Abstract
To determine the excretion dynamics and genotypic characteristics of rotavirus A (RVA), a longitudinal observational study was performed in 10 pigs from 3 litters at a farrow-to-finish farm. A total of 400 fecal samples were directly collected from the rectums of individual pigs (aged 7 to 217 days) at 3- to 14-day intervals. Seventy-one samples (17.5%) were positive for RVA by reverse transcription-PCR designed to detect the VP7 and VP4 genes. At least 13 combinations of 5 G (G2, G4, G5, G9, and G11) and 6 P (P[6], P[7], P[13], P[23], P[27], and P[34]) genotypes were identified by direct sequencing of the PCR products. We were able to detect RVA VP7 sequences from each pig 4 to 6 times with intervals of 7 to 52 days (from 7 to 119 days of age). Each pig harbored RVAs with at least 3 to 6 different combinations of G and P genotypes, while repeated excretions of RVAs carrying the same combinations of G and P genotypes were also observed. Virus shedding and changes in G and P genotypes appeared to be associated with movement of the pigs into weaning, growing, and finishing barns. These results indicated that, over their lifetimes, pigs raised for meat frequently and intermittently excrete genetically diverse RVAs.
INTRODUCTION
Rotavirus A (RVA) is a major cause of severe diarrhea in children worldwide and is estimated to cause more than 453,000 deaths each year among children aged 5 years or younger (9, 40). Similar to findings in humans, RVA is also one of the most frequently detected enteropathogens associated with acute enteritis among young farm animals such as piglets and calves. RVA infections pose an economic threat to the livestock industry due to increased morbidity and mortality rates, and poor growth performance (8, 20, 35). Although implementing and maintaining appropriate health management practices are obviously crucial for controlling RVA-induced diarrhea in piglets and calves (8, 43), there is a lack of understanding of the viral ecology within these meat animals from birth to harvest.
RVA is a member of the genus Rotavirus, within the family Reoviridae. Its genome consists of 11 segments of double-stranded RNAs encased in a triple-layered capsid. These segments encode six structural (VP1 to VP4, VP6, VP7) and five or sometimes six nonstructural (NSP1 to NSP5) proteins (9). Since the two outer capsid proteins VP7 and VP4 independently induce neutralizing antibodies, they define G and P serotypes, respectively, and play an important role in protective immunity (9). Recently, a new rotavirus classification system, which defines different genotypes for all the 11 genome segments based on the percentage nucleotide (nt) sequence identity in each segment, was proposed (27, 28). According to this scheme, 27 G and 35 P genotypes have been described for RVAs of humans and animals (27). To date, 12 G genotypes (G1 to G6, G8 to G12, and G26) and 13 P genotypes (P[1], P[5] to P[8], P[11], P[13], P[19], P[23], P[26], P[27], P[32], and P[34]) have been detected in pigs (5, 16, 23, 25, 27, 34, 37).
Although five G genotypes (G1 to G4, and G9) and three P genotypes (P[4], P[6], and P[8]) represent the majority of clinically important RVA strains in humans (36), the intensive RVA surveillance associated with introduction of the RVA vaccine into human populations has resulted in the detection of RVAs with unusual genotypes, including those commonly detected in pigs and cattle (14, 19, 21, 29, 38). Animal RVAs are therefore regarded as a potential reservoir for the genetic diversity of human RVAs, and consequently their ecology has been of great concern (24). Further, an increase was been noted in the incidence of porcine RVA diarrhea on large-scale pig farms (7, 22). Understanding the viral ecology within a pig population is crucial for preventing or reducing the incidence of porcine RVA diarrhea as well as transmission of porcine RVAs to humans. However, most studies of porcine RVAs have focused on the genotypes present in suckling and weaned pigs (6, 16, 41), which is not informative about viral ecology and genotypic characteristics when animals are raised on farms over an extended period.
Here, to understand the ecology of RVA infection within a farm over an extended period, we investigated the excretion dynamics and genotypic characteristics of RVAs isolated from animals over their lifetimes on a farrow-to-finish swine farm in Japan.
MATERIALS AND METHODS
Study design.
Pigs were kept at a 900-sow farrow-to-finish farm located in Tochigi Prefecture, Japan, from November 2002 to June 2003. The farm was managed by a continuous-flow production system, which means that the farrowing, weaning, growing, and finishing barns are never been depopulated and that pigs of different ages are sometimes housed together in different pens in the same building. Since pigs are always present, there is no opportunity for total cleaning and disinfecting of the facility (17). Pigs were nursed with their sows in each farrowing pen until weaning at 28 days of age. They were comingled with those from other litters at the time of weaning, transferred into a weaning barn, and distributed into pens in a weaning barn (10 to 12 pigs per pen). Pigs were next transferred into growing and finishing barns at 59 and 101 days of age without rearranging the group or commingling with other pigs. Fences that allowed nose-to-nose contact separated the pens in each barn. To optimize growth and nutrition, diets were formulated to meet the requirements published in the Japanese Feeding Standard for Swine (33) and were modified based on age period (approximate, in days) as follows: early suckling (5 to 28 days), mid-suckling to early weaning (10 to 40 days), late weaning (35 to 65 days), growing (60 to 130 days), and finishing (greater than 120 days). All food was purchased from a commercial source. Pigs had constant access to drinking water.
Ten piglets from three litters (three or four piglets per litter) born on the same day were randomly selected and ear tagged for identification. They were housed together as one group after weaning without commingling of other pigs. Fecal samples were directly collected from the rectum of each pig twice weekly (from 7 to 119 days of age) or once every 2 weeks (after 119 days of age). The samples were stored and shipped at 4°C. After recording fecal consistency, the samples were stored at −80°C until RNA extraction. The Animal Ethical Committee and the Animal Care and Use Committee of National Institute of Animal Health approved all animal experimentation.
Preparation of RNA and RT-PCR.
Total RNA was extracted from fecal suspensions using TRIzol-LS (Invitrogen Corp., Carlsbad, CA) and subjected to reverse transcription-PCR (RT-PCR). A negative control was included for each RNA extraction. The VP7 gene and the VP8* fragments of the VP4 gene were amplified using a Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA) with primer pairs Beg9/End9 and Con2/Con3, respectively (13, 15). A negative control from each RNA extraction, a negative control (Milli-Q water), and a positive control (porcine RVA OSU strain RNA) were included in each run. The amplicons were analyzed by 2% agarose gel electrophoresis and visualized under UV light after ethidium bromide staining. Although the VP4 genes were further reamplified using the TaKaRa Ex Taq kit (TaKaRa Bio Inc., Shiga, Japan) and 1 μl of the RT-PCR product as the template, the VP4 sequences were able to be determined in only 24 of the 71 samples in which VP7 sequences were detected. Therefore, the samples were further analyzed for the VP4 gene using a different primer pair, VP4-13F (5′-GCT TCG CTC ATT TAY AGA C-3′) and VP4-821R (5′-TCT CTA TTA TAT TGC ATT TCT TTC C-3′), which was designed to detect porcine RVAs. RT-PCR conditions were as follows: 50°C for 30 min, 95°C for 15 min, followed by 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min with a final extension at 72°C for 7 min. Reamplification was performed using 1 μl of the RT-PCR product as template as follows: 94°C for 5 min, followed by 30 cycles at 90°C for 30 s, 50°C for 30 s, and 72°C for 1 min with a final extension at 72°C for 7 min.
Nucleotide sequence analysis.
VP7 and VP4 PCR products were purified using MicroSpin S-400 HR columns (GE Healthcare, Uppsala, Sweden) or a QIAquick gel extraction kit (Qiagen, Valencia, CA) and sequenced in both directions on an Applied Biosystems 3100 automated DNA sequencer using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequences were assembled, edited, and analyzed using MEGA 5 software (39). The nucleotide sequences of the VP7 or VP4 genes from the fecal samples were compared with those of reference strains available from GenBank (27). Alignments of multiple sequences were performed using the CLUSTAL W algorithm. Genetic distances were calculated using the Kimura-2 correction parameter, and phylogenetic dendrograms were constructed via the neighbor-joining method with 1,000 bootstrap replications (26).
Statistical analyses.
Statistical analyses for comparison of RVA detection rate in solid feces and soft-to-watery feces were carried out using the chi-square test.
Nucleotide sequence accession numbers.
The VP7 or VP4 genes of RVAs detected in the present study shared more than 98.7% nucleotide identity with those belonging to the same genotype, except for P[13]. We have therefore submitted the nucleotide sequences of 15 representative isolates to GenBank/EMBL/DDBJ under the following strain names and accession numbers (in parentheses): RVA/Pig-wt/JPN/pig2-98d/2003/G11P[7] (VP7, AB701777; VP4, AB701781), RVA/Pig-wt/JPN/pig5-31d/2002/G9P[23] (VP7, AB690412; VP4, AB701782), RVA/Pig-wt/JPN/pig5-88d/2003/G5P[27] (VP7, AB690410; VP4, AB690421), RVA/pig-wt/JPN/pig6-7d/2002/G4P[6] (VP7, AB690413; VP4, AB701783), RVA/Pig-wt/JPN/pig8-115d/2003/G2P[34] (VP7, AB690411; VP4, AB690422), RVA/Pig-wt/JPN/pig8-31d/2002/G9P[6] (VP7, AB701778; VP4, AB701784), RVA/Pig-wt/JPN/pig8-63d/2003/G2P[27] (VP7, AB701779; VP4, AB701785), RVA/Pig-wt/JPN/pig9-28d/2002/G5P[6] (VP7, AB690403; VP4, AB690414), RVA/Pig-wt/JPN/pig9-42d/2002/G5P[13] (VP7, AB690404; VP4, AB690415), RVA/Pig-wt/JPN/pig9-49d/2002/G5P[7] (VP7, AB690405; VP4, AB690416), RVA/Pig-wt/JPN/pig9-59d/2003/G11P[27] (VP7, AB690406; VP4, AB690417), RVA/Pig-wt/JPN/pig9-94d/2003/G11P[13] (VP7, AB690407; VP4, AB690418), RVA/Pig-wt/JPN/pig9-98d/2003/G5P[13] (VP7, AB690408; VP4, AB690419), RVA/Pig-wt/JPN/pig9-112d/2003/G11P[13] (VP7, AB701780; VP4, AB701786), and RVA/Pig-wt/JPN/pig9-115d/2003/G11P[34] (VP7, AB690409; VP4, AB690420).
RESULTS
Detection rate of RVAs.
Of the 400 fecal samples collected from 10 individual pigs from 7 to 217 days of age, 71 (17.8%) were positive for the RVA VP7 gene. The detection rate was highest during the weaning period (37.8%), followed by the growing (20.0%), finishing (6.7%), and suckling (3.3%) periods. The relationship between RVA detection and fecal consistency was evaluated in 340 samples collected during the weaning, growing, and finishing periods. One hundred eleven samples (32.6%) were soft or pasty to watery as shown in Table 1. The highest incidence of soft-to-watery feces was observed during the growing period (57.7%, 75 of 130) followed by the weaning (17.8%, 16 of 90) and finishing (16.7%, 20 of 120) periods. The detection rate of RVA nucleotide sequences was significantly higher in soft-to-watery (20.0%, 4 of 20) than in solid (4.0%, 4 of 100) feces in the finishing period (P < 0.01). RVAs were also detected at higher rates in soft-to-watery than in solid feces in the weaning (56.3%, 9 of 16 versus 33.8%, 25 of 74) and growing (22.7%, 17 of 75 versus 18.2%, 10 of 55) periods, although the differences were not judged to be statistically significant. The relationship between RVA detection and fecal consistency could not be evaluated during the suckling period because the records of fecal consistency were not available for feces collected from 7 to 17 days of age, and only two samples were positive for RVA sequences (Table 1).
Table 1.
Fecal consistency and G and P genotypes of detected RVAs: sampling of 10 pigs from 7 to 217 days of age
| Stage | Age (days) | Piga |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
||||||||||||
| Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | Fecal consist | Genotype | ||
| Suckling | 7 | nr | — | nr | — | nr | — | nr | — | nr | — | nr | G4P[6] | nr | — | nr | — | nr | — | nr | — |
| 10 | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | |
| 14 | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | |
| 17 | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | nr | — | |
| 21 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 24 | N | — | S | GndP[6] | N | — | N | — | N | — | S | — | N | — | N | — | N | — | N | — | |
| Weaning | 28 | S | G9P[6] | N | — | N | G5P[6] | D | — | N | — | D | G5P[6] | N | — | N | — | N | G5P[6] | N | G5P[13] |
| 31 | S | G9P[23] | N | GxP[x] | N | G5P[6] | N | — | N | G9P[23] | S | — | N | — | N | G9P[6] | N | GxP[x] | N | — | |
| 35 | N | — | N | — | N | — | N | — | N | — | N | — | D | G5P[13] | N | — | N | — | D | — | |
| 38 | N | G5P[7] | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 42 | N | G5P[7] | D | G5P[7] | N | G5P[13] | N | G5P[7] | D | — | N | G5P[7] | S | G5P[7] | N | — | N | G5P[13] | S | G5P[13] | |
| 45 | N | GxP[7] | N | G5P[7] | N | GxP[13] | N | G5P[7] | D | GxP[7] | N | G5P[7] | N | G5P[13] | N | G5P[7] | N | — | D | G5P[13] | |
| 49 | N | — | N | — | N | — | S | — | N | — | N | — | N | — | N | G5P[x] | N | G5P[7] | N | — | |
| 52 | N | — | N | — | N | — | N | — | S | — | N | — | N | — | N | — | N | — | N | — | |
| 56 | N | — | N | — | N | G11P[13] | N | — | S | — | N | — | N | — | N | — | N | — | N | G11P[13] | |
| Growing | 59 | N | G11P[34] | N | G11P[ND] | N | G11P[13] | N | G11P[27] | S | — | S | G2P[27] | D | G11P[x] | N | — | N | G11P[27] | D | G11P[27] |
| 63 | N | — | N | — | N | — | D | — | D | — | N | — | S | — | S | G2P[27] | N | — | D | — | |
| 66 | N | — | N | — | N | — | D | G2P[27] | D | — | N | — | N | — | N | — | N | — | N | — | |
| 70 | N | — | N | — | N | — | D | — | D | — | S | — | S | — | S | — | N | — | D | — | |
| 73 | D | — | D | G2P[27] | N | — | S | — | D | — | N | — | S | — | D | — | N | — | D | — | |
| 77 | D | — | D | — | N | — | D | — | D | — | D | — | D | — | D | — | N | — | D | — | |
| 80 | D | — | D | — | N | — | D | — | D | — | D | — | D | — | N | — | N | — | D | — | |
| 84 | N | — | D | — | N | — | D | — | D | — | D | — | D | — | S | — | N | — | D | — | |
| 88 | D | — | N | — | N | — | N | — | D | G5P[27] | D | — | N | — | N | — | D | — | D | — | |
| 92 | D | — | S | — | N | — | D | — | S | — | S | G5P[7] | S | — | D | — | N | — | D | — | |
| 94 | D | — | N | GndP[7] | N | — | N | — | N | — | N | G5P[7] | D | G5P[7] | D | — | N | G11P[13] | D | — | |
| 98 | D | GxP[27] | D | G11P[7] | N | G5P[7] | S | G5P[7] | D | — | D | G5P[7] | D | GxP[7] | D | — | S | G5P[13] | D | GxP[13] | |
| 101 | D | — | N | — | N | — | D | G5P[7] | D | — | N | G5P[7] | N | — | N | — | N | — | N | — | |
| Finishing | 105 | N | — | N | — | D | — | D | G5P[7] | N | — | N | — | N | — | N | — | N | — | N | — |
| 108 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 112 | N | — | N | — | N | — | D | — | D | — | N | — | D | — | D | — | N | G11P[13] | D | — | |
| 115 | N | — | N | — | N | G11P[13] | D | — | D | — | N | — | N | — | D | G2P[34] | N | G11P[34] | N | GxP[ND] | |
| 119 | S | — | S | — | N | — | S | G11P[7] | S | G2P[34] | N | — | N | — | S | — | N | — | S | — | |
| 133 | N | — | N | — | N | — | N | — | D | — | N | — | N | — | S | — | N | — | N | — | |
| 148 | N | — | N | — | N | — | N | — | S | — | N | — | N | — | N | — | N | — | N | — | |
| 161 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 175 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 189 | N | — | N | — | N | — | N | — | D | — | N | — | N | — | N | — | N | — | N | — | |
| 203 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
| 217 | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | N | — | |
Fecal consist, fecal consistency of the sample. Genotype, RVA G and P genotypes. —, Samples were negative for RT-PCR targeting the VP7 and VP4 genes. Gx and P[x], VP7 and VP4 sequences could not be determined because of mixed infection with more than one strain of RVAs. Gnd and P[ND], the VP7 and VP4 nucleotide sequences could not be determined because of unsatisfactory quality of the sequence trace data. Other abbreviations: N, normal; S, soft (boldface); D, pasty to watery (shaded); nr, record unavailable.
Genetic analysis of RVAs.
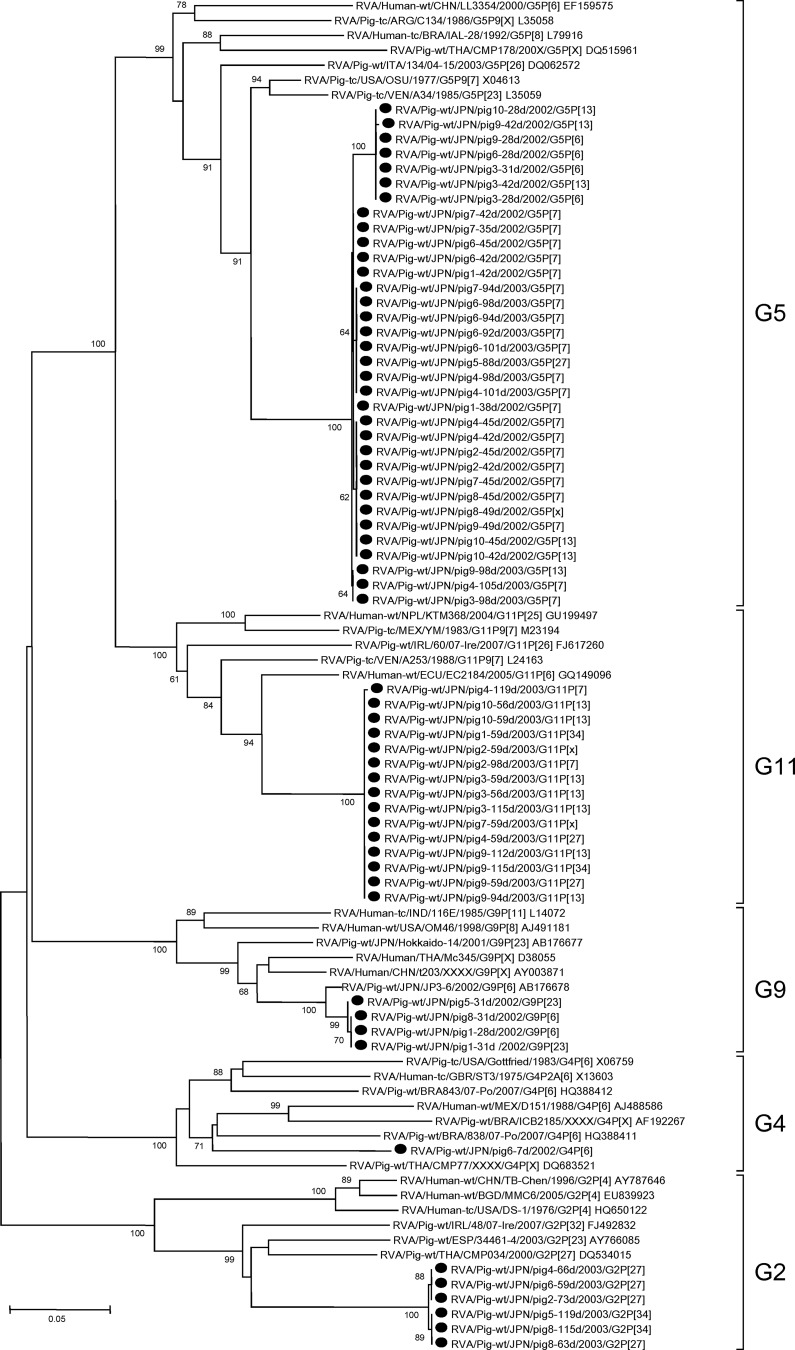
The nucleotide sequences of the VP7 genes (710 nucleotides, corresponding to nucleotides 172 to 881 in the VP7 gene of the porcine G5 OSU strain) were successfully determined for 60 samples. In the remaining samples, VP7 sequences could not be determined due to mixed infection with more than one strain of RVAs (eight samples) or unsatisfactory quality of sequence trace data (three samples). Phylogenic analysis allowed us to classify the 60 RVA sequences as one of five G genotypes (G2, G4, G5, G9, and G11) of 27 established G genotypes (Fig. 1). Nucleotide sequences corresponding to the G5 genotype were detected most frequently (47.9%, n = 34), followed by those with G11 (21.1%, n = 15), G2 (8.5%, n = 6), G9 (5.6%, n = 4), and G4 (1.4%, n = 1) genotypes. The VP7 genes of the isolates belonging to the same G genotype were genetically closely related to each other with more than 99.7% nucleotide (nt) identities to each other, except those of the G5 isolates, which shared 98.7% to 100% nt identities to each other.
Fig 1.
Phylogenetic dendrograms based on partial nucleotide sequences of the VP7 genes from RVA isolates identified in this study and those of reference and selected strains of each G genotype. Bootstrap values above 60% are shown at the branch nodes. The strains identified in this study are shown with black circles. The sequences obtained from GenBank are shown with strain names followed by GenBank accession numbers.
The nucleotide sequences of G5 isolates were most closely related to those of prototype porcine G5 strains OSU and A34 (90.5 to 92.9% nt identities) (Fig. 1). In contrast, the nucleotide sequences of G11 isolates were genetically distant from that of prototype porcine G11 strain YM and were assigned to a separate branch more closely related to the genomes of the human EC2184 and porcine A253 strains (88.6% to 91.7% nt identities) (Fig. 1). The nucleotide sequences of G2 isolates were genetically distant from those of prototype human G2 strains DS-1 and TB-Chen and were assigned to a separate branch related to porcine G2 strains including CMP034 (87.0% to 87.3% nt identities) (Fig. 1). Similarly, the nucleotide sequences of G9 isolates were distant from those of human prototype G9 strains 116E and OM46 and were closely related to those of porcine G9 strain JP3-6 (98.0% to 98.2% nt identity). The nucleotide sequence of the G4 isolate was genetically distant from that of porcine prototype G4 strain Gottfried, and it belonged to a lineage composed of porcine strains BRA838/07-Po and ICB2185 and human strain D151 (83.6% to 85.0% nt identities) (Fig. 1).
Among the 71 samples, the VP8* fragments of VP4 gene were detected using the primer pair Con2/Con3 in 19 samples by RT-PCR and in 7 samples by reamplification of the RT-PCR product. Among them, the VP4 nucleotide sequences could be determined in only 24 samples. The remaining 47 samples were further analyzed using the primer pair VP4-13F/VP4-821R. VP4 genes were detected in 26 samples via RT-PCR and in 21 samples by reamplification of the RT-PCR products. VP4 nucleotide sequences were able to be determined in 41 samples. VP4 nucleotide sequences (663 to 672 nt) corresponding to nt 88 to 762 of the porcine P[7] OSU strain were determined in 65 of the 71 samples. In the remaining six samples, VP4 sequences could not be determined due either to mixed infection with more than one strain of RVA (four samples) or to unsatisfactory quality of the sequence trace data (two samples).
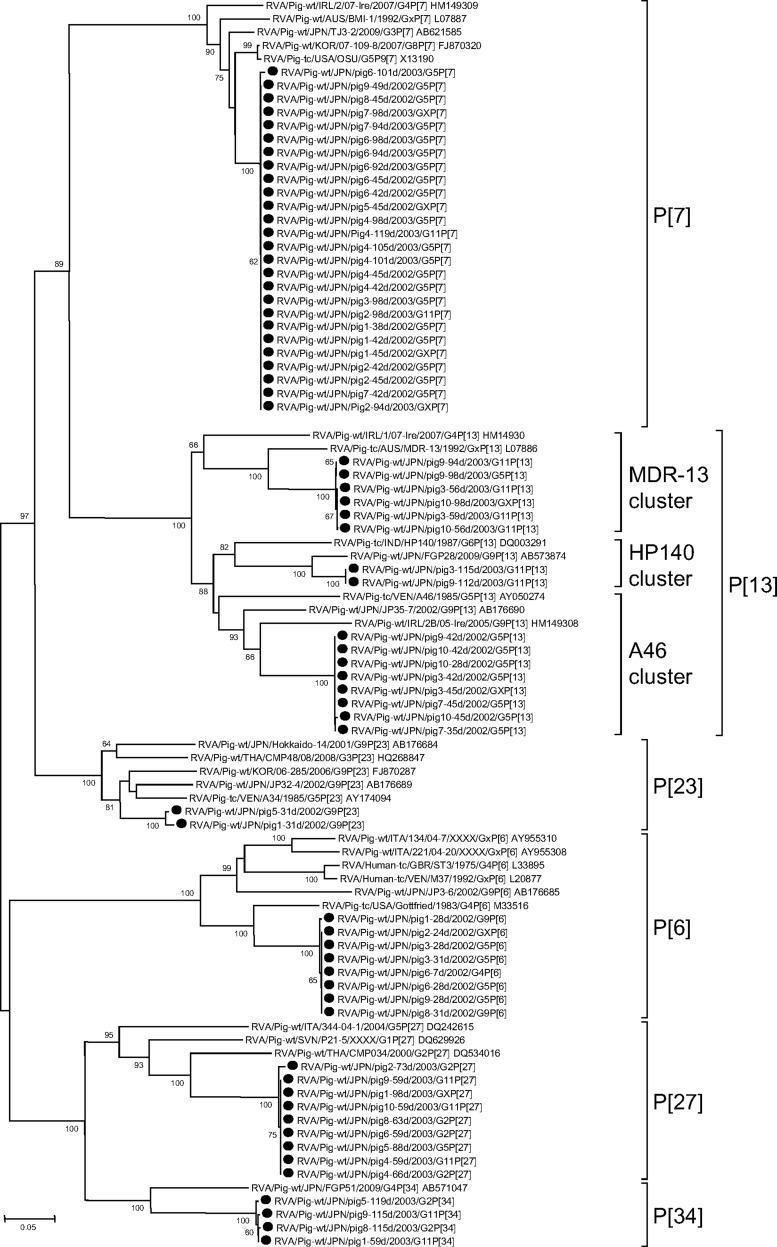
Phylogenic analysis enabled us to classify the 65 RVA VP4 sequences as one of six P genotypes (P[6], P[7], P[13], P[23], P[27], and P[34]) of 35 established P genotypes (Fig. 2). P[7] genotypes were detected most frequently (39.4%, n = 28), followed by P[13] (22.5%, n = 16), P[27] (14.1%, n = 10), P[6] (7.0%, n = 5), P[34] (5.6%, n = 4), and P[23] (2.8%, n = 2). The nucleotide sequences corresponding to the P[6], P[7], P[23], P[27], and P[34] genotypes shared greater than 99.0% nt identities, and each genotype was able to be assigned to a single branch (Fig. 2). In contrast, the P[13] sequences were able to be easily separated into three clusters (MDR-13, HP140, and A46), with nucleotide sequence identities within and between clusters ranging from 99.9% to 100% and from 77.7% to 78.9%, respectively (Fig. 2). The P[13] sequences contained a 6-nt insertion after nt 564 of the porcine OSU VP4 gene. Nucleotides 415 to 417 of the OSU VP4 gene were deleted from P[6] and P[27] sequences (data not shown). None of the VP4 sequences displayed a close genetic relationship with those of human strains (Fig. 2).
Fig 2.
Phylogenetic dendrogram based on the partial nucleotide sequences of the VP8* fragment of VP4 genes from RVA isolates identified in this study and those of reference and selected strains of each genotype. Bootstrap values above 60% are shown at the branch nodes. The strains identified in this study are shown with black circles. The sequences obtained from GenBank are shown with strain names followed by GenBank accession numbers.
Shedding pattern of RVAs.
Each of the pigs started to excrete RVAs at an average age of 28 days old (ranging from 7 to 42 days old) and ended at an average age of 109 days (range: 98 to 119 days). RVA shedding could be detected repeatedly at least 4 to 6 times within 7 to 52 days. The G and P genotypes detected in each pig are summarized in Table 1.
In the suckling period, virus shedding was detected in only one pig at 7 (pig 6, G4P[6]) and 24 (pig 2, P[6] genotype) days of age, respectively.
Within 1 week after weaning, RVA shedding was detected in the feces of seven pigs (pigs 1, 3, 5, and 7 to 10) for the first time and in two pigs (pigs 2 and 6) for the second time. This peak in RVA shedding included combinations of two G (G5 and G9) and three P (P[6], P[13], and P[23]) genotypes. Mixed infection with more than one RVA strain was observed in two pigs (pigs 2 and 9 at 31 days).
We detected a second peak of RVA shedding (from 42 to 45 days of age) soon after the first peak in the same weaning barn. In nine pigs, RVAs had been detected on previous occasions, and in one pig (pig 4), this was the first instance. We detected RVA sequences of G5 together with P[7] or P[13] genotypes in the second peak. Notably, the same G5 genotype RVAs were detected at the first and second peaks in five pigs (pigs 3, 6, 7, 9, and 10). Among them, the same combinations of G and P genotypes were detected in two pigs (G5P[13] in pigs 7 and 10). The P[13] sequences detected in the first and second peaks all belonged to the A46 cluster (Fig. 2). The mixed infection was also observed at this time in four pigs (pigs 1, 3, and 5 at 45 days, and pig 8 at 49 days).
The third peak was characterized by predominant detection of G11 in combination with P[13], P[27], or P[34] genotypes starting from 56 days and peaked at 59 days, which were just before and after their movement into the growing barn. P[13] sequences were detected both at the second and third peaks in pigs 3 and 10, although the P[13] sequences belonged to the MDR-13 cluster and were genetically distant from those detected in the first and second peaks (Fig. 2). G2P[27] genotype RVAs were detected sporadically in the first half of the growing period (59 to 73 days). The mixed infection was observed again at this peak in pig 7 at 59 days.
The fourth peak was characterized by predominant detection of G5P[7] genotypes starting from 88 days and peaking at 98 days—one week before their transfer into the finishing barn. G5P[7] genotypes were detected consistently in the feces of pigs 4 and 6 before they entered the finishing barn and in pig 4 even after its entry. P[7] sequences sharing a close genetic relationship with those identified at the second peak were detected in pigs 2, 4, 6, and 7 again at this peak. Re-excretion of the same G5P[7] genotype was observed in pigs 4, 6, and 7. Other genotypes were identified in pigs 2, 5, and 9 and included G5P[13], G5P[27], G11P[7], and G11P[13]. P[13] sequences were identified in pigs 9 and 10 for the second and fourth times. These P[13] sequences, together with those identified at the third peak, represent a branch in the MDR-13 cluster (Fig. 2). Mixed infection was also observed at this peak in pigs 7 and 10 at 98 days of age.
During the finishing period, G2 and G11 combined with P[7], P[13], and P[34] genotypes were detected sporadically from 112 to 119 days. The P[13] sequences identified in pigs 3 and 9 at these times were genetically distant from those identified at the excretion peaks and represented a separate branch in the HP140 cluster (Fig. 2).
A total of 13 different combinations of G and P genotypes (G2P[27], G2P[34], G4P[6], G5P[6], G5P[7], G5P[13], G5P[27], G9P[6], G9P[23], G11P[7], G11P[13], G11P[27], and G11P[34]) were identified among pigs aged 7 to 119 days. We identified RVAs with at least three to six different combinations of two or three G and two to five P genotypes in individual pigs. Repeated excretion of RVAs with the same G or P genotypes occurred in eight pigs (pigs 2 to 4 and 6 to 10). Among them, six pigs (pigs 3, 4, 6, 7, 9, and 10) excreted RVAs twice with the same combinations of G and P genotypes with intervals ranging from 10 to 56 days.
DISCUSSION
The zoonotic potential of porcine RVAs has been a great concern (19, 24, 30, 31). Together with continuous monitoring of genotypic characteristics of porcine RVAs, understanding the viral ecology within a pig population is crucial for preventing or reducing the incidence of porcine RVA diarrhea as well as transmission of porcine RVAs to humans. Here, we demonstrate the repeated and intermittent lifetime shedding of genetically diverse RVAs into feces by individual pigs raised for meat production in a longitudinal observational study.
In the present study, virus shedding, as indicated by detection of the nucleotide sequences of RVAs, was observed not only in the suckling and weaning periods but also in the growing and finishing periods. These results differ from those of Fu and Hampson (10, 11), which indicate that all pigs become infected with RVA before 40 days of age, although viral antigens are not detected in pigs over 2 months old. This discrepancy in findings may be due to a number of factors, such as differences in the sensitivity of the methods used for virus detection. RT-PCR detection of the RVA genome has been reported to be 100 to 1,000 times more sensitive than a commercial enzyme-linked immunosorbent assay (ELISA) (18). We analyzed the virus excretion using the RT-PCR targeting the VP7 gene, whereas ELISA was employed by Fu and Hampson (10, 11). Steyer et al. reported that RVA could be detected in 18.9% of fattening pigs aged more than 70 days using RT-PCR (38), and Benfield et al. demonstrated RVA excretion by isolating viruses from the feces of sows before and after farrowing (3). Taken together, the present and previous findings indicate that RVA infections can indeed occur in pigs throughout their lifetimes.
Note that we were able to detect VP4 genes via RT-PCR using the primer pair Con2/Con3 in only 26 of the 71 samples in which VP7 genes were detected. Although the reamplification of the RT-PCR product using the same primer pair increased the detection rate, VP4 genes could not be amplified in 45 of the 71 samples, and VP4 nucleotide sequences could not be determined in 47 samples. By designing new primer pair VP4-13F/VP4-821R targeting porcine RVAs and reamplifying the RT-PCR products, we could amplify VP4 genes in all 47 samples, which enabled us to determine the nucleotide sequences of 41 additional samples. Because the Con2/Con3 primer pair was designed to detect and genotype human RVAs by heminested PCR (13), it might not be suitable for detecting porcine RVAs in which a variety of P genotypes have been identified.
Here, our genetic analysis of the excreted RVAs identified a variety of genotypes, including at least 5 G (G2, G4, G5, G9, and G11) and 6 P (P[6], P[7], P[13], P[23], P[27], and P[34]) genotypes and 13 different combinations (G2P[27], G2P[34], G4P[6], G5P[6], G5P[7], G5P[13], G5P[27], G9P[6], G9P[23], G11P[7], G11P[13], G11P[27], and G11P[34]). Our results, as well as those of others (2, 6, 32), clearly indicate significant genetic diversity of RVAs within a pig herd. Although such diversity may have resulted from the introduction of new RVA strains into a farm, our previous report indicated the additional possibility of genetic reassortment between RVA strains within the herd (32). In the present study, the excreted RVA strains belonging to the same G or P genotypes had highly similar VP7 or VP4 genes (except P[13]), while some of them combined with different P or G genotypes. These results suggest that gene reassortment occurred within the farm. However, we were not able to rule out the possibility of mixed infection with more than one RVA strain. Indeed, the samples in which the VP7 or VP4 nucleotide sequences or both could not be determined because of mixed infections were observed concurrently with the RVA excretion peaks. Further, each excretion peak involved RVAs consisting of several combinations of G and P genotypes. These findings suggest that mixed infection occurs frequently throughout the lifetime of the pigs and that the genetic diversity of porcine RVAs within a herd might be maintained thorough mixed infections and subsequent genome reassortment events.
Although recurrent excretion of RVA from pigs has been reported (4, 10), whether or not this represents reinfection or persistent infection at undetectable titers remains unclear. Repeated and intermittent excretion was also observed in the present study, and our genetic analysis of excreted RVAs indicated that this could have been caused by ingestion of RVAs consisting of different combinations of G and P genotypes. Further, virus excretion and changes in the G and P genotypes tended to occur in association with movement of the pigs into different barns (weaning, growing, and finishing barns), suggesting that such reinfection and changes in G and P genotypes may have resulted from exposure to different RVA strains that persist in each barn (12, 42). The farm described in the present study was managed by a continuous-flow system, and consequently not every pen in the barns could be washed and disinfected to eliminate excreted viruses. Further, viruses could also be transmitted from neighboring pigs, because pens were separated by fences that allowed the pigs to come into direct contact. The frequency of exposure and excretion of RVAs within a pig herd might therefore be reduced by changing the management system and structure of the pens.
Recently, asymptomatic RVA infections in pigs and cattle have caused concern, as RVAs are excreted from subclinically infected animals and can cause a diarrheal outbreak in the herd (1, 6, 38). Further, such RVAs may also represent a source of zoonotic transmission (38). Our present findings indicated that asymptomatic infections could occur in a pig population more frequently than we estimated based on the studies by Fu and Hampson (10, 11). Although we did not isolate viruses and assess their infectivity or pathogenicity in our present study, Abe et al. (1) and Collins et al. (6) successfully isolated RVAs from asymptomatic animals. Taken together, these findings indicate the need for greater focus on preventing direct transmission of RVAs from animals to humans that are in contact with livestock to reduce the occurrence of gene reassortment events that could result in efficiently replicating animal-human reassortant strains in humans. In addition, as indicated by Steyer et al. (38), regarding the wide use of livestock manure as fertilizer and the stability of RVAs in the environment, transmission of animal RVAs via environmental samples such as water sources and raw food should also be assessed.
In conclusion, we demonstrate here the repeated and intermittent lifetime shedding of genetically diverse RVAs into feces by individual pigs raised for meat production. We believe that our data illuminate one set of mechanisms influencing the distribution of RVA in a swine population.
ACKNOWLEDGMENTS
We thank Hidetoshi Ikeda and Koshi Yamamoto for providing the fecal samples and information about the farm's history. We also thank Itsuro Yamane for advice on statistical analysis and Hiroko Kunifuda for technical support.
This research was supported in part by a research grant from the National Institute of Animal Health, Japan.
Footnotes
Published ahead of print 4 April 2012
REFERENCES
- 1. Abe M, et al. 2009. Molecular epidemiology of rotaviruses among healthy calves in Japan: isolation of a novel bovine rotavirus bearing new P and G genotypes. Virus Res. 144:250–257 [DOI] [PubMed] [Google Scholar]
- 2. Barreiros M, Alfieri A, Alfieri A, Medici K, Leite J. 2003. An outbreak of diarrhoea in one-week-old piglets caused by group A rotavirus genotypes P[7],G3 and P[7],G5. Vet. Res. Commun. 27:505–512 [DOI] [PubMed] [Google Scholar]
- 3. Benfield DA, Stotz I, Moore R, McAdaragh JP. 1982. Shedding of rotavirus in feces of sows before and after farrowing. J. Clin. Microbiol. 16:186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernard S, Jestin A. 1985. Rotavirus infections in conventional pigs-kinetics excretion in feces of rotavirus antigens, antibodies and immune-complexes by pigs from birth up to 3 months of age. Zentralbl. Veterinarmed. B 32:306–315 [DOI] [PubMed] [Google Scholar]
- 5. Collins PJ, Martella V, Buonavoglia C, O'Shea H. 2010. Identification of a G2-like porcine rotavirus bearing a novel VP4 type, P[32]. Vet. Res. 41:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins PJ, Martella V, Sleator RD, Fanning S, O'Shea H. 2010. Detection and characterisation of group A rotavirus in asymptomatic piglets in southern Ireland. Arch. Virol. 155:1247–1259 [DOI] [PubMed] [Google Scholar]
- 7. Dewey C, Carman S, Pasma T, Josephson G, McEwen B. 2003. Relationship between group A porcine rotavirus and management practices in swine herds in Ontario. Can. Vet. J. 44:649–653 [PMC free article] [PubMed] [Google Scholar]
- 8. Dhama K, Chauhan RS, Mahendran M, Malik SVS. 2009. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 33:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Estes MK, Kapikian AZ. 2007. Rotaviruses, p 1917–1973 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 10. Fu Z, Hampson D. 1987. Group-A rotavirus excretion patterns in naturally infected-pigs. Res. Vet. Sci. 43:297–300 [PubMed] [Google Scholar]
- 11. Fu Z, Hampson D. 1989. Natural transmission of group-A rotavirus within a pig-population. Res. Vet. Sci. 46:312–317 [PubMed] [Google Scholar]
- 12. Fu Z, Hampson D, Blackmore D. 1989. Detection and survival of group a rotavirus in a piggery. Vet. Rec. 125:576–578 [PubMed] [Google Scholar]
- 13. Gentsch JR, et al. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gentsch JR, et al. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis. 192(Suppl. 1):S146–S159 [DOI] [PubMed] [Google Scholar]
- 15. Gouvea V, et al. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halaihel N, et al. 2010. Enteric calicivirus and rotavirus infections in domestic pigs. Epidemiol. Infect. 138:542–548 [DOI] [PubMed] [Google Scholar]
- 17. Harris DL. 2000. Introduction, p 3–36 In Multi-site pig production, 1st ed Iowa State University Press, Ames, IA [Google Scholar]
- 18. Husain M, Seth P, Broor S. 1995. Detection of group-A rotavirus by reverse-transcriptase and polymerase chain-reaction in feces from children with acute gastroenteritis. Arch. Virol. 140:1225–1233 [DOI] [PubMed] [Google Scholar]
- 19. Iturriza-Gómara M, et al. 2011. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol. Infect. 139:895–909 [DOI] [PubMed] [Google Scholar]
- 20. Katsuda K, Kohmoto M, Kawashima K, Tsunemitsu H. 2006. Frequency of enteropathogen detection in suckling and weaned pigs with diarrhea in Japan. J. Vet. Diagn. Invest. 18:350–354 [DOI] [PubMed] [Google Scholar]
- 21. Khoury H, Ogilvie I, El Khoury AC, Duan Y, Goetghebeur MM. 2011. Burden of rotavirus gastroenteritis in the Middle Eastern and North African pediatric population. BMC Infect. Dis. 11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linares RD, et al. 2009. Frequency of group A rotavirus in piglet stool samples from non-vaccinated Brazilian pig herds. Braz. Arch. Biol. Technol. 52:63–68 [Google Scholar]
- 23. Maneekarn N, et al. 2006. Detection of rare G3P[19] porcine rotavirus strains in Chiang Mai, Thailand, provides evidence for origin of the VP4 genes of Mc323 and Mc345 human rotaviruses. J. Clin. Microbiol. 44:4113–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet. Microbiol. 140:246–255 [DOI] [PubMed] [Google Scholar]
- 25. Martella V, et al. 2007. Identification of group A porcine rotavirus strains bearing a novel VP4 (P) genotype in Italian swine herds. J. Clin. Microbiol. 45:577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matthijnssens J, et al. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 82:3204–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matthijnssens J, et al. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156:1397–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matthijnssens J, et al. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 153:1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matthijnssens J, et al. 2009. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J. Virol. 83:2917–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matthijnssens J, et al. 2010. Reassortment of human rotavirus gene segments into G11 rotavirus strains. Emerg. Infect. Dis. 16:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Midgley SE, et al. 2011. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet. Microbiol. doi:10.1016/j.vetmic.2011.10.027 [DOI] [PubMed] [Google Scholar]
- 32. Miyazaki A, et al. 2011. Genetic diversity of group A rotaviruses associated with repeated outbreaks of diarrhea in a farrow-to-finish farm: identification of a porcine rotavirus strain bearing a novel VP7 genotype, G26. Vet. Res. 42:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. National Agriculture and Bio-oriented Research Organization 2005. Japanese feeding standard for swine. (In Japanese.) Japan Livestock Industry Association, Tokyo, Japan [Google Scholar]
- 34. Parra GI, et al. 2008. Phylogenetic analysis of porcine rotavirus in Argentina: increasing diversity of G4 strains and evidence of interspecies transmission. Vet. Microbiol. 126:243–250 [DOI] [PubMed] [Google Scholar]
- 35. Saif LJ, Fernandez FM. 1996. Group A rotavirus veterinary vaccines. J. Infect. Dis. 174(Suppl. 1):S98–S106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29–56 [DOI] [PubMed] [Google Scholar]
- 37. Steyer A, et al. 2007. Molecular characterization of a new porcine rotavirus P genotype found in an asymptomatic pig in Slovenia. Virology 359:275–282 [DOI] [PubMed] [Google Scholar]
- 38. Steyer A, Poljsak-Prijatelj M, Barlic-Maganja D, Marin J. 2008. Human, porcine and bovine rotaviruses in Slovenia: evidence of interspecies transmission and genome reassortment. J. Gen. Virol. 89:1690–1698 [DOI] [PubMed] [Google Scholar]
- 39. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tate JE, et al. 2012. The WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 12:136–141 [DOI] [PubMed] [Google Scholar]
- 41. Teodoroff TA, et al. 2005. Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains. J. Clin. Microbiol. 43:1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Woode G. 1978. Epizootiology of bovine rotavirus infection. Vet. Rec. 103:44–46 [DOI] [PubMed] [Google Scholar]
- 43. Yuan L, Stevenson WG, Saif LJ. 2006. Rotavirus and reovirus, p 435–454 In Straw EB, Zimmerman JJ, D'Allaire S, Taylor JD. (ed), Diseases of swine, 9th ed Blackwell Publishing Professional, Ames, IA [Google Scholar]