Abstract
H2B monoubiquitylation (H2Bub1), which is required for multiple methylations of both H3K4 and H3K79, has been implicated in gene expression in numerous organisms ranging from yeast to human. However, the molecular crosstalk between H2Bub1 and other modifications, especially the methylations of H3K4 and H3K79, remains unclear in vertebrates. To better understand the functional role of H2Bub1, we measured genome-wide histone modification patterns in human cells. Our results suggest that H2Bub1 has dual roles, one that is H3 methylation dependent, and another that is H3 methylation independent. First, H2Bub1 is a 5′-enriched active transcription mark and co-occupies with H3K79 methylations in actively transcribed regions. Second, this study shows for the first time that H2Bub1 plays a histone H3 methylations-independent role in chromatin architecture. Furthermore, the results of this work indicate that H2Bub1 is largely positioned at the exon–intron boundaries of highly expressed exons, and it demonstrates increased occupancy in skipped exons compared with flanking exons in the human and mouse genomes. Our findings collectively suggest that a potentiating mechanism links H2Bub1 to both H3K79 methylations in actively transcribed regions and the exon–intron structure of highly expressed exons via the regulation of nucleosome dynamics during transcription elongation.
Post-translational modification of the four core histones is a ubiquitous process during the regulation of gene activation and repression. Histone modifications are also involved in various cellular processes, including alternative splicing (Kolasinska-Zwierz et al. 2009; Luco et al. 2010). Several histone modifications can engage in crosstalk with other histone modifications, in either cis or trans. One well-known example of histone crosstalk in trans is the requirement of H2B C terminus monoubiquitylation (H2B monoubiquitylation, H2Bub1) for the multiple methylations of H3K4 and H3K79 (Dover et al. 2002; Sun and Allis 2002; Shahbazian et al. 2005), which is evolutionarily conserved from yeast to human. In Saccharomyces cerevisiae, monoubiquitylation of lysine residue 123 in histone H2B was shown to play a positive role in H3K4 and H3K79 methylations, which are involved in transcription initiation and elongation, respectively (Briggs et al. 2002; Sun and Allis 2002). In mammals, H2Bub1 coupled to RAD6-mediated transcription has been reported to directly stimulate H3K4 methylation (Kim et al. 2009), and synthetically ubiquitylated H2B stimulates DOT1L-mediated H3K79 methylations in vitro (McGinty et al. 2008; Kim et al. 2009; Oh et al. 2010). Despite many efforts to infer the roles of histone modifications, however, the crosstalk between H2Bub1 and the multiple methylations of H3K4 and H3K79 is not fully understood, especially in mammals.
The signaling between H2Bub1 and histone H3 methylation is clearly linked to chromatin dynamics during transcription elongation, but the manner with which H2Bub1-containing nucleosomes regulate chromatin architecture independently of histone H3 methylation during transcription elongation is unclear. In this regard, Fleming et al. (2008) found that H2Bub1 plays specific roles in GAL1/10 gene expression in budding yeast, and these roles are independent of its regulation of H3 methylation. In addition, Pavri et al. (2006) demonstrated that H2Bub1 directly stimulates FACT (FAcilitates Chromatin Transcription)-mediated transcription using an in vitro chromatin-based transcription system. The ubiquitin moiety of histone H2B is removed by the deubiquitylases, UBP8 and UBP10 (Weake and Workman 2008). Loss of the H2B E2/E3 ubiquitin ligase RAD6/BRE1 or substitution of the H2B C-terminal lysine residue with arginine leads to transcription defects, suggesting that the dynamic turnover of H2Bub1 is critical to both transcription and gene silencing. Recent studies have proposed that H2Bub1 regulates chromatin dynamics by enhancing nucleosome stability (Chandrasekharan et al. 2009) and disrupts local and higher-order chromatin compaction (Fierz et al. 2011). These findings strongly suggest that H2Bub1 is involved in additional regulatory pathways beside trans-crosstalk with the methylations of histone H3. To understand the genome-wide characteristics of H2Bub1 in mammals, Minsky et al. (2008) developed a mammalian H2Bub1-specific monoclonal antibody. Subsequent use of ChIP-chip and ChIP-seq analysis showed that H2Bub1 is associated with the transcribed regions of highly expressed genes in HeLa cells (Shema et al. 2008). However, the prior data are insufficient for an investigation of the genome-wide properties and unique roles of H2Bub1 in cooperation with other modifications. Therefore, we herein generated high-throughput sequencing (ChIP-seq) data for the genome-wide occupancies of H2Bub1, nucleosome, H3K4me3, H3K36me3, H3K79me1/2/3, H3ac, and mRNA-seq in human embryonic carcinoma (NCCIT) cells. Furthermore, to investigate the effects of H2Bub1 reduction, we performed mRNA-seq and ChIP-seq of H2Bub1 in NCCIT cells transfected with RNF20-small interfering RNA (siRNF20), which decreased the H2Bub1 signal. We found that H2Bub1 is a 5′-enriched active transcription mark and that the methylations of H3K79 co-occupied with H2Bub1 in highly expressed genes. Notably, H2Bub1 was strongly enriched at the exon–intron boundaries of highly expressed exons and appeared to correlate with exon skipping.
Results
H2Bub1 is a 5′-enriched active transcription mark
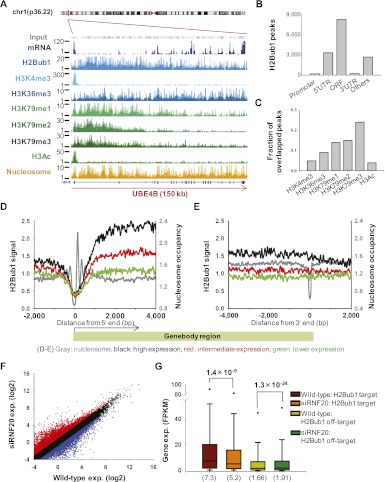
To determine genome-wide patterns of histone modifications, including histone H2B monoubiquitylation, we carried out ChIP-seq analysis using undifferentiated human teratocarcinoma NCCIT cells. NCCIT cells were established from a mediastinal mixed germ cell tumor (Teshima et al. 1988) at an intermediate stage between seminoma and embryonal carcinoma (Damjanov 1993). NCCIT is a developmentally pluripotent cell line that can differentiate into derivatives of all three embryonic germ layers and extra-embryonic cell lineages (Damjanov et al. 1993). H2Bub1 demonstrated relatively strong signals near the 5′ regions of actively transcribed genes, and then gradually decreased along the transcribed region; this is shown for the UBE4B gene (ubiquitin conjugation factor E4 B) in Figure 1A, as an example. Similar patterns were also observed for H3K79 methylations, while H3K4me3 and H3ac were enriched in the regions surrounding the transcriptional start sites (TSS). These patterns, as well as all other observed modification patterns, were consistent with previously reported results in human cells (Barski et al. 2007; Wang et al. 2008).
Figure 1.
Varying genomic properties of the H2Bub1-enriched regions. (A) The ChIP-seq results for UBE4B (ubiquitin conjugation factor E4B) are shown. (Y-axis) Read counts of the sequence tags in 30-bp increments. H2Bub1 was enriched at the 5′ end and gradually decreased along the transcribed region, similar to the methylation pattern of H3K79. (B) MACS analysis recognized 14,679 H2Bub1 peaks and assigned them to 4173 genes. (C) The H2Bub1 peaks overlapped with the H3K79me3 peaks (24% of the H3K79me3 peaks) in 2765 genes. (D,E) H2Bub1 signals in the regions surrounding the transcription start site (TSS) and transcription end site (TES). (Y-axis) H2Bub1 signal; (x-axis) relative position (bp). Nucleosomes were well positioned in the regions surrounding the TSS, while H2Bub1 signals were depleted in these regions. The gene expression levels were divided into three groups: highly expressed (black lines, top 30%), lower expressed (green lines, bottom 30%), and intermediately expressed (red lines, middle 40%). (F) Comparison of gene expression changes between wild-type and siRNF20-transfected NCCIT cells. (Red and blue) Genes demonstrating twofold or more up-regulation and down-regulation, respectively. (G) Gene expression changes among the H2Bub1-targeted genes. Among ∼25,000 genes, 3824 were H2Bub1-targeted genes, and the remaining 21,310 genes were defined as H2Bub1 off-targeted genes. P-values were calculated using the Kolmogorov-Smirnov (KS) test (Stephens 1970). Parentheses indicate median values.
To infer the genomic regions that showed H2Bub1 enrichment, we detected H2Bub1 peaks using the MACS peak detection software (see Supplemental Methods). In agreement with the 5′-enriched pattern we had observed for H2Bub1, most of the H2Bub1 peaks were located in the 5′ UTR and open reading frame (ORF) regions (Fig. 1B). This result suggests that the specific function of H2Bub1 is localized to transcribed regions, not promoter regions, further indicating that H2Bub1 may play a role in potentiating transcriptional elongation. The H2Bub1 peaks highly overlapped with the H3K79 methylation peaks; ∼24% of the H3K79me3 peaks and H2Bub1 peaks matched, while H3ac and H3K4me3 had similar numbers of peaks (Fig. 1C). Gene set analysis using DAVID indicated that genes doubly targeted by both H2Bub1 and H3K79 methylation peaks were enriched in protein modification, metabolism, and mRNA-related biological functions (Supplemental Fig. 1A).
Since previous studies have suggested that H2Bub1 is associated with highly expressed genes (Minsky et al. 2008; Shema et al. 2008), we investigated whether H2Bub1 is a transcription-dependent active mark in human cells. H2Bub1 demonstrated strong signals in highly expressed genes (Fig. 1D, black line), and these signals decreased along the transcribed regions (Fig. 1E). The H2Bub1 signal showed a more than twofold enrichment in highly expressed genes compared with lower-expressed genes. To examine whether the strong H2Bub1 signal was associated with increased gene activation, we performed mRNA-seq for siRNF20-transfected NCCIT cells. Our results revealed that H2Bub1, H3K4me3, and H3K79me3 levels decreased in the RNF20 knockdown cells (Supplemental Fig. 2). Since the H3K4me3 and H3K79 methylations, which are known to function as gene activation signals, decreased with the reduction of H2Bub1 signal, we hypothesized that the highly expressed genes targeted by H2Bub1 were expected to demonstrate down-regulation. However, using a twofold expressional change as a cutoff value, we found that although 4654 genes were down-regulated in siRNF20-transfected NCCIT cells, a comparable number (5304) were up-regulated (Fig. 1F).
To investigate the biological functions of specifically regulated genes in siRNF20-transfected NCCIT cells, we conducted gene set analysis (Supplemental Fig. 1B). According to the GO (Gene Ontology), the down-regulated genes were enriched for cell cycle-related functions related to tumor suppression. This result is consistent with the previous suggestion that H2Bub1 may function as a tumor suppressor (Shema et al. 2008). The up-regulated genes were also enriched for developmental regulation-related genes, suggesting a new possible role for H2Bub1 in developmental processes. The number of up-regulated genes observed in siRNF20-transfected NCCIT cells also raises the possibility that H2Bub1 may have an additional role in gene regulation beyond its crosstalk with H3 lysine methylations. However, although many genes were up-regulated in siRNF20-transfected NCCIT cells, the H2Bub1-targeted genes tended to be significantly decreased (Fig. 1G, brown and orange), while those not targeted by H2Bub1 tended to be slightly increased (Fig. 1G, yellow–green and green). The presence of gene expression-dependent H2Bub1 patterns and more significant changes in H2Bub1-targeted genes in the siRNF20-transfected NCCIT cells suggest that H2Bub1 is a 5′-enriched gene activation mark.
Trans-crosstalk between H2Bub1 and H3K79 methylations is associated with gene expression
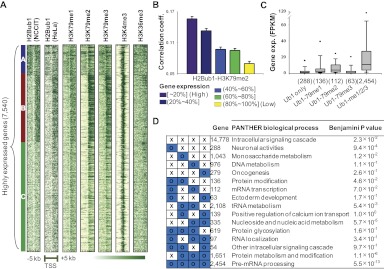
H2Bub1 has demonstrated trans-crosstalk with the methylations of H3K4 and K79 in a variety of organisms ranging from yeast to human (Dover et al. 2002; Sun and Allis 2002; Kim et al. 2005, 2009; Shahbazian et al. 2005). We verified these relationships in NCCIT cells (Supplemental Fig. 2), and then set out to investigate whether this trans-crosstalk generated genome-wide co-occupation patterns and/or was associated with gene activation. We first clustered the H2Bub1 patterns in the regions surrounding the TSS (including 5 kb upstream/downstream from the TSS) using k-means clustering (see Supplemental Methods). We found three distinct patterns (Fig. 2A). After sorting genes according to the H2Bub1-clustered results in NCCIT cells, we compared our results with those for H2Bub1 in HeLa cells (Shema et al. 2008) as well as those for H3K79me1/2/3, H3K4me3, and H3K36me3 in NCCIT cells (Fig. 2A). The H2Bub1 clustering patterns were fairly consistent between NCCIT and HeLa cells, indicating that these cluster patterns are unlikely to be cell-type specific. The methylations of H3K79 demonstrated genome-wide correlation patterns with H2Bub1. However, although H2Bub1 is a prerequisite for H3K4me3 (Kim et al. 2009), H3K4me3 did not highly overlap with H2Bub1. The trimethylation patterns of H3K36 (used here as a control) were independent of H2Bub1. These results were consistent with the low number of overlapping peaks between H2Bub1 and H3K4me3/H3K36me3 (Fig. 1C). The discrepancy between H3K4me3 and H3K79me1/2/3 suggests that H3K4me3 is affected by other pathways in addition to an H2Bub1-dependent pathway; these might include crosstalk with the lysine acetylations of H3 (Hung et al. 2009), counter-correlation with H3R2 methylation (Guccione et al. 2007), and several other related pathways. Clearly, the bidirectional pattern of H3K4me3 in the regions surrounding the TSS and the broader distribution of H2Bub1 in transcribed regions provide strong evidence that there is a discrepancy between H2Bub1 and H3K4me3. However, the hDOT1L-mediated H3K79 methylations are tightly regulated by H2Bub1 in the same nucleosome (McGinty et al. 2008). In agreement with these clustering patterns, the H3K79 methylations and H2Bub1, but not H3K4me3, showed strongly correlated patterns of average modification signals in the gene body regions (Supplemental Fig. 3A–E).
Figure 2.
Genome-wide correlation patterns between H2Bub1 and H3K79 methylations. (A) Three distinct H2Bub1 patterns were identified for highly expressed genes (top 30%, 7540) in the regions surrounding the TSS, as assessed by k-means clustering. Other modification patterns are presented in the gene order shown for H2Bub1. Cluster A shows strongly enriched bidirectional patterns, while both clusters B and C were enriched downstream from the TSS. (B) The correlation patterns between H3K79me2 and H2Bub1 affected gene activation. Genes were divided into five groups according to their gene expression levels (purple: highly expressed genes; yellow: lower expressed genes). Pearson correlation coefficients were calculated between H3K79 methylations and H2Bub1 within each gene. (Y-axis) Average correlation coefficient of each group. (C) Boxplots of gene expression levels in FPKM scores for different combinations of H2Bub1- and H3K79-methylation-targeted genes. Modification-targeted genes were defined as those containing at least one corresponding modification peak. (Values in parentheses) Number of genes. (D) Gene set analysis for all combinations of H2Bub1- and H3K79 methylation-targeted genes using DAVID. PANTHER biological processes are presented with corrected P-values.
Since H2Bub1 demonstrated genome-wide correlations with the methylations of H3K79, which are regarded as gene activation marks, we investigated whether this genome-wide correlation pattern was associated with changes in gene activation. If trans-crosstalk between H2Bub1 and H3K79 methylations is important in gene activation, H3K79 methylations should show strong correlations with H2Bub1 in highly expressed genes. Thus, gene expression levels and correlation coefficients were compared between H2Bub1 and the methylations of H3K79 in the gene body regions. Among the mono-, di-, and trimethylations of H3K79, dimethylation showed the most significant relationship, with highly expressed genes (purple, top 20%) demonstrating more than twofold higher correlation coefficients with these genes compared with the lower-expressed genes (Fig. 2B). This tendency was maintained for the mono- and trimethylations, suggesting that trans-crosstalk between H2Bub1 and the methylations of H3K79 is related to gene activation (Supplemental Fig. 3F,G).
To test the effect of trans-crosstalk on gene activation, we extended our analysis to genes that are cotargeted by H2Bub1 and the methylations of H3K79. Using the modification peaks defined in Figure 1, B and C, we identified genes that were solely targeted by H2Bub1, those that were doubly targeted (H2Bub1-H3K79me1, H2Bub1-H3K79me2, and H2Bub1-H3K79me3), and those that were simultaneously targeted (H2Bub1-H3K79me1/2/3). We found that the H2Bub1-targeted genes tended to be simultaneously cotargeted by H3K79 mono-, di-, and trimethylations (around 60% of the 4173 H2Bub1-targeted genes). Furthermore, the genes that were simultaneously targeted (H2Bub1-H3K79me1/2/3) were highly activated compared with H2Bub1 solely or doubly targeted genes (Fig. 2C). In agreement with this result, high signals for both H2Bub1 and H3K79 methylations in the gene body regions were associated with high gene expression levels (red spots in Supplemental Fig. 3C–E).
Despite this apparent trans-crosstalk between H2Bub1 and H3K79 methylations, however, it was clear that genes were targeted by different combinations of these modifications. In this regard, we wondered what properties might be associated with different combinatorial patterns of these modifications. All 16 possible gene groups were generated according to the presence of each modification and gene set analysis was conducted. Each combinatorial pattern was characterized by distinct biological functions (Fig. 2D).
H2Bub1 is highly enriched at the exon–intron boundaries of highly expressed exons
Recent yeast studies showed that C-terminal mutants of H2B (K123R in budding yeast and K119R in fission yeast) demonstrated more severe phenotypes and modulated transcription among more genes compared with H3K4R or H3K79R mutants, indicating that H2Bub1 plays one or more roles in transcription elongation independent of its regulation of histone H3 methylation in yeast (Tanny et al. 2007; Fleming et al. 2008). Since several papers have emphasized the role of H2Bub1 in modulating nucleosome architecture (Chandrasekharan et al. 2009; Batta et al. 2011; Fierz et al. 2011; Schulze et al. 2011), we herein sought to identify correlation patterns between H2Bub1 and nucleosome architecture.
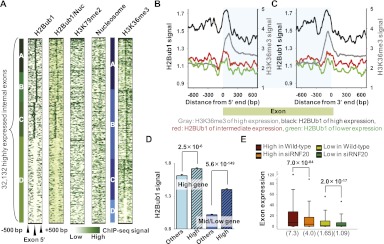
First, we focused on the relationship between H2Bub1 signals and exon–intron structure. To investigate the various modification patterns in the regions surrounding the exon–intron boundaries, we used k-means clustering (see Supplemental Methods). As shown in Figure 3A, four distinct patterns were clearly identified in the exon boundary regions (encompassing 500 bp upstream of and downstream from the exon 5′ end): high density in the central region (cluster A), slightly biased to the exonic region (cluster B), slightly biased to the intronic region (cluster C), and low density in the central region (cluster D).
Figure 3.
H2Bub1 is enriched at the exon boundaries. Internal exons separated from flanking exons by at least 500 bp (n = 107,107) were considered. (A) Clustered exon boundary patterns 500 bp upstream of and downstream from the 5′ end of the exons are shown. H2Bub1/Nuc indicates the H2Bub1 patterns obtained following normalization with respect to nucleosome occupancy. (B,C) Average H2Bub1 signals at the exon 5′ and 3′ ends according to exon expression levels. Similar to the H3K36me3 signals of highly expressed exons (gray lines), H2Bub1 was enriched at the exon boundaries of highly expressed exons (black lines). (X-axis) Relative position (bp) from each 5′ and 3′ end. (D) Average H2Bub1 signals of highly expressed exons in both highly expressed genes and middle/lower expressed genes are shown with standard deviation error bars (y-axis). (High) Highly expressed exons (top 30%) in each gene expression group; (Others) remaining exons. P-values were calculated using the Student's t-test. (E) Boxplots of exon expression changes between wild-type and siRNF20-transfected NCCIT cells. The H2Bub1-enriched (top 30%, n = 32,132) and H2Bu1-depleted (bottom 30%, n = 32,132) exons were defined according to the H2Bub1 signal for each exon. The parentheses indicate median values.
This property of H2Bub1 was also observed in HeLa cells (Supplemental Fig. 4A). Because exon boundaries are also nucleosome enriched (Andersson et al. 2009; Tilgner et al. 2009), H2Bub1 density was normalized with respect to nucleosome occupancy (see Supplemental Methods). H2Bub1 was still enriched at the exon boundaries after this normalization (Fig. 3A, H2Bub1/Nuc), implying that this enrichment was not attributable to nucleosome occupancy. Unlike H2Bub1, no patterns were recognized in H3K4me3 and H3K79 methylations (Fig. 3A; Supplemental Fig. 4A), demonstrating that the enrichment of H2Bub1 at the exon boundaries was independent of crosstalk with H3 methylations. We also confirmed that H3K36me3 and nucleosome were enriched at the exon boundaries (Fig. 3A) as previously reported (Kolasinska-Zwierz et al. 2009; Schwartz and Ast 2010).
Next, we examined whether the degree of H2Bub1 enrichment depended on the expression levels of exons. Indeed, highly expressed exons showed stronger enrichment at both the 5′ and 3′ ends of the exon boundaries (Fig. 3B,C, black lines). However, in intermediately or lower-expressed exons, H2Bub1 demonstrated only very slight enrichment or no specific patterns at the exon boundaries (Fig. 3B,C, red and green lines). Similar results were obtained in HeLa cells (Supplemental Fig. 4B,C) and mouse myoblast cells (Vethantham et al. 2012; Supplemental Fig. 4D,E). Interestingly, Pearson correlation coefficient showed a slight positive correlation between the H2Bub1 signals and exon expression levels (r = 0.05, P = 4.2 × 10−53). However, this was less of a correlation than that of H3K36me3 (r = 0.19, P = 0) (data not shown), suggesting that H2Bub1 is important in highly expressed exons, but that it is less prominent than H3K36me3 in determining exon expression levels.
It is possible that the high H2Bub1 densities at the boundaries of highly expressed exons are just the result of the overall higher level of H2Bub1 throughout the bodies of highly expressed genes compared with intermediately or lower expressed genes (Fig. 3D). However, regardless of the gene expression levels, highly expressed exons showed stronger H2Bub1 signals (Fig. 3D, High) compared with the remaining exons (Fig. 3D, Others). This tendency was more significant in the intermediately or lower-expressed genes (Fig. 3D, blue bars), supporting the notion that H2Bub1 enrichment plays a unique potentiating role in highly expressed exonic regions.
To validate the role of H2Bub1 in highly expressed exons, we used wild-type and siRNF20-transfected NCCIT cells, and compared the expression levels among exons with high H2Bub1 signals (top 30% of exons according to H2Bub1 signal) and those with low H2Bub1 signals (bottom 30% of exons). The expression levels of exons with high H2Bub1 signals significantly decreased when H2Bub1 expression was reduced by siRNF20 transfection (Fig. 3E, brown and orange). In contrast, the expression levels of exons with low H2Bub1 signals decreased less significantly in siRNF20-trasnfected NCCIT cells (Fig. 3E, yellow-green and green). Thus, our findings collectively suggest that H2Bub1 is strongly enriched at the exon boundaries of highly expressed exons and that this property is evolutionarily well conserved in mammals.
We next examined whether distinct H2Bub1 patterns at the exon boundaries (Fig. 3A) could be associated with exon–intron structure. When patterns of exon length, gene strand, upstream intron length, and exon position were analyzed according to clusters, only upstream/downstream intron length and exon position showed cluster-specific patterns. Exons located following short upstream/downstream introns tended to be included in cluster A or D compared with cluster B and C (Supplemental Fig. 4F,G). Additionally, more exons near the 3′ end tended to be assigned to cluster D (Supplemental Fig. 4H). This tendency may be associated with the 5′-bias of the H2Bub1 pattern. Most of the exons in cluster D were marked by H3K36me3 (84% of the exons in cluster D were included in clusters A–C of H3K36me3). These data indicate that H3K36me3 may play a complementary role in exons unmarked by H2Bub1 (Supplemental Fig. 4I).
H2Bub1 is highly enriched in skipped exons in mammals
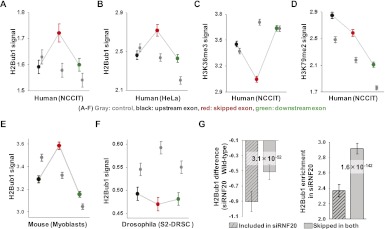
Previous studies reported that two exon determination marks, nucleosome and H3K36me3, are less prominent in skipped exons (Hon et al. 2009; Choi 2010), and an association between histone modifications and alternative splicing has been experimentally validated (Luco et al. 2010). Since H2Bub1 is also enriched at exon boundaries, we compared H2Bub1 signals between skipped exons and flanking exons (see Methods) to determine whether H2Bub1 was correlated with skipped exons. Interestingly, H2Bub1 signals were higher for skipped exons in human cells (Fig. 4A,B). Consistent with previous results, reduced nucleosome and H3K36me3 signals were also observed in the skipped exons (Fig. 4C; Supplemental Fig. 5A), whereas no specific pattern was observed for the control, H3K79me2 (Fig. 4D), or H3K4me3/H3K79me1/H3K79me3 (data not shown). To test whether this result was specific to human cells, genome-wide H2Bub1 data from mouse and Drosophila cells were also analyzed. Notably, an evolutionarily conserved pattern was observed in the mouse (Fig. 4E), while an opposing result (i.e., reduced signals in skipped exons) was found in Drosophila cells (Fig. 4F). Exon skipping was also correlated with the clustered patterns of H2Bub1 (Fig. 3A), as skipped exons were slightly enriched in clusters A and B compared with clusters C and D (Supplemental Fig. 5B). Interestingly, the tendency of H2Bub1 specificity in skipped exons was stronger in clusters A and B compared with clusters C and D (Supplemental Fig. 5C). These cluster-dependent patterns support a putative H2Bub1 role in exon skipping, since clusters A and B include exons enriched by H2Bub1 in their exonic regions.
Figure 4.
Skipped exons have a stronger H2Bub1 signal than flanking exons. (A–F) H2Bub1, H3K36me3, and H3K79me2 signals in skipped exons and their flanking exons. (Gray) Three constitutive exons used as controls. In the control, H2Bub1 shows decreased patterns from 5′ to 3′, except in Drosophila (n = 16,724 in NCCIT). (Red, black, green) Skipped exon, upstream of the skipped exon, and downstream from the skipped exon, respectively. (Y-axis) Average modification signals with standard deviation error bars. In NCCIT (n = 12,337), HeLa (n = 9722), and myoblast (n = 12,356) cells, the H2Bub1 signals were higher in skipped exons than in flanking exons. In Drosophila, however, the H2Bub1 signal was lower in skipped exons than in flanking exons (n = 3201). The H3K36me3 signal was lower in skipped exons and H3K79me2 did not show any specific pattern in the skipped exons. (G) Differences in H2Bub1 between wild-type and siRNF20-transfected NCCIT cells for persistently skipped exons and included exons with standard deviation error bars (left). H2Bub1 enrichment is shown for both persistently skipped exons and included exons in siRFN20-transfected cells with standard deviation error bars (right). H2Bub1 enrichment was determined by dividing the H2Bub1 tag density by the average tag density across the whole genome (see Methods). P-values were calculated using the KS-test.
To validate the association between H2Bub1 and exon skipping, the number of skipped exons was measured in siRNF20-transfected NCCIT cells. Of the 34,763 exons skipped in wild-type cells, 21,804 were persistently skipped in siRNF20-transfected NCCIT cells, while the remaining 12,959 exons were included. Thus, the knockdown of RNF20 expression elicited changes to 37% of the skipped exons in wild-type cells, and this effect may be attributable to a reduction of H2Bub1 signals. When we compared H2Bub1 signals between wild-type and siRNF20-transfected cells, the persistently skipped exons showed lower degrees of H2Bub1 reduction (Fig. 4G, left). A similar pattern was observed when we compared H2Bub1 signals between siEGFP-transfected NCCIT cells (control) and siRNF20-trnasfected NCCIT cells (Supplemental Fig. 5D). In the siRNF20-transfected NCCIT cells, the included exons showed relatively lower H2Bub1 signal enrichments compared with the persistently skipped exons, suggesting that increased H2Bub1 signals are correlated with exon skipping (Fig. 4G, right).
To explain the persistent skipping of exons despite reductions in the H2Bub1 signal, we hypothesized that other modifications, such as nucleosome occupancy and H3K36me3, may compensate in exon skipping. The patterns of H2Bub1 and nucleosome in wild-type cells seemed to be independent of whether the exons were persistently skipped or not (Supplemental Fig. 5F,G). However, the H3K36me3 signals seemed to correlate more strongly with the persistently skipped exons (green lines in Supplemental Fig. 5E, 15% decreased H3K36 signals compared with flanking exons) compared with included exons (blue lines in Supplemental Fig. 5E, 8% decreased H3K36me3 signals compared with flanking exons), suggesting that H3K36me3 may compensate for the reduction of H2Bub1. Therefore, various chromatin features should be considered together when we seek to elucidate the effect of H2Bub1 on exon skipping. Although there are many limitations to this analysis, our results strongly support the idea that H2Bub1 plays a potentiating role in exon skipping.
Specific H2Bub1 properties in mammals are associated with exon skipping
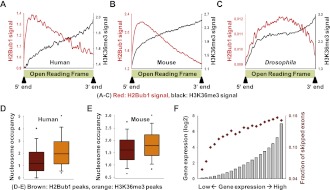
The correlative patterns between H2Bub1 signals and exon skipping raised many interesting questions. To examine the discrepancy of H2Bub1 patterns in skipped exons between mammals and Drosophila, we investigated H2Bub1 patterns in ORF regions in human, mouse, and fly cells. In NCCIT cells, H2Bub1 and H3K36me3 demonstrated opposing patterns, in that H2Bub1 was biased to the 5′ end and H3K36me3 was biased to the 3′ end (Fig. 5A). A similar pattern was observed in mouse cells (Fig. 5B). In Drosophila cells, however, H3K36me3 demonstrated a pattern similar to that in human and mouse cells (Gan et al. 2010), while H2Bub1 was enriched in central ORF regions (Fig. 5C; Kharchenko et al. 2010). The central enrichment of H2Bub1 has also been reported in Arabidopsis (Roudier et al. 2011). This phenomenon may suggest that the 5′ bias of H2Bub1 in mammals results from a gain-of-function during evolution. In light of the observed discrepancy in exon skipping between mammals and fly, we suspect that different mechanisms underlie the enrichment of H2Bub1 in ORF regions, leading to different correlation patterns with exon skipping.
Figure 5.
Unique properties of H2Bub1 in mammals. (A–C) Average H2Bub1 and H3K36me3 patterns of ORF regions in various organisms are shown. In mammals, H2Bub1 density was high at the 5′ end regions and rapidly decreased along the transcribed regions (red lines). In Drosophila cells, in contrast, H2Bub1 was enriched in the central ORF regions. (D,E) Boxplots of nucleosome occupancy for H2Bub1 and H3K36me3 peaked regions. The P-values between H2Bub1 and H3K36me3 were zero for both human and mouse, as assessed using the KS test. (F) Average gene expression levels (bar graph) and fractions of skipped exons (red diamonds) are shown for 20 subgroups equally divided according to their gene expression levels.
With regard to the opposing correlative patterns between H2Bub1 and H3K36me3 in skipped exons, we hypothesized that these may arise through differences in the regulation of the nucleosome architecture. A recent study reported that H2Bub1 disrupts local and higher-order chromatin compaction, indicating that it plays significant roles in nucleosome architecture (Fierz et al. 2011). In yeast, a functional interplay between SPT16 and H2Bub1 regulates nucleosome reassembly in GAL1 gene (Fleming et al. 2008), and dynamic regulation of H2B ubiquitylation/deubiquitylation could be equally important for transcription (Henry et al. 2003; Daniel et al. 2004; Weake and Workman 2008; Song and Ahn 2010). Notably, our data indicate that the H2Bub1 peak-containing regions demonstrated reduced nucleosome occupancy compared with H3K36me3 (Fig. 5D; see Supplemental Methods). The mouse genome also demonstrated a similar pattern (Fig. 5E). Thus, in the wake of transcription, H2Bub1 and H3K36me3 may affect nucleosome dynamics in different ways, potentially leading to opposing correlative patterns of exon skipping.
The enrichment of H2Bub1 in skipped exons (Fig. 4A,B,E) seemed to be counterintuitive, given that the H2Bub1 signals were enriched in highly expressed exons (Fig. 3B,C). A specific set of histone modifications (including H2Bub1) may cause a well-defined “exonic nucleosome” during transcription elongation (Keren et al. 2010). The more frequent occurrence of exon skipping in highly expressed genes (Fig. 5F) may be one reason for the seemingly counterintuitive pattern observed herein. We expect that the more complex functions of H2Bub1 may be linked to exon skipping in a context-dependent manner.
Discussion
The findings in this study suggest that H2B monoubiquitylation plays several roles during transcription, acting in both H3 methylation-dependent and -independent fashions. First, H2Bub1 is a 5′-enriched active transcription mark in the human genome. Second, H2Bub1 has genome-wide correlation patterns with the H3K79 methylations and co-occupancies between H2Bub1, and the methylations of H3K79 are associated with highly expressed genes. Third, H2Bub1 is strongly enriched at the exon boundaries of highly expressed exons. Fourth, H2Bub1 displays an increased density in skipped exons. Together, these results define additional functions for H2B monoubiquitylation during active transcription in humans.
One interesting finding of this study is that combinatorial patterns of H2Bub1 and H3K79 methylations are characterized by distinct biological functions. Although co-occupancies between H2Bub1 and the methylations of H3K79 seemed to be strongly associated with pre-mRNA processing-related genes (Supplemental Fig. 1A), the functions of these genes differed according to the combinatorial patterns (Fig. 2D). With respect to gene expression levels, H3K79me2 showed a stronger tendency for cooperation with H2Bub1 compared with H3K79 mono- and trimethylations (Fig. 2B; Supplemental Fig. 3F,G). Although DOT1L has been established to modify all H3K79 methylations, our results suggest that the multiple methylations of H3K79 may play distinct roles in different contexts.
In terms of genome-wide occupancy, the ability of H2Bub1 to stimulate H3K79 methylations is evolutionarily conserved from yeast to human. Therefore, like H2Bub1, the methylations of H3K79 are expected to be enriched surrounding the intron–exon boundaries. However, our present results suggest that H2Bub1 has a more unique role in determining exon–intron structure. A recent study suggested that the effects of H2Bub1 in nucleosome stability and structure may be related to the enrichment of H2Bub1 in intron–exon boundaries (Chandrasekharan et al. 2009). Given all of the available evidence, the present study sheds light on the role of H2Bub1 in exon–intron structure, suggesting that it helps mediate complex splicing mechanisms in higher eukaryotes, independent of the crosstalk between H2Bub1 and the methylations of H3K79.
Two models explain the mechanism of exon and intron selection: intron definition and exon definition (Keren et al. 2010). Placing the basal splicing machinery across introns is regarded as the ancient mechanism. In mammals, however, although the length of the exons has remained relatively constant, intron length has gradually expanded. As opposed to fungi, in which introns have remained short and are thought to be the recognized unit for selection (i.e., intron definition), higher eukaryotes use exon definition as their major mechanism for selection. In this regard, H2Bub1 can be hypothesized to have achieved an uncharacterized function in exon definition due to evolutionary pressure. Further studies will be required to validate our hypothesis through comparative genomic approaches.
Ubiquitylation, which results in the addition of a bulky moiety to lysine residues within proteins, exerts a dramatic effect on protein conformation and function. Monoubiquitylation of the H2B C terminus affects chromatin structure in vivo, thereby modulating transcription elongation (Weake and Workman 2008). Here, we found that histone H2Bub1 is enriched at exon boundaries (Fig. 3; Supplemental Fig. 4) and correlates with exon skipping (Fig. 4). According to the “speed bump” model, the speed of RNA PolII determines the binding of splicing factors during transcription. In this model, the speed of PolII is regulated by various barriers, including high-nucleosome density, DNA methylation, and several types of histone modifications (Keren et al. 2010; Schwartz and Ast 2010). All of the present correlative findings reinforce the notion that H2Bub1 potentiates alternative splicing. Moreover, we found that the methylations of H3K4 and H3K79 are not tightly related to exon–intron structure or exon skipping in human cells, suggesting that H2Bub1 plays a unique role in chromatin architecture, independent of the methylations of histone H3.
Methods
Antibodies
Polyclonal rabbit antibodies recognizing H3ac, H3K4me3, H3K36me3, H3K79me1, H3K79me2, and H3K79me3 were purchased from Abcam or produced in-house as previously described (Oh et al. 2010). Antibodies against H2Bub1 were obtained from Millipore (17-650) and Cell Signaling (5546). The antibody specificity of the anti-H2Bub1 from Millipore is shown in Supplemental Figure 6. The anti-H2Bub1 from Cell Signaling was also confirmed (data not shown).
Cell culture, chromatin immunoprecipitation (ChIP) assay, and mononucleosome purification
The NCCIT embryonic carcinoma cell line was cultured on 10-cm dishes in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% antibiotic-antimycotic solution (Hyclone) at 37°C in a humidified atmosphere containing of 5% CO2.
Cross-linking was performed by incubating cells in 1% formaldehyde (Sigma) for 20 min at 25°C and then quenching cells with 125 mM glycine. For ChIP, phosphate buffered saline (PBS)-washed cells were lysed in 400 μL of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.0), and DNA was sonicated to create fragments 400–500 nt in length using a Sonic Dismembrator (Model 500, Fisher). The chromatin was cleared by centrifugation for 15 min at 13,000 rpm and 4°C and subjected to immunoprecipitation for 2 h at 4°C using the indicated antibodies and a mixture of protein A and G agarose (GE Healthcare). The protein A and G agarose/antibody/chromatin complexes were washed for 10 min each with the following solutions: low-salt wash buffer (20 mM Tris-Cl at pH 8.0, 0.1% SDS, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl), high-salt wash buffer (20 mM Tris-Cl at pH 8.0, 0.1% SDS, 2 mM EDTA, 1% Triton X-100, 500 mM NaCl), LiCl buffer (10 mM Tris-Cl at pH 8.0, 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholic acid), and TE buffer (10 mM Tris-Cl at pH 8.0, 1 mM EDTA). The chromatin complexes were eluted twice in 250 μL of elution buffer (1% SDS, 0.1 M NaHCO3) by incubation at 25°C for 15 min with rotation, and then reverse cross-linked by heating overnight at 68°C with 400 mM NaCl.
For mononucleosome purification, PBS-washed cells were lysed in 400 μL of lysis buffer (18% Ficoll-400 [Sigma F2637], 10 mM KH2PO4, 10 mM K2PO4, 1 mM MgCl2, 0.25 mM EGTA, and 0.1 mM PMSF) and centrifuged at 4°C for 15 min at 13,000 rpm. The nuclear pellets were resuspended with 1 mL of Buffer A (10 mM Tris-Cl at pH 7.4, 150 mM NaCl, 5 mM KCl, 1 mM EDTA, 0.1 mM PMSF), and then mixed with 5 μL of CaCl2 (final concentration, 5 mM) and 1.5 μL of MNase (500 U/2.5 μL, NEB). After incubation at 37°C for 5 min, the reactions were stopped by adding 100 μL of 1% SDS and 50 μL of 0.5 mM EDTA. The MNase-digested nuclei were reverse cross-linked by heating overnight at 68°C with 350 mM NaCl. Finally, the reverse cross-linked samples were treated with RNase A and proteinase K (Sigma), isolated by phenol/chloroform extraction, and precipitated using 3 M sodium acetate (pH 5.2) with absolute ethanol.
siRNA and transfection
A 21-nt synthetic siRNA duplex designed to inhibit expression of RNF20 (sense, 5′-ACAUCCGUAUCAUCCUUAAUU-3′, and antisense, 5′-UUAAGGAUGAUACGGAUGUUU-3′) was synthesized and purified by Samchully Pharm. The day before transfection, NCCIT cells were seeded at a density of 2 × 105 cells/well in 6-well plates. siRNF20 was then transfected into the cells using FuGene HD (Roche) according to the manufacturer's instructions. Three days after transfection, the samples were separated by SDS-PAGE and subjected to immunoblot analysis using the indicated antibodies.
Illumina library construction and sequencing
The purified ChIP DNA fragments were ligated to a pair of adaptors designed to enable sequencing on the Illumina Genome Analyzer. The ligation products were size-fractionated on an agarose gel, and the 200–300-bp fragments were subjected to 18 cycles of PCR amplification. Cluster generation and 72 cycles of single-read sequencing were performed.
The mRNA sequencing library was generated using the mRNA-seq sample prep kit (Illumina). Polyadenylated mRNA was purified from 10 μg of total RNA using Sera-mag oligo(dT) beads (Illumina), and then fragmented by incubation with fragmentation buffer (Illumina) at 94°C for 5 min. First-strand cDNA was synthesized using SuperScript II (Invitrogen) and random primers (Illumina), and second-strand cDNA was synthesized using RNaseH and DNA polymerase I (Illumina). The resulting cDNA fragments were ligated to a pair of adaptors designed for sequencing on the Illumina Genome Analyzer. The ligation products were size-fractionated to obtain 200-bp fragments on a 2% agarose gel and subjected to 15 cycles of PCR amplification. Cluster generation and 72 cycles of single-read sequencing were performed.
Data processing
The sequence tags were mapped to the human genome (UCSC hg18) using Bowtie (Langmead et al. 2009) with default options. The mapped sequence reads were extended to the average fragment length (200 bp for ChIP-seq and 150 bp for MNase-seq), and the number of overlapping sequence tags was determined using 30-bp windows with 15-bp steps across the human genome. The ChIP-seq signal was defined as the ratio of target read count/target size divided by total read count/genome size (Jung and Kim 2009). The mRNA-seq data were mapped to the human genome using TopHat (Trapnell et al. 2009) with default parameters, and the expression level of each gene was defined using the FPKM (fragment per kilobase of exon per million fragments mapped) scores generated by Cufflinks (Trapnell et al. 2010). Genes with FPKM scores <0.05 were removed, as they could represent unexpressed or unsequenced regions. The expression level of each exon was defined as exon read count/exon size divided by total read count/genome size.
Data analysis
Genome annotations were downloaded from the UCSC Genome Browser, human genome Build 36.1 (hg18 assembly). Gene definitions were obtained from Refseq Gene track in the UCSC Genome Browser (Karolchik et al. 2009). Exons were defined using Refseq Gene track and only internal exons separated from flanking exons by at least 500 bp were analyzed.
For HeLa cells, the H2Bub1 ChIP-seq data were kindly provided by Shema et al. (2008). The mRNA-seq data were downloaded from the GEO database with accession number GSM546927 (Guo et al. 2010).
For our analysis of Drosophila melanogaster, H2Bub1 ChIP-chip and mRNA-seq for the S2-DRSC cell line (late embryonic stage embryo-derived cell line) were downloaded from the GEO database using accession numbers GSE20773 (Roy et al. 2011) and GSM461176 (Brooks et al. 2011), respectively. The H3K36me3 ChIP-seq data were downloaded from the GEO database with accession number GSM480158 (Gan et al. 2010). Gene definitions were given by their Refseq gene track (dm3) in the UCSC Genome Browser and other processes were the same as those used for the human genome data.
For mouse myoblast cells, the H2Bub1 and H3K36me3 ChIP-seq data were downloaded from ArrayExpress with accession number E-GEOD-25308 (Asp et al. 2011; Vethantham et al. 2012), and the RNA-seq data were downloaded from the GEO database with accession number GSM521256 (Trapnell et al. 2010). Gene definitions were given by their Refseq gene track (mm9) in the UCSC Genome Browser, and the other processes were the same as those used for the human genome data.
Calculating modification signals in skipped exons
Alternatively spliced exons were defined by more than threefold repression of cassette exons compared with flanking exons after the removal of the first and last two exons from the analysis. To precisely define skipped exons, only exons of highly expressed genes (the top 30%) were considered; however, the same tendency was achieved when all genes were used. In NCCIT cells, HeLa cells, siRNF20-transfected NCCIT cells, mouse myoblast cells, and Drosophila melanogaster cells, we observed 12337, 9722, 11496, 12356, and 3201 skipped exons, respectively. As a control, we selected three constitutive exons that were separated from skipped exons by at least two exons. NCCIT cells contained 16,724 control exons. All exon definitions were provided by the Refseq gene tracks for all organisms, as obtained from the UCSC Genome Browser.
When we compared exon skipping between wild-type and siRNF20-transfected cells, we selected skipped exons with H2Bub1-specific patterns in order to infer the effect of RNF20 knockdown. Among the skipped exons, we selected those showing a more than or less than 1.5-fold enrichment of H2Bub1 signals compared with flanking exons. We found that 5821 exons were persistently skipped in both cell types, while 2749 exons were skipped only in wild-type cells. To check the effect of our definition of exon skipping, we changed the threshold value for exon expression levels from threefold to twofold. Using this parameter, we observed a similar tendency in our results. The H2Bub1 signal difference between wild-type and siRNF20-transfected cells was calculated by comparing the fold enrichment of H2Bub1 signals versus the average H2Bub1 signal (total read count/genome size).
Data access
The ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE25882.
Acknowledgments
Inkyung Jung thanks the members of the Bioinformatics and Computational Biology Laboratory and Yoo-Sun Lee for helpful discussions. We also thank the modENCODE project for allowing us to use their data. This work is also supported by grants from the Epigenomic Research Program for Human Stem Cells (2007-2004134), the Future-Based Technology Development Program of the National Research Foundation of Korea (2010-0020590), and the KAIST Future Systems Healthcare Project, Ministry of Education, Science and Technology, South Korea. This work is also supported by the CHUNG Moon Soul Center for BioInformation and BioElectronics (CMSC), and by the Ministry of Education, Science and Technology, South Korea (Bioinformatics Pipeline for Stem Cell Epigenomics).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.120634.111.
References
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD 2011. PNAS Plus: Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci 108: E149–E158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF 2011. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25: 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498 doi: 10.1038/nature00970 [DOI] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci 106: 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK 2010. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol 11: R70 doi: 10.1186/gb-2010-11-7-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanov I 1993. Pathogenesis of testicular germ cell tumours. Eur Urol 23: 2–5 [DOI] [PubMed] [Google Scholar]
- Damjanov I, Horvat B, Gibas Z 1993. Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab Invest 68: 220–232 [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR 3rd, Grant PA 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871 [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW 2011. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66 [DOI] [PubMed] [Google Scholar]
- Gan Q, Schones DE, Ho Eun S, Wei G, Cui K, Zhao K, Chen X 2010. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol 11: R42 doi: 10.1186/gb-2010-11-4-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937 [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B 2009. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol 5: e1000566 doi: 10.1371/journal.pcbi.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O 2009. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 33: 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung I, Kim D 2009. Regulatory patterns of histone modifications to control the DNA methylation status at CpG islands. IBC 1: 1–8 [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ 2009. The UCSC Genome Browser. Curr Protoc Bioinformatics 28: 1.4.1–1.4.26 [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2010. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG 2005. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20: 759–770 [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG 2009. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10: 483–488 [DOI] [PubMed] [Google Scholar]
- Oh S, Jeong K, Kim H, Kwon CS, Lee D 2010. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun 399: 512–517 [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2011. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS 2011. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev 25: 2242–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Ast G 2010. Chromatin density and splicing destiny: On the cross-talk between chromatin structure and splicing. EMBO J 29: 1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 19: 271–277 [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ahn SH 2010. A Bre1-associated protein, large 1 (Lge1), promotes H2B ubiquitylation during the early stages of transcription elongation. J Biol Chem 285: 2361–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens MA 1970. Use of the Kolmogorov-Smirnov, Cramer-Von Mises and related statistics without extensive tables. JR Stat Soc 32: 115–122 [Google Scholar]
- Sun ZW, Allis CD 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD 2007. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev 21: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima T, Chatani M, Hata K, Inoue T 1988. High-dose rate intracavitary therapy for carcinoma of the uterine cervix: II. Risk factors for rectal complication. Int J Radiat Oncol Biol Phys 14: 281–286 [DOI] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Yang Y, Bowman C, Asp P, Lee JH, Skalnik DG, Dynlacht BD 2012. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol Cell Biol 32: 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2008. Histone ubiquitination: Triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]