Abstract
We analyzed the cycle threshold (CT) of PCR surveillance MRSA swabs obtained from veterans. Lower CT on admission was associated with a positive culture from nasal swabs at discharge. Compared to PCR, direct plating of nasal swabs performed poorly, especially for patients with an elevated CT. The CT is strongly correlated with quantitative nasal cultures. Clinical and infection control applications of the CT have yet to be defined and warrant further evaluation.
TEXT
Screening for methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization is done in an attempt to control the spread of MRSA within health care settings and prevent subsequent MRSA infection. The Veterans Health Administration (VHA) was an early adopter of active universal nasal MRSA colonization surveillance for all patients admitted to acute-care facilities, with many hospitals and states subsequently following this course (8, 13).
Most active surveillance programs rely on rapid detection methods using direct chromogenic agar cultures and/or real-time PCR to minimize delays in the identification and isolation of carriers in an attempt to prevent nosocomial transmission of MRSA. Although they are less costly, direct chromogenic agar cultures are insensitive compared to PCR (15, 22). Broth enrichment in tryptic soy broth (TSB) prior to plating has been shown to increase the sensitivity of chromogenic techniques but also increases the workload and the time to result, making this method problematic for routine use in the clinical microbiology laboratory (11). Therefore, PCR has become the gold standard in determining colonization status.
Few studies have evaluated the impact of nasal MRSA colonization burden and results of agar based screening tests. In 2009, Wolk et al. demonstrated that a lower MRSA colonization burden is associated with discordant screening tests (positive PCR test and negative agar test), and the MRSA cycle threshold (CT) values for the concordant samples in the Xpert MRSA assay (Cepheid, Sunnyvale, CA) were statistically lower than those for discordant samples (21). In our study, we examined the variation between PCR and direct culture screening methods for nasal MRSA colonization among hospitalized veterans, compared the CT values for patients with discordant screening results at admission and discharge, and evaluated the quantitative abilities of the Xpert MRSA PCR assay.
In October of 2007, the VHA and the Centers for Disease Control and Prevention developed a MRSA bundle to be instituted in all United States VHA medical centers. The MRSA bundle utilized, in addition to other measures, active surveillance of nasal MRSA colonization for all patients admitted to the hospital, transferred between units, and discharged from the hospital. The Atlanta VA Medical center (AVAMC) is a large tertiary-care medical center serving over 80,000 veterans. The patients admitted to the AVAMC are primarily male (∼95%) and either Caucasian (∼54%) or African American (∼44%) with significant medical and surgical comorbidities. Like many VHA medical centers, admission and transfer screening is done with a PCR-based test, while discharge screening is done by directly plating nasal swabs onto chromogenic agar. Extranasal sites are not routinely screened for MRSA, and MRSA decolonization is rarely implemented.
MRSA screening at the AVAMC is performed using a double swab, one swab with two swab heads. (Copan swabs with liquid Stuart medium; Cepheid). Both swab heads are inserted (together) 1 cm into each nasal vestibule and rotated 4 revolutions while maintaining even contact with the nasal mucosa. Swabs are sent directly to the microbiology laboratory for immediate testing, with one swab being used for PCR or culture and the partner swab being saved as a backup. Admission and transfer screening is performed by the Xpert MRSA PCR assay according to the manufacturer's instructions (which includes initial vortexing of the swab in the PCR cassette), and a CT from 15 to 36 is considered positive. Discharge screening cultures are performed by direct inoculation (without initial vortexing) onto Spectra MRSA chromogenic agar (Remel, Lenexa, KS) (16), as rapid detection is unnecessary upon discharge and culture is significantly less costly.
From October 2007 through January 2008, 2,237 admission or transfer MRSA nasal screens were performed at the AVAMC, and 369 (16.5%) were positive, corresponding to 272 patients with a positive admission and/or transfer screen (many admissions had multiple positive nasal screens). The CT value for all positive MRSA admission or transfer screens was recorded. A corresponding discharge nasal surveillance culture was performed for 181 of the 272 patients (the remaining 91 patients did not have a discharge nasal swab performed due to poor compliance with obtaining discharge swabs early in the implementation of the VHA MRSA Directive). Of the 181 patients with a positive admission or transfer screen, 62 (34.3%) had a negative discharge culture for MRSA. For these 62 MRSA-discordant patients (positive PCR test and negative agar test), 39 partner discharge swabs were available for additional PCR testing by the Xpert MRSA assay. Of the 39 partner nasal discharge swabs, 16 (41.0%) were PCR positive and 23 (59.0%) were PCR negative. The 23 unavailable partner swabs either had been used by the clinical lab for retesting or had been inadvertently discarded prior to retesting. These results demonstrate both the poor testing characteristics of direct plating without prior broth enrichment or vortexing (41.0% false-negative rate among discordant samples) and the intermittent nature of MRSA nasal colonization (59% negative on repeat PCR testing) in a subgroup of patients.
The mean admission CT of the 62 patients with PCR-positive admission swabs and negative discharge cultures was compared to the mean admission CT of the 119 patients with PCR-positive admission swabs and discharge cultures with a two-sample t test (SAS version 9.2; SAS, Cary, NC). The mean CT was significantly lower in those that remained positive at discharge than in those that were negative at discharge (26.1 versus 30.1; P < 0.0001). This is consistent with previous studies demonstrating higher nasal bacterial burden in patients persistently colonized with S. aureus (14).
Five hospital-acquired MRSA infections (HAI) occurred during hospitalization of the 272 patient (1.84% of admissions). The admission CT of patients that developed a HAI was not different from that of patients that did not develop a HAI (25.5 versus 27.2; P = 0.4674). The very few HAI in this cohort limits our power to detect a significant difference between the two groups and warrants further evaluation with a dedicated case-control study.
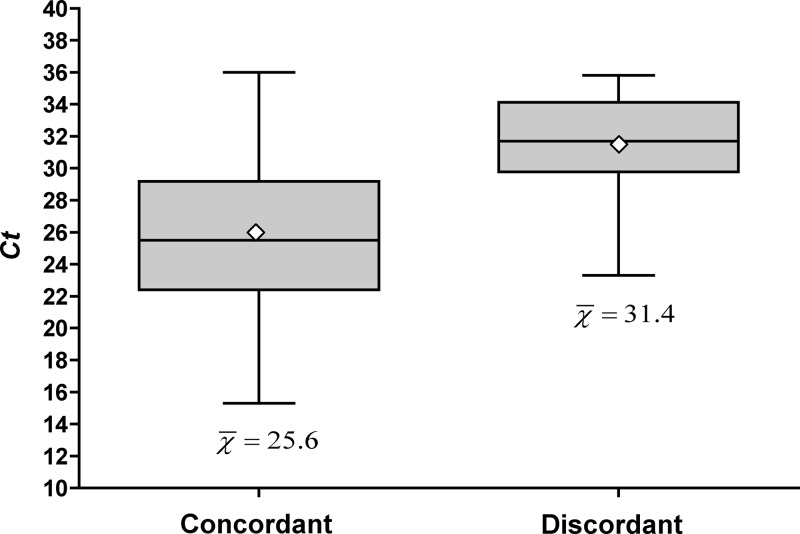
To ensure that clinical factors (i.e., antibiotics) did not contribute to the discrepancies seen between admission/transfer PCR and discharge culture results, we analyzed partner nasal swabs by PCR and culture simultaneously. Partner swabs for 204 consecutive positive admission or transfer PCR screens were collected from January to May 2011 and directly inoculated onto Spectra MRSA agar by cross-streaking in four quadrants to yield semiquantitative culture results (1+ to 4+), identical to the method used for discharge nasal screens (23). All swabs were inoculated onto Spectra agar within 48 h of a positive Xpert assay. The mean CT was 27.3 for all positive swabs, with a standard deviation of 5.0. Only 146/204 (71.6%) were positive by direct inoculation to Spectra MRSA agar. The mean CT of swabs that were positive by both PCR and direct agar culture was 25.6, compared to a CT of 31.4 for those that were positive by PCR and negative on direct inoculation (P < 0.005) (Fig. 1).
Fig 1.
Box plot of cycle thresholds (Xpert MRSA assay) among patients with a positive Xpert nasal screen and simultaneous positive direct agar screen (concordant) and those with a positive Xpert nasal screen and a simultaneous negative direct agar screen (discordant). The shaded boxes represent the 25th percentile to the 75th percentile. The central horizontal line represents the 50th percentile, and vertical lines represent the maximum and minimum values. The open diamond reflects the mean. t test for comparing two means, P = <0.005.
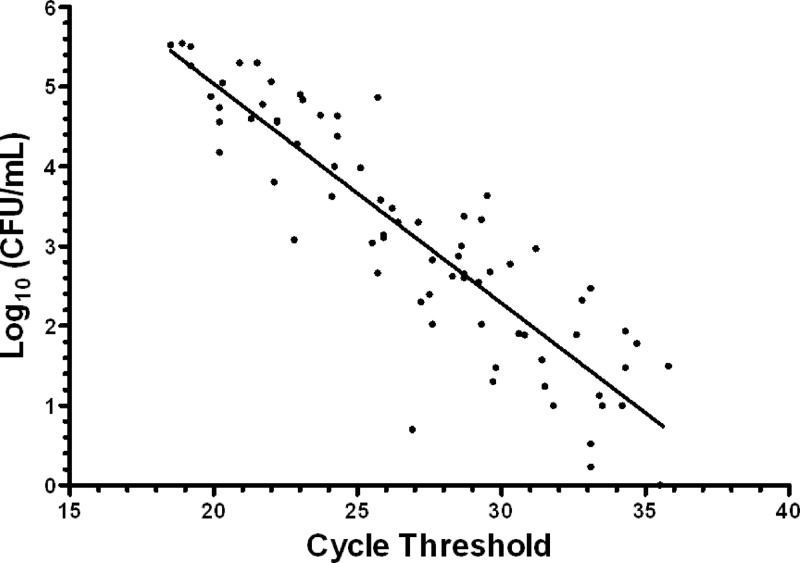
In an attempt to assess the quantitative capabilities of the Xpert MRSA assay, 76 PCR-positive admission partner swabs were placed into 500 μl of tryptic soy broth (Becton Dickinson Co., Sparks, MD) and vortexed for 10 s to simulate the initial step in the Xpert assay. Serial dilutions of the broth were spiral plated onto Spectra MRSA agar; colonies were counted at 24 h and converted to log10 CFU/ml (12). Using this method, 73/76 PCR-positive partner swabs grew MRSA. A strong correlation between CT and log10 CFU/ml was observed, with a Pearson's correlation coefficient of −0.89 (P < 0.0001) (Fig. 2). The following linear regression line was fit to the data: log10 (CFU/ml) = 10.54 − 0.26CT.
Fig 2.
Relationship of cycle threshold (Xpert MRSA assay) and quantitative cultures with the corresponding fitted line (Pearson's correlation coefficient, r = −0.89, P < 0.0001). The regression equation is as follows: log10(CFU/ml) = 10.54 − 0.26CT.
Using a preexisting MRSA surveillance system, we have demonstrated that the MRSA CT on the Xpert MRSA assay is a reliable marker of nasal MRSA colonization burden, that direct plating of nasal swabs is a less reliable test for patients with a low MRSA nasal burden, and that a higher MRSA nasal burden is associated with subsequent positive nasal screens.
Nasal S. aureus colonization clearly increases the risk of developing subsequent S. aureus infections (3, 4, 6, 7, 18). Nasal colonization is now commonly being determined by PCR-based testing (1, 8, 9) due to its ease of implementation, reliability, and rapid results. An additional advantage of PCR-based tests is an easily obtained measure of quantification in the CT. Quantification of nasal S. aureus colonization was described over 50 years ago and has been linked to the persistently colonized state (14, 20), an increased risk of contamination of the environment (5, 17), an increased risk of infection (10, 19), and a higher likelihood that other body sites (besides the nares) are colonized (12). In those studies, S. aureus carriage was quantified by labor- and time-intensive techniques not easily implemented in a clinical practice. Now, with an easily obtained and reliable measure of nasal quantification, the clinical and infection control implications of nasal S. aureus burden can be further evaluated.
The Xpert MRSA assay targets the staphylococcal cassette chromosome mec (SCCmec)-orfX junction and does not specifically target the mecA gene. Because of this, methicillin-sensitive S. aureus (MSSA) isolates with empty cassettes may test positive with the Xpert MRSA assay. A recent report evaluated this phenomenon and reported a false-positive rate of 7.7% in positive nasal MRSA screens (2). We did not attempt to isolate MSSA from swabs that were PCR positive and culture negative, and this is a limitation of our study. However, the large discrepancy seen between PCR testing modalities and direct plating is not likely all due to this phenomenon. As our data demonstrate, negative results for direct culturing are most likely due to the inability of MRSA to transfer from swab to agar and not a limitation of the agar itself. This failure of transmission likely causes the majority of discordant results. Vortexing nasal swabs prior to plating greatly enhanced culture results.
Direct plating, used by the AVAMC for all discharge cultures in an attempt to measure MRSA acquisition in the hospital, likely misses a significant proportion of patients with low-level MRSA nasal colonization, which calls into question the reliability of this protocol and its cost savings. The potential role of nasal MRSA quantification in infection control and clinical care is unknown and deserves further study.
ACKNOWLEDGMENTS
We thank Priscilla Maldonado-Estrada, Henry K. Lowery, and the Atlanta VAMC clinical microbiology laboratory for their assistance in specimen collection and handling.
This study was supported by CDC Interagency Agreement 07FED706507 and PHS grant UL RR025008 from the Clinical and Translational Science Award program, National Institutes of Health, National Center for Research Resources.
We have no potential conflicts of interest to declare.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Aldeyab MA, et al. 2009. Can. the use of a rapid polymerase chain screening method decrease the incidence of nosocomial meticillin-resistant Staphylococcus aureus? J. Hosp. Infect. 71:22–28 [DOI] [PubMed] [Google Scholar]
- 2. Arbefeville SS, et al. 2011. Prevalence and genetic relatedness of methicillin-susceptible Staphylococcus aureus isolates detected by the Xpert MRSA nasal assay. J. Clin. Microbiol. 49:2996–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Datta R, Huang SS. 2008. Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin. Infect. Dis. 47:176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. 2004. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin. Infect. Dis. 39:776–782 [DOI] [PubMed] [Google Scholar]
- 5. Ehrenkranz NJ. 1964. Person-to-person transmission of Staphylococcus aureus. Quantitative characterization of nasal carriers spreading infection. N. Engl. J. Med. 271:225–230 [DOI] [PubMed] [Google Scholar]
- 6. Honda H, et al. 2010. Staphylococcus aureus nasal colonization and subsequent infection in intensive care unit patients: does methicillin resistance matter? Infect. Control Hosp. Epidemiol. 31:584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang SS, Platt R. 2003. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin. Infect. Dis. 36:281–285 [DOI] [PubMed] [Google Scholar]
- 8. Jain R, et al. 2011. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 364:1419–1430 [DOI] [PubMed] [Google Scholar]
- 9. Jeyaratnam D, et al. 2008. Impact of rapid screening tests on acquisition of meticillin resistant Staphylococcus aureus: cluster randomised crossover trial. BMJ 336:927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalmeijer MD, van Nieuwland-Bollen E, Bogaers-Hofman D, de Baere GA. 2000. Nasal carriage of Staphylococcus aureus is a major risk factor for surgical-site infections in orthopedic surgery. Infect. Control Hosp. Epidemiol. 21:319–323 [DOI] [PubMed] [Google Scholar]
- 11. Lee S, et al. 2008. Comparison of culture screening protocols for methicillin-resistant Staphylococcus aureus (MRSA) using a chromogenic agar (MRSA-Select). Ann. Clin. Lab Sci. 38:254–257 [PubMed] [Google Scholar]
- 12. Mermel LA, Cartony JM, Covington P, Maxey G, Morse D. 2011. Methicillin-resistant Staphylococcus aureus colonization at different body sites: a prospective, quantitative analysis. J. Clin. Microbiol. 49:1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muder RR, et al. 2008. Implementation of an industrial systems-engineering approach to reduce the incidence of methicillin-resistant Staphylococcus aureus infection. Infect. Control Hosp. Epidemiol. 29:702–708 [DOI] [PubMed] [Google Scholar]
- 14. Nouwen JL, et al. 2004. Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a “culture rule.” Clin. Infect. Dis. 39:806–811 [DOI] [PubMed] [Google Scholar]
- 15. Paule SM, et al. 2009. Chromogenic media vs real-time PCR for nasal surveillance of methicillin-resistant Staphylococcus aureus: impact on detection of MRSA-positive persons. Am. J. Clin. Pathol. 131:532–539 [DOI] [PubMed] [Google Scholar]
- 16. Peterson JF, et al. 2010. Spectra MRSA, a new chromogenic agar medium to screen for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 48:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Solberg CO. 1965. A study of carriers of Staphylococcus aureus with special regard to quantitative bacterial estimations. Acta Med. Scand. Suppl. 436:1–96 [PubMed] [Google Scholar]
- 18. Wertheim HF, et al. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705 [DOI] [PubMed] [Google Scholar]
- 19. White A. 1963. Increased infection rates in heavy nasal carriers of coagulase-positive staphylococci. Antimicrob. Agents Chemother. 161:667–670 [PubMed] [Google Scholar]
- 20. White A. 1961. Quantitative studies of nasal carriers of staphylococci among hospitalized patients. J. Clin. Invest. 40:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolk DM, Marx JL, Dominguez L, Driscoll D, Schifman RB. 2009. Comparison of MRSASelect agar, CHROMagar methicillin-resistant Staphylococcus aureus (MRSA) medium, and Xpert MRSA PCR for detection of MRSA in nares: diagnostic accuracy for surveillance samples with various bacterial densities. J. Clin. Microbiol. 47:3933–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolk DM, et al. 2009. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J. Clin. Microbiol. 47:758–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. York MK. 2004. Paratechnical processing of specimens for aerobic bacteriology, p 3.3.1.1–3.3.1.9 In Isenberg HD. (ed), Clinical microbiology procedures handbook, 2nd ed, vol 1 ASM Press, Washington, DC [Google Scholar]