Abstract
An important role in the treatment regimens for Mycoplasma pneumoniae infections is played by macrolide (ML) antibiotics. In the past few years, however, a steady increase has been detected in the worldwide prevalence of ML-resistant (MLr) M. pneumoniae strains. It is obvious that this increase necessitates a continuous monitoring of MLr and, when detected, modification of antibiotic treatment modalities. Previously, we developed a pyrosequencing-based assay system for the genetic determination of MLr as well as molecular typing of M. pneumoniae. In this study, the sensitivity of this system was improved by the inclusion of a nested-PCR protocol. The modified system was applied to 114 M. pneumoniae-positive specimens that were obtained from a collection of 4,390 samples from patients with acute respiratory tract infections. These samples were collected between 1997 and 2008 in The Netherlands. The pyrosequencing system produced reliable data in 86% of the specimens that contained >500 M. pneumoniae genome copies/ml of patient sample. Each of these samples contained DNA of the ML-sensitive genotype. While 43% of the samples were found to harbor the M. pneumoniae subtype 1 genotype, 57% contained the subtype 2 genotype. We conclude that the pyrosequencing-based assay system is a useful tool for MLr determination and molecular typing of M. pneumoniae in patient samples. MLr-associated M. pneumoniae genotypes, however, were not found in the current study population.
INTRODUCTION
Mycoplasma pneumoniae is a pathogen of the human respiratory tract and one of the most prevalent causes of community-acquired pneumonia. General treatment options for M. pneumoniae infections include macrolides (MLs), tetracyclines, and fluoroquinolones. In young children, however, tetracyclines and fluoroquinolones are contraindicated due to unfavorable effects, such as discoloration of teeth by tetracyclines and putative cartilage damage induced by fluoroquinolones. The antibiotics of choice for this patient group are therefore ML antibiotics, such as clarithromycin and azithromycin. In the past decade, however, increasing numbers of M. pneumoniae clinical isolates were found to be resistant against these antibiotics. ML resistance (MLr) was first reported in Japan in 2002 (14), and since then it has also been detected in the United States (10, 26), Europe (3, 7, 17, 18), eastern Asia (13), and the Middle East (1). The prevalence of MLr varies widely, from 1 to 2% in Denmark (18) to as high as 90% in China (11). Clearly, the emergence of MLr poses a significant threat to the use of MLs in the treatment of M. pneumoniae infections. It is therefore important that MLr among M. pneumoniae isolates is rapidly and efficiently monitored in order to allow effective antibiotic treatment.
Although the sensitivity of M. pneumoniae to antibiotics can be determined unambiguously on cultured bacterial isolates, the culturing of M. pneumoniae is time-consuming and precludes the timely identification of antibiotic-resistant isolates. The monitoring of MLr in this species is therefore most efficiently performed using molecular assays. These assays are aimed at the detection of specific point mutations in the bacterial 23S rRNA gene, which determine the MLr phenotype. Since the first report of MLr in M. pneumoniae, several molecular assays have been developed to detect the MLr-associated point mutations. In most cases, these assays rely either on time-consuming Sanger sequencing of PCR products or on real-time PCR followed by high-resolution melting analysis, which requires a high level of expertise in interpretation of the results (17, 26).
Previously, we described a pyrosequencing system to assess MLr and to determine the genotype (i.e., subtype 1 or subtype 2) of cultured M. pneumoniae strains (20). This system consists of four separate assays (two assays to assess MLr and two assays to determine genotype) and was found to be highly useful when applied to cultures of M. pneumoniae. In order to develop a culture-independent pyrosequencing system that is suitable for the determination of both MLr and the genotype of M. pneumoniae in clinical specimens, we modified the original pyrosequencing system by increasing its sensitivity. In the present study, we applied this modified system to M. pneumoniae-positive clinical specimens collected during a 12-year surveillance period in The Netherlands.
Here, we demonstrate that the pyrosequencing system is highly convenient for molecular characterization of M. pneumoniae directly on patient material. The system can be used in parallel with a PCR-based surveillance system and can be applied to the monitoring of epidemics. In addition, the pyrosequencing system can serve as a guide for clinicians in the diagnosis and treatment of acute respiratory infections (ARIs) caused by M. pneumoniae. Finally, we show that all M. pneumoniae-positive specimens from the collection contained bacterial isolates with an ML-sensitive (MLs) genotype. Thus, MLr was not detected among M. pneumoniae strains circulating in The Netherlands between 1997 and 2008.
(Part of this work was presented at the spring scientific meeting of the Dutch Society for Medical Microbiology, 19 April 2011, and at the annual meeting of the Dutch Society for Pediatrics, 5 November 2011.)
MATERIALS AND METHODS
Surveillance network, patients, and clinical specimens.
The nationwide Continuous Morbidity Registration at the Dutch Sentinel General Practice Network, coordinated by NIVEL (the Netherlands Institute for Health Service Research), continuously monitors consultations for influenza-like illness (ILI) in The Netherlands. In addition, this network aims to monitor the circulation of influenza viruses and other pathogens, including M. pneumoniae, among patients with ILI or other ARIs. ARI was defined as a respiratory infection with acute onset plus one of the following symptoms: coryza, sore throat, cough, frontal headache, retrosternal pain, or myalgia. ILI was defined as an ARI accompanied by fever (rectal temperature > 38°C).
Sixty-one general practitioners (GPs) from 45 general practices participate in this surveillance network. Together, they serve about 0.8% of the Dutch population, nationally representative by age, gender, geographic distribution, and population density (5). The GPs weekly report the number of patients consulting their practice with ILI and are asked to collect a nose swab and a throat swab on a weekly basis from two patients with ILI. If the GPs do not encounter patients with ILI, they are asked to sample two patients with another ARI (4, 22, 23). From 2001 until 2003, healthy controls were asked to participate in the surveillance as part of a case-control study on environmental risk factors for ARI (23).
In this study, we used the data and all M. pneumoniae-positive specimens collected in this surveillance network from 1997 to 2008. M. pneumoniae was monitored during each year of the surveillance, except for weeks 40 to 52 in 2003, the entire year in 2004, and weeks 1 to 36 in 2005. For each patient, a questionnaire was completed, which included age, gender, time of onset of illness, clinical symptoms, and diagnosis. The nose and the throat swab were combined and transported in 4 ml of gelatin-lactoalbumin-yeast (GLY) medium containing 0.1 mg/ml pimaricin and 0.2 mg/ml gentamicin at ambient temperature. The specimens were sent by regular mail to the Virology Department of the Laboratory for Infectious Diseases and Perinatal Screening of the National Institute for Public Health and the Environment for analysis.
M. pneumoniae strains.
All M. pneumoniae strains used in this study, including reference strains M129 (ATCC no. 29342), PI 1428 (ATCC no. 29085), FH (ATCC no. 15531), and MAC (ATCC no. 15492), as well as clinical strains R035, P05/132, M688/98, and T79 (20), were cultured in Mycoplasma medium, as described previously (12).
Detection and quantification of M. pneumoniae DNA.
Patient specimens were analyzed prospectively for the presence of M. pneumoniae DNA by nested PCR, as described by Dorigo-Zetsma et al. (6). Residual material from the original specimen was stored at −80°C for future purposes. For this study, all stored M. pneumoniae-positive specimens were retrieved. DNA was extracted from each original specimen (200 μl) using the QIAamp DNA minikit (Qiagen, The Netherlands) according to the manufacturer's recommendations. Phocine herpesvirus was added before isolation as an internal control for real-time PCR, as described before (15). The DNA was eluted in a final volume of 50 μl. A quantitative real-time (TaqMan) PCR assay was used to quantify the M. pneumoniae DNA load, as previously described (20).
MLr genotype identification and subtype determination by pyrosequencing.
To identify genotypes (point mutations) associated with either MLr or ML sensitivity (MLS), two pyrosequencing assays targeting the M. pneumoniae 23S rRNA gene were used (20). These assays can determine the presence of the MLr-associated mutations at positions 2063, 2064, and 2067 (assay 1) and at position 2617 (assay 2) of the 23S rRNA gene (20). Initially, the assays were designed to be performed on cultured bacteria. In this study, we aimed to use these assays directly on DNA isolated from clinical specimens. We therefore designed a nested-PCR protocol to improve the detection limit and specificity of the original protocol (20). PCR and sequence primers for both assays are listed in Table 1. The first (external) PCR was identical for both assays. The PCR mixtures (25 μl) contained 0.4 μM each primer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 2 mM MgSO4, 0.02 U/μl Pfu DNA polymerase (Fermentas), 1× Pfu buffer (Fermentas), and 5 μl template DNA. The following cycling conditions were used: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. The (internal) PCRs were performed as previously described (20), using 1 μl of template from the first PCR. A negative control was taken along in each PCR run. The resulting (biotinylated) PCR products from the second PCR were immobilized to streptavidin-Sepharose high-performance beads (GE Healthcare) and processed to yield high-quality, single-stranded DNA using the PyroMark vacuum prep workstation (20). The pyrosequencing reactions and sequence analyses were performed using the PyroMarkQ96MD sequencer (Qiagen) and accompanying software.
Table 1.
Oligonucleotide primers used in this study
| Assay/target(s) and primer typea | Orientation | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|
| MPN141 | |||
| External | Forward | GAA ACA CCT CCT CCA CCA AC | 151 |
| Reverse | CCA TCT AAC AGT TCA GCG AG | ||
| Internal | Forward | GAA TGA TGT GGT GGG GGT TG | 84 |
| Reverse | Biotin-GGG GTG CGT ACA ATA CCA TCA A | ||
| Sequencing | CAA CGC CGC AAA GAT | ||
| MPN528a | |||
| External | Forward | TAC GAC TAG GAG ATC ATC CAG | 232 |
| Reverse | TTA AGG TGT TTG AGT TGG TGT | ||
| Internal | Forward | ATC TAC CGA TTC AAC CAA CTG CT | 166 |
| Reverse | Biotin-GCT AAC TGC GCT AGA GCA AAA T | ||
| Sequencing | AGA AAT CGA AAA CTG ACT AT | ||
| 23S rRNA | |||
| Assay 1 | |||
| External | Forward | ACC ATC TCT TGA CTG TCT CG | 183 |
| Reverse | CAT CAA CAA GTC CTA GCG AAC | ||
| Internal | Forward | TCG GTG AAA TCC AGG TAC G | 130 |
| Reverse | Biotin-CAT CGA TTG CTC CTA CCT ATT CTC | ||
| Sequencing | AGG CGC AAC GGG ACG | ||
| Assay 2 | |||
| External | Forward | GAT TAA AGA GAT ACG TGA GTT G | 175 |
| Reverse | ATA GAC ACG TTA CTA CCC AG | ||
| Internal | Forward | Biotin-TTC AAA CCG TCG TGA GAC AG | 113 |
| Reverse | AAC TGG AGC ATA AGA GGT GTC CT | ||
| Sequencing | CTA CGG GCA CAA TAG AT |
The sequences of the internal and sequencing primers have previously been described by Spuesens et al. (20).
To discriminate between the two major M. pneumoniae subtypes (i.e., subtypes 1 and 2), we previously developed two different pyrosequencing assays (20). The first assay targets a subtype-specific single-nucleotide polymorphism (SNP) located in a conserved region near the 3′ end of the MPN141 gene (which encodes the major cytadhesin, P1, of M. pneumoniae), whereas the second targets a subtype-specific SNP in the MPN528a gene (which encodes a homologue of RecU Holliday junction resolving enzymes) (19, 20). Both assays were improved by a nested protocol in a similar fashion to that described above for the MLr and MLs assays, using similar conditions. The primers used in each assay are listed in Table 1.
Statistical analysis.
The analysis of a possible association between genomic copy load and either patient characteristics or subtype was performed using the Mann-Whitney U test. To analyze the relationship between subtype and age groups or the presence of clinical symptoms, we used the chi-square test. Significance was set at P = 0.05. All statistical analyses were performed using SPSS statistical software version 16.0.1.
RESULTS
Prevalence of M. pneumoniae in The Netherlands between 1997 and 2008 among patients with ARI.
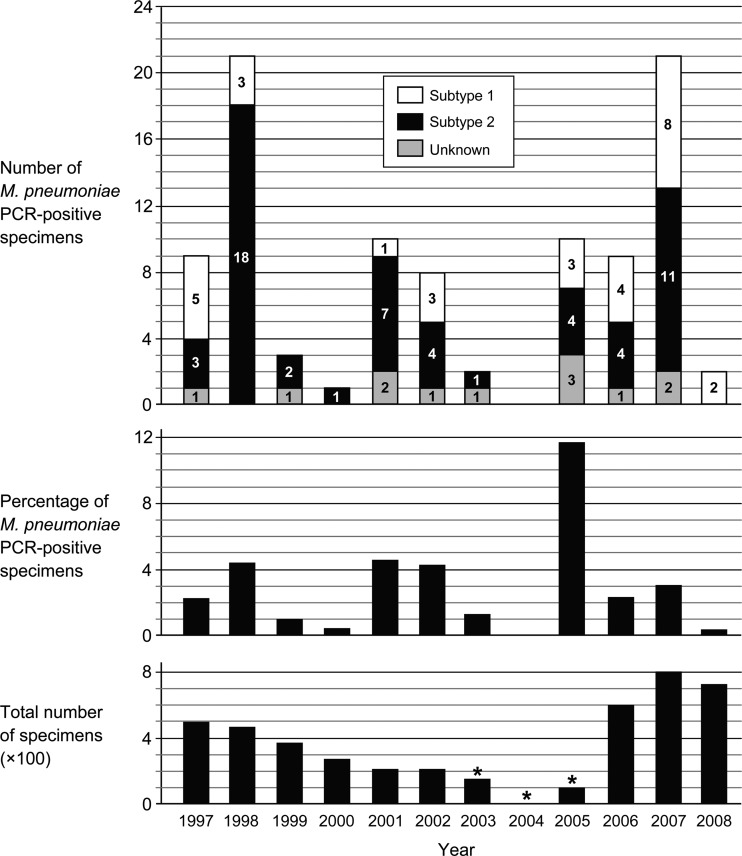
Of the 4,390 specimens that were collected between 1997 and 2008 from patients with ILI or another ARI, 114 (2.6%) tested positive for M. pneumoniae using a conventional, nested-PCR assay. Of 588 specimens that were collected from healthy individuals, 5 were positive for M. pneumoniae; these specimens were not included in this study. From 3 of the 114 M. pneumoniae-positive specimens (each originating from 1997), original material was no longer available. The M. pneumoniae DNA load in the remaining 111 M. pneumoniae-positive specimens was determined using a quantitative, real-time PCR assay (20). In 15 of these specimens, the real-time PCR assay was negative. These specimens were therefore not analyzed further. In the other 96 specimens, the M. pneumoniae genomic DNA could be quantified (Table 2). The genomic copy loads in these specimens ranged from 250 genomic copies/ml up to 1.2 × 106 copies/ml of original patient sample. As shown in Fig. 1, the prevalence of M. pneumoniae fluctuated over the years and was highest in 2005 (11.5%). Most M. pneumoniae cases were observed in two groups of patients—i.e., patients 0 to 15 years of age (32.3%) and patients 31 to 45 years of age (31.3%) (Table 2). The symptom that was reported most frequently was coughing. An association between the bacterial DNA load, patient age, and clinical symptoms was not found. There was also no correlation between the bacterial DNA load and the delay between the date of onset of illness and the moment of specimen collection.
Table 2.
Characteristics of 96 confirmed M. pneumoniae PCR-positive patients
| Variable | Total no. (%) of patients (n = 96)a | Distribution per genomic copy load (%)b |
||
|---|---|---|---|---|
| <500 (n = 12) | 500-5,000 (n = 37) | >5,000 (n = 47) | ||
| Age (yr) | ||||
| 0–15 | 31 (32.3) | 7 (58.3) | 13 (35.1) | 11 (23.4) |
| 16–30 | 16 (16.7) | 2 (16.6) | 5 (13.5) | 9 (19.1) |
| 31–45 | 30 (31.3) | 1 (8.3) | 12 (32.4) | 17 (36.2) |
| 46–60 | 15 (15.6) | 1 (8.3) | 4 (10.8) | 10 (21.3) |
| >60 | 4 (4.2) | 1 (8.3) | 3 (8.1) | 0 (0.0) |
| Gender | ||||
| Female | 55 (57.3) | 7 (58.3) | 20 (54.1) | 28 (59.6) |
| Male | 41 (42.7) | 5 (41.7) | 17 (45.9) | 19 (40.4) |
| Duration of Illness at time of sampling | ||||
| ≤7 days | 83 (88.3) | 9 (75.0) | 31 (86.1) | 43 (93.5) |
| >7 days | 11 (11.7) | 3 (25.0) | 5 (13.9) | 3 (6.5) |
| Symptomsc | ||||
| Fever | 60 (69.8) | 8 (72.7) | 27 (77.1) | 25 (62.5) |
| Malaise | 50 (58.1) | 6 (54.5) | 22 (62.9) | 22 (55.0) |
| Headache | 22 (33.8) | 0 (0.0) | 10 (38.5) | 12 (38.7) |
| Muscle pains | 31 (36.0) | 2 (18.2) | 13 (37.1) | 16 (40.0) |
| Sore throat | 31 (36.0) | 6 (54.5) | 16 (45.7) | 9 (22.5) |
| Cough | 76 (88.4) | 9 (81.1) | 32 (91.4) | 35 (87.5) |
| Shortness of breath | 2 (8.7) | 0 (0.0) | 2 (25.0) | 0 (0.0) |
| Rhinorrhea | 17 (27.0) | 2 (22.2) | 12 (44.4) | 3 (11.1) |
| Diagnosis | ||||
| ARI | 43 (44.8) | 8 (66.7) | 16 (43.2) | 19 (40.4) |
| ILI | 53 (55.2) | 4 (33.3) | 21 (56.8) | 28 (59.6) |
Questionnaires were not complete for all patients.
Bacterial genomic copy loads per ml of the original patient specimen, as determined by real-time PCR. There was no association between the load of M. pneumoniae DNA and specific patient characteristics.
The symptom shortness of breath was only recorded from 1997 through 1999. The symptom frontal headache was not recorded from 2000 through 2003. The symptom rhinorrhea was recorded from 2000 onwards.
Fig 1.
M. pneumoniae-positive specimens (n = 96) and M. pneumoniae subtype detected per year. (Top panel) Absolute numbers of M. pneumoniae PCR-positive specimens. The numbers of subtype 1 and subtype 2 genotypes are represented in each bar by the white and black areas, respectively; the numbers are also indicated within these areas. The gray areas depict the number of specimens in which a subtype could not be determined (“Unknown”). In these cases, insufficient amounts of PCR product were produced, which precluded the generation of pyrosequencing results. (Middle panel) Prevalence (%) of M. pneumoniae PCR-positive specimens per year in the sentinel surveillance network. (Bottom panel) The total number of collected specimens each year. No specimens were tested for M. pneumoniae in weeks 40 to 52 of 2003, the entire year 2004, and in weeks 1 to 36 of 2005 (as indicated by the asterisks).
Generation and evaluation of a highly sensitive pyrosequencing system.
Previously, we described the design and use of four pyrosequencing assays for determination of both MLr and genotype of M. pneumoniae strains (20). A major drawback of these assays, however, is that they can only be applied to cultured bacterial isolates, as their lower detection limit is relatively high (approximately 5,000 copies per ml of the original sample). We therefore set out to improve the detection limit of the four assays, which will be referred to here as the “pyrosequencing system,” such that they can be performed directly on clinical specimens from M. pneumoniae-positive patients. For this purpose, the initial PCRs of the pyrosequencing system were converted into nested reactions (as described in Materials and Methods).
To determine the lower detection limit of the system on patient specimens, we performed experiments in which throat swabs from healthy persons were spiked with known quantities of M. pneumoniae DNA. The swabs were rinsed in 1 ml phosphate-buffered saline, and after DNA extraction, the M. pneumoniae DNA load was measured by real-time PCR. The detection limit for each of the four assays from the pyrosequencing system was found to be ∼5 × 102 genomic copies per ml of sample (which is ∼10-fold more sensitive than the original protocol). At copy numbers below 5 × 102 genomic copies per ml, the assays failed to provide reliable results. In these cases, the amount of generated PCR product was too low to allow the generation of high-quality, reproducible pyrosequencing data.
To test whether the modified pyrosequencing system is able to detect the genotypes associated with either ML resistance (MLr) or ML sensitivity (MLs) and to discriminate between the two different subtypes of M. pneumoniae, we applied the system to different isolates from our M. pneumoniae strain collection. These isolates included four strains with an MLr genotype (the subtype 1 strains R035 and P05/132 and subtype 2 strains M688/98 and T79) and four strains with an MLs genotype (subtype 1 strains M129 and PI1428 and subtype 2 strains MAC and FH). Similar to the original pyrosequencing system, the modified system correctly determined the subtypes as well as the MLr/MLs genotype of each of the tested strains (data not shown).
MLr/MLs and M. pneumoniae subtype determination in clinical specimens.
The modified pyrosequencing system was applied directly to DNA isolated from the 96 M. pneumoniae real-time PCR-positive patient samples. Seventy-seven of these specimens (80.2%) yielded a reliable result in the MLr and MLs assays from the pyrosequencing system. As expected from the spiking experiments, the performance of the tests was highly dependent on the genomic load in the original sample. In specimens with a load above 5 × 102 genomic copies/ml, the MLr and MLs assays generated reliable data in 86% of cases. At genomic copy numbers lower than 5 × 102 genomic copies/ml, however, results were obtained in only 41% of the cases. All specimens that produced reliable data in the MLr and MLs assays were found to carry the MLs genotype. Thus, the MLr genotype was not detected in the current sample collection.
The two M. pneumoniae subtyping assays from the pyrosequencing system were slightly more sensitive than the MLr and MLs assays: in 85 of the 96 M. pneumoniae DNA-positive specimens (88.5%), the subtype could be determined. Similar to what was reported for the MLr and MLs assays, the success of the subtyping assays was highly dependent on the M. pneumoniae genomic load in the original specimens. In specimens with a load above 5 × 102 per ml, the genotype could be reliably determined in 92% of the cases. In specimens with loads of 5 × 102 genomic copies/ml or lower, the subtype was determined in 67% of the cases. The results from the two subtyping assays corresponded for all specimens. The majority of the specimens obtained during the entire sample period contained M. pneumoniae subtype 2 sequences (57%). This percentage varied over the years and was highest in 1998 and 2001 (Fig. 1). We did not find an association between bacterial subtype, patient age, and clinical symptoms or between subtype and bacterial DNA load.
DISCUSSION
In this study, a modified pyrosequencing system was used for MLr determination and molecular typing of M. pneumoniae directly in clinical specimens. We found this modified system to generate reliable results in 86% of the samples that carried >500 M. pneumoniae genome copies/ml. This lower detection limit was determined by spiking patient samples with known quantities of purified M. pneumoniae DNA. The main modification made to the original pyrosequencing protocol was the inclusion of a nested PCR instead of a single PCR (20). Clearly, the use of a nested-PCR protocol may also have drawbacks. First, a higher PCR sensitivity may lead to increased problems related to contamination. However, strict guidelines were followed in the handling and containment of PCR products: i.e., each step of the pyrosequencing system was performed in a separate room. As a consequence, we have not experienced problems with DNA contamination during the course of this study. Second, a nested-PCR step may increase the risk of generating PCR errors, which may lead to incorrect sequencing results and incorrect assignment of either subtype or MLr/MLs genotype. To avoid this problem, a heat-stable DNA polymerase with proofreading activity was used throughout this study, and we have not experienced any (PCR-induced) sequence variation in any of the analyzed amplicons.
Although ML-resistant M. pneumoniae isolates have recently been detected in many European countries (such as Germany, Denmark, France, and Italy), we did not detect any genotypes associated with MLr in the Dutch patient cohort that we studied. This apparent inconsistency may be explained by the nature of our study population, which consists of ambulant patients. It might be that the prevalence of MLr is higher among (hospitalized) patients that are subjected to antibiotic treatment. It is also possible that MLr among M. pneumoniae strains in The Netherlands emerges more slowly than in other countries due to the restricted use of antibiotics in The Netherlands (2). Thus, the selection for strains with an MLr genotype may not be as strong in this country as in neighboring countries, in which antibiotics are administered with a higher frequency. However, the easy administration and the favorable pharmacokinetic profile of newer MLs, like clarithromycin and azithromycin, will likely stimulate their prescription. It should therefore be considered that MLr will also emerge in The Netherlands in the near future.
In our study population, a remarkable fluctuation in the prevalence of M. pneumoniae was observed over time. Similar fluctuations have been reported in epidemiological studies from other countries (8, 9, 18). The highest percentage of M. pneumoniae-positive specimens was detected in the year 2005 (Fig. 1). Interestingly, during the same period (i.e., the years 2004 and 2005), an epidemic of M. pneumoniae infections occurred in Denmark (18). A possible explanation for the periodic fluctuations in the incidence of M. pneumoniae infections is antigenic variation of bacterial surface-exposed proteins, such as the P1 protein (21). This antigenic variation may result in a temporary reduction of the immunity against M. pneumoniae in the population (24). It is also possible that the specific immunological memory against M. pneumoniae wanes rapidly, allowing the bacteria to reinfect individuals within 1 to 2 years after the previous infection (9).
Another notable epidemiological observation in this study was the relatively low prevalence of M. pneumoniae in our patient population. This may, at least in part, be explained by the age distribution of the population. More specifically, the burden of M. pneumoniae infections lies primarily in childhood (25), whereas the specimens in the surveillance network were collected regardless of age. In addition, the primary aim of the surveillance network is to monitor influenza virus infections, and those patients were selected that were suspected of an influenza virus infection.
As a consequence of the relatively low prevalence of M. pneumoniae in the group of patients that was studied, it was difficult to analyze a putative correlation between the presence of either subtype 1 or subtype 2 M. pneumoniae isolates in the Dutch population. Shifts in the prevalence of these subtypes have been reported to occur, such that the subtype that predominates in the human population is replaced by the other subtype every 8 to 10 years (9). In this study, however, such shifts were not observed, which may, at least in part, be due to the length of the sampling period being too short. Regardless, the clinical implications of the detection of either subtype 1 or subtype 2 strains are still unclear. In this study, we did not find any association between the subtype of the infecting M. pneumoniae strain, the pathogenesis of infection, and bacterial DNA load. A similar conclusion was drawn by Nilsson and coworkers in a Swedish study, in which 45 M. pneumoniae PCR-positive patients were included (16). As a consequence, molecular (sub)typing of M. pneumoniae isolates is currently only useful for epidemiological purposes.
In conclusion, pyrosequencing is a valuable tool for the molecular characterization of M. pneumoniae. Using this system, we did not detect any MLr-associated M. pneumoniae genotypes in patient specimens collected between 1997 and 2008 in The Netherlands. However, as MLr is increasing in neighboring countries, either due to local antibiotic pressure or due to actual spread, it is important that MLr be monitored, not only in countries with a high prevalence of MLr, but also in countries with a low prevalence of antibiotic resistance, such as The Netherlands.
ACKNOWLEDGMENTS
We are thankful to Y. Hu for retrieving all specimens used in this study and to S. Jenny and the many other technicians involved in the diagnostics of the surveillance specimens.
A.M.C.V.R. is supported by grants from Erasmus MC, the European Society for Pediatric Infectious Diseases, and the Netherlands Organization for Health Research and Development.
Footnotes
Published ahead of print 11 April 2012
REFERENCES
- 1. Averbuch D, Hidalgo-Grass C, Moses AE, Engelhard D, Nir-Paz R. 2011. Macrolide resistance in Mycoplasma pneumoniae, Israel, 2010. Emerg. Infect. Dis. 17:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Butler CC, et al. 2009. Variation in antibiotic prescribing and its impact on recovery in patients with acute cough in primary care: prospective study in 13 countries. BMJ 338:b2242 doi:10.1136/bmj.b2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chironna M, et al. 2011. Emergence of macrolide-resistant strains during an outbreak of Mycoplasma pneumoniae infections in children. J. Antimicrob. Chemother. 66:734–737 [DOI] [PubMed] [Google Scholar]
- 4. Dijkstra F, Donker GA, Wilbrink B, Van Gageldonk-Lafeber AB, Van Der Sande MA. 2009. Long time trends in influenza-like illness and associated determinants in The Netherlands. Epidemiol. Infect. 137:473–479 [DOI] [PubMed] [Google Scholar]
- 5. Donker G. 2009. Continuous Morbidity Registration Dutch Sentinel General Practice Network. Annual report. http//:www.nivel.nl/peilstations
- 6. Dorigo-Zetsma JW, Zaat SA, Vriesema AJ, Dankert J. 1999. Demonstration by a nested PCR for Mycoplasma pneumoniae that M. pneumoniae load in the throat is higher in patients hospitalised for M. pneumoniae infection than in non-hospitalised subjects. J. Med. Microbiol. 48:1115–1122 [DOI] [PubMed] [Google Scholar]
- 7. Dumke R, von Baum H, Luck PC, Jacobs E. 2010. Occurrence of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Clin. Microbiol. Infect. 16:613–616 [DOI] [PubMed] [Google Scholar]
- 8. Dumke R, Von Baum H, Luck PC, Jacobs E. 2010. Subtypes and variants of Mycoplasma pneumoniae: local and temporal changes in Germany 2003–2006 and absence of a correlation between the genotype in the respiratory tract and the occurrence of genotype-specific antibodies in the sera of infected patients. Epidemiol. Infect. 138:1829–1837 [DOI] [PubMed] [Google Scholar]
- 9. Kenri T, et al. 2008. Genotyping analysis of Mycoplasma pneumoniae clinical strains in Japan between 1995 and 2005: type shift phenomenon of M. pneumoniae clinical strains. J. Med. Microbiol. 57:469–475 [DOI] [PubMed] [Google Scholar]
- 10. Li X, et al. 2009. Emerging macrolide resistance in Mycoplasma pneumoniae in children: detection and characterization of resistant isolates. Pediatr. Infect. Dis. J. 28:693–696 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, et al. 2010. Characterization of macrolide resistance in Mycoplasma pneumoniae isolated from children in Shanghai, China. Diagn. Microbiol. Infect. Dis. 67:355–358 [DOI] [PubMed] [Google Scholar]
- 12. Maquelin K, et al. 2009. Raman spectroscopic typing reveals the presence of carotenoids in Mycoplasma pneumoniae. Microbiology 155:2068–2077 [DOI] [PubMed] [Google Scholar]
- 13. Matsubara K, et al. 2009. A comparative clinical study of macrolide-sensitive and macrolide-resistant Mycoplasma pneumoniae infections in pediatric patients. J. Infect. Chemother. 15:380–383 [DOI] [PubMed] [Google Scholar]
- 14. Matsuoka M, et al. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob. Agents Chemother. 48:4624–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niesters HG. 2001. Quantitation of viral load using real-time amplification techniques. Methods 25:419–429 [DOI] [PubMed] [Google Scholar]
- 16. Nilsson AC, Bjorkman P, Welinder-Olsson C, Widell A, Persson K. 2010. Clinical severity of Mycoplasma pneumoniae (MP) infection is associated with bacterial load in oropharyngeal secretions but not with MP genotype. BMC Infect. Dis. 10:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peuchant O, et al. 2009. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J. Antimicrob. Chemother. 64:52–58 [DOI] [PubMed] [Google Scholar]
- 18. Rasmussen JN, et al. 2010. Increased incidence of Mycoplasma pneumoniae infections detected by laboratory-based surveillance in Denmark in 2010. Euro Surveill. 15:19708. [PubMed] [Google Scholar]
- 19. Sluijter M, et al. 2010. The Mycoplasma genitalium MG352-encoded protein is a Holliday junction resolvase that has a non-functional orthologue in Mycoplasma pneumoniae. Mol. Microbiol. 77:1261–1277 [DOI] [PubMed] [Google Scholar]
- 20. Spuesens EB, et al. 2010. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae by pyrosequencing. J. Microbiol. Methods 82:214–222 [DOI] [PubMed] [Google Scholar]
- 21. Spuesens EB, et al. 2009. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155:2182–2196 [DOI] [PubMed] [Google Scholar]
- 22. van Gageldonk-Lafeber AB, et al. 2005. A case-control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin. Infect. Dis. 41:490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Gageldonk-Lafeber AB, et al. 2007. Risk factors for acute respiratory tract infections in general practitioner patients in The Netherlands: a case-control study. BMC Infect. Dis. 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vink C, Rudenko G, Seifert HS. 25 December 2011. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol. Rev. [Epub ahead of print.] doi:10.1111/j.1574–6976.2011.00321.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolff BJ, Thacker WL, Schwartz SB, Winchell JM. 2008. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob. Agents Chemother. 52:3542–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]