Abstract
Motivation: As many complex disease and expression phenotypes are the outcome of intricate perturbation of molecular networks underlying gene regulation resulted from interdependent genome variations, association mapping of causal QTLs or expression quantitative trait loci must consider both additive and epistatic effects of multiple candidate genotypes. This problem poses a significant challenge to contemporary genome-wide-association (GWA) mapping technologies because of its computational complexity. Fortunately, a plethora of recent developments in biological network community, especially the availability of genetic interaction networks, make it possible to construct informative priors of complex interactions between genotypes, which can substantially reduce the complexity and increase the statistical power of GWA inference.
Results: In this article, we consider the problem of learning a multitask regression model while taking advantage of the prior information on structures on both the inputs (genetic variations) and outputs (expression levels). We propose a novel regularization scheme over multitask regression called jointly structured input–output lasso based on an ℓ1/ℓ2 norm, which allows shared sparsity patterns for related inputs and outputs to be optimally estimated. Such patterns capture multiple related single nucleotide polymorphisms (SNPs) that jointly influence multiple-related expression traits. In addition, we generalize this new multitask regression to structurally regularized polynomial regression to detect epistatic interactions with manageable complexity by exploiting the prior knowledge on candidate SNPs for epistatic effects from biological experiments. We demonstrate our method on simulated and yeast eQTL datasets.
Availability: Software is available at http://www.sailing.cs.cmu.edu/.
Contact: epxing@cs.cmu.edu
1 INTRODUCTION
One of the fundamental problems in computational biology is to understand associations between genomic variations such as single nucleotide polymorphisms (SNPs) and phenotypic variations. Complex phenotypes (e.g. disease syndromes or pathological signatures) usually consist of a large number of quantitative traits such as clinical and molecular (e.g. gene expression) signals. Differences between these phenotypes involve the complex interplay of a large number of SNPs that perturb the function of disease-related genes in the context of a regulatory or interaction network, rather than these SNPs acting individually (Brem et al., 2005). Thus, unraveling the causal genetic variations and understanding the mechanisms of consequent cell and tissue transformation requires an analysis that jointly considers the epistatic and marginal effects of genomic locations that affect the phenotypic variations. Specifically, genomic locations that influence the expression levels of genes or mRNAs are called expression quantitative trait loci (eQTLs).
Previously, linear marginal effects of eQTLs have been studied extensively in the past decade. To increase the power of detecting causal genetic variants reliably, many different approaches have been proposed that take advantage of the correlation structures in the form of either physical or inferred molecular networks in the genome and phenome, and other prior knowledge of such structures from previous studies. For example, graph-guided fused lasso analyzed multiple traits simultaneously by considering a network of multiple traits to find genetic markers with pleiotropic effects that affect multiple-correlated traits jointly (Kim and Xing, 2009). Another approach, Lirnet, was proposed to make use of prior knowledge on regulatory features, such as conservation scores for a more informed search of association SNPs (Lee et al., 2009).
Unlike linear effects of eQTLs, detecting non-linear SNP–SNP interactions is still in its infancy due to a very large number of possible interactions between SNPs [we refer readers to (Phillips, 2008) for the meanings of SNP–SNP interactions or epistasis]. For example, a typical association analysis often involves up to millions of SNPs; with J~ 106 SNPs, the number of candidates of SNP pairs for pairwise interactions becomes J2 which is clearly pushing the limit of practical computing resources. More importantly, it also carries a serious statistical issue that is the multiple testing problem (Bendera and Langeb, 2001). If we test all SNP pairs to find epistatic effects, only a handful of SNP pairs may be left after correcting for multiple hypothesis testing. To cope with the problem, previous methods attempted to reduce the number of SNP pairs for hypothesis testing in various ways. For example, Devlin et al. (2003) proposed a two-step approach. It first fits a linear regression model with only marginal effects and then considers only the SNPs with significant marginal effects for epistatic interactions. This approach reduces the candidate interaction pairs but it significantly limits the scope of the analysis since genetic loci with epistatic effects may not show any marginal effects. The two-step approach will completely miss such interactions. Another proposed approach is sequential search (Storey et al., 2005). It chooses a primary SNP by finding the SNP with the largest marginal effect, and then the secondary SNP is selected in such a way that residual sum of squares is minimized in a regression setting with the two chosen SNPs. Even though it has been shown that this approach is more powerful than exhaustive 2D search, it still suffers from the cases where interacting SNPs do not have significant marginal effects. Furthermore, Emily et al. proposed a different approach where the candidates of SNP pairs for epistatic effects are chosen using biology networks (Emily et al., 2009). In particular, under the guidance of protein–protein interaction network they reported significant interacting SNP pairs for susceptibility to diseases such as hypertension and bipolar disorder.
Apparently, there is a growing need for a scalable but mathematically principled approach to make effective use of structures in both the genome and the transcriptome; to capture higher order interactions between the genetic variations; and to enable consistent, optimal, and computationally efficient high-dimensional inference for large-scale genome-wide-association (GWA) mapping. In this article, we propose jointly structured input-output lasso (SIOL) model for multitask regression that systematically addresses these challenges.
Specifically, we propose ‘struct i/o multitask regression’, a novel regression method with structured reguralizers which incorporates genome and transcriptome structures into a linear regression model and detects first-order effects of SNPs in the genome. Here, genome (input) structure refers to the phenomenon where multiple-related SNPs are associated with a single trait; transcriptome (output) structure corresponds to pleiotropic effect that is a single SNP is associated with multiple related traits. Note that traits refer to diverse biological outputs such as eye color, onset of diseases and expression levels of genes. Taking into account the structures of the datasets, we can significantly improve GWA inference. First, the genome structure enables us to capture correlated SNPs jointly. When SNPs are linked with the genes with the same biological functions via pathways or biological networks, they are likely to be jointly associated with a trait. Thus, it would be desirable to choose the correlated SNPs all together as eQTLs. Second, output structure allows us to find SNPs which are associated with multiple-related traits jointly. The pleiotropy is well-known phenomenon where a single mutation affects multiple related traits (Dudley et al., 2005). Therefore, we induce this effect over multiple-related traits in our model for better GWA inference.
Extending the model ‘struct i/o multi-task regression’, we propose ‘structured polynomial multitask regression’ to detect epistatic effects as well as marginal effects of SNPs in the genome. In this model, we adopt polynomial regression (Gavrilets and Scheiner, 1993) and include additional regressors for higher order terms. However, in genome-wide association studies, considering all pairs of SNPs is infeasible even for second-order polynomial regression because given a typical human genome with ~105 SNPs, we need to include ~1010 regressors which is both computationally and statistically unmanageable. To find a reasonable set of candidates of SNP pairs, we exploit prior knowledge to guide the search for plausible SNP pairs with epistatic effects. Specifically, we will use a synthetic genetic interaction network (Costanzo et al., 2010) in our eQTL analysis of yeast. The network was constructed using large-scale synthetic genetic array (SGA) analysis (Tong et al., 2004), where query mutations are crossed to the array of viable gene deletion mutants to generate double mutants [see (Boone et al., 2007) for a review]. If two separate genes with mutations that are viable in a single mutant cause a cell death or sickness, then we call the situation a synthetic lethal or sick interaction and edges in the network are created based on this information. As this network contains information on pairs of genes whose mutations affect the phenotype only when the mutations on both genes are present, this represents a set of ground-truth epistatic interactions. Furthermore, it is reasonable to expect that SNPs that lie in cis to the two genes with a synthetic interaction are likely to interact epistatically as well. Thus, in our approach, we use the synthetic genetic interaction network to suggest those SNPs that lie in cis to the epistatically interacting genes as candidate SNPs for epistatic effects on the trait in question. In addition, one can use any other available resources to determine the candidate SNP pairs. For example, it would be possible to include pairs of SNPs having marginal effects to expand the search space without blowing up the number of candidates. In our experiments, we also included SNP pairs that were statistically significant by two-locus epistasis test (P-value <10−5).
Given our proposed models, we need to solve convex optimization problems. To optimize our models, we developed a simple and efficient algorithm called hierarchical group-thresholding method. As we shall see later, in our regularizer, we have non-separable penalties due to the overlap between input and output groups. Thus, traditional methods such as a coordinate descent method cannot be directly applied to our problems. Our optimization technique efficiently solved our problems by checking possible sparsity patterns using optimality conditions and updating non-zero regression coefficients using a coordinate descent method.
Our experimental results confirmed the efficacy of our approach. In our simulation study, our method significantly outperformed other competitors in terms of recall and precision rates in finding true eQTLs with marginal and epistatic effects. Also, applying our model with the genetic interaction network to yeast eQTL dataset (Brem and Kruglyak, 2005), we detected SNPs having marginal and epistatic effects in yeast genome. Interestingly, we found a novel SNP pair (chr1:154328 and chr5:350744) with interaction effects that affects >400 traits related to the same GO category of ribosome biogenesis (corrected P-value for enrichment =1.2×10−36). It turns out that these SNPs are very closely located to NUP60 and RAD51 (within 500 bp), both of which interact with each other (P-value for interaction =3×10−7) (Costanzo et al., 2010). Two-locus epistasis test was not able to detect this epistatic effect with P-value cutoff (<10−5). The SNP pair was also not reported in Storey et al. (2005). The rest of this article is organized as follows. We first present background of linear regression model with structured sparsity in Section 2. Then, in Section 3, we explain struct i/o multitask regression demonstrating how input and output structures can be incorporated into a regression model. Then, as an extension of the previous model, we propose structured polynomial multitask regression that considers both marginal and epistatic effects. Finally, we confirm the benefits of the use of input/output structures using simulated datasets, followed by the analysis of eQTLs with epistatic and marginal effects in yeast.
2 BACKGROUND: LINEAR REGRESSION WITH STRUCTURED SPARSITY
We begin with a brief review on regularized regression approaches including lasso (Tibshirani, 1996), group lasso (Yuan and Lin, 2006) and multitask lasso (Obozinski et al., 2006).
2.1 Notation for matrix operations
Given a matrix B∈ℝK×J, we denote the k-th row by βk, the j-th column by βj, and the (k, j) element by βkj. ‖·‖F denotes the matrix Frobenius norm, ‖·‖1 denotes an ℓ1 norm (entry−wise matrix ℓ1 norm for a matrix argument), and ‖·‖2 represents an ℓ2 norm. Given the set of groups 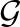 ={g1,…, g|
={g1,…, g|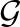 |} defined as a subset of the power set of {1,…, J}, βkg represents the vector with elements {βkj:j∈g, g∈
|} defined as a subset of the power set of {1,…, J}, βkg represents the vector with elements {βkj:j∈g, g∈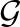 }, or equivalently a subvector of βk for group g. Similarly, for the set of groups
}, or equivalently a subvector of βk for group g. Similarly, for the set of groups 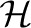 ={h1,…, h|
={h1,…, h|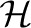 |} over K rows of matrix B, we denote by βhj the vector with elements {βkj:k∈h, h∈
|} over K rows of matrix B, we denote by βhj the vector with elements {βkj:k∈h, h∈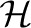 }. We also define the submatrix of Bhg as a |h|×|g| matrix with elements {βkj:k∈h, j∈g, h∈
}. We also define the submatrix of Bhg as a |h|×|g| matrix with elements {βkj:k∈h, j∈g, h∈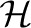 , g∈
, g∈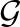 }.
}.
2.2 Lasso, group lasso and multi-task lasso
Assuming that data are collected for N samples at J inputs and K outputs, we let xji denote the observation for the i-th sample and the j-th input. In the problem of genetic association mapping, for haploid organisms, xji takes values from {0, 1}, and for diploid organisms, the value of xji is set to the number of minor alleles at the j-th genetic locus. Let yki denote the observation of k-th output for the i-th individual. Then, we use a regression model that combines a linear model for the marginal effects of individual inputs as follows:
where B∈ℝK×J represents the regression coefficient matrix and E∈ℝK×N is a matrix of noise terms whose elements are assumed to be identically and independently distributed as Gaussian with zero mean and constant variance. In this model, we have only marginal effects. However, one can extend the model to include higher order interactions as we shall see later in Section 4. Throughout this article, we assume that X and Y are standardized. Then, we consider a model without an intercept.
2.2.1 Lasso and group lasso
Lasso is a widely used technique for obtaining a sparse estimate of the regression coefficients. Especially, it has been popular in genome-wide association studies as it is known that it works well even when J>>N (Tibshirani, 1996). The estimates of lasso can be obtained by optimizing the residual sum of squares along with ℓ1 norm as follows:
| (1) |
where λ is the tuning parameter that determines the amount of penalization. A larger value of λ tends to encourage a greater number of the βkj's to be set exactly to zero. The optimal value of λ can be determined by cross validation on regression error, or via an information-theoretic test based on BIC.
In the problem in equation (1), the input variables are independently considered. We consider the grouping of SNPs and apply the group-lasso penalty to B (Yuan and Lin, 2006):
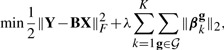 |
(2) |
where 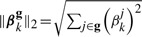 and g∈
and g∈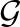 represents a group of SNPs (inputs). The penalty term in equation (2) encourages shrinkage of groups of regression coefficients βkg, ∀g∈
represents a group of SNPs (inputs). The penalty term in equation (2) encourages shrinkage of groups of regression coefficients βkg, ∀g∈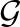 . Note that g can be given by prior domain knowledge or computational algorithms. For example, g can be a cluster of SNPs in genetic interaction networks as these SNPs might influence on the same traits jointly. Also, SNPs in the same pathways can be a reasonable group g (Wang et al., 2007). If such domain knowledge is unavailable, we can use computational methods [e.g. graphical lasso Friedman et al. (2008) and clustering algorithms] to infer meaningful groups of SNPs.
. Note that g can be given by prior domain knowledge or computational algorithms. For example, g can be a cluster of SNPs in genetic interaction networks as these SNPs might influence on the same traits jointly. Also, SNPs in the same pathways can be a reasonable group g (Wang et al., 2007). If such domain knowledge is unavailable, we can use computational methods [e.g. graphical lasso Friedman et al. (2008) and clustering algorithms] to infer meaningful groups of SNPs.
2.2.2 Multitask regression with ℓ1/ℓ2 regularization
A multitask regression with ℓ1/ℓ2-regularization was proposed to learn a joint sparsity pattern across multiple tasks (Obozinski et al., 2006). The ℓ1/ℓ2 penalty allows us to borrow information across multiple regression tasks. The ℓ1/ℓ2-regularized multitask regression is defined as follows:
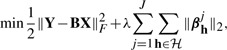 |
(3) |
where 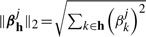 and h∈
and h∈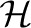 is a group of traits (outputs). The penalty term in equation (3) encourages the outputs in group h to have a common set of relevant inputs. Notice that h can also be learned from prior knowledge or computational techniques. For example, one can group the genes sharing the same biological functions to form h. It is reasonable as the genes with the same functions are likely to be affected by common SNPs. If such domain knowledge is unavailable, one can find
is a group of traits (outputs). The penalty term in equation (3) encourages the outputs in group h to have a common set of relevant inputs. Notice that h can also be learned from prior knowledge or computational techniques. For example, one can group the genes sharing the same biological functions to form h. It is reasonable as the genes with the same functions are likely to be affected by common SNPs. If such domain knowledge is unavailable, one can find 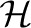 using computational methods. For example, one can estimate a trait network using a graph inference algorithm, and then find the clusters of traits in the network using a clustering algorithm.
using computational methods. For example, one can estimate a trait network using a graph inference algorithm, and then find the clusters of traits in the network using a clustering algorithm.
3 STRUCT I/O MULTITASK REGRESSION
We propose our method (SIOL) that incorporates structural constraints on both the inputs and outputs. The model combines the mixed-norm regularizers for the groups of inputs and outputs, which leads to the following optimization problem:
| (4a) |
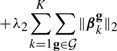 |
(4b) |
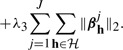 |
(4c) |
The term in equation (4b) incorporates the groupings of the inputs 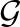 ={g1,…, g|
={g1,…, g|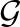 |}, where go represents the o-th group of correlated inputs, and the term in equation (4c) incorporates the groupings of the outputs
|}, where go represents the o-th group of correlated inputs, and the term in equation (4c) incorporates the groupings of the outputs 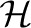 ={h1,…, h|
={h1,…, h|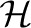 |}, where hm represents the m-th group of correlated outputs.
|}, where hm represents the m-th group of correlated outputs.
Although our proposed model is simply convex combination of ℓ1 norm, ℓ1/ℓ2 norm for input groups and ℓ1/ℓ2 norm for output groups, it is non-trivial to characterize the behavior of the model. Here, we will demonstrate the grouping effects induced by our model, and their benefits in terms of rich structured sparsity in B. Recall that we denote by Bhg the block of coefficients for input group g and output group h. We start with Karush–Kuhn–Tucker (KKT) condition for equation (4):
| (5) |
where skj, ckj and dkj are the subgradient of ℓ1 norm, ℓ1/ℓ2 norm for input groups, and ℓ1/ℓ2 norm for output groups with respect to βkj, respectively. We also define rkj=yk−∑l≠j βkl xl.
First, we consider the case where all coefficients in Bhg become zero simultaneously, that is Bhg=0. Using KKT condition in equation (5), we can see that Bhg=0 if and only if
| (6) |
This condition is due to Cauchy–Schwarz inequality, ∑j∈g(ckj)2≤1, and ∑k∈h(dkj)2≤1. Here, if λ1, λ2 and λ3 are large, Bhg is likely to be zero jointly. This structural sparsity is useful to filter out a large number of irrelevant covariates as it considers both the group of correlated inputs g and the group of correlated outputs h simultaneously.
Our model also inherits grouping effects for only input (or output) groups. For the analysis of such grouping effects, we fix the groups of zero coefficients that overlap with, say, an input group βkg. Formally speaking, let us define ξ={j: (βh′j=0, j∈g, h′∈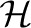 ) ∨ (βkg′=0, j∈g′ ∧ g)}, and fix βkjs for all j∈ξ. Using the KKT condition in equation (6), βkg=0 if
) ∨ (βkg′=0, j∈g′ ∧ g)}, and fix βkjs for all j∈ξ. Using the KKT condition in equation (6), βkg=0 if
| (7) |
Here, we know that dkj=0 for j∈g − ξ (βkj=0 and βhj≠0) and λ2 ∑j∈g (βkj)2=λ2 ∑j∈g−ξ (βkj)2, and hence ∑j∈g−ξ ( λ2 ckj + λ3 dkj)2≤λ22. This technique was introduced in (Yuan et al., 2011) to handle overlapping group lasso penalty. One can see that if the size of ξ is large, βkg tends to be zero together because it reduces the left-hand side of equation (7). This behavior explains the correlation effects between input and output group structures. When a group of coefficients (βkg, βhj) corresponding to an input group or an output group become zero, they affect other groups of coefficients that overlap with them; and the overlapped coefficients are more likely to be zero. These correlation effects between overlapping groups are desirable for inducing appropriate structured sparsity as it allows us to share information across different inputs and different outputs simultaneously. We skip the analysis of the grouping effects for output groups as the argument is the same except that the input and output group are reversed.
Finally, we also have individual sparsity due to ℓ1 penalty. Suppose that we have βkg≠0 and βhj≠0. Using the KKT condition, βkj=0 if and only if
| (8) |
It is equivalent to the condition of lasso that sets an individual regression coefficient to zero. Note that if λ2=λ3=0, we have only individual sparsity, and our model is the same as lasso. When input and output groups should contain both zero and non-zero entries, we can handle the situations using equation (8).
When applied to GWA mapping of eQTLs, our model offers a number of desirable properties. It is likely that our model can detect association SNPs with low signal-to-noise ratio by taking advantage of rich structural information. In GWA studies, one of the main challenges is to detect SNPs having weak signals with limited sample size. In complex diseases such as cancer and diabetes, biologists believe that multiple SNPs are jointly responsible for diseases but not necessarily with strong marginal effects (McCarthy et al., 2008). Even though they do not have strong effects on phenotypic traits individually, it is important to detect them because they might cause significant consequences collectively. However, such causal SNPs are hard to detect mainly due to insufficient number of samples. Our model deals with this challenge by taking advantage of both input and output group structures. First, by grouping inputs (or SNPs), we can increase the signal-to-noise ratio. Suppose each SNP has small signal marginally, if a group of coefficients is relevant, their joint strength will be increased, and it is unlikely that they are jointly set to zero. Conversely, if a group of coefficients is irrelevant, their joint strength will still be small, and it is likely that they are set to zero. Second, taking advantage of the output groups, we can share information across the correlated outputs, and it decreases the sample size required for successful support recovery (Negahban and Wainwright, 2011). Overall, to detect causal SNPs having small effects, our model increases signal-to-noise ratio by grouping the SNPs, and simultaneously decreases the required number of samples by grouping phenotypic traits.
4 STRUCTURED POLYNOMIAL MULTITASK REGRESSION
In addition to marginal effects, we are also interested in detecting interaction effects where multiple SNPs affect phenotypic traits through their interactions. Let us explain interaction effects using an example. Suppose that there are two variants A/a and B/b for an organism. Uppercase and lowercase letters represent major and minor genotypes in population, respectively. Assuming that there is an interaction effect between the two variants, the following scenario can be possible. If an individual has A(major) and B(major) at two genomic locations, there are no effects on the sample. Similarly, genotypes (A and b) or (a and B) cannot affect any traits of an individual. However, if an individual has two minor genotypes a and b, his/her traits can be changed accordingly. Therefore, to detect pairwise interaction effects, we should consider all pairs of SNPs instead of considering each SNP individually. However, it is often infeasible to test all SNP pairs in eQTL mappings or association studies. For example, for human genomes with ~105 SNPs, we should take into account ~1010 candidates of SNP pairs. It is clearly computational demanding and statistically challenging due to multiple hypothesis testing problems. Here, we will show how to generate good and relatively small number of candidates of SNP pairs using genetic interaction networks (Costanzo et al., 2010). Having individual and interaction terms in our model, our model will be able to detect marginal and interaction effects in the genome simultaneously.
Following common practice in GWA literature, here, we consider only pairwise interactions between SNP pairs. Instead of including all SNP pairs as regressors, we use a synthetic genetic interaction network (Costanzo et al., 2010) to define a relatively small candidate set U of interacting SNP pairs. A synthetic genetic interaction network is derived from biological evidence of pairwise functional interactions between genes, such as double knockout experiments (Boone et al., 2007; Costanzo et al., 2010; Koh et al., 2009; Tong et al., 2004). It contains information about pairs of genes whose mutations affect the phenotype only when the mutations are present on both genes, and this represents a set of ground-truth interaction effects. Given such a network, we consider only those pairs of SNPs that are physically located in the genome near the genes that interact in the network within a certain distance. A set of SNP pairs U generated by this scheme is not only much smaller than an exhaustive pair-set but also biologically more plausible. It should also be noted that it is possible to include other sets of SNP pairs from other resources in our candidate set. For example, in our experiments, we also added SNP pairs that passed two-locus epistasis test with P-value <10−5 into the set U.
We generate the group of SNPs or interacting SNP pairs in two steps. In the first step, we find highly interconnected subgraphs (or clusters) from the genetic interaction network using any graph clustering algorithms. In our experiments, we used MCODE algorithm (Bader and Hogue, 2003) for clustering the network. The clusters consist of genes, and the members in each cluster are likely to interact with each other. In the second step, we group all the SNPs or SNP pairs that are linked to the genes in a cluster. We linked the genes and SNPs based on physical locations in the genome. For example, if a SNP is located nearby a gene within a certain distance (e.g. <500 bp), they are linked together. Finally, we define individual SNPs in the mth group as gm∈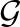 and SNP pairs in the m-th group as lm∈
and SNP pairs in the m-th group as lm∈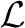 .
.
We then look for associations between inputs/input-pairs and outputs via equation (9) which is equivalent to equation (4) except that it includes additional interaction terms:
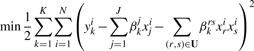 |
(9a) |
| (9b) |
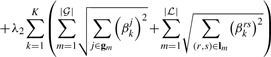 |
(9c) |
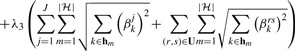 |
(9d) |
| (9e) |
In equation (9), we explicitly show two different tuning parameters for ℓ1 penalty depending on whether a covariate is modeling an individual effect (λ1) or interaction effect (λ4) because they might need different level of sparsity. Note that this problem is the same as equation (4) if we treat interaction terms xri xsi as additional covariates. However, as we have an additional tuning parameter λ4, it requires more computation to determine optimal tuning parameters using crossvalidation procedure.
5 OPTIMIZATION
Unfortunately, the optimization problem resultant from equations (4), (9) is non-trivial. One may find out that each βkj appears in all three regularization terms. Thus, our structured regularizer is non-separable, and simple coordinate descent optimization is not applicable. To solve this challenging problem, we developed a highly efficient optimization technique called hierarchical group-thresholding, and we expect that our method can be scaled to very large datasets such as human eQTL datasets. In our experiments, it took < 3 s for our method to optimize equation (9) when λ1=λ4=0.05, λ2=λ3=0.1, N≤3500, J≤600, and K=10 on a desktop with 2.83 GHz CPU and 4 GB RAM. Here, we briefly describe our optimization algorithm. We start with non-zero regression coefficients of B initialized by other methods [e.g. ridge regression Hoerl and Kennard (1970)]. We then iterate the following two procedures until our model converges. First, we set the groups of (or individual) coefficients to zero according to optimality conditions. Second, we update non-zero coefficients using a coordinate descent method. In this article, however, we are unable to elaborate the optimization technique due to the lack of space.
6 EXPERIMENTS
In this section, we apply our method to both simulation datasets and yeast eQTL datasets (Brem and Kruglyak, 2005) to demonstrate the performance of our method. In our simulations, for comparison, we selected methods including sparse group lasso (Friedman et al., 2010), sparse multitask lasso, lasso (Tibshirani, 1996) and single SNP analysis (PLINK) (Purcell et al., 2007). Note that sparse group lasso is a linear regression model with ℓ1 and ℓ1/ℓ2 penalty for the input groups, and sparse multitask lasso has regularizer that combines ℓ1 and ℓ1/ℓ2 penalty for the output groups. For real data analysis, using yeast genetic interaction network (Costanzo et al., 2010), we show our analysis of eQTLs having marginal and epistatic effects. In all our experiments, we use the following encoding for SNPs. We set xji=0 if the j-th SNP from the i-th sample is a major genotype and xji=1 if it is a minor genotype.
6.1 Simulation study
As the ground-truth associations between SNPs and gene expressions for yeast are unknown, to systematically evaluate the performance of different methods, we perform a simulation study using the yeast genotypes with output values simulated from the known regression coefficients. For generating X, we first selected 60 SNPs from the yeast genome sample (N=100) as the original input covariates. We then simulated 60 pairwise interaction terms (xji×xj′i) by randomly selecting input-pairs from the 60 SNPs mentioned above. Pooling the 60 marginal terms and 60 pairwise interaction terms resulted in a input space of 120 dimensions. We simulated B matrix which reflects the true associations. We used different association strengths of 0.2 and 0.4 when simulating true B. Specifically, we set the coefficients {βk6…, βk10}, {βk31…, βk35}, {βk66…, βk70} and {βk86…, βk90} for all k=1,…, 10 to the non-zero values of association strengths. Given the extracted yeast genotype X and simulated B, we made output variables (or traits) by Y=BX with Gaussian noise with zero mean and unit variance. For the definition of input and output groups, we grouped 5 consecutive input variables, and grouped 10 output variables assuming that all the output variables belong to the same group. For each parameter setting, we generated 20 datasets which have different 100 samples randomly selected from 114 samples in the yeast eQTL dataset and randomly chosen 60 SNP pairs. Based on the datasets, we report the average performance using precision recall curves.
In Figure 1(a and b), we show the performance of our method (SIOL) and other methods including sparse group lasso (S-Group Lasso), sparse multitask lasso (S-Multi-task Lasso), lasso and single SNP analysis performed by PLINK (Purcell et al., 2007) for different association strengths of 0.2 and 0.4. For the results of single SNP analysis, we discarded SNPs with large P-values (>0.001) which is equivalent to the P-value cutoff 0.1 with Bonferroni correction. For all the methods, the tuning parameters were learned using cross validation. From the simulation results, we observed the following behaviors of various methods.
SIOL significantly outperformed all the other competitors for all association strengths persistently. In particular, when the problem is difficult (e.g. association strength 0.2), the performance gap between our method and others was more substantial.
The performances of lasso and single SNP were comparable when association strength was small (e.g. 0.2). With high recall rate, single SNP outperformed lasso but lasso performed better than single SNP with high precision rate.
All sparse learning techniques improved performance significantly as the association strength increased from 0.2 to 0.4. However, single SNP could not take advantage of the high association strength effectively. This result supports that multivariate analysis should be preferred over single variate analysis such as single SNP when there are multiple causal SNPs.
Fig. 1.
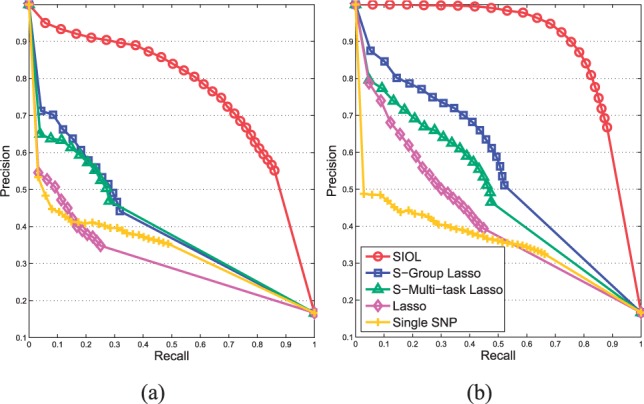
Precision recall curves on the recovery of true non-zero coefficients by changing the threshold of relevant covariates for our proposed method (SIOL) and other methods including sparse group lasso (S-Group Lasso), sparse multi-task lasso (S-Multi-task Lasso), Lasso and single SNP analysis under different association strengths of (a) 0.2 and (b) 0.4
In our simulation study, we verified that our method truly takes advantage of input and output structures effectively when there exists meaningful prior information on both input and output sides. Also, it should be noted that the performance of our method can be in par with that of sparse group lasso or sparse multitask lasso even when one of group structures is unreliable. Our method can adjust tuning parameters to ignore any incorrect group structures.
6.2 Yeast eQTL dataset
We apply SIOL to budding yeast (Saccharomyces cerevisiae) data (Brem and Kruglyak, 2005) with 1260 unique SNPs (out of 2956 SNPs) and the observed gene-expression levels of 5637 genes. As network prior knowledge, we used genetic interaction network reported in (Costanzo et al., 2010) with stringent cutoff to construct the set of candidates of SNP pairs U. We follow the procedure in Section 4 to make U with an additional set of significant SNP pairs with P-value <10−5 computed from two-locus epistasis test. When determining the set U, we assumed that a SNP is linked to a gene if the distance between them is <500 bp. We consider it a reasonable choice for cis-effect as the size of intergene regions for S. cerevisiae is 515 bp on average (Sunnerhagen and Piskur, 2006). As a result, we included 982 interaction terms from the interaction network in X with 1260 individual SNPs. The number SNP pairs from two-locus epistasis test was different depending on the trait. For generating input structures, we processed the network data as follows. We started with genetic interaction data which include 74 984 interactions between gene pairs. We then extracted genetic interactions with low P-values (<0.001). Given 44 056 significant interactions, using MCODE clustering algorithm, we found 55 gene clusters. Using the gene clusters, we generated the groups of individual SNPs and pairs of SNPs according to the scheme in Section 4. For generating output structures, we applied hierarchical clustering to the yeast gene expression data with cutoff 0.8, resulting in 2233 trait clusters.
6.2.1 Marginal effects in yeast eQTL dataset
We briefly analyze eQTLs having marginal effects in the yeast dataset as the focus of our analysis will be on epistatic interactions. In general, the association results for marginal effects by our method, lasso and single SNP analysis showed similar patterns for strong associations. However, we observed differences for SNPs with small or medium sized signals. For example, our results had fewer non-zero regression coefficients compared with lasso. One possible explanation would be that the grouping effects induced by our model might have removed false predictions with small or medium sized effects. To illustrate eQTLs with marginal effects, we show some examples of association SNPs using GenAMap (Curtis et al., 2012). Figure 2 demonstrates a Manhattan plot on Chromosome 7 for two genes including YER160C and YJR029W. Both genes have the same GO category ‘transposition’. As both genes share the same GO category, it is likely that they are affected by the same SNPs if there exist any association SNPs for both genes. In our results, we could see that the same SNPs on Chromosome 7 are associated with both genes as shown in Figure 2. However, single SNP analysis did not find any significant association SNPs in the region. Lasso detected association SNPs in the region but they were associated with only YER160C rather than both of them (lasso plot is not shown to avoid cluttered plots).
Fig. 2.
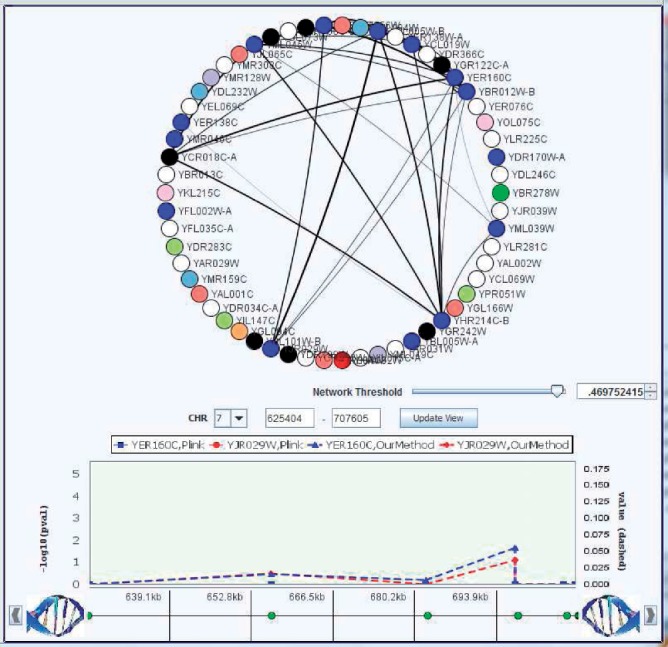
Manhattan plot for association between (YER160C and YJR029W) and SNPs on Chromosome 7. The two genes YER160C and YJR029W share the same GO category ‘transposition’. Our method detected SNPs which affect both two genes in this region. However, single SNP analysis did not find any association SNPs and lasso found SNPs associated only with YER160C in this region. This figure was generated using GenAMap software (Curtis et al., 2012)
6.2.2 Epistatic effects in yeast eQTL dataset
As we analyze a large number of genes (5637), it is interesting to find SNP pairs that affect a large number of traits, which are often called hotspots. Here, we analyze the hotspots having epistatic interactions, and compare our results with the results of two-locus epistasis test for all SNP pairs performed by PLINK (Purcell et al., 2007). Recall that two-locus epistasis test is the widely used statistical technique for detecting interaction effects between a SNP pair (r, s) based on the following model: yki ~ b0+b1 xri+b2 xsi+b3 xri xsi, ∀i, k. It computes P-value for the association of the SNP pair (r, s) by testing the significance of the interaction term b3. In the following analysis, we discarded all SNP pairs if the correlation coefficient between the pairs >0.5 to avoid trivial interaction effects.
We first identified the most significant hotspots that affect >100 gene traits. To make sure that we include only significant interactions, we considered interaction terms if their absolute value of regression coefficients are >0.05. For the results of two-locus epistasis test, we considered all SNP pairs with P-value <10−5. Figure 3(a and b) shows the hotspots found by our method and two-locus epistasis test. The rings in the figure represent the yeast genome from Chromosome 1 (located at the top of each circle) to 16 clockwise. The lines show epistatic interactions between the two genomic locations at both ends. Interestingly, our method detected 11 hotspots, and two-locus epistasis test found only two hotspots with epistatic interactions. In Table 1, we summarized the hotspots with epistatic effects identified by our method. Notably, hotspot 1 (epistatic interaction between chr1:154328 and chr5:350744) affects 455 genes which are enriched with the GO category of ribosome biogenesis with the corrected P-value for enrichment <10−35 (multiple testing correction is performed by false discovery rate (Maere et al., 2005)). This SNP pair was included in our candidates from the genetic interaction network. There is a significant genetic interaction between NUP60 and RAD51 with P-value 3×10−7 (Costanzo et al., 2010), and both genes are located at chr1:152257-153877 and chr5:349975-351178, respectively. As both SNPs are closely located to NUP60 and RAD51, it is reasonable to hypothesize that two SNPs at chr1:154328 and chr5:350744 affected the two genes, and their genetic interaction in turn acted on a large number of genes related to ribosome biogenesis. We further investigated the mechanism of this significant SNP–SNP interaction. In our literature survey, RAD51 (RADiation sensitive) is strand exchange protein involved in DNA repair system (Sung, 1994), and NUP60 (NUclear Pore) is the subunit of the nuclear pore complex involved in nuclear export system (Denning et al., 2001). Also, it has been reported that yeast cells are excessively sensitive to DNA damaging agents if there exist mutations in NUP60 (Nagai et al., 2008). In our results, we also found out that the SNP close to NUP60 did not have significant marginal effects, and the SNP in RAD51 had marginal effects. According to these facts, it would be possible to hypothesize as follows. When there are no mutations in RAD51, the point mutation in NUP60 cannot affect other traits as the single mutation is not strong enough and if there exist DNA damaging agents in the environment, DNA repair system would be able to handle them. However, when there exists the point mutation in RAD51 involved in DNA repair system, DNA damaging agents would severely harm yeast cells with the point mutation in NUP60 as DNA repair system might not work properly due to the mutation in RAD51 (recall that the SNP in RAD51 had marginal effects). As a result, both mutations in NUP60 and RAD51 could make a large impact on many gene traits.
Fig. 3.
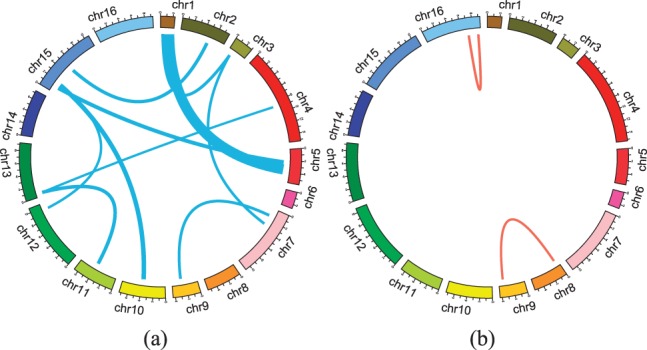
Hotspots of SNP pairs with epistatic effects identified by (a) our method and (b) two-locus epistasis test. This figure represents the yeast genome in a circular format generated using Circos software (Krzywinski et al., 2009). In clockwise direction, from the top of the circles, we show 16 chromosomes. Lines indicate interaction effects between two connected locations in the genome. Thickness of the lines is proportional to the number of traits affected by the interaction effects. Here we show interaction effects which influence > 100 gene traits. The hotspots for (a) are shown in Table 1. In (b), two SNP pairs are found including chr16:718892-chr16:890898 (affected genes are enriched with the GO category of ribosome biogenesis with corrected P-value 1.6×10−36), and chr8:56246-chr9:362631 (affected genes are enriched with the GO category of vacuolar protein catabolic process with corrected P-value 1.6×10−14)
Table 1.
Hotspots of SNP pairs having epistatic effects in yeast identified by our method
| Hotspot label | SNP1 location | SNP2 location | Number of affected traits | GO category of affected traits | Corrected P-value of GO category |
|---|---|---|---|---|---|
| 1 | chr1:154328 | chr5:350744 | 455 | Ribosome biogenesis | 1.2×10−36 |
| 2 | chr10:380085 | chr15:170945 | 195 | Ribosome biogenesis | 1.6×10−12 |
| 3 | chr10:380085 | chr15:175594 | 185 | Ribosome biogenesis | 4.1×10−12 |
| 4 | chr5:222998 | chr15:108577 | 170 | Response to temperature stimulus | 2.9×10−6 |
| 5 | chr11:388373 | chr13:64970 | 155 | Regulation of translation | 1.8×10−32 |
| 6 | chr2:499012 | chr15:519764 | 145 | Vacuolar protein catabolic process | 1.4×10−7 |
| 7 | chr1:41483 | chr3:64311 | 130 | ||
| 8 | chr7:141949 | chr9:277908 | 125 | ||
| 9 | chr3:64311 | chr7:312740 | 115 | Glycoprotein metabolic process | 1.5×10−4 |
| 10 | chr12:957108 | chr15:170945 | 110 | Vacuolar protein catabolic process | 7.8×10−16 |
| 11 | chr4:864542 | chr13:64970 | 105 | Ribonucleoprotein complex biogenesis | 3.7×10−6 |
Furthermore, we looked at the hotspots which affect >10 gene traits. Figure 4a and 4b shows epistatic interactions identified by our method and two-locus epistasis test, respectively. In this figure, we show significant interactions with regression coefficient cutoff >0.1 for our method, and P-value cutoff <10−6 for two-locus epistasis test. These cutoffs are arbitrarily chosen to make the number of hotspots found by both methods similar. Surprisingly, two methods showed very different hotspots with epistatic interactions. Figure 4a was very similar to Figure 3a but in Figure 4b several hotspots emerged which were absent in Figure 3b. We will analyze these hotspots in two ways. First, we will look at the hotspots with epistatic effects which appeared in both Figure 4a and 4b. Then, we will investigate the differences between the two results. First, we observed that both methods found significant epistatic effects between Chromosomes 1 and 5. Recall that in our previous analysis of the hotspots, this interaction was discussed (see hotspot 1 in Table 1). Among all significant SNP pairs found by two-locus epistasis test, there was no SNP pair identical to hotspot 1 but there were 30 SNP pairs close to it (within <50 kb). Also, it turns out that these 30 SNP pairs had very strong correlation with hotspot 1.
Fig. 4.
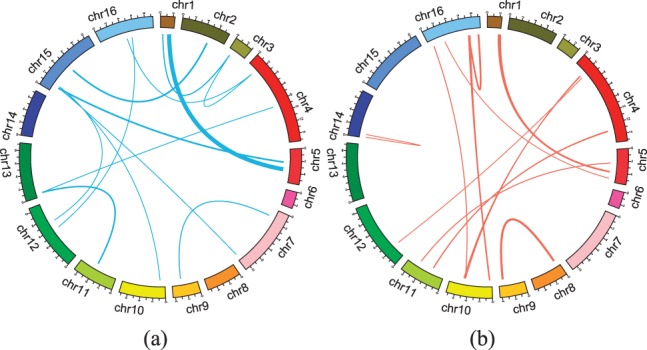
Hotspots of SNP pairs with epistatic effects which influence > 10 gene traits found by (a) our method and (b) two-locus epistasis test by PLINK (Purcell et al., 2007). This figure was generated using Circos software (Krzywinski et al., 2009)
In Figure 6, we show scatter plot to illustrate the strong correlations between hotspot 1 and these 30 SNP pairs. More interestingly, the total number of genes affected by these 30 SNP pairs was 416, and it is very similar to 455, that is, the number of genes affected by hotspot 1. According to these facts and our previous analysis for the mechanism of hotspot 1, it seems that hotspot 1 is truly significant, and two-locus epistasis test found significant SNP pairs that are close to the true location but failed to find the exact location of hotspot 1. It supports that our algorithm could find such a significant hotspot affecting >400 genes by detecting exact SNP pairs. Second, we investigated the differences between the two results in Figure 4a and 4b. As we cannot report all the results in this article, we focused on a SNP pair (chr10:87113-chr15:141621) in Figure 4a, and another SNP pair (chr8:63314-chr9:362631) in Figure 4b. Figure 5a and 5b shows the average gene expression levels for each SNP pair. In this figure, x-axis represents the genotype ∈{0, 1} which is the multiplication of two SNPs (SNP1 × SNP2, where SNP1, SNP2 ∈{0, 1}), and y-axis represents the average gene expression levels of individuals with given genotype. Each line in Figure 5a and 5b shows how the average gene expression level changes as the genotype changes from 0 to 1 for each trait affected by the SNP pairs with error bars of one SD. Interestingly, in Figure 5a, we could see that there is a consistent pattern, where for most gene traits, the expression levels decreased as the genotype changed from 0 to 1. However, as shown in Figure 5b, for the SNP pair found by two-locus epistasis test, we could not find such a coherent pattern. It seems that we found consistent gene expression patterns for the SNP pair as our model finds SNPs using input and output group structures. Conversely, it is possible that two-locus epistasis test found the SNP pair, which affected the expression levels of multiple genes with different patterns as it analyzed each SNP pair separately.
Fig. 6.
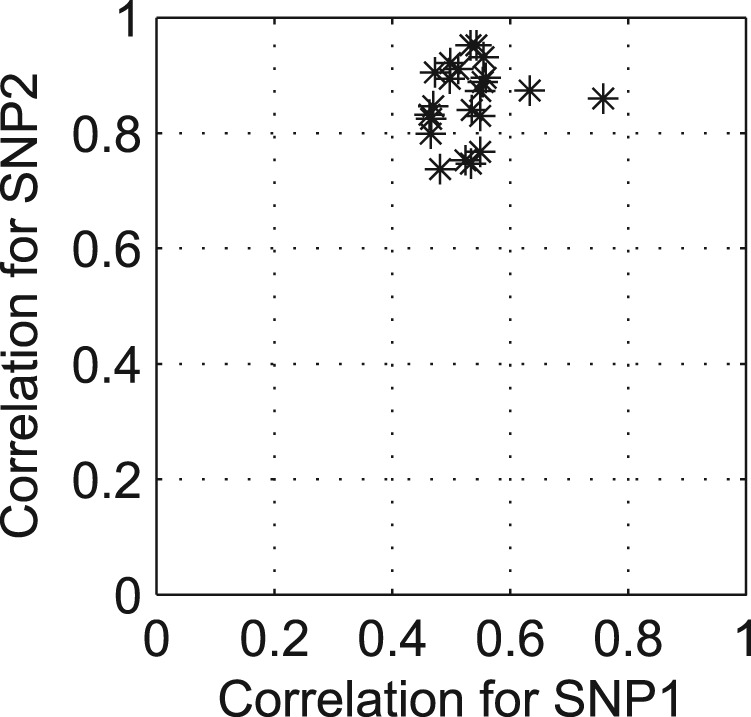
The scatter plot for illustrating the correlations between the SNP pair at hotspot 1 (SNP1, SNP2) and SNP pairs detected by two-locus epistasis test (P-value <10−6) close to hotspot 1 (within <50 kb). Each dot represents a SNP pair found by two-locus epistasis test, and it was perturbed by a small amount of random noise to avoid overlapping dots
Fig. 5.
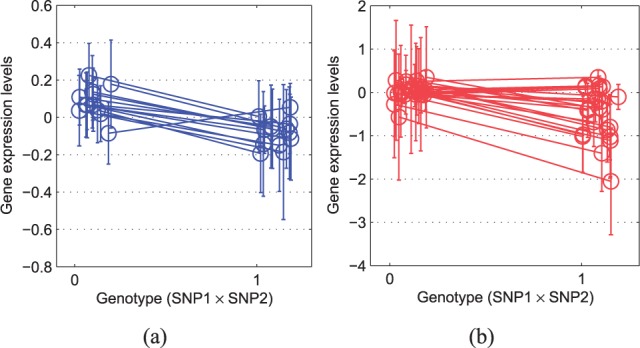
Variations of gene expression levels according to the genotypes of (a) a SNP pair (chr10:87113-chr15:141621) found by our method, and (b) a different SNP pair (chr8:63314-chr9:362631) found by two-locus epistasis test. Here, x-axis represents genotypes (SNP1 × SNP2 ∈{0, 1}) and y-axis shows the average expression levels of the genes affected by the corresponding SNP pairs. There are multiple lines in each penal as both SNP pairs are associated with multiple traits. A small noise was added to the genotypes to avoid overlapping of the error bars (±1 SD from the mean)
7 DISCUSSIONS
We proposed a novel sparse learning technique called jointly–SIOL. We introduced a structured regularizer that includes ℓ1 penalty, ℓ1/ℓ2 penalty for input and output groups simultaneously. Using the rich structured regularizer, we made it possible to use input and output group structures in a single framework. Our experiments showed that our method can effectively use structural information and improve the accuracy for detecting association SNPs.
7.1 P-value computation
Note that our method gives biased regression coefficients. Thus, it would be desirable to compute P-values to estimate the significance of the non-zero coefficients and control false discovery rate. For high-dimensional regression problems, a few approaches have been proposed to compute P-values (Meinshausen et al., 2009; Wasserman and Roeder, 2009). We can use these techniques to compute P-values for the covariates (SNPs or SNP pairs) selected by our method. Here, we briefly describe ‘screen and clean’ procedure proposed by (Wasserman and Roeder, 2009). It starts with randomly dividing the data (X, Y) into two equal-sized groups, Ψ1=(X1:⌊N/2⌋, Y1:⌊N/2⌋) and Ψ2=(X⌊N/2⌋+1:N, Y⌊N/2⌋+1:N). First, we apply our method with tuning parameters determined by cross validation to the first group of data, Ψ1, and find non-zero coefficients. We denote by Qk={j: βkj≠0, j=1,…, J}, k=1,…, K, the set of covariates chosen by our method. Second, for all k, we calculate P-values of the covariates in Qk based on the second group of data, Ψ2, using ordinary least-squares estimates. We then adjust the P-values by multiplication with |Qk|. For the covariates not in Qk, we assign P-value of 1. Recently, ‘multisplit’ method (Meinshausen et al., 2009) was proposed for computing P-values based on aggregate results of the above procedure. Using P-values computed by ‘multisplit’ method, the authors showed that family-wise error and false discovery rate can be controlled.
7.2 Comparison between SIOL and other models
A unique contribution of our model is to use both input structures (groups of SNPs) and output structures (groups of traits) simultaneously in a highly general setting. Note that we can deal with overlapping groups, and hence, a SNP or a trait can be involved in multiple groups. Unlike our model, most previous models considered only input or only output structure. For example, composite absolute penalties are introduced to incorporate grouping and hierarchical structures on input sides (Zhao et al., 2009). Adaptive multitask lasso is proposed to consider the groups of traits with many regulatory features in the genome (Lee et al., 2010). Graph-guided fused lasso (GFlasso) (Kim and Xing, 2009) and tree-guided group lasso (Kim and Xing, 2010) are developed to incorporate graph and tree structures on output sides, respectively. Recently, Curtis et al. (2012) proposed graph–graph-guided fused lasso (gGFlasso) which attempted to use input structure on transcriptome and output structure on phenome. However, it is different from our model as it is based on graph structures rather than group structures.
7.3 Application of our method to Other eQTL data
To apply our method to human eQTL datasets or eQTL datasets for other species, it is important to find reliable groups of SNPs or groups of traits. In yeast eQTL studies, we have experimentally validated genetic interaction networks. However, such reliable genetic interaction networks may not be available for other domains. Instead, we have abundant biological knowledge such as regulatory features, LD (linkage disequilibrium) structures, pathway databases and protein–protein interaction networks. One needs to find meaningful groups of SNPs or groups of traits using such biological information.
7.4 Effects of unreliable grouping information
Given unreliable grouping information, it is likely that our method finds only a few SNPs that are strongly associated with traits. Note that our method shrink the regression coefficient matrix B based on the predefined groups of SNPs and traits. Ideally, coefficients should be grouped together if they can be jointly shrunk rather than jointly selected. Suppose there is a group of SNPs which contains a few relevant SNPs and many irrelevant SNPs. In that case, it is likely that the coefficients corresponding to the group are jointly shrunk to zero, and we might fail to capture the relevant SNPs within the group. Conversely, if a group has no relevant SNPs, our method can still shrink the group of coefficients effectively. Overall, when using unreliable grouping information, our model might select SNPs having strong association but miss many SNPs having weak association.
7.5 Future work
We still have many challenging issues for association mapping problems. For example, it is non-trivial to find biologically meaningful and reliable input and output group structures. One of possible approaches would be to combine GWA mapping with the inference of grouping information to improve the performance of both tasks synergistically. Also, we want to apply our method to human disease data as well. Detection of SNPs with true epistatic and marginal effects will shed light on the better understanding of genetic factors of complex diseases.
Funding: This work was done under a support from NIH 1 R01 GM087694-01; NIH 1RC2HL101487-01 (ARRA); AFOSR FA9550010247; ONR N0001140910758; NSF Career DBI-0546594; NSF IIS-0713379; and Alfred P. Sloan Fellowship awarded to E.P.X.
Conflict of Interest: none declared.
REFERENCES
- Bader G., Hogue C. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendera R., Langeb S. Adjusting for multiple testing – when and how? J. Clin. Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- Boone C., et al. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Brem R., Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. PNAS. 2005;102:1572. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R., et al. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., et al. The genetic landscape of a cell. Science. 2010;327:425. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis R., et al. Pacific Symposium on Biocomputing. Hawaii, USA: 2012. Finding genome-transcriptome-phenome association with structured association mapping and visualization in genamap. [PubMed] [Google Scholar]
- Denning D., et al. The nucleoporin Nup60p functions as a Gsp1p–GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 2001;154:937–950. doi: 10.1083/jcb.200101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., et al. Analysis of multilocus models of association. Genet. Epidemiol. 2003;25:36–47. doi: 10.1002/gepi.10237. [DOI] [PubMed] [Google Scholar]
- Dudley A., et al. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 2005;1:2005.0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emily M., et al. Using biological networks to search for interacting loci in genome-wide association studies. Eur. J. Hum. Genet. 2009;17:1231–1240. doi: 10.1038/ejhg.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., et al. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., et al. A note on the group Lasso and a sparse group Lasso. 2010 arXiv:1001.0736v1 [math.ST] [Google Scholar]
- Gavrilets S., Scheiner S. The genetics of phenotypic plasticity. VI. theoretical predictions for directional selection. J. Evolut. Biol. 1993;6:49–68. [Google Scholar]
- Hoerl A., Kennard R. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12:55–67. [Google Scholar]
- Kim S., Xing E. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 2009;5:e1000587. doi: 10.1371/journal.pgen.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Xing E. Proceedings of the 27th Annual International Conference on Machine Learning. Haifa, Israel: 2010. Tree-guided group lasso for multi-task regression with structured sparsity. [Google Scholar]
- Koh J., et al. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucl. Acids Res.. 2009;38:D502–D507. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., et al. Learning a prior on regulatory potential from eQTL data. PLoS Genetics. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., et al. Adaptive multi-task lasso: with application to eQTL detection. Adv. Neural Inform. Process. Syst. 2010;23:1306–1314. [Google Scholar]
- Maere S., et al. Bingo: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- McCarthy M., et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Meinshausen N., et al. P-values for high-dimensional regression. J. Am. Stat. Assoc. 2009;104:1671–1681. [Google Scholar]
- Nagai S., et al. Functional targeting of DNA damage to a nuclear pore-associated sumo-dependent ubiquitin ligase. Science. 2008;322:597. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negahban S., Wainwright M. Simultaneous support recovery in high dimensions: Benefits and perils of block ℓ1/ℓ∞-regularization. IEEE Trans. Inform. Theory. 2011;57:3841–3863. [Google Scholar]
- Obozinski G., et al. Joint covariate selection for grouped classification. Vol. 743. Berkeley: Technical Report. Department of Statistics, University of California; 2006. [Google Scholar]
- Phillips P. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J., et al. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3:1380. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sunnerhagen P., Piskur J. Comparative genomics: using fungi as models. Vol. 15. Heidelberg, Germany: Springer; 2006. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. Roy. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- Tong A., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wang K., et al. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L., Roeder K. High dimensional variable selection. Ann. stat. 2009;37:2178. doi: 10.1214/08-aos646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Lin Y. Model selection and estimation in regression with grouped variables. J. Roy. Stat. Soc. B (Stat. Methodol.) 2006;68:49–67. [Google Scholar]
- Yuan L., et al. Adv. Neural Inform. Process. Syst. Granada, Spain: 2011. Efficient methods for overlapping group lasso. [Google Scholar]
- Zhao P., et al. The composite absolute penalties family for grouped and hierarchical variable selection. Ann. Stat. 2009;37:3468–3497. [Google Scholar]


