The zinc finger transcription factor Zbtb46 specifically marks cDCs and their committed precursors and, when overexpressed in BM progenitors, promotes cDC development at the expense of granulocytes.
Abstract
Distinguishing dendritic cells (DCs) from other cells of the mononuclear phagocyte system is complicated by the shared expression of cell surface markers such as CD11c. In this study, we identified Zbtb46 (BTBD4) as a transcription factor selectively expressed by classical DCs (cDCs) and their committed progenitors but not by plasmacytoid DCs (pDCs), monocytes, macrophages, or other lymphoid or myeloid lineages. Using homologous recombination, we replaced the first coding exon of Zbtb46 with GFP to inactivate the locus while allowing detection of Zbtb46 expression. GFP expression in Zbtb46gfp/+ mice recapitulated the cDC-specific expression of the native locus, being restricted to cDC precursors (pre-cDCs) and lymphoid organ– and tissue-resident cDCs. GFP+ pre-cDCs had restricted developmental potential, generating cDCs but not pDCs, monocytes, or macrophages. Outside the immune system, Zbtb46 was expressed in committed erythroid progenitors and endothelial cell populations. Zbtb46 overexpression in bone marrow progenitor cells inhibited granulocyte potential and promoted cDC development, and although cDCs developed in Zbtb46gfp/gfp (Zbtb46 deficient) mice, they maintained expression of granulocyte colony-stimulating factor and leukemia inhibitory factor receptors, which are normally down-regulated in cDCs. Thus, Zbtb46 may help enforce cDC identity by restricting responsiveness to non-DC growth factors and may serve as a useful marker to identify rare cDC progenitors and distinguish between cDCs and other mononuclear phagocyte lineages.
DCs are immune accessory cells critical for both innate and adaptive responses against pathogens (Steinman, 2012). Multiple subtypes of DCs have been identified that have distinct functions and molecular characteristics (Shortman and Naik, 2007). The types of DCs that appear to be present in both human and mouse include plasmacytoid DCs (pDCs), which generate type-1 interferon, and antigen-presenting classical DCs (cDCs), both of which are present in lymphoid and nonlymphoid tissues. At steady-state, cDCs can be further divided into CD8α+ and CD8α− subsets, which possess differential capacities for promoting responses to pathogens as a result of differences in their ability to cross-present antigens and secrete cytokines (den Haan et al., 2000; Hildner et al., 2008). Moreover, upon infection, additional subsets of cDCs can be induced from monocytes and share many of the distinguishing markers of homeostatic cDCs but may carry out distinct functions (Cheong et al., 2010).
Distinguishing these various DC subsets from closely related lineages such as monocytes and macrophages remains problematic. First, distinctions are based on patterns and expression levels of many surface markers rather than a single unique identifier. Second, expression levels for these markers may be altered by cellular activation through cytokines and innate sensing pathways or anatomical localization (Geissmann et al., 2010; Hume, 2011). In analyzing the physiological role of DCs, CD11c has frequently been used as a surrogate marker to identify the lineage. As such, the CD11c locus has been modified to express fluorescent reporters, Cre recombinase, or the diphtheria toxin receptor (DTR) for the purpose of allowing tracking of DCs or to allow the inducible depletion of DCs for functional studies in vivo (Jung et al., 2002; Lindquist et al., 2004; Caton et al., 2007). However, the interpretation of studies using these reagents has been confounded by the expression of CD11c by other lineages (Probst et al., 2005; Murphy, 2011). For example, in CD11c-DTR mice, the depletion of DCs by administration of diphtheria toxin A also leads to depletion of other cell types, including tissue-resident, marginal zone, and metallophilic macrophages, NK cells, and NKT cells, as well as some CD11c+ B and T cells (van Rijt et al., 2005; Bennett and Clausen, 2007; Hume, 2011). Other loci, including LysM, Csf1r, Cx3cr1, and CD11b, have been modified to express fluorescent proteins, but these are also expressed not only in DCs but in additional myeloid populations as well (Hume, 2011). No single marker has been identified to uniquely distinguish cDCs from other myeloid or lymphoid lineages, prompting the suggestion that identification of bona-fide cDCs may require the simultaneous analysis of surface phenotype, cellular derivation, function, and anatomical location (Geissmann et al., 2010).
DCs are derived from BM myeloid progenitors that undergo progressive lineage restriction. The common myeloid progenitor (CMP) generates the common DC progenitor (CDP), which has lost the ability to give rise to granulocytes and macrophages (derived from the downstream granulocyte macrophage precursor [GMP]) but still retains both pDC and cDC potential (Onai et al., 2007; Naik et al., 2007). The CDP appears to be the immediate precursor of both pre-pDCs (Cisse et al., 2008) and pre-cDCs (Naik et al., 2006; Liu et al., 2009), which are restricted to their respective lineage but not yet fully mature. Terminal cDC development occurs in peripheral lymphoid and nonlymphoid organs after Flt3L (fms-like tyrosine kinase 3 ligand)-dependent expansion (Waskow et al., 2008).
We previously suggested that accurately defining the cDC lineage might benefit from methods based on lineage-specific transcription factors induced during these early stages of commitment (Satpathy et al., 2011). Several transcription factors have already been found to regulate DC development (Satpathy et al., 2011) but are not suitable for the purpose of specifically marking the lineage in vivo. For example, Irf8−/− mice lack pDC as well as CD8α+ cDC lineages but also have additional defects in other immune cell types (Schiavoni et al., 2002; Wang and Morse, 2009). Likewise, the transcription factor Batf3, which is required for the development of the CD8α+ cDC lineage, is expressed in cDCs but also in other myeloid cells (Heng and Painter, 2008) and differentiated TH1 cells (Hildner et al., 2008).
In this study, we identify a member of the BTB-ZF (Broad complex, Tramtrack, Bric-à-brac, and Zinc finger) family (Kelly and Daniel, 2006), Zbtb46 (BTBD4), as specifically expressed in cDCs among immune lineages. By targeting the locus to express GFP, we are able to distinguish cDCs from all other lineages within the immune system, including closely related myeloid lineages. Furthermore, Zbtb46 expression identifies the earliest committed precursor of cDCs. Zbtb46 is not required for cDC development in vivo but regulates silencing of G-CSF and leukemia inhibitory factor receptors that normally occurs during cDC differentiation.
RESULTS
Zbtb46 is selectively expressed in cDCs and committed cDC progenitors
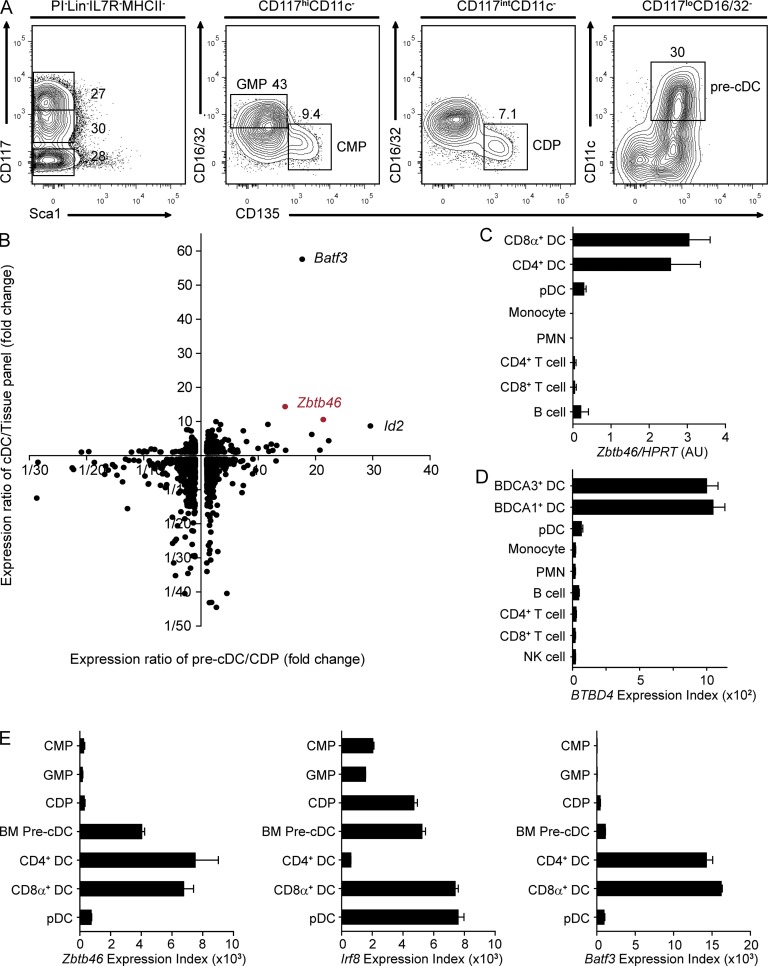
To identify transcription factors expressed in early cDC progenitors but not in other myeloid precursors, we purified CMPs, GMPs, CDPs, and BM and splenic pre-cDCs (Fig. 1 A) and performed gene expression analysis (Fig. 1 B). Transcription factors were compared for their induction in splenic pre-cDCs compared with BM CDPs and for their lineage specificity in cDCs compared with a broad panel of hematopoietic and nonhematopoietic cells (Fig. 1 B; Lattin et al., 2008). This method identified previously studied cDC genes, Batf3 and Id2, as well as the novel factor Zbtb46. Zbtb46 was induced nearly 20-fold in splenic pre-cDCs relative to CDPs and was highly cDC specific across the tissue panel (Fig. 1 B).
Figure 1.
Zbtb46 is expressed in BM pre-cDCs and cDCs in both mouse and human. (A) BM cells from WT mice were stained for expression of the indicated markers. Two-color histograms are shown for live cells pregated as indicated above the diagram. Numbers represent the percentage of cells within the indicated gate. Data are representative of five independent experiments with at least two mice each. (B) Sorted populations in A were analyzed by gene expression microarray as described in Materials and methods. Gene expression levels were determined for a list of all transcription factors using dChip software (Li and Wong, 2001a,b) for the splenic pre-cDC, BM CDP, CD8α+ and CD4+ cDCs, and a broad panel of tissues (Lattin et al., 2008). For each transcription factor, the horizontal axis indicates the ratio of gene expression in the pre-cDC compared with the CDP, and the vertical axis indicates the ratio of the mean expression in CD8α+ and CD4+ DCs compared with the mean expression in the tissue panel excluding DCs. Each dot indicates an individual probe set. (C) Shown is the normalized expression value of Zbtb46 mRNA determined by quantitative RT-PCR for the indicated cell populations. Data represent three independently sorted replicates from three mice. (D) Shown is the expression value of BTBD4 mRNA in the indicated cells derived from Lindstedt et al. (2005) and Du et al. (2006). (E) Shown are the expression values for mouse Zbtb46, Irf8, and Batf3 mRNA transcripts in the indicated cell populations determined by expression microarray. Microarray analysis in B and E represents either two or three independently obtained arrays from progenitors sorted from three to five pooled WT mice. cDC arrays in E are derived from Robbins et al. (2008). (C–E) Bars represent the mean ± SEM.
Zbtb46 was expressed in both the CD8α+ and CD4+ subsets of cDCs but at very low levels in pDCs, monocytes, granulocytes, and T and B cells (Fig. 1 C). Zbtb46 is conserved as BTBD4 in humans, where it was also specifically expressed in both major subsets of cDCs but not pDCs, monocytes, granulocytes, or T, B, or NK cells (Fig. 1 D). In mouse, Zbtb46 was first expressed in the BM pre-cDC and increased during differentiation into mature cDC subsets (Fig. 1 E). In contrast, Irf8 was expressed in the earlier CMP and GMP stages, highly induced in the CDP, and maintained in the pre-cDC and subsequent CD8α+ cDC and pDC. Expression of Batf3 was low in BM progenitors and was induced beginning in the later splenic pre-cDC stage and maintained in mature cDCs (Fig. 1 E; Edelson et al., 2010).
Zbtb46-driven GFP expression identifies cDCs in lymphoid organs
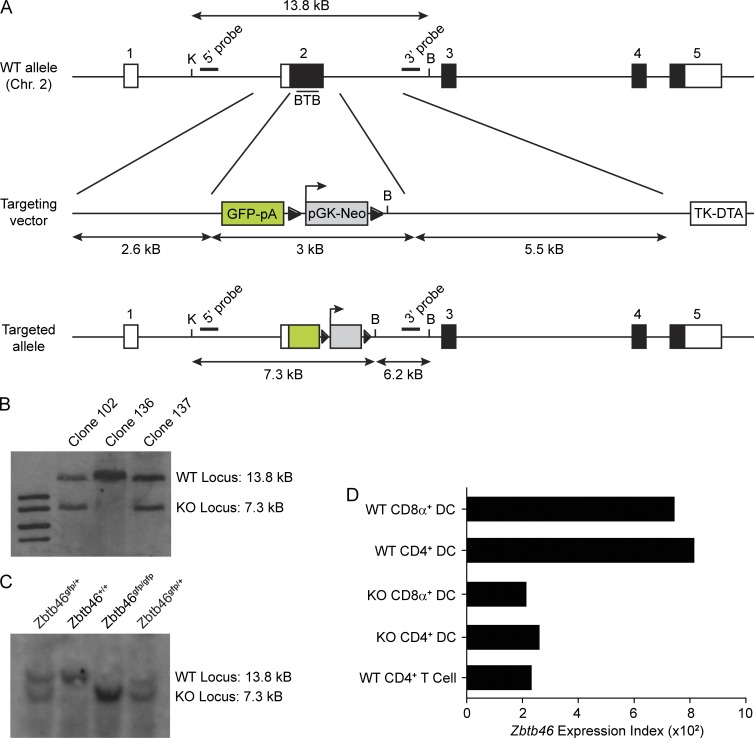
Zbtb46 encodes a 601-aa member of the BTB-ZF family of transcriptional repressors (Kelly and Daniel, 2006) for which no studies have been reported. To examine its expression pattern and function in vivo, we designed a targeting strategy to inactivate the locus while also allowing for detection of its expression. We inserted a GFP reporter cassette into exon 2 beginning at the first coding methionine of Zbtb46, thus removing the native translational start site and completely eliminating exon 2, which encodes the Zbtb46 BTB domain (Fig. 2 A). Two ES cell clones harboring the correctly targeted Zbtb46gfp allele were obtained (Fig. 2 B) and used to generate germline-targeted mice on the pure 129SvEv genetic background (Fig. 2 C). Both of these correctly targeted clones were successfully transmitted through the germline, and progeny derived from each clone exhibited the same phenotype and patterns of GFP expression. Data in this study are exclusively from descendants of clone 102. Mice of genotypes Zbtb46gfp/+ and Zbtb46gfp/gfp are viable and are produced at normal Mendelian frequencies from heterozygous breeding pairs. Zbtb46 transcripts were reduced in DCs isolated from Zbtb46gfp/gfp mice to background levels, indicating that this targeting strategy interrupted normal gene transcription as intended (Fig. 2 D).
Figure 2.
Generation of the Zbtb46gfp locus. (A) The endogenous mouse Zbtb46 locus (top) contains five exons, with the first exon entirely noncoding and the second exon containing the methionine initiation codon and encoding the BTB domain of Zbtb46 (line). The Zbtb46 locus, the targeting vector, and mutant allele with retention of the neomycin (pGK-Neo) selection cassette are shown. Exons are displayed as boxes, with coding regions shaded. Double digestion of the germline locus with KpnI (K) and BamHI (B) yields a restriction fragment of 13.8 kB, which is detected by both the 5′ probe and 3′ probe. Correctly targeted clones show an additional 7.3-kB band detected by the 5′ probe and a 6.2-kB band detected by the 3′ probe. Triangles indicate LoxP sequences. DTA, diphtheria toxin A; pA, polyadenylation signal; TK, thymidine kinase promoter. (B) Southern blot analysis of targeted clones. KpnI–BamHI double digests of clones 102, 136, and 137 were electrophoresed and hybridized with 5′ probe as previously described (Kohyama et al., 2009). (C) Southern blot analysis of tail DNA. Tail biopsies of germline-transmitted descendents of clone 102 obtained from breeding of heterozygous Zbtb46gfp/+ mice were double digested with KpnI and BamHI, electrophoresed, and hybridized with 5′ probe. Shown above the gel are the interpreted genotypes. (D) Splenic CD4+ cDCs and CD8α+ cDCs were isolated from Zbtb46+/+ (WT) and Zbtb46gfp/gfp (KO) mice, and gene expression microarray analysis was performed in comparison with WT CD4+ T cells. Shown is the expression value of the Zbtb46 probe set from the Affymetrix mouse Gene 1.0 array for the indicated cell types. Data are assembled from one to two replicate arrays obtained in one experiment.
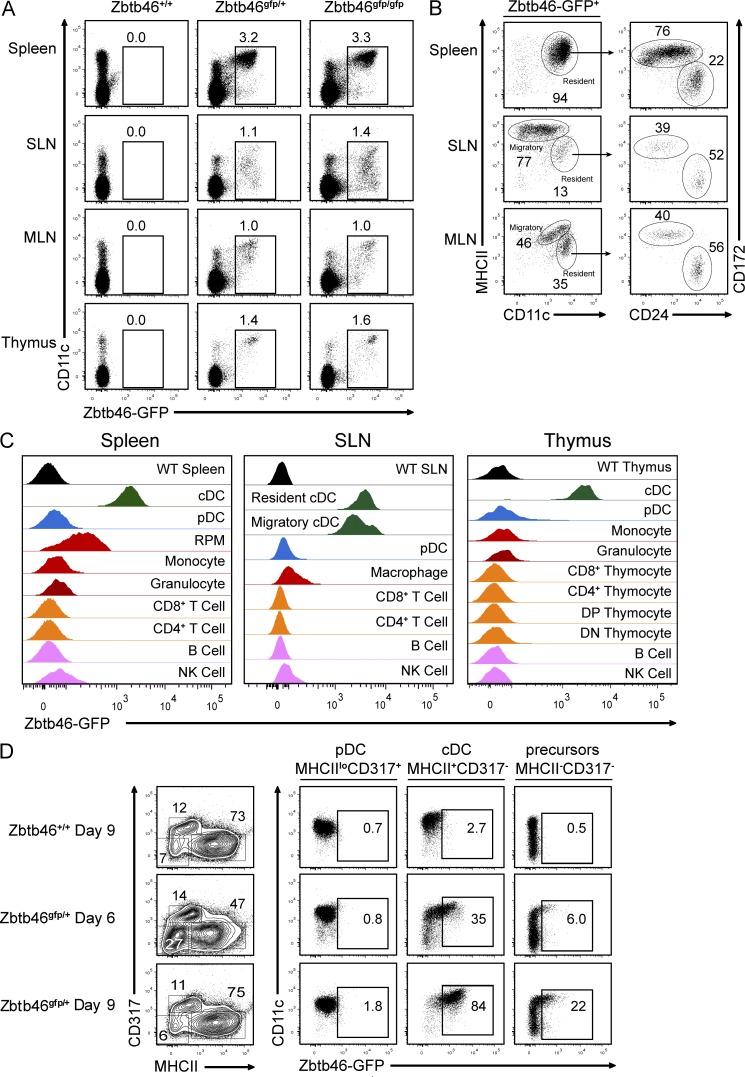
We first analyzed Zbtb46 expression in lymphoid organs, including spleen, skin-draining LNs (SLNs), mesenteric LNs (MLNs), and thymus (Fig. 3 A). In CD45+ cells analyzed by FACS, GFP-expressing cells were present at equivalent frequencies in both Zbtb46gfp/+ and Zbtb46gfp/gfp mice, indicating that no major populations of cells failed to develop in the absence of Zbtb46 (Fig. 3 A). In spleen, GFP+ cells were overwhelmingly CD11c+MHCII+ and comprised both CD172+CD24− and CD172−CD24+ populations (Fig. 3 B), indicating Zbtb46 expression in both major subsets of cDCs. In LNs, GFP was not only expressed by cells with high levels of CD11c, but also by migratory cDCs that expressed reduced levels of CD11c. Here, CD11chi cells expressed intermediate levels of MHCII, representing lymphoid-resident DCs, whereas CD11cint/lo cells expressed high levels of MHCII, representing migratory DCs (Fig. 3 B; Shortman and Naik, 2007). Again, GFP+ resident DCs in SLNs and MLNs contained both CD172+CD24− and CD172−CD24+ populations (Fig. 3 B), indicating that Zbtb46 is expressed in all cDC subsets in peripheral lymphoid organs as well.
Figure 3.
Zbtb46gfp specifically identifies cDCs in the mouse. Cells from Zbtb46+/+, Zbtb46gfp/+, and Zbtb46gfp/gfp were harvested from the spleen, SLNs, MLNs, and thymus as indicated and stained for CD11c, MHCII, CD172, CD24, and CD45. (A) Shown are two-color histograms for the indicated markers after gating for CD45 expression. Numbers represent the percentage of cells in the indicated gate. (B) Shown are two-color histograms for the indicated markers derived from a GFP+ gate as indicated in A from Zbtb46gfp/gfp mice. (C) Shown are single-color histograms for GFP expression of cells from Zbtb46+/+ mice (WT) or cells from Zbtb46gfp/+ mice stained to identify cDCs (MHCIIhiCD11chi), resident cDCs (MHCIIintCD11chi), migratory cDCs (MHCIIhiCD11cint/lo), pDCs (CD317+B220+), RPMs (F4/80+ autofluorescent), monocytes (CD11bhiCD11c−MHCII−Ly6G−), granulocytes (Ly6G+CD11b+), CD8+ T cells (CD8α+CD11c−), CD4+ T cells (CD4+CD11c−), B cells (B220+MHCII+), NK cells (NKp46+), DP thymocytes (CD4+CD8+Lin−), or DN thymocytes (CD4−CD8−Lin−). (D) BM cells from Zbtb46+/+ or Zbtb46gfp/+ mice were cultured with 100 ng/ml Flt3L for 6–9 d as indicated, harvested, and stained for CD11c, MHCII, and CD317. Gates identifying pDCs, cDCs, and precursors are indicated in two-color histograms for CD317 and MHCII expression on the left. Shown on the right are two-color histograms for CD11c and GFP expression for pDCs, cDCs, and precursor cells. All data in this figure are representative of three to five independent experiments using at least two mice per group.
Notably, GFP expression was not detected in any other myeloid or lymphoid lineages (Fig. 3 C). In particular, pDCs were devoid of GFP expression in spleen, SLNs, and thymus, as were monocytes, granulocytes, and NK cells. Red pulp macrophages (RPMs) showed a low level of autofluorescence that did not represent authentic GFP expression, as similar levels of autofluorescence were observed in RPMs of Zbtb46+/+ (WT) mice (not depicted) and anti-GFP staining was not detected in RPMs by immunofluorescence microscopy (see Fig. 8 D, bottom). This restricted GFP expression in cDCs but not pDCs was also observed in Flt3L-treated BM cultures (Fig. 3 D). Here, GFP expression was observed in both committed cDCs and pre-cDCs but not pDCs at both 6 and 9 d of differentiation.
Figure 8.
Zbtb46 is expressed in definitive erythroid precursors and endothelial cells. (A) BM cells from Zbtb46+/+ (WT) or Zbtb46gfp/gfp (KO) mice were stained for expression of lineage markers B220, NKp461, and CD3 (Lin), Sca-1, CD16/32, CD41, CD105, CD117, CD135, and CD150 and analyzed using a FACSAria. Shown are two-color histograms for the indicated markers on cells previously gated for expression as indicated above the diagram. Numbers represent the percentage of cells within the indicated gate. The gates used to identify MkP, GMP, CMP, CDP, CFU-E, pre–CFU-E, pre-MegE, and pre-GM populations for later GFP expression analysis are indicated. Data are representative of five independent experiments with at least two mice per group. (B) Shown are single-color histograms for GFP expression of cells stained in A for each indicated gate. (C) Shown are mean expression values of Zbtb46 (±SEM) for each indicated cell population derived from Pronk et al. (2007). Data are assembled from two to four replicate arrays. (D) Spleen sections from Zbtb46gfp/+ (left) or Zbtb46gfp/+ → WT BM chimera (right) mice were stained for CD11c, B220, F4/80, and GFP. Arrows indicate GFP+CD11c− endothelial cells. Data are representative of three independent experiments with one or two mice per group. Bars, 200 µm. (E) Splenocytes were stained for expression of CD11c, CD31, MADCAM-1, and Flk1. Shown are two-color histograms of cells from Zbtb46gfp/+ (left and middle) or a Zbtb46gfp/+ → WT BM chimera (right) for the indicated markers of cells previously gated as indicated above the diagram. Gates are labeled by cellular identity. Data are representative of three independent experiments with one or two mice each.
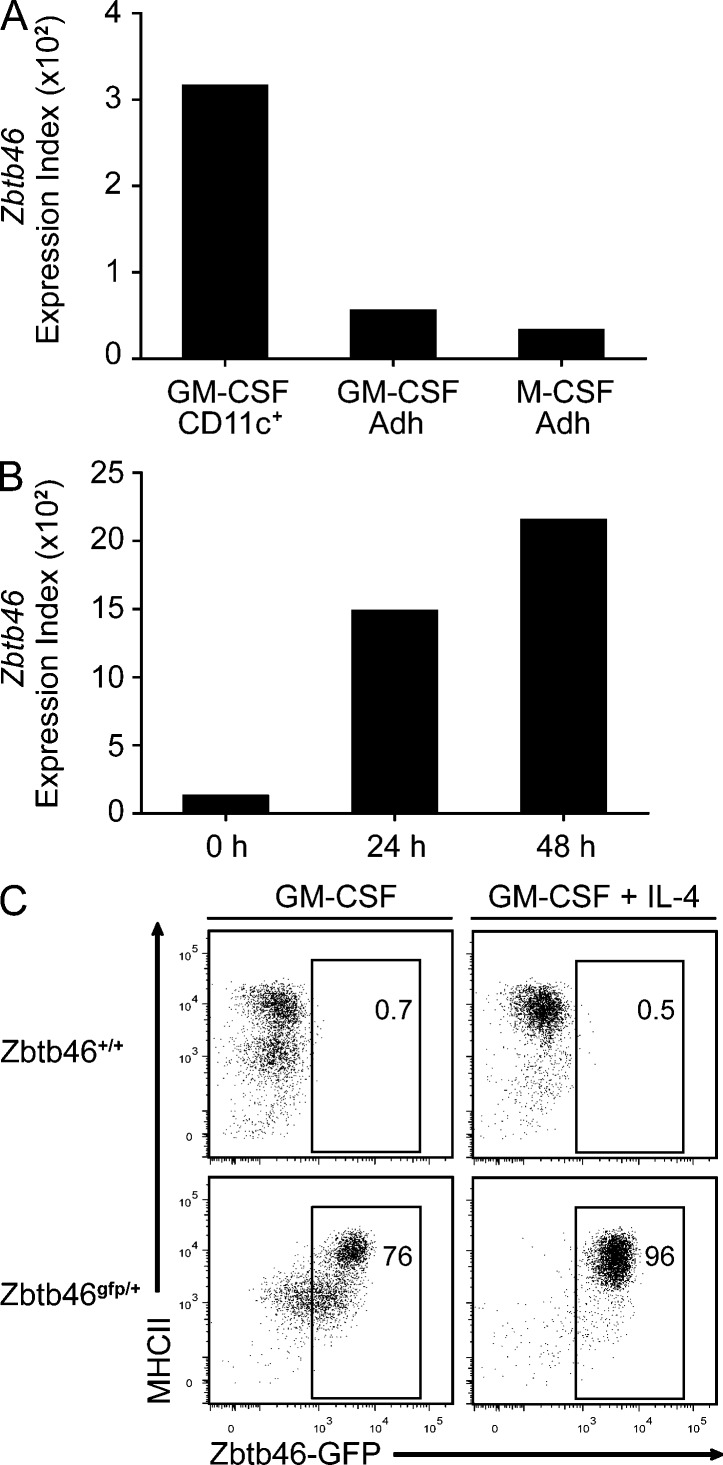
Interestingly, Zbtb46 was also expressed in nonadherent CD11c+ cDCs generated in BM cultures treated with GM-CSF (Fig. 4 A; Sallusto and Lanzavecchia, 1994). Adherent cells either from GM-CSF–treated cultures, which represent macrophages and granulocytes, or from M-CSF–treated cultures, which represent macrophages, did not express Zbtb46 (Fig. 4 A). Purified monocytes cultured with GM-CSF induced the expression of Zbtb46 after 24 h (Fig. 4 B) and induced high levels of Zbtb46gfp after 4 d in culture (Fig. 4 C). Notably, after 4 d of treatment with GM-CSF alone, Zbtb46gfp was induced in ∼75% of cells, whereas with the addition of both GM-CSF and IL-4, they became uniformly MHCII high and >95% GFP positive, indicating that IL-4 may enhance DC differentiation of monocytes. These results demonstrate that both Flt3L- and GM-CSF–dependent derivations of cDCs express Zbtb46, whereas macrophages and granulocytes do not.
Figure 4.
GM-CSF–derived cDCs express Zbtb46gfp. (A) BM cells from C57BL/6 mice were treated with 20 ng/ml GM-CSF or M-CSF, and cultures were harvested on day 7 and divided into adherent and nonadherent fractions. Nonadherent CD11c+ cDCs from the GM-CSF–treated cultures were purified by positive selection, and microarray analysis was performed on cDCs (GM-CSF CD11c+) and both adherent populations (Adh) for the indicated cultures. Shown is the expression value of the Zbtb46 probe set. (B) BM monocytes were purified by negative selection using antibodies for Ly6G, B220, CD4, CD8α, DX5, CD11c, Ter119, and CD117, resulting in a fraction of cells that were >90% pure for expression of CD11b and Ly6C. Cells were cultured with 20 ng/ml GM-CSF for the indicated number of hours and harvested, and microarray analysis was performed. Shown is the expression value of the Zbtb46 probe set. Data in A and B are from one experimental replicate obtained from cells pooled from three mice. (C) Monocytes were purified from BM by cell sorting as CD11b+Ly6C+Ly6G−CD11c−MHCII−Siglec-H− cells and cultured in 20 ng/ml GM-CSF with or without 20 ng/ml IL-4 as indicated and analyzed after 4 d for expression of Zbtb46gfp and MHCII. Data are representative of one experiment in which each sample was performed in triplicate. Numbers represent the percentage of cells in the indicated gate.
Zbtb46gfp expression distinguishes cDCs from macrophages in peripheral tissues
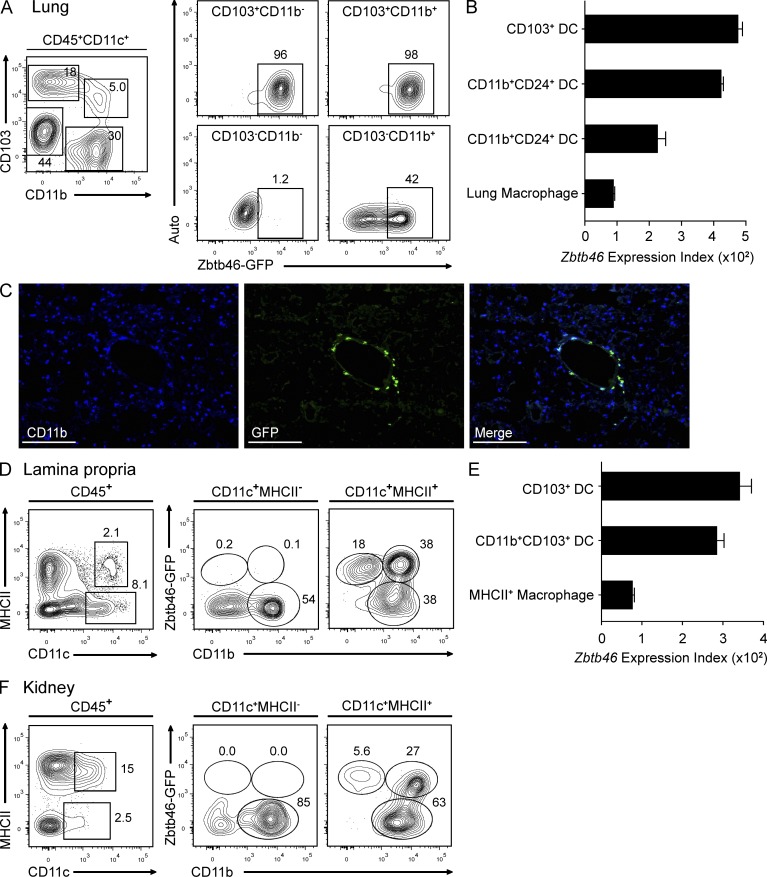
A particular problem arises in using CD11c to distinguish cDCs from macrophages in peripheral nonlymphoid tissues such as lung (GeurtsvanKessel and Lambrecht, 2008), intestine (Manicassamy et al., 2010; Murphy, 2011), or kidney (Krüger et al., 2004). In the lung, cDCs and macrophages both express CD11c but differ in expression of CD103 and CD11b, with cDCs comprising CD103+CD11b−, CD103+CD11b+, and CD103−CD11b+ populations and macrophages residing within a CD103−CD11blo/−autofluorescent+ gate (Edelson et al., 2010). Consistent with this, we found that Zbtb46gfp was uniformly expressed in CD103+CD11b− and CD103+CD11b+ populations of cDCs and not expressed in CD11c+CD103−CD11blo/− macrophages (Fig. 5 A). Notably, the CD11c+CD103−CD11b+ cells were heterogeneous for GFP expression, and although considered to represent cDCs (Edelson et al., 2010), our result suggests that this heterogeneous population likely includes GFP− macrophages that resemble cDCs in marker expression but not transcriptional profile. This agrees with the suggestion of a recent study that peripheral CD11c+CD103−CD11b+ cells contain macrophages as they show partial dependence on M-CSF (Ginhoux et al., 2009). Furthermore, Zbtb46 expression determined by microarray was high in both the CD103+CD11b− and CD24+CD11b+ cDC populations and low in lung macrophages (Fig. 5 B). The CD24−CD11b+ lung cDCs expressed Zbtb46 at an intermediate level, consistent with the notion of heterogeneity. Accordingly, analysis of lung tissue sections by immunofluorescence microscopy showed that only a subset of CD11b+ cells (Fig. 5 C) or CD11c+ cells (not depicted) were positive for expression of Zbtb46gfp. These GFP-positive cells were largely found to surround large vessels and conducting airways.
Figure 5.
Zbtb46gfp distinguishes cDCs from macrophages in peripheral tissues. (A) Cells were harvested from perfused lungs of Zbtb46gfp/gfp mice and analyzed for expression of CD11c, MHCII, CD45, CD103, CD11b, GFP, and autofluorescence (centered on 700 nm). Shown (right) are two-color histograms for GFP expression and autofluorescence for the indicated populations. Numbers represent the percentage of cells in the indicated gate. Data are representative of three independent experiments with three mice per group. (B) Shown are the mean expression values of Zbtb46 (±SEM) determined by microarray for the indicated cell populations derived from the ImmGen Database (Heng and Painter, 2008). Data are assembled from two to four replicate arrays. (C) Lung sections from Zbtb46gfp/+ BM chimeras were stained for CD11b and anti-GFP. Data are representative of five independent experiments. Bars, 200 µm. (D) Cells were harvested from small intestinal lamina propria of Zbtb46gfp/+ mice and analyzed for expression of CD11c, MHCII, CD45, CD103, CD11b, and GFP. Shown (right) are two-color histograms for CD11b and GFP expression for the indicated populations. Data are representative of three independent experiments with two mice each. (E) Shown are the mean expression values of Zbtb46 (±SEM) in the indicated cells derived from Heng and Painter (2008). Data are assembled from four to seven replicate arrays. (F) Cells harvested from the kidney from Zbtb46gfp/gfp mice were stained as in D. Data are representative of three independent experiments with two mice each.
Another peripheral tissue in which the distinction of cDCs and macrophages is problematic is the lamina propria of the intestinal tract (Bogunovic et al., 2009; Varol et al., 2009), where both macrophages and DCs can express CD11c (Manicassamy et al., 2010; Murphy, 2011). In the lamina propria, we found that the majority of CD11c+ cells lacked expression of MHCII; these classically have been suggested to represent macrophages (Pabst and Bernhardt, 2010). In agreement, we found that none of these CD11c+MHCII− cells expressed Zbtb46gfp (Fig. 5 D). Within CD11c+MHCII+ cells, Zbtb46gfp was uniformly expressed in the CD11b− (CD103+) subset (Fig. 5 D), previously shown to be derived from CDPs (Bogunovic et al., 2009; Varol et al., 2009). However, among the CD11b+ subset of CD11c+MHCII+ cells, only half expressed Zbtb46gfp (Fig. 5 D). Again, this is consistent with previous suggestions that some CD11c+MHCII+CD11b+ cells in the lamina propria arise from monocytes and represent MHCII+ macrophages (Bogunovic et al., 2009; Varol et al., 2009). Consistently, Zbtb46 expression by microarray analysis was high in both of the CD103+ cDC subsets but low in lamina propria macrophages (Fig. 5 E).
In the kidney, Zbtb46 expression can similarly distinguish between cDCs and macrophages as in lamina propria and lung (Fig. 5 F). CD11c+ cells in the kidney have also been recognized to contain both cDCs and macrophages (Krüger et al., 2004). As in the lamina propria, a large fraction of CD11c+MHCII− cells coexpress CD11b and have been identified as macrophages (Krüger et al., 2004). We found that this population of macrophages was uniformly negative for GFP expression. In contrast, kidney cells that have been previously characterized to represent CD103+ cDCs, which are CD11c+MHCII+CD11b− (Ginhoux et al., 2009), were uniformly positive for GFP expression. Finally, the CD11c+MHCII+CD11b+ cells, which have been suggested to represent CD11b+ cDCs, were heterogeneous for GFP expression, suggesting that some of these cells may represent an MHCII+ macrophage population as well.
Zbtb46 expression indicates the identity of unclassified myeloid cell types
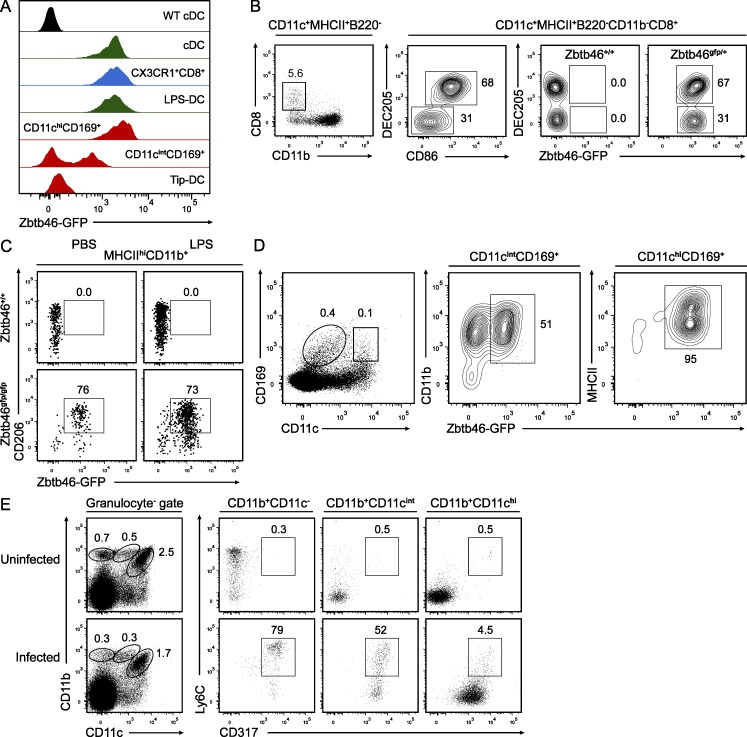
Zbtb46 expression also clarifies the identity of several recently described myeloid cell types that have characteristics of both cDCs and other myeloid lineages. The recently described CX3CR1+CD8α+ splenic population was initially thought to represent a pDC-related lineage based on its E2-2 expression and immunoglobulin gene rearrangements but also resembles cDCs by its high expression of CD11c and MHCII (Bar-On et al., 2010). LPS-induced CD206+ DCs arise from monocytes during inflammation, and so may represent macrophages or DCs but were interpreted as DCs based on their similarity to GM-CSF–derived DCs (Cheong et al., 2010). Notably, both of these types of cells were uniformly positive for Zbtb46gfp expression (Fig. 6, A–C), helping to exclude molecular heterogeneity within these populations and supporting the interpretation that they represent cDC lineages.
Figure 6.
Zbtb46gfp is expressed in CX3CR1+CD8α+, LPS-induced, and CD169+ DCs. (A) Shown are single-color histograms of GFP expression from Zbtb46+/+ mice (WT) or Zbtb46gfp/+ mice and stained to identify cDCs (MHCIIhiCD11chi), CX3CR1+CD8α+ DCs (CD8α+MHCII+CD11chiDEC205−), LPS-DCs (MHCIIhiCD11b+CD206+), CD11chiCD169+ cells, CD11cintCD169+ cells, or Tip-DCs (CD11cintCD11bhiLy6C+) according to the gating strategies shown in B–E. Data are representative of one to three independent experiments with at least three mice per group. (B) Splenocytes from Zbtb46+/+ and Zbtb46gfp/+ mice were stained for expression of the indicated markers. Cells within the indicated gate of the far left panel were analyzed for expression of DEC205 and CD86 (middle left), and DEC205 and Zbtb46gfp (right panels) for either Zbtb46+/+ or Zbtb46gfp/+ mice. Data are representative of three independent experiments with three mice each. Numbers represent the percentage of cells in the indicated gate. (C) SLNs from Zbtb46+/+ and Zbtb46gfp/gfp mice treated i.v. with either PBS or 5 µg LPS were analyzed after 18 h for expression of the indicated markers for cells previously gated as indicated above the histogram. Data are representative of two independent experiments involving three mice per group. (D) Cells from MLNs of Zbtb46gfp/+ mice were analyzed for expression of the indicated markers on cells previously gated as indicated above the histogram. Data are representative of three independent experiments with three mice per group. (E) Splenocytes from Zbtb46gfp/gfp mice infected with L. monocytogenes for 2 d were analyzed for expression of CD11b and CD11c (left) and gates established for monocytes, Tip-DCs, and cDCs. Cells within each gate were analyzed for expression of Ly6C and CD317.
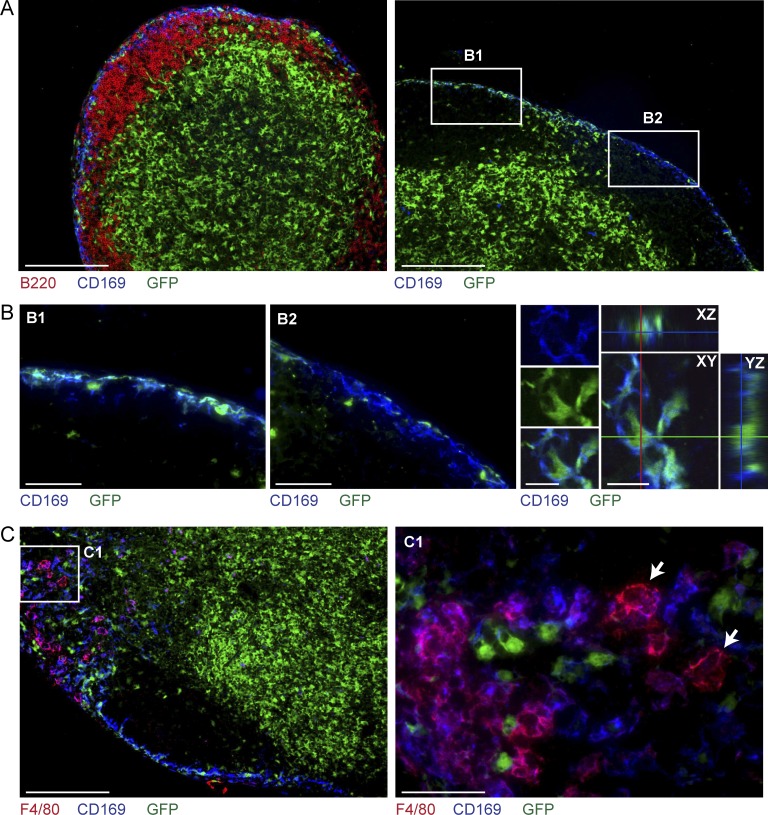
In the past few years, a CD169+ population of cells located in the subcapsular region of LNs was interpreted to represent a novel subtype of macrophage that captures and preserves extracellular antigen for presentation to B cells (Junt et al., 2007; Phan et al., 2009). Analysis of Zbtb46gfp/+ mice by FACS (Fig. 6, A and D) and immunofluorescence microscopy (Fig. 7, A and B) provides the first evidence that these cells may be molecularly heterogeneous. Approximately half of CD169+ subcapsular cells in both SLNs and MLNs were positive for Zbtb46gfp expression. In contrast, medullary macrophages, defined by their coexpression of CD169 and F4/80 (Phan et al., 2009), were uniformly GFP negative (Fig. 7 C). Another recent study identified a separate subset of CD169+ cells that expresses high levels of CD11c and MHCII (Asano et al., 2011), which by gene expression pattern resemble CD8α+ cDCs (Heng and Painter, 2008) rather than macrophages. We found that this population of cells was uniformly positive for Zbtb46gfp expression (Fig. 6 D), favoring the interpretation that this subset of CD169+CD11chi cells represents cDCs.
Figure 7.
Zbtb46gfp is expressed in a subset of CD169+ subcapsular cells but not in medullary macrophages. (A) SLN sections from Zbtb46gfp/+ → WT BM chimeras were analyzed by fluorescence microscopy for expression of B220, CD169, and GFP. (B) Areas B1 and B2 from A (right) are shown at higher magnification as indicated (first and second panels). Confocal images along three planes (x, y, and z; right) confirm GFP expression within CD169+ cells. (C) MLN sections from Zbtb46gfp/+ → WT BM chimeras were analyzed for expression of F4/80, CD169, and GFP. The area indicated as C1 (left) is shown at higher magnification (right) to demonstrate that medullary macrophages are negative for GFP expression (arrows). Immunofluorescence images are representative of three to four independent experiments with one or two mice each. Confocal images are representative of one independent experiment in which three individual mice were analyzed. Bars: (A and C [left]) 200 µm; (B [left and middle] and C [right]) 50 µm; (B, right) 10 µm.
Finally, TNF/iNOS-producing DCs (Tip-DCs), which are of monocyte origin and are induced after infection with Listeria monocytogenes (Serbina et al., 2003), were negative for expression of Zbtb46gfp (Fig. 6 A). This finding may indicate that Tip-DCs are more highly related to a form of activated monocyte than to DCs, as recently suggested (Geissmann et al., 2010). Accordingly, these cells expressed Ly6C and CD317 similar to activated monocytes (Fig. 6 E). Of note, during L. monocytogenes infection or LPS-induced inflammation, Zbtb46gfp expression remained restricted to cDCs and loss of Zbtb46 did not affect the induction or maturation of cDCs (not depicted).
Zbtb46gfp is expressed in definitive erythroid precursors and endothelial cells
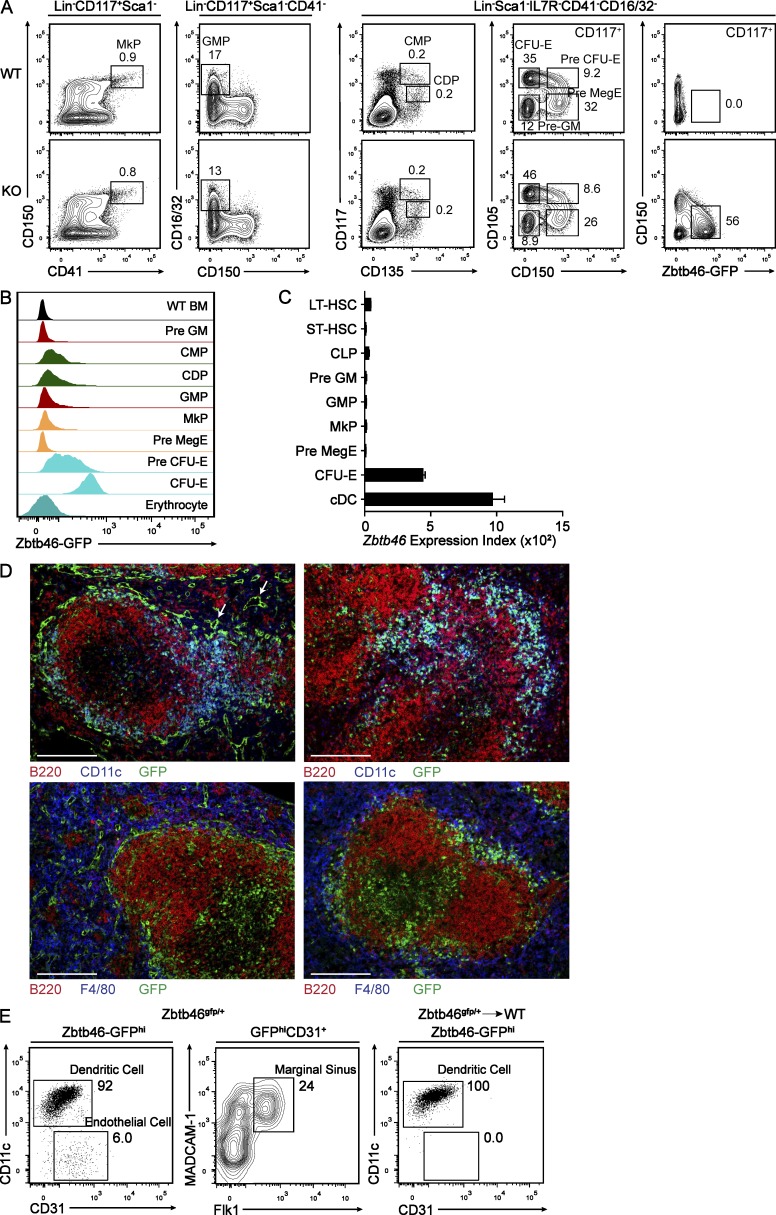
Above we observed that a small fraction of Zbtb46gfp-expressing cells were negative for CD11c and MHCII expression in lymphoid organs (Fig. 3 B). To determine the identity of these GFP-expressing cells, we first examined BM for indications of GFP expression in progenitor populations (Fig. 8 A). First, we noted that there were no changes in the frequency of progenitor populations between Zbtb46+/+ control and Zbtb46gfp/gfp mice (Fig. 8 A). Within these identified progenitor populations, GFP was expressed only within the committed erythroid precursor populations: pre–CFU-erythroid (CFU-E) and CFU-E (Fig. 8, A and B; Pronk et al., 2007). However, unlike the persistent expression of GFP in mature DCs, GFP was not detectable in mature erythrocytes (Fig. 8 B). Consistently, among these populations, Zbtb46 messenger RNA (mRNA) was only observed in committed erythroid progenitors at ∼50% of the level of cDCs by microarray analysis (Fig. 8 C). To determine whether expression of Zbtb46 in erythroid progenitors impacted gene expression, we purified pre-MegE, pre–CFU-E, and CFU-E populations from Zbtb46+/+, Zbtb46gfp/+, and Zbtb46gfp/gfp mice and performed microarray analysis. Notably, there was very little change in gene expression caused by the loss of Zbtb46 in these progenitors (Fig. S1 A).
We next examined Zbtb46gfp expression in the spleen using immunofluorescence (Fig. 8 D, left). As expected, GFP was observed in a subset of cells coexpressing CD11c, consistent with its expression by cDCs as determined by FACS. However, in addition, we observed that GFP was also expressed by cells associated with the vasculature of the splenic marginal zone and red pulp (Fig. 8 D), suggesting expression by endothelium. To test this, we reexamined splenic stromal cells by FACS for GFP expression using markers to identify endothelium and distinguish between these cells and DCs (Fig. 8 E). In spleen, the majority of GFP-expressing cells expressed CD11c, but 6% were negative for CD11c and were positive for CD31 expression. Of these cells, 24% expressed MADCAM-1 and Flk1, markers of endothelial cells associated with the marginal sinus (Zindl et al., 2009). Because endothelial cells in adult mice are not of hematopoietic origin, we generated chimeras by transferring Zbtb46gfp/+ BM into lethally irradiated WT mice. In these chimeras, Zbtb46gfp expression remained specific but was no longer found in CD11c−CD31+ endothelial cells either by FACS or by immunofluorescence (Fig. 8, D [right] and E). The use of radiation chimeras, either Zbtb46gfp/+ into WT or WT into Zbtb46gfp/+, could be used to isolate the GFP expression for functional studies of either DCs or endothelium, respectively. In summary, outside of its selective expression in cDCs within immune lineages, Zbtb46 is also expressed transiently by erythroid progenitors and constitutively by endothelial cells.
Zbtb46gfp identifies cDC-committed progenitors in BM and peripheral lymphoid organs
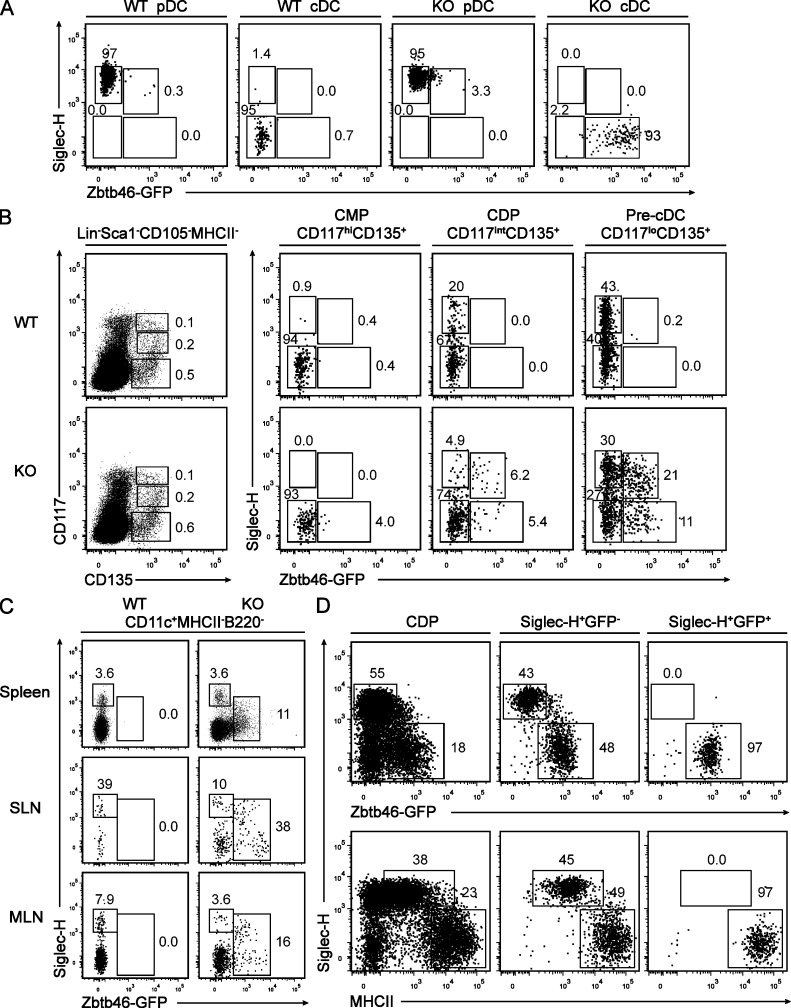
The lack of truly cDC-specific markers has confounded the purification of committed cDC progenitors away from cells capable of pDC development. The most successful purifications of a committed pre-cDC relied on CD11c and CD135, but these retained some pDC potential (Fig. 1 A; Liu et al., 2009), perhaps because of the expression of these markers on pre-pDCs as well. Furthermore, the in vivo expansion of pre-cDCs by Flt3L administration hinders their subsequent identification because of the down-regulation of Flt3 (CD135) itself (Waskow et al., 2008). Finally, at the single cell level, a fraction of cells identified as CDPs lacked pDC potential (Naik et al., 2007), suggesting the need for additional factors in defining progenitors of DC lineages.
To ask whether Zbtb46 expression could distinguish between committed progenitors of cDCs and pDCs, we analyzed its expression in BM DC progenitors in Zbtb46+/+ and Zbtb46gfp/gfp mice. First, as in the spleen, mature BM pDCs did not express GFP in Zbtb46gfp/gfp mice, and mature BM MHCII+ cDCs were positive for GFP expression (Fig. 9 A). We examined GFP expression using levels of CD117 (c-kit) to distinguish CMPs (CD117hi), CDPs (CD117int), and the fraction containing pre-cDCs (CD117lo; Fig. 9 B, left). CMPs were completely negative for both GFP and Siglec-H, a marker expressed on pDCs (Zhang et al., 2006), as expected (Fig. 9 B, middle left). The current definition of CDPs uses Lin−Sca-1−IL-7R−CD117intCD135+ gating (Naik et al., 2007; Onai et al., 2007), but it has been noted that a fraction of these cells are already committed to either pDC or cDC lineages (Naik et al., 2007). We noted both in Zbtb46+/+ and Zbtb46gfp/gfp mice that a small fraction of CDPs expressed Siglec-H. Importantly, these cells were heterogeneous for Zbtb46 expression. In Zbtb46gfp/gfp mice, GFP was expressed in approximately half of these Siglec-H+ CDPs (Fig. 9 B, middle right). In the CD117loCD135+ fraction containing pre-cDCs, a greater number of cells expressed Siglec-H, and again approximately half expressed GFP (Fig. 9 B, right). This population of Siglec-H+Zbtb46+ cells could also be observed in peripheral lymphoid organs (Fig. 9 C).
Figure 9.
Zbtb46 expression identifies committed pre-cDCs in BM and peripheral lymphoid tissues. (A) BM from Zbtb46+/+ (WT) and Zbtb46gfp/gfp (KO) mice was stained for expression of Siglec-H, CD317, B220, CD11c, and MHCII. Shown are two-color histograms for Siglec-H and GFP expression for cells previously gated for positive expression of B220, CD317, and Siglec-H (pDC) or for positive expression of CD11c and MHCII (cDC) as indicated above the diagram. Numbers represent the percentage of cells within the indicated gate. Data are representative of three independent experiments with three mice each. (B) BM cells from Zbtb46+/+ (WT) and Zbtb46gfp/gfp (KO) mice were stained for expression of B220 and NKp46 (Lin), Sca-1, CD105, MHCII, CD117, CD135, CD11c, and Siglec-H. Shown are two-color histograms for expression of the indicated markers for cells previously gated as indicated above the diagram. Data are representative of five independent experiments with two mice each. (C) Splenocytes, SLNs, and MLNs from Zbtb46+/+ (WT) and Zbtb46gfp/gfp (KO) mice were analyzed for expression of CD11c, MHCII, B220, Siglec-H, and GFP. Shown are histograms depicting Siglec-H and GFP expression for cells previously gated as indicated above the diagram. Data are representative of two independent experiments with one or two mice each. (D) BM cells from Zbtb46gfp/gfp mice were sorted into populations of Lin−Sca-1−CD105−CD117intCD135+MHCII− (CDP; left), Lin−Sca-1−CD105−MHCII−CD117loCD135+Siglec-H+GFP− (Siglec-H+GFP−; middle), or Lin−Sca-1−CD105−MHCII−CD117loCD135+Siglec-H+ and GFP+ (Siglec-H+GFP+; right) as indicated above the diagram. Cells were cultured in 100 ng/ml Flt3L for 4 d and analyzed by FACS for expression of Siglec-H, GFP, and MHCII. Data are representative of three independent experiments with two to three mice each.
We wondered whether GFP expression would mark definitive commitment to the cDC lineage independent of Siglec-H expression. For this, we sorted the CD117lo cells into populations that were either Siglec-H+ Zbtb46gfp-negative or Siglec-H+ Zbtb46gfp-positive and compared them with CDPs for their ability to generate pDCs and cDCs after in vitro culture with Flt3L (Fig. 9 D). Although CDPs and Siglec-H+ Zbtb46gfp-negative cells could generate both cDCs and pDCs, the Siglec-H+ Zbtb46gfp-positive cells were able to generate only cDCs but not pDCs (Fig. 9 D). This indicates that expression of Zbtb46 identifies a committed progenitor of the cDC lineage. Furthermore, this commitment step does not require a functional Zbtb46 protein because cDCs developed from both Zbtb46+/+ and Zbtb46gfp/gfp progenitors. It will be interesting to determine whether the divergence of pDCs and cDCs takes place at the Siglec-H+Zbtb46− stage of CDP development.
Zbtb46 represses alternate myeloid potential in BM progenitor cells
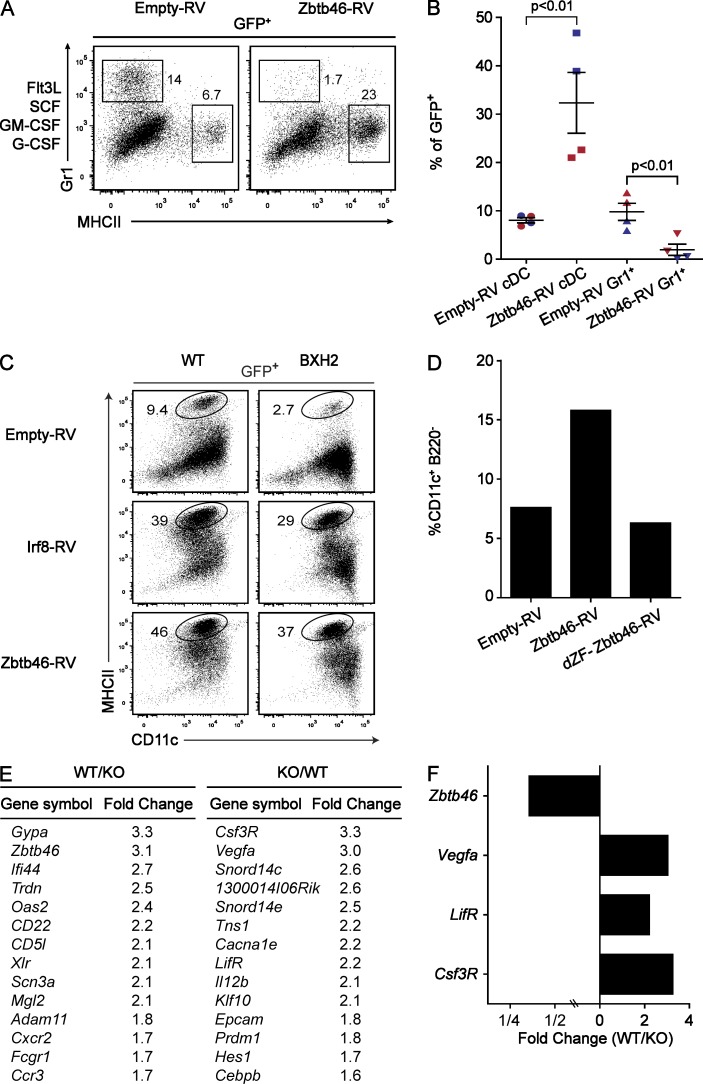
Because Zbtb46 expression first became apparent at the pre-cDC stage, we wondered whether it might act in regulating lineage restriction of cDCs. We expressed Zbtb46 by retrovirus in BM cells and examined its effect on myeloid differentiation in a system using a mixture of factors supporting both DC and alternate myeloid lineage development (Fig. 10, A and B). Although infection of progenitors with control retroviruses supported development of both granulocytes and cDCs in this setting, expression of Zbtb46 led to a 10-fold reduction in the growth of granulocytes and an enhanced outgrowth of cDCs. Notably, this effect was even evident when Zbtb46 was expressed in BM progenitors from Irf8 mutant mice (either Irf8-deficient [not depicted] or BXH2 recombinant inbred mice, harboring a point mutation in the Irf8 gene [Fig. 10 C; Tailor et al., 2008]), which normally do not support the development of cDCs. The effects of Zbtb46 are specific because deletion of its zinc finger domains abrogated the enhancement of cDC development (Fig. 10 D).
Figure 10.
Zbtb46 inhibits granulocyte potential, enhances cDC development, and regulates expression of non-DC growth factor receptors. (A) GFP-RV (Empty-RV; Ranganath et al., 1998) or a retrovirus encoding mouse Zbtb46 (Zbtb46-RV) was used to infect BM cells and cultured with growth factors as described in Materials and methods. On day 5, cells were harvested and analyzed for expression of Gr1, MHCII, and GFP. Shown are two-color histograms for the retroviral constructs indicated above the diagram for the indicated markers for cells gated on positive expression of GFP (GFP+). Numbers represent the percentage of cells in the indicated gates. Data are representative of three independent experiments using at least two mice per group. (B) Replicates from two experiments (each experiment is uniquely colored) described in A are shown. Numbers represent the percentage of cells on day 5 that are present within the gates for cDCs or granulocytes (Gr1+) as indicated below the diagram. Bars represent the mean ± SEM. (C) BM from WT or BXH2 recombinant inbred mice (Tailor et al., 2008) was infected with GFP-RV, a retrovirus encoding full-length mouse Irf8 (Irf8-RV) or Zbtb46-RV as described in A and analyzed on day 5 for expression of CD11c and MHCII. Similar diminishment of cDC development was seen with Irf8−/− BM, which was similarly restored by Zbtb46-RV (not depicted). Data are representative of two independent experiments with two mice per group. (D) GFP-RV vector (Empty-RV) or retroviruses expressing full-length Zbtb46 (Zbtb46-RV) or a truncated Zbtb46 in which the region of cDNA encoding the C-terminal zinc finger domains (amino acids 404–601) were removed (dZF-Zbtb46-RV) and were used to infect BM progenitors as described in A, and cells were transferred into sublethally irradiated CD45.1 recipients. After 14 d, splenocytes were harvested from recipient mice, and CD45.2+GFP+ cells were analyzed for expression of CD11c, B220, and MHCII. Shown are the percentage of cells within the B220−CD11c+ gate for cells previously gated as CD45.2+GFP+ for the indicated retroviral construct. Data are representative of three independent experiments with one mouse per group. (E) Sorted steady-state splenic CD172+CD4+ cDCs from Zbtb46+/+ (WT) and Zbtb46gfp/gfp (KO) mice were purified, and gene expression microarrays were performed. Genes that are increased in the WT sample relative to the Zbtb46gfp/gfp (KO) sample (left) are listed with the associated fold induction. Genes that are increased in Zbtb46gfp/gfp (KO) mice relative to WT are listed with their fold induction (right). (F) Fold changes of the indicated genes derived from E are shown for their expression in WT compared with Zbtb46gfp/gfp (KO) mice.
Because Zbtb46 belongs to a family of transcriptional repressors, some of which are known to participate in lineage fate decisions within the immune system (Wildt et al., 2007), we analyzed gene expression microarrays from WT or Zbtb46gfp/gfp CD4+ splenic cDCs to identify genes that may be induced or repressed by the loss of Zbtb46 (Fig. 10 E and Fig. S1 B). In these steady-state DCs, it appeared that Zbtb46 regulated relatively few target genes. Notably, among the few genes altered were the receptors for G-CSF (Csf3R) and leukemia inhibitory factor (Fig. 10 F), which were increased in their relative expression in Zbtb46-deficient CD4+ cDCs. These results suggest that Zbtb46 may enforce cDC lineage restriction by extinguishing the expression of alternate myeloid growth factor receptors.
DISCUSSION
The precise assignment of cellular identity can be hampered by the fact that many lineages share expression of common surface markers. This is particularly true for cDCs, which have overlapping expression of various markers in common with pDCs, monocytes, and macrophages. The hypothesis that transcription factors define distinct lineages prompted our efforts to generate a Zbtb46-knockin reporter to help distinguish between these closely related myeloid subsets. We find that within the immune system, Zbtb46 expression specifically identifies the cDC lineage, both in lymphoid and nonlymphoid tissues. Because Zbtb46 is also expressed in endothelial cells and transiently during erythroid differentiation, studies such as intravital microscopy or lineage ablation using the Zbtb46 locus may benefit from the use of BM chimeras to eliminate effects of expression by non-DCs. In examining Zbtb46-deficient mice, it seems that cDCs, vasculature, and erythrocytes each develop without major abnormalities, suggesting that potential defects will reside in induced responses or that there is compensation by an unidentified factor.
Our data indicate that Zbtb46 expression at the stage of the pre-DC is a marker for cells committed to the cDC lineage that have lost potential for pDCs and other myeloid cells. This is a useful distinction because it has not been precisely clear at which point potential for pDC development is extinguished (Liu and Nussenzweig, 2010; Belz and Nutt, 2012). By combining Zbtb46gfp expression with analysis of other markers, we have determined that although Siglec-H is specific for pDCs in peripheral organs, BM progenitors expressing Siglec-H actually retain potential for cDC development. This result might explain discrepancies between two recent studies that attempted to deplete pDCs based either on Siglec-H–DTR (Takagi et al., 2011) or BDCA2-DTR (Swiecki et al., 2010) because the former would likely cause the loss of some cDC populations not depleted by the latter.
Besides cDCs in lymphoid and peripheral tissues, several unconventional myeloid subsets have been described whose precise origin and identity have been unclear. In these cell types, examination of Zbtb46 expression seems to aid in their classification into either cDC or macrophage lineages. For example, Tip-DCs were originally described as a type of induced inflammatory DC lineage (Serbina et al., 2003) but recently were suggested to more closely resemble human M1 macrophages (Geissmann et al., 2010). The lack of Zbtb46 expression in Tip-DCs may support this latter interpretation. In addition, subcapsular macrophages expressing CD169 were considered to represent an F4/80-negative macrophage lineage (Junt et al., 2007; Phan et al., 2009). Our results suggest that this population is heterogeneous with respect to Zbtb46 expression and could indicate that these cells may be more closely related to cDCs than previously thought.
Although current results indicate that Zbtb46 is dispensable for development of the recognized types of cDCs, our analysis has not yet examined the functional properties of these cells in settings of immune challenge. We may conclude that specification and commitment in vivo appears normal, but we have uncovered some evidence for subtle changes in gene expression in CD4+ cDCs. These findings will require further studies to clarify the functional consequences of such changes. Importantly, heterozygous Zbtb46gfp/+ mice or BM chimeras should benefit studies in which the identification of cDCs or committed pre-cDCs from monocytes and macrophages is necessary.
MATERIALS AND METHODS
Mice.
Zbtb46gfp mice were maintained on the pure 129SvEv background by mating germline progeny from the EDJ22 ESC line with mice purchased from Taconic. Experiments were performed with sex-matched littermate mice at 6–10 wk of age. Irf8−/− mice were obtained from the European Mutant Mouse Archive and maintained on the C57BL/6 background. BXH2/TyJ mice were purchased from the Jackson Laboratory and maintained on the C57BL/6 background. Mice were bred and maintained in our specific pathogen–free animal facility according to institutional guidelines and with protocols approved by the Animal Studies Committee of Washington University in St. Louis.
Generation of the Zbtb46gfp targeting construct.
The Zbtb46gfp targeting construct was assembled with the Gateway recombination cloning system (Invitrogen). To construct pENTR-lox-GFP.Neo, a 1,017-bp PCR product containing eGFP followed by the SV40-pA terminator was amplified from gDNA of CX3CR1gfp/+ mice (Jung et al., 2000) using primers f666-pEGFP-N, 5′-AATAGTCGACCGGTCGCCACCATGGT-3′, and r1635-pEGFP-N1, 5′-AATCTCGAGATTAACGCTTACAATTTACGCCTTAAGATAC-3′, and cloned into pGEM-T Easy (Promega). The resulting plasmid was digested using SalI and XhoI, and the 1,004-bp eGFP-pA cassette was cloned into the SalI site 5′ of the PGK–neor (phosphoglycerate kinase promoter–neomycin phosphotransferase) gene cassette of the pLNTK targeting vector (Gorman et al., 1996) to generate pLNTK-EGFP (8,313 bp). Finally, the 2,825-bp eGFP-PGK–neor cassette from pLNTK-EGFP was released using SalI and XhoI and ligated bluntly into the 2,554-bp backbone of NotI-digested pENTR-lox-Puro (Iiizumi et al., 2006). To construct pENTR-Zbtb46-5HA, the 5′ homology arm was amplified by PCR from 129SvEv-EDJ22 ESC genomic DNA using the following oligonucleotides, which contain attB4 and attB1 sites: 5′-GGGGACAACTTTGTATAGAAAAGTTGCTGGTTTGAATTAGGGTACTTGAAG-3′ and 5′-GGGGACTGCTTTTTTGTACAAACTTGCTGGGAGTCTTGCTGCTGTGT-3′. The attB4-attB1 PCR fragment was inserted into the pDONR (P4-P1R) plasmid (Invitrogen) by the BP recombination reaction generating pENTR-Zbtb46-5HA. To construct pENTR-Zbtb46-3HA, the 3′ homology arm was amplified by PCR from genomic DNA using the following oligonucleotides, which contain attB2 and attB3 sites: 5′-GGGGACAGCTTTCTTGTACAAAGTGGCGCTGACTCAGAAGAATGTC-3′ and 5′-GGGGACAACTTTGTATAATAAAGTTGAATAATTCTCTAATAGAATAT-3′. The attB2-attB3 PCR fragment was inserted into pDONR(P2R-P3) plasmid (Invitrogen) by the BP recombination reaction generating pENTR-Zbtb46-3HA. LR recombination reaction was performed using pENTR-Zbtb46-5HA, pENTR-Zbtb46-3HA, pENTR-lox-GFP.Neo, and pDEST DTA-MLS to generate the final targeting construct. The linearized vector was electroporated into EDJ22 embryonic stem cells (129SvEv background), and targeted clones were identified by Southern blot analysis with the 5′ probe and confirmed with the 3′ probe. Probes were amplified from genomic DNA using the following primers: Zbtb465P forward, 5′-TTGTAGTCAGCTTCTCACTTGTC-3′; Zbtb465P reverse, 5′-CAGTATGTTCGTAGGTTTTGTGG-3′; Zbtb463P forward, 5′-CAATTAAGGAAAAGTTGAAG-3′; and Zbtb463P reverse, 5′-TTATCCATTGAGCTGTCTCT-3′. Blastocyst injections were performed, and male chimeras were bred to female 129SvEv mice. Genotyping PCR was conducted to confirm germline transmission and further determine the genotype of progeny mice using the following primers: ScreenGFP reverse, 5′-AACTTGTGGCCGTTTACGTC-3′; Screen 5HA forward, 5′-GACCTCATACCTCCTTAGCA-3′; and Screen WT reverse, 5′-GGCGGAGTACATGAAGTCAA-3′.
DC preparation.
Lymphoid organ and nonlymphoid organ DCs were harvested and prepared as described previously (Edelson et al., 2010). In brief, spleens, MLNs, SLNs (inguinal), and kidneys were minced and digested in 5 ml Iscove’s modified Dulbecco’s media + 10% FCS (cIMDM) with 250 µg/ml collagenase B (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 30 min at 37°C with stirring. Cells were passed through a 70-µm strainer before red blood cells were lysed with ACK lysis buffer. Cells were counted on a Vi-CELL analyzer (Beckman Coulter), and 5–10 × 106 cells were used per antibody staining reaction. Lung cell suspensions were prepared after perfusion with 10 ml Dulbecco’s PBS (DPBS) via injections into the right ventricle after transection of the lower aorta. Dissected and minced lungs were digested in 5 ml cIMDM with 4 mg/ml collagenase D (Roche) for 1 h at 37°C with stirring. SI cell suspensions were prepared after removal of Peyer’s patches and fat. Intestines were opened longitudinally, washed of fecal contents, cut into 0.5-cm pieces, and incubated in HBSS medium (Life Technologies) + 5% FBS + 2 mM EDTA at 37°C for 20 min while shaking at 250 rpm. This process was repeated three times. The tissue suspensions were passed through a 100-µm filter, and the remaining tissue pieces were minced and incubated in HBSS medium + 5% FBS with collagenase B and DNaseI at 37°C for 30 min with shaking.
Antibodies and flow cytometry.
Staining was performed at 4°C in the presence of Fc Block (clone 2.4G2; BD) in FACS buffer (DPBS + 0.5% BSA + 2 mm EDTA). The following antibodies were purchased from BD: APC anti-CD4 (RM4-5), V450 anti-GR1 (RB6-8C5), PE-Cy7 anti-CD8α (53–6.7), PE-Cy7 anti-CD24 (M1/69), and APC anti-CD172a/SIRPα (P84). These were purchased from eBioscience: PE anti-NKp46 (29A1.4), PerCP-Cy5.5 anti-CD11b (M1/70), APC eFluor780 anti-CD11c (N418), PE anti-CD103 (2E7), APC anti-CD317/BST2 (eBio927), eFluor450 anti-MHCII (I-A/I-E; M5/114.15.2), APC anti-CD45.2 (104), PerCP-Cy5.5 anti-CD16/32 (93), PE anti-CD41 (eBioMWReg30), APC anti-Flk1 (Avas12a1), APC anti-CD150 (mShad150), APC-eFluor780 anti-CD117 (2B8), V500 anti-B220 (RA3-6B2), eFluor450 anti-CD105 (MJ7/18), PE anti-CD135 (A2F10), biotin anti–MADCAM-1 (MECA-367), APC anti-F4/80 (BM8), Alexa Fluor 700 anti–Sca-1 (D7), PerCP-eFluor710 anti–Siglec-H (eBio440C), eFluor450 anti-CD31 (390), eFluor450 anti-Ter119 (Ter119), and PERCPCy5.5 anti-CD14 (Sa2-8). These reagents were purchased from Miltenyi Biotec: PE anti-CD205/DEC205 (NLDC-145) and APC anti-CD205/DEC205 (NLDC-145). APC anti-CD206 (MR5D3) was purchased from BioLegend. In general, all antibodies were used at a 1:200 dilution. Anti-DEC205 was used at a 1:20 dilution. Cells were analyzed on FACSCanto II or FACSAria II flow cytometers (BD), and data were analyzed with FlowJo software (Tree Star). For immunofluorescence experiments, the following antibodies were purchased from Invitrogen: rabbit polyclonal anti-GFP, Alexa Fluor 488 anti–rabbit IgG (A11034), streptavidin Alexa Fluor 555, and biotin anti-F4/80 (BM8); from BioLegend: Alexa Fluor 647 anti-CD11c (N418); from AbD Serotec: APC anti-CD169 (3D6.112); and from eBioscience: biotin anti-B220 (RA3-6B2).
Progenitor isolation and cell culture.
BM was harvested from femur, tibia, humerus, and pelvis. Bones were fragmented by mortar and pestle, and debris was removed by gradient centrifugation using Histopaque 1119 (Sigma-Aldrich). Cells were passed through a 70-µm strainer, and red blood cells were lysed with ACK lysis buffer. Cells were counted on a Vi-CELL analyzer, and 5–10 × 106 were stained for analysis or entire BM stained for sorting. Gates used to define GMP, CMP, CDP, and pre-cDC are based on previous studies (Akashi et al., 2000; Onai et al., 2007). CMPs were identified as Lin−c-kithiSca-1−CD11c−CD135+CD16/32− and IL-7R−. CDPs were identified as Lin−c-kitintSca-1−CD135+CD16/32− and IL-7R−. GMPs were identified as Lin−CD135−c-kithiSca-1−CD16/32+ and IL-7R−. For cell purification by sorting we used a FACSAria. Cells were sorted into PBS supplemented with 0.5% BSA and 2 mM EDTA. Cell purities of at least 95% were confirmed by postsort analysis. For Flt3L culture experiments, whole BM (2 × 106 cells/ml in 4 ml cIMDM for 6 or 9 d) or sorted cells (1–5 × 104 cells/200 µl cIMDM for 4 d) were cultured at 37°C in 100 ng/ml Flt3L (PeproTech). For GM-CSF and M-CSF culture experiments, whole BM (106 cells/ml in 4 ml cIMDM for 7 d) was cultured at 37°C in 20 ng/ml GM-CSF or M-CSF (PeproTech). For monocyte cultures, BM monocytes were identified as Ly6C+CD11b+Ly6G−CD11c−MHCII−Siglec-H−, and 5 × 104 cells were sorted and cultured in 20 ng/ml GM-CSF with or without 20 ng/ml IL-4 for 4 d (PeproTech).
Expression microarray analysis.
Total RNA was isolated from cells using the RNAqueous-Micro kit (Invitrogen). For Mouse Genome 430 2.0 arrays, RNA was amplified, labeled, fragmented, and hybridized using the 3′ IVT Express kit (Affymetrix). Data were normalized within experiments, and expression values were modeled using DNA-Chip analyzer (dChip) software (http://www.dChip.org; Li and Wong, 2001a). For Mouse Gene 1.0 ST arrays, RNA was amplified with the WT Expression kit (Invitrogen) and labeled, fragmented, and hybridized with the WT Terminal Labeling and Hybridization kit (Affymetrix). Data were processed using RMA (robust multichip average) quantile normalization, and expression values were modeled using ArrayStar software (DNASTAR). All original microarray data have been deposited in GEO DataSets under accession no. GSE37030.
Quantitative RT-PCR.
Gene expression analysis used cells isolated to >95% purity from WT 129SvEv spleens using a FACSAria II. RNA and cDNAs were prepared with RNeasy Micro kit (QIAGEN) and Superscript III reverse transcription (Invitrogen). Real-time PCR and a StepOnePlus Real-Time PCR system (Applied Biosystems) were used according to the manufacturer’s instructions, with the Quantitation Standard-Curve method and HotStart-IT SYBR Green qPCR Master Mix (Affymetrix/USB). PCR conditions were 10 min at 95°C, followed by 40 two-step cycles consisting of 15 s at 95°C and 1 min at 60°C. Primers used for measurement of Zbtb46 expression were as follows: Zbtb46 qPCR forward, 5′-AGAGAGCACATGAAGCGACA-3′; Zbtb46 qPCR reverse, 5′-CTGGCTGCAGACATGAACAC-3′; HPRT forward, 5′-TCAGTCAACGGGGGACATAAA-3′; and HPRT reverse, 5′-GGGGCTGTACTGCTTAACCAG-3′.
Immunofluorescence microscopy.
Organs were fixed in 2% paraformaldehyde for 8 h followed by incubation in 30% sucrose/H2O overnight, embedded in OCT compound, and 7-µm cryosections were produced. After two washes in DPBS, sections were blocked in CAS Block (Invitrogen) containing 0.2% Triton X-100 for 10 min at room temperature. Sections were stained with primary and secondary reagents diluted in CAS Block containing 0.2% Triton X-100 and mounted with ProLong Gold Antifade reagent containing DAPI (Invitrogen). Four-color epifluorescence microscopy was performed using an AxioCam MRn microscope equipped with an AX10 camera (Carl Zeiss). Monochrome images were acquired with AxioVision software (Carl Zeiss) using either 10× or 40× objectives and exported into ImageJ software (National Institutes of Health) for subsequent color balancing and overlaying. Confocal microscopy was performed using an LSM 510 META confocal laser-scanning microscope and analyzed with LSM Image Examiner software (Carl Zeiss). Images were taken using a water immersion C-Apochromat 40× objective, and z-stack images were acquired at 1-µm intervals.
BM chimeras.
BM cells from Zbtb46gfp/+ were collected as described in Progenitor isolation and cell culture, and 2 × 106 total BM cells were transplanted by retroorbital i.v. injection into WT 129SvEv recipients that had received 1,200 rads of whole body irradiation on the day of transplant. Chimeras were analyzed at 6–8 wk after transplant. Sublethal irradiation was accomplished with 600 rads of whole body irradiation.
LPS and L. monocytogenes.
Mice were infected with 104 live L. monocytogenes (WT strain EGD). L. monocytogenes was stored as glycerol stocks at −80°C and diluted into pyrogen-free saline for i.v. injection into mice, and analysis was performed 2 d later. 5 µg LPS E. coli 055:B5 (Sigma-Aldrich) diluted in 200 µl pyrogen-free saline was injected i.v. to induce CD206+ DCs, and analysis was performed 18 h later in SLNs. Intracellular staining for CD206 was performed after cells were fixed in 4% paraformaldehyde in DPBS and permeabilized in 0.1% saponin/DPBS.
Retrovirus infection.
The Zbtb46 full-length cDNA was cloned from splenic cDNA using RT-PCR with the following primers: Zbtb46isoform1 reverse, 5′-ATTACTCGAGCTAGGAGATCCAGGCAAAGTCT-3′; and Zbtb46isoform1 forward, 5′-AAAAGGATCCATCGATACCATGAACAACCGAAAGGAAGATA-3′. The resulting 1.8-kB PCR fragment was digested with BamHI and XhoI and cloned into the BglII and XhoI sites of the GFP-RV (Ranganath et al., 1998). Retroviral vectors were transfected into Phoenix E cells as described previously (Sedy et al., 2005), and viral supernatants were collected 2 d later. BM cells were infected with viral supernatants in the presence of 2 µg/ml polybrene by spin infection at 2,500 rpm for 30 min. Cells were cultured in the presence of 10 ng/ml of each of the factors: Flt3L, stem cell factor (SCF), GM-CSF, and G-CSF (PeproTech). Cells were harvested on day 5 and analyzed by FACS.
Statistical analysis.
Differences between groups were analyzed by an unpaired, two-tailed Student’s t test, with p-values ≤0.05 considered significant (Prism; GraphPad Software).
Online supplemental material.
Fig. S1 shows gene expression analysis of erythroid precursors and CD4+ cDCs from WT and Zbtb46 KO mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120030/DC1.
Supplementary Material
Acknowledgments
We thank the ImmGen consortium for use of the ImmGen Database (Heng and Painter, 2008), Mike White for blastocyst injections and generation of mouse chimeras, and the Alvin J. Siteman Cancer Center at Washington University in St. Louis School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility.
This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (AI076427-02), and Department of Defense (W81XWH-09-1-0185; to K.M. Murphy); the American Heart Association (12PRE8610005; to A.T. Satpathy); the German Research Foundation (AL 1038/1-1; to J.C. Albring); and the Burroughs Wellcome Fund Career Award for Medical Scientists, Jack H. Ladenson Fellowship in Experimental Clinical Pathology at Washington University in St. Louis School of Medicine, and American Society of Hematology Scholar Award (to B.T. Edelson). The Alvin J. Siteman Cancer Center is supported in part by National Cancer Institute Cancer Center Support Grant #P30 CA91842.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- cDC
- classical DC
- CDP
- common DC progenitor
- CFU-E
- CFU-erythroid
- CMP
- common myeloid progenitor
- DTR
- diphtheria toxin receptor
- GMP
- granulocyte macrophage precursor
- MLN
- mesenteric LN
- mRNA
- messenger RNA
- pDC
- plasmacytoid DC
- RPM
- red pulp macrophage
- SLN
- skin-draining LN
- Tip-DC
- TNF/iNOS-producing DC
References
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Asano K., Nabeyama A., Miyake Y., Qiu C.H., Kurita A., Tomura M., Kanagawa O., Fujii S., Tanaka M. 2011. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity. 34:85–95 10.1016/j.immuni.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Bar-On L., Birnberg T., Lewis K.L., Edelson B.T., Bruder D., Hildner K., Buer J., Murphy K.M., Reizis B., Jung S. 2010. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 107:14745–14750 10.1073/pnas.1001562107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz G.T., Nutt S.L. 2012. Transcriptional programming of the dendritic cell network. Nat. Rev. Immunol. 12:101–113 10.1038/nri3149 [DOI] [PubMed] [Google Scholar]
- Bennett C.L., Clausen B.E. 2007. DC ablation in mice: promises, pitfalls, and challenges. Trends Immunol. 28:525–531 10.1016/j.it.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. 2007. Notch–RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G., et al. 2010. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 143:416–429 10.1016/j.cell.2010.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N.S., Kant S.G., et al. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 135:37–48 10.1016/j.cell.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan J.M., Lehar S.M., Bevan M.J. 2000. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Tang Y., Xu H., Lit L., Walker W., Ashwood P., Gregg J.P., Sharp F.R. 2006. Genomic profiles for human peripheral blood T cells, B cells, natural killer cells, monocytes, and polymorphonuclear cells: comparisons to ischemic stroke, migraine, and Tourette syndrome. Genomics. 87:693–703 10.1016/j.ygeno.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207:823–836 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F., Gordon S., Hume D.A., Mowat A.M., Randolph G.J. 2010. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 10:453–460 10.1038/nri2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Lambrecht B.N. 2008. Division of labor between dendritic cell subsets of the lung. Mucosal Immunol. 1:442–450 10.1038/mi.2008.39 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.R., van der Stoep N., Monroe R., Cogne M., Davidson L., Alt F.W. 1996. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 5:241–252 10.1016/S1074-7613(00)80319-2 [DOI] [PubMed] [Google Scholar]
- Heng T.S., Painter M.W.; Immunological Genome Project Consortium 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A. 2011. Applications of myeloid-specific promoters in transgenic mice support in vivo imaging and functional genomics but do not support the concept of distinct macrophage and dendritic cell lineages or roles in immunity. J. Leukoc. Biol. 89:525–538 10.1189/jlb.0810472 [DOI] [PubMed] [Google Scholar]
- Iiizumi S., Nomura Y., So S., Uegaki K., Aoki K., Shibahara K., Adachi N., Koyama H. 2006. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 41:311–316 10.2144/000112233 [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., Littman D.R. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Unutmaz D., Wong P., Sano G., De los Santos K., Sparwasser T., Wu S., Vuthoori S., Ko K., Zavala F., et al. 2002. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 17:211–220 10.1016/S1074-7613(02)00365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junt T., Moseman E.A., Iannacone M., Massberg S., Lang P.A., Boes M., Fink K., Henrickson S.E., Shayakhmetov D.M., Di Paolo N.C., et al. 2007. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 450:110–114 10.1038/nature06287 [DOI] [PubMed] [Google Scholar]
- Kelly K.F., Daniel J.M. 2006. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 16:578–587 10.1016/j.tcb.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Kohyama M., Ise W., Edelson B.T., Wilker P.R., Hildner K., Mejia C., Frazier W.A., Murphy T.L., Murphy K.M. 2009. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 457:318–321 10.1038/nature07472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger T., Benke D., Eitner F., Lang A., Wirtz M., Hamilton-Williams E.E., Engel D., Giese B., Müller-Newen G., Floege J., Kurts C. 2004. Identification and functional characterization of dendritic cells in the healthy murine kidney and in experimental glomerulonephritis. J. Am. Soc. Nephrol. 15:613–621 10.1097/01.ASN.0000114553.36258.91 [DOI] [PubMed] [Google Scholar]
- Lattin J.E., Schroder K., Su A.I., Walker J.R., Zhang J., Wiltshire T., Saijo K., Glass C.K., Hume D.A., Kellie S., Sweet M.J. 2008. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 4:5 10.1186/1745-7580-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. 2001a. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 98:31–36 10.1073/pnas.011404098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. 2001b. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:H0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist R.L., Shakhar G., Dudziak D., Wardemann H., Eisenreich T., Dustin M.L., Nussenzweig M.C. 2004. Visualizing dendritic cell networks in vivo. Nat. Immunol. 5:1243–1250 10.1038/ni1139 [DOI] [PubMed] [Google Scholar]
- Lindstedt M., Lundberg K., Borrebaeck C.A. 2005. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175:4839–4846 [DOI] [PubMed] [Google Scholar]
- Liu K., Nussenzweig M.C. 2010. Origin and development of dendritic cells. Immunol. Rev. 234:45–54 10.1111/j.0105-2896.2009.00879.x [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 10.1126/science.1170540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy S., Reizis B., Ravindran R., Nakaya H., Salazar-Gonzalez R.M., Wang Y.C., Pulendran B. 2010. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 329:849–853 10.1126/science.1188510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M. 2011. Comment on “Activation of β-catenin in dendritic cells regulates immunity versus tolerance in the intestine”. Science. 333:405, author reply :405 10.1126/science.1198277 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Metcalf D., van Nieuwenhuijze A., Wicks I., Wu L., O’Keeffe M., Shortman K. 2006. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 7:663–671 10.1038/ni1340 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Pabst O., Bernhardt G. 2010. The puzzle of intestinal lamina propria dendritic cells and macrophages. Eur. J. Immunol. 40:2107–2111 10.1002/eji.201040557 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Green J.A., Gray E.E., Xu Y., Cyster J.G. 2009. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 10:786–793 10.1038/ni.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst H.C., Tschannen K., Odermatt B., Schwendener R., Zinkernagel R.M., Van Den Broek M. 2005. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin. Exp. Immunol. 141:398–404 10.1111/j.1365-2249.2005.02868.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. 2007. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 1:428–442 10.1016/j.stem.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Ranganath S., Ouyang W., Bhattarcharya D., Sha W.C., Grupe A., Peltz G., Murphy K.M. 1998. GATA-3-dependent enhancer activity in IL-4 gene regulation. J. Immunol. 161:3822–3826 [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109–1118 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Murphy K.M., Kc W. 2011. Transcription factor networks in dendritic cell development. Semin. Immunol. 23:388–397 10.1016/j.smim.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G., Mattei F., Sestili P., Borghi P., Venditti M., Morse H.C., III, Belardelli F., Gabriele L. 2002. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med. 196:1415–1425 10.1084/jem.20021263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedy J.R., Gavrieli M., Potter K.G., Hurchla M.A., Lindsley R.C., Hildner K., Scheu S., Pfeffer K., Ware C.F., Murphy T.L., Murphy K.M. 2005. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat. Immunol. 6:90–98 10.1038/ni1144 [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. 2003. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 19:59–70 10.1016/S1074-7613(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Shortman K., Naik S.H. 2007. Steady-state and inflammatory dendritic-cell development. Nat. Rev. Immunol. 7:19–30 10.1038/nri1996 [DOI] [PubMed] [Google Scholar]
- Steinman R.M. 2012. Decisions about dendritic cells: past, present, and future. Annu. Rev. Immunol. 30:1–22 10.1146/annurev-immunol-100311-102839 [DOI] [PubMed] [Google Scholar]
- Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M. 2010. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 33:955–966 10.1016/j.immuni.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Morse H.C., III, Ozato K. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 111:1942–1945 10.1182/blood-2007-07-100750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., Fukaya T., Eizumi K., Sato Y., Sato K., Shibazaki A., Otsuka H., Hijikata A., Watanabe T., Ohara O., et al. 2011. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity. 35:958–971 10.1016/j.immuni.2011.10.014 [DOI] [PubMed] [Google Scholar]
- van Rijt L.S., Jung S., Kleinjan A., Vos N., Willart M., Duez C., Hoogsteden H.C., Lambrecht B.N. 2005. In vivo depletion of lung CD11c+ dendritic cells during allergen challenge abrogates the characteristic features of asthma. J. Exp. Med. 201:981–991 10.1084/jem.20042311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.D., Shakhar G., Jung S. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Wang H., Morse H.C., III 2009. IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol. Res. 43:109–117 10.1007/s12026-008-8055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. 2008. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9:676–683 10.1038/ni.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt K.F., Sun G., Grueter B., Fischer M., Zamisch M., Ehlers M., Bosselut R. 2007. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 179:4405–4414 [DOI] [PubMed] [Google Scholar]
- Zhang J., Raper A., Sugita N., Hingorani R., Salio M., Palmowski M.J., Cerundolo V., Crocker P.R. 2006. Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood. 107:3600–3608 10.1182/blood-2005-09-3842 [DOI] [PubMed] [Google Scholar]
- Zindl C.L., Kim T.H., Zeng M., Archambault A.S., Grayson M.H., Choi K., Schreiber R.D., Chaplin D.D. 2009. The lymphotoxin LTalpha(1)beta(2) controls postnatal and adult spleen marginal sinus vascular structure and function. Immunity. 30:408–420 10.1016/j.immuni.2009.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.