Abstract
This article describes PCR fingerprinting using the universal primer T3B to distinguish among species of the Sporothrix complex, S. brasiliensis, S. globosa, S. mexicana, and S. schenckii. This methodology generated distinct banding patterns, allowing the correct identification of all 35 clinical isolates at the species level, confirmed by partial calmodulin (CAL) gene sequence analyses. This methodology is simple, reliable, rapid, and cheap, making it an ideal routine identification system for clinical mycology laboratories.
TEXT
Sporotrichosis is a globally distributed subcutaneous mycosis with areas of high endemicity (11, 21) that is caused by the dimorphic fungus Sporothrix schenckii (22). Sporotrichosis has been regarded as a job-related disease occurring as isolated cases or small outbreaks affecting people exposed to plants or soil (1, 6, 7). Rio de Janeiro State, Brazil, is a region of sporotrichosis hyperendemicity where several human and animal cases have been described since 1998 (5, 23).
The diagnosis of sporotrichosis is attained by clinical, epidemiological, and laboratorial data, including culture and analysis of phenotypic characteristics. The first description of PCR for sporotrichosis' diagnosis was reported in 2001 (9). A diagnostic nested PCR assay targeting the S. schenckii 18S rRNA gene was further evaluated and showed high sensitivity and specificity, indicating that PCR may be clinically useful for diagnosis (8).
S. schenckii was long considered a single taxon, although great genetic variation within this species has been described (10). By associating phenotypic and genotypic features, Marimon et al. (13) recognized three new species, Sporothrix brasiliensis, Sporothrix globosa, and Sporothrix mexicana, and proposed an identification key for these Sporothrix species. S. globosa has worldwide distribution (12, 18), whereas S. brasiliensis is apparently restricted to Brazil (13) and S. mexicana to Mexican environmental samples (13), although the latter was recently identified in Portugal (3). Additionally, these authors have proposed the promotion of S. schenckii var. luriei to the status of a new species, Sporothrix luriei (14). However, identification based only on phenotypic characteristics is often inconclusive due to phenotypic variability within these species. Therefore, new rapid and reliable identification strategies are necessary (19).
Analysis of tRNA intergenic spacers was first used to distinguish Streptococcus species (15). It has also been applied successfully for Candida identification (2, 16, 24). Here, we evaluate T3B PCR fingerprinting to differentiate clinical Sporothrix strains at the species level in comparison to analysis of partial calmodulin (CAL) gene sequences (19).
Thirty-five Sporothrix spp. isolates from the Fungal Culture Collection of IPEC/Fiocruz were included in this study approved by the Ethics Commission of the same institution. Among them, S. brasiliensis type strain CBS 120339 (IPEC16490), S. globosa IPEC27135 (18), S. schenckii IPEC29334 (IOC1226) (19), and S. mexicana (MUM11.02) (3) were used as controls. All 35 strains were previously phenotypically characterized (3, 19), and 15 isolates could not be identified (Table 1).
Table 1.
Comparison of tools applied in the characterization of strains of the Sporothrix complex
| Strain | Phenotypic identificationa | Genotypic characterization |
||
|---|---|---|---|---|
| Final identificationb | GenBank accession no.c | Referenced | ||
| IPEC16490 | S. brasiliensis | S. brasiliensis | AM116899 | 19 |
| IPEC27445-3 | S. brasiliensis | S. brasiliensis | HQ426950 | 19 |
| IPEC27052 | Sporothrix sp.* | S. brasiliensis | HQ426941 | 19 |
| IPEC27135 | Sporothrix sp.* | S. globosa | GU456632 | 3 |
| IPEC27387 | Sporothrix sp.* | S. brasiliensis | HQ426948 | 19 |
| IPEC34067 | Sporothrix sp.* | S. brasiliensis | HQ426952 | 19 |
| IPEC27372 | Sporothrix sp.* | S. brasiliensis | HQ426947 | 19 |
| IPEC25011 | S. brasiliensis | S. brasiliensis | HQ426935 | 19 |
| IPEC33605 | Sporothrix sp.* | S. brasiliensis | HQ426957 | 19 |
| IPEC27930 | Sporothrix sp.* | S. brasiliensis | HQ426951 | 19 |
| IPEC28772 | Sporothrix sp.* | S. brasiliensis | HQ426955 | 19 |
| IPEC28457 | S. brasiliensis | S. brasiliensis | JN995607 | This study |
| IPEC34007 | Sporothrix sp.* | S. brasiliensis | HQ426959 | This study |
| IPEC27177-2 | Sporothrix sp.* | S. brasiliensis | HQ426944 | 19 |
| IPEC27087 | S. brasiliensis | S. brasiliensis | HQ426942 | 19 |
| IPEC27288 | Sporothrix sp.* | S. brasiliensis | HQ426945 | 19 |
| IPEC27209 | Sporothrix sp.* | S. brasiliensis | HQ426946 | 19 |
| IPEC28604 | S. brasiliensis | S. brasiliensis | HQ426953 | 19 |
| IPEC26945 | Sporothrix sp.* | S. brasiliensis | HQ426939 | 19 |
| IPEC27130 | Sporothrix sp.* | S. brasiliensis | HQ426943 | 19 |
| IPEC25521 | Sporothrix sp.* | S. brasiliensis | HQ426936 | 19 |
| IPEC16919 | S. brasiliensis | S. brasiliensis | HQ426930 | 19 |
| IPEC18782A | S. brasiliensis | S. brasiliensis | HQ426933 | This study |
| IPEC28329 | S. schenckii* | S. brasiliensis | JN995610 | This study |
| IPEC27022 | S. brasiliensis | S. brasiliensis | HQ426940 | 19 |
| IPEC28487 | S. brasiliensis | S. brasiliensis | HQ426928 | 19 |
| IPEC28665 | S. brasiliensis | S. brasiliensis | JN995606 | This study |
| IPEC28790 | S. brasiliensis | S. brasiliensis | HQ426956 | 19 |
| IPEC29334 | S. schenckii | S. schenckii | HQ426962 | 19 |
| IPEC26961 | S. schenckii | S. schenckii | JN995605 | This study |
| IPEC27157-1 | S. schenckii | S. schenckii | JN995604 | This study |
| IPEC27100 | S. schenckii* | S. brasiliensis | JN995609 | This study |
| IPEC27133 | S. schenckii* | S. brasiliensis | JN995608 | This study |
| MUM 11.02 | S. mexicana or S. schenckii* | S. mexicana | JF970258 | 4 |
| IPEC27722 | S. mexicana* | S. schenckii | HQ426961 | 19 |
Performed according to reference 19. Asterisks indicate incorrect phenotypic identifications.
Calmodulin sequencing and T3B concordant identification.
Of strain used for characterization.
Reference from which the partial gene calmodulin sequencing result was obtained.
Genomic DNA was extracted from the mycelial phase (25), and PCR was performed with the primer T3B (5′-AGGTCGCGGGTTCGAATCC-3′) (24). T3B PCR fingerprinting reproducibility was confirmed by repeating the assays at least 3 times under the same conditions at two different laboratories. The T3B profiles were analyzed using the software Bionumerics 5.1 (Applied Maths BVBA, Saint-Martens-Latem, Belgium). Similarity coefficients were calculated using the Dice algorithm, and cluster analysis was performed by means of the unweighted-pair group method using average linkages (UPGMA). Sequencing of the CAL gene was performed as previously described (17) using the sequencing platform at Fiocruz, Brazil (20). Sequences were edited with Sequencer 4.6 (Genes Codes Corporation), aligned with MEGA 4.0.2 software, and compared with sequences available from NCBI GenBank by BLAST. Phylogenetic analyses were performed using MEGA with 1,000 bootstrap replicates (4) (http://www.megasoftware.net/).
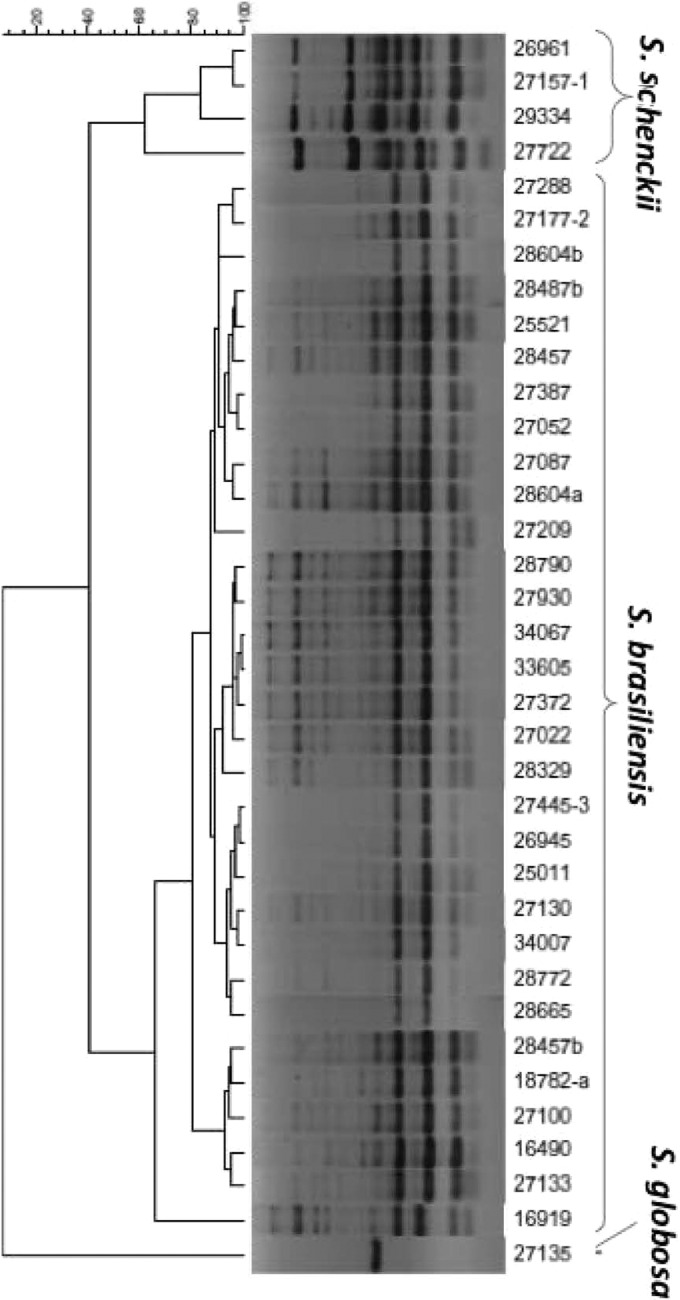
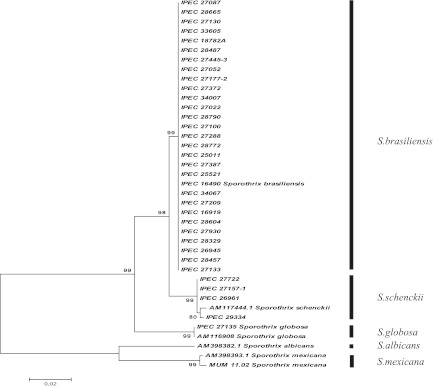
T3B PCR fingerprinting of control strains showed profiles with DNA fragments ranging from 300 to 1,500 bp that allowed a clear distinction of S. brasiliensis, S. globosa, S. mexicana, and S. schenckii (Fig. 1). A dendrogram derived from analysis of the T3B profiles of all isolates splits Sporothrix strains into three groups, not the expected four, with IPEC27722, the only strain phenotypically characterized as S. mexicana, grouping into the S. schenckii cluster. Regardless of this discrepancy, the T3B profiling showed a high correspondence between clusters and Sporothrix species, with all isolates clustering with their respective control strain (Fig. 2). The T3B fingerprinting identification was confirmed by comparison with the CAL gene partial sequences obtained along with sequences from the NCBI database, AM398393.1 (S. mexicana), AM398382.1 (Sporothrix albicans), AM116908 (S. globosa), and AM117444.1 (S. schenckii). The phylogenetic tree of the CAL locus analyzed by neighbor joining revealed five distinct clades represented by the five species (Fig. 3). Overall, there was 100% agreement between T3B PCR fingerprinting and CAL locus sequencing on species identification. Comparing these results with phenotypic analysis, 14 of the 15 isolates with inconclusive phenotypes grouped within the S. brasiliensis cluster and IPEC27135 grouped within the S. globosa cluster. The genotypic analyses also allowed the correction of phenotypic misidentifications, such as for the isolate IPEC27722, which was phenotypically characterized as S. mexicana but grouped within the S. schenckii cluster, as well as for the isolates IPEC27133, IPEC27100, and IPEC28329, which identified as S. schenckii but clustered with S. brasiliensis.
Fig 1.
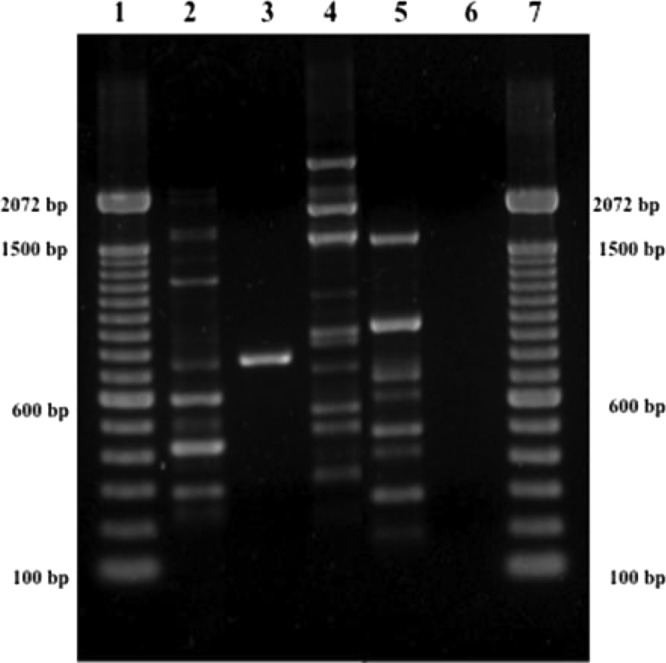
Representative T3B PCR fingerprinting profiles of the Sporothrix complex. (1 and 7) Molecular marker DNA ladder, 100 bp (Invitrogen). (2) S. brasiliensis (IPEC 16490). (3) S. globosa (IPEC 27135). (4) S. mexicana (MUM 11.02). (5) S. schenckii (IPEC27722). (6) Negative control.
Fig 2.
Dendrogram showing the degrees of similarity of T3B fingerprinting profiles among the Sporothrix isolates by using the Dice coefficient and the UPGMA cluster method. The cophenetic correlation coefficient (0.97) indicates a very good fit for this analysis.
Fig 3.
Consensus tree of Sporothrix based on partial calmodulin (CAL) gene sequences of 35 strains and the NCBI public GenBank sequences AM398393.1 (S. mexicana), AM398382.1 (S. albicans), and AM117444.1 (S. schenckii) that was constructed with MEGA version 4.0.2 and 1,000 bootstrap replicates.
Identification of the Sporothrix species complex has been based on a polyphasic approach using a combination of phenotypic methodologies and sequencing (13, 14, 18, 19). The proposed identification key based on phenotypic tests (13) was reported as easy and reliable for species differentiation without the need of molecular techniques. However, the results are often inconclusive or ambiguous, and some species are too closely related to show phenotypic differences. This work describes the assessment of DNA polymorphism within the Sporothrix complex by genomic DNA amplification with the single nonspecific primer T3B. The T3B PCR profiles were highly informative and generated clearly distinct banding patterns for each species, allowing their differentiation. S. brasiliensis strains showed bands sharing similarity higher than 80%, but intraspecies variation was observed. This variation was previously demonstrated for Candida (2), and band-sharing values observed are within the variation range (70 to 85%) expected for strains within the same species (16).
Comparison of the inconclusive phenotypic identification of isolates with the results obtained by T3B PCR fingerprinting demonstrated the sensitivity of the latter, since species of those strains could all be determined. This proposed identification technique is simple, reliable, rapid, and cheaper and requires less technical expertise than sequencing. The generated, computer-scanned PCR profiles can form the basis of a database which can be used for future identifications of atypical or unidentifiable Sporothrix isolates. Furthermore, these advantages are especially valuable in a laboratory with limited facilities, making it an ideal identification methodology for clinical mycology laboratories.
Nucleotide sequence accession numbers.
All sequences were deposited in the GenBank database under accession numbers GU456632, HQ426928 to HQ426962, and JN995604 to JN995610.
ACKNOWLEDGMENTS
Financial support was provided by FAPERJ (grant proc. E-26/111.619/2008). R.M.Z.-O. is supported in part by CNPq 350338/2000-0. M.M.E.D.O. was supported in part by a Cyted Grant (RED MICOMOL_CYTED CIFG8231770) for his work at CBMA, Universidade do Minho, Braga, Portugal.
We thank M. M. Muniz and R. Franco-Duarte for their assistance and N. Lima for kindly supplying the S. mexicana isolate.
Automated sequencing was done using the genomic platform/DNA sequencing platform at Fundação Oswaldo Cruz—PDTIS/FIOCRUZ (RPT01A), Brazil.
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. Cooper CR, Dixon DM, Salkin IF. 1992. Laboratory-acquired sporotrichosis. J. Med. Vet. Mycol. 30:169–171 [DOI] [PubMed] [Google Scholar]
- 2. Correia A, Sampaio P, Almeida J, Pais C. 2004. Study of molecular epidemiology of candidiasis in Portugal by PCR fingerprinting of Candida clinical isolates. J. Clin. Microbiol. 42:5899–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dias NM, Oliveira MME, Santos C, Zancope-Oliveira RM, Lima N. 2011. Sporotrichosis caused by Sporothrix mexicana, Portugal. Emerg. Infect. Dis. 17:1975–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 5. Freitas DF, Valle ACF, Almeida-Paes R, Bastos FI, Gutierrez-Galhardo MC. 2010. Zoonotic sporotrichosis in Rio de Janeiro, Brazil: a protracted epidemic yet to be curbed. Clin. Infect. Dis. 50:453. [DOI] [PubMed] [Google Scholar]
- 6. Hajjeh R, et al. 1997. Outbreak of sporotrichosis among tree nursery workers. J. Infect. Dis. 176:499–504 [DOI] [PubMed] [Google Scholar]
- 7. Hay RJ, Morris-Jones R. 2008. Outbreaks of sporotrichosis. Curr. Opin. Infect. Dis. 21:119–121 [DOI] [PubMed] [Google Scholar]
- 8. Hu S, et al. 2003. Detection of Sporothrix schenckii in clinical samples by a nested PCR assay. J. Clin. Microbiol. 41:1414–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kano R, Nakamura Y, Watanabe S, Tsujimoto H, Hasegawa A. 2001. Identification of Sporothrix schenckii based on sequences of the chitin synthase 1 gene. Mycoses 44:261–265 [PubMed] [Google Scholar]
- 10. Liu X, et al. 2003. Characterization of Sporothrix schenckii by random amplification of polymorphic DNA assay. Chin. Med. J. 116:239–242 [PubMed] [Google Scholar]
- 11. Lopez-Romero E, et al. 2011. Sporothrix schenckii complex and sporotrichosis, an emerging health problem. Future Microbiol. 6:85–102 [DOI] [PubMed] [Google Scholar]
- 12. Madrid H, et al. 2009. Sporothrix globosa, a pathogenic fungus with widespread geographical distribution. Rev. Iberoam. Micol. 26:218–222 [DOI] [PubMed] [Google Scholar]
- 13. Marimon R, et al. 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J. Clin. Microbiol. 45:3198–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marimon R, Gene J, Cano J, Guarro J. 2008. Sporothrix luriei: a rare fungus from clinical origin. Med. Mycol. 46:621–625 [DOI] [PubMed] [Google Scholar]
- 15. McClelland M, Petersen C, Welsh J. 1992. Length polymorphisms in tRNA intergenic spacers detected by using the polymerase chain reaction can distinguish streptococcal strains and species. J. Clin. Microbiol. 30:1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyer W, Maszewska K, Sorrell TC. 2001. PCR fingerprinting: a convenient molecular tool to distinguish between Candida dubliniensis and Candida albicans. Med. Mycol. 39:185–193 [DOI] [PubMed] [Google Scholar]
- 17. O'Donnell K, Aoki NHT, Cigelnik E. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41:61–78 [Google Scholar]
- 18. Oliveira MME, et al. 2010. Sporotrichosis caused by Sporothrix globosa in Rio de Janeiro, Brazil: case report. Mycopathologia 169:359–363 [DOI] [PubMed] [Google Scholar]
- 19. Oliveira MME, Almeida-Paes R, Muniz MM, Gutierrez-Galhardo MC, Zancope-Oliveira RM. 2011. Phenotypic and molecular identification of Sporothrix isolates from an epidemic area of sporotrichosis in Brazil. Mycopathologia 172:257–267 [DOI] [PubMed] [Google Scholar]
- 20. Otto TD, et al. 2008. ChromaPipe: a pipeline for analysis, quality control and management for a DNA sequencing facility. Genet. Mol. Res. 7:861–871 [DOI] [PubMed] [Google Scholar]
- 21. Queiroz-Telles F, Nucci M, Colombo AL, Tobon A, Restrepo A. 2011. Mycoses of implantation in Latin America: an overview of epidemiology, clinical manifestations, diagnosis and treatment. Med. Mycol. 49:225–236 [DOI] [PubMed] [Google Scholar]
- 22. Ramos-e-Silva M, Vasconcelos C, Carneiro S, Cestari T. 2007. Sporotrichosis. Clin. Dermatol. 25:181–187 [DOI] [PubMed] [Google Scholar]
- 23. Schubach A, Barros MB, Wanke B. 2008. Epidemic sporotrichosis. Curr. Opin. Infect. Dis. 21:129–133 [DOI] [PubMed] [Google Scholar]
- 24. Thanos M, et al. 1996. Rapid identification of Candida species by DNA fingerprinting with PCR. J. Clin. Microbiol. 34:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woods JP, Kersulyte D, Goldman WE, Berg DE. 1993. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J. Clin. Microbiol. 31:463–464 [DOI] [PMC free article] [PubMed] [Google Scholar]