Abstract
As for other chronic viral diseases, quantification of hepatitis delta virus (HDV) loads may be useful for patient management. We describe a one-step quantitative reverse transcription-PCR assay that is reliable and automatable and meets the regulatory authorities' standards for accurate quantification of the major HDV genotypes. It includes an internal control and uses in vitro-transcribed RNAs as standards. Its linearity range is 500 to 1.7 × 1011 copies/ml, its sensitivity is around 150 copies/ml, its repeatability is around 15%, and its reproducibility is below 0.25 log10 copies/ml.
TEXT
Hepatitis delta virus (HDV), a satellite of hepatitis B virus, chronically infects more than 15 million people (8, 18). It causes the most difficult-to-treat form of viral hepatitis (1, 13). As for other chronic viral infections, quantification of HDV loads may improve patient management and help clarify the pathophysiology of HDV infection. Commercial kits accurately quantify only HDV type 1 (HDV-1) genotypes (2), and other previously published in-house methods do not meet the required criteria for diagnostic method accreditation, especially because of the lack of an internal control (IC) (7, 9, 10, 11, 19) or the validation of only HDV-1 genotypes. Here we describe a one-step quantitative reverse transcription-PCR (qRT-PCR) assay that can be automated for the accurate quantification of all of the HDV genotypes that circulate in Europe in the presence of an encapsulated heterologous RNA used as an IC. According to the manufacturer's instructions, the IC is added to each sample before extraction and thus monitors the overall performance of the assay.
Nucleic acids were extracted from 500 μl EDTA-plasma or serum and eluted in 25 μl using NucliSENS easyMAG (bioMérieux, Marcy l'Etoile, France) by following the Generic 2.0.1 protocol. A one-step qRT-PCR was performed with the Quantitect Virus kit (Qiagen, Courtaboeuf, France) as described in the supplemental material on a Rotor-Gene 6000 device (Qiagen). Coamplification of IC and HDV RNAs occurred in the same tube. Detection of the IC was done with the Quasar 670-labeled probe and primers provided in the kit (Simplexa Extraction and Amplification Control Set—RNA; Focus Diagnostics, Cypress, CA, and Eurobio, Les Ulis, France). Two forward primers (AgD-F1, AgD-F2) and a reverse primer (AgD-R) were designed (Table 1) to bind conserved parts of the gene encoding the delta antigen, resulting in a PCR product of 129 bp (1158 to 1287). Design of an appropriate probe proved to be difficult due to the high variability (4) and GC content of the HDV genome. Detection was done with an LNA-Black Hole Quencher 1 (BHQ1) probe from Eurogentec (17) and compared to the result obtained with a TaqMan-minor groove binder (MGB) probe (Applied Biosystems) binding to the same site. Better sensitivity, accuracy, and fluorescence ratios were obtained with the LNA probe than with the TaqMan probe (see Fig. S2 in the supplemental material). Dually labeled probes like TaqMan can work with a few mismatches (6). However, we had to introduce a degenerate position to cover the high diversity of HDV genotypes. LNA nucleotides, chosen to be directed to highly conserved positions of the HDV target, further allowed some polymorphism in less conserved positions (Table 1; see Fig. S1 in the supplemental material). The cycle threshold values for the IC were very stable, at around 26.2 ± 1.7, even for high viral loads. Its presence did not significantly affect HDV quantification compared to that with the assay without an IC (difference of 0.34 ± 0.23 log10 copies/ml). The majority of the previously published qRT-PCR assays for HDV RNA quantification did not include an IC and employed plasmids or cDNAs as calibrators (7, 9, 10, 11, 14). Plasmid-based standard curves usually underrate RNA samples (16). In vitro-transcribed RNAs (HDV-1, -5, -6, -7, and -8) were thus serially diluted in QuantiTect nucleic acid dilution buffer (Qiagen, Courtaboeuf, France) and used as standards. The standard curve generated with HDV-1 standards is shown in Fig. 1. We found a 1-log difference when plasmid standards were used instead of RNA standards (data not shown), a difference also described by Terlizzi et al. (16). Indeed, the use of in vitro-synthesized RNA standards provides the control for both the RT and PCR steps and is thus more reliable than other methods using DNA-based standards.
Table 1.
Sequences of the primers and probes used in this study
| Primer or probe | Sequencea | Locationb |
|---|---|---|
| Primers | ||
| AgD-F1 | 5′CGGGCCGGCTGTTCTTCT3′ | 1158–1175 |
| AgD-F2 | 5′CGGGCCGGCTACTCTTCT3′ | 1158–1175 |
| AgD-R | 5′AAGGAAGGCCCTCGAGAACA3′ | 1287–1268 |
| Probes | ||
| TaqMan-MGB | 5′CTCTTCTTCCTCCYTGCTGA3′ | 1215–1234 |
| LNA-BHQc | 5′CTTCTTCCTCCYTGCTGA3′ | 1217–1234 |
| dLy86 | 5′CTCTTCCTCCTCCGCGCTGA3′ | 1215–1234 |
| dLy20 | 5′CTCTTCTTCTTCCTTGCTCA3′ | 1215–1234 |
| dLy131 | 5′CTCTTCCTCTTCCTTGCTGA3′ | 1215–1234 |
TaqMan-MGB and LNA-BHQ probe sequences were aligned with patients' sequences correctly detected by the LNA-BHQ probe but not by the TaqMan-MGB probe. Boldface letters in the two forward primer sequences indicate differences. Boldface letters in the probe sequences show the differences between the probe and patient sequences.
Numbering refers to the HDV-1 genome (accession number M21012).
LNA nucleotides are underlined.
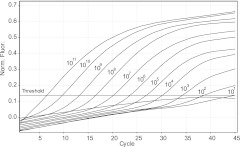
Fig 1.
Typical real-time amplification plot generated from a 10-fold dilution series of the short RNA HDV-1 standard. From left to right, the curves represent 2.7 × 1011 to 2.7 × 101 HDV RNA copies/reaction. The curve that stayed below the threshold corresponds to 2.7 copies/reaction. Norm. Fluor., normalized fluorescence.
A specificity of 100% was achieved (verified with plasma from HDV-negative patients replicating hepatitis B virus [HBV], HIV, hepatitis C virus [HCV], cytomegalovirus, or hepatitis E virus and apparently uninfected patients).
HDV-1 standard RNA at extreme concentrations (3.38 × 100 and 3.38 × 1010 copies/μl) were detected but out of the linear range. Taking into account the dilution factor inherent to the extraction technique (1/50), the dynamic range of HDV RNA quantification (linearity) determined with both RNA transcripts and serum samples extended from 500 to 1.7 × 1011 copies/ml of serum for clinical samples. Standard curves generated within the dynamic range in 15 independent experiments showed a mean R2 value of 0.999 and a PCR efficiency of 97%.
The lower limit of detection (LOD) was established from triplicate analyses of 2 10-fold dilutions of the standard RNA (27 and 2.7 copies/reaction mixture) and determined to be 168 copies/ml. Dilutions of clinical samples confirmed the LOD to be around 150 copies/ml (see Fig. S2 in the supplemental material).
The linearity and accuracy of the assay were verified by using dilutions of patient plasma (see Fig. S2 in the supplemental material) and of HDV transcripts of the HDV-5, -6, -7, and -8 genotypes (see Fig. S3 in the supplemental material). Their theoretical concentrations were compared with those measured based on the HDV-1 standard curve. Measured and expected HDV RNA levels were well correlated (R2 values of 0.999, 0.999, 0.988, and 0.998, respectively) with differences of 0.00 to 0.46 log10 copies/ml. Due to the influx of migrants, more than 30% of the HDV carriers in France were born in Africa and are likely infected with an African strain (HDV-5, -6, -7, or -8) or an African HDV-1 variant. Accurate detection and quantification of all genotypes are mandatory for a gold standard qRT-PCR assay.
Repeatability, evaluated with 20 extractions of serum samples from two patients, yielded coefficients of variation (CVs) of 15 and 15.3% (7.41 ± 0.07 and 5.63 ± 0.07 log10 copies/ml), respectively, with a variation of only 0.07 log10. Results from 15 independent assays of the low-, medium-, and high-level RNA transcripts and one patient's serum varied from 0.03 log for the high-level standard to 0.11 log for the low-level standard (CVs of 21.9, 15.5, 7.4, and 14.7, respectively). This is in the range of commercially available assays for HBV, HCV, or HIV and below the maximum allowed analytical variability of 0.25 log (3).
Samples from 80 patients were quantified using the qRT-PCR with the LNA probe. Viral loads ranged from 2.7 to 10.4 log10 copies/ml, with a median viral load of 6.0 log10 copies/ml. The four highest viral loads corresponded to sera from two young patients and two HIV-positive patients, in accordance with previous observations (15). Two samples had 150 and 500 copies/ml. Twenty-nine patient samples (28.6%) were found to be negative (below the LOD). Among these samples, 25 were also found to be negative with the qualitative assay and were HDV IgM negative. The other four samples were also found to be negative with the qualitative assay but were still positive for anti-HDV-IgM antibodies; two of them had very low hepatitis B surface antigen levels.
In the context of new emerging antiviral molecules, it is crucial to achieve the most sensitive results. Here we found 18 samples out of 76 with quantitative results between the quantification limit (5 × 102 copies/ml) and 105 copies/ml, while the method of Ferns et al., the other in-house assay that includes an IC, found only 1 sample out of 44 below 105 copies/ml (5), placing its sensitivity in question. The target amplified, the antigen delta gene in the present assay versus the highly structured ribozyme in the other one, might explain this discrepancy.
The duplex one-step qRT-PCR assay presented here allows accurate quantification of the major HDV genotypes with a single primer-probe mixture and provides essential features mandatory for diagnostic use (12). These characteristics are wide dynamic range encompassing the viral range of HDV-infected patients, good sensitivity and variability, inclusion of an IC, and a reduced contamination risk due to closed-tube procedures and a one-step reaction. Hepatitis delta is still an orphan hepatitis in terms of knowledge of its pathogenicity, diagnostic tools, and antiviral treatment. The present method could help improve understanding of the pathophysiology and clinical evolution of HDV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Frédérique Fégé, Christine Garrigou, Delphine Laurençot, and Valérie Nevers for excellent technical assistance. We thank Frederic Le Gal, Emmanuel Gordien, Si Ahmed Si Nataf, and Ségolène Brichler for helpful discussions.
We have no conflict of interest concerning this study.
This work was supported by INSERM U851 and U1052, the French National Agency for Research on AIDS and Viral Hepatitis (ANRS), and Hospices Civils de Lyon. P.D. was supported by Assistance Publique—Hôpitaux de Paris and Université Paris 13 (Interface INSERM contract 2007-2011).
Footnotes
Published ahead of print 14 March 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Buti M, et al. 2011. Clinical outcome of acute and chronic hepatitis delta over time: a long-term follow-up study. J. Viral Hepat. 18:434–442 [DOI] [PubMed] [Google Scholar]
- 2. Butt A, Le Gal F, Brichler S, Gordien E. 2011. Hepatitis delta virus RNA quantification by real time rt-PCR techniques: performances of two commercial kits compared to an “in house” consensus technique. International Meeting on the Molecular Biology of Hepatitis B Viruses European Association for the Study of the Liver, Geneva, Switzerland [Google Scholar]
- 3. Ciotti M, Marcuccilli F, Guenci T, Prignano MG, Perno CF. 2008. Evaluation of the Abbott RealTime HBV DNA assay and comparison to the Cobas AmpliPrep/Cobas TaqMan 48 assay in monitoring patients with chronic cases of hepatitis B. J. Clin. Microbiol. 46:1517–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dény P. 2006. Hepatitis delta virus genetic variability: from genotypes I, II, III to eight major clades? Curr. Top. Microbiol. Immunol. 307:151–171 [DOI] [PubMed] [Google Scholar]
- 5. Ferns RB, Nastouli E, Garson JA. 2011. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J. Virol. Methods 179:189–194 [DOI] [PubMed] [Google Scholar]
- 6. Gunson RN, Collins TC, Carman WF. 2006. Practical experience of high throughput real time PCR in the routine diagnostic virology setting. J. Clin. Virol. 35:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofmann J, et al. 2010. Quantitative detection and typing of hepatitis D virus in human serum by real-time polymerase chain reaction and melting curve analysis. Diagn. Microbiol. Infect. Dis. 67:172–179 [DOI] [PubMed] [Google Scholar]
- 8. Hughes SA, Wedemeyer H, Harrison PM. 2011. Hepatitis delta virus. Lancet 378:73–85 [DOI] [PubMed] [Google Scholar]
- 9. Le Gal F, et al. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J. Clin. Microbiol. 43:2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mederacke I, et al. 2010. Establishment of a novel quantitative hepatitis D virus (HDV) RNA assay using the Cobas TaqMan platform to study HDV RNA kinetics. J. Clin. Microbiol. 48:2022–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pollicino T, et al. 2011. Replicative and transcriptional activities of hepatitis B virus in patients coinfected with hepatitis B and hepatitis delta viruses. J. Virol. 85:432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raymaekers M, Smets R, Maes B, Cartuyvels R. 2009. Checklist for optimization and validation of real-time PCR assays. J. Clin. Lab Anal. 23:145–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romeo R, Colombo M. 2011. Treatment of chronic hepatitis delta: mission impossible? Gastroenterology 141:2268–2271 [DOI] [PubMed] [Google Scholar]
- 14. Schaper M, et al. 2010. Quantitative longitudinal evaluations of hepatitis delta virus RNA and hepatitis B virus DNA shows a dynamic, complex replicative profile in chronic hepatitis B and D. J. Hepatol. 52:658–664 [DOI] [PubMed] [Google Scholar]
- 15. Soriano V, et al. 2011. Hepatitis delta in HIV-infected individuals in Europe. AIDS 25:1987–1992 [DOI] [PubMed] [Google Scholar]
- 16. Terlizzi ME, et al. 2011. Improvement of HRV quantification using cRNA-based standards for real time RT-PCR. Mol. Biotechnol. 48:15–18 [DOI] [PubMed] [Google Scholar]
- 17. Vester B, Wengel J. 2004. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43:13233–13241 [DOI] [PubMed] [Google Scholar]
- 18. Wedemeyer H. 2010. Re-emerging interest in hepatitis delta: new insights into the dynamic interplay between HBV and HDV. J. Hepatol. 52:627–629 [DOI] [PubMed] [Google Scholar]
- 19. Yamashiro T, et al. 2004. Quantitation of the level of hepatitis delta virus RNA in serum, by real-time polymerase chain reaction—and its possible correlation with the clinical stage of liver disease. J. Infect. Dis. 189:1151–1157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.