Abstract
Chimpanzee and gorilla chromosomes differ from human chromosomes by the presence of large blocks of subterminal heterochromatin thought to be composed primarily of arrays of tandem satellite sequence. We explore their sequence composition and organization and show a complex organization composed of specific sets of segmental duplications that have hyperexpanded in concert with the formation of subterminal satellites. These regions are highly copy number polymorphic between and within species, and copy number differences involving hundreds of copies can be accurately estimated by assaying read-depth of next-generation sequencing data sets. Phylogenetic and comparative genomic analyses suggest that the structures have arisen largely independently in the two lineages with the exception of a few seed sequences present in the common ancestor of humans and African apes. We propose a model where an ancestral human-chimpanzee pericentric inversion and the ancestral chromosome 2 fusion both predisposed and protected the chimpanzee and human genomes, respectively, to the formation of subtelomeric heterochromatin. Our findings highlight the complex interplay between duplicated sequences and chromosomal rearrangements that rapidly alter the cytogenetic landscape in a short period of evolutionary time.
Chimpanzee and gorilla chromosomes differ cytogenetically from human chromosomes by 11 large-scale rearrangements (nine paracentric and pericentric inversions, one translocation, and one fusion) and by the presence of additional terminal G-bands adjacent to the telomere. These subtelomeric caps are heterochromatic in nature and are completely absent from the karyotype of human and orangutan (Haaf and Schmid 1987; IJdo et al. 1991; Ventura et al. 2011). Subterminal heterochromatin has been thought to be composed primarily of a tandem array of a 32-bp subterminal satellite (StSat) creating large subterminal blocks of constitutive heterochromatic regions (Royle et al. 1994; Koga et al. 2011) adjacent to the canonical telomeric TTAGGG sequence (Greider and Blackburn 1989). While almost all gorilla chromosomes show the presence of subterminal caps, only half of chimpanzee chromosomes possess such structures (Fan et al. 2002). In both chimpanzee and pygmy chimpanzee, this process has also created islands of interstitial heterochromatin on both chromosomes VII and XIII (Royle et al. 1994).
Due to the high-copy repetitive nature of these regions, the subtelomeric heterochromatin, like centromeric and secondary constriction on acrocentric chromosomes, is not represented in the existing genome assemblies of the chimpanzee (The Chimpanzee Sequencing and Analysis Consortium 2005) and gorilla. Unlike centromeric satellite sequences, which were well-characterized prior to the era of full genome sequencing (Rudd and Willard 2004), there are relatively few detailed molecular studies of the organization or evolution of the subterminal caps (Royle et al. 1994). Conservatively, it has been estimated by dot-blot analysis that the subterminal heterochromatin constitutes >3 Mbp (0.1%) of the total genomic DNA of each species (Yunis and Prakash 1982; Koga et al. 2011).
Subtelomeric regions are more generally recognized as extremely dynamic regions of chromosomes (Mefford and Trask 2002; Prieto et al. 2004; Gonzalez-Garcia et al. 2006; Carreto et al. 2008; Nieves et al. 2011). In humans, large complex blocks of duplicated sequences—zones of subtelomeric duplication—typically define the last 50–150 kbp of human chromosomes (Mefford and Trask 2002; Riethman et al. 2005). It has been postulated that this genomic dynamism occurs during meiotic prophase when all the chromatids interconnect, allowing for nonhomologous chromosome exchange of chromosome ends (Wallace and Hulten 1985). This contact may explain why specific subtelomeric regions preferentially associate and share a high degree of sequence identity despite mapping to nonhomologous chromosomes (Hirai et al. 2005). The high sequence identity among subtelomeric regions has been an important consideration in understanding changes in the chromatin environment related to some human genomic disorders (Gerber et al. 2011; Kudo et al. 2011; van der Maarel et al. 2011).
Recently, we identified the presence of specific segmental duplications that have hyperexpanded within the subterminal cap of both gorilla and chimpanzee (Cheng et al. 2005; Marques-Bonet et al. 2009; Ventura et al. 2011). In this study, we perform a detailed investigation into the organization and evolution of subterminal caps using molecular cytogenetics, targeted clone-based sequencing, and phylogenetic-based analyses. Based on the evolutionary history of hominid chromosomal rearrangements, we develop a model to explain the distribution and organization of subtelomeric caps in gorilla and chimpanzee, as well as their absence in humans. We hypothesize that ancestral chromosomal rearrangements were important in catalyzing the spread of these chromosome structures and that the fusion of human chromosome 2 squelched this process in humans by eliminating the source of the StSat sequence shortly after human speciation.
Results
Subtelomeric cap distribution among gorilla and chimpanzee chromosomes
In order to test the localization of telomeric TTAGGG sequences with respect to the 32-bp StSat, we began by performing bicolor fluorescent in situ hybridization (FISH) experiments using biotinylated oligonucleotides designed to the satellite sequence in conjunction with a commercial PNA (peptide nucleic acid) corresponding to telomeric TTAGGG sequence (Methods). As expected, the telomeric satellite colocalizes with “caps” in both species, mapping immediately proximal to the PNA telomere probe (Supplemental Fig. S1). We were able to identify all chromosome arms carrying StSat in chimpanzee (42/96 chromosome tips and interstitial signals on chromosomes VII and XIII) and in gorilla (80/96 chromosome tips). Cap signals were detected on short arms of IIp and IIq chromosomes in chimpanzee and on both arms of IIq in gorilla. No traces of signals were observed on the long arms of chimpanzee IIp and IIq or on the short arm of gorilla IIp. For gorilla, in particular, we did not detect signals on any of the p arms of the acrocentric chromosomes (IIp, IX, XIII, XIV, XV, XVIII, XXI, and XXII), except for chromosome IIq showing signals on the p and q arms.
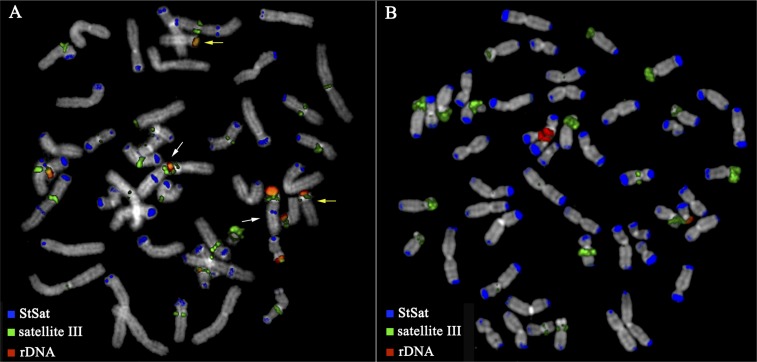
Satellite III (SatIII) DNA and ribosomal DNA (rDNA) have been reported to localize to the short arm of acrocentric chromosomes in chimpanzee and gorilla (Jarmuz et al. 2007). In order to understand the chromosomal distribution of these high-copy repeats, we performed three-color FISH experiments in chimpanzee and gorilla using StSat, rDNA, and SatIII (group 1 and group 2) (Bandyopadhyay et al. 2001b) specific probes. In gorilla, we found that all chromosome termini or arms hybridized strongly to one of these probes, but never did these probes colocalize to the same arm (Fig. 1). In chimpanzee, not all chromosomes showed hybridization signals for satellite sequences and, as in gorilla, StSat probes never colocalized with either rDNA or SatIII probes. Both rDNA and SatIII probes, however, did colocalize on chromosomes XIIIpter and XIVpter. These results and the reduced signal intensity suggest a less pronounced spreading of satellite sequences to the tips of chimpanzee chromosomes when compared to gorilla (Fig. 1). In chimpanzee, we note that not all chromosomal ends are capped by StSat or another form of satellite. These chromosomal ends that stain G-negative are more similar to the terminal chromosomal structures found in human and orangutan.
Figure 1.
Three-color FISH experiment using StSat, SatIII, and rDNA on chimpanzee and gorilla chromosomes. SatIII and rDNA never colocalize with the StSat in both species. (A) In chimpanzee, however, rDNA and SatIII probes strongly colocalize on chromosomes XIIIpter (white arrows) and XIVpter (yellow arrows). (B) In gorilla, all chromosomal tips are positive for one of the three forms of the satellite.
Molecular cytogenetic analysis of subterminal regions
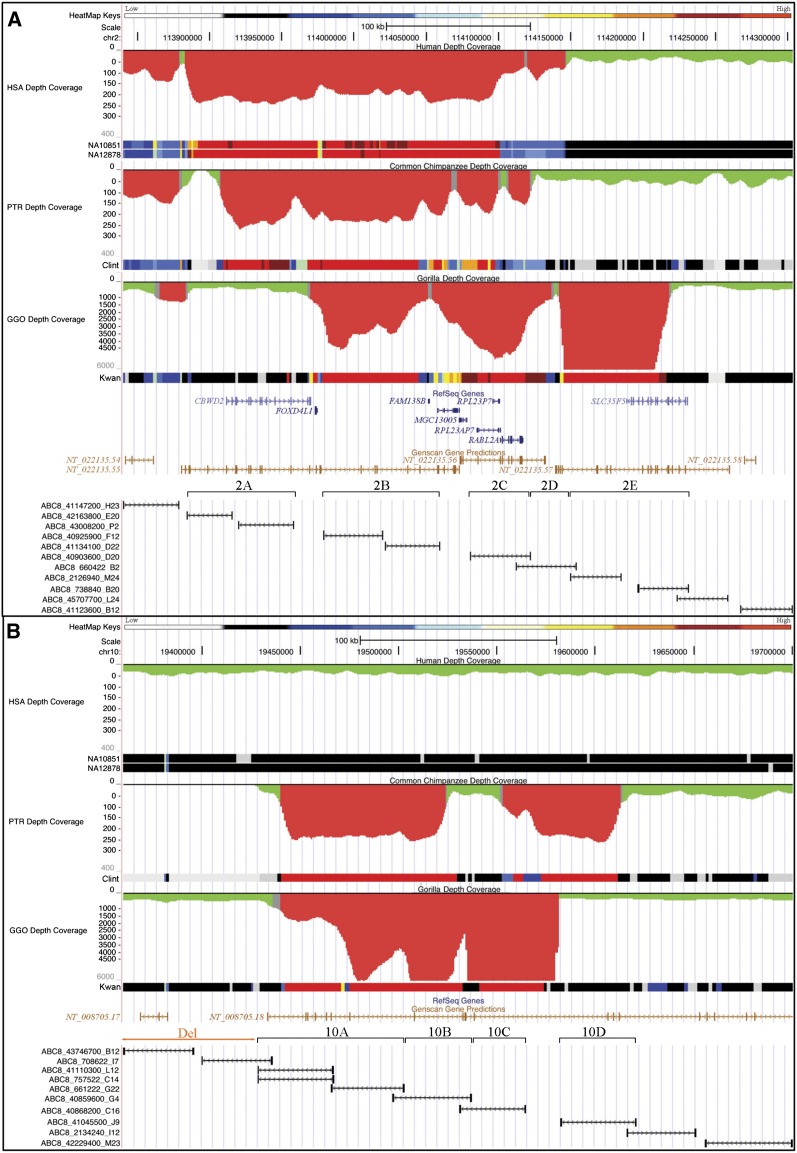
We previously identified two regions of segmental duplication that mapped to the subterminal caps and had hyperexpanded to hundreds of copies in both chimpanzee and gorilla. This included a 36-kbp chimpanzee-hyperexpanded segment homologous to human chromosome 2, contained in the chimpanzee clone AC150905, and a 45-kbp gorilla-hyperexpanded segment homologous to human chromosome 10 (19534885-19579478 NCBI35) (Fan et al. 2002; Cheng et al. 2005; Marques-Bonet et al. 2009; Ventura et al. 2011). To better define the cap structure organization in both apes and delineate the full extent of these duplications, we performed a series of comparative FISH experiments using a set of overlapping human fosmid clones (Kidd et al. 2010) mapping to a 463-kbp region of human chromosome 2 (113840263-114303469) and a 341-kbp region mapping to chromosome 10 (19359773-19700390) (Table 1; Fig. 2). Signals were distinguished as mapping to the subterminal cap or immediately proximal to the heterochromatic cap, termed “subcap” for the purpose of this study.
Table 1.
Comparative hybridization of chromosome 2 and chromosome 10 hyperexpansions
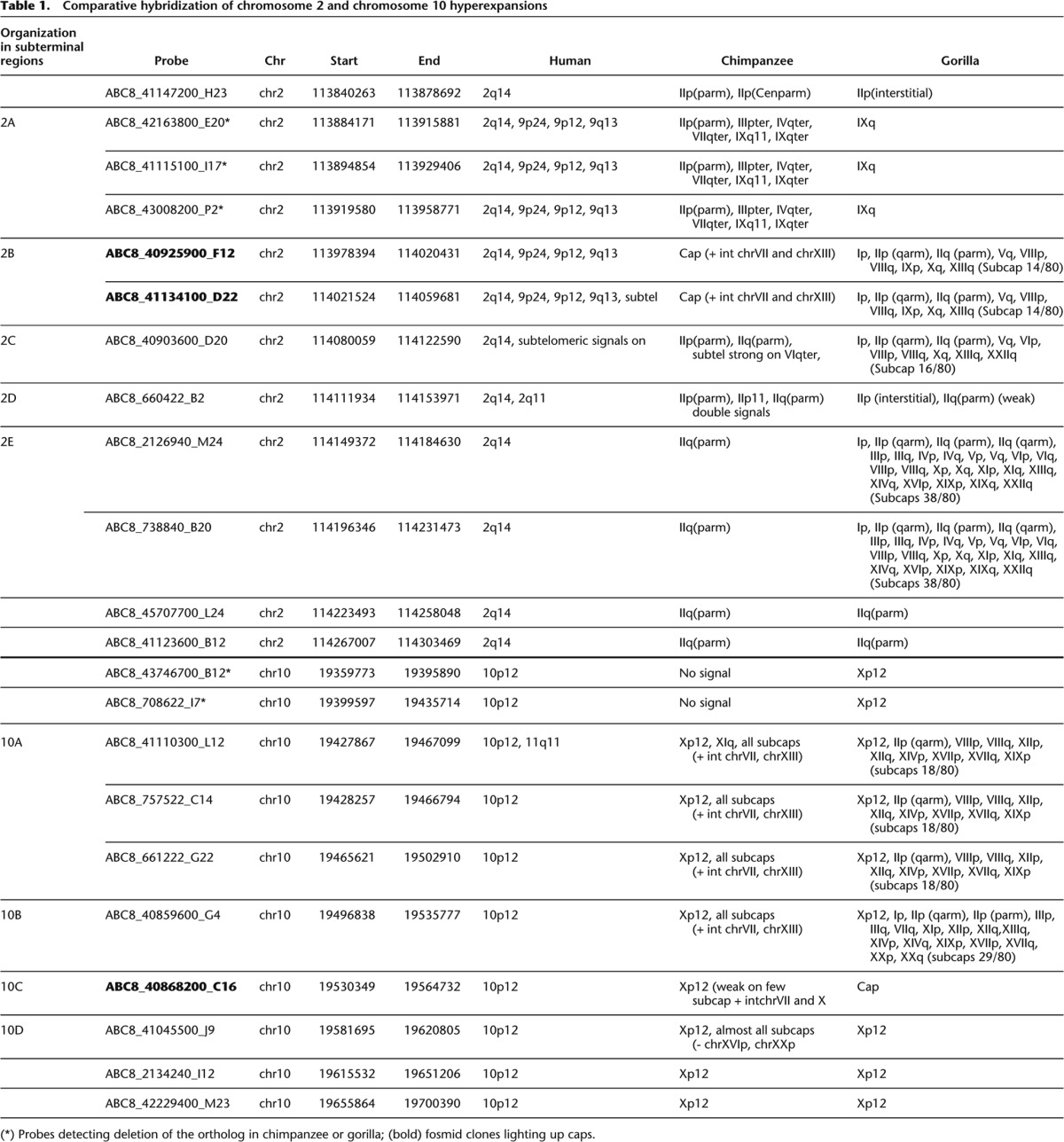
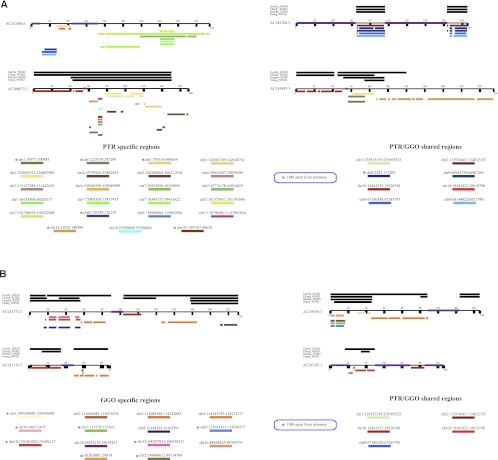
Figure 2.
Duplication and fosmid probe map. The pattern of segmental duplications is shown for (A) a 463-kbp region of human chromosome 2 (chr2:113840263-114303469) and (B) a 341-kbp region mapping to chromosome 10 (chr10:19359773-19700390) based on human genome annotation (NCBI build35). Computationally predicted human, chimpanzee, and gorilla duplications (red = excess depth-of-coverage of aligned whole-genome shotgun sequence) as well as a heat map indicating the copy number are displayed. Fosmid clone contigs are reported below each genomic region. Human fosmid probes underlying each region are shown and grouped (2A–E and 10A–D) based on the pattern of hybridization to chimpanzee and gorilla subtelomeric cap and subcap regions. (HSA) Homo sapiens, (PTR) Pan troglodytes, (GGO) Gorilla gorilla.
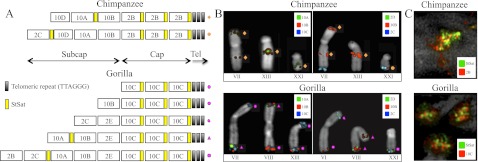
Based on differences in the pattern of hybridization, we defined three main areas from human chromosome 2 (2B, C, and E) in chimpanzee and four regions from human chromosome 10 (10A–D) in gorilla as part of the subterminal regions. We note that fosmid clones corresponding to region 2D did not show any signal to either cap or subcap regions (Table 1; Fig. 3A,B). Due to the close proximity of the regions and the compact nature of subterminal heterochromatin, FISH experiments were generally uninformative in resolving the order of the probes. Thus, we defined the organization of these regions assuming that higher-copy probes mapped more distally than lower-copy-number probes based on studies of the organization of human subtelomeric regions (Mefford and Trask 2002; Linardopoulou et al. 2005; Rudd et al. 2009). This assumption was subsequently validated by BAC-based clone sequencing (see below). FISH experiments on metaphase and interphase nuclei and stretched chromosomes using StSat and human cap-specific fosmid probes (2B and 10C) showed a clear interdigitation of satellite sequences and chromosome 2 and 10 probes in subtelomeric cap-associated regions of chimpanzee and gorilla, respectively (Fig. 3C).
Figure 3.
Schematic representation of the organization of subtelomeric regions. (A) Cap and subcap regions have been reported according to FISH experiments and copy number count in chimpanzee (top lines) and gorilla (bottom lines). (B) Extracted chromosomes representing examples of all the different organizations observed in the subterminal regions of chimpanzee and gorilla. (C) FISH on stretched chromosomes showing the interdigitation between StSat and regions 2B and 10C in chimpanzee and gorilla, respectively.
There were three important observations. First, we found that probes mapping to the subterminal caps hybridized much more intensely than probes mapping to the subcap, irrespective of the species analyzed. We interpreted this as representing increases in copy number of these regions in the cap when compared to the subcap of African great ape chromosomes. Second, we observed a remarkable reciprocity in cap and subcap locations between gorilla and chimpanzee. For example, probes ABC8_40925900_F12 and ABC8_41134100_D22 (Fig. 2, region 2B) hybridized exclusively to the caps in chimpanzee but only to the subcaps in gorilla. Similarly, probe ABC8_40868200_C16 (Fig. 2, region 10C) showed strong hybridization signals to the caps in gorilla but very weak signals to most chimpanzee subcaps (Table 1). Third, the subcap hyperexpansions were accompanied by fixed, large-scale deletions of the ancestral locus where the duplicated sequences originated. We identified a 75-kbp deleted segment in gorilla mapping to human chromosome 2 (Fig. 2, region 2A) and a 76-kbp region in chimpanzee mapping to human chromosome 10 (chr10: 19359773-19435714) (Table 1). Both deleted regions showed no traces of RefSeq genes but displayed predictions of genes in the area (Fig. 2).
In order to understand the fate of the most terminal portions of the p-arm of chromosomes IIp and IIq in chimpanzee and gorilla following the fusion, we performed additional three-color hybridization experiments using BAC probes corresponding to human chromosome 2p and 2q (RP11-301C20: chr2:35,539,435-35,655,555 and RP11-361O8: chr2:165,546,718-165,724,972, respectively) along with fosmid probes corresponding to the region of interest. This approach was essential to discriminate the acrocentric and almost cytogenetically indistinguishable chromosomes IIp and IIq in both species. We observed no signals to the chimpanzee subcap locations using fosmid clones from the chromosome 2 region (Fig. 2), while gorilla displayed subcap signals using both chromosomes 2 and 10 fosmid clones (Supplemental Fig. S2; Supplemental Table S1). It is noteworthy that region 2A showed signals on the orthologous IIp and pericentromeric regions on chromosome IX in chimpanzee, while no signals were observed at the orthologous gorilla chromosome IIp. Gorilla showed only signals on chromosome IX and likewise for macaque, which was used as representative of Old World monkeys (data not shown). These results date the duplication from chromosome IX to chromosome II after the divergence of gorilla from the chimpanzee-human ancestor.
Chimpanzee interstitial heterochromatin
We performed additional analyses to localize the interstitial heterochromatic regions in the chimpanzee genome. We selected a set of overlapping human fosmid clones corresponding to the previously reported region of insertion (Gross et al. 2006) and used FISH to more precisely localize the integration sites of the heterochromatic chimpanzee blocks on chromosomes VII and XIII. We localized the insertion breakpoint of the chromosome VII heterochromatic block to a 12.3-kbp interval (as defined by probes ABC8_40890400_C6 at chr7: 101930395-101963680 and ABC8_720140_F22 at chr7: 101951324-101986595) and the chromosome XIII breakpoint to a 5.9-kbp interval (as defined by probes ABC8_2114240_I22 at chr13: 45906763-45941160 and ABC8_43215900_F22 at chr13: 45935232-45970371) (Supplemental Table S2; Supplemental Fig. S3). While the organization of these interstitial heterochromatic blocks was virtually identical in composition to chimpanzee subterminal caps, none of these chromosome 7 or 13 probes produced any signals in the caps or subcaps of the chimpanzee chromosomes, suggesting an independent and unidirectional transfer of sequence from the subterminal cap region to euchromatin. Notably, the integration sites correspond to human-chimpanzee and human-chimpanzee-gorilla segmentally duplicated regions in chromosome VII and chromosome XIII, respectively. Further, in the case of chromosome VII, the insertion point maps within 1 Mbp from an inversion breakpoint specific to the ancestral lineage of human and chimpanzee (Ventura et al. 2011).
Copy number variation between and within species
We performed two sets of experiments to assess the extent of variation between and within species. To assess qualitative differences in the chromosomal distribution, we first performed cohybridization experiments on metaphase chromosomal spreads from two gorillas, two chimpanzees, and one bonobo. Marked differences in the distribution and signal intensity were observed, especially when comparing homologous chromosomes in gorilla (Figs. 3B, 4), where subterminal regions showed differences in the organization of the cap (see chromosomes XIX and XXII). The two common chimpanzees and the bonobo showed a much more uniform distribution of probes, with the exception of the interstitial chromosome VII heterochromatic block, which appears less complex in bonobo when compared to the two common chimpanzees (Figs. 3B, 4).
Figure 4.
Summary of FISH experiments in human, two common chimpanzees, one pigmy chimpanzee, and two gorillas. Colored circles represent locations of the probes in great apes chromosomes; half circles represent heterozygous signals. Difference in interstitial heterochromatin on chromosome VII is displayed between common and pygmy chimpanzees. (PPA) Pan paniscus.
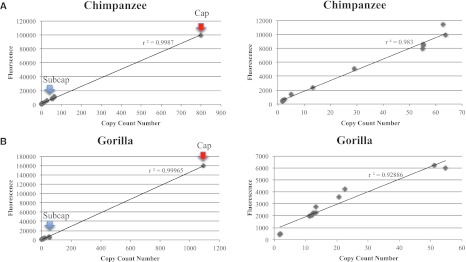
To obtain a quantitative assessment of copy number variation, we performed read-depth-based copy number predictions as previously described in Sudmant et al. (2010). Briefly, next-generation sequence reads were mapped to reference sequences and read-depth was used to estimate copy number after correcting for GC-bias. While the efficacy of this approach has been validated for low- to middle-copy duplications, the dynamic range response of high-copy repeats had not been previously tested. To assess this more formally, we initially estimated the median copy number for various subterminal cap regions in three species (human, chimpanzee, and gorilla) and compared these values to the number and relative fluorescence intensity normalized by the value of maximum fluorescence (VMF = 255). The relative fluorescence intensity and read-depth copy number for each locus in each species were highly correlated (r2 = 0.93–0.99) (Supplemental Table S3; Fig. 5; Supplemental Fig. S4), suggesting that copy number, even for high-copy repeats, may be accurately estimated.
Figure 5.
Distribution copy number counts versus fluorescence. Copy number estimates of sequence read-depth for each genomic region correlate strongly (r2 = 0.99) with measured fluorescence intensity for every probe distributed on chromosome 2 and chromosome 10 (see Supplemental Table S3). Red arrows show the location of the single probe positive for cap, while blue arrows display the group of probes positive for subcap in (A) chimpanzee and (B) gorilla (left panels). The correlation rates reduce when hyperexpanded segments are excluded from the analysis (right panels).
We estimate the median species copy number for various subcap regions ranges from 29 to 64 in chimpanzee with greater diversity and lower copy number (12–55) than observed in gorilla (Supplemental Table S3). We estimate the median copy of the hyperexpanded segments within the subterminal cap to be 798 in chimpanzee compared to 1092 in gorilla. In contrast, all analyzed human genomes, as well as the Neanderthal, were either predicted to be diploid for the corresponding segments or showed only modest evidence of duplication (<14 copies). Adjusting for the size of the probes and differences in copy number for each segment, the amount of segmental duplication added to the cap and subcap regions is strikingly similar (∼46 Mbp) in both gorilla and chimpanzee (Supplemental Table S3). This value exceeds the initial estimates of StSat sequence by more than 10-fold.
Sequence analyses
Since the subcap and cap regions of gorilla and chimpanzee chromosomes are missing from the corresponding primate genome assemblies, we identified large-insert clones by hybridization and sequenced the inserts using capillary-based sequencing methods. We also used BLAST to search for sequence similarity with StSat (contained in the pCht7 and pCht13 clones) (Royle et al. 1994) for GenBank accessions corresponding to BAC clones carrying these sequences within the nucleotide (nr/nt) and high-throughput genomic sequence (HTGS) collections. We identified 17 chimpanzee and nine gorilla clones whose inserts had been sequenced and had tracts of StSat sequence (Supplemental Table S4). The corresponding sequences were annotated as StSat, the presence of segmental duplications in human, chimpanzee, and gorilla, and homology with the human reference genome using a suite of computational programs (Methods).
Our analysis showed that all 26 BAC sequences were highly duplicated in chimpanzee and gorilla genomes and carried both species-specific as well as ancestrally shared duplicated sequences (Fig. 6A,B). None of the BACs, however, was composed solely of satellite sequences, but rather they showed a patchwork of segmental duplications mixed with tracts of StSat sequences. While variable in size, most of the satellite tracts were >20 kbp, with some exceeding 60 kbp within a given BAC (Fig. 6A,B; Supplemental Fig. S5A,B). It should be noted, in this regard, that nine out of the 26 BACs failed to completely assemble or assembled into sequence contigs substantially shorter than the typical BAC insert length for these libraries, indicating difficulty in cloning and sequence assembly of these regions.
Figure 6.
Genomic organization based on large-insert clone sequencing. Sequence organization shown for large-insert BAC clones that carry StSat sequences in (A) chimpanzee and (B) gorilla. Sequences were annotated for the presence of duplications (whole-genome shotgun sequence detection [WSSD]) in each species (black bars above line), StSat sequence (purple), and the location of segmental duplications in the human genome based on BLAST sequence similarity searches (colored bars). Patchworks of segmental duplications are mixed with tracts of StSat sequences in all the clones analyzed.
Four chimpanzee BAC clones showed a relatively unique architecture—segmental duplications from human chromosome 10 as well as several locations on chromosome 7, including chr7: 101708105-102032998. The latter region was previously defined as flanking the interstitial heterochromatic block specific to the chimpanzee genome. Copy number sequence read-depth analysis predicts a dramatic increase in copy number transitioning from the chromosome VII to chromosome X regions consistent with the transition point from euchromatin to the interstitial heterochromatin in the chimpanzee genome (Supplemental Fig. S6).
Although we identified between 20 and 30 distinct segmental duplications associated with StSats, several were observed much more frequently. Eight out of 17 chimpanzee BAC clones carried segmental duplications corresponding to the human chromosome 2 region 2B, while four of those eight showed similarity to the chromosome 10 region 10B (Fig. 2). Similarly, seven out of nine gorilla BAC clones showed similarity to region 10C, flanked by StSat consistent with previous FISH and copy number estimates. Such shared paralogous sequences are particularly valuable because they provide an opportunity to assess the phylogenetic relationship among various copies between and within species.
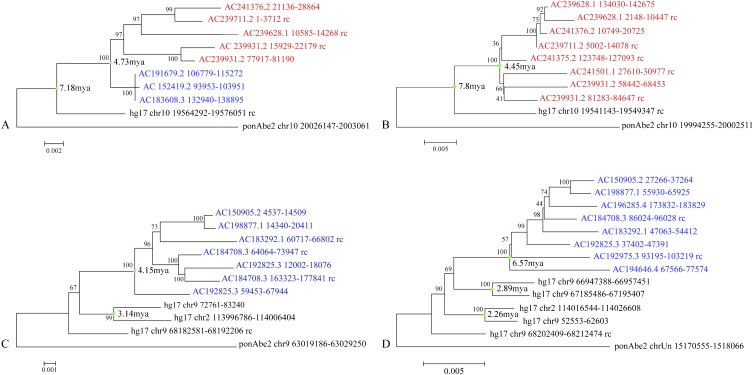
We specifically searched for shared sequences located within 10 kbp of StSat sequences in an effort to date the expansion of StSat sequences during hominid evolution. We identified four regions: two mapping to human chromosome 2 (39.6 and 10.0 kbp in length) and two mapping to chromosome 10 (11.8 and 8.2 kbp in length) where we had successfully resolved seven to nine distinct copies among the chimpanzee and gorilla clone sequences. Homologous sequences from these BACs and the human and orangutan reference genomes were extracted, aligned (ClustalW), and phylogenetic trees were constructed (MEGA4 maximum likelihood). We used orangutan as an outgroup because these loci were single-copy in this species and mapped, in most cases, to the ancestral locus.
The phylogenetic analyses generally support independent expansions of the duplicated sequences in chimpanzee and gorilla. Using an estimated orangutan divergence of 14 million years ago (mya) from the human lineage, we can approximate these expansions to have initiated between 5 and 7 mya. The topology of the trees suggests that the spread of the subterminal segmental duplications was continuous within each lineage (as indicated by the varying branch length and terminal subclades) and accumulated over a long period of evolutionary time. Interestingly, only one of the paralogous segments (a chromosome 10 segment) was shared at the sequence level between both gorilla and chimpanzee subterminal clones. Phylogenetic analysis (Fig. 7; Supplemental Table S5) provides compelling evidence that this segment was duplicated in the common ancestor of chimpanzee and gorilla (7–8 mya) but then subsequently and gradually expanded to the subterminal caps of both chimpanzee and gorilla. These duplications are not present in contemporary humans, potentially due to subsequent loss in the ancestral lineage.
Figure 7.
Phylogenetic analysis. Sequences shared among distinct subterminal genomic loci in chimpanzee (blue) and gorilla (red) were aligned to human (NCBI build35) and orangutan (ponAbe2) orthologous sequences (see text for details). Neighbor-joining phylogenetic trees (bootstrap values at each node) were constructed from four different genomic regions (A–D). Only the chromosome 10 segment (A) is shared between gorilla and chimpanzee cap sequences. We estimate the evolutionary time (green dots = mya) of duplications and expansions assuming a human-orangutan divergence of 14 mya.
Discussion
In this study, we characterize the genomic organization and evolution of the chromosomal caps of gorilla and chimpanzee chromosomes. We operationally distinguish two genomic architectures—cap regions, consisting of hyperexpanded arrays of human chromosome 2 and 10 segmental duplications (chimpanzee and gorilla, respectively) interspersed with StSat (Fig. 2), and subcap regions composed of lower-copy segmental duplications admixed with occasional tracts of StSat. This subterminal heterochromatin and the corresponding transition regions are absent from the genomes of both human and orangutan. Phylogenetic and cytogenetic data suggest that the genomic architecture of these regions began to form in the common ancestor of human, chimpanzee, and gorilla ∼7–8 mya (Fig. 7A). In contrast, we estimate that the expansion and spread of cap sequences to multiple chromosomes occurred soon after speciation in gorilla and chimpanzee (4–5 mya), albeit independently, with a reciprocal definition of cap and subcap sequences in each lineage. It is unclear why the progenitor cap and subcap sequences were quiescent for 3 million years, but the emergence of the StSat sequence must have been important in generating this instability since it, along with the chromosome 2 and 10 segmental duplications, are the only common sequence elements. We hypothesize that chromosome II rearrangements played a central role in triggering this instability and specifying differences in the evolutionary trajectory in human, chimpanzee, and gorilla.
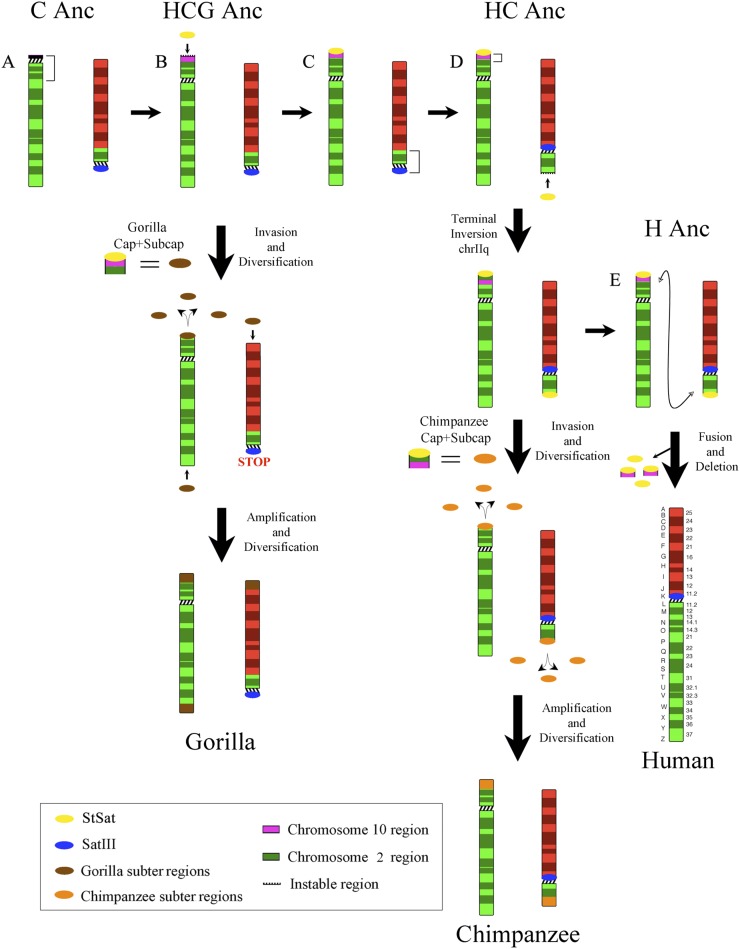
Three evolutionary large-scale chromosomal rearrangements involving chromosome II over the last 12 million years of hominid evolution have been previously described (Fig. 8), namely: (1) a pericentric inversion within acrocentric chromosome IIq in the gorilla-chimpanzee ancestor after separation from orangutan (Yunis and Prakash 1982; Roberto et al. 2008; Ventura et al. 2011), (2) a pericentric inversion on chromosome IIp in the human-chimpanzee ancestor after divergence from gorilla lineage (Wienberg et al. 1994; Roberto et al. 2008), and (3) a fusion of IIp and IIq specifically in the ancestral lineage of human to create chromosome 2 (Yunis and Prakash 1982; Wienberg et al. 1994).
Figure 8.
Model of subterminal heterochromatin evolution in chimpanzee and gorilla genomes. Shown is a hypothetical mechanism of evolution of subterminal satellite (StSat), chromosome 2, and chromosome 10 regions that have contributed to the subtelomeric chromosomal regions in chimpanzee and gorilla. (C Anc) Catarrhine ancestor, (HCG Anc) human-chimpanzee-gorilla ancestor, (HC Anc) human-chimpanzee ancestor, (H Anc) human ancestor. (A) A pericentric inversion of chromosome IIq in the gorilla-chimpanzee ancestor (Yunis and Prakash 1982; Roberto et al. 2008; Ventura et al. 2011). (B) A segment of chromosome 10 was duplicatively transposed to the short arm of chromosome IIq in the common ancestor of human-chimpanzee-gorilla, placing it in close proximity to centromeric satellite sequences. (C) A pericentric inversion that occurred on chromosome IIp in the human-chimpanzee ancestor (Wienberg et al. 1994; Roberto et al. 2008) internalizing the SatIII sequences. (D) A smaller local inversion in ancestral human-chimpanzee IIq placed the chromosome 2 segment adjacent to the StSat; StSat and potentially duplicated sequences spread to the distal end of the short arm of chromosome IIp in the human and chimpanzee ancestor. (E) A fusion of IIp and IIq human ancestor created human chromosome 2 (Yunis and Prakash 1982; Wienberg et al. 1994) and squelched the spreading of StSat to human subtelomeric regions.
To explain our results, we propose the following possible model (Fig. 8). A segment of chromosome X was duplicatively transposed to the short arm of chromosome IIq in the common ancestor of human-chimpanzee-gorilla. The initial IIq inversion placed centromeric satellite sequences in close proximity to this segment potentiating the emergence of StSat sequences. The second inversion of chromosome IIp occurred in the ancestor of human and chimpanzee after separation from the gorilla ancestor. This event internalized the protective SatIII sequence (Fig. 8) and effectively rendered chromosome IIp “exposed” to the spread of duplicated sequences. (Note: Sequence similarity searches detect vestigial SatIII sequence homology specifically at the centromeric breakpoint of this large-scale inversion in human chromosome 2p.) We hypothesize that the existence of a smaller local inversion in ancestral human-chimpanzee IIq altered the order of chromosome 2 and 10 segments, placing the chromosome 2 segment adjacent to the StSat and, as a result, susceptible to hyperexpansion. A key step was the spread of the StSat and potentially duplicated sequences to the distal end of the short arm of chromosome IIp in the human and chimpanzee ancestor—where additional events occurred, such as the duplicative transposition of a chromosome IX segment (segment 2A) (Fig. 8; Table 1). In this model, both IIp and IIq could serve as sources for the spread of these sequences to other telomeric regions and the formation of subterminal heterochromatin. In the human ancestor, subterminal heterochromatin was circumvented by the fusion of chromosome IIp and IIq leading to the deletion of this unstable sequence from the human lineage. It is possible that the presence of StSat sequence along with sequence homology may have, in fact, played a role in triggering this event.
This scenario would explain (1) the absence of subterminal heterochromatin in humans, (2) the differential distribution of subterminal heterochromatin in chimpanzee and gorilla chromosomes, and (3) differences in the composition of the cap and subcap sequences—in particular with respect to the hyperexpansion of chromosome 2 and 10 sequences in gorilla and chimpanzee, respectively (Cheng et al. 2005; Marques-Bonet et al. 2009). In our model, both the evolution and spread of subterminal heterochromatin was inextricably linked to large-scale chromosomal rearrangements that catalyzed its formation. In contrast, the fusion of chromosome IIp and IIq squelched this process in humans by eliminating the source of the StSat sequence. Since the phylogenetic analyses of both human and chimpanzee suggest the spread of these segments occurred >4 mya, it follows that the fusion of chromosome IIp and IIq occurred early during evolution; otherwise, subterminal heterochromatin would have evolved within our lineage.
We recognize, however, that this is only one of many possible explanations that might account for the observed differences among human, chimpanzee, and gorilla subterminal regions. It may be also possible that functional constraints, such as the expression or suppression of critical genes in humans, may have contributed to these dramatic differences between humans and great apes. Human diseases such as fascioscapulohumeral muscular dystrophy (FSHD), for example, arise from chromatin deregulation (Lemmers et al. 2010) near the telomere of human chromosome 4. From an evolutionary perspective, it is known that specific gene families have expanded immediately proximal to the telomeres of human chromosomes. Of particular relevance in this regard is the Wiskott-Aldrich Syndrome Protein and SCAR Homolog (WASH) gene family (Linardopoulou et al. 2007). This gene family has both expanded within the subtelomeric regions, with a copy located specifically at the site of the chromosome 2 fusion. The function of the specific family members is not known, but as a class, they are thought to be important in the reorganization of the actin cytoskeleton in filopodia protrusions (Linardopoulou et al. 2007).
Among hominids, chimpanzees are the only lineage wherein heterochromatic blocks developed interstitially (chromosomes VII and XIII). Our sequence and FISH analyses indicate that the composition of these regions is indistinguishable from subterminal heterochromatin. Refined mapping of the breakpoints reveals that the interstitial heterochromatin formed within segmentally duplicated regions shared between human and chimpanzee for chromosome VII and between human, chimpanzee, and gorilla for chromosome XIII. Thus, it is quite likely that homology between these interstitial blocks of segmental duplication and the subtelomeric region provided a gateway for the movement of StSat and chromosome 2 segments internally (IJdo et al. 1991; Hirai et al. 2005). Once present, these sequences amplified, creating heterochromatic interstitial G-band positive regions in this species. The fact that the chromosome VII interstitial heterochromatin has not fixed in the chimpanzee population may indicate that these interstitial “colonizations” occurred relatively recently (Hirai et al. 2005) or, alternatively, are being deleted.
Hirai and colleagues (2005) described a retrotransposable compound repeat DNA organization (RCRO) associated with subterminal heterochromatin. The authors hypothesized that this sequence was important in inducing and prolonging bouquet formation during meiotic prophase specifically in chimpanzee when compared to human and other primate chromosomes. Our data reveal a much more complex organization of subterminal heterochromatin that involved segmental duplication as opposed to retrotransposition—although it is possible that the RCRO originally described is part of this architecture. Our copy number estimates and sequence analysis (Fig. 5) suggest that StSat and segmental duplication expansions have added ∼45–46 Mbp of highly identical (1%–2% divergent) sequences to the ends of many gorilla and chimpanzee chromosomes.
It has been previously reported that the presence of large blocks of heterochromatin at the ends of chromosomes may alter patterns of meiotic recombination (Miklos and Nankivell 1976). In chimpanzee, it is, thus, possible that this architecture directly interferes with synapsis during meiosis, resulting in an overall reduction of the total number of chiasma and a prolonged association of telomeric regions during zygotene and pachytene. Indeed, preliminary findings by Hirai and colleagues reported a mean lower chiasma frequency in chimpanzee when compared to human (Hirai et al. 2005). While it is now known that the pattern of fine-scale recombination differs significantly between human and chimpanzee (Ptak et al. 2005), it will be interesting to determine whether this difference is most pronounced in close proximity to regions of subterminal heterochromatin. This will require both higher quality sequence and fine-scale mapping of meiotic crossover events in these complex regions of human and great ape chromosomes.
Methods
Fluorescent in situ hybridization
Gorilla and chimpanzee BAC and human fosmid clones were used as probes in FISH assays on human and great ape metaphase spreads. Metaphases from nonhuman primates (common chimpanzee, gorilla, bonobo, and rhesus monkey) were obtained from lymphoblastoid or fibroblast cell lines; human metaphase spreads were obtained from PHA-stimulated peripheral lymphocytes of normal donors by standard procedures. DNA extraction from BACs and fosmids was performed as already reported (Ventura et al. 2001). FISH experiments were essentially performed as previously described (Ventura et al. 2004). Briefly, DNA probes were directly labeled with Cy3-dUTP, Cy5-dUTP (GE Healthcare), or Fluorescein-dUTP (Invitrogen) by nick translation. Two hundred ng of labeled probe were hybridized on metaphases spreads; hybridization was performed overnight at 37°C in 2× SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, 5 μg COT1 DNA (Roche), and 3 μg sonicated salmon sperm DNA, in a volume of 10 μL. Post-hybridization washing was performed at 60°C in 0.1× SSC (three times, high stringency). Washes of interspecies hybridization experiments were performed at lower stringency: 37°C in 2× SSC, 50% formamide (× 3), followed by washes at 42°C in 2× SSC (× 3). Chromosome identification was obtained by DAPI staining, producing a Q-banding pattern. Digital images were obtained using a Leica DMRXA epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments). Cy3, Cy5, Fluorescein, and DAPI fluorescence signals, detected with specific filters, were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Stretched chromosomes
Chimpanzee and gorilla chromosomes were stretched mechanically. Colcemid-treated lymphoblastoid cells were washed in phosphate-buffered saline (PBS), counted, and resuspended in a hypotonic solution (75 mM KCl, Cf: 60,000 cells/mL) for 15 min. Then, 0.5 mL of the suspension was cytocentrifuged (Shandon Cytospin 3 centrifuge) onto coated glass slides (Thermo Shandon Double Cytoslide) at 800 rpm for 4 min and fixed in methanol at −20°C for 15 min and in methanol:acetic acid 3:1 for 30 min. The slides were aged at 80°C for 1 h before hybridization.
Oligo-FISH and PNA probes
Biotinylated primers were designed on the StSat sequence (5′TCCATGTTTATACAGATAGCGGTG3′-Bio) and directly used as probes (0.001 μmol) in FISH experiments on chimpanzee and gorilla metaphase spreads. Probe and target were denaturated at 74°C for 6 min and hybridized o/n at 37°C in 2× SSC, 50% (v/v) formamide, 10% (w/v) dextran sulphate, and 3 μg sonicated salmon sperm DNA, in a volume of 10 μL. Slides were quickly washed in 2× SSC, 50% formamide at room temperature, and signals were detected (detection buffer: 1% BSA/1× SSC0/0.1 Tween 20) with avidin-FITC at 37°C for 1 h. Excess fluorochrome was washed away in 4× SSC/0.1 Tween 20 at room temperature, and chromosomes were DAPI stained.
PNA probes hybridized to denatured telomeric sequences in cells permeabilized in hot formamide. Two μL of ready-to-use Cy3-conjugated telomere PNA probe (Dako, Telomere PNA FISH Kits) were used as probes in fluorescent in situ cohybridization assays to evaluate the respective localization of telomeric sequences and the StSat on gorilla and chimpanzee chromosomes.
Satellite III and rDNA chromosomal distribution
To determine the chromosomal distribution, primer sets were designed from the known sequences of each SatIII subfamily (Bandyopadhyay et al. 2001a,b) (pE-1: GATTCGATTCCATTGCACTCG-GGACTGAAACAAAATGGAGACC; pE-2: ATGCAGCCTGGGTGACCT-AAGAATCCATACCACACC; pR-1: TGTGCCTCTGTGTTACAT-ACTGCCATCCTTTCCACC; pR-2: ACGCTGGGTGATGGAGTGAAATAC-ACTCCATTTCATTCCGCCGC; pR-4: TAAGCGTGGAATGGGTTTGAGC-CATCCGATTCCATTTCACTAC; pK-1: ATCGAATGGATTCCTAATTG-CGATATCTTCTGTTACACG; pW-1: AATGGGATGGAACCGAGTGG-CCTTTCATTTCAAGTCCCTTCGC) and used to amplify genomic DNA by polymerase chain reactions (PCR) on human, chimpanzee, and gorilla genomic DNA. Amplified products were then directly labeled with Fluorescein-dUTP by PCR labeling and used as probes in FISH assays. The use of PCR labeling avoids the possible contamination from genomic DNA by nick translation labeling of PCR products. PCR labeling was carried out in a final volume of 20 μL that contained 100 ng PCR product, 2 μL 10× reaction buffer (Invitrogen), 2 μL 50 mM MgCl2, 0.5 μL each primer (10 μM), 0.5 2 mM μL dACG, 2.5 μL 1 mM Fluorescein-dUTP, 5 μL 2% BSA, and 0.3 μL Taq polymerase (5 U/μL). For both amplification reactions, the cycling parameters used were as follows: 3 min initial denaturation at 94°C, followed by 30 cycles of: 94°C for 30 sec, 56°C for 30 sec, and 72°C for 30 sec. Final extension was at 72°C for 10 min.
In order to assess rDNA localization in great apes, clones containing rDNA were extracted by a BLAST sequence similarity search of the human rDNA complete repeating unit (accession: U13369.1) against the HTGS database. Several human BAC clones were selected and tested; the clone giving the most consistent and reliable hybridization signals was chosen as a probe for the cohybridization experiments: CH507-159O11.
BAC sequencing
Chimpanzee and gorilla clones were selected for complete insert sequencing using capillary sequencing methods (McPherson et al. 2001) in order to obtain high-quality finished sequence within duplicated regions. Rearrangements were visualized using Miropeats (Parsons 1995) and previously described in-house visualization tools (Kidd et al. 2010).
Data access
A subset of clones was selected for complete insert sequencing. The sequences have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession nos. AC198877.1, AC183608.3, AC192975.3, AC097005.1, AC194646.4, AC200072.3, AC191679.2, AC183292.1, AC150905.2, AC152419.2, AC204739.3, AC192628.1, AC196285.4, AC213004.6, AC184708.3, AC192825.3, AC145493.3, AC239281.3, AC241375.2, AC239628.1, AC239393.3, AC239711.2, AC241376.2, AC241501.1, AC239640.2, AC239931.2.
Acknowledgments
We thank Tonia Brown for valuable comments in the preparation of this manuscript. The authors are grateful to Lisa Faust, Mollie Herget, and the staff of the Lincoln Park Zoo for gorilla material used in this study. We are also indebted to the large genome sequencing centers for early access to BAC end sequence data for targeted analysis of structural rearrangements. This work was supported, in part, by NIH grants HG002385 and GM058815 to E.E.E. and NIH grant U54 HG003079 to R.K.W. P.H.S. is supported by a Howard Hughes Medical Institute International Student Fellowship. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.136556.111.
References
- Bandyopadhyay R, Berend SA, Page SL, Choo KH, Shaffer LG 2001a. Satellite III sequences on 14p and their relevance to Robertsonian translocation formation. Chromosome Res 9: 235–242 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay R, McQuillan C, Page SL, Choo KH, Shaffer LG 2001b. Identification and characterization of satellite III subfamilies to the acrocentric chromosomes. Chromosome Res 9: 223–233 [DOI] [PubMed] [Google Scholar]
- Carreto L, Eiriz MF, Gomes AC, Pereira PM, Schuller D, Santos MA 2008. Comparative genomics of wild type yeast strains unveils important genome diversity. BMC Genomics 9: 524 doi: 10.1186/1471-2164-9-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93 [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Fan Y, Linardopoulou E, Friedman C, Williams E, Trask BJ 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res 12: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber JC, Neuhann TM, Tyshchenko N, Smitka M, Hackmann K 2011. Expanding the clinical and neuroradiological phenotype of 6q27 microdeletion: Olfactory bulb aplasia and anosmia. Am J Med Genet A 155: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia M, Gonzalez-Sanchez M, Puertas MJ 2006. The high variability of subtelomeric heterochromatin and connections between nonhomologous chromosomes, suggest frequent ectopic recombination in rye meiocytes. Cytogenet Genome Res 115: 179–185 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Gross M, Starke H, Trifonov V, Claussen U, Liehr T, Weise A 2006. A molecular cytogenetic study of chromosome evolution in chimpanzee. Cytogenet Genome Res 112: 67–75 [DOI] [PubMed] [Google Scholar]
- Haaf T, Schmid M 1987. Chromosome heteromorphisms in the gorilla karyotype. Analyses with distamycin A/DAPI, quinacrine and 5-azacytidine. J Hered 78: 287–292 [DOI] [PubMed] [Google Scholar]
- Hirai H, Matsubayashi K, Kumazaki K, Kato A, Maeda N, Kim HS 2005. Chimpanzee chromosomes: Retrotransposable compound repeat DNA organization (RCRO) and its influence on meiotic prophase and crossing-over. Cytogenet Genome Res 108: 248–254 [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA 1991. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc Natl Acad Sci 88: 9051–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz M, Glotzbach CD, Bailey KA, Bandyopadhyay R, Shaffer LG 2007. The evolution of satellite III DNA subfamilies among primates. Am J Hum Genet 80: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, Kallicki J, Kaul R, Wilson RK, Eichler EE 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Notohara M, Hirai H 2011. Evolution of subterminal satellite (StSat) repeats in hominids. Genetica 139: 167–175 [DOI] [PubMed] [Google Scholar]
- Kudo H, Emi M, Ishigaki Y, Tsunoda U, Hinokio Y, Ishii M, Sato H, Yamada T, Katagiri H, Oka Y 2011. Frequent loss of genome gap region in 4p16.3 subtelomere in early-onset type 2 diabetes mellitus. Exp Diabetes Res 2011: 498460 doi: 10.1155/2011/498460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. 2010. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329: 1650–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ 2007. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet 3: e237 doi: 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JD, Marra M, Hillier L, Waterston RH, Chinwalla A, Wallis J, Sekhon M, Wylie K, Mardis ER, Wilson RK, et al. 2001. A physical map of the human genome. Nature 409: 934–941 [DOI] [PubMed] [Google Scholar]
- Mefford HC, Trask BJ 2002. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3: 91–102 [DOI] [PubMed] [Google Scholar]
- Miklos GL, Nankivell RN 1976. Telomeric satellite DNA functions in regulating recombination. Chromosoma 56: 143–167 [DOI] [PubMed] [Google Scholar]
- Nieves M, De Oliveira EH, Amaral PJ, Nagamachi CY, Pieczarka JC, Muhlmann MC, Mudry MD 2011. Analysis of the heterochromatin of Cebus (Primates, Platyrrhini) by micro-FISH and banding pattern comparisons. J Genet 90: 111–117 [DOI] [PubMed] [Google Scholar]
- Parsons JD 1995. Miropeats: Graphical DNA sequence comparisons. Comput Appl Biosci 11: 615–619 [DOI] [PubMed] [Google Scholar]
- Prieto P, Martin A, Cabrera A 2004. Chromosomal distribution of telomeric and telomeric-associated sequences in Hordeum chilense by in situ hybridization. Hereditas 141: 122–127 [DOI] [PubMed] [Google Scholar]
- Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Paabo S 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet 37: 429–434 [DOI] [PubMed] [Google Scholar]
- Riethman H, Ambrosini A, Paul S 2005. Human subtelomere structure and variation. Chromosome Res 13: 505–515 [DOI] [PubMed] [Google Scholar]
- Roberto R, Misceo D, D'Addabbo P, Archidiacono N, Rocchi M 2008. Refinement of macaque synteny arrangement with respect to the official rheMac2 macaque sequence assembly. Chromosome Res 16: 977–985 [DOI] [PubMed] [Google Scholar]
- Royle NJ, Baird DM, Jeffreys AJ 1994. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat Genet 6: 52–56 [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet 20: 529–533 [DOI] [PubMed] [Google Scholar]
- Rudd MK, Endicott RM, Friedman C, Walker M, Young JM, Osoegawa K, de Jong PJ, Green ED, Trask BJ 2009. Comparative sequence analysis of primate subtelomeres originating from a chromosome fission event. Genome Res 19: 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Tawil R, Tapscott SJ 2011. Facioscapulohumeral muscular dystrophy and DUX4: Breaking the silence. Trends Mol Med 17: 252–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M 2001. Centromere emergence in evolution. Genome Res 11: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D'Addabbo P, Wandall A, Bjorck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res 14: 1696–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Catacchio CR, Alkan C, Marques-Bonet T, Sajjadian S, Graves TA, Hormozdiari F, Navarro A, Malig M, Baker C, et al. 2011. Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res 21: 1640–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BM, Hulten MA 1985. Meiotic chromosome pairing in the normal human female. Ann Hum Genet 49: 215–226 [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Ludecke HJ, Senger G, Horsthemke B, Claussen U, Cremer T, Arnold N, Lengauer C 1994. The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosome Res 2: 405–410 [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525–1530 [DOI] [PubMed] [Google Scholar]