Abstract
Calmodulin is a prototypical and versatile Ca2+ sensor with EF-hands as its high-affinity Ca2+ binding domains. Calmodulin is present in all eukaryotic cells, mediating Ca2+-dependent signaling. Upon binding Ca2+, calmodulin changes its conformation to form complexes with a diverse array of target proteins. Despite a wealth of knowledge on calmodulin, little is known on how target proteins regulate calmodulin’s ability to bind Ca2+. Here, we take advantage of two splice variants of SK2 channels, which are activated by Ca2+-bound calmodulin, but show different sensitivity to Ca2+ for their activation. Protein crystal structures and other experiments show that depending on which SK2 splice variant it binds to calmodulin adopts drastically different conformations with different affinities for Ca2+ at its C-lobe. Such target protein induced conformational changes make calmodulin a dynamic Ca2+ sensor, capable of responding to different Ca2+ concentrations in cellular Ca2+ signaling.
Introduction
As the prototypical Ca2+ sensor, calmodulin (CaM) is widely expressed in all eukaryotic cells, mediating a variety of cellular signaling processes, including regulation of enzymatic activities, modulation of ion channel activities, synaptic transmission and plasticity, and regulation of gene expression (Clapham, 2007; Deisseroth et al., 1998; Dick et al., 2008; Drum et al., 2002; Halling et al., 2005; McLaughlin and Murray, 2005; Wayman et al., 2008; Xia et al., 1998; Zuhlke et al., 1999). A diverse array of target proteins has been identified, such as CaM kinases and ion channels, which are known to interact with CaM with or without Ca2+. CaM is also a versatile Ca2+ sensor, capable of responding to a wide range of Ca2+ concentrations (10−12 M – 10−6 M) in Ca2+-dependent signal transduction (Chin and Means, 2000). Four canonical EF-hands, two located at the CaM N terminus (N-lobe) and the other two at the C terminus (C-lobe), serve as the high affinity Ca2+ binding motifs. (Meador et al., 1992; Meador et al., 1993). The N- and C-lobes are connected by an extremely flexible central linker region. EF-hands at the C-lobe are generally thought to have a higher affinity for Ca2+ than those in the N-lobe (Andersson et al., 1983; Crouch and Klee, 1980). Upon binding Ca2+, CaM changes its conformation from the closed configuration to the open one, exposing the hydrophobic surfaces within the N- and C-lobes for Ca2+-dependent interactions with the target proteins (Chin and Means, 2000; Halling et al., 2005; Hoeflich and Ikura, 2002; Ikura et al., 1992; Ishida and Vogel, 2006; Kranz et al., 2002; Meador et al., 1992; Meador et al., 1993; Schumacher et al., 2004). Additional conformational changes, most noticeably unwinding of the α-helix of the CaM linker region (R74 to E83) to various degrees, allow CaM to adopt different conformations in its interactions with different target proteins, ranging from the compact (collapsed) conformation to its full extended conformation (Drum et al., 2002; Fallon and Quiocho, 2003; Ikura et al., 1992; Meador et al., 1992; Meador et al., 1993; Mori et al., 2008; Van Petegem et al., 2005).
Such structural flexibility explains how CaM is capable of interacting with target proteins with distinct structural features (Halling et al., 2005; Ishida and Vogel, 2006). So far, most structural studies have focused on how binding of Ca2+ to CaM exposes the hydrophobic surfaces for interaction with target proteins. Despite a wealth of knowledge on CaM, much less is known about how CaM’s affinity for Ca2+ is determined, in particular, how binding of CaMBDs can reciprocally affect CaM’s conformations and consequently change CaM’s affinities for Ca2+. It is generally thought that formation of CaM-target protein complexes increases CaM’s affinity for Ca2+. Furthermore, it is also less clear how regulation of CaM’s affinity for Ca2+ by target proteins is achieved at the molecular level. This knowledge gap is in part due to technical difficulties, such as the size limitation of proteins used in NMR experiments or the challenges of getting protein crystals of CaM complexed with bigger target proteins. Consequently, short CaMBD peptides (typically 15–30 mers) are often used in structural studies, and they are often too short to produce any significant impact on CaM’s conformations.
Small-conductance Ca2+-activated potassium channels (SK) are widely expressed in brain and play pivotal roles in regulating neuronal excitability, dendritic integration and synaptic transmission (Faber, 2009; Kohler et al., 1996; Stocker, 2004). Activation of SK is achieved exclusively by intracellular Ca2+ (Xia et al., 1998). CaM, constitutively tethered to SK, serves as the high-affinity Ca2+ sensor. Binding of Ca2+ to CaM, particularly at the N-lobe, changes the channel conformation and opens the channel (Schumacher et al., 2004; Schumacher et al., 2001). Activation of SKs is highly sensitive to Ca2+ with EC50 around 300 – 700 nM (Kohler et al., 1996; Xia et al., 1998). Three SK genes have been identified, KCNN1, KCNN2 and KCNN3 (Bond et al., 2005). We have recently identified an SK2 splice variant, SK2-b, which is less sensitive to Ca2+ for its activation, compared to the original SK2 channel, SK2-a (unpublished data). These two SK2 splice variants are virtually identical in their primary amino acid sequence with the exception that SK2-b has three additional amino acid residues, A463, R464 and K465 (ARK), in its CaMBD.
Since SK channels are exclusively activated by Ca2+-bound CaM, this pair of SK2 splice variants provides a unique experimental model for us to test how target proteins may regulate the CaM’s ability to bind Ca2+. We have determined the X-ray crystal structure of CaM complexed with CaMBD from SK2-b (CaMBD2-b) at a resolution of 1.9Å and compared to the previously determined structure of CaM-CaMBD2-a (Schumacher et al., 2001). Along with biochemical, biophysical and electrophysiological data, we demonstrate that the structural flexibility of CaM occurs not only at the CaM linker region, but also extensively in its hydrophobic interfaces, induced by these two CaMBDs. Our results provide direct experimental evidence supporting the notion that target proteins can change the conformation of CaM and more importantly regulate the affinity of CaM for Ca2+, thus making CaM a dynamic Ca2+ sensor capable of responding to a wide range of Ca2+ concentrations in cellular Ca2+ signal transduction.
Results
Structure of the CaM-CaMBD2-b complex
To explore how target proteins may affect the affinity of CaM for Ca2+, we determined the X-ray crystal structure of the CaM-CaMBD2-b complex in the presence of Ca2+ at a resolution of 1.9Å (Figures 1 & S1, Table 1). The CaM-CaMBD2-b complex shows a 2×2 architecture, with two CaMs (vertical) and two CaMBD2-b peptides (horizontal) in an antiparallel arrangement (Figures 1A & 1C). Both CaMBD2-b and CaMBD2-a adopt the same hair-pin α-helical structure (Figures 1A & 1C). Insertion of ARK does not disrupt the propensity of CaMBD2-b to form α-helix (Figures 1A & S1D). Nor are there any direct contacts between ARK and the CaM hydrophobic interfaces (Figure 1A), where Ca2+-dependent interactions between CaM and its target proteins typically take place, although R464 forms a salt bridge with E120 of CaM outside of the hydrophobic interfaces. In both complexes, the CaM N-lobe interacts with the C-terminal fragment of the CaMBD peptide, whereas the C-lobe interacts with the N-terminal fragment of the CaMBD peptide. Compared to CaM-CaMBD2-a, the structure of the CaM-CaMBD2-b complex shows distinct features, particularly the CaM conformation (Figure 1).
Figure 1. A distinct conformation of the CaM-CaMBD2-b complex.
(A & B) Structure of the CaM-CaMBD2-b complex. A 2×2 complex is formed with two horizontal CaMBD2-b peptides, green, and two CaM molecules, salmon (A). Also shown are side chains of the three residues A463, R464 and K465 in CaMBD2-b. A 90°-turn of the structure shows that CaM in CaMBD2-b adopts an “S”-shaped configuration (B).
(C & D) Structure of the CaM-CaMBD2-a complex (1G4Y). Two CaMBD2-a peptides, gold, and two CaM molecules, blue, form the 2×2 complex (C). CaM in the CaM-CaMBD2-a complex displays a “C”-like structure (D).
(E & F) Comparison of the structures of CaM from CaM-CaMBD2-b (salmon) and CaM-CaMBD-2a (blue). The CaM structures are aligned at the N-lobe. Notice the difference at the linker region (R74-E82).
See also Figure S1
Table 1.
Crystallographic statistics
| Data Collectiona | |
|---|---|
| CaM-CaMBD2-b Complex | |
| Space Group Unit Cell Dimensions |
P1 a = 47.7 Å, b = 65.6 Å, c = 66.1 Å α = 90.7°, β = 110.5°, γ = 111.0° |
| Wavelength (Å) | 1.075 |
| Resolution range (Å) | 30 – 1.9 (2.0 – 1.9) |
| Completeness (%) | 97.9 (96.1) |
| Total Observations | 620,836 (77,935) |
| Unique Observations | 53,499 (7,715) |
| Mean Redundancy | 11.6 (10.1) |
| Mean I/σ(I) | 15.6 (2.6) |
| Rmergeb | 0.093 (0.936) |
| Rpimc | 0.030 (0.325) |
| Model Refinementa | |
| Resolution Range (Å) | 20–1.9 (1.97–1.9) |
| Number of reflections | 53,458 (4,945) |
| Rworkd | 0.170 (0.242) |
| Rfreed | 0.195 (0.288) |
| Number of atoms/Average B-factor (Å2) | 4,123/43.9 |
| Protein | 3,687/42.0 |
| Calcium ions | 8/31.0 |
| Solvent | 318/49.6 |
| Ligands (glycerol, sulfate, phenylurea) | 110/58.2 |
| Phi/Psi, most favored (%) | 99.3 |
| R.m.s.d. bond angles (°) | 0.885 |
| R.m.s.d. bond lengths (Å) | 0.013 |
Values in parentheses refer to data in the highest resolution shell.
Rmerge = ΣhklΣj|Ij − 〈I〉| / ΣhklΣj Ij. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents.
Rpim (precision-indicating merge) = Σhkl (1/nhkl − 1)1/2 Σj|Ij − 〈I〉| / ΣhklΣj Ij. n is the number of observations of reflection hkl.
Rcryst = Σhkl|Fobs − kFcalc|/ Σhkl|Fobs|. Rfree = Rcryst for 5% of reflections excluded from crystallographic refinement.
Overall, significant differences exist in the CaM structure between CaM-CaMBD2-b and CaM-CaMBD2-a (r.m.s.d. = 12.16 Å). Such differences, however, are not spread uniformly across the entire CaM. The CaM N-lobe from both complexes displays nearly identical structure, with r.m.s.d = 0.86 Å. In contrast, the CaM C-lobe shows dramatic differences between the two complexes (r.m.s.d. = 2.53 Å). A significant difference is also seen in the CaM linker region (r.m.s.d. = 4.83 Å, Figures 1E & 1F). The CaM linker region in our model maintains a rigid α-helix, leading to approximately a 180-degree rotation of the C-lobe (Figures 1E & 1F). Consequently, CaM in our structure adopts an “S”-like configuration (Figures 1B & 1F). In comparison, the CaM linker region in CaM-CaMBD2-a unfolds from R74 to E82, resulting in a moderate collapse of CaM and thus a “C”-like configuration (Figures 1D & 1F). Comparison was also made between our structure and that of the CaM-edema-factor complex (1K90, Figure S1E). In the latter complex, the CaM linker region unwinds within a much shorter span of the α-helix. Another obvious difference is in how CaMBDs are positioned in these two complexes. In CaM-CaMBD2-b, the two CaMBD2-b peptides are seen at both sides of the S-shaped CaM and they are completely separated from each other (Figures 1A & 1B). In contrast, the two CaMBD2-a peptides are much closer to each other and are wrapped around by CaM (Figures 1C & 1D, also see Figure 3).
Figure 3. Different thermodynamic profiles for CaM-CaMBD2-b and CaM-CaMBD2-a.
(A) Surface representation of the CaM-CaMBD2-b complex. CaMBD2-b (green) interacts with CaM (salmon) only at both the N- and C-lobes. There is no physical contact between the CaMBD2-b peptides.
(B) Surface representation of the CaM-CaMBD2-a complex. The CaMBD2-a peptides (gold) form extensive contacts with CaM (blue) as well as between themselves.
(C) Results of ITC experiments. Ca2+-bound CaM was titrated into either CaMBD2-a or CaMBD2-b. For CaM-CaMBD2-a, a two-site model is required to fit the data (the smooth line). On the other hand, a single-site model can adequately fit the data and use of the two-site model did not statistically improve the fitting.
(D) Thermodynamic profiles for formation of the CaM-CaMBD2-a and CaM-CaMBD2-b complexes. Plotted are the Gibbs free energy (ΔG), enthalpy (ΔH) and entropy (−TΔS) for formation of both CaM-CaMBD2-a and CaM-CaMBD2-b. ΔG is calculated from ΔH and TΔS (Table S1), which were determined by ITC at 20°C (ΔG = ΔH − TΔS).
See also Figure S2 and Table S1
The most intriguing and unexpected observation is that all four EF-hands of CaM in the CaM-CaMBD2-b complex are occupied by Ca2+ ions (Figure 1). This is in striking contrast to that of the CaM-CaMBD2-a complex, in which the CaM C-lobe is Ca2+-free (Figure 1) (Schumacher et al., 2001). It has been suggested that the CaM C-lobe in the CaM-CaMBD2-a complex might adopt a semi-open configuration, which could contribute to the failure of the C-lobe to bind Ca2+ (Schumacher et al., 2004). However, structural comparison shows that the CaM C-lobe from CaM-CaMBD2-a is more likely in an open configuration, in which helices VI and VII are completely separated from helices V and VIII, similar to the CaM C-lobe from either CaM-CaMBD2- b or CaM-edema factor, both of which are Ca2+-bound in their CaM C-lobes (Figures S1F & S1G). While differences can be seen in the four helices of the CaM C-lobe between CaM-CaMBD2-b and CaM-CaMBD2-a, more significant changes are observed in their loop regions, where the EF-hands reside (Figure S1F). Failure to bind Ca2+ is also reported for the EF-hands of the CaM N-lobe of the CaM-edema factor complex (Figure S1E).
CaM-CaMBD2-b differs from CaM-CaMBD2-a in solution
We first sought to determine whether, like CaM-CaMBD2-a (Schumacher et al., 2001), the CaM-CaMBD2-b complex would adopt a 2×2 configuration in solution in the presence of Ca2+. Sedimentation equilibrium (SE) experiments were performed for both CaM-CaMBD2-b and CaM-CaMBD2-a with or without Ca2+. Without Ca2+ (Figures 2A & 2B), the molecular mass is 28.5 ± 0.7 kDa (mean ± sd) for CaM-CaMBD2-b and 24.9 ± 2.0 kDa CaM-CaMBD2-a very close to the predicted molecular mass of a 1×1 complex, 29.3 kDa and 28.9 kDa respectively. In the presence of Ca2+, the molecular mass is doubled to 61.1 ± 1.6 kDa for CaM-CaMBD2-b and 55.8 ± 1.0 kDa for CaM-CaMBD2-a, indicating that Ca2+ promotes formation of a 2×2 complex in solution (Figures 2C & 2D). The SE results agree with previous reports for CaM-CaMBD2-a, which also show that without Ca2+ the CaM-CaMBD interaction occurs at an interface different from that in the presence of Ca2+ (Schumacher et al., 2004; Schumacher et al., 2001).
Figure 2. Ca2+ promotes formation of a 2×2 complex for both CaM-CaMBD2-a and CaM-CaMBD2-b in solution.
(A & B) A 1×1 complex of CaM-CaMBD2-a and CaM-CaMBD2-b in the absence of Ca2+. Upper panels are raw SE data (symbols) at different centrifugation speeds as indicated. Curves are fit of the raw data to a single-component model using Sedphat. Lower panels are residuals of the fit. Without Ca2+ both CaM-CaMBD2-a and CaM-CaMBD2-b are 1×1, with a predicted molecular mass of 28.9 kDa and 29.3 kDa respectively.
(C & D) A 2×2 complex of CaM-CaMBD2-a and CaM-CaMBD2-b in the presence of Ca2+. Upper panels are raw SE data (symbols) with fit to a single-component model (curves), and lower panels are residuals of the fit. Ca2+ promotes formation of a 2×2 complex for both CaM-CaMBD2-a and CaM-CaMBD2-b with a predicted molecular mass of 57.8 kDa and 58.6 kDa respectively.
We then turned our attention to whether both CaM complexes might be different from each other when in solution. The structures predict that formation of the CaM-CaMBD2-b complex is a much simpler process, involving primarily the interaction between CaM and the CaMBD2-b peptide (Figure 3A). In contrast, formation of the CaM-CaMBD2-a complex includes not only the CaM-CaMBD2-a interaction but also the extensive interactions along the longer helices of two CaMBD2-a (Figure 3B). The interactions between two CaMBD2-a peptides are quite substantial, with the total surface area of 1208Å2, compared to 1174Å2 for the CaM-CaMBD2-a interaction at the N-lobe and 1149Å2 for the CaM-CaMBD2-a interaction at the C-lobe. Thus, formation of the CaM-CaMBD2-a complex in the presence of Ca2+ undergoes a thermodynamically more complex process than that of the CaM-CaMBD2-b complex.
Isothermal titration calorimetry (ITC) was used to examine the thermodynamic profiles associated with formation of the 2×2 complex of CaM-CaMBD2-b or CaM-CaMBD2-a. For both complexes, changes in heats become minuscule when the molar ratio of CaM/CaMBD approaches 1, indicating that formation of the 2×2 complex has been completed (Figures 3C & S2). Formation of the CaM-CaMBD2-b complex shows a sigmoidal relationship of the heat evolved versus the molar ratio of CaM/CaMBD2-b, which is well modeled by a single-binding-site isotherm (smooth curve, Figures 3C & S2, and Table S1). In contrast, formation of the CaM-CaMBD2-a complex produces a biphasic change in heats, which is best fitted by a two-binding-site model (smooth curve, Figure 3C & S2, and Table S1). Analyses show that for CaM-CaMBD2-a the rising phase is ethalpically favored (ΔH1 = −34.73 ± 0.96 Kcal/mol), whereas the declining phase is entropy-driven (ΔS2 = 87.2 ± 1.98 cal/mol/K). Despite dramatic differences in ΔH and TΔS, the Gibbs free energy (ΔG) for formation of CaM-CaMBD2-a is essentially the same as that of CaM-CaMBD2-b (Fig. 3D). Previous studies on other CaM complexes have shown that CaM has the same affinity for different CaM-target proteins despite differences in their thermodynamics for formation of these CaM complexes (e.g. Frederick et al., 2007). Thus, in the presence of Ca2+, formation of CaM-CaMBD2-a and CaM-CaMBD2-b undergoes different thermodynamic processes (ΔH and −TΔS in Figure 3D), indicating that in solution these two CaM complexes are different from each other. Furthermore, in the presence of Ca2+, CaM has essentially the same affinities for both CaMBD2-a and CaMBD2-b (ΔG in Figure 3D).
Structural plasticity in the CaM hydrophobic binding interfaces
In CaM-CaMBD2-b, the three additional amino acids, ARK, do not directly interact with CaM at its hydrophobic interfaces (Figure 1A). How can insertion of ARK in CaMBD2-b produce such a dramatic impact on the CaM conformation? Typically, a complete α-helical turn requires 3.6 amino acid residues. Insertion of ARK produces less than one full α-helical turn (60 degrees less), and therefore alters how CaM interacts with CaMBD2-b. Upon binding Ca2+, CaM undergoes conformational changes and exposes the hydrophobic surfaces in both its N- and C-lobes that interact with target proteins (Chin and Means, 2000; Halling et al., 2005; Hoeflich and Ikura, 2002; Ishida and Vogel, 2006). Extensive structural analysis was performed to identify amino acid residues involved in formation of the CaM-CaMBD2-b and CaM-CaMBD2-a complexes.
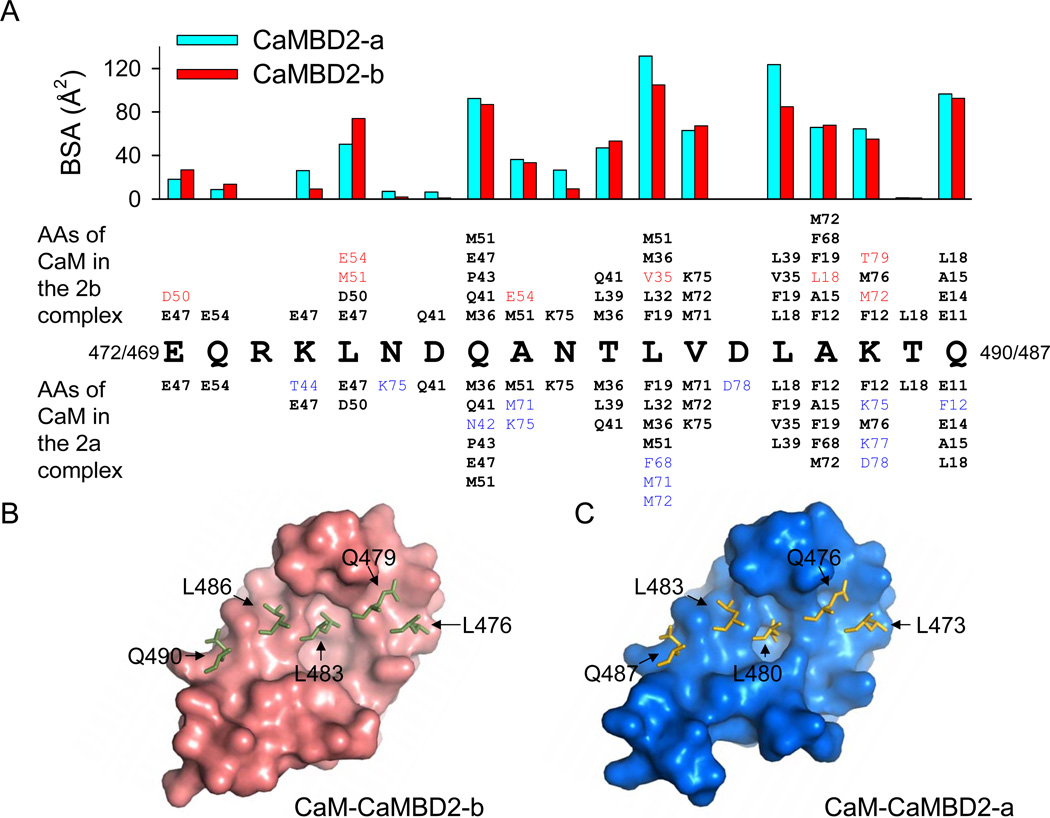
Two peptide fragments in both CaMBD2-a and CaMBD2-b interact with the CaM hydrophobic surfaces, fragment E469/E472 to L488/L491 (Insertion of ARK shifts numbering by 3 in CaMBD2-b) interacts with the CaM N-lobe and fragment R419 to L440 forms contacts with the CaM C-lobe. Figures 4 and 5 show the plots of buried surface area (BSA) for CaMBD residues in their interactions with CaM (bar graphs), as well as lists of CaM residues, within 5 Å radius of each CaMBD residues, that are in contacts with these CaMBD residues. L480/L483, with the highest BSA, serves as the anchor residue, which interacts with the hydrophobic pocket in the N-lobe. The corresponding anchor residue interacting with the CaM C-lobe is L428. Clearly, the same set of key residues in CaMBD interacts with CaM in formation of both CaM-CaMBD2-b and CaM-CaMBD2-a.
Figure 4. Interactions of CaM and CaMBDs at the CaM N-lobe.
(A) Amino acid residues involved in formation of CaM-CaMBD2-b and CaM-CaMBD2-a with Ca2+. Buried surface area (BSA, bar graphs) identifies key residues in CaMBD2-b and CaMBD2-a which interact with CaM. A C-terminal fragment, from E472/E469 to L491/L488, interacts with the CaM N-lobe. Insertion of ARK shifts the numbering by three for CaMBD2-b. Also listed are CaM residues, in a smaller type face, that are in contacts (within 5 Å radius) with individual CaMBD residues. Shown in red are the CaM residues which only interact with key residues in CaMBD2-b. CaM residues in cyan form contacts only with CaMBD2-a. CaM residues in a black bold type face interact with both.
(B & C) Hydrophobic interfaces of the CaM N-lobe in complex with CaMBD2-b (B) or CaMBD2-a (C). Overlaid are key residues from CaMBD2-b or CaMBD2-a. Both N-lobes are aligned from L4 to R74, with r.m.s.d. = 0.86 Å, to create the graphs.
See also Figure S3
Figure 5. Interactions of CaM and CaMBDs at the CaM C-lobe.
(A) Amino acid residues involved in formation of CaM-CaMBD2-b and CaM-CaMBD2-a in the presence of Ca2+, similar to descriptions in Fig. 3. An N-terminal fragment, from R419 to L440, interacts with the CaM C-lobe.
(B & C) Hydrophobic interfaces of the CaM C-lobe in complex with CaMBD2-b (B) or CaMBD2-a (C). Both C-lobes are aligned from I85 to T146, with r.m.s.d. = 2.53 Å, to create the graphs.
See also Figure S4
The hydrophobic interfaces in the CaM N- and C-lobes that interact with target proteins are formed by amino acid residues, particularly methionines, from all four helices of each lobe. Hydrophobic pockets, which harbor the anchor residues from CaMBD, are thought to consist primarily of FLMM, F19, L32, M51 and M71 at the N-lobe, and F92, L105, M124 and M144 at the C-lobe (Ataman et al., 2007). In both CaM-CaMBD2-a and CaM-CaMBD2-b, the hydrophobic pocket at the N-lobe is made primarily of F19, L32, M36 and M51 (Figure 3A). Overall, a similar set of CaM residues contributes to formation of the hydrophobic interface at the N-lobe to interact with CaMBD2-b or CaMBD2-a in the presence of Ca2+. The CaM N-lobe hydrophobic interfaces from CaM-CaMBD2-b and CaM-CaMBD2-a share similar surface topology (Figures 4B, 4C & S3).
The hydrophobic surfaces in the CaM C-lobe vary greatly between the CaM-CaMBD2-b and CaM-CaMBD2-a complexes, and there is very little overlap between the CaM residues that contact a given target amino acid residues from CaMBD2-a or CaMBD2-b (Figure 5A). In the CaM-CaMBD2-b complex, the CaM C-lobe hydrophobic pocket includes F92, L105, F141, M144 and M145, while the corresponding hydrophobic pocket in CaM-CaMBD2-a is made up of I85, A88, F89, F92, L105, V108, M109 and M124 (Figures S4A & S4B). Consequently, the location of the hydrophobic pocket at the C-lobe changes significantly (Figures 5B & 5C). Another example is the hydrophobic surface for W432 from CaMBD, which interacts primarily with helix V in CaM-CaMBD2-b instead of helix VII in CaM-CaMBD2-a (Figures S4C & S4D). Thus, substantial rearrangement of the CaM C-lobe hydrophobic interface takes place, depending on which CaMBD is complexed with CaM.
Effects of CaM target proteins on the conformation of individual EF-hands in CaM
Comparison of the structures of two CaM-complexes demonstrates that both CaMBD2-b and CaMBD2-a produce significant impact on the conformation of CaM, in particular, rearrangement of the hydrophobic interface at the C-lobe. In contrast, such rearrangement of the hydrophobic interface is minimal at the N-lobe. We next sought to determine whether rearrangement of the hydrophobic interfaces in the CaM C-lobe affects the conformation of the EF-hands, which might provide explanations to why the CaM C-lobe of the CaM-CaMBD2-a complex fails to bind Ca2+. A canonical EF-hand includes six Ca2+-coordinating amino acid residues, located in the loop region and arranged in: 1(+X), 3(+Y), 5(+Z), 7(−X), 9(−Y) and 12(−Z) (Figure S5) (Gifford et al., 2007).
In Figure 6, the loop region of each CaM EF-hand in our structure is superimposed and compared to that of the CaM-CaMBD2-a or CaM-edema factor complex. For the N-lobe, where EF-hands bind Ca2+ ions in both CaM-CaMBD2-a and CaM-CaMBD2-b, the loop structure matches well for both EF-hand 1 (r.m.s.d. = 0.26 Å) and EF-hand 2 (r.m.s.d. = 0.46 Å, Figure 6A). On the other hand, EF-hand 3 and EF-hand 4 of the CaM C-lobe display significant structural differences between CaM-CaMBD2b and CaM-CaMBD2-a, with r.m.s.d. of 1.28 Å and 1.67 Å respectively (Figure 6B). If the comparison includes side chains, the differences become even more significant, with r.m.s.d. of 1.96 Å for EF-hand 3 and 2.46 Å for EF-hand 4. The most significant differences lie with the glutamate residue at position 12(−Z) of the EF-hand, E104 and E140 (Figure 6B). In CaM-CaMBD2-a both E104 and E140 point away from the Ca2+ coordination sphere. Significant differences are also observed at position 1(+X), D93 and D129, and position 3(+Z), D95 and D131. Analysis shows that the structural differences at EF-hand 3 and EF-hand 4 are not caused by crystal packing.
Figure 6. Conformational plasticity of the CaM EF-hands.
(A & B) Structures of EF-hands from CaM-CaMBD2-b (salmon) and CaM-CaMBD2-a (blue). EF-hands 1 and 2 are aligned from D20 to E31 and D56 to E67 respectively. EF-hands 3 and 4 are aligned from D93 to E104 and D129 to E140 respectively. Side chains at positions 7 and 9 are not shown.
(C & D) Structures of EF-hands from CaM-CaMBD2-b (salmon) and the CaM-edema factor (pale cyan, 1K90). Significant differences exist in the structures of EF-hands 1 and 2, but not EF-hands 3 and 4. Side chains at positions 7 and 9 are not shown.
See also Figure S5
We further compared the structures of EF-hands in CaM-CaMBD2-b to those of the CaM-edema factor complex (1K90), which fail to bind Ca2+ at the CaM N-lobe (Drum et al., 2002). As expected, the conformation of EF-hand 1 and EF-hand 2 in 1K90 is dramatically different from that of our structure with r.m.s.d. of 2.08 Å and 1.68 Å respectively (Figure 6C). Several key residues in both EF-hand 1 and EF-hand 2 of 1K90 show significantly altered spatial orientations, including D24, E31, D56 and D58. On the other hand, there is little difference at EF-hand 3 and EF-hand 4 of the CaM C-lobe between CaM-CaMBD2-b and the CaM-edema factor complex (Figure 6D). Thus, in CaM-CaMBD2-a, failure of binding to Ca2+ at the CaM C-lobe results primarily from altered conformations of EF-hand 3 and EF-hand 4 as a consequence of rearrangement of helices V, VI, VII and VIII. CaMBD2-b, on the other hand, interacts with a different hydrophobic interface at the CaM C-lobe and restores the ability of EF-hand 3 and EF-hand 4 to bind Ca2+.
CaM as a dynamic Ca2+ sensor
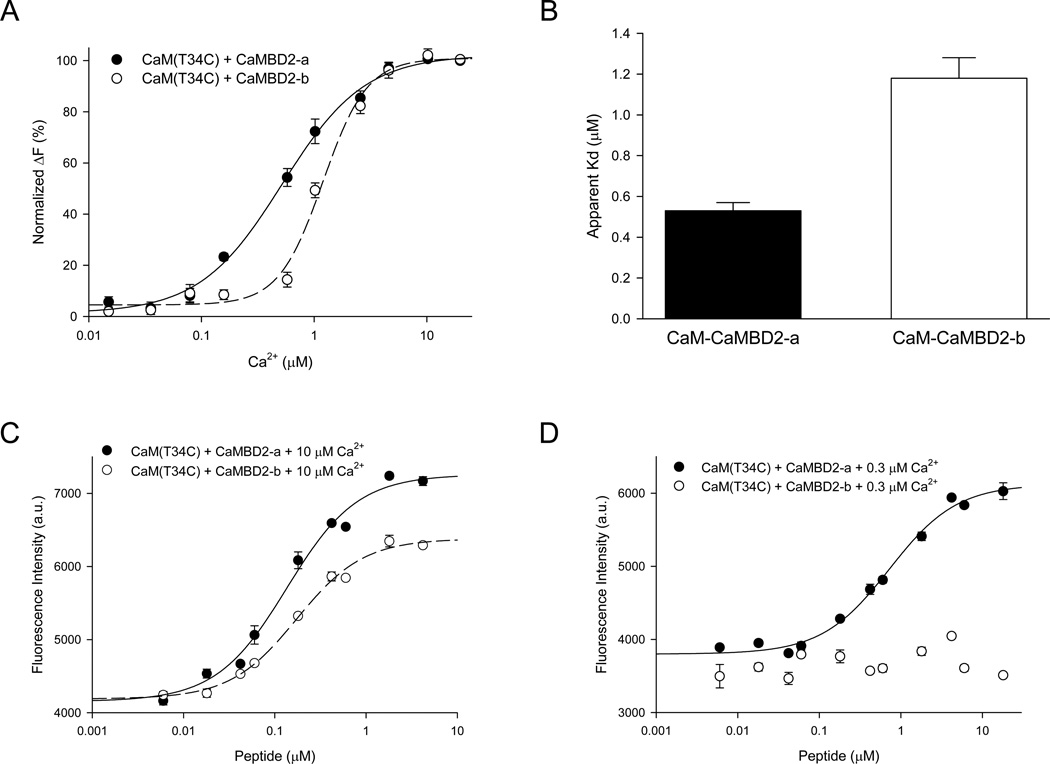
Our recent work has shown that SK2-b is less sensitive to Ca2+ for its activation (unpublished data). While differences in structures between the two CaM complexes may provide a plausible explanation, structures alone cannot rule out other potential scenarios. Binding assays were performed using AEDANS-labeled CaM(T34C or T110C) and CaMBD2-a or CaMBD2-b at different Ca2+ concentrations to examine the interactions between CaM and CaMBDs. The fluorescence emission intensity increases with Ca2+ concentrations in formation of both the CaM-CaMBD2-a and CaM-CaMBD2-b complexes (Figure S6). There is, however, a significant rightward shift of the dose-response curve for the Ca2+-dependent formation of the CaM-CaMBD2-b complex (Figure 7A). The apparent Kd for Ca2+ is increased from 0.53 ± 0.04 µM (n = 7) for formation of CaM-CaMBD2-a to 1.18 ± 0.10 µM (n = 4) for that of CaM-CaMBD2-b (p < 0.001, Figure 7B), suggesting that CaM, when complexed with CaMBD2-b, may have reduced its affinity for Ca2+. The Hill coefficient is 1.13 ± 0.04 (n = 7) for CaM-CaMBD2-a and 2.05 ± 0.18 (n = 4) for CaM-CaMBD2-b. Identical results were obtained with CaM(T110C).
Figure 7. The CaM-CaMBD2-b complex has a reduced apparent affinity for Ca2+.
(A & B) Formation of CaM-CaMBD2-b (n = 4) complex is less sensitive to Ca2+, compared to that of CaM-CaMBD2-a (n = 7). Increases in fluorescence intensity (normalized) are plotted against free Ca2+ concentrations (A). There is a significant reduction in the apparent Kd for Ca2+ (p < 0.001, B).
(C & D) Dose-dependent binding of CaMBD2-b (n = 3) and CaMBD2-a (n = 3) to AEDANS labeled CaM(T34C) in the presence of 10 µM Ca2+ (C) or 0.3 µM Ca2+ (D). CaM(T110C) yielded the same results.
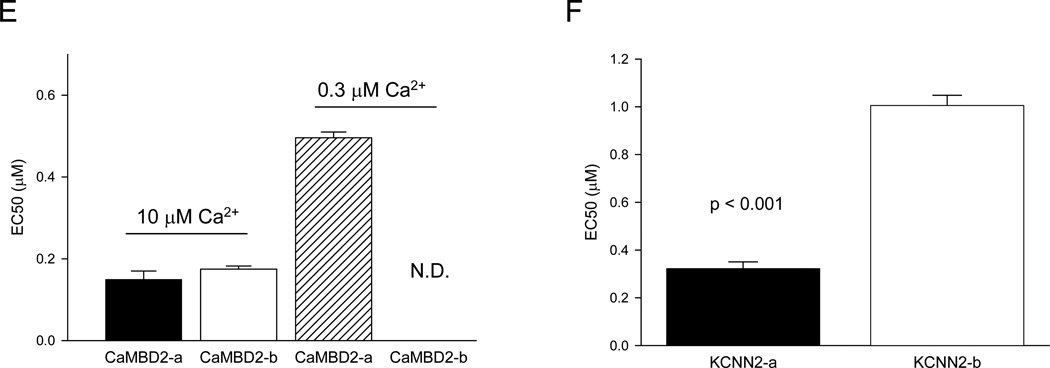
(E) The EC50 of CaM for CaMBDs obtained at 10 µM or 0.3 µM Ca2+. The EC50 of CaM for CaMBD2-b at 0.3 µM Ca2+ could not be determined (N.D.).
(F) The EC50 for activation of SK2-b (n=10) and SK2-a (n=8) channels by Ca2+ from electrophysiology experiments, (P<0.001).
See also Figure S6
However, such a shift in Ca2+-dependent formation of the CaM-CaMBD2-b complex could also result from a reduction of the affinity of Ca2+-bound CaM for CaMBD2-b (Figure S6A). Additional binding assays were performed to test such a possibility with or without Ca2+. At a saturating Ca2+ concentration of 10 µM (Figure 7A), the fluorescence intensity of CaM(T34C) increases with the peptide concentrations of both CaMBD2-a and CaMBD2-b (Figure 7C). There is no difference in the EC50 for formation of either CaM-CaMBD2-a (0.15 ± 0.02 µM) or CaM-CaMBD2-b (0.18 ± 0.01 µM, n = 3, p = 0.318), suggesting that CaM, once Ca2+-bound, has the same affinity for both CaMBD2-a and CaMBD2-b (Figure 7E). Identical results were obtained with CaM(T110C) instead of CaM(T34C) (data not shown). The results are consistent with our ITC data that with saturating Ca2+ CaM has the same affinity for CaMBD2-a and CaMBD2-b. Additional binding assays were performed at the Ca2+ concentration of 0.3 µM to further evaluate the affinity of CaM for CaMBD2-b. The fluorescence intensity of CaM(T34C) increases with CaMBD2-a, with an EC50 of 0.72 ± 0.09 µM, (n = 3, Figure 7E). In contrast, the CaMBD2-b peptide produces little change in the fluorescence intensity, as though there is little or no interaction between CaM and CaMBD2-b (Figure 7D). Identical results were obtained CaM (T110C). Finally, when the same binding assays were repeated without Ca2+, dose-dependent changes in the fluorescence intensity with CaMBD2-b were observed with CaM(T110C), but not CaM(T34C). CaM has the same EC50 for both CaMBD2-a and CaMBD2-b (data not shown). In the presence of Ca2+, we never observed differential results using CaM(T34C) or CaM(T110C). This led us to conclude that increased fluorescence intensity in the presence of Ca2+ results from formation of a stable 2×2 complex, consistent with the SE data (Figure 2).
Collectively, the results of binding assays are consistent with the model that CaM, when complexed with CaMBD2-b, has a reduced affinity for Ca2+. In principle, a reduction in Ca2+ affinity at either the N- or C-lobe will cause a right-shift of Ca2+ dependent formation of the CaM-CaMBD2-b complex (Figure 7A). Computer modeling using molecular dynamics (MD) was performed to test whether the CaM N- and C-lobes in CaM-CaMBD2-b might have different affinities for Ca2+. Thermodynamic integration calculations for the annihilation of bound Ca2+ were performed using all-atom, explicit-solvent MD simulations. The mean ΔΔG associated with annihilation of Ca2+ (N-lobe vs. C-lobe) is 18.39 ± 6.35 kcal/mol (2.72 – 32.88 kcal/mol, Table S2), indicating that the CaM N-lobe more stably binds Ca2+ than does C-lobe in the CaM-CaMBD2-b complex.
Electrophysiology experiments again confirm that SK2-b is less sensitive to Ca2+ for its activation compared to SK2-a. The EC50 for activation of SK2-b by Ca2+ is 1.01 ± 0.04 µM (n = 12), approximately three times higher than the EC50 for activation of SK2-a (0.32 ± 0.03 µM, n = 8, p < 0.001, Figure 7F). The magnitude of the change in EC50 for Ca2+-dependent channel activation is comparable to that of Ca2+-dependent interaction between CaM and CaMBDs (Figure 7D). The Hill coefficient is 3.64 ± 0.28 (n = 12) for SK2-b and 3.58 ± 0.29 (n = 8) for SK2-a. This is likely due to the fact that a functional SK channel is a tetramer, and each SK channel will have four CaM molecules attached. These results show that CaM, when in complex with CaMBD2-b, has a lower affinity for Ca2+, which is the primary reason that SK2-b becomes less sensitive to Ca2+ for its activation. Depending on which SK2 splice variant CaM binds to, CaM changes its conformation and its affinity for Ca2+, capable of responding to a much wider range of Ca2+ concentrations in Ca2+-dependent signaling.
Discussion
In this study, we show that insertion of three additional amino acid residues, ARK, in CaMBD2-b drastically alters the conformation of CaM as well as CaM's affinity for Ca2+ in CaM-CaMBD2-b compared to CaM-CaMBD2-a. Several key structural features contribute to such significant changes. (1) Both CaMBD2-a and CaMBD2-b form a 2×2 complex with CaM in solution (Figure 2). Without the constraint of the 2×2 configuration, CaM in CaM-CaMBD2-b will not adopt a conformation different from that of CaM-CaMBD2-a and nor will it change its affinity for Ca2+. (2) Insertion of ARK rotates the downstream residues of CaMBD2-b by less than one full α-helical turn (~300°). This effectively changes the relative spatial orientations of the two fragments in CaMBD2-b which interact with the CaM N- and C-lobes. (3) The conformation of the CaM N-lobe is virtually identical in both CaM-CaMBD2-a and CaM-CaMBD2-b (Figures 1, 4 & 6). Therefore, the affinity of the CaM N-lobe for Ca2+ will be similar, if not identical, between the two CaM complexes. (4) The structural flexibility allows substantial conformational changes of CaM in the hydrophobic interface at the CaM C-lobe (Figures 1, 5 & 6). One important consequence is that such conformational changes restore the ability of the CaM C-lobe to bind Ca2+ in CaM-CaMBD2-b. Thus, the CaM C-lobe can be Ca2+-bound or Ca2+-free (a very low affinity for Ca2+), depending on which SK2 variant peptide it interacts with (Figure 6).
Formation of the CaM-CaMBD2-b complex (2×2) becomes Ca2+-dependent, not only at the CaM N-lobe, but also at the CaM C-lobe (Figure 8A). In contrast, for CaM-CaMBD2-a, exposure of the hydrophobic interface at the CaM C-lobe is induced, primarily, by CaMBD2-a, independent of Ca2+ (Figure 8B). It is generally thought that the CaM C-lobe has a higher affinity for Ca2+ than the N-lobe (Andersson et al., 1983; Crouch and Klee, 1980). When CaM is associated with its target proteins, the affinity of CaM for Ca2+ becomes even higher (Peersen et al., 1997). Our results show that the opposite can happen as well. The CaM C-lobe in the CaM-CaMBD2-b complex has a lower affinity for Ca2+ than the N-lobe, effectively determining the overall reduced Ca2+ sensitivity in formation of the 2×2 CaM-CaMBD2-b complex (a rightward shift of the dose-response curve in Figure 7A).
Figure 8. A molecular model for changes in CaM’s affinity for Ca2+, induced by CaMBD2-b or CaMBD2-a.
In the presence of Ca2+, CaM forms a 2×2 complex with both CaMBD2-b and CaMBD2-a in solution. Formation of the CaM-CaMBD2-b complex in the presence of Ca2+ requires that both the CaM N- and C-lobes become Ca2+-bound before they can interact with CaMBD2-b (A). The CaM C-lobe has a lower affinity for Ca2+ than the N-lobe (K1C < K1N), effectively determining the overall reduced Ca2+ sensitivity in formation of the 2×2 CaM-CaMBD2-b complex. In contrast, formation of the CaM-CaMBD2-a complex is Ca2+-dependent at the CaM N-lobe and becomes Ca2+-independent at the CaM C-lobe. (B). The open symbols represent the Ca2+-free EF-hands, while the filled circles and squares represent Ca2+-bound EF-hands. The open triangles represent EF-hands which are no longer able to bind Ca2+.
Structural flexibility has been proposed as a mechanism through which CaM is able to interact with a variety of target proteins, even though these target proteins do not share common structural features. Globally, conformational changes of CaM include at least two major processes: (1) unwinding of the α-helix in the CaM linker region (R74 to E83), and (2) exposure of the hydrophobic interfaces at both the N- and C-lobes of when CaM becomes Ca2+-bound (Chin and Means, 2000; Drum et al., 2002; Fallon and Quiocho, 2003; Halling et al., 2005; Hoeflich and Ikura, 2002; Ikura et al., 1992; Ishida and Vogel, 2006; Meador et al., 1992; Meador et al., 1993; Mori et al., 2008; Van Petegem et al., 2005). The CaM linker region is extremely flexible. When CaM is complexed with its target proteins, the linker region often unwinds and allows CaM to wrap around the target proteins with its N- and C-lobes (Meador et al., 1992; Meador et al., 1993; Mori et al., 2008; Osawa et al., 1999; Van Petegem et al., 2005). Different from these previously reported structures, CaM in the CaM-CaMBD2-b complex adopts a rigid α-helical conformation in its linker region (Figure 1). Such an extended conformation of CaM has been reported in other CaM complexes (Larsson et al., 2001; Rodriguez-Castaneda et al., 2010).
Once Ca2+-bound, CaM undergoes significant conformational changes in the N- and C-lobes, adopting an open mode configuration which exposes the hydrophobic interfaces for interactions with the target proteins (Chin and Means, 2000; Halling et al., 2005; Hoeflich and Ikura, 2002; Ishida and Vogel, 2006; Meador et al., 1992; Meador et al., 1993; Schumacher et al., 2004). This Ca2+-dependent transition from the closed mode to the open mode is key to Ca2+-dependent interaction between CaM and CaM target proteins. The consensus is that exposure of the CaM hydrophobic interface results from binding of Ca2+ to the EF-hands at both the CaM N- and C-lobes. On the other hand, the CaM C-lobe in CaM-CaMBD2-a fails to bind Ca2+, but nevertheless is able to expose its hydrophobic interface for its interaction with CaMBD2-a. This Ca2+-independent interaction is achieved through the semi-open conformation, particularly at the CaM C-lobe (Chagot and Chazin, 2011; Swindells and Ikura, 1996; Urbauer et al., 1995). Our work shows that the hydrophobic interface at the CaM C-lobe can be made of almost entirely different sets of amino acid residues, even though the interacting amino acids on CaMBD remain the same (Figures 4 & 5), again demonstrating the structure plasticity of CaM in mediating Ca2+-dependent signaling.
CaM is dynamic Ca2+ sensor, capable of responding to a wide range of Ca2+ concentrations, e.g. 10−12 M – 10−6 M, in Ca2+-dependent cellular signaling (Chin and Means, 2000). Previous work on CaM-target protein complexes often used short CaMBD peptides. While valuable information has been obtained, one disadvantage of using short CaMBD peptides is that the interactions between CaM and CaMBD are greatly simplified, especially the lack of the effects of the target proteins on the conformation of CaM. The results of this study, together with the work on CaM-CaMBD2-a (1G4Y) and CaM-edema factor (1K90), demonstrate that target proteins have significant impact not only on the CaM’s structure, but, more importantly, on CaM’s affinity for Ca2+. For instance, CaM in the CaM-CaMBD2-a complex has lost its ability to bind Ca2+ at its C-lobe, while CaM in the CaM-CaMBD2-b has restored its ability to bind Ca2+ at its C-lobe. CaM in the CaM-edema factor complex fails to bind Ca2+ at its N-lobe (Drum et al., 2002). Thus, target proteins can drastically decrease, not just increase, the affinity of CaM for Ca2+. Thus, changes in CaM’s conformations, induced by target proteins, provide an additional mechanism that allows CaM to respond to Ca2+ signals of different strength, as observed in local vs. global Ca2+ sensing at sites of Ca2+ entry into the cytoplasm (Dick et al., 2008; Tadross et al., 2008).
Experimental Procedures
Protein Expression and Purification
Rat CaM cDNA was cloned into pET-28b (Novagen), expressed in E. coli strain Rosetta2(DE3) (Novagen) and purified using a low substitution phenyl sepharose fast flow column and an AKTA purifier (GE Healthcare). The codons of the CaMBD2-b were optimized for expression in E. coli and the synthetic gene was cloned into pET-28b. The sequence of the CaMBD2-b (including the His-tag) used for expression is: MDTQLTKRVKNAAANVLRETWLIYKNTKLVKKIDHAKVRKHQRKFLQAIHQARKLRSVKMEQRKLNDQANTLVDLAKTQLEHHHHHH. This C-terminal His-tag fusion protein fragment was expressed, solubilized with 0.2% w/v sarkosyl, and purified on a nickel column (Qiagen). Both CaM and CaMBD were subsequently purified using Sephacryl S-100 high resolution gel filtration column (GE Healthcare). The protein complex was formed by slowly adding the CaMBD2-b to CaM. The complex was then purified using the gel filtration column (GE Healthcare) pre-equilibrated in a solution with (in mM) Tris-HCl 10, NaCl 50, and CaCl2 10 (pH 7.5). Fractions were collected and concentrated to 3 mM. Protein concentrations were determined by predicted extinction coefficients.
Crystallization and Structure Determination
Please see the Supplemental Experimental Procedures for details.
Sedimentation Equilibrium
Sedimentation equilibrium (SE) experiments were performed as previously described (Schumacher et al., 2001). Briefly, for SE analysis of the complexes in the presence of Ca2+, both protein complexes were dialyzed into a solution with (in mM) NaCl 50, Tris 10, and CaCl2 10, pH 7.5. For SE analysis of the complexes in the absence of Ca2+, both protein complexes were dialyzed into a solution with (in mM) NaCl 50, Tris 10, and EGTA 5, pH 7.5. Protein complexes at 30 µM loading concentration were sedimented to equilibrium at 10000, 13000, 18000, and 25000 rpm in a Beckman XL-I Analytical Ultracentrifuge using an An 50 Ti rotor. Radial absorbance scans measured at 280 nm determined the distribution of the complexes in the centrifugal field. Data analysis was performed using SEDFIT and SEDPHAT (Schuck et al., 2002). A single component model yielded the best fit to the data.
Isothermal Titration Calorimetry (ITC)
For ITC experiments, CaM, CAMBD2-a, and CaMBD2-b were expressed and purified using affinity columns followed by gel filtration with the following buffer (in mM): HEPES 20, NaCl 100, pH 7.5. Both CaM and CaMBDs were dialyzed into the same buffer supplemented with 10 mM CaCl2. CaM (90 µM) was titrated into CaMBD2-a (10 µM) or CaMBD2-b (10 µM) at 20°C using a microcalorimeter (MicroCal/GE). The injection volume was 4 µl and the initial cell volume of CaMBDs was 1.43 ml. For data analysis, the dilution heat generated from titration of CaM into the buffer without CaMBD peptides was measured and subtracted from that with either CaMBD2-b or CaMBD2-a. There were no noticeable changes in the dilution heat. Data were fitted to standard equations in Origin 7.0 supplied by MicroCal. Identical results were obtained in three independent experiments with different batches of the purified proteins.
Fluorescence measurements with fluorophore-labeled CaM
Dansyl-labeled CaM is often used to quantify the interaction between CaM and its target proteins (e.g. Zuhlke et al., 1999). Dansyl-labeling, however, will modify the Lys residues in CaM and potentially alter the interaction between CaM and CaMBD, since K75, K77 and K115 in CaM are clearly involved in interacting with CaMBD (Figures 4 & 5). Instead, fluorophore-labeled CaM was created by introducing a cysteine mutation in CaM (T34C or T110C), as previously described (Halling et al., 2009; Spratt et al., 2007). The fluorophore changes its fluorescence intensity in response to changes in the local environment near the fluorophore. The mutant CaM was expressed and purified as describe above (plus 0.5 mM TCEP) and was labeled with 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (AEDANS, Invitrogen). All proteins were dialyzed into the solution containing (in mM) HEPES 20, NaCl 100, EGTA 10, HEDTA 10, TCEP 0.2, pH 7.0. Binding assays were performed by mixing 0.2 µM of AEDANS-labeled CaM(T34C) or CaM(T110C) with 3 µM of either CaMBD2-a or CaMBD2-b and adding Ca2+ to its final free concentrations calculated with the software by Chris Patton of Stanford University (http://www.stanford.edu/~cpatton/maxc.html). The mixture was incubated for 3 h at room temperature in dark. The fluorescence was measured using a Safire II microplate reader (Texan) with excitation at 340 nm (10 nm bandwidth) and emission between 400 and 600 nm (scanned with 20 nm bandwidth) at 28°C. The increase of fluorescence intensity, at emission of 510 nm, was plotted as a function of the free Ca2+ concentrations and fitted with a standard dose-response curve. In a separate experiment, AEDANS labeled CaM (0.2 µM) was mixed with an increasing amount of CaMBDs at Ca2+ concentrations as indicated. Changes in the fluorescence intensity were plotted as a function of the CaMBD concentrations and fitted with a standard dose-response curve.
Electrophysiology
Please see the Supplemental Experimental Procedures for details.
Calculation of relative binding affinities of the CaM N- and C-lobes for Ca2+
The CaM-CaMBD2-b crystal structure was subject to 18 ns of explicit solvent MD simulation using NAMD v2.8 (Phillips et al., 2005) and the CHARMM force field (Mackerell et al., 2004), at 298 K and 1 bar to provide an ensemble of configurations. Configurations were sampled at 4, 8, 12, and 16 ns to provide initial configurations for thermodynamic integration (TI) calculations (Frenkel and Smit, 2002). TI was performed along a discrete alchemical pathway connecting the Ca2+-bound state to a Ca2+-free state for two particular Ca2+: that bound to EF-hands in the CaM N-lobe (chain A), and that bound to EF-hands in the CaM C-lobe (chain A). The TI order parameters for van der Waals and electrostatic interactions were varied from 1.0 (Ca2+ fully bound) to 0.0 (Ca2+ fully absent) according to a schedule that turns electrostatics off first before completely turning off van der Waals (See Supplemental Methods). Numerical integration was performed using the trapezoidal rule.
Statistics
Where applicable, data are expressed as mean ± sem. Student’s-tests are used for data comparison.
Supplementary Material
Highlights.
We show structure of CaM-CaMBD2-b, a SK2 splice variant with reduced Ca2+ sensitivity
Dramatic changes in the CaM structure in CaM-CaMBD2-b compared to CaM in CaM-CaMBD2- a
Conformational changes at the CaM C-lobe result in its reduced affinity for Ca2+
Results demonstrate CaM alters its affinity for Ca2+ induced by the target proteins
Acknowledgements
We would like to thank Drs. Horn, Aldrich, Levitan, Covarrubias, Root, Scott, Milev, Horne and Armen for their encouragement and helpful discussions; Drs. Yue, Tang and Leppla for providing plasmids of CaM mutants and edema factor; Drs. Eto and Butler for their help with fluorescence measurement; Jefferson Kimmel Center structural facility for access of equipment in initial protein crystal screening, initial in-house X-ray diffraction and ITC experiments; staff at the Beamline facility (X29A) of the Brookhaven National labs for assistance with collection of X-ray diffraction data. The coordinates of the CaM-CaMBD2-b complex have been deposited in the Protein Data Bank under accession code 3SJQ. The accession number of the SK2-b gene is JN857942. The work is supported by grants from NIH to JFZ (R01MH073060 & R01NS39355).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflict of interests for the authors involved in this work.
References
- Andersson A, Forsén S, Thulin E, HJ V. Cadmium-113 nuclear magnetic resonance studies of proteolytic fragments of calmodulin: assignment of strong and weak cation binding sites. Biochem. 1983;22:2309–2313. doi: 10.1021/bi00279a001. [DOI] [PubMed] [Google Scholar]
- Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA. The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains. Structure. 2007;15:1603–1617. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CT, Maylie J, Adelman JP. SK channels in excitability, pacemaking and synaptic integration. Curr. Opin. Neurobiol. 2005;15:305–311. doi: 10.1016/j.conb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- Chagot B, Chazin WJ. Solution NMR Structure of Apo-Calmodulin in Complex with the IQ Motif of Human Cardiac Sodium Channel NaV1.5. J. Mol. Biol. 2011;406:106–119. doi: 10.1016/j.jmb.2010.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. Calcium Signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Crouch TH, Klee CB. Positive cooperative binding of calcium to bovine brain calmodulin. Biochem. 1980;19:3692–3698. doi: 10.1021/bi00557a009. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- Dick IE, Tadross MR, Liang H, Tay LH, Yang W, Yue DT. A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature. 2008;451:830–834. doi: 10.1038/nature06529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drum CL, Yan S-Z, Bard J, Shen Y-Q, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang W-J. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- Faber ESL. Functions and Modulation of Neuronal SK Channels. Cell Biochem Biophys. 2009;55:127–139. doi: 10.1007/s12013-009-9062-7. [DOI] [PubMed] [Google Scholar]
- Fallon JL, Quiocho FA. A closed compact structure of native Ca2+-calmodulin. Structure. 2003;11:1303–1307. doi: 10.1016/j.str.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Frenkel D, Smit B. Understanding Molecular Simulation. San Diego: Academic Press; 2002. [Google Scholar]
- Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- Halling DB, Aracena-Parks P, Hamilton SL. Regulation of voltage-gated Ca2+ channels by calmodulin. Sci. STKE. 2005;2005 doi: 10.1126/stke.3152005re15. re15- [DOI] [PubMed] [Google Scholar]
- Halling DB, Georgiou DK, Black DJ, Yang G, Fallon JL, Quiocho FA, Pedersen SE, Hamilton SL. Determinants in CaV1 channels that regulate the Ca2+ sensitivity of bound calmodulin. J. Biol. Chem. 2009;284:20041–20051. doi: 10.1074/jbc.M109.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Ikura M. Calmodulin in Action: Diversity in Target Recognition and Activation Mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Ishida H, Vogel HJ. Protein-peptide interaction studies demonstrate the versatility of calmodulin target protein binding. Protein Pept Lett. 2006;13:455–465. doi: 10.2174/092986606776819600. [DOI] [PubMed] [Google Scholar]
- Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- Kranz JK, Lee EK, Nairn AC, Wand AJ. A derect test of the reductionist approach to structural studies of calmodulin activity. J Biol Chem. 2002;277:16351–16354. doi: 10.1074/jbc.C200139200. [DOI] [PubMed] [Google Scholar]
- Larsson G, Schleucher J, Onions J, Hermann S, Grundström T, Wijmenga SS. A novel target recognition revealed by calmodulin in complex with the basic helix–loop–helix transcription factor SEF2-1/E2-2. Protein Sci. 2001;10:169–186. doi: 10.1110/ps.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- Mori MX, Vander Kooi CW, Leahy DJ, Yue DT. Crystal Structure of the CaV2 IQ Domain in Complex with Ca2+/Calmodulin: High-Resolution Mechanistic Implications for Channel Regulation by Ca2+ Structure. 2008;16:607–620. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. A novel target recognition revealed by calmodulin in complex with Ca2+- calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- Peersen OB, Madsen TS, Falke JJ. Intermolecular tuning of calmodulin by target peptides and proteins: Differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 1997;6:794–807. doi: 10.1002/pro.5560060406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Castaneda F, Maestre-Martinez M, Coudevylle N, Dimova K, Junge H, Lipstein N, Lee D, Becker S, Brose N, Jahn O, et al. Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity. Embo J. 2010;29:680–691. doi: 10.1038/emboj.2009.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size- Distribution Analysis of Proteins by Analytical Ultracentrifugation: Strategies and Application to Model Systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Crum M, Miller MC. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12:849–860. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a CaCa2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- Spratt DE, Taiakina V, Palmer M, Guillemette JG. Differential binding of calmodulin domains to constitutive and inducible nitric oxide synthase enzymes. Biochem. 2007;46:8288–8300. doi: 10.1021/bi062130b. [DOI] [PubMed] [Google Scholar]
- Stocker M. Ca2+-activated K+ channels: molecular determinants and function of the SK family. Nat Rev Neurosci. 2004;5:758–770. doi: 10.1038/nrn1516. [DOI] [PubMed] [Google Scholar]
- Swindells MB, Ikura M. Pre-formation of the semi-open conformation by the apo-calmodulin C-terminal domain and implications for binding IQ-motifs. Nat Struct Mol Biol. 1996;3:501–504. doi: 10.1038/nsb0696-501. [DOI] [PubMed] [Google Scholar]
- Tadross MR, Dick IE, Yue DT. Mechanism of Local and Global Ca2+ Sensing by Calmodulin in Complex with a Ca2+ Channel. Cell. 2008;133:1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Petegem F, Chatelain FC, Minor DL. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat Struct Mol Biol. 2005;12:1108–1115. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Lee Y-S, Tokumitsu H, Silva A, Soderling TR. Calmodulin-Kinases: Modulators of Neuronal Development and Plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XM, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen JE, Ishii T, Hirschberg B, Bond CT, Lutsenko S, et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature. 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- Urbauer JL, Short JH, Dow LK, Wand AJ. Structural Analysis of a Novel Interaction by Calmodulin: High-Affinity Binding of a Peptide in the Absence of Calcium. Biochem. 1995;34:8099–8109. doi: 10.1021/bi00025a016. [DOI] [PubMed] [Google Scholar]
- Zuhlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.