Abstract
A 49-year-old renal transplant recipient was admitted to our hospital due to abundant liquid diarrhea and dehydration. Parasitological investigations, including genotyping, led to the diagnosis of intestinal microsporidiosis due to a new and highly divergent internal transcribed spacer (ITS) genotype of Enterocytozoon bieneusi. The potential route of transmission through horse stools is discussed.
CASE REPORT
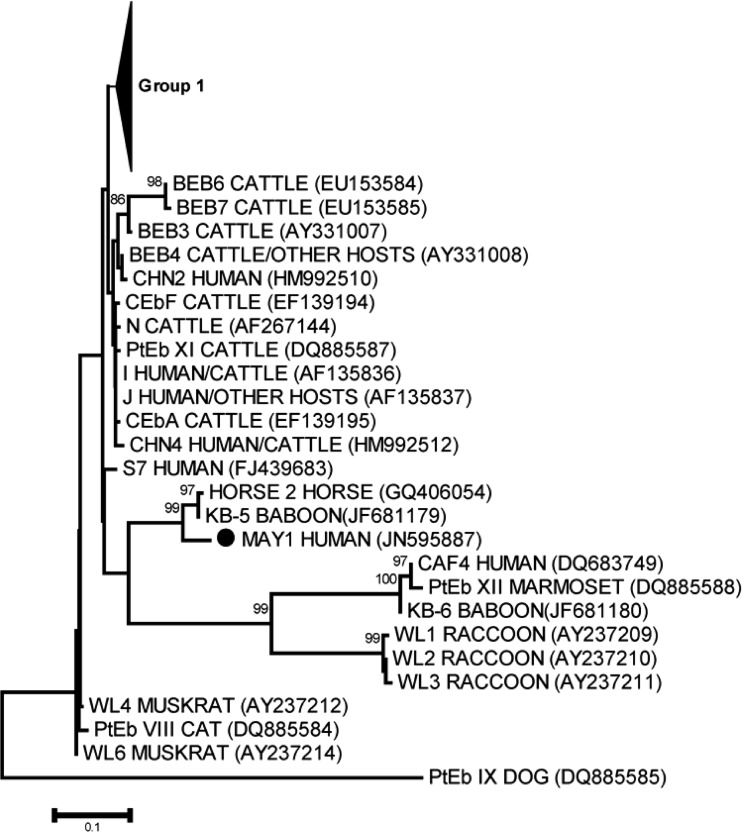
A49-year-old man who received a renal transplant in November 1998 was admitted in December 2005 to the renal transplant unit at Nice University Hospital (France) because of abundant liquid diarrhea and dehydration leading to acute renal failure. His medical history included chronic renal failure secondary to polycystic kidney disease. The follow-up after kidney transplantation revealed no underlying problem. At the time of diarrhea, he was given prednisolone (5 mg once daily). Stool samples were performed and sent to the laboratory for microbiological investigations. Bacterial and viral investigations were negative. However, several structures consistent with microsporidial spores were found after microscopic examination of both Uvitex and modified trichrome stains. The patient was successfully treated with nitazoxanide (1,000 mg twice a day for 60 consecutive days), and the clinical symptoms disappeared. A stool sample performed after nitazoxanide withdrawal was negative for microsporidia. Enterocytozoon bieneusi was identified by a species-specific real-time PCR assay as described previously (6). Briefly, DNA was extracted using the QIAamp DNA stool kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. This E. bieneusi specific real-time PCR assay targeting the small-subunit rRNA (SSU rRNA) was applied using a Rotor Gene 3000 platform (Corbett Life Science, Mortlake, Sydney, Australia). Specific detection of E. bieneusi was performed using primers Eb1 (CGACAGCCTGTGTGTGAGAATAC) and Eb5 (CAAACGAATGACTTGACCCTGTAA). Detection of the 180-bp product was ensured by use of the specific TaqMan probe EbS2 (FAM-TGCTTAATTTAACTCAACGCGGGAAAA-TAMRA [where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine]). To provide further insight into the transmission route (zoonotic or human-to-human transmission) of E. bieneusi in our patient, the internal transcribed spacer rRNA (ITS rRNA) region was amplified and sequenced with the primer set MSP3 (GGAATTCACACCGCCCGTCRYTAT) and MSP4B (CCAAGCTTATGCTTAAGTCCAGGGAG) (6). PCR products were purified, and sequencing was performed using a BigDye Terminator sequencing kit on an ABI Prism 3130 genetic analyzer (Applied Biosystems). Nucleotide sequences were analyzed using Seqscape software (Applied Biosystems). Surprisingly, comparison of the nucleotide sequences of this isolate to the GenBank database using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed only a weak similarity to previously reported E. bieneusi genotypes. The most similar genotypes (94% homology, 241 bp) were genotypes Horse 2 and KB-5 (GenBank accession numbers GQ406054 and JF681179, respectively), which have been recently described in horses and in captive baboons, respectively (9, 14). Importantly, a phylogenetic analysis of the ITS rRNA nucleotide sequences of our E. bieneusi isolate (named MAY1), together with all ITS rRNA sequences previously reported from humans or human and animal hosts, confirmed the close genetic relationship between our isolate and these highly divergent genotypes (Fig. 1). Epidemiological investigation by questioning the spouse and the patient revealed that the week before the symptoms appeared the patient worked on his farm, where he breeds horses. Indeed, he had to clean a well in which water was contaminated with horse stool. The main hypothesis is that our patient probably got infected during that episode.
Fig 1.
Phylogenetic relationships among Enterocytozoon bieneusi genotype group 1 and all other genotypes reported and available in GenBank as well as the nucleotide sequence identified in this study (MAY1), inferred by a neighbor-joining analysis of the ITS rRNA gene sequence, based on genetic distances calculated by the Kimura two-parameter model. ●, nucleotide sequence determined in this study (MAY1). Bootstrap values of less than 75% are not shown. The complete phylogenetic tree, including all E. bieneusi ITS genotypes published at the time this report was prepared, is available upon request.
Microsporidia are a diverse group of obligate intracellular parasites currently classified as fungi (10). They infect a wide range of eukaryotic cells in numerous invertebrate and vertebrate hosts, including humans and domestic and wild animals (10). Enterocytozoon bieneusi and the Encephalitozoon spp. are the major species infecting humans, with E. bieneusi being the most prevalent (10, 15). Transmission occurs mainly through fecal-oral routes, with sources of infection including other infected humans and animals, contaminated water, and, as illustrated recently, food (1, 4). Microsporidia have emerged as an important cause of opportunistic infection in patients with AIDS, being predominantly associated with wasting and diarrhea. However, recent studies have provided clear evidence that these infections are not restricted to AIDS and are also common in immunocompromised non-HIV-infected individuals, such as solid-organ transplant recipients (5, 7, 8). Moreover, Sak et al. have recently provided new insights into the understanding of microsporidiosis, highlighting a high prevalence of microsporidia in healthy subjects (11, 12). These findings suggest that the real distribution of microsporidiosis in humans is probably underestimated (1).
Enterocytozoon bieneusi is probably the species in the genus with the most extensive genetic diversity (13). This genetic diversity of E. bieneusi relies on molecular methods, genetically distinct isolates having similar morphological characteristics (13). Presently, sequencing analysis of the ITS rRNA region is still considered the gold standard for genotyping and epidemiological studies of E. bieneusi (13, 15). Until now, more than 90 E. bieneusi genotypes have been reported, with new genotypes being regularly described (9, 13). It is now clear that both host-adapted E. bieneusi genotypes with narrow host ranges and potentially zoonotic genotypes with wide host specificity have been identified (13). Analysis of nucleotide sequences of ITS rRNA provides valuable information about the transmission and pathogenic potential of E. bieneusi because it allows determination of the genotype in human and animal isolates. The most striking finding of our report is the discovery of a new and highly divergent E. bieneusi genotype in a human host. Indeed, nearly all of the genotypes that have been shown to infect humans so far belong to group 1 (3). To the best of our knowledge, human infections by E. bieneusi isolates that cluster outside group 1 are exceptional and have been described in a single recent study performed in Central Africa (3). Taken together, E. bieneusi genotype MAY1 and the one recently reported by Breton et al. in Gabon and Cameroon (genotype CAF4) are the only descriptions of a highly divergent E. bieneusi genotype infecting a human host (3). Genotype MAY1 can be grouped with genotype Horse 2, recently identified in an equid species (14). Interestingly, in our patient, symptoms began after contact with water contaminated with horse stool. Genetic similarity between our new genotype and the one described in an equid species, as well as the close contact with horse stools, suggest a zoonotic transmission from horses to our patient. Unfortunately, stools from horses were not available for analysis.
In the present case, E. bieneusi infection appeared 7 years after transplantation. This finding is in agreement with previous data from literature showing reported cases occurring from 19 days to up to 15 years after kidney transplantation (7). At the time of diagnosis, the patient was given immunosuppressive therapy that was maintained while nitazoxanide was introduced. Albendazole and fumagillin are the main drugs used to treat E. bieneusi infections (7). However, relapse is often observed after albendazole withdrawal and the efficacy of fumagillin is counterbalanced by its adverse effects, with fumagillin exhibiting bone marrow toxicity leading to thrombocytopenia and neutropenia (1, 7). Here, a complete recovery was obtained with nitazoxanide. This drug is not considered the first-line therapy but has been already used with success in an HIV-infected patient (2).
In conclusion, we report a new and highly divergent genotype of E. bieneusi that is also the first described E. bieneusi genotype with close proximity to one recently described in horses (14). Whereas the real prevalence of microsporidiosis in equid species has been poorly investigated so far, our data suggest that horses could act as potential sources of human microsporidial infections as suggested recently (14). These findings must be considered for the management of immunocompromised patients such as HIV-infected patients and solid-organ transplant recipients. In high-risk patients, giving advice such as avoiding close contact with animals and following prophylactic measures will probably reduce the burden of this neglected disease.
Nucleotide sequence accession number.
ITS rRNA nucleotide sequences of our E. bieneusi isolate have been deposited in the GenBank database under accession number JN595887.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Anane S, Attouchi H. 2010. Microsporidiosis: epidemiology, clinical data and therapy. Gastroenterol. Clin. Biol. 34:450–464 [DOI] [PubMed] [Google Scholar]
- 2. Bicart-Sée A, Massip P, Linas MD, Datry A. 2000. Successful treatment with nitazoxanide of Enterocytozoon bieneusi microsporidiosis in a patient with AIDS. Antimicrob. Agents Chemother. 44:167–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breton J, et al. 2007. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 45:2580–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Decraene V, Lebbad M, Botero-Kleiven S, Gustavsson AM, Löfdahl M. 2011. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 9:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Didier ES, Weiss LM. 2011. Microsporidiosis: not just in AIDS patients. Curr. Opin. Infect. Dis. 24:490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espern A, et al. 2007. Molecular study of microsporidiosis due to Enterocytozoon bieneusi and Encephalitozoon intestinalis among human immunodeficiency virus-infected patients from two geographical areas: Niamey, Niger, and Hanoi, Vietnam. J. Clin. Microbiol. 45:2999–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galván AL, et al. 2011. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 49:1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanternier F, et al. 2009. Microsporidiosis in solid organ transplant recipients: two Enterocytozoon bieneusi cases and review. Transpl. Infect. Dis. 11:83–88 [DOI] [PubMed] [Google Scholar]
- 9. Li W, et al. 2011. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J. Clin. Microbiol. 49:4326–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mathis A, Weber R, Deplazes P. 2005. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 18:423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sak B, Kváč M, Kučerová Z, Květoňová D, Saková K. 2011. Latent microsporidial infection in immunocompetent individuals—a longitudinal study. PLoS Negl. Trop. Dis. 5:e1162 doi:10.1371/journal.pntd.0001162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sak B, et al. 2011. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J. Clin. Microbiol. 49:1064–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santín M, Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 56:34–38 [DOI] [PubMed] [Google Scholar]
- 14. Santín M, Vecino JA, Fayer R. 2010. A zoonotic genotype of Enterocytozoon bieneusi in horses. J. Parasitol. 96:157–161 [DOI] [PubMed] [Google Scholar]
- 15. Santín M, Fayer R. 2011. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 90:363–371 [DOI] [PubMed] [Google Scholar]