Abstract
A new hepatitis C virus (HCV) core antigen (HCV Ag) assay was thought to have a good correlation with HCV RNA. The aim was to elucidate the usefulness of this HCV Ag assay in community screening. In a township where HCV is endemic, 405 residents aged 58 years or older responded to a follow-up community screening. All subjects were tested for anti-HCV (AxSYM, version 3.0; Abbott Diagnostics) and HCV Ag (Architect HCV Ag test; Abbott Diagnostics). For subjects with anti-HCV signal-to-cutoff ratios (S/CO) > 10 and/or HCV Ag > 3 fmol/liter, HCV RNA data (Taqman HCV RNA; Roche Diagnostics) were further checked. A total of 115 (28.4%) subjects had their serum HCV RNA levels measured, and 93 were HCV RNA positive. The other 290 subjects were supposed to be HCV RNA negative. HCV Ag was significantly correlated with HCV RNA according to the following equation: (log HCV RNA) = 2.08 + 1.03 (log HCV Ag) (R2 = 0.94; P < 0.001). As determined using a combination of the values for anti-HCV (S/CO > 40) and HCV Ag (>3 fmol/liter) as a cutoff to predict viremia, the sensitivity, specificity, accuracy, positive predictive value, and negative predictive value were 96.8%, 100%, 99.3%, 100%, and 99%, respectively. In conclusion, for a community study, HCV Ag showed good correlation with HCV RNA. In addition, anti-HCV or HCV Ag can predict HCV viremia well, while a combination of anti-HCV (>40 S/CO) and HCV Ag (>3 fmol/liter) can provide the best result validity.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is a common etiology of liver cirrhosis and hepatocellular carcinoma, with an estimated 170 million chronic carriers worldwide (9, 22). Successful eradication of HCV has been shown to improve the prognosis of HCV-induced liver disease and reduce the associated mortality (23, 27). Hence, to adequately screen individuals with an active infection is a crucial issue in areas where HCV is endemic (24).
In clinical practice, diagnosis of HCV infection in hospitals is usually based on the detection of anti-HCV antibodies in the serum. Several anti-HCV assays have been used as the common serological marker for HCV infection for more than 20 years. However, most assays cannot distinguish infected individuals with an ongoing active infection from those who have recovered from acute infection. Unlike anti-HCV antibodies, serum HCV RNA is a reliable marker for the diagnosis of an ongoing HCV infection and is usually used for monitoring anti-HCV treatment. But its high cost, and the requirement for considerable technical skill and related equipment, limit its routine use in community screening (16).
The HCV core antigen (HCV Ag) possibly exists in both complete HCV virions and RNA-free core protein structures and has been detected in the serum of infected individuals (8, 13, 20). Several HCV Ag assays developed in the last decade have been shown to have good correlation with HCV RNA assays (3, 21, 25, 28). Hence, these assays were used as an alternative to HCV RNA for the diagnosis of active HCV infection as well as for the monitoring of the response to antiviral therapy (2, 4). A sensitive quantitative immunoassay (Architect HCV Ag test; Abbott Diagnostics) was launched recently (15, 17) and was also reported to have excellent correlation with HCV screening for viremia in special groups such as hemodialysis patients (14).
In community screening, although anti-HCV assays have been used as a first-line screening test for decades, individuals with an ongoing active HCV infection were not identified unless by checking their serum HCV RNA levels. Since HCV Ag assays showed good correlation with HCV RNA and might be used as an economical substitute for HCV RNA testing in the hospital (2, 4), it is interesting to survey the role of HCV Ag for HCV screening in the community.
The aims of the community study conducted in an area where HCV is endemic were to elucidate the utility of the new HCV Ag assay for HCV screening compared with that of the anti-HCV assay and the correlation with HCV viremia.
MATERIALS AND METHODS
Tzukuan township is located in southern Taiwan and has a total population of about 40,000 residents. It has been reported to be an area where HCV is endemic (11), with an estimated anti-HCV prevalence of 41.6%. In 1997, all 9,632 residents in this township who were aged 45 or older were invited for HCV screening by telephone and mail. Of the residents who responded to the invitations, a total of 2,909 (30.6% of the age group) were screened for anti-HCV with blood tests and ultrasound examination (10). A follow-up study was conducted in 2005, and 1,002 participants responded (6, 26). In 2010, we conducted follow-up community screening with this cohort, and 405 of the 1,002 residents responded.
All participating subjects were tested for anti-HCV and HCV Ag. Since the lower limit of positive-detection levels for HCV reactions in the HCV Ag kit was reported to be 3 fmol/liter, for participants with anti-HCV titers detected at a signal-to-cutoff ratio (S/CO) > 10 and/or HCV Ag > 3 fmol/liter, HCV RNA was further checked. The others were considered to be HCV RNA negative.
Anti-HCV enzyme immunoassay.
A third-generation anti-HCV enzyme immunoassay (EIA) (AxSYM, version 3.0; Abbott, Chicago, IL) was used for anti-HCV detection. The presence of anti-HCV can be expressed as a signal-to-cutoff ratio (S/CO). An S/CO of >1.0 was interpreted as reactive. Our previous study (5) showed that viremia was present for 95% (114/120) of results at S/CO ratios above 40, 51% (26/51) at S/CO ratios between 10 and 40, and none (0/21) at S/CO ratios between 1 and 10. The U.S. Centers for Disease Control and Prevention (CDC) also suggests that the S/CO ratio of this kit with a true predictive rate > 95% is above 10 (1).
Architect HCV Ag assay.
The HCV Ag assay is a two-step immunoassay, using chemiluminescent microparticle immunoassay technology for quantitative measurement of the HCV core Ag. Specimens with concentration values < 3.00 fmol/liter are considered nonreactive for HCV Ag, whereas values ≥ 3.00 fmol/liter are considered to represent reactivity. The upper limit of linearity is 20,000 fmol/liter.
HCV RNA assay.
The HCV RNA concentrations were detected using a Cobas AmpliPrep/Cobas TaqMan HCV kit (Amplicor, Roche Diagnostics, Branchburg, NJ). The lower limit of detection of this assay was 15 IU/ml, with a linear range of between 43 × 106 and 69 × 106 IU/ml.
Statistical analysis.
Lineal regression analysis was used to assess the linear association between the HCV Ag and HCV RNA concentrations as well as anti-HCV and HCV RNA concentrations in log scales. Sensitivity, specificity, positive predictive values (PPV) and negative predictive values (NPV), and accuracy of prediction of HCV viremia for different cutoff values of anti-HCV and HCV Ag tests were computed. All data were analyzed by using SPSS software (SPSS; version 15.0). Statistical significance was set at P < 0.05.
RESULTS
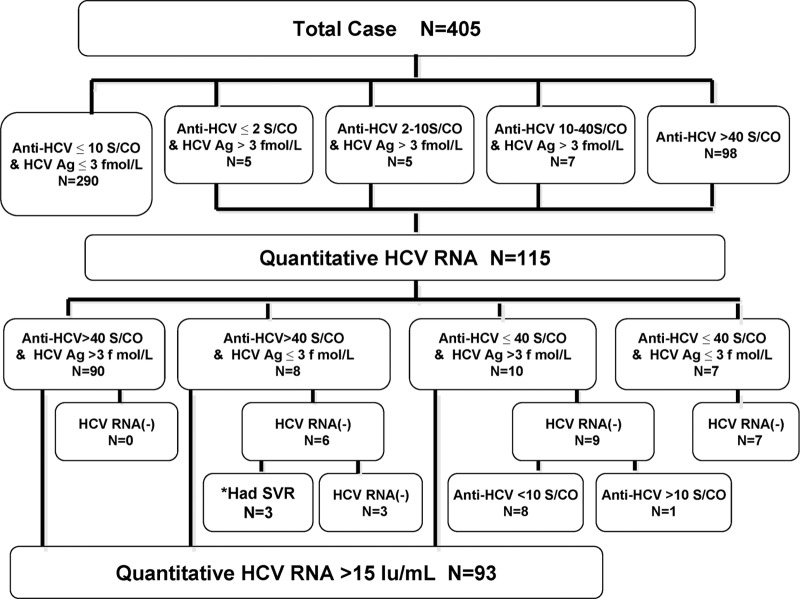
Among 405 respondents in this study, there were 192 (47.4%) men and 213 (52.6%) women, with a mean age of 69.6 ± 7.7 years. Figure 1 shows the flow chart of the variations among 405 subjects according to Anti-HCV, HCV Ag, and HCV RNA data. A total of 115 (28.4%) subjects with anti-HCV > 10 S/CO and/or HCV Ag > 3 fmol/liter had their serum HCV RNA levels measured, and 93 of them were positive for HCV RNA. We further combined anti-HCV = 40 S/CO and HCV Ag = 3 fmol/liter as two cutoff points; all 90 subjects with anti-HCV > 40 S/CO and HCV Ag > 3 fmol/liter were HCV RNA positive. In contrast, all 7 subjects with anti-HCV ≤ 40 S/CO and HCV Ag ≤ 3 fmol/liter were HCV RNA negative. Of the 8 subjects with anti-HCV > 40 S/CO and HCV Ag ≤ 3 fmol/liter, 6 were HCV RNA negative and 3 of them had received anti-HCV treatment since the last screening, with sustained virological response (SVR) achievement. Of the other 10 subjects with anti-HCV ≤ 40 S/CO and HCV Ag > 3 fmol/liter, 9 were HCV RNA negative and 8 of those had anti-HCV < 10 S/CO.
Fig 1.
Flow chart of the variations among 405 participants according to anti-HCV, HCV Ag, and HCV RNA. *Had SVR, 3 subjects received anti-HCV treatment between the two screenings and achieved sustained virological response.
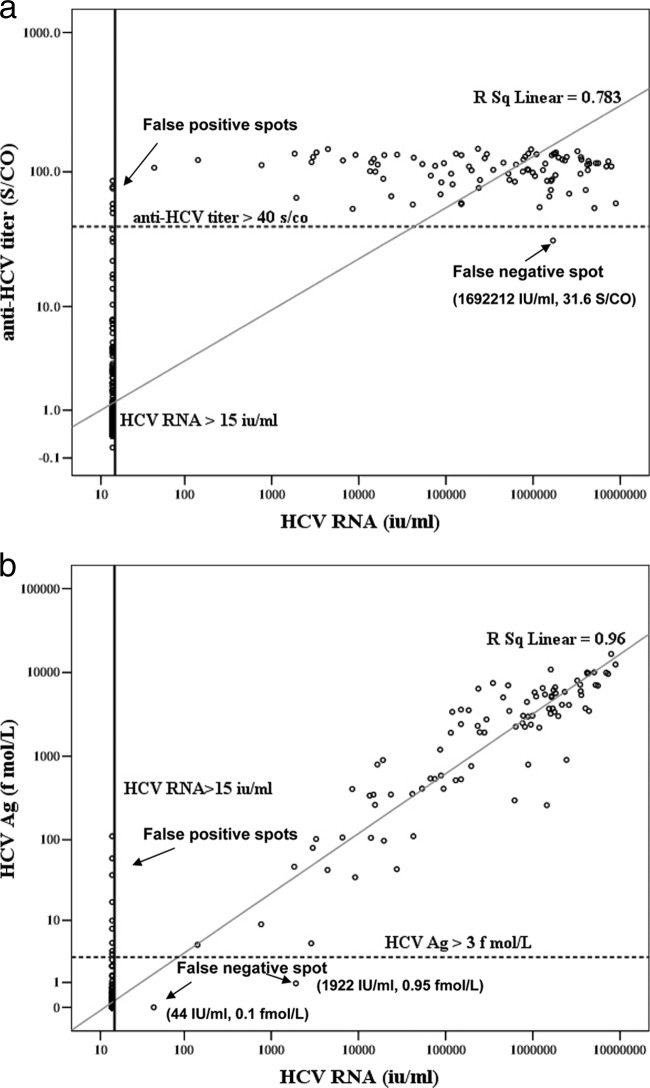
We compared anti-HCV and HCV RNA as well as HCV Ag and HCV RNA levels in log scales. Figure 2a shows that anti-HCV had a significant correlation with HCV RNA in 405 participants (log HCV RNA = 1.45 + 1.67 [log anti-HCV], R2 = 0.75, P < 0.001). Using anti-HCV = 40 S/CO as the cutoff line, we identified 6 false-positive (FP) cases. Only 1 false-negative (FN) case with 31.6 S/CO and 1,692,212 IU/ml was identified. With respect to HCV Ag and HCV RNA, we identified 9 FP cases when we drew the cutoff line with HCV Ag = 3 fmol/liter. Only 2 FN cases were identified, with levels of 0.1 fmol/liter and 44 IU/ml and levels of 0.95 fmol/liter and 1,922 IU/ml, respectively (Fig. 2b).
Fig 2.
(a) Correlation between anti-HCV and HCV RNA results (n = 405). Anti-HCV and HCV RNA concentrations were log transformed prior to analysis. The lower-limit detection line for HCV RNA was 15 IU/ml. Using anti-HCV = 40 S/CO as the cutoff line, we identified 6 false-positive cases and 1 false-negative case. A significant linear regressive correlation between anti-HCV and HCV RNA was noted. (Log HCV RNA = 1.45 + 1.67 [log anti-HCV], R2 = 0.75, P < 0.001). (b) Correlation between HCV Ag and HCV RNA results (n = 405). HCV Ag and HCV RNA concentrations were log transformed prior to analysis. The lower-limit detection line for HCV RNA was 15 IU/ml. Using HCV Ag = 3 fmol/liter as the cutoff line, we identified 9 false-positive cases and 2 false-negative cases. A significant linear regressive correlation between HCV Ag and HCV RNA was noted (log HCV RNA = 2.08 + 1.03 [log HCV Ag], R2 = 0.94, P < 0.001).
HCV Ag had a better correlation with HCV RNA in this study (log HCV RNA = 2.08 + 1.03 [log HCV Ag], R2 = 0.94, P < 0.001 in 405 subjects). We further limited the study sample to 115 subjects, having actually measured their HCV RNA levels, and HCV Ag was still well correlated with HCV viremia (log HCV RNA = 1.78 + 1.14 [log HCV Ag], R2 = 0.85, P < 0.001), with a better correlation than that seen with anti-HCV.
Table 1 shows the HCV RNA results according to different cutoff values for anti-HCV and HCV Ag among the study subjects. Using anti-HCV to predict HCV viremia, S/CO equaling 40 was the best cutoff, with 1 FN and 6 FP results. Using HCV Ag = 3 fmol/liter as the cutoff, we had 2 FN and 9 FP results whereas 10 fmol/liter had 5 FN and 4 FP results. Using combined anti-HCV (S/CO > 40) and HCV Ag (>3 fmol/liter) to predict HCV viremia, there were only 3 FN and no FP. Table 2 shows the prediction results for HCV viremia as determined using different cutoff values for the different serological tests. Among them, using anti-HCV = 40 S/CO as the cutoff, we identified the best sensitivity (98.9%) and negative predictive value (NPV) (99.7%) for the prediction of HCV viremia, while combining anti-HCV = 40 S/CO and HCV Ag = 3 fmol/liter obtained the best specificity (100%), accuracy (99.3%), and positive predict value (PPV) (100%).
Table 1.
Comparisons of anti-HCV, HCV Ag, and HCV RNA test results in 405 participantsa
| HCV RNA test resultb | No. of participants |
|||||||
|---|---|---|---|---|---|---|---|---|
| Anti-HCV (S/CO) |
HCV Ag (fmol/liter) |
HCV Ag (S/CO) |
Anti-HCV (S/CO)/HCV Ag (fmol/liter) |
|||||
| >40 | ≤40 | >3 | ≤3 | >10 | ≤10 | >40/>3 | Other | |
| P | 92 | 1 | 91 | 2 | 88 | 5 | 90 | 3 |
| N | 6 | 306 | 9 | 303 | 4 | 308 | 0 | 312 |
Abbreviations: anti-HCV, hepatitis C virus antibody; HCV Ag, hepatitis C virus core antigen; P, positive; N, negative.
HCV RNA > 15 IU/ml was defined as representing an HCV RNA-positive result and HCV RNA < 15 IU/ml as representing an HCV RNA-negative result.
Table 2.
Comparisons of anti-HCV and HCV-Ag and a combination of anti-HCV plus HCV Ag tests to predict HCV viremia in a community screening (n = 405)a
| Test(s) | % Sen (95% CI) | % Spe (95% CI) | % Acc (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|
| Anti-HCV (S/CO = 40) | 98.9 (95.1 ∼ 99.9) | 98.1 (96.3 ∼ 99.2) | 98.3 (97.1 ∼ 99.5) | 93.9 (88.3 ∼ 98.7) | 99.7 (98.4 ∼ 100) |
| HCV Ag (3 fmol/liter) | 97.8 (93.4 ∼ 99.5) | 97.1 (95.3 ∼ 99.4) | 97.3 (95.3 ∼ 99.2) | 91.0 (84.5 ∼ 96.1) | 99.3 (98.5 ∼ 100) |
| HCV Ag (10 fmol/liter) | 94.6 (89.8 ∼ 98.3) | 98.7 (97.4 ∼ 99.6) | 97.8 (96.2 ∼ 99.1) | 95.7 (90.9 ∼ 99.3) | 98.4 (97.5 ∼ 99.4) |
| Anti-HCV (S/CO = 40) + HCVAg (3 fmol/liter) | 96.8 (92.5 ∼ 99.4) | 100 (99.3 ∼ 100) | 99.3 (98.5 ∼ 100) | 100 (97.7 ∼ 100) | 99.0 (98.4 ∼ 100) |
Abbreviations: Anti-HCV, anti-hepatitis C virus antibody; HCV Ag, hepatitis C virus core antigen; Sen, sensitivity; Spe, specificity; Acc, accuracy; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
DISCUSSION
Successful eradication of HCV could improve the prognosis of HCV-induced liver disease and reduce the associated mortality (23, 27). Hence, in areas where HCV is endemic, it is important to identify the individuals with an ongoing active infection to administer further anti-HCV treatment. Anti-HCV assays are the most common serological markers for the diagnosis of HCV infection and are usually used as the first-line screening test in the community. However, most anti-HCV assays do not correlate well with HCV viremia (18).
Unlike anti-HCV, HCV RNA in the sera of the subjects indicates active viral replication. The detection of HCV viremia is currently regarded as the method of choice for the identification of an active infection. However, its high cost and time-consuming nature and the need for sophisticated technical equipment and highly trained personnel limit its routine use (16). Hence, in community screening, it is used as a second-line tool to confirm ongoing HCV infection among anti-HCV antibody-positive individuals.
Previous studies demonstrated that detection of HCV Ag in serum or plasma is useful as an indirect marker of HCV replication due to the excellent correlation between HCV Ag and HCV RNA concentrations (3, 21, 25, 28). In addition, HCV Ag assays, which are easier to perform than reverse transcription-PCR (RT-PCR), also save time and are less expensive (2, 4). They are used currently to monitor patients undergoing antiviral therapy and to determine the clinical efficacy of such treatment.
In this community-based study, all participants living in the township of Tzukuan, in a southern Taiwan area where HCV is endemic, were members of the aged population. We used the third-generation anti-HCV assay and HCV Ag for HCV screening. Our previous studies (5, 7, 12) reported that this third-generation anti-HCV kit could predict an active HCV infection by selection of an appropriate cutoff. The results of the present study also suggest that anti-HCV had a fair correlation with HCV RNA. Using anti-HCV to predict HCV viremia, S/CO equaling 40 was the best cutoff, with 6 false-positive (FP) cases and only 1 false-negative (FN) case.
Regarding the HCV Ag assay in this study, it seemed to have shown better correlation with HCV RNA than anti-HCV for 405 subjects (log HCV RNA = 2.08 + 1.03 [log HCV Ag], R2 = 0.94, P < 0.001). Even limiting the study sample to 115 subjects, having actually measured their HCV RNA levels, HCV Ag was still well correlated with HCV viremia (log HCV RNA = 1.78 + 1.14 [log HCV Ag], R2 = 0.85, P < 0.001). Although we might overestimate the correlation between HCV Ag and HCV RNA, in real practice, this assay still could have a good quantitative prediction of viremia.
Using HCV Ag to predict HCV viremia with a cutoff of 3 fmol/liter, 9 FP and 2 FN were obtained, whereas, using 10 fmol/liter as the cutoff, the number of FP decreased to 4 but the number of FN increased to 5. Comparing the two cutoffs, 3 fmol/liter has better sensitivity and a better NPV than 10 fmol/liter but poorer specificity, accuracy, and PPV.
If we ignore the budget for screening tools, using combined anti-HCV = 40 S/CO and HCV Ag = 3 fmol/liter as the predictive markers, we were able to obtain excellent sensitivity, specificity, accuracy, PPV, and NPV for viremia prediction at 96.8%, 100%, 99.3%, 100%, and 99%. Using this algorithm of two tests, all 90 subjects with anti-HCV > 40 S/CO and HCV Ag > 3 fmol/liter gave positive HCV RNA results. In contrast, all 7 subjects with anti-HCV ≤ 40 S/CO and HCV Ag ≤ 3 fmol/liter gave negative HCV RNA results. If we were to apply this algorithm with the same sensitivity and specificity to populations with low HCV prevalence, where the prevalence is only 5%, for example, the PPV and NPV would be 100% and 99.7%, respectively. It seemed that this algorithm might still be robust even in populations with low HCV prevalence. More large-scale community studies are needed to validate this algorithm in populations with very low HCV prevalence.
In this age cohort, the third-generation anti-HCV assay was shown to have fair linear correlation but good predictive indices for HCV viremia by using the cutoff as an S/CO of 40. Although the Architect HCV Ag assay was slightly more expensive than the anti-HCV assay, after selecting an adequate cutoff, we could also acquire the ideal predictive indices for HCV viremia, which were comparable to those of anti-HCV. Moreover, it showed an excellent and much better correlation with HCV RNA than the anti-HCV assay. It seemed that both the anti-HCV and HCV Ag assays would be appropriate for use as first-line screening tools due to their good qualitative prediction of HCV viremia. Compared with the anti-HCV assay, HCV Ag could even predict HCV RNA quantitatively to distinguish between an ongoing infection and a recovered phase of acute infection. If we had a limited budget for screening, HCV Ag might offer us more information associated with HCV infection.
This study focused mainly on the evaluation of qualitative and quantitative prediction of HCV RNA by analysis of anti-HCV and HCV Ag in a community screening. With a limited budget, only those subjects who had a possibility of active infection were further checked for their viremia levels. The others were classified as HCV-RNA negative without actual examinations, which might overestimate the correlation between HCV Ag or anti-HCV and HCV RNA. In addition, this age cohort was the first community cohort analyzed using a combination of anti-HCV and HCV Ag tests for HCV screening; more study cohorts are needed to validate the generalizability of this testing algorithm in the future. Finally, HCV genotypes are important for antiviral treatment; however, the early identification of different genotypes is not necessary in a community screening. Hence, the influence of different genotypes on the correlations among anti-HCV, HCV Ag, and HCV RNA (19) was not elucidated. Further longitudinal community-based studies are needed to clarify this issue.
For community screening in the general population, the use of anti-HCV S/CO > 40 or HCV Ag > 3 fmol/liter had a good qualitative prediction for HCV viremia and was affordable as a first-line screening tool. Compared with anti-HCV, HCV Ag was slightly more expensive but showed a better quantitative prediction for HCV RNA. If we were to conduct a community screening with a limited budget in an area where HCV is endemic, using HCV Ag assays could offer us more information associated with active HCV infection. Finally, the results of the present study suggest that, with a sufficient budget, a combination of anti-HCV (>40 S/CO) and HCV Ag (>3 fmol/liter) should be the most valid screening predictor for HCV viremia, with PPV and NPV of 100% and 99%, respectively. In conclusion, to better understand the characteristics of the screening population and assays for diagnoses, we would choose to perform adequately tested assays with an appropriate budget.
ACKNOWLEDGMENTS
This study was supported by a grant from Chang Gung Memorial Hospital (CMRPG:890611) to Yuan-Hung Kuo and was funded by Abbott Diagnostics for providing reagents.
We have no conflicts of interest or funding source.
Footnotes
Published ahead of print 29 March 2012
REFERENCES
- 1. Centers for Disease Control and Prevention Hepatitis C information for health professionals: laboratory testing. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/hepatitis/HCV/LabTesting.htm [Google Scholar]
- 2. Enomoto M, et al. 2005. Chemiluminescence enzyme immunoassay for monitoring hepatitis C virus core protein during interferon-alpha 2b and ribavirin therapy in patients with genotype 1 and high viral loads. J. Med. Virol. 77:77–82 [DOI] [PubMed] [Google Scholar]
- 3. Fabrizi F, et al. 2005. Novel assay using total hepatitis C virus (HCV) core antigen quantification for diagnosis of HCV infection in dialysis patients. J. Clin. Microbiol. 43:414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayashi K, et al. 2005. Usefulness of a new immuno-radiometric assay to detect hepatitis C core antigen in a community-based population. J. Viral Hepat. 12:106–110 [DOI] [PubMed] [Google Scholar]
- 5. Huang WS, et al. 2005. Prediction of viremia for cases of hepatitis C virus (HCV) infection using a third-generation anti-HCV enzyme immunoassay test. Hepatogastroenterology 52:893–896 [PubMed] [Google Scholar]
- 6. Huang YC, et al. 2011. Community-based screening for hepatocellular carcinoma in elderly residents in a hepatitis B- and C-endemic area. J. Gastroenterol. Hepatol. 26:129–134 [DOI] [PubMed] [Google Scholar]
- 7. Hung CH, et al. 2008. Identified cases of acute hepatitis C from computerized laboratory database: a hospital-based epidemiological and clinical study. J. Infect. 56:274–280 [DOI] [PubMed] [Google Scholar]
- 8. Kaiser T, et al. 2008. Kinetics of hepatitis C viral RNA and HCV-antigen during dialysis sessions: evidence for differential viral load reduction on dialysis. J. Med. Virol. 80:1195–1201 [DOI] [PubMed] [Google Scholar]
- 9. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74–81 [DOI] [PubMed] [Google Scholar]
- 10. Lu SN, et al. 2003. Molecular epidemiological and clinical aspects of hepatitis D virus in a unique triple hepatitis viruses (B, C, D) endemic community in Taiwan. J. Med. Virol. 70:74–80 [DOI] [PubMed] [Google Scholar]
- 11. Lu SN, et al. 1997. Different viral aetiology of hepatocellular carcinoma between two hepatitis B and C endemic townships in Taiwan. J. Gastroenterol. Hepatol. 12:547–550 [DOI] [PubMed] [Google Scholar]
- 12. Lu SN, et al. 2004. Is it possible to diagnose acute hepatitis C virus (HCV) infection by a rising anti-HCV titre rather than by seroconversion? J. Viral Hepat. 11:563–570 [DOI] [PubMed] [Google Scholar]
- 13. Maillard P, et al. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miedouge M, Saune K, Kamar N, Rieu M. 2010. Analytical evaluation of HCV core antigen and interest for HCV screening in haemodialysis patients. J. Clin. Virol. 48:18–21 [DOI] [PubMed] [Google Scholar]
- 15. Morota K, et al. 2009. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J. Virol. Methods 157:8–14 [DOI] [PubMed] [Google Scholar]
- 16. National Institutes of Health 2002. NIH consensus statement on management of hepatitis C. NIH Consens. State Sci. Statements 19:1–46 [PubMed] [Google Scholar]
- 17. Park Y, et al. 2010. New automated hepatitis C virus (HCV) core antigen assay as an alternative to real-time PCR for HCV RNA quantification. J. Clin. Microbiol. 48:2253–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richter SS. 2002. Laboratory assays for diagnosis and management of hepatitis C virus infection. J. Clin. Microbiol. 40:4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ross RS, et al. 2010. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J. Clin. Microbiol. 48:1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schüttler CG, et al. 2004. Variable v ratio of hepatitis C virus RNA to viral core antigen in patient sera. J. Clin. Microbiol. 42:1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seme K, Poljak M, Babic DZ, Mocilnik T, Vince A. 2005. The role of core antigen detection in management of hepatitis C: a critical review. J. Clin. Virol. 32:92–101 [DOI] [PubMed] [Google Scholar]
- 22. Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 23. Singal AK, et al. 2010. Antiviral therapy reduces risk of hepatocellular carcinoma in patients with hepatitis C virus-related cirrhosis. Clin. Gastroenterol. Hepatol. 8:192–199 [DOI] [PubMed] [Google Scholar]
- 24. Sroczynski G, et al. 2009. Long-term effectiveness and cost-effectiveness of screening for hepatitis C virus infection. Eur. J. Public Health 19:245–253 [DOI] [PubMed] [Google Scholar]
- 25. Tanaka E, et al. 2000. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology 32:388–393 [DOI] [PubMed] [Google Scholar]
- 26. Tsai PS, et al. 2011. Acquirement and disappearance of HBsAg and anti-HCV in an aged population: a follow-up study in an endemic township. Liver Int. 31:971–979 [DOI] [PubMed] [Google Scholar]
- 27. Wang JH, et al. 2010. Liver stiffness decrease after effective antiviral therapy in patients with chronic hepatitis C: longitudinal study using FibroScan. J. Gastroenterol. Hepatol. 25:964–969 [DOI] [PubMed] [Google Scholar]
- 28. Zanetti AR, et al. 2003. Total HCV core antigen assay: a new marker of hepatitis C viremia for monitoring the progress of therapy. J. Med. Virol. 70:27–30 [DOI] [PubMed] [Google Scholar]