Abstract
Segmented filamentous bacteria (SFB) are host-specific intestinal symbionts that comprise a distinct clade within the Clostridiaceae, designated Candidatus Arthromitus. SFB display a unique life cycle within the host, involving differentiation into multiple cell types. The latter include filaments that attach intimately to intestinal epithelial cells, and from which “holdfasts” and spores develop. SFB induce a multifaceted immune response, leading to host protection from intestinal pathogens. Cultivation resistance has hindered characterization of these enigmatic bacteria. In the present study, we isolated five SFB filaments from a mouse using a microfluidic device equipped with laser tweezers, generated genome sequences from each, and compared these sequences with each other, as well as to recently published SFB genome sequences. Based on the resulting analyses, SFB appear to be dependent on the host for a variety of essential nutrients. SFB have a relatively high abundance of predicted proteins devoted to cell cycle control and to envelope biogenesis, and have a group of SFB-specific autolysins and a dynamin-like protein. Among the five filament genomes, an average of 8.6% of predicted proteins were novel, including a family of secreted SFB-specific proteins. Four ADP-ribosyltransferase (ADPRT) sequence types, and a myosin-cross-reactive antigen (MCRA) protein were discovered; we hypothesize that they are involved in modulation of host responses. The presence of polymorphisms among mouse SFB genomes suggests the evolution of distinct SFB lineages. Overall, our results reveal several aspects of SFB adaptation to the mammalian intestinal tract.
Symbiotic relationships—the intimate living together of genetically distinct organisms—are a universal phenomenon characterizing life on Earth (Portier 1918). Segmented filamentous bacteria (SFB) are intestinal symbionts of a variety of vertebrates including monkey, mouse, rat, dog, chicken, and trout (Hampton 1962; Klaasen et al. 1993a; Snel et al. 1995; Umesaki et al. 1995; Imaoka et al. 1997). SFB comprise a distinct clade of phylogenetically closely related, uncultivated clostridia within the family Clostridiaceae 1, designated Candidatus Arthromitus (Supplemental Fig. S1). The term “arthromitus” (Greek arthron, joint; mitos, thread) was originally coined by Joseph Leidy in 1849, summarizing the variety of thread-like microorganisms he observed though meticulous microscopic inspections of animal intestinal microbial communities (Leidy 1849).
One of the most intriguing and unifying features of SFB is their intimate interaction with their hosts, most notably, their firm attachment to epithelial cells of the distal ileum by one end of the filament (Chase and Erlandsen 1976). SFB differentiate during their life cycle to form multicellular filaments, ranging in length from 50 to 1000 μm, as well as single “holdfasts” and spores, i.e., vegetative and dormant reproductive cells, respectively (Chase and Erlandsen 1976; Klaasen et al. 1992). Holdfasts are teardrop-shaped cells; their pointed end contains a finely textured matrix from which ribosomes are excluded, and with this end they attach to host cells (Chase and Erlandsen 1976). Both holdfast and spore cells attach to host intestinal epithelial cells, and through continuous growth and septa formation produce a chain of attached, clonal progeny, known as segment cells. Within older segments, holdfasts and spores differentiate and are eventually released (Chase and Erlandsen 1976; Klaasen et al. 1992).
Attempts to transplant SFB from one animal host species into the germ-free intestine of another host species have failed, suggesting host specificity among the members of the SFB clade (Tannock et al. 1984). Significantly, within their respective hosts, SFB induce a multifaceted immune response. The immune response can involve components of the innate as well as adaptive immune system, including IgA production, fine-tuning of the homeostatic profile of pro-inflammatory helper T cells (Th17) and anti-inflammatory regulatory T cells, and induction of epithelial cell-derived antimicrobial peptides (Klaasen et al. 1993b; Umesaki et al. 1999; Gaboriau-Routhiau et al. 2009; Ivanov et al. 2009; Salzman et al. 2010). Depending on the model system, colonization by SFB can either confer protection against intestinal pathogens (Garland et al. 1982; Ivanov et al. 2009) or promote autoimmune disorders such as autoimmune arthritis and experimental autoimmune encephalomyelitis through induction of specific T-cell subsets (Wu et al. 2010; Lee et al. 2011). Until recently, we lacked information about the SFB-associated genetic determinants that might facilitate host–microbe interactions.
The first SFB genome sequences have recently been described (Kuwahara et al. 2011; Prakash et al. 2011; Sczesnak et al. 2011), although their resistance to cultivation ex vivo continues to hamper more detailed studies of these peculiar bacteria. In the present study, we isolated five individual mouse SFB filaments and explored their genome sequences separately. We took advantage of recent technological advances in microfluidics and genomics that make it now possible to study low-abundance uncultivated microbes at the single-cell level (Marcy et al. 2007; Harrington et al. 2010). The ability to treat single bacterial structures as individual units of study provides insights into microevolution and microdiversity that may be lost in genome analyses based on consensus sequences from a large population of cells.
We identified several genetic determinants that may be important for SFB niche adaptation, host–microbe interaction, and cell differentiation, and for distinguishing SFB from other clostridia that inhabit the intestinal tract of animals. We discovered four variant sequences for a novel ADP-ribosyltransferase (ADPRT) family and a new predicted myosin-cross-reactive antigen (MCRA) family protein. In addition, we identified sequence polymorphisms among the SFB genomes, suggesting underlying genetic heterogeneity, and distinct lineages. Differences among our sequences, recently published mouse SFB genome sequences, and a rat SFB genome sequence may provide clues about the basis of SFB host specificity.
Results
Single filament isolation and recovery of genomic sequences
To characterize SFB genomic repertoire, we separately isolated five individual filaments from fecal material of an SFB-monocolonized mouse using laser tweezers in concert with a microfluidic device (Supplemental Material; Supplemental Fig. S2; Supplemental Movies 1–5). Their genomic DNA was amplified and subjected to multiplex pyrosequencing. Between 38 and 67 Mb was obtained for each of the five individual filaments. Assemblies were obtained for each individual SFB filament (SFB-1 to SFB-5) (Table 1; Supplemental Material; Supplemental Table S1). In addition, two coassemblies were obtained by combining the reads from the five individual SFB filaments, resulting in “SFB-co,” which was assembled de novo, and in “SFB-mouse-SU,” which was assembled through the use of a recently published, complete mouse SFB genome sequence, SFB-mouse-Yit (AP012209) (Prakash et al. 2011), which is closely related to our SFB (see below) (Table 1; Supplemental Material; Supplemental Table S1).
Table 1.
Genome information for the individual SFB assemblies (SFB-1 to SFB-5) and the coassemblies (SFB-co, SFB-mouse-SU)
The majority of the SFB genome sequence was recovered, as estimated by (1) rarefaction analysis (Supplemental Material; Supplemental Fig. S3); (2) shared sequence analysis of the five individual SFB genomes (Supplemental Material); (3) presence of the 36 universal distributed clusters of orthologous genes (COGs) (Supplemental Material; Supplemental Fig. S4; Ciccarelli et al. 2006); (4) number of total COGs compared with other clostridia (Supplemental Fig. S4); and (5) number of tRNAs compared with other clostridia (Supplemental Material; Supplemental Fig. S3). We recovered 98.7% of the genome sequence using the five SFB filaments, when compared with the genome of SFB-mouse-Yit (1.586 Mb) (Prakash et al. 2011). The mean GC content was 27.9%–28.4% for the seven SFB genome assemblies, which is typical for taxa belonging to the family Clostridiaceae 1 (Supplemental Fig. S3; Table 1; Supplemental Table S1). Seventy-eight percent of the protein-coding genes had high similarity to sequences in other clostridia or Firmicutes (Table 1; Supplemental Table S1), but on average, 8.6% of the predicted proteins showed no significant sequence similarity.
Comparative genomics
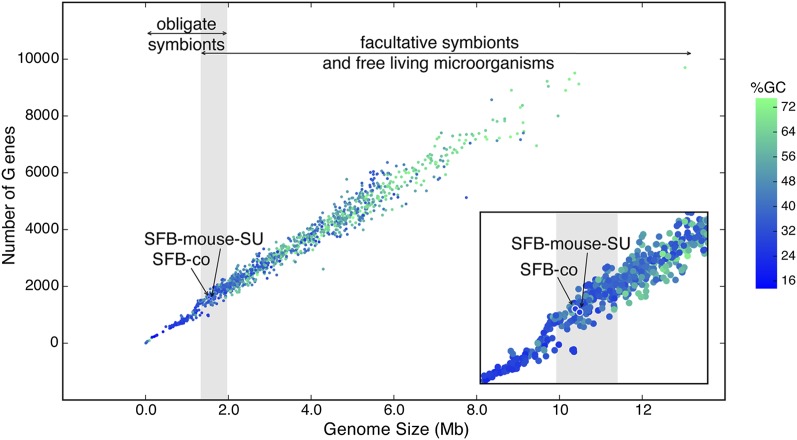
In Bacteria and Archaea, genome size and number of genes are directly proportional to each other (Konstantinidis and Tiedje 2004). The ratio of gene number to assembly size for SFB-co and for SFB-mouse-SU is roughly similar to that of 1247 complete microbial genomes (Fig. 1), further suggesting that we recovered nearly complete genome sequences for SFB. Although there is no strict relationship between genome size and phylogenetic affiliation, smaller genomes tend to have a lower mean GC content compared with larger genomes (Fig. 1; Supplemental Fig. S5). Obligate symbionts, such as endosymbionts of insects and nematodes, have smaller genomes compared with free-living and facultative symbionts (Fig. 1; Supplemental Fig. S5; Wu et al. 2004; McCutcheon and Moran 2007). Based on the SFB-co and SFB-mouse-SU assembly sizes, predicted number of genes, and GC content, SFB is positioned between obligate symbionts and facultative symbionts, reflecting its close association with the animal host.
Figure 1.
Relationship between genome size and number of genes for SFB-co, SFB-mouse-SU, and 1247 complete microbial genomes. Microbial strains are color-coded by mean GC content (%). (Inset) Close-up of the region of the plotted data that includes SFB-co and SFB-mouse-SU.
Phylogenetic analysis of 16S rRNA sequences has previously suggested that SFB are affiliated with the family Clostridiaceae 1, in which SFB from different animal hosts form a distinct clade, designated Candidatus Arthromitus (Supplemental Fig. S1; Snel et al. 1994, 1995; Umesaki et al. 1995). We confirmed this family affiliation with the predicted protein sequences of five highly conserved genes (polC, tufA, recA, pheT, and secA). In most cases, the SFB lineage arises close to the root of the family, suggesting divergence early in the evolution of Clostridiaceae 1. The longer branches observed for some SFB sequences may be an indication of a faster evolutionary rate (Supplemental Fig. S6); the predicted sequences for these five proteins from each of the individual SFB genomes exhibited 100% sequence identity among each other, arguing against the possibility of sequence errors.
To obtain a more detailed view of shared as well as distinct features of SFB compared with other members of the Clostridiaceae 1, we performed unsupervised clustering of the predicted proteomes based on sequence similarity, manually annotated the resulting clusters, and used principal coordinates analysis to explore the variation in cluster abundance between species (Supplemental Material). The individual Clostridiaceae 1 family members can each be distinguished by the presence of specific sequence clusters (Fig. 2; Supplemental Fig. S7). Both Clostridium perfringens SM101 and Clostridium novyi NT are characterized by the presence of specific transposases, respectively. Clostridium tetani E88 is characterized by several protein clusters, including clusters of surface-located N-acetylmuramoyl-L-alanine amidases (cell wall hydrolases, autolysins), transposases, and additional surface proteins with putative adhesion and enzymatic properties (Fig. 2; Supplemental Fig. S7). SFB are distinguished by four clusters of SFB-specific predicted proteins: Cluster.1, putative secreted hypothetical proteins; Cluster.2, N-acetylmuramoyl-L-alanine amidases; Cluster.3, putative secreted hypothetical proteins; and Cluster.4, hypothetical proteins (Fig. 2; Supplemental Fig. S7; Supplemental Table S2; see below).
Figure 2.
Principal component analysis (PCA) of protein clusters from the predicted proteomes from SFB-co and four other members of the Clostridiaceae 1. A vector in close proximity of a clostridia strain indicates that the protein cluster is highly enriched in that particular strain. The length of a vector indicates the magnitude of influence of that particular protein cluster in relation to all clusters, in explaining differences in clostridia proteome composition. See Supplemental Figure S7 for PC3 versus PC4.
SFB-specific proteins
The SFB protein Cluster.1 is composed of 19 predicted proteins, which have limited similarity to known bacterial proteins and are predicted to be secreted (Fig. 2; Supplemental Figs. S7, S8; Supplemental Table S2). Although these proteins have a low overall sequence similarity among themselves, their unifying features are four conserved regions at the C terminus of the protein (Supplemental Fig S8; Supplemental Table S2) that together have similarity to protein domain PF13946 (DUF4214). Tandem alpha-helices are predicted at the C terminus of some of these proteins (Supplemental Fig. S8C). The three-dimensional structure of the proteins is similar to that of the phycobilisome linker polypeptide (PDB ID 2KY4). The proteins that compose Cluster.3 exhibit sequence similarity in the C-terminal region to the C terminus of Cluster.1 proteins; they are also putatively secreted; and one protein has a predicted fibronectin type III domain (Supplemental Table S2). Based on their predicted extracellular location and the limited number of catalytic domains usually found in secreted degradative enzymes, Cluster.1 and Cluster.3 proteins may be components of surface structures, possibly relevant to niche adaptation.
SFB-specific predicted protein polymorphisms
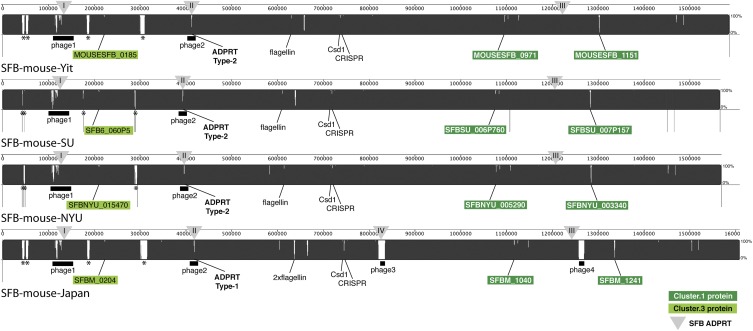
All SFB-specific proteins described here were also discovered in the genomes of other SFB populations (Supplemental Table S3). Interestingly, polymorphisms were detected in some Cluster.1 and Cluster.3 proteins, suggesting the existence of distinct genetic lineages (Fig. 3; Supplemental Fig. S9). Protein Cluster.1 and Cluster.3 homologs from SFB-mouse-Yit and SFB-mouse-NYU exhibit high sequence identity (Supplemental Fig. S9; Supplemental Table S3). The proteins from SFB-mouse-SU have high similarity to the homologs from SFB-mouse-Yit and SFB-mouse-NYU as well but exhibit occasional distinct amino acid residues in some proteins (Supplemental Fig. S9; Supplemental Table S3). Several Cluster.1 and Cluster.3 homologs from SFB-mouse-Japan exhibit striking differences compared with the respective proteins from SFB-mouse-Yit, SFB-mouse-NYU, and SFB-mouse-SU (Supplemental Fig. S9; Supplemental Table S3). The SFB-specific proteins described here are also encoded in the SFB-rat-Yit genome. On average, the overall predicted protein sequence identity between SFB-mouse-Yit, and SFB-rat-Yit genomes is 83.4% (Prakash et al. 2011). SFB-specific proteins have an overall predicted sequence identity of 75.2% ± 2.0% (SEM) between mouse-SFB and rat-SFB. Six SFB-specific proteins from rat had a sequence identity below 65% compared with their respective homolog in mouse-SFB (Supplemental Fig. S9; Supplemental Table S3). Together, this suggests that SFB encode proteins that are predicted to be secreted, not found in any other bacteria described to date, and that some of these predicted proteins exhibit polymorphisms that are suggestive of distinct SFB-mouse lineages, and may be relevant to host specificity.
Figure 3.
Genome sequence comparison of whole and nearly complete SFB genomes, and SFB-specific proteins. The genome sequences of SFB-mouse-SU, SFB-mouse-Yit (AP012209) (Prakash et al. 2011), SFB-mouse-NYU (AGAG01000000) (Sczesnak et al. 2011), and SFB-mouse-Japan (AP012202) (Kuwahara et al. 2011) were aligned using Mauve. (Black-filled areas) High sequence identity; (white-filled areas) differences among some or all genomes. SFB-specific proteins that exhibit extensive polymorphisms among genomes are labeled: Cluster.1 proteins (dark green box); Cluster.3 proteins (light green box). (Triangles) The genome locations of the four types of ADP-ribosyltransferases. (*) Ribosomal RNA operons that are missing from some genomes. Thin vertical black lines for SFB-mouse-SU, and SFB-mouse-NYU indicate the borders of contigs. Two similar flagellins (FliC3, and FliC4) in SFB-mouse-Japan are encoded at a position where only one copy is observed in the other genomes; sequence fragments from single cell SFB suggest the presence of two flagellins as well (this study). See Supplemental Figure S9 and Supplemental Table S3 for further sequence comparisons of SFB-specific proteins among genomes.
Metabolism
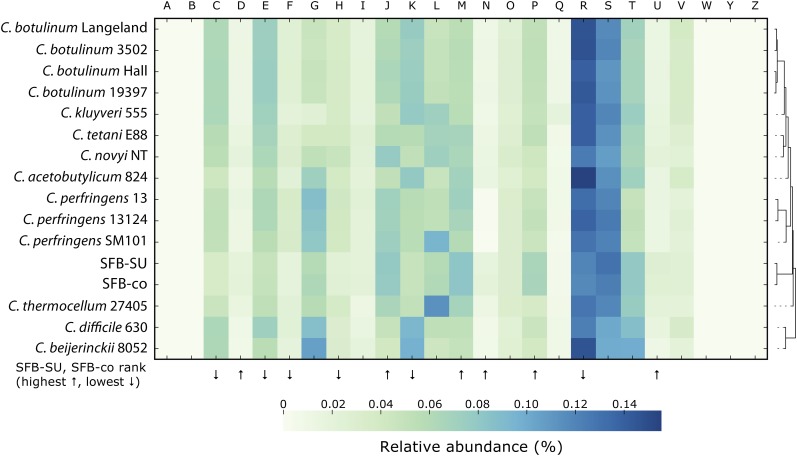
As is the case for many Clostridia, SFB appear to have a heterotrophic lifestyle. Glycolysis and pentose phosphate pathways were identified in the SFB data sets and found to be nearly complete. Genes for fructose and mannose metabolism were identified and were similar to those in C. perfringens SM101 and Clostridium acetobutylicum ATCC 824. We did not find genes encoding components of the electron transport chain. Substrate-level phosphorylation may be performed in SFB by phosphoglycerate kinase (SFBSU_003P17), pyruvate kinase (SFBSU_002P48), and acetate kinase (SFBSU_006P356). Molecular hydrogen may be produced via a pyruvate ferredoxin oxidoreductase (SFBSU_006P288). A number of predicted dehydrogenases, including alcohol, aldehyde, and lactate dehydrogenases, may facilitate the oxidation of substrates and generate fermentation products such as lactate, acetate, and ethanol. In contrast, only a small fraction of the enzymes required for the synthesis of amino acids, vitamins, and cofactors were found in the SFB data sets (Fig. 4; Supplemental Fig. S10). In particular, pathways for riboflavin (vitamin B2), porphyrin (including vitamin B12), and thiamine (vitamin B1) biosynthesis as well as pathways for biosynthesis of the amino acids alanine, threonine, glycine, methionine, arginine, and proline appeared to be incomplete, as compared with corresponding gene sets in other clostridia.
Figure 4.
Functional categories of cluster orthologous groups (COGs). Analysis of the predicted proteome of SFB-co and SFB-mouse-SU in comparison to other clostridia. Relative abundance levels of COG functional categories were clustered using a Euclidean distance metric and displayed as a heatmap, with low abundance (light green) and high abundance (dark blue). (A) RNA processing and modification. (B) Chromatin structure and dynamics. (C) Energy production and conversion. (D) Cell cycle control, cell division, chromosome partitioning. (E) Amino acid transport and metabolism. (F) Nucleotide transport and metabolism. (G) Carbohydrate transport and metabolism. (H) Coenzyme transport and metabolism. (I) Lipid transport and metabolism. (J) Translation, ribosomal structure and biogenesis. (K) Transcription. (L) Replication, recombination and repair. (M) Cell wall/membrane/envelope biogenesis. (N) Cell motility. (O) Post-translational modification, protein turnover, chaperones. (P) Inorganic ion transport and metabolism. (Q) Secondary metabolites biosynthesis, transport, and catabolism. (R) General function prediction only. (S) Function unknown. (T) Signal transduction mechanisms. (U) Intracellular trafficking, secretion, vesicular transport. (V) Defense mechanisms. (W) Extracellular structures. (Y) Nuclear structure. (Z) Cytoskeleton.
SFB encode proteins predicted to be involved in the transport of various small molecules and ions, such as amino acids, oligopeptide/dipeptides, manganese, zinc, iron, and phosphate. SFB are also predicted to express phosphonate and phosphate transporters, as well as several PTS systems for the uptake of sugars. In addition, a variety of predicted extracellular proteases, including a potential mucin-degrading alpha-N-acetylglucosaminidase and a zinc-dependent phospholipase (Table 2), were found in the SFB gene inventory that could facilitate the degradation of higher-molecular-weight compounds. We hypothesize that SFB have an anaerobic metabolism similar to that of other proteolytic clostridia that break down larger peptides, take up amino acids, and ferment them into products that may include short-chain fatty acids (SCFA). In contrast to other clostridia, SFB appear to require vitamins, cofactors, and amino acids from the environment to sustain cellular processes.
Table 2.
Selected gene products potentially involved in host–microbe interaction and tolerance to antimicrobial action
Cell differentiation
The relative abundance of genes predicted to encode functions related to cell cycle control (COG category D) and envelope biogenesis (COG category M), as well as trafficking (COG category U) in SFB-co and SFB-mouse-SU, was higher than in other clostridia (Fig. 4) and may reflect the complex cell differentiation processes during the SFB life cycle. We found eight N-acetylmuramoyl-L-alanine amidases (cell wall hydrolases, PF01510, PF05105, PF01520) in SFB-mouse-SU, four of which formed a cluster that was unique to SFB (Supplemental Table S2; see above). These four predicted proteins contain several amino acid residues that have been previously reported to be involved in zinc binding, substrate binding, and catalysis (Low et al. 2005; Zoll et al. 2010). We hypothesize that the four predicted SFB enzymes may be important in the formation of the different SFB cell morphotypes, analogous to the role of autolysins in cell separation in Bacillus subtilis (Chai et al. 2010). They may also be responsible for the release of holdfasts and spores from the filament and facilitate excretion of cytoplasmic proteins and potential effectors (Pasztor et al. 2010).
Many predicted proteins were found in SFB that are homologous to proteins that control cell cycle, cell differentiation, and cell shape in B. subtilis and other bacteria, such as FtsZ, FtsW, FtsEX, SepF, MreBCD, YvcK, Spo0A, SpoIIIE, GerE, SpoVT, GerAA-AC, DivIVA, and CinA. In addition, we found a protein with similarity to bacterial dynamin-like proteins (BDLP; SFBSU_006P653). Dynamins are a family of proteins that mediate membrane remodeling and promote membrane curvature in eukaryotic endocytosis, scission of a range of organelles, cell division, and maintenance of cell shape (Praefcke and McMahon 2004). The crystal structure of the BDLP of the filamentous cyanobacterium Nostoc punctiforme has been described; it localizes to the cell membrane in a punctate pattern (Low and Lowe 2006). The predicted three-dimensional structure of the BDLP of SFB exhibits striking similarity to the BDLP of N. punctiforme (Supplemental Fig. S11). It is tempting to speculate that the SFB BDLP may play an important role in formation of SFB cell morphotypes, including scission processes during the development of progeny cells, and formation of the nipple-like curved structure of holdfast cells important for establishing firm host–microbe interaction.
Host–microbe interactions
We identified several genetic determinants that might facilitate SFB interactions with the host (Table 2). For example, SFB appear to encode flagella, although these structures have not been previously observed. Four proteins are predicted to harbor flagellin-N and flagellin-C domains, of which three are similar to flagellin proteins previously observed in other clostridia or Firmicutes. Flagella may allow SFB to swim through the mucus layers adjacent to the epithelium and respond to chemotactic signals; motility in the mucus layers may be facilitated by SFB-encoded mucin-degrading enzymes such as the alpha-N-acetylglucosaminidase. Flagella might also facilitate adhesion of SFB to surfaces, and flagellin itself may be recognized by the host and trigger immune responses (Vijay-Kumar et al. 2006; Uematsu et al. 2008; Van Maele et al. 2010).
The SFB genomes are predicted to contain a homolog of the myosin-cross-reactive antigen (MCRA) that may be involved in host–cell interactions and survival in the host environment. MCRA was originally identified as an antigen in individuals with acute rheumatic fever (ARF), and other forms of autoimmune disease caused by Streptococcus pyogenes (Kil et al. 1994). Streptococcal MCRA is thought to play a role in these conditions through its elicitation of antibodies that cross-react with host myosin (Kil et al. 1994). Intriguingly, SFB have been associated with the induction of autoimmune disease in some animal models (Wu et al. 2010; Lee et al. 2011). The MCRA-homolog protein from SFB has 73.9% and 50.2% sequence identities to the MCRAs from Streptococcus mutans UA159, and S. pyogenes SSI-1, respectively. Other homologs of MCRA have recently been identified in other bacteria, such as Staphylococcus aureus and Lactobacillus acidophilus, where they appear to contribute to resistance to oxidative stress, host survival, and adherence to host cells (O'Flaherty and Klaenhammer 2010; Malachowa et al. 2011).
SFB ADP-ribosyltransferases (ADPRTs)
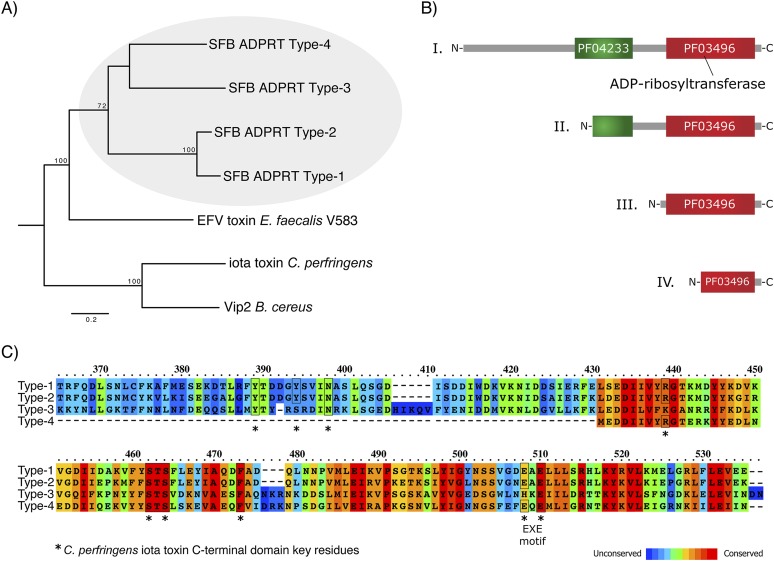
In total, we discovered four ADP-ribosyltransferase sequence types in SFB genomes, referred to hereafter as ADPRT Type-1 to Type-4 (Figs. 3, 5; Supplemental Fig. S12). Three ADPRT types (Type-1, -2, and -3) were found in the SFB genomes of the present study, as well as in the SFB-mouse-Yit and SFB-mouse-NYU genomes. SFB-mouse-Japan encodes complete and truncated versions of Type-1, a Type-3 ADPRT, and a Type-4 ADPRT (Figs. 3, 5; Supplemental Fig. S12). Type-1 ADPRTs are encoded in phage element 1 and are composed of a C-terminal ADP-ribosyltransferase domain (PF03496) and an N-terminal portion containing a phage protein-like domain (PF04233) (Figs. 3, 5; Supplemental Fig. S12). Type-2 ADPRTs are encoded in phage element 2 and are composed of a C-terminal ADPRT domain (PF03496) and a truncated PF04233 domain. The Type-1 ADPRT in phage element 2 in SFB-mouse-Japan has the same predicted length as the Type-2 in other SFB genomes (Supplemental Fig. S12). ADPRT Type-3 is composed of an ADPRT-domain only, and ADPRT Type-4 is composed of a ADPRT-domain that is truncated at the N terminus (Fig. 5; Supplemental Fig. S12). ADPRT Type-3 is located in the chromosomal neighborhood of genes encoding a protein with a cupin-like domain (PF07883), a catalase, a rubrerythrin, and hypothetical proteins. ADPRT Type-4 in SFB-mouse-Japan is located in phage element 3 (Figs. 3, 5; Supplemental Fig. S12).
Figure 5.
ADP-ribosyltransferases (ADPRTs) in segmented filamentous bacteria (SFB). (A) Phylogenetic tree based on four ADPRT sequence types from SFB and ADPRTs from other Firmicutes. Proteins were aligned using MUSCLE, all columns with gaps removed, and phylogeny inferred using a maximum likelihood method. Bootstrap statistical support for branchings with values ≥70 are shown. SFB ADPRT Type-1, SFBSU_002P79; SFB ADPRT Type-2, SFBSU_006P112; SFB ADPRT Type-3, SFBSU_007P84; SFB ADPRT Type-4, SFBM_0768; EFV toxin Enterococcus faecalis V583, NP_814127; Iota toxin Clostridium perfringens, Q46220; Vip2 Bacillus cereus, 1QS1_A. (B) Protein domain organization of the four ADPRTs in SFB. (PF03496) ADP-ribosyltransferase domain; (PF04233) phage protein-like domain. See Figure 3, and Supplemental Figure S12 for information about their localization in SFB genomes, respectively. (C) Amino acid sequence conservation among SFB ADPRT sequence types. (*) Key amino acid residues, involved in catalysis as described for the C. perfringens iota toxin (Y246, Y251, N255, R295, S338-S340, F349, E378, E380) (Tsuge et al. 2008).
Type-1 and Type-2 ADPRT-domains share predicted sequence and structural similarities with the C-terminal domains of the C. perfringens iota toxin and the vegetative insecticidal protein VIP2 of Bacillus cereus (Fig. 5; Tsuge et al. 2008). Key residues described for the iota toxin of C. perfringens in catalyzing ADP-ribosylation of G-actin at Arg 177, including the EXE-motif, are present in both Type-1 and Type-2 SFB ADPRT-domains (Fig. 5). Type-3 and Type-4 SFB ADPRT-domains have some but not all of the key residues (Fig. 5). Proteins with both PF04233 and PF03496 (ADPRT) domains can be found in other Firmicutes, such as Anaerotruncus colihominis DSM 17241 (Uniprot B0P707), Lactobacillus reuteri 100-23 (B3XNE3), Streptococcus mitis B6 (D3H775), Streptococcus pyogenes MGAS9429 (Q1CQR7), and Enterococcus faecalis V583 (Q838U8). Interestingly, the ADPRT from E. faecalis V583, referred to as EFV toxin, was recently identified in silico and its toxicity demonstrated in a yeast expression system (Fieldhouse et al. 2010). The predicted SFB ADPRT Type-1 has structural similarity to the EFV toxin in E. faecalis V583 (Supplemental Fig. S12C) (Fieldhouse et al. 2010). In E. faecalis V583, the ADPRT gene is also found in the neighborhood of phage-related genes, suggesting that these genes may have been originally acquired through phage infection (Fieldhouse et al. 2010). A bacteriophage-encoded toxin has recently been found to protect against wasp parasitism in a symbiotic bacterium–aphid mutualism (Oliver et al. 2009). It is tempting to speculate that the SFB ADPRTs observed here may target host cells and modulate symbiotic interactions.
Host environment survival
To prevent potential antimicrobial compounds from reaching their targets, SFB may use proteins with efflux functions (e.g., MATE proteins) (Table 2). Bile acids can have antimicrobial and immunomodulatory activities (Stacey and Webb 1947; Inagaki et al. 2006); SFB possess a predicted choloylglycine hydrolase, which may catalyze the deconjugation of conjugated bile acids, as has been described for other gut community members (Jones et al. 2008; Lambert et al. 2008). The action of lysozyme, expressed by Paneth cells as an integral component of the innate immune defense, may be diminished by modifications performed by SFB O-acetyltransferases and polysaccharide deacetylase (Rosenthal et al. 1983; Vollmer and Tomasz 2000; Bera et al. 2006; Milani et al. 2010). SFB contain several genes predicted to encode rubrerythrin, as well as two catalases, which may be involved in oxidative stress protection. SFB encode an arginase that for several microorganisms facilitates immune evasion through catabolism of arginine which is the precursor for nitric oxide (NO) synthesis (Das et al. 2010). The NO-generating macrophage enzyme nitric oxide synthase 2 (Nos2) was up-regulated in the intestines of ex germ-free mice upon colonization by SFB (Ivanov et al. 2009). SFB may also produce an extracellular polysaccharide capsule, which in other organisms serves as a protective shield against antimicrobial attack, mediates adhesion, and facilitates recognition by the host. There are homologs in SFB of genes that encode critical participants in capsule biosynthesis in other organisms, such as O-antigen ligase (SFBSU_006P638), UDP-N-acetylglucosamine 2-epimerase (SFBSU_007P192), and several glycosyl transferases, GT1 (PF00534) (e.g., SFBSU_006P639) and GT2 (PF00535) (e.g., SFBSU_006P601) (Bruggemann and Gottschalk 2008). It has been reported that SFB are surrounded by a “fuzzy,” electron-dense material, which may be a polysaccharide (Koopman et al. 1987; Sanford 1991).
Sequence variation between individual SFB filaments
We used three approaches to explore the individual SFB filaments, SFB-1 to SFB-5, for the presence of genome sequence variation and detected a number of SNPs (Supplemental Fig. S13; Table 3; Supplemental Table S4; Supplemental Material). Among the loci exhibiting polymorphisms that were identified by at least two approaches were oxaloacetate decarboxylase subunit alpha (OadA), pyruvate kinase (PK), and flagellar motor switch protein (FliN) (Supplemental Fig. S14). OadA is part of an oxaloacetate decarboxylase Na+ pump that generates pyruvate from oxaloacetate (Studer et al. 2007). In filament SFB-5 (Supplemental Fig. S2B5), a conserved lysine (K) residue in OadA appears to be substituted by a threonine (T) residue. Mutation of the homologous lysine residue in Vibrio cholerae renders the enzyme inactive, and an oadA mutant of Legionella pneumophila, a facultative intracellular symbiont, exhibits a growth defect in eukaryotic host cells (Jain et al. 1996; Studer et al. 2007). Interestingly, in the same filament, SFB-5, a nucleotide polymorphism was also found in the pyruvate kinase-encoding gene, leading to a valine (V) to alanine (A) change in the protein (Supplemental Fig. S14). PK is a key enzyme in cellular energy metabolism and catalyzes the conversion of phosphoenolpyruvate to pyruvate. The polymorphism in SFB-5 is located in the C-terminal PK_C domain and is not among key catalytic residues described for PKs in other organisms.
Table 3.
Single nucleotide polymorphisms among individual SFB genomes
In SFB-4 we detected a nucleotide polymorphism in the flagellar motor switch protein (FliN)–encoding gene, resulting in a predicted alanine (A) to threonine (T) change in the C-terminal SPOA-domain (Supplemental Fig. S14). FliN, together with FliG and FliM, forms the switch complex that controls the direction of flagella rotation. The alanine residue in the C-terminal domain is highly conserved among bacteria and appears to be part of the structural core of FliN (Brown et al. 2005). In bacteria, mutations in FliN can cause a range of effects, including defects in flagella export and failure in flagella rotation (Irikura et al. 1993; Brown et al. 2005).
Sequence variation between SFB populations
The genome sequences of SFB-mouse-SU and SFB-co are based on the genomes from five SFB filaments, while SFB-mouse-Yit, SFB-mouse-NYU, and SFB-mouse-Japan are each based on the genomes from a few thousand SFB; thus, these latter consensus sequences provide an estimate of the dominant alleles in a given SFB population. The OadA, PK, and FliN variants observed here (see above) in individual SFB filaments (Supplemental Fig. S14) may be examples of minor variants coexisting in a population of SFB cells.
To explore sequence variation between SFB-mouse-SU, SFB-mouse-Yit, SFB-mouse-NYU, and SFB-rat-Yit, we performed multiple genome sequence comparisons (Supplemental Fig. S15; Supplemental Material). Overall, all SFB genomes appear highly syntenic (Supplemental Fig. S15). SFB-mouse-Yit, SFB-mouse-SU, and SFB-mouse-NYU exhibit >99% sequence identity. SFB-mouse-SU is slightly distinct from SFB-mouse-Yit and SFB-mouse-NYU. SFB-mouse-Japan has >98% sequence identity to the other SFB-mouse genomes. Among the chromosomal features that exhibit highest variability among the SFB-mouse genomes are the CRISPR-arrays, some phage-related genes, and hypothetical proteins, including Cluster.1 and Cluster.3 SFB-specific proteins (Fig. 3; Supplemental Material; see above).
Discussion
Several important phylogenetic lineages of bacteria and key members of complex, host-associated microbial communities remain poorly characterized because of their resistance to cultivation in the laboratory. We used a microfluidic device to isolate individual segmented filamentous bacteria (SFB) from a mouse and explore their genomic repertoire, in order to gain insight into the biology of this enigmatic group of bacteria. We obtained nearly 99% coverage of a complete SFB genome sequence from each of five individual SFB filaments. An average of 8.6% of the predicted proteins from the filaments were novel, including a family of secreted SFB-specific proteins with hitherto unknown function.
Based on careful inspection by electron microscopy, a unique life cycle has been proposed for SFB, involving multicellular filamentous morphotypes that attach to intestinal epithelial cells, as well as individual progeny cells, i.e., “holdfasts” and spores, that differentiate within the segment cells of filaments (Chase and Erlandsen 1976; Klaasen et al. 1992). We propose that as part of its symbiotic relationship with the host, the host provides SFB with nutrients including various amino acids as well as vitamins and cofactors (Fig. 6). Intimate host–SFB contact may facilitate the direct transport of essential nutrients from host to SFB. In conventional mice with a complex gut microbiota, SFB may also be able to obtain nutrients from other microbes; the fact that SFB carry out a complex life cycle within monocolonized mice indicates that the host alone is sufficient for this aspect of SFB biology. Filament growth following host attachment suggests that nutrients may be transported from the host-attached segment cell to the descending segment cells within a filament, because SFB appear to be unable to synthesize many metabolic building blocks de novo but are able to perform complex and energy-consuming cell differentiation processes within older segment cells. Differentiation into holdfasts and spores may be induced by early nutrient limitation in the segments most distant from the host. Growth as a filament may provide SFB with some level of protection against phagocytosis due to their large size, as has been observed for other filamentous bacteria (Hahn et al. 1999; Rosenberger and Finlay 2002; Justice et al. 2006). In contrast, smaller morphotypes, such as holdfasts, may be phagocytosed under some conditions (Yamauchi and Snel 2000).
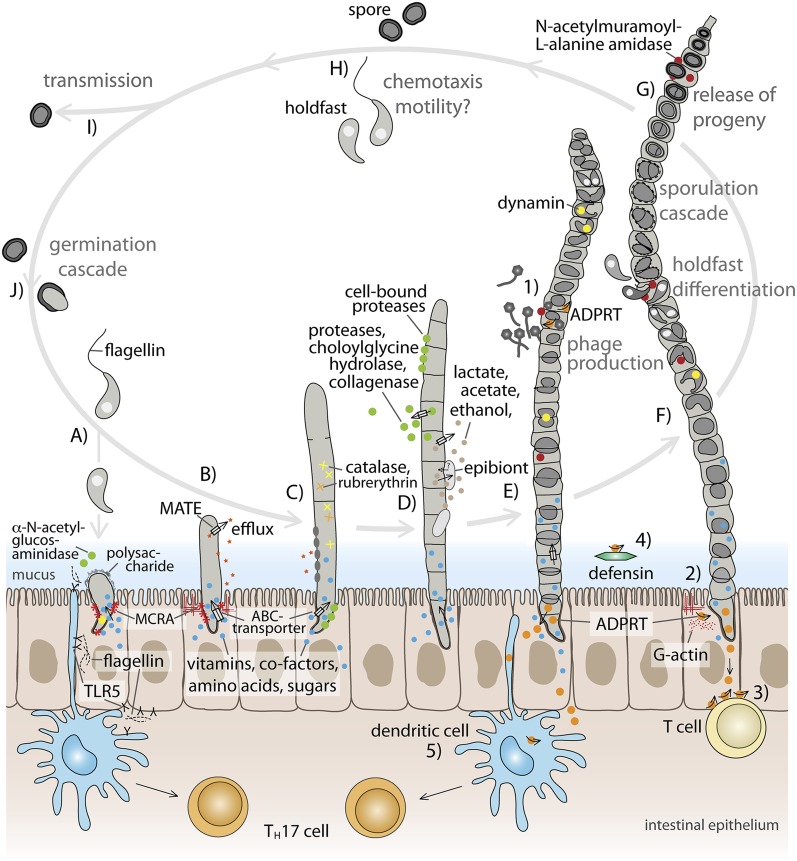
Figure 6.
Proposed life cycle of segmented filamentous bacteria (SFB) in the murine intestine, and putative targets of SFB ADP-ribosyltransferases (ADPRTs). This schematic is based on the present genome sequence study, previous microscopy studies (e.g., Davis and Savage 1974; Chase and Erlandsen 1976; Koopman et al. 1987; Klaasen et al. 1992), and other SFB-related literature. (A) Migration of SFB through the mucus layer may be facilitated by chemotaxis/flagella-driven motility, and degradation of mucin by excreted alpha-N-acetylglucosaminidase. Attachment of holdfast cell to intestinal epithelial cell in the small intestine. Flagellin may be recognized by host determinants, trigger immune responses, and protect epithelial cells from apoptosis. SFB may obtain essential nutrients from the host and import them using a variety of transport systems. (B–D) Undifferentiated filaments; (E,F) differentiated filaments. (C–F) Filaments may be protected from phagocytosis by host cells due to their size. (B) Growth of holdfast segment cell. (C) Early stage of filament development. Anchored holdfast segment, formation of primary segment cell, and secondary segments via new transverse septa. (D) Filament growth through division of segment cells. At various stages during the life cycle, fermentation products and extracellular enzymes may be released. Released products may also be used by other microorganisms, such as “epibionts” of unknown affiliation that have been observed to be associated with SFB. (E) Initiation of differentiation of reproductive cells. Scissions and cell-shape modulation processes may be facilitated by dynamin. Tertiary traverse septum formation, and production of spherical cells, that subsequently form C-shaped cells. Differentiation of one C-shaped cell into two holdfast cells. Phage production may occur at certain stages during the SFB life cycle. (F) Differentiation of holdfasts and spores. Formation of holdfasts with condensed nuclei, and release into the environment, likely facilitated by the action of autolysins. Nonreleased holdfasts can differentiate further into spores. Two holdfasts develop into one spore. Precipitation of cortex and spore coat material around holdfasts as plaques. The size of the holdfasts decrease and they are drawn together. Cortex and spore coat increase in thickness. Release of spores, facilitated by autolysins. (G–I) Released progeny may be dispersed within the same host as well as transmitted to new hosts. Migration of holdfasts may be facilitated by flagella-driven motility and chemotaxis. (J) Induction of germination cascade in spores within the host through unknown signal (possibly, bile). (1–5) Potential targets of ADP-ribosylation by SFB ADPRT. (1) Regulation of phage production through modulation of bacterial RNA polymerase activity. (2) Modification of G-actin, and inhibition of actin polymerization. (3) ADP-ribosylation of T-cell surface proteins, and modulation of T-cell homeostasis. (4) ADP-ribosylation of defensins, and modulation of their activity. (5) ADP-ribosylation of G protein in dendritic cells, leading to up-regulation of cAMP production, and secretion of effector molecules that induce Th17 cells. (Blue circle) Essential nutrients, e.g., amino acids, vitamins, co-factors, sugars. (Gray circle) Fermentation products, such as lactate, acetate, ethanol, H2, CO2. (Red asterisk) Myosin-cross-reactive antigen (MCRA). (Gray box with arrow) Transporters for import and export of small molecules, e.g., MATE-efflux, ABC-transporters, PTS system, ion transporters. (Green circle) Extracellular and surface-located enzymes, such as proteases, bile acid hydrolase (choloylglycine hydrolase), collagenase, alpha-N-acetyl-glucosaminidase, fibronectin binding protein. (Red circle) Autolysin (N-acetylmuramoyl-L-alanine amidase). (Yellow circle) Dynamin. (Orange circle) SFB ADP-ribosyltransferase. (Gray oval) Surface modifications, such as O-acetylation, and polysaccharide deacetylation. (Yellow cross) Catalase. (Orange cross) Ruberythrin. (Red star) Antimicrobials, such as NO, antimicrobial peptides, lysozyme. (Gray irregular shape) Polysaccharide.
We identified a group of autolysins that may play a role in cell morphotype differentiation during the SFB life cycle, as well as facilitate the release of progeny cells from the filament (Fig. 6). SFB dynamin may be involved in differentiation of the SFB morphotypes, e.g., by facilitating scission, as well as formation of the nipple-like curved structure of holdfast cells. Although not previously discussed, the presence of flagella- and chemotaxis-related genes suggests a motile cell type. One wonders whether the tail-like structures associated with some holdfasts are, in fact, flagella (Chase and Erlandsen 1976, Fig. 6). Newly differentiated holdfast cells may express flagella, and upon release from the segment cell, may migrate and colonize new intestinal epithelial cells within the same host. Spore cells, in particular, may play an important role in SFB transmission between hosts, because they may be able to survive outside of the host for prolonged periods of time. Bile in the small intestine may act as a trigger for germination (Wilson 1983). Ensuring a constant generation of progeny cells may be a particular challenge for SFB, since their lifestyle involves tight adherence to the most highly regenerative tissue within mammals (Barker et al. 2007). In return for provision of nutrients, SFB may, among other things, protect their host against colonization by intestinal pathogens (Garland et al. 1982; Ivanov et al. 2009).
We discovered four novel predicted ADP-ribosyl transferases (ADPRTs), which have, to varying degrees, sequence similarities to known ADPRTs that ribosylate actin and thereby inhibit its polymerization. A previous study reported circular localization of polymerized actin in epithelial cells around the attachment sites of SFB and proposed a role for this phenomenon in the stability of SFB–host adherence (Jepson et al. 1993). The SFB ADPRTs may either target actin or other proteins (Fig. 6). SFB–host attachment may, in addition or instead, involve the SFB myosin-cross-reactive antigen (MCRA). The MCRA homolog in S. pyogenes is cell-membrane-associated and has been described to have actin-like properties and fatty acid double-bond hydratase activity, and to be located at the cell surface (MCRA homolog in S. aureus), where it facilitates adherence to host tissue (Fig. 6; Barnett and Cunningham 1992; Kil et al. 1994; Volkov et al. 2010; Malachowa et al. 2011). SFB ADPRTs may be involved in regulating phage replication, similar to the regulation of bacteriophage T4 replication by enterobacteria (Tiemann et al. 2004). Mammals, including mice and humans, possess several endogenic ADPRTs that have sequence similarity with bacterial ADPRTs, including the EXE-motif (Glowacki et al. 2002). They appear to be expressed in a tissue-specific manner and may be involved in regulating cellular processes, such as T-cell functions through ADP-ribosylation of T-cell surface proteins, such as LFA-1, CD8, CD27, CD43, CD44, and CD45 (Nemoto et al. 1996; Okamoto et al. 1998; Liu et al. 1999; Glowacki et al. 2002; Adriouch et al. 2007). Another possible role for SFB ADPRTs may be the ADP-ribosylation of defensins, similar to the ADP-ribosylation of alpha-defensin-1 by a human endogenic ADPRT (Paone et al. 2002). Alternatively, the SFB ADPRTs may induce Th17 cells through modulation of dendritic cell activity via ADP-ribosylation of G-protein, as described for the ADP-ribosyltransferase cholera toxin (CT) (Datta et al. 2010). It has been hypothesized previously that SFB may stimulate Th17 cell induction via dendritic cells (Ivanov et al. 2009). The presence of three to four ADPRT sequence variants in SFB lineages suggests multiple, differential roles for these protein domains.
The generation of five independent genome data sets gave us the opportunity to examine similarity as well as variability between five individual SFB filaments that originate from the same mouse fecal sample. SNPs detected among the five SFB filaments provide a snapshot of genetic variability at a given time in a given mouse-associated SFB population. Sequence variation could be the result of spontaneous mutations that may either be eliminated over time or become fixed in the population. Population diversification may be driven by competition or coevolution (Kassen and Rainey 2004; Yoder and Nuismer 2010). The mutation of a conserved residue in OadA (synthesizes pyruvate from oxaloacetate) in filament SFB-5 may have a negative effect on its growth, because mutations of the same residue in the OadA of other bacteria are deleterious (Jain et al. 1996; Studer et al. 2007). Intriguingly, filament SFB-5 exhibits dwarfism compared with the other SFB filaments (Supplemental Fig. S2). A second mutation in filament SFB-5, namely, in PK (synthesizes pyruvate from phosphoenolpyruvate), suggests an epistatic interaction (i.e., the effect of one gene is modified by the effect of another gene) (Lehner 2011). More specifically, it may be an example of a compensatory adaptation and, at least in part, alleviate the phenotypic consequences of the mutated OadA in SFB-5 (Moore et al. 2000). Alternatively, it may be an initial step in a path of sequential mutations guiding this subpopulation toward an adaptive peak, alleviating the negative effects of mutated OadA (Poelwijk et al. 2007; Meyer et al. 2012).
Other recently published SFB genome sequences gave us the opportunity to explore differences between different SFB populations. SFB-mouse-SU, SFB-mouse-Yit, and SFB-mouse-NYU exhibited high sequence similarity, which is not surprising given that the mice from which those SFB were derived all originated from the same laboratory. We identified, however, several sequence polymorphisms among those genomes that suggest the presence of distinct SFB lineages within the same mouse colony. Distinct SFB lineages could contribute to the fitness of SFB and may be responsible for reported differences in SFB prevalence, as well as abundance in apparently identical animals, housed under the same conditions (Garland et al. 1982; Klaasen et al. 1992). In addition, there are striking differences in SFB colonization between various mouse strains that may be reflected in differences we identified between SFB-mouse-Japan and the other SFB-mouse genomes (SFB-mouse-SU, SFB-mouse-Yit, and SFB-mouse-NYU) (Klaasen et al. 1993a; Ohashi et al. 2006; Ivanov et al. 2009).
The observations described in this study offer starting points for future research on the ecology, evolution, cell biology, and physiology of SFB and their interactions with their hosts.
Methods
Microfluidic device cell sorting and genome amplification
Microfluidic 48-channel cell-sorting devices were produced by the Stanford Microfluidics Foundry. Individual SFB filaments were isolated from a fecal sample from an SFB-monocolonized mouse (gift from Y. Umesaki, Yakult). No other bacterial morphotypes or 16S rDNA sequence types, other than those characteristic of SFB, were found in the fecal material as examined by fluorescent in situ hybridization (FISH) and broad-range 16S rDNA sequencing. Individual cells were separated from the bulk sample using a laser trap (Supplemental Fig. S2A). The Repli-G midi MDA reagents (QIAGEN) were used to amplify genomes from individual SFB in 60-nL chambers on the device. Reaction products were examined for the presence and identity of 16S rDNA sequences through bacterial broad-range and SFB-specific PCR, and sequencing. For details about the microfluidic device, cell sorting, and genome amplification, see the Supplemental Material.
DNA extraction, 16S rDNA amplification, and fluorescent in situ hybridization (FISH)
DNA was extracted from fecal samples using the QIAamp DNA Stool Mini Kit (QIAGEN), according to the manufacturer's recommendations. The 16S rRNA gene was amplified using broad-range bacterial-specific primers Bact8FM and Bact1391R (Palmer et al. 2007). PCR products were cloned and inserts sequenced on both strands. To examine multiple displacement amplification (MDA) products subsequent to cell sorting for the presence of SFB-specific, as well as bacterial 16S rDNA sequences, PCR was performed using SFB-specific primers SFB747F and SFB1266R and bacterial broad-range primers Bact8FM and Bact1391R. Five samples, from which only SFB-specific 16S rDNA sequences were obtained, were selected for pyrosequencing. For FISH analysis, bacterial cells were fixed according to a protocol designed for fixation of Gram-positive bacteria (Roller et al. 1994). Cy3-labeled SFB-specific probe SFB1266R and Cy5-labeled probe EUB338 (Amann et al. 1990) were used to detect SFB and bacteria, respectively. For details about DNA extraction, primer design, 16S rDNA amplification, cloning and insert sequencing, and FISH analysis, see the Supplemental Material.
Sequencing library preparation and shotgun sequencing
Aliquots of each first-round MDA reaction product were reamplified using the Repli-G midi kit (QIAGEN). Multiplexed shotgun libraries were prepared according to the GS-FLX Titanium general library protocol (Roche-454 Life Sciences) using custom bar-coded adaptor oligos. Sequencing libraries were quantified by digital PCR (Fluidigm), using amplification primers complementary to the titanium adaptor. DNA pyrosequencing of the shotgun library was performed on a Roche 454 Genome Sequencer FLX instrument using titanium chemistry. For details about sequencing library preparation, quantification, and shotgun sequencing, see the Supplemental Material.
Assembly, gene prediction, annotation, and genome size prediction
Reads from the shotgun pyrosequencing runs were binned according to individual SFB filament and trimmed using the sfffile tool (Roche). Reads from each SFB filament were individually assembled (SFB-1 to SFB-5) and coassembled (SFB-co) de novo using Newbler version 2.5.3 (Roche) with an increased value for the expected read depth to account for the increased variation in read depth as a result of MDA. In addition, the reads from all five cells were coassembled by mapping against the complete genome sequence from SFB-mouse-Yit (AP012209) (Prakash et al. 2011); the resulting assembly is referred to as SFB-mouse-SU. Protein-coding genes were predicted and annotated. To determine the approximate genome size, rarefaction and shared sequence analyses were performed. For details about assembly, removal of chimeric and contaminant reads, gene prediction, annotation, and genome size prediction, see the Supplemental Material.
Comparative genomics and cluster analysis
Comparative genomic analyses of gene and protein sequences were based on a local copy of NCBI's Complete Microbial Genomes (downloaded 2010-02-01). For the cluster analysis, predicted proteins from the SFB coassembly and members of the Clostridiaceae 1 were compared with each other using BLASTp, and the resulting hits were then filtered and used to create an adjacency list representation with the proteins as nodes and the percent bitscore as the edge weight. The network list was analyzed using the Markov cluster (MCL) algorithm to produce a list of protein clusters approximating protein families (http://micans.org/mcl/). A matrix was constructed and analyzed using Principal Component Analysis (PCA). Comparative whole and nearly complete SFB genome sequence analysis was performed using Mauve (Darling et al. 2004), Artemis Comparison Tool (ACT) (Carver et al. 2008), and Kalign2 (Lassmann et al. 2009). For details about comparative genomics and cluster analysis, see the Supplemental Material.
Sequence analysis, phylogenetic tree construction, and SNP detection
Proteins were aligned using MUSCLE (Edgar 2004), 16S rDNA sequences were aligned using SILVA INcremental Aligner, and maximum likelihood phylogenetic analyses were performed. Protein structures were predicted using PHYRE2. Amino acid conservations were examined through PRALINE. Via three independent approaches, the presence of sequence variation between individual SFB (SFB-1 to SFB-5) was explored. First, BLAST pairwise comparisons of nucleotide and amino acid sequences from all protein-coding genes between and among the individual SFB assemblies were performed, and polymorphisms were recorded for best reciprocal hit (BRH) pairs between genomes (Supplemental Table S4; Supplemental Material). In a second approach, SNPs were identified as discrepancies between aligned individual SFB genome assemblies and the coassembly (Table 3; Supplemental Material). In a third approach, SNPs were identified as discrepancies between individual SFB whose reads were mapped against the complete genome SFB-mouse-Yit (Supplemental Fig. S13; Supplemental Material). For more information about sequence analysis, including single nucleotide polymorphism (SNP) detection and phylogenetic tree construction, see the Supplemental Material.
Data access
The Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under the umbrella project PRJNA70149. The genome sequences described in this paper are the first versions, AFXR01000000 (SFB-1), AFXS01000000 (SFB-2), AFXT01000000 (SFB-3), AFXU01000000 (SFB-4), AFXV01000000 (SFB-5), AFXW01000000 (SFB-co), and AGVP01000000 (SFB-mouse-SU).
Acknowledgments
We thank Y. Umesaki (Yakult, Japan) and D. Littman (New York University) for providing fecal samples from SFB-monocolonized mice; S.D. Pedersen (Technical University of Denmark) for assistance with the FISH assay; M.A. Fischbach (UCSF) for helpful suggestions; P. Coggill (Welcome Trust Sanger Institute) for support with Pfam domain PF13946; S. Kulkarni (Stanford University, California) for providing fecal samples from conventional laboratory mice; and J. Haagensen (Stanford University, California) for providing Imaris software. This work was supported by National Institutes of Health Director's Pioneer Award DP1OD000964 to D.A.R. and R01 HG004863 to D.A.R. and S.R.Q. D.A.R. is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University. S.J.P. was supported by the Danish Council for Independent Research/Natural Sciences and is currently supported by Lundbeckfonden. E.D.H. was supported by a Human Frontiers Science Program LTF.
Authors' contributions: S.J.P. and D.A.R. conceived the study; S.J.P., E.D.H., S.R.Q., P.C.B., and D.A.R. designed the research; S.J.P. and P.C.B. performed the research; S.J.P., E.D.H., P.C.B., S.R.Q., and D.A.R. contributed new reagents/analytic tools; S.J.P., E.D.H., P.C.B., S.R.Q., and D.A.R. analyzed the data; S.J.P. and D.A.R. wrote the manuscript; and E.D.H., P.C.B., and S.R.Q. edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131482.111.
Freely available online through the Genome Research Open Access option.
References
- Adriouch S, Hubert S, Pechberty S, Koch-Nolte F, Haag F, Seman M 2007. NAD+ released during inflammation participates in T cell homeostasis by inducing ART2-mediated death of naive T cells in vivo. J Immunol 179: 186–194 [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56: 1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Barnett LA, Cunningham MW 1992. Evidence for actinlike proteins in an M protein-negative strain of Streptococcus pyogenes. Infect Immun 60: 3932–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A, Biswas R, Herbert S, Gotz F 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect Immun 74: 4598–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol 187: 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Gottschalk G 2008. Comparative genomics of clostridia: Link between the ecological niche and cell surface properties. Ann NY Acad Sci 1125: 73–81 [DOI] [PubMed] [Google Scholar]
- Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA 2008. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24: 2672–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R 2010. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev 24: 754–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase DG, Erlandsen SL 1976. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J Bacteriol 127: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT 2004. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Lahiri A, Chakravortty D 2010. Modulation of the arginase pathway in the context of microbial pathogenesis: A metabolic enzyme moonlighting as an immune modulator. PLoS Pathog 6: e1000899 doi: 10.1371/journal.ppat.1000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SK, Sabet M, Nguyen KP, Valdez PA, Gonzalez-Navajas JM, Islam S, Mihajlov I, Fierer J, Insel PA, Webster NJ, et al. 2010. Mucosal adjuvant activity of cholera toxin requires Th17 cells and protects against inhalation anthrax. Proc Natl Acad Sci 107: 10638–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CP, Savage DC 1974. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun 10: 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieldhouse RJ, Turgeon Z, White D, Merrill AR 2010. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput Biol 6: e1001029 doi: 10.1371/journal.pcbi.1001029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31: 677–689 [DOI] [PubMed] [Google Scholar]
- Garland CD, Lee A, Dickson MR 1982. Segmented filamentous bacteria in the rodent small intestine: Their colonization of growing animals and possible role in host resistance to Salmonella. Microb Ecol 8: 181–190 [DOI] [PubMed] [Google Scholar]
- Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, et al. 2002. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci 11: 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Moore ER, Hofle MG 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl Environ Microbiol 65: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton JC. 1962. Observations on the relations between intestinal epithelial cells and cellular components of luminal contents in the distal ileum of the mouse, pp. LL9–LLI0. Academic Press, New York. [Google Scholar]
- Harrington ED, Arumugam M, Raes J, Bork P, Relman DA 2010. SmashCell: A software framework for the analysis of single-cell amplified genome sequences. Bioinformatics 26: 2979–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka A, Okada Y, Matsumoto S, Setoyama H, Umesaki Y 1997. 16S ribosomal DNA sequence divergence of segmented filamentous bacteria with special reference to inter-species and within-species variation of host animals. Syst Appl Microbiol 20: 418–422 [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. 2006. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci 103: 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol 175: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain B, Brand BC, Luck PC, Di Berardino M, Dimroth P, Hacker J 1996. An oxaloacetate decarboxylase homologue protein influences the intracellular survival of Legionella pneumophila. FEMS Microbiol Lett 145: 273–279 [DOI] [PubMed] [Google Scholar]
- Jepson MA, Clark MA, Simmons NL, Hirst BH 1993. Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infect Immun 61: 4001–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci 105: 13580–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Seed PC, Hultgren SJ 2006. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci 103: 19884–19889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R, Rainey PB 2004. The ecology and genetics of microbial diversity. Annu Rev Microbiol 58: 207–231 [DOI] [PubMed] [Google Scholar]
- Kil KS, Cunningham MW, Barnett LA 1994. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect Immun 62: 2440–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Poelma FG, Beynen AC 1992. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev 8: 165–180 [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC 1993a. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim 27: 141–150 [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC 1993b. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun 61: 303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci 101: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JP, Stadhouders AM, Kennis HM, De Boer H 1987. The attachment of filamentous segmented micro-organisms to the distal ileum wall of the mouse: A scanning and transmission electron microscopy study. Lab Anim 21: 48–52 [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, et al. 2011. The lifestyle of the segmented filamentous bacterium: A non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res 18: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JM, Bongers RS, de Vos WM, Kleerebezem M 2008. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl Environ Microbiol 74: 4719–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Frings O, Sonnhammer EL 2009. Kalign2: High-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res 37: 858–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci (Suppl 1) 108: 4615–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B 2011. Molecular mechanisms of epistasis within and between genes. Trends Genet 27: 323–331 [DOI] [PubMed] [Google Scholar]
- Leidy J 1849. October 9th. Proc Acad Nat Sci Philad 4: 227 [Google Scholar]
- Liu ZX, Yu Y, Dennert G 1999. A cell surface ADP-ribosyltransferase modulates T cell receptor association and signaling. J Biol Chem 274: 17399–17401 [DOI] [PubMed] [Google Scholar]
- Low HH, Lowe J 2006. A bacterial dynamin-like protein. Nature 444: 766–769 [DOI] [PubMed] [Google Scholar]
- Low LY, Yang C, Perego M, Osterman A, Liddington RC 2005. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J Biol Chem 280: 35433–35439 [DOI] [PubMed] [Google Scholar]
- Malachowa N, Kohler PL, Schlievert PM, Chuang ON, Dunny GM, Kobayashi SD, Miedzobrodzki J, Bohach GA, Seo KS 2011. Characterization of a Staphylococcus aureus surface virulence factor that promotes resistance to oxidative killing and infectious endocarditis. Infect Immun 79: 342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, et al. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci 104: 11889–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci 104: 19392–19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE 2012. Repeatability and contingency in the evolution of a key innovation in phage λ. Science 335: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani CJ, Aziz RK, Locke JB, Dahesh S, Nizet V, Buchanan JT 2010. The novel polysaccharide deacetylase homologue Pdi contributes to virulence of the aquatic pathogen Streptococcus iniae. Microbiology 156: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore FB, Rozen DE, Lenski RE 2000. Pervasive compensatory adaptation in Escherichia coli. Proc Biol Sci 267: 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto E, Yu Y, Dennert G 1996. Cell surface ADP-ribosyltransferase regulates lymphocyte function-associated molecule-1 (LFA-1) function in T cells. J Immunol 157: 3341–3349 [PubMed] [Google Scholar]
- O'Flaherty SJ, Klaenhammer TR 2010. Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology 156: 3360–3367 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Hiraguchi M, Ushida K 2006. The composition of intestinal bacteria affects the level of luminal IgA. Biosci Biotechnol Biochem 70: 3031–3035 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Azhipa O, Yu Y, Russo E, Dennert G 1998. Expression of ADP-ribosyltransferase on normal T lymphocytes and effects of nicotinamide adenine dinucleotide on their function. J Immunol 160: 4190–4198 [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Hunter MS, Moran NA 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325: 992–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO 2007. Development of the human infant intestinal microbiota. PLoS Biol 5: e177 doi: 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J 2002. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci 99: 8231–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasztor L, Ziebandt AK, Nega M, Schlag M, Haase S, Franz-Wachtel M, Madlung J, Nordheim A, Heinrichs DE, Gotz F 2010. Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J Biol Chem 285: 36794–36803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ 2007. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445: 383–386 [DOI] [PubMed] [Google Scholar]
- Portier P 1918. Les symbiotes Masson, Paris. [Google Scholar]
- Praefcke GJ, McMahon HT 2004. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, et al. 2011. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe 10: 273–284 [DOI] [PubMed] [Google Scholar]
- Roller C, Wagner M, Amann R, Ludwig W, Schleifer KH 1994. In situ probing of gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiology 140: 2849–2858 [DOI] [PubMed] [Google Scholar]
- Rosenberger CM, Finlay BB 2002. Macrophages inhibit Salmonella typhimurium replication through MEK/ERK kinase and phagocyte NADPH oxidase activities. J Biol Chem 277: 18753–18762 [DOI] [PubMed] [Google Scholar]
- Rosenthal RS, Folkening WJ, Miller DR, Swim SC 1983. Resistance of O-acetylated gonococcal peptidoglycan to human peptidoglycan-degrading enzymes. Infect Immun 40: 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford SE 1991. Light and electron microscopic observations of a segmented filamentous bacterium attached to the mucosa of the terminal ileum of pigs. J Vet Diagn Invest 3: 328–333 [DOI] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II 2011. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel J, Blok HJ, Kengen HMP, Ludwig W, Poelma FGJ, Koopman JP, Akkermans ADL 1994. Phylogenetic characterization of clostridium related segmented filamentous bacteria in mice based on 16S ribosomal-RNA analysis. Syst Appl Microbiol 17: 172–179 [Google Scholar]
- Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD 1995. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus.” Int J Syst Bacteriol 45: 780–782 [DOI] [PubMed] [Google Scholar]
- Stacey M, Webb M 1947. Studies on the antibacterial properties of the bile acids and some compounds derived from cholanic acid. Proc R Soc Med 134: 523–537 [DOI] [PubMed] [Google Scholar]
- Studer R, Dahinden P, Wang WW, Auchli Y, Li XD, Dimroth P 2007. Crystal structure of the carboxyltransferase domain of the oxaloacetate decarboxylase Na+ pump from Vibrio cholerae. J Mol Biol 367: 547–557 [DOI] [PubMed] [Google Scholar]
- Tannock GW, Miller JR, Savage DC 1984. Host specificity of filamentous, segmented microorganisms adherent to the small bowel epithelium in mice and rats. Appl Environ Microbiol 47: 441–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann B, Depping R, Gineikiene E, Kaliniene L, Nivinskas R, Ruger W 2004. ModA and ModB, two ADP-ribosyltransferases encoded by bacteriophage T4: Catalytic properties and mutation analysis. J Bacteriol 186: 7262–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge H, Nagahama M, Oda M, Iwamoto S, Utsunomiya H, Marquez VE, Katunuma N, Nishizawa M, Sakurai J 2008. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proc Natl Acad Sci 105: 7399–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, et al. 2008. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 9: 769–776 [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H 1995. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol 39: 555–562 [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K 1999. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun 67: 3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maele L, Carnoy C, Cayet D, Songhet P, Dumoutier L, Ferrero I, Janot L, Erard F, Bertout J, Leger H, et al. 2010. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3negCD127+ immune cells in spleen and mucosa. J Immunol 185: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Wu H, Jones R, Grant G, Babbin B, King TP, Kelly D, Gewirtz AT, Neish AS 2006. Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am J Pathol 169: 1686–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov A, Liavonchanka A, Kamneva O, Fiedler T, Goebel C, Kreikemeyer B, Feussner I 2010. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J Biol Chem 285: 10353–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Tomasz A 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J Biol Chem 275: 20496–20501 [DOI] [PubMed] [Google Scholar]
- Wilson KH 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18: 1017–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol 2: e69 doi: 10.1371/journal.pbio.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32: 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi KE, Snel J 2000. Transmission electron microscopic demonstration of phagocytosis and intracellular processing of segmented filamentous bacteria by intestinal epithelial cells of the chick ileum. Infect Immun 68: 6496–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JB, Nuismer SL 2010. When does coevolution promote diversification? Am Nat 176: 802–817 [DOI] [PubMed] [Google Scholar]
- Zoll S, Patzold B, Schlag M, Gotz F, Kalbacher H, Stehle T 2010. Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS Pathog 6: e1000807 doi: 10.1371/journal.ppat.1000807 [DOI] [PMC free article] [PubMed] [Google Scholar]