Abstract
Rapid assays are still needed to detect rifabutin (RFB) susceptibility for proper tuberculosis treatment. To assess the use of the GenoType MTBDRplus assay and subsequent rpoB gene sequencing on detection of RFB susceptibility, we analyzed 800 multidrug-resistant Mycobacterium tuberculosis isolates, and 13% (104/800) were RFB susceptible. Of the 104 RFB-susceptible isolates, 71 (68.3%) isolates were rapidly identified using two molecular assays, while the remaining isolates could be determined using conventional drug-susceptibility testing according to the clinician's decision.
TEXT
Rifamycins are a group of antibiotics that belong to a subclass of the large family ansamycin. They interact with the β-subunit of the bacterial RNA polymerase encoded by the rpoB gene, inhibit DNA-dependent RNA synthesis, and are particularly effective against mycobacteria. Two potent rifamycin derivatives, rifampin (RIF) and rifabutin (RFB), are used as effective antibiotics for treatment of tuberculosis (TB). The cross-resistance rate between the two rifamycin derivatives is high (1, 10, 11, 12). However, RFB might be still potent against certain multidrug-resistant (MDR) Mycobacterium tuberculosis strains (with MDR defined as resistant to at least isoniazid [INH] and RIF) and is an alternative to RIF for TB patients with serious side effects during treatment or coinfected with HIV (8).
RFB is not included in a routine conventional first-line drug susceptibility testing (DST) for M. tuberculosis isolates, and subsequent RFB susceptibility testing of MDR isolates needs an additional 3 to 4 weeks. Several molecular tests have been applied to detect RIF resistance, such as the GenoType MTBDRplus assay and the INNO-LiPA Rif assay (3, 7). The GenoType MTBDRplus assay, a line-probe assay endorsed by World Health Organization in 2008, can detect M. tuberculosis complex and resistance to INH and RIF (5). The GenoType MTBDRplus assay has been successfully applied to detection of culture samples since 2007 and clinical specimens since 2010 in Taiwan. Moreover, D516V mutation of the rpoB gene has been confirmed to confer resistance specifically to RIF in the commercial INNO-LiPA Rif assay (11, 13). The GenoType MTBDRplus assay also has a probe targeting D516V that might be used for identification of RFB susceptibility. However, the feasibility of the GenoType MTBDRplus assay to simultaneously identify RFB-susceptible isolates was not known and was thus evaluated in this study.
MDR isolates.
We analyzed 800 MDR M. tuberculosis isolates collected from clinical mycobacteriology laboratories from January 2006 to December 2010. RFB and first-line DST were performed using the agar proportion method on either Middlebrook 7H10 or 7H11 according to standardized protocols (9).
rpoB gene sequencing.
The rpoB gene was amplified with primers rpoB-F (5′-TCG GCG AGC CCA TCACGT CG-3′) and rpoB-R (5′-GCG TAC ACC GAC AGC GAG CC-3′), which yielded a 541-bp fragment containing the hot spot region (6). In addition, A 365-bp fragment targeting the V146F mutation (V176F according to the M. tuberculosis numbering system) was amplified and sequenced with the primers TB-176-F (5′-CTT CTC CGG GTC GAT GTC GTT G-3′) and TB-176-R (5′-CGC GCT TGT CGA CGT CAA ACT C-3′) (4). PCR products were sequenced, and data were assembled and edited thereafter.
The GenoType MTBDRplus assay.
The GenoType MTBDRplus assay was performed according to the instructions provided by the manufacturer (Hain Lifescience GmbH, Nehren, Germany) (2). Eight wild-type (wt) probes and four mutation (mut) probes were used to determine the resistance to RIF. The probes for detecting target sequences were as follows: wt1 (codons 505 to 509), wt2 (codons 510 to 513), wt3 (codons 513 to 517), wt4 (codons 516 to 519), wt5 (codons 518 to 522), wt6 (codons 522 to 525), wt7 (codons 526 to 529), wt8 (codons 530 to 533), mut1 (codon D516V), mut2A (codon H526Y), mut2B (codon H526D), and mut3 (codon S531L). Due to overlapping design of probes for detection of mutations at codons 513, 516, and 522, continuous absence of each of the wt2 and wt3, wt3 and wt4, and wt5 and wt6 pairs could represent a single mutation. To clarify the specific pattern and its usage as a marker for detecting RFB susceptibility, we defined group 1 isolates as those missing only wt2, only wt3 and wt4, or only wt5 and wt6, while group 2 was designated as containing isolates simultaneously missing wt7, mut2A, and mut2B (Table 1).
Table 1.
Correlations between specific mutations of the rpoB genes and patterns of the GenoType MTBDRplus assay for identification of RFB-susceptible isolatesa
| Mutation codon no. | Codon | Amino acid change | No. of isolates | No. (%) of RFB-resistant isolates | Pattern by GenoType MTBDRplus assay |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt1 | wt2 | wt3 | wt4 | wt5 | wt6 | wt7 | wt8 | mut1 | mut2 | mut3 | mut4 | |||||
| 143 | CGT/TGT | R→C | 1 | 0 (0) | ||||||||||||
| 146 | GTC/TTC | V→F | 6 | 6 (100) | ||||||||||||
| 511 | CTG/CCG | L→P | 3 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
| 513 | CAA/AAA | Q→K | 10 | 10 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAA/CTA | Q→L | 4 | 4 (100) | ■ | ■ | ■ | ■ | ■ | ■ | |||||||
| CAA/GAA | Q→E | 1 | 1 (100) | ■ | ■ | ■ | ■ | ■ | ■ | |||||||
| CAA/CCA | Q→P | 7 | 7 (100) | ■ | ■ | ■ | ■ | ■ | ■ | |||||||
| 516 | GAC/TAC | D→Y | 14 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| GAC/GTC | D→V | 22 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| GAC/TTC | D→F | 8 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | |||||||
| 522 | TCG/TTG | S→L | 5 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| 526 | CAC/CGC | H→R | 18 | 18 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
| CAC/TAC | H→Y | 53 | 53 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
| CAC/GAC | H→D | 32 | 32 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
| CAC/CAA | H→Q | 2 | 2 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAC/CCC | H→P | 1 | 1 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAC/TGC | H→C | 2 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAC/CTC | H→L | 11 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAC/ACC | H→T | 1 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| CAC/AAC | H→N | 4 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| 529 | CGA/CTA | R→L | 1 | 0 (0) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
| 531 | TCG/TTG | S→L | 491 | 491 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||
| TCG/TGG | S→W | 16 | 16 (100) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||||
| 533 | CTG/CCG | L→P | 27 | 8 (29.6) | ■ | ■ | ■ | ■ | ■ | ■ | ■ | |||||
Shading highlights mutations that confer both RIF and RFB resistance.
Of the 800 MDR M. tuberculosis isolates, 104 (13%) RIF-resistant but RFB-susceptible isolates were identified. In addition, we identified 740 isolates with a single mutation, 41 with multiple mutations, 6 with deletions, and 13 without mutation at the beginning region or at the 81-bp hot spot region of the rpoB gene. Of the 740 MDR M. tuberculosis isolates with a single mutation in the rpoB gene, 91 (12.3%) were susceptible to RFB. Of the 91 RFB-susceptible isolates, 52 (57.1%) isolates with single mutations at codons 511, 516, and 522 can be confirmed directly using the GenoType MTBDRplus assay (Table 1). However, isolates with multiple mutations or deletions might be incorrectly interpreted as RFB susceptible (Table 2). Of the 47 isolates with multiple mutations and deletions, 21 isolates would not be misinterpreted as isolates with only a single mutation, including 12 isolates that had an absence of multiple probes at a discontinuous position, four isolates that were missing probes wt7 and wt8, two isolates missing probes wt3 to wt6, one isolate missing probes wt1 and wt2, one isolate missing probes wt1 to wt4, and one isolate missing probes wt4 and wt5. Nevertheless, 24 isolates were misinterpreted as isolates with single mutations and two isolates, including one with V144A and V146F and another with V146F and S164P, were misinterpreted as wild-type isolates (Table 2).
Table 2.
Multidrug-resistant Mycobacterium tuberculosis isolates with triple mutations, double mutations, or deletion in the rpoB gene, their patterns of the GenoType MTBDRplus assay, and possibility of misinterpretation as patterns for single mutation
| Mutated codons | No. of MDR isolates | No. of RFB-susceptible isolates | Pattern of the GenoType MTBDRplus assay |
Misinterpretation of mutation site(s) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt1 | wt2 | wt3 | wt4 | wt5 | wt6 | wt7 | wt8 | mut1 | mut2A | mut2B | mut3 | ||||
| Triple mutations | |||||||||||||||
| A501T, H526R, R529Q | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 526 or 529 only | ||||||
| Double mutations | |||||||||||||||
| V146A, L533P | 3 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 531 or 533 only | ||||||
| T480I, S531L | 3 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 531 or 533 only | ||||||
| L533P, E562A | 2 | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 531 or 533 only | |||||
| D444V, S531L | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 531 or 533 only | ||||||
| S531L, I561V | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 531 or 533 only | ||||||
| Q513E, M558K | 2 | ■ | ■ | ■ | ■ | ■ | ■ | 513 only | |||||||
| E458K, Q513L | 1 | ■ | ■ | ■ | ■ | ■ | ■ | 513 only | |||||||
| L511P, M515L | 1 | 1 | ■ | ■ | ■ | ■ | ■ | ■ | 513 only | ||||||
| D516Y, Q148R | 2 | ■ | ■ | ■ | ■ | ■ | ■ | 516 only | |||||||
| D516G, I572F | 1 | ■ | ■ | ■ | ■ | ■ | ■ | 516 only | |||||||
| S164P, H526N | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 526 only | ||||||
| L511P, S512G | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 511 only | ||||||
| H526N, L533P | 4 | ■ | ■ | ■ | ■ | ■ | ■ | Direct IDa | |||||||
| N518S, H526P | 1 | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | |||||||
| D516Y, T525I | 1 | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||||
| D516G, L533P | 2 | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||||
| D516Y, L533P | 1 | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||||
| D516N, H526D | 2 | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||||
| D516Y, H526D | 1 | ■ | ■ | ■ | ■ | Direct ID | |||||||||
| D516G, S522W | 2 | 2 | ■ | ■ | ■ | ■ | Direct ID | ||||||||
| L511 silent, H526Y | 1 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||
| L511P, H526N | 2 | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | |||||||
| L511R, D516A | 1 | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | |||||||
| L511P, F505L | 1 | 1 | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||
| V144A, V146F | 1 | None | |||||||||||||
| V146F, S164P | 1 | None | |||||||||||||
| Deletion | |||||||||||||||
| 509–511 (or 510–512) | 2 | 2 | ■ | ■ | ■ | ■ | ■ | ■ | ■ | 511 only | |||||
| 517 | 2 | 2 | ■ | ■ | ■ | ■ | ■ | ■ | 516 only | ||||||
| 9 bp at 513–516 | 1 | ■ | ■ | ■ | ■ | Direct ID | |||||||||
| 518 (or 519) | 1 | 1 | ■ | ■ | ■ | ■ | ■ | ■ | Direct ID | ||||||
| Total | 47 | 10 (21.3%) | |||||||||||||
Direct ID, isolates with multiple mutations or deletions could be identified directly by the GenoType MTBDRplus assay.
Only 1% of isolates with codon 531 mutations have other concurrent mutations, while the percentages of isolates with codon 511 or codon 146 were 70% and 45.5%, respectively (Table 3). In addition, 71.4% and 84.6% of isolates with either codon 511 or codon 516 mutations changed their susceptibilities to RFB. With the high multiple-mutation rate and high proportion of the above-mentioned isolates changing their susceptibilities, confirmation of RFB susceptibility by rpoB gene sequencing was recommended.
Table 3.
MDR Mycobacterium tuberculosis isolates with multiple mutations in the rpoB gene and the change in their resistance to RFB
| Mutated codon | No. (%) of isolates with multiple mutations | No. (%) of isolates changing resistance to RFBa |
|---|---|---|
| 146 | 5 (45.5) | 0 |
| 511 | 7 (70.0) | 5 (71.4) |
| 513 | 3 (12.0) | 0 |
| 516 | 13 (22.8) | 11 (84.6) |
| 526 | 13 (9.5) | 7 (53.8) |
| 531 | 5 (1.0) | 0 |
| 533 | 12 (30.8) | ND |
| Total | 58 (7.5) |
All changes shown were from susceptible to resistant. ND, not determined.
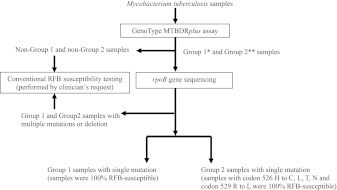
Consequently, we proposed an algorithm for identification of RFB-susceptible M. tuberculosis (Fig. 1). For group 1 samples with a single mutation and group 2 samples with specific single mutations at either codon 526 or 529, 100% of the samples were RFB susceptible. In this study, of the 104 RFB-susceptible isolates, 50% and 18.3% were identified from group 1 samples with a single mutation and group 2 samples with a single mutation at codon 526 (H to C, L, T, or N) and codon 529 (R to L), respectively (Fig. 1). Furthermore, according to clinical decisions or the patient's status, determination of the remaining 31.7% RFB-susceptible isolates could be optionally performed using conventional RFB DST. We have thus demonstrated that a line-probe assay in combination of rpoB gene sequencing could simultaneously identify TB, resistance to INH and RIF, and susceptibility to RFB.
Fig 1.
Proposed flowchart for identification of rifabutin (RFB) susceptibility. In this study, 104 rifabutin-susceptible isolates were identified among 800 multidrug-resistant Mycobacterium tuberculosis isolates. Shown is a suggested algorithm for identification of rifabutin susceptibility. *, group 1 represents samples missing only wt2, only wt3 and wt4, or only wt5 and wt6; **, group 2 represents samples simultaneously missing wt7, mut2A, and mut2B.
ACKNOWLEDGMENTS
This work was supported by grants DOH98-DC-2025 and 101CTP1004-2 from the Centers for Disease Control, Department of Health, Taiwan.
We thank a number of clinical mycobacterial laboratories in Taiwan for submitting Mycobacterium tuberculosis isolates.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Chikamatsu K, Mizuno K, Yamada H, Mitarai S. 2009. Cross-resistance between rifampicin and rifabutin among multi-drug resistant Mycobacterium tuberculosis strains. Kekkaku 84:631–633 [PubMed] [Google Scholar]
- 2. Hain Lifescience GmbH 2009. GenoType MTBDRplus: instruction manual. Hain Lifescience GmbH, Nehren, Germany [Google Scholar]
- 3. Hauck Y, Fabre M, Vergnaud G, Soler C, Pourcel C. 2009. Comparison of two commercial assays for the characterization of rpoB mutations in Mycobacterium tuberculosis and description of new mutations conferring weak resistance to rifampicin. J. Antimicrob. Chemother. 64:259–262 [DOI] [PubMed] [Google Scholar]
- 4. Heep M, Rieger U, Beck D, Lehn N. 2000. Mutations in the beginning of the rpoB gene can induce resistance to rifamycins in both Helicobacter pylori and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:1075–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang WL, Chen HY, Kuo YM, Jou R. 2009. Performance assessment of the GenoType MTBDRplus test and DNA sequencing in detection of multidrug-resistant Mycobacterium tuberculosis. J. Clin. Microbiol. 47:2520–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jou R, Chen HY, Chiang CY, Yu MC, Su IJ. 2005. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis isolates and identification of 11 novel rpoB alleles in Taiwan. J. Clin. Microbiol. 43:1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ling DI, Zwerling AA, Pai M. 2008. Rapid diagnosis of drug-resistant TB using line probe assays: from evidence to policy. Expert Rev. Respir. Med. 2:583–588 [DOI] [PubMed] [Google Scholar]
- 8. Luna-Herrera J, Reddy MV, Gangadharam PR. 1995. In-vitro and intracellular activity of rifabutin on drug-susceptible and multiple drug-resistant (MDR) tubercle bacilli. J. Antimicrob. Chemother. 36:355–363 [DOI] [PubMed] [Google Scholar]
- 9. National Committee for Clinical Laboratory Standards 2000. Susceptibilty testing of mycobacteria, nocardia, and other aerobic actinomycetes, vol 23, no 18 Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA: [PubMed] [Google Scholar]
- 10. Senol G, Erbaycu A, Ozsöz A. 2005. Incidence of cross resistance between rifampicin and rifabutin in Mycobacterium tuberculosis strains in Izmir, Turkey. J. Chemother. 17:380–384 [DOI] [PubMed] [Google Scholar]
- 11. Sintchenko V, Chew WK, Jelfs PJ, Gilbert GL. 1999. Mutations in rpoB gene and rifabutin susceptibility of multidrug-resistant Mycobacterium tuberculosis strains isolated in Australia. Pathology 31:257–260 [DOI] [PubMed] [Google Scholar]
- 12. Uzun M, Erturan Z, Anğ O. 2002. Investigation of cross-resistance between rifampin and rifabutin in Mycobacterium tuberculosis complex strains. Int. J. Tuberc. Lung Dis. 6:164–165 [PubMed] [Google Scholar]
- 13. Yoshida S, et al. 2010. Comparison of rifabutin susceptibility and rpoB mutations in multi-drug-resistant Mycobacterium tuberculosis strains by DNA sequencing and the line probe assay. J. Infect. Chemother. 16:360–363 [DOI] [PubMed] [Google Scholar]