Abstract
Summary: The two main functions of bioinformatics are the organization and analysis of biological data using computational resources. Geneious Basic has been designed to be an easy-to-use and flexible desktop software application framework for the organization and analysis of biological data, with a focus on molecular sequences and related data types. It integrates numerous industry-standard discovery analysis tools, with interactive visualizations to generate publication-ready images. One key contribution to researchers in the life sciences is the Geneious public application programming interface (API) that affords the ability to leverage the existing framework of the Geneious Basic software platform for virtually unlimited extension and customization. The result is an increase in the speed and quality of development of computation tools for the life sciences, due to the functionality and graphical user interface available to the developer through the public API. Geneious Basic represents an ideal platform for the bioinformatics community to leverage existing components and to integrate their own specific requirements for the discovery, analysis and visualization of biological data.
Availability and implementation: Binaries and public API freely available for download at http://www.geneious.com/basic, implemented in Java and supported on Linux, Apple OSX and MS Windows. The software is also available from the Bio-Linux package repository at http://nebc.nerc.ac.uk/news/geneiousonbl.
Contact: peter@biomatters.com
1 INTRODUCTION
Research in the biosciences increasingly depends upon bioinformatics for the effective organization and analysis of biological data and experimental results. Having access to the appropriate bioinformatics tools is crucial to the success of any research project. The field of bioinformatics itself has previously been segregated into the parallel realms of data organization (databases, access, integration, web services and search tools) and data analysis (efficient algorithms and statistical modeling). As a result, a large proportion of bioinformatics work involves the chaperoning of data from a variety of sources through a pipeline of analysis tools. This approach typically involves a number of ad hoc labor-intensive steps to convert file formats and create transitional file types. Geneious Basic was created to provide a general framework for research-focused bioinformatics tasks to overcome these challenges and take advantage of modern computing trends. Highlights of the platform include automated database searching, data backup functionality and an extensible API for the integration of novel bioinformatics analysis tools.
2 METHODS
Geneious Basic is written in Java Swing to maximize interoperability among all commonly used operating systems. It is compiled under and requires Java 5 to run. The application provides core modules to enable the visualization, manipulation and transfer of DNA sequences (linear, circular and short oligos such as primers and probes), amino acid sequences, pair-wise and multiple alignments, phylogenetic trees, 3D structures, sequence chromatograms, contig assemblies, microsatellite electropherograms and statistical graphs.
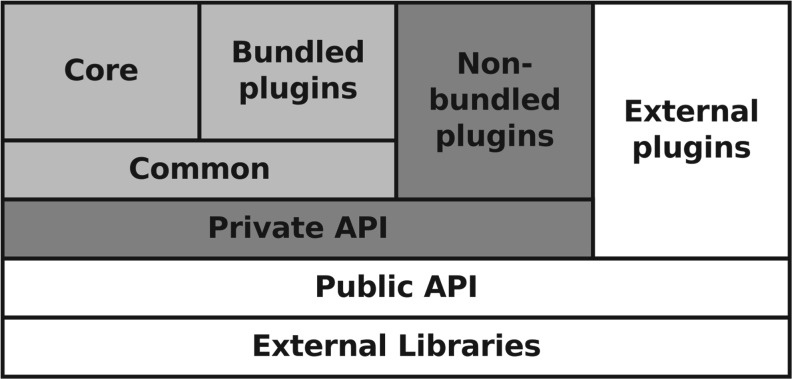
The underlying software framework for Geneious Basic is modular and multi-tiered with a focus on handling bioinformatics data and tools (Fig. 1). It integrates a comprehensive plugin system which is grounded in the extensible Geneious API. The public API component allows plugin developers to leverage the functionality and user interface of the Geneious platform while concentrating on the development of processes and algorithms. The API download from the Geneious website provides a number of skeleton plugin examples to use as a basis for new plugins, allowing developers with a basic level of Java knowledge to develop fully functional plugins that greatly extend the functionality of Geneious Basic.
Fig. 1.
Modular overview of the Geneious Basic software stack. Top-most modules have dependencies on lower modules. The unshaded modules represent the publicly accessible modules for plugin development.
The public API allows developers to leverage online sequence search web services such as NCBI BLAST. Also using the public API, developers can implement plugins that exploit external binaries and even online computational resources, a growing trend in the field of Bioinformatics (Schatz et al., 2010).
3 RESULTS
The Geneious Basic graphical user interface comprises of three main panels, a variety of pull-down menus and many right-click functions for common bioinformatics analyses. Geneious Basic displays selected file(s) in a variety of different ways including sequence view (linear and circular), dotplot view, query-centric alignment view, protein domain view, 3D structure view, text view and notes.
A number of analysis algorithms are implemented within Geneious Basic. For a given document, the user can select from among the appropriate analysis tools to process the data; for example, in the case of a sequence document, the user may choose to perform a BLAST (Altschul et al., 1990) search for a given query sequence against a specific online repository, align against other sequences or generate protein translations. The combinations of available tools and document types allow for the implementation of complex workflows in a single, consistent environment (Fig. 2).
Fig. 2.
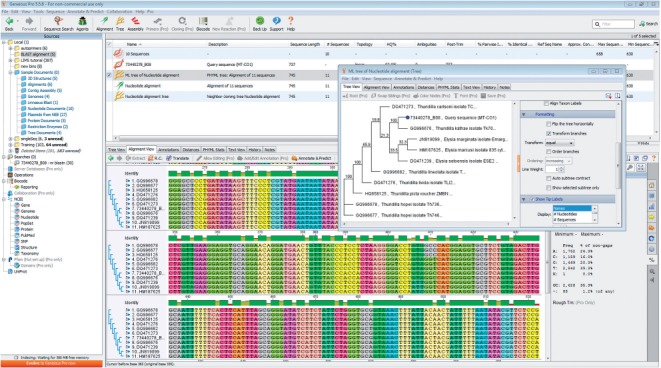
The phylogenetic tree showing the relationship of a MT-CO1 sequence from an unknown source with similar public sequence. Sequences were identified from Genbank using BLAST. Sequences were downloaded then aligned using the Geneious Aligner and a phylogenetic tree built using PhyML. All of these steps were performed within Geneious Basic.
Geneious Basic has an active community of plugin developers, using the public API to contribute third-party plugins and additional functionality within Geneious. The Species Delimitation Plugin, a plugin for summarizing measures of phylogenetic support for user-selected collections of taxa on user-supplied trees (Masters et al., 2011), is one example of scientific research leveraging the Geneious public API for publishable academic research. At the time of the writing of this manuscript, there were nine available plugins for Geneious Basic (Table 1), from plugins wrapping industry standard tools such as Phobos Tandem Repeat Finder (Mayer 2006–2010) to tools that exploit online compute resources such as Green Button.
Table 1.
Summary of available plugins for Geneious Basic
| Plugin name | Category | Author |
|---|---|---|
| DualBrothers Recombination Detect | Phylogenetics | Marc Suchard |
| Green Button | Supercomputing | Biomatters Ltd. |
| Heterozygotes | Sequencing | Biomatters Ltd. |
| InterProScan | Protein | Michael Thon |
| MrBayes | Phylogenetics | Marc Suchard |
| Phobos Tandem Repeat Finder | Nucleotide | Christoph Mayer |
| PhyML | Phylogenetics | Vincent Lefort |
| Species Delimitation | Protein | Masters and Ross |
| Transmembrane Prediction | Protein | Marc Suchard |
4 DISCUSSION
The functionality in Geneious Basic can be compared with a number of other desktop software packages, such as VectorNTI (Lu and Moriyama, 2004), CLC Bio, Sequencher (GeneCodes), Lasergene (DNAstar) and MEGA4 (Tamura et al., 2007). In comparison to the above, the focus of Geneious Basic is to provide an extensible desktop platform in which existing components can be leveraged by third-party plugin developers. This is a unique and important contribution to bioinformatics, allowing users to customize and extend core to their needs without significant manual intervention, matching the success of extensible platforms in parallel spaces such as network analysis and workflow management (Goecks et al., 2010; Smoot et al., 2010). Also comparable is Geneious Pro (Drummond et al., 2010), a commercial extension to Geneious Basic. It shares the base functionality of Geneious Basic and additionally provides tools focused on data sharing, collaboration and advanced next-generation sequencing functionality.
Geneious Basic brings together a large and disparate number of complementary data sources, analysis methods and visualization tools. It is the intention of the authors that by encouraging development to the public API new analysis methods and visualization tools will continue to grow and diversify the range of bioinformatics tasks available to all researchers.
Geneious Basic is cross-platform and is available for Linux, Apple OSX and MS Windows as a standalone download. It is also available as a preconfigured package in the comprehensive Bio-Linux operating system (Field et al., 2006).
Conflict of Interest: MK is currently employed by Biomatters Ltd, the makers of Geneious and hold share options as part of the company share options scheme.
Footnotes
†Current affiliation: Google Inc.
REFERENCES
- Altschul S.F., et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Drummond, A.J. et al. (2010) Geneious v5.5. Available at http://www.geneious.com (last accessed 12 March 2012)
- Field D., et al. Open software for biologists: from famine to feast. Nat. Biotechnol. 2006;24:801–803. doi: 10.1038/nbt0706-801. [DOI] [PubMed] [Google Scholar]
- Goecks J., et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Moriyama E.N. Vector NTI, a balanced all-in-one sequence analysis suite. Brief. Bioinform. 2004;5:378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- Masters B.C., et al. Species Delimitation - a Geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 2011;11:154–157. doi: 10.1111/j.1755-0998.2010.02896.x. [DOI] [PubMed] [Google Scholar]
- Mayer,C. (2006–2010) Phobos 3.3.11. Available at http://www.rub.de/spezzoo/cm/cm_phobos.htm (last accessed 12 March 2012)
- Schatz M.C., et al. Cloud computing and the DNA data race. Nat. Biotechnol. 2010;28:691–693. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]