Abstract
High-risk human papillomavirus (HPV) DNA/RNA testing provides higher sensitivity but lower specificity than cytology for the identification of high-grade cervical intraepithelial neoplasia (CIN). Several new HPV tests are now available for this purpose, and a direct comparison of their properties is needed. Seven tests were evaluated with samples in liquid PreservCyt transport medium from 1,099 women referred for colposcopy: the Hybrid Capture 2 (Qiagen), Cobas (Roche), PreTect HPV-Proofer (NorChip), Aptima HPV (Gen-Probe), and Abbott RealTime assays, the BD HPV test, and CINtec p16INK4a cytology (mtm laboratories) immunocytochemistry. Sensitivity, specificity, and positive predictive value (PPV) were based on the worst histology found on either the biopsy or the treatment specimen after central review. Three hundred fifty-nine women (32.7%) had CIN grade 2+ (CIN2+), with 224 (20.4%) having CIN3+. For detection of CIN2+, Hybrid Capture 2 had 96.3% sensitivity, 19.5% specificity, and 37.4% PPV. Cobas had 95.2% sensitivity, 24.0% specificity, and 37.6% PPV. The BD HPV test had 95.0% sensitivity, 24.2% specificity, and 37.8% PPV. Abbott RealTime had 93.3% sensitivity, 27.3% specificity, and 38.2% PPV. Aptima had 95.3% sensitivity, 28.8% specificity, and 39.3% PPV. PreTect HPV-Proofer had 74.1% sensitivity, 70.8% specificity, and 55.4% PPV. CINtec p16INK4a cytology had 85.7% sensitivity, 54.7% specificity, and 49.1% PPV. Cytology of a specimen taken at colposcopy (mild dyskaryosis or worse) had 88.9% sensitivity, 58.1% specificity, and 50.7% PPV. Our study confirms that, in a referral setting, HPV testing by a number of different tests provides high sensitivity for high-grade disease. Further work is needed to confirm these findings in a routine screening setting.
INTRODUCTION
High-risk (HR) human papillomavirus (HPV) is a necessary factor for the development of cervical cancer (31), but the presence of HR HPV DNA does not invariably lead to disease. We have shown that the detection of HR HPV DNA provides high sensitivity but has lower specificity than cytology for the identification of high-grade cervical lesions in a screening population in the United Kingdom (9), and this finding has been replicated in several other studies (1, 3, 8, 15–17, 21, 22, 26, 27). In addition, prospective studies have shown that HPV DNA-positive women who do not have disease initially are significantly more likely to develop high-grade squamous intraepithelial lesions (SILs) within 10 years than women with a negative HPV DNA test (4, 12, 14, 18). If testing for HR HPV DNA is to be used as a primary cervical screening test, refinements or additional tests are highly desirable to improve its specificity while retaining its very high sensitivity. Candidates include HPV typing, detection of HPV E6/E7 mRNA tests, and p16INK4a cytology, which have shown higher specificity than HPV DNA testing (10, 19).
The introduction of a liquid-based medium for collection of cytological specimens has allowed other molecular techniques to be evaluated more easily as adjunctive or triage tests (2). We have previously compared a number of different molecular tests in a population referred for colposcopy because of abnormal cytology (7, 29). However, there are now a number of newer tests, which also require evaluation. A number of studies have compared two tests against each other (usually the comparator was the Hybrid Capture 2 [HC2] assay), but as far as we are aware, apart from our previous study, none have simultaneously compared such a wide range of tests using the same sample. The aim of this study was to compare directly the sensitivity and specificity of several tests for the detection of high-grade cervical intraepithelial neoplasia (CIN) using aliquots from the same sample from the same women in a population referred for colposcopy because of abnormal cytology. This population was chosen because the high disease rate allows comparison of the sensitivities of the different tests within a modest sample size. Use of a screening population would require a sample size at least 20 times larger in order to have similar power. All the tests were compared against the “gold standard” of centrally reviewed histopathology.
MATERIALS AND METHODS
The study population comprised 1,099 women who had been referred to the colposcopy clinics at the Hammersmith and St. Mary's Hospitals in London, United Kingdom, between September 2007 and October 2009 because of abnormal screening smears (Fig. 1). In England, women are referred for colposcopy if they have cytology showing mild dyskaryosis or worse or three smears showing borderline dyskaryosis. While not a screening population, the advantage was a broad range of outcomes and a high disease rate, which would enable accurate comparisons of sensitivity and specificity in a relatively small sample. Women were eligible if they had been referred as a result of one or more abnormal cervical smears, were not pregnant, had not been treated previously for CIN, and had not had a hysterectomy. All women received a patient information sheet explaining the study and provided written consent. Approvals were obtained from the relevant local research ethics committees.
Fig 1.
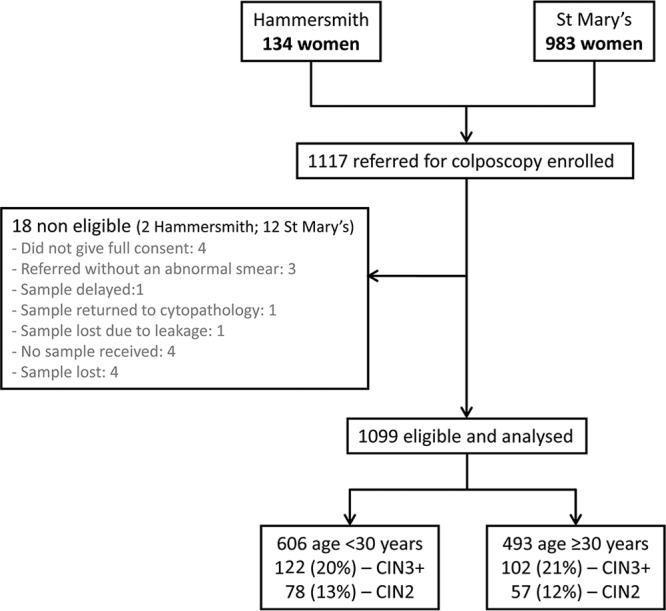
Study flowchart for women included in the study. Numbers shown are numbers of women.
Prior to colposcopy, two cervical samples were obtained using a Cervex broom and placed in separate 20-ml vials of PreservCyt transport medium. Two samples were taken because the number of tests proposed required more aliquots than could be obtained from a single sample. Testing from two samples has been shown to have good concordance for molecular testing (11). Colposcopy was then performed in the usual manner. The liquid-based cytology (LBC) samples were transported to The Doctors Laboratory (TDL), where an aliquot from the first sample was removed for cytology, processed using the ThinPrep system, and returned to the relevant cytopathology departments for reporting. Cytopathology reporting was carried out blindly, although the cytopathologists were inevitably aware that the sample came from a colposcopy clinic. The remaining material from the first sample was used for NorChip PreTect HPV-Proofer testing (which was also performed by TDL).
Both samples were then sent to the laboratory at Queen Mary, University of London (QMUL), where aliquots were removed for the other tests. The Qiagen Hybrid Capture 2 assay and mtm laboratories CINtec p16INK4a cytology were carried out using material from the first sample. The second sample was used for the Abbott RealTime, Roche Cobas, Gen-Probe Aptima, and BD HPV tests. Tests were carried out in the QMUL laboratory according to standard manufacturers' instructions. The molecular testing laboratories were blinded to the cytology and histopathology results.
Neither the women nor their clinicians were informed of the HPV test results, and the results were not used to influence patient management.
Histopathology was first reported locally and then centrally reviewed by M.Y., who was blinded to all study test results. Where discrepant readings occurred, further review was undertaken by M.S., and the majority opinion was taken; where all three readings were discrepant (one each of CIN less severe than grade 2 [<CIN2], CIN2, and CIN3), CIN2 was assigned. All results are presented on the basis of the reviewed histopathology, and the highest grade of abnormality seen in the biopsy or treatment specimen was used.
Laboratory methods.
In this study, the following assays were carried out and scored in strict accordance with the manufacturers' protocols.
(i) DNA-based detection and genotyping assays. (a) Hybrid Capture 2 assay.
The Hybrid Capture 2 assay (Qiagen) detects 13 HR HPV genotypes collectively and is based on the hybridization of HPV DNA to a 13-HR-HPV-type RNA probe cocktail. The DNA-RNA hybrid is captured by an anti-DNA-RNA antibody and detected by chemiluminescence. Readings over 1 relative light unit (RLU) are considered positive.
(b) Cobas 4800 test.
The Cobas 4800 (Roche Diagnostics) test is a qualitative in vitro test for the detection of 14 HR HPV types. The test specifically identifies HPV type 16 (HPV-16) and HPV-18 while concurrently detecting the rest of the high-risk types as a group (types 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68).
(c) Abbott RealTime High Risk HPV assay.
The Abbott RealTime High Risk HPV assay (Abbott Molecular, Wiesbaden, Germany) is a qualitative multiplex real-time test that also specifically identifies HPV-16 and HPV-18 while concurrently detecting the rest of the high-risk types as a group (types 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68).
(d) BD HPV test.
The BD HPV test (BD Diagnostics) a real-time PCR (at the time of writing, not yet commercially available) which detects 14 HR HPV types. Typing is provided for types 16, 18, 31, 45, 51, 52, and 59. The remaining HPV types are grouped into the type 33, 56, 58, and 66 group and the type 35, 39, and 68 group.
(ii) RNA-based detection assays. (a) PreTect HPV-Proofer assay.
The PreTect HPV-Proofer assay (NorChip) was performed by TDL. PreTect HPV-Proofer is a real-time multiplex nucleic acid sequence-based amplification assay for isothermal amplification of E6/E7 mRNA expressed by five high-risk HPV types (types 16, 18, 31, 33, and 45) using proprietary primer sets (19).
(b) Aptima (Gen-Probe) test.
The Aptima (Gen-Probe) assay is based on target capture, transcription-mediated amplification, and hybridization protection for the detection of E7 mRNA expression of 14 HR HPV types. Although not commercially available as yet, in this study, typing was also performed at Gen-Probe Laboratories for HPV types 16, 31, and 33 and HPV types 18 and 45 combined (13).
(iii) p16INK4a cytology.
p16INK4a cytology uses immunocytochemical detection of overexpression of the p16INK4a tumor suppressor gene (CINtec cytology; mtm laboratories) and scoring using nuclear morphology.
Immunostaining was done at mtm laboratories using monoclonal antibody to p16INK4a with antimouse as secondary antibody and detected by diaminobenzidine chromogen. The p16INK4a score was based on nuclear assessment of brown-stained cells by four criteria (32): A, increased size; B, granular or hyperchromatic chromatin; C, irregular shape; D, variable morphology from cell to cell.
Cells positive for any one of these criteria were scored as 2. Cells positive for criterion A and one other criterion were scored as 3, and cells positive for criterion A and more than one other criterion were scored as 4. The sample score was the highest score observed. Reporting of CINtec p16INK4a cytology was carried out blindly by C.B. according to a scoring method recently described (32, 33).
Sample adequacy was assessed by a cellular DNA or RNA marker for all molecular assays except HC2. Assay variability was controlled by including a pooled positive control and a pooled negative control in each run for each test. Reagents from the same batch were used throughout, and the controls provided in the kits showed little assay variability over time.
Data entry and all statistical analysis were carried out at QMUL.
Statistical methods.
The main outcome measures were sensitivity, specificity, and positive predictive value (PPV). Confidence intervals for these were based on binomial statistics. Comparisons between tests were conducted by McNemar's test for matched pairs and giving odds ratios and 95% confidence intervals for discordant samples. The study was designed to detect with 80% power a difference in sensitivity of 95% versus 90%. This assumed 300 CIN2+ cases and a discordant difference of 20 (positive/negative) versus 5 (negative/positive). Assays were considered to be significantly different if the two-sided P value was less than 0.05. Additional calculations of sensitivity, specificity, and PPV were carried out for women aged ≤30 and women aged >30 years. Full details of type-specific results will appear elsewhere, but positivity results for HPV-16 and HPV-18 are shown.
We considered the worst histology within 9 months of the initial baseline visit. Histologically confirmed CIN2+ and CIN3+ were taken as the primary endpoints. We also reported CIN2 separately. Specificity is reported only for <CIN2, as we do not consider the detection of CIN2 to be a false positive. Where appropriate, we also computed receiver operating characteristic (ROC) curves to further compare sensitivity and specificity at different cutoffs for Hybrid Capture 2, BD HPV, Abbott RealTime, Roche Cobas, Gen-Probe Aptima, and mtm laboratories p16INK4a.
All statistical analyses were carried out using the Stata (version 10.1) program (StataCorp).
RESULTS
Figure 1 (flowchart) shows the study recruitment. A total of 1,117 women consented to participate in the study, and the tests from 1,099 women were analyzed. The median length of time between the referral smear and colposcopy was 1.8 months (interquartile ratio [IQR], 1.4 to 2.6 months; range, 0.3 to 59 months). The median age of the women was 29 years (IQR, 26.6 to 34.3 years). Over three-quarters of the women were under 35 years of age, with 55% being under the age of 29 years.
The referral population comprised approximately 24% of women with high-grade dyskaryosis, and 76% had low-grade disease (16.7% borderline, 44.2% with a single mildly dyskaryotic cytology, and 15.4% with mild dyskaryosis and one or more borderline smears or worse). Over 20% of the concurrent smears were borderline, 32% were mild dyskaryosis, and approximately 26% were high grade. Just under one-fifth (19%) of the concurrent smear specimens taken in the colposcopy clinic were negative. This may reflect sampling variability in the smear or regression of disease.
Table 1 tabulates referral cytology against worst histology. Twenty-seven percent of the women either had a normal colposcopy with no biopsy or a negative biopsy, 37% of women had CIN1 or other minor abnormalities, 12% had CIN2, and 20% had CIN3 or worse.
Table 1.
Results of referral smear and worst-reviewed consensus histology within 9 months of baseline cytology
| Referral smear | No. (%) of women with the following histology result: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Not taken | Unsatisfactory | Normal | HPV only | CIN1 | CIN2 | CIN3 | CGIN,a high grade | Invasive squamous carcinoma | Total | |
| Normal | 1 | 1 | 2 (0.2) | |||||||
| Borderline | 49 | 3 | 29 | 5 | 75 | 8 | 10 | 3 | 1 | 183 (16.7) |
| Mild dyskaryosis (1 single mild) | 94 | 7 | 71 | 9 | 210 | 60 | 35 | 486 (44.2) | ||
| Mild dyskaryosis (1 mild + 1 or more borderline or worse) | 25 | 2 | 16 | 4 | 79 | 25 | 18 | 169 (15.4) | ||
| Moderate dyskaryosis | 3 | 2 | 9 | 2 | 35 | 35 | 86 | 1 | 1 | 174 (15.8) |
| Severe dyskaryosis or worse | 2 | 2 | 5 | 7 | 61 | 1 | 7 | 85 (7.7) | ||
| Total | 172 (15.7) | 16 (1.5) | 128 (11.6) | 20 (1.8) | 404 (36.8) | 135 (12.3) | 210 (19.1) | 5 (0.5) | 9 (0.8) | 1,099 (100.0) |
CGIN, cervical glandular intraepithelial neoplasia.
Cytology of samples taken at colposcopy (mild dyskaryosis or worse) had 88.9% sensitivity, 58.1% specificity, and 50.7% PPV. The cytology was read with the knowledge that the patient was having colposcopy and so may not reflect normal screening practice.
The overall HPV positivity of the different tests is shown in Table 2 and ranged from 79% to 86%, apart from p16INK4a and PreTect HPV-Proofer, at 58.9% and 43.9%, respectively. The proportion of women positive for HPV type 16 and/or 18 was similar across tests (for HPV type 16, the positivity ranged from 26.2% to 31.9%, and for HPV type 18, it ranged from 7.8% to 11.8%; Table 2).
Table 2.
HPV positivity and type-specific results of different tests
| Test | No. of women tested (no. with single mild or less smear) | % of women positive for: |
|||
|---|---|---|---|---|---|
| HPV | HPV-16 | HPV-18 | Other HPVa | ||
| Qiagen Hybrid Capture 2 | 1,067 (649) | 85.8 | |||
| Roche Cobas | 1,095 (670) | 82.3 | 31.9 | 10.0 | 71.1 |
| Abbott RealTime PCR | 1,095 (670) | 79.4 | 30.5 | 8.5 | 67.6 |
| BD HPV | 1,097 (670) | 82.0 | 30.5 | 9.4 | 71.4 |
| Gen-Probe Aptima | 1,097 (670) | 79.0 | 30.0 | 11.8b | |
| NorChip PreTect HPV-Proofer | 1,057 (641) | 43.9 | 26.2 | 7.8 | 42.8 |
| mtm laboratories p16INK4a | 974 (591) | 58.9 | |||
Other HPV only for those negative for HPV-16 and HPV-18.
Tested HPV-18 and HPV-45 positive combined.
Sensitivity, specificity, and PPV for CIN2+, CIN2 alone, and CIN3+ of the different tests (all samples taken concurrently at colposcopy) are reported in Table 3. We have chosen to report separately the results for CIN2+ and CIN3+ because CIN2 is less likely to progress and has greater variability in diagnosis. CIN3 has been shown to have greater reproducibility.
Table 3.
Sensitivity, specificity, and PPV of different tests for detection of high-grade disease
| Test and grade assessed | All women |
Women with referral less than or equal to single mild dyskaryosis |
||||
|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | Sensitivity (%) | Specificity (%) | PPV (%) | |
| Qiagen Hybrid Capture 2 | ||||||
| CIN3+ | 98.7 (96.1–99.7)a | 24.0 (21.3–26.9) | 100.0 (92.7–100.0) | 9.2 (6.9–11.9) | ||
| CIN2+ | 96.3 (93.8–98.0) | 19.5 (16.7–22.6) | 37.4 (34.2–40.6) | 93.0 (86.8–96.9) | 19.9 (16.5–23.5) | 20.0 (16.7–23.6) |
| CIN2 | 92.4 (86.5–96.3) | 87.9 (77.5–94.6) | ||||
| Roche Cobas | ||||||
| CIN3+ | 97.3 (94.2–99.0) | 23.9 (21.1–26.8) | 100.0 (92.7–100.0) | 9.3 (7.0–12.2) | ||
| CIN2+ | 95.2 (92.5–97.2) | 24.0 (20.9–27.2) | 37.6 (34.5–40.9) | 94.9 (89.2–98.1) | 25.1 (21.6–29.0) | 21.1 (17.7–24.9) |
| CIN2 | 91.9 (85.9–95.9) | 91.2 (81.8–96.7) | ||||
| Abbott RealTime PCR | ||||||
| CIN3+ | 97.3 (94.2–99.0) | 24.7 (21.9–27.8) | 100.0 (92.7–100.0) | 9.8 (7.3–12.7) | ||
| CIN2+ | 93.3 (90.1–95.6) | 27.3 (24.1–30.7) | 38.2 (35.0–41.5) | 92.3 (85.9–96.4) | 28.9 (25.2–32.9) | 21.6 (18.0–25.4) |
| CIN2 | 86.7 (79.7–91.9) | 86.8 (76.4–93.8) | ||||
| BD HPV | ||||||
| CIN3+ | 97.8 (94.8–99.3) | 24.2 (21.5–27.2) | 100.0 (92.7–100.0) | 9.4 (7.0–12.2) | ||
| CIN2+ | 95.0 (92.2–97.0) | 24.2 (21.2–27.5) | 37.8 (34.6–41.0) | 94.0 (88.1–97.6) | 25.3 (21.7–29.2) | 21.0 (17.6–24.8) |
| CIN2 | 90.4 (84.1–94.8) | 89.7 (79.9–95.8) | ||||
| Gen-Probe Aptima | ||||||
| CIN3+ | 97.8 (94.8–99.3) | 25.1 (22.3–28.2) | 100.0 (92.7–100.0) | 9.8 (7.3–12.7) | ||
| CIN2+ | 95.3 (92.5–97.2) | 28.8 (25.6–32.2) | 39.3 (36.1–42.7) | 92.3 (85.9–96.4) | 29.1 (25.4–33.1) | 21.6 (18.1–25.5) |
| CIN2 | 91.1 (85.0–95.3) | 86.8 (76.4–93.8) | ||||
| mtm laboratories p16INK4a | ||||||
| CIN3+ | 90.2 (85.3–93.9) | 32.2 (28.4–36.2) | 83.7 (69.3–93.2) | 12.5 (8.9–16.9) | ||
| CIN2+ | 85.7 (81.5–89.3) | 54.7 (50.8–58.6) | 49.1 (45.0–53.3) | 76.2 (66.9–84.0) | 57.2 (52.7–61.6) | 27.8 (22.7–33.3) |
| CIN2 | 78.2 (69.9–85.1) | 71.0 (58.1–81.8) | ||||
| NorChip PreTect HPV-Proofer | ||||||
| CIN3+ | 80.3 (74.4–85.3) | 37.7 (33.3–42.3) | 80.9 (66.7–90.9) | 16.6 (12.0–22.1) | ||
| CIN2+ | 74.1 (69.1–78.6) | 70.8 (67.3–74.2) | 55.4 (50.7–60.0) | 73.6 (64.4–81.6) | 72.1 (68.1–75.9) | 35.4 (29.2–41.9) |
| CIN2 | 63.6 (54.6–71.9) | 68.3 (55.3–79.4) | ||||
| Cytology (mild or worse) | ||||||
| CIN3+ | 92.9 (88.7–95.9) | 33.1 (29.4–36.9) | ||||
| CIN2+ | 88.9 (85.1–91.9) | 58.1 (54.5–61.7) | 50.7 (46.7–54.7) | NAb | ||
| CIN2 | 82.2 (74.7–88.3) | |||||
Data in parentheses represent 95% confidence intervals.
NA, not available.
Five adjunctive tests had a sensitivity greater than 95% for CIN3+: Hybrid Capture 2, Cobas 4800, Abbott RealTime, BD HPV, and Aptima. The Abbott RealTime test was significantly less sensitive for CIN2+ than Hybrid Capture 2, BD HPV, and Aptima and had a marginally but not significantly lower sensitivity than Cobas 4800. All these tests were significantly more sensitive than p16INK4a or PreTect HPV-Proofer.
There were seven cancers, of which all but one (a microinvasive stage 1A cancer missed by PreTect HPV-Proofer only) were detected by all the HPV tests. This sample tested positive for HPV-52 by BD HPV and for other HPV types by Abbott RealTime and Cobas 4800.
Overall, the highest specificity was achieved with the PreTect HPV-Proofer (70.8%, for CIN2+), but this test had relatively low sensitivity (Table 3). The CINtec p16INK4a cytology test had a significantly lower specificity (54.7% and 49.4% for CIN2+ and CIN3+, respectively) than PreTect HPV-Proofer but a higher specificity than the other tests. Of the five highly sensitive tests, Hybrid Capture 2 showed significantly lower specificity than the other four tests (McNemar's test). Aptima and Abbott RealTime were also significantly more specific than Cobas 4800 and BD HPV. When focusing on CIN3+, overall, sensitivity is slightly improved; however, the ordering of the tests remains similar (Table 3). Because there is uncertainty about the progressive potential of CIN2, we provide sensitivity estimates for both CIN2+ and CIN3+; however, few would consider CIN2 to be a false positive, so we do not report specificity for CIN3+.
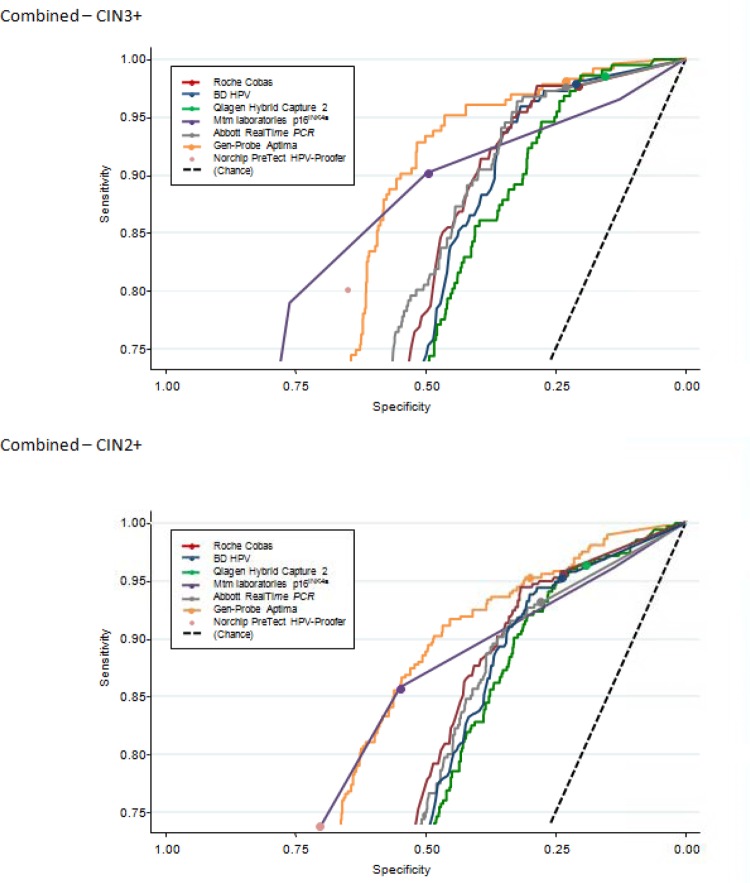
Figure 2 shows the effects on sensitivity and specificity of using different cutoffs for Hybrid Capture 2, Cobas, Abbott RealTime, Aptima, BD HPV, and p16INK4a in predicting histologically confirmed high-grade disease (CIN2+ and CIN3+). The ROC curves for the highly sensitive tests are very similar. If the cutoff for Hybrid Capture 2 was raised (from ≥1 RLU to ≥2 RLU), the sensitivity remained relatively unchanged, while the specificity slightly improved. Using a cutoff ≥3 instead of ≥2 (see Materials and Methods) for p16INK4a reduced the sensitivity for CIN3+ (from 90.2% to 79.0%), although it increased the specificity from 49.4% to 75.9%. For the BD HPV test, lowering the cutoff to a cycle threshold (CT) value of ≤33 from the nominal ≤36.2 would result in a small decrease in sensitivity for CIN3+ (from 97.8% to 97.3%) and for CIN2+ (from 95% to 93.8%), while it would result in an increase in specificity for CIN3+ (from 21.9% to 26.7%) and for CIN2+ (from 24.1% to 29.4%).
Fig 2.
ROC curves for tests to detect CIN2+ and CIN3+. Sensitivity plotted against specificity calculated at different cutoffs for each test is shown. Plots are presented separately for CIN3+ and CIN2+, with a dashed line used to represent the line of chance.
Sensitivity was slightly higher for all tests in younger women (age <30 years), being (for CIN3+) between 3.0% and 5.3% higher for all tests except PreTect HPV-Proofer, whose sensitivity was 88.9% in women under age 30 years and 70.3% in those over age 30 years. All tests showed higher specificity in the older age group (data not shown).
To more accurately reflect the use of these tests in a situation where HPV triage has been recommended, we also considered the restricted population of women who had a borderline or mildly dyskaryotic referral smear. The results were generally similar (Table 3). For the five tests that showed the highest sensitivity (Hybrid Capture 2, Abbott RealTime, Cobas 4800, BD HPV, and Aptima), specificities were similar when only women with borderline or mildly dyskaryotic smears were considered, and the relative performance (i.e., the order) of the tests was unchanged in these lower-risk cytology categories.
It should be noted that the tests did not all miss the same cases, making comparisons more complex. There were 120 CIN2+ individuals missed by at least one test: 87 individuals were negative by one test only (60 by PreTect HPV-Proofer, 26 by CINtec p16INK4a cytology, and 1 by Hybrid Capture 2); 10 individuals were negative by two tests (7 by PreTect HPV-Proofer and CINtec p16INK4a cytology, 3 by PreTect HPV-Proofer and Abbott RealTime); 23 individuals were negative by three or more tests. Data on individual HPV types were complicated by the large number of multiple infections and will be reported separately.
DISCUSSION
This study's strength is that it is possible to compare at the same time a wide range of adjunctive tests with samples taken from the same woman. All the women in the population studied were referred with abnormal cytology, but they had a wide range of histological outcomes, which facilitated the ability to look at relative sensitivity. One other study (23) has compared several commercially available tests in the same cohort. The study was performed in 281 women referred with an abnormal smear, and the tests compared were HC2, PreTect HPV-Proofer, Roche's linear array, and the DR.HPV IVD kit (DR.Chip Biotech, Inc.). Sensitivity from CIN3+ was highest for HC2 and linear array (100%), whereas NorChip PreText HPV-Proofer showed a high specificity (88%). Another study based in a screening setting (5) compared Abbott RealTime with Hybrid Capture 2. The study was performed in a random sample of 998 specimens and showed sensitivities for CIN2+ of 96.4% and 97.6% for Abbott RealTime and HC2, respectively. Specificity was 92.3% for Abbott RealTime and 92.6% for HC2.
A recent population-based study in France (20) which compared HC2 with Aptima showed sensitivities similar to those from our study for CIN2+ of 92% for Aptima and 96.4% for HC2. Specificities for CIN2+ were much higher in the FASE study (91.8% for Aptima versus 86.4% for HC2) than our study (28.8% for Aptima versus 19.5% for HC2). This difference in specificity is due to the differences in a screening population versus a referral population. A second population-based study from Slovenia (25) compared Abbott RealTime with Hybrid Capture 2. In that study, Abbott RealTime showed an overall sensitivity of 98.2%, compared with 93.3% in our study, while Hybrid Capture 2 showed a sensitivity of 94.7%, compared with 96.3% in our study. As with the FASE study, specificity was much higher for both tests, for similar reasons.
A recent study (6) compared the performance of the BD HPV assay to that of HC2 in an archived subset of ALTS clinical trial specimens among women referred with either atypical squamous cells of undetermined significance (ASCUS) or low-grade squamous intraepithelial lesion (LSILs; borderline or mild dyskaryosis). The clinical performance of the BD assay (sensitivity = 90.0%; specificity = 43.9%; PPV = 12.9%) for detection of a 2-year worst histologic outcome of CIN3 was comparable to that of HC2 (sensitivity = 87.5%; specificity = 41.8%; PPV = 12.2%). In addition, specimens that tested HC2 negative and BD HPV positive were more likely to be positive for oncogenic HPV genotypes, whereas those that tested HC2 positive and BD HPV negative were more likely to be positive for low-risk HPV genotypes.
A study conducted in the United States (28) compared HC2 with Cobas 4800 among 1,578 women with ASCUS cytology. This study showed a slightly lower sensitivity for CIN2+ for both tests (87.2% for HC2, 90.0% for Cobas 4800). However, specificity in the study by Stoler et al. (28) was considerably higher (71.1% for HC2, 70.5% for Cobas 4800). It should be noted that in the United Kingdom, women are referred for colposcopy only after three smears showing borderline dyskaryosis, whereas in the study by Stoler et al. (28), referral for colposcopy took place after a single ASCUS result.
Five tests demonstrated very high sensitivity, indicating that they are unlikely to miss significant disease, and of these, the Aptima and Abbott RealTime tests had the best specificity, suggesting that these tests may lead to fewer unnecessary colposcopy referrals. Sample adequacy was not an issue for these samples, as they were collected by trained health care professionals in referral clinics. Sample adequacy can be a problem when using material after cytology slides have been prepared, but since we had two 20-ml vials, this was not an issue here.
The results for CINtec p16INK4a cytology were less favorable than those in other recent publications (33), which found a sensitivity of 100% in an LSIL population, with a specificity of 81.7%. However, there is a wide variation in the literature, which may be at least partly related to changes in and a lack of standardization in reporting of p16INK4a cytology results (10, 30). The results for CINtec p16INK4a cytology are similar to those of the concurrent cytology. However, the concurrent cytology was based on samples taken at colposcopy, where sampling and reading may be better than those for routine screening samples.
High sensitivity is clearly important. However, it is known that many CIN2 and some CIN3 lesions will regress spontaneously, and it is possible that tests with lower sensitivity still identify the lesions which are destined to progress to cancer. Among individuals with CIN2+ in our study, 13 tested positive by HC2 but were missed by Abbott RealTime, whereas only 2 tested positive by Abbott RealTime but were missed by HC2. These results differ from those of Poljak et al. (24) in the comparison of HC2 with Abbott RealTime, where 7 individuals with CIN3+ tested positive by HC2 but were missed by Abbott RealTime and 15 were positive by Abbott RealTime but negative by HC2.
Triage is most needed for women with a single mildly dyskaryotic or borderline dyskaryosis on smear. For this group, the absolute sensitivity and the relative performance of the tests were similar to those for the whole cohort, but specificity was higher, especially in the case of borderline dyskaryotic smears, indicating the value of a triage test for these women. These data provide useful relative comparisons of the performances of these tests in such a situation. However, a limitation of this study in terms of triage is that the HPV tests were not done on the referral sample, as would normally be the case when using liquid-based cytology.
It should be noted that this study was not designed to provide specific algorithms for cervical screening or triage but was designed to compare a range of HPV tests in a sample of women with a high disease rate, with a particular focus on evaluating the test sensitivities relative to each other. The very high sensitivity in this situation has provided reassurance that disease is unlikely to be missed in women with abnormal cytology. It also suggests that sensitivity will be very high in cytology-negative women, although this requires direct verification. However, there is already some evidence to support the relative performances of the tests in a screening setting, although only in pairwise comparisons (5, 6, 20, 25).
Compared with the population for our previous publication (29), this population contained a somewhat greater proportion of women aged under 29 years and fewer women aged over 45 years, which may account for the overall higher HPV positivity rates. This may also contribute to the higher proportion of individuals with CIN2 and the lower specificity observed compared to the previous study.
Our study confirms that, in a referral setting, HPV testing by a number of different tests offers high sensitivity for high-grade disease. However, this finding may not be extrapolated to a screening setting, and further work in unselected screening populations is needed.
ACKNOWLEDGMENTS
We thank the staff in the Departments of Cytopathology and Colposcopy at the Hammersmith Hospital and St. Mary's Hospital, London, for their help with this study.
Jack Cuzick is on advisory boards for Gen-Probe, Roche, Qiagen, Abbott, and BD; Mark Stoler is a consultant to MSD, Roche, BD, Gen-Probe, Qiagen, and mtm laboratories; Christine Bergeron is on advisory boards for Gen-Probe, Roche, and mtm laboratories.
This study was supported by Cancer Research United Kingdom Programme grants C569/A10404 and C8162/A10406 and supplemented with financial contributions and/or assay kits from Qiagen, Gen-Probe Incorporated, Abbott Molecular, BD Diagnostics, mtm laboratories, NorChip, and Roche Diagnostics.
Footnotes
Published ahead of print 14 March 2012
REFERENCES
- 1. Almonte M, et al. 2007. Cervical screening by visual inspection, HPV testing, liquid-based and conventional cytology in Amazonian Peru. Int. J. Cancer 121:796–802 [DOI] [PubMed] [Google Scholar]
- 2. Arbyn M, Ronco G, Cuzick J, Wentzensen N, Castle PE. 2009. How to evaluate emerging technologies in cervical cancer screening? Int. J. Cancer 125:2489–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bulkmans NW, et al. 2007. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet 370:1764–1772 [DOI] [PubMed] [Google Scholar]
- 4. Bulkmans NW, Rozendaal L, Voorhorst FJ, Snijders PJ, Meijer CJ. 2005. Long-term protective effect of high-risk human papillomavirus testing in population-based cervical screening. Br. J. Cancer 92:1800–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carozzi FM, et al. 2011. Comparison of clinical performance of Abbott RealTime High Risk HPV test with that of Hybrid Capture 2 assay in a screening setting. J. Clin. Microbiol. 49:1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castle PE, et al. 2011. Evaluation of a new DNA test for detection of carcinogenic human papillomavirus. J. Clin. Microbiol. 49:3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuzick J, et al. 2010. Performance of the Abbott RealTime high-risk HPV test in women with abnormal cervical cytology smears. J. Med. Virol. 82:1186–1191 [DOI] [PubMed] [Google Scholar]
- 8. Cuzick J, et al. 2006. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int. J. Cancer 119:1095–1101 [DOI] [PubMed] [Google Scholar]
- 9. Cuzick J, et al. 2003. Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet 362:1871–1876 [DOI] [PubMed] [Google Scholar]
- 10. Denton KJ, et al. 2010. The sensitivity and specificity of p16(INK4a) cytology versus HPV testing for detecting high-grade cervical disease in the triage of ASC-US and LSIL Pap cytology results. Am. J. Clin. Pathol. 134:12–21 [DOI] [PubMed] [Google Scholar]
- 11. de Sanjose S, et al. 1999. Screening for genital human papillomavirus: results from an international validation study on human papillomavirus sampling techniques. Diagn. Mol. Pathol. 8:26–31 [DOI] [PubMed] [Google Scholar]
- 12. Dillner J, et al. 2008. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ 337:a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joo S, Salva N, Nater T, Dockter J. 2010. Preliminary evaluation of the Aptima HPV 16 18/45 genotype assay for the detection of human papillomavirus types 16, 18 and 45 in liquid Pap specimens, poster 252. International Papillomavirus Conference, Montreal, Quebec, Canada [Google Scholar]
- 14. Khan MJ, et al. 2005. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 97:1072–1079 [DOI] [PubMed] [Google Scholar]
- 15. Kitchener HC, et al. 2009. ARTISTIC: a randomised trial of human papillomavirus (HPV) testing in primary cervical screening. Health Technol. Assess. 13(51):1–150, iii-iv [DOI] [PubMed] [Google Scholar]
- 16. Kotaniemi-Talonen L, et al. 2008. Screening with a primary human papillomavirus test does not increase detection of cervical cancer and intraepithelial neoplasia 3. Eur. J. Cancer 44:565–571 [DOI] [PubMed] [Google Scholar]
- 17. Mayrand MH, et al. 2007. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N. Engl. J. Med. 357:1579–1588 [DOI] [PubMed] [Google Scholar]
- 18. Mesher D, et al. 2010. Long-term follow-up of cervical disease in women screened by cytology and HPV testing: results from the HART study. Br. J. Cancer 102:1405–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Molden T, Kraus I, Skomedal H, Nordstrom T, Karlsen F. 2007. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J. Virol. Methods 142:204–212 [DOI] [PubMed] [Google Scholar]
- 20. Monsonego J, et al. 2011. Evaluation of oncogenic human papillomavirus RNA and DNA tests with liquid-based cytology in primary cervical cancer screening: the FASE study. Int. J. Cancer 129:691–701 [DOI] [PubMed] [Google Scholar]
- 21. Naucler P, et al. 2009. Efficacy of HPV DNA testing with cytology triage and/or repeat HPV DNA testing in primary cervical cancer screening. J. Natl. Cancer Inst. 101:88–99 [DOI] [PubMed] [Google Scholar]
- 22. Naucler P, et al. 2007. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N. Engl. J. Med. 357:1589–1597 [DOI] [PubMed] [Google Scholar]
- 23. Paolini F, et al. 2011. High risk human papillomavirus genotyping in clinical samples: evaluation of different commercial tests. Int. J. Immunopathol. Pharmacol. 24:127–138 [DOI] [PubMed] [Google Scholar]
- 24. Poljak M, et al. 2009. The Abbott RealTime High Risk HPV test: comparative evaluation of analytical specificity and clinical sensitivity for cervical carcinoma and CIN 3 lesions with the Hybrid Capture 2 HPV DNA test. Acta Dermatovenerol. Alp. Panonica Adriat. 18:94–103 [PubMed] [Google Scholar]
- 25. Poljak M, et al. 2011. Comparison of clinical and analytical performance of the Abbott RealTime High Risk HPV test to the performance of Hybrid Capture 2 in population-based cervical cancer screening. J. Clin. Microbiol. 49:1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ronco G, et al. 2010. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 11:249–257 [DOI] [PubMed] [Google Scholar]
- 27. Sankaranarayanan R, et al. 2009. HPV screening for cervical cancer in rural India. N. Engl. J. Med. 360:1385–1394 [DOI] [PubMed] [Google Scholar]
- 28. Stoler MH, et al. 2011. High-risk human papillomavirus testing in women with ASC-US cytology: results from the ATHENA HPV study. Am. J. Clin. Pathol. 135:468–475 [DOI] [PubMed] [Google Scholar]
- 29. Szarewski A, et al. 2008. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol. Biomarkers Prev. 17:3033–3042 [DOI] [PubMed] [Google Scholar]
- 30. Tsoumpou I, et al. 2009. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat. Rev. 35:210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walboomers JM, et al. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12–19 [DOI] [PubMed] [Google Scholar]
- 32. Wentzensen N, et al. 2005. Evaluation of a nuclear score for p16INK4a-stained cervical squamous cells in liquid-based cytology samples. Cancer 105:461–467 [DOI] [PubMed] [Google Scholar]
- 33. Wentzensen N, Bergeron C, Cas F, Vinokurova S, von Knebel Doeberitz M. 2007. Triage of women with ASCUS and LSIL cytology: use of qualitative assessment of p16INK4a positive cells to identify patients with high-grade cervical intraepithelial neoplasia. Cancer 111:58–66 [DOI] [PubMed] [Google Scholar]