuracy (Kong et al. 2007). Positive CPC scores indicate positive protein-coding potential, whereas negative CPC scores indicate low coding-potential. In agreement with this, of 1500 randomly chosen annotated ncRNA transcripts analyzed, most yielded negative CPC scores, whereas 1500 randomly selected protein-coding genes yielded positive CPC scores (Fig. 3A). Newly discovered BRAF-regulated transcripts returned a similar CPC profile to the control ncRNA population, suggesting that many of these transcripts may be lncRNAs (Fig. 3A). Next, to search for biologically important, highly expressed, BRAF-regulated lncRNAs, we assessed expression levels of up-regulated novel transcripts with negative CPC scores, in a publicly available melanoma RNA-seq data set published recently (Berger et al. 2010). This study did not search for lncRNAs but instead focused on identifying novel gene fusions and other events driven by underlying aberrations in genomic DNA. Re-mapping the RNA-seq data from Berger et al. (2010; including intergenic regions) allowed us to derive expression values (using Cuffdiff) for the novel lncRNAs identified in our study. Clustering of these data revealed recurrent, overexpression of a novel lncRNA that we named BANCR (Fig. 3B–H; Supplemental Fig. 4).
Figure 3.
Oncogenic BRAFV600E regulates expression of many novel transcripts. (A) Frequency distribution plot showing coding potential analysis (CPC scores) of novel transcripts discovered by de novo assembly. One thousand five hundred random annotated ncRNAs and 1500 random protein-coding genes were also analyzed for reference. (B) Analysis of BRAF up-regulated novel lncRNAs in a publicly available melanoma RNA-seq data set. We remapped and reanalyzed Berger et al. (2010) data and derived expression values for novel transcripts up-regulated in all three of our samples. Robustly up-regulated novel transcripts (with negative CPC scores) were clustered with Berger et al. (2010) data revealing recurrently highly expressed lncRNAs. (C) Histograms of raw RNA-seq data in control sample, in melanocytes overexpressing BRAFV600E and in both primary melanomas (Mel. 1, Mel. 2). Scripture and Trinity assembly for BANCR is also shown. y-axis is number of RNA-seq reads normalized for mapping variation. (D) CPC score for BANCR. Examples of lncRNAs HOTAIR and XIST and protein-coding genes BRAF and TP53 are shown for reference. (E) BANCR expression in primary melanomas validated by qRT-PCR. (F) BANCR expression in melanocytes overexpressing BRAFV600E or NRASG12V measured by qRT-PCR. Data are means from three experiments ± SD. (G) BANCR Northern blot using RNA from control melanocytes (MCs), melanocytes overexpressing BRAFV600E, or sk-mel-5 melanoma cells. (H) Schematic of BANCR locus.
RNA-seq data histograms and de novo assembly using Scripture indicated that BANCR is a four-exon transcript of 693 bp that is highly induced by BRAFV600E in melanocytes and is overexpressed in primary tumors (Fig. 3C). We also detected the BANCR transcript using the Trinity algorithm (Grabherr et al. 2011), which does not rely on mapping of RNA-seq reads to a reference genome sequence. This complementary approach provided orthogonal validation of BANCR transcript architecture, and in further support of this, there are spliced ESTs that overlap with our BANCR gene prediction (e.g., EST W58600). De novo assembled transcripts also align faithfully with RNA-seq reads in the independent melanoma data set of Berger et al. (2010; Supplemental Fig. 4B). The CPC score for BANCR is similar to that derived for the well-characterized lncRNAs HOTAIR (Rinn et al. 2007) and XIST (Borsani et al. 1991), whereas control protein-coding transcripts such as BRAF and TP53 yield positive CPC scores (Fig. 3D). BANCR expression levels were increased in 4/5 additional BRAFV600E melanoma samples, to levels as high as 20- to 40-fold (Fig. 3E). We also validated up-regulation of BANCR by both BRAFV600E and mutant active NRAS in melanocytes by qPCR (Fig. 3F). Northern blot analysis revealed up-regulation of a single band corresponding to BANCR RNA in melanocytes expressing BRAFV600E and in the BRAF-mutant melanoma cell line sk-mel-5 (Fig. 3G). BANCR is transcribed at a locus on chromosome 9 and lies in a gene desert 40 kb downstream from TJP2 and 20 kb upstream of FAM189A2 (Fig. 3H). Expression of the most proximal genes upstream of and downstream from BANCR was not significantly changed (Supplemental Table 8), suggesting that BANCR is not a cis-regulator of neighboring genes as has been described for a subset of lncRNAs (Pasmant et al. 2007; Yap et al. 2010). By exploring multi-tissue RNA-seq data from ENCODE, we found that BANCR expression is largely melanocyte/melanoma-specific (Fig. 2A, marked with an asterisk), but we did detect low level expression in K562 leukemia cells and H1ES cells (Fig. 2A; Supplemental Fig. 5). In addition, the BANCR locus is enriched for chromatin marks associated with active transcription, and interestingly, the BANCR promoter was shown to be enriched for STAT1 transcription factor binding in HeLa-s3 cells (ENCODE ChIP-seq data) (Supplemental Fig. 5).
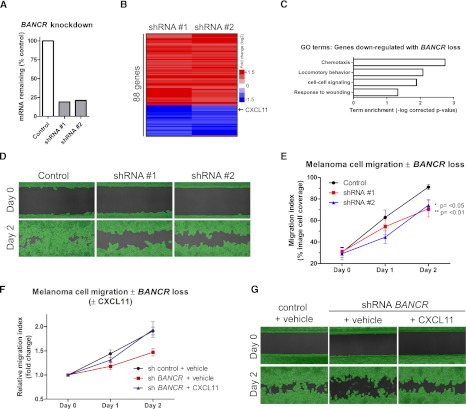
To explore if BANCR may be relevant to BRAFV600E-driven oncogenic impacts, we undertook shRNA-mediated BANCR depletion in melanoma cells. First, gene expression profiling was performed using complementary DNA (cDNA) microarrays on Colo829 BRAFV600E melanoma cells in which BANCR expression had been reduced to <25% using shRNAs (Fig. 4A). Eighty-eight genes changed significantly following BANCR knockdown using independent BANCR shRNA sequences (Fig. 4B). Genes repressed by BANCR depletion were enriched for GO terms, including locomotory behavior and chemotaxis (Fig. 4C), suggesting a potential role for BANCR in melanoma cell motility. Consistent with this, BANCR depletion impaired migration of melanoma cells without affecting viability and proliferation (Fig. 4D,E; Supplemental Fig. 6A), indicating that BANCR is at least partially necessary for melanoma cell migration in vitro. We next tested if factors down-regulated by BANCR loss were able to rescue defective cell migration. We found that the chemokine CXCL11, which is down-regulated by BANCR knockdown (Supplemental Fig. 6B), was able to rescue the migration defect without significantly altering baseline migration (Fig. 4F,G; Supplemental Fig. 6C). These data indicate that BANCR is a novel BRAFV600E-induced lncRNA that regulates melanoma cell migration in vitro in part by regulating expression of CXCL11.
Figure 4.
BANCR loss impairs melanoma cell migration. (A) Quantification of BANCR knockdown by qPCR using two independent shRNAs. (B) Microarray analysis of Colo829 melanoma cells following shRNA knockdown of BANCR. (C) Significant gene ontology (GO) terms associated with genes repressed by BANCR loss. (D) Representative images showing amount of cell migration at day 0 and day 2. Melanoma cells were infected with lentivirus expressing either nontargeting control shRNA (control) or one of two duplicate independent shRNA sequences targeting BANCR (shRNA 1, shRNA 2). Green highlight, shown for clarity, is a mask applied by the analysis software for accurate, unbiased migration quantification. (E) Quantification of replicate images (n = 6 per timepoint) taken over migration assay timecourse. Values are means from two independent experiments ± SD. Statistical analysis performed by one-way ANOVA, **P ≤ 0.01, *P ≤ 0.05 versus control. (E) Quantification of replicate images (n = 3 per timepoint) taken over migration assay timecourse ± CXCL11. Cells were infected with shRNA control lentivirus and treated with vehicle (PBS), infected with BANCR shRNA, and treated with vehicle or infected with BANCR shRNA and treated with CXCL11 (10 ng/mL). (G) Representative images showing amount of cell migration at day 0 and day 2.
Discussion
In this study, we performed RNA-seq of BRAFV600E-expressing primary human melanocytes to characterize how this oncogene remodels the melanocyte transcriptome. To enhance cancer relevance, these data were filtered through RNA-seq data from duplicate BRAFV600E mutant human melanomas to characterize BRAFV600E-regulated RNA transcripts that are also recurrently altered in melanoma. By use of this approach, we identified 39 annotated lncRNAs regulated by BRAFV600E that are also concordantly expressed in melanoma. By using de novo transcript assembly, we identified 70 additional un-annotated BRAFV600E-regulated transcripts that were also concordantly expressed in melanoma. By comparing these data to a reanalyzed, publicly available melanoma RNA-seq data set, we identified a list of priority BRAF target transcripts that are recurrently altered in cancer. From these transcripts, the putative lncRNA, BANCR, was identified as a potential regulator of melanoma cell migration.
Large-scale transcriptome analyses indicate that ∼50% of transcribed RNA has no protein-coding potential (Claverie 2005; Kapranov et al. 2007; Mercer et al. 2009). How this “dark matter” of the genome is altered in cancer, and the functional importance of such alterations, is only beginning to be explored. Whereas ncRNA species such as microRNAs are established as being important in cancer, more recently described lncRNAs are less well studied in this context. Altered expression of lncRNAs has been reported in several cancers: For example, HOTAIR is up-regulated in primary and metastatic breast tumors, and high levels of HOTAIR were associated with metastasis and poor survival (Gupta et al. 2010); lincRNAp21 is regulated by TP53 and mediates the TP53 response (Huarte et al. 2010); SPRY4-IT1 is overexpressed in melanoma (Khaitan et al. 2011); the lncRNA PANDA is aberrantly expressed in breast cancer and is implicated in regulating the DNA damage response (Hung et al. 2011); MALAT-1 is associated with metastasis in non-small-cell lung cancer (Ji et al. 2003); and ANRIL is involved in epigenetic silencing of p15 in leukemia (Yu et al. 2008). Whether altered lncRNA expression represents noise resulting from tumor heterogeneity and genomic instability or whether this is driven by oncogenic driver events has not been determined systematically. Employing RNA-seq and using BRAFV600E melanoma as a system to interrogate oncogene-regulated lncRNAs allowed visualization as to how an oncogene transforms the transcriptional landscape in normal cells. It also facilitated the identification of several novel transcripts and lncRNAs that are frequently dysregulated in melanoma, many of which are also expressed in other tissues.
Recent RNA sequencing of eight short-term melanoma cultures originating from stage 4 tumors and two melanoma cell lines identified 11 novel melanoma gene fusions and 12 novel read-through transcripts (Berger et al. 2010). This insightful study, however, did not seek to discover novel RNA transcripts, which may have been technically challenging due to the considerably lower sequencing depth compared with that of our present study. Berger et al. (2010) obtained an average of 14.2 million total reads and 8.4 million mapped reads per sample, whereas our present study achieved an average of 151 million total reads and 132 million mapped reads per sample representing 10×–15× increased coverage (Supplemental Table 9). We subsequently remapped and reassembled Berger et al. (2010) data and included intergenic regions and our de novo assembled novel transcripts, which increased mapped reads from 8.4 million to around 20 million (Supplemental Table 10). This allowed interrogation of the expression of the newly discovered novel transcripts described here and revealed that BANCR is recurrently overexpressed in melanoma. Since BANCR is overexpressed in both our primary melanomas and metastatic samples from Berger et al. (2010), BANCR may play a potential role throughout the spectrum of disease progression. Consistent with this, knockdown of BANCR in metastatic melanoma cell lines inhibited melanoma cell migration (Fig. 4).
Our findings that CXCL11 was down-regulated by BANCR loss in migration-deficient melanoma cells and that migration could be rescued by reintroduction of recombinant CXCL11 are consistent with previous findings. For example, it has been shown that the anti-tumor drugs dimethylfumarate (DMF) and dacarbazine (DTIC) reduced migration and CXCL11 expression in melanoma cells in vitro (Valero et al. 2010). In addition, melanoma cells lacking the CXCL11 receptor CXCR3 exhibited reduced migration in a murine model of melanoma metastasis (Kawada et al. 2004). Our data further implicate CXCL11 as being an important mediator of melanoma cell migration and identify BANCR as an important regulator.
In conclusion, we have used RNA-seq to identify annotated and novel transcripts regulated by oncogenic BRAFV600E. Interrogation of novel lncRNAs up-regulated by BRAFV600E that were also recurrently up-regulated in melanoma identified BANCR, a novel lncRNA that regulates melanoma cell migration. The combination of RNA-seq of normal cells expressing relevant oncogenes with RNA-seq of the corresponding oncogene-expressing human cancers may provide a useful general approach to discover new oncogene-regulated RNA transcripts of possible clinical relevance to human cancer.
Methods
Cell culture
Primary human melanocytes were isolated from fresh, discarded surgical foreskin specimens obtained from Lucile Packard Children's Hospital at Stanford through and Institutional Review Board–approved protocol. Isolation performed as previously described (Chudnovsky et al. 2005). Primary melanocytes were cultured in melanocyte medium 254 (Invitrogen). 293T and sk-mel-5 cells were cultured in DMEM + 10% FCS (Invitrogen). All cells were cultured in a 37°C, 5% CO2 humidified incubator. Colo-829 cells were cultured as above using RPMI + 10% FCS (Invitrogen).
Lentiviral infections and shRNA vector construction
BRAFV600E and NRASG12V were cloned into the lentiviral vector pRRL-sin-cPPT-hPGK-WPRE. Helper packaging plasmids used were pucMDG, and pCMVΔ8.91. 293T cells were transfected overnight with virus production vectors using calcium phosphate transfection, and the cell medium was replaced with DMEM +10% FCS the following day. Transfected cells were transferred to a 32°C, 5% CO2 incubator overnight for virus production. Melanocytes were infected with diluted high titer lentivirus by centrifugation in 10-cm culture plates at 32°C in the presence of 5 μg/mL polybrene (Sigma). As a control, a population of normal melanocytes pooled from three donors was infected with virus transmitting RFP. Custom-made shRNAs were cloned into the lentiviral vector pGIPZ (Openbiosystems) and were designed using RNAi Central shRNA des. tool (http://cancan.cshl.edu/RNAi_central/RNAi.cgi?type=shRNA). Cells were infected twice, 24 h apart as above to achieve best knockdown. All shRNA sequences are shown in Supplemental Table 2.
Western blotting
Cells were lysed in cell lysis buffer (no. 9803, Cell Signaling); protein concentration was determined using Bio-Rad protein assay dye (no. 500-0006, BioRad); and polyacrylaminde gel electrophoresis was performed using NuPAGE 10% Bis-Tris gels (Invitrogen). Proteins were transferred to a Amersham Hybond-P PVDF membrane (GE Healthcare). Antibodies used were as follows: BRAF (sc-5284, Santa Cruz Biotechnology), pERK (no. 4377, Cell Signaling), total ERK (no. 9102, Cell Signaling), beta-actin (A2668, Sigma), anti-Mouse HRP (NA931), and anti-Rabbit HRP (NA934) secondary antibodies (GE healthcare). Membranes were developed using ECL reagent (GE Healthcare).
Northern blotting
RNA was separated by RNA gel electrophoreses and transferred to Hybond-N+ nylon membrane (Amersham) overnight using 20× SSC buffer. The membrane was cross-linked using a UV Stratalinker. The Northern probe was made by PCR-cloning BANCR cDNA followed by labeling with α-32P-labeled dCTP (Perkin Elmer). Northern membrane and probe were incubated overnight at 60°C in a rotating incubator. The membrane was then washed with 2× SSC buffer +0.1% SDS and exposed overnight at −80°C for visualization.
RNA extraction, cDNA library construction, and cDNA sequencing
Total RNA was isolated with an RNeasy mini kit according to the manufacturer's instructions (Qiagen). cDNA sequencing libraries were prepared using the Illumina mRNA sequencing prep kit (RS-100-0801), which selects for poly-A containing mRNAs. Two micrograms of total RNA from each sample was used for library preparation. High-throughput, 101-bp paired-end cDNA sequencing was performed by Elimbio.
RNA-seq data analysis
Sequencing reads were mapped to NCBI build 36.1/hg18 using “TopHat,” and mapped sequences were aligned to RefSeq and Gencode transcriptomes using “Cufflinks” (Trapnell et al. 2010). Transcripts were also assembled using “Scripture,” which performs unbiased de novo transcript assembly without using a reference transcriptome (Guttman et al. 2010). For fold change calculations, RNA-seq data were compared with data from a population of normal melanocytes pooled from three donors, infected with lentivirus transmitting RFP. Transcript coding potential calculation was performed using “CPC calculator” (Kong et al. 2007). GO terms associated with significantly changed protein-coding genes were calculated using DAVID (Huang da et al. 2009).
ENCODE data analysis
For multi-tissue expression analysis, paired-end RNA-seq data for H1ESC, K562, HSMM, NHLF, HUVEC, GM12878, HELA, HEPG2, MCF7, and NHEK cells (all ENCODE project) were downloaded from the UCSC Genome Browser. Expression was quantified with Cufflinks using a reference transcriptome composed of RefSeq, GENCODE, and predicted Scripture-derived transcripts. FPKM values were hierarchically clustered and visualized using Java Treeview.
Transcription factor binding
Binding sites for 77 transcription factors were obtained from the UCSC table browser (ENCODE transcription factor ChIP-seq peak files). Transcript promoter regions were defined as 3 kb upstream of to 1 kb downstream from the TSS. With this, we generated a regulatory matrix of transcripts as rows and 77 transcription factors as columns. A 0 indicates no binding; 1, positive binding detected.
ChIP-seq data for K562 and H1ES cells from the ENCODE project (Birney et al. 2007) for H3K4me3 and RNA pol II (significant called ChIP-seq peaks) were downloaded from the UCSC Genome Browser. We also downloaded data for DNaseI hypersensitive regions for primary human melanocytes, K562, and H1ES cells. Wiggle files were converted to bedGraph format, and the average signal was calculated beginning upstream of the TSS by a distance of 50% of the gene body (TSS to transcription stop site) and continuing downstream from the transcript by the same 50% distance. These data were sorted into 100 bins normalized to the transcript length.
Reverse transcription real-time qPCR
RNA was reverse transcribed using the iScript Reverse transcription kit (BioRad). qPCR was performed using the Maxima SYBR Green/ROX qPCR master mix (Thermo Scientific). Primers for qPCR were designed using “OligoPerfect” primer design software (Invitrogen), and the primer specificity and efficiency/dynamic range was confirmed to be linear over four orders of magnitude during optimization prior to use in experiments. All qPCR primers used are listed in Supplemental Table 1.
Microarray analysis
Microarrays were performed using GeneChip Human Genome U133 Plus 2.0 array chips (Affymetrix) on RNA extracted from Colo829 melanoma cells ± BANCR knockdown. cDNA synthesis, labeling, and array procedures were conducted at the Stanford PAN facility.
Melanoma RNA
RNA from primary human melanomas was purchased from Asterand. H and E histological images of melanoma samples and tumor grading information was also provided by Asterand.
BRAF and NRAS genotyping
The BRAFV600 locus and NRASQ61 locus were amplified and purified from melanoma cDNA. BRAF locus primers were as follows: forward, agcttgtatcaccatctccatatca; reverse, accataaaaattatctggtccctgt. NRAS locus primers were as follows: forward, ctgagtacaaactggtggtggtt; reverse, ggcttgttttgtatcaactgtcc. Products (BRAF 401 bp, NRAS 386 bp) were resolved by gel electrophoresis, purified, and sequenced by Sanger sequencing.
Migration assays
A modified “scratch” migration assay that does not involve variable, physical wounding of cell cultures was used to assess cell migration (Ibidi, see http://www.ibidi.de). Cells were seeded at 40,000 cells per chamber and allowed to attach overnight. The following day, culture inserts were removed and light microscopy images acquired (three per condition). Cells were cultured during the migration assay under normal conditions, and images were subsequently acquired 24 h and 48 h later. Images were analyzed using automated image analysis software (http://ibidi.wimasis.com/). For CXCL11 experiments, CXCL11 (R&D Systems) diluted in PBS was applied at 10 ng/mL.
Data access
All RNA-seq data and cDNA microarray data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE33092.
Acknowledgments
This work was supported by the United States Veterans Affairs Office of Research and Development and by NIH/NIAMS grant AR49737 to P.A.K. We thank Leszek Lisowski, PhD, for technical advice with lentiviral infections; Shiying Tao for primary melanocyte isolations; and Todd Ridky, MD, PhD, for cloning assistance.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.140061.112.
References
- Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, Johnson LA, Robinson J, Verhaak RG, Sougnez C, et al. 2010. Integrative analysis of the melanoma transcriptome. Genome Res 20: 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. 2010. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467: 596–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329 [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA 2005. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet 37: 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie JM 2005. Fewer genes, more noncoding RNA. Science 309: 1529–1530 [DOI] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, You MJ, DePinho RA, McMahon M, Bosenberg M 2009. BrafV600E cooperates with Pten loss to induce metastatic melanoma. Nat Genet 41: 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954 [DOI] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R 2009. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15: 294–303 [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. 2011. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. 2010. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468: 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannengiesser C, Spatz A, Michiels S, Eychene A, Dessen P, Lazar V, Winnepenninckx V, Lesueur F, Druillennec S, Robert C, et al. 2008. Gene expression signature associated with BRAF mutations in human primary cutaneous melanomas. Mol Oncol 1: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T, Inaba K, Minato N, Oshima M, Taketo MM 2004. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res 64: 4010–4017 [DOI] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ 2011. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 71: 3852–3862 [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G 2007. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. 2010. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Michailidou C, Jones M, Walker P, Kamarashev J, Kelly A, Hurlstone AF 2009. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. Dis Model Mech 2: 399–411 [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer LM, East P, Reis-Filho JS, Marais R 2009. Identification of direct transcriptional targets of (V600E)BRAF/MEK signalling in melanoma. Pigment Cell Melanoma Res 22: 785–798 [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I 2007. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67: 3963–3969 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V, Gartner J, Jiang J, Cherukuri PF, Molinolo A, et al. 2011. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet 43: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein JC, Sznol M, Pavlick AC, Ariyan S, Cheng E, Bacchiocchi A, Kluger HM, Narayan D, Halaban R 2010. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med 8: 67 doi: 10.1186/1479-5876-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero T, Steele S, Neumuller K, Bracher A, Niederleithner H, Pehamberger H, Petzelbauer P, Loewe R 2010. Combination of dacarbazine and dimethylfumarate efficiently reduces melanoma lymph node metastasis. J Invest Dermatol 130: 1087–1094 [DOI] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. 2010. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18: 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 29: 3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, et al. 2011. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 43: 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H 2008. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]