Abstract
Candida pseudorugosa is a novel species closely related to Candida rugosa for which only one case has been reported. We report the first case of a bloodstream infection in humans caused by a Candida sp. closely related to C. pseudorugosa. We contribute evidence to show this organism as a potential human pathogen that may be misidentified by conventional methods, also pointing out its lower sensitivity to azoles and other antifungal agents.
CASE REPORT
The patient was a 49-year-old woman from Buenos Aires, Argentina. She was diagnosed with glioblastoma multiforme and underwent surgery twice in 2007. On 20 June 2008, the patient was operated on for a new tumor at the Hospital de Clínicas “José de San Martín.” During surgery, purulent material over and under the dura mater was observed. This material was collected and sent to the microbiology laboratory to be cultured. Four days later, the patient got a central venous catheter, and empirical antibiotic treatment with a combination of vancomycin (1 g every 12 h) and meropenem (2 g every 8 h) was initiated. Propionibacterium acnes was detected in the purulent material, and the antibiotic treatment was changed to ampicillin, 3 g every 12 h. On 7 July, 17 days after surgery, the patient developed a fever, so two blood samples were taken by venipuncture and the catheter was removed, and all these samples were sent to the microbiology laboratory. Yeasts were detected in the blood samples cultured in Bact/ALERT 3D (bioMérieux, Inc., Durham, NC) and in the catheter. Therefore, empirical antifungal treatment was initiated with 400 mg/day of fluconazole. After 48 h of incubation on CHROMagar Candida (CHROMagar Company, Paris, France), two different colonies were isolated from each sample. One of the colonies was pink, and the other one was blue (Table 1). All isolates were identified as Candida rugosa by ID32C (bioMérieux, Marcy l'Etoile, France). Code numbers for the pink and blue colony isolates were 1300300015 and 1100300015, respectively. In vitro antifungal susceptibilities to fluconazole and voriconazole were determined using an agar diffusion test (Neo-Sensitabs, Rosco, Denmark). The blue isolates were dose-dependent susceptible to fluconazole (17 to 19 mm); the pink isolates were susceptible (24 to 27 mm). All isolates were susceptible to voriconazole (23 to 30 mm). In view of these results, after a 1-week treatment with fluconazole, amphotericin B deoxycholate was administered at doses of 0.7 mg/kg/day for 14 days. After four and ten days of treatment, blood cultures were negative. The patient improved and was discharged. In September 2008, the patient's condition recurred and she died as a result of the new tumor.
Table 1.
Strain features and GenBank accession numbers
| Strain (DMic no.) | Sample | Color in CHROMagar |
GenBank accession no. |
||
|---|---|---|---|---|---|
| First subculture | Successive subcultures at 48 h/after 48 ha | ITS1-5.8S-ITS2 | D1/D2 domain | ||
| 103837 | Catheter | Pink | Green/blue | JF345209 | JF345215 |
| 103838 | Catheter | Blue | Green/blue | JF345210 | JF345216 |
| 103839 | Blood | Pink | Green/blue | JF345211 | JF345217 |
| 103840 | Blood | Blue | Green/blue | JF345212 | JF345218 |
| 103841 | Blood | Pink | Green/blue | JF345213 | JF345219 |
| 103842 | Blood | Blue | Green/blue | JF345214 | JF345220 |
All the green subcultures turned blue after 48 h of incubation.
All yeast isolates were sent to the National Reference Centre, the Mycology Department of the National Institute of Infectious Diseases “Dr. Carlos G. Malbrán,” to be further studied. Isolates were reisolated on CHROMagar Candida to ensure purity and viability. All isolates turned green after incubation at 28°C for 24 to 48 h, but after 48 h they turned blue (Table 1). Morphological, physiological, and biochemical tests were performed according to the standard methodology (42). These characteristics were similar for all isolates, and they were identified as Candida rugosa. The results of the tests were as follows. For morphological characteristics, on YM agar (42) after 10 days at 28°C, the colonies were butyrous, cream colored, wrinkled, and had a wavy margin; on malt extract broth (42) after 3 days at 28°C, the cells became ovoid, ellipsoid, or cylindrical and occurred singly, in pairs, or in chains; in Dalmau plate culture on cornmeal agar (42), abundant pseudohyphae developed after 3 days at 28°C. For physiological and biochemical characteristics, fermentation was absent; glucose, galactose, d-xylose, d-glucosamine, N-acetyl-d-glucosamine, glycerol, manitol, glucitol, dl-lactate, and succinate were assimilated; l-arabinose was strongly or weakly assimilated; maltose and trehalose were not assimilated or weakly assimilated; l-sorbose, sucrose, cellobiose, lactose, melibiose, raffinose, d-arabinose, d-ribose, l-rhamnose, erythritol, galactitol, methyl-α-d-glucoside, salicin, citrate, and myo-inositol were not assimilated; ribitol was variably assimilated; nitrate was not assimilated. The urease test was negative. Isolates grew well at 28, 35, 37, and 42°C but failed to grow at 45°C.
Molecular characterization was performed by sequencing two different variable regions of ribosomal DNA widely used in the identification of yeast (4, 5, 12, 17, 18, 20, 23, 32): the ITS1-5.8s-ITS2 region with primers ITS1/4 (40) and the D1/D2 domain of the 26S rRNA gene with primers NL1/4 (40). Sequences were identical for all isolates (for GenBank accession numbers, see Table 1). The isolates were compared to sequences published in the GenBank database (National Center of Biotechnology Information, National Library of Medicine, Bethesda, MD) using the BLASTN program. The isolates showed 94% similarity for the ITS1-5.8s-ITS2 region (24 nucleotide differences) and 99% similarity for the 26S D1/D2 domain with the XH1164 strain (3 nucleotide differences), GenBank numbers DQ234792.1 and DQ234791.1, respectively. Strain XH1164 (CBS10433) is the type strain of the novel species Candida pseudorugosa, recently described by Li et al. (22). Phenotypic characteristics of this type strain were very similar to those observed in the isolates presented in this study. The isolates also showed 100% and 99% similarity (1 gap difference) for the 26S D1/D2 domain with the C. pseudorugosa-like strains MZKI K-259 and MZKI K-269, which were isolated from subglacial ice by Butinar et al. (3), GenBank numbers EU056285 and EU056286, respectively. On the basis of these data, all isolates were finally identified as Candida spp. closely related to C. pseudorugosa and were introduced in the Culture Collection of the Mycology Department (DMic) (Table 1).
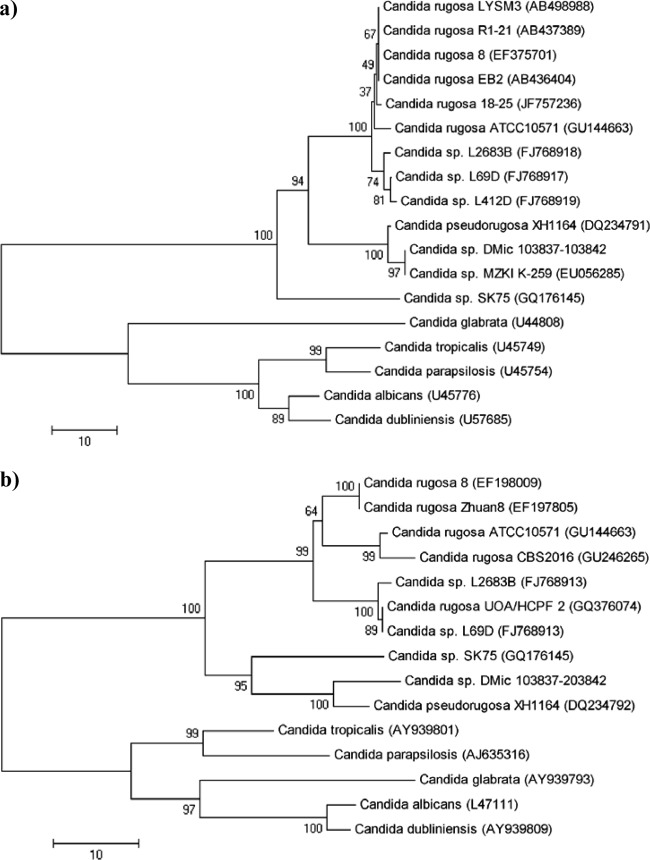
Phylogenetics analyses based on the ITS1-5.8S-ITS2 region and the 26S D1/D2 domain sequences (Fig. 1) were performed by neighbor-joining methodology using the MEGA 5.0 program. The analysis of both regions showed that all our isolates fell in the same group as the C. pseudorugosa XH1164 type strain and the close relationship between this species and C. rugosa.
Fig 1.
Neighbor-joining tree based on the 26S rRNA gene D1/D2 domain (a) and ITS1-5.8S-ITS2 region (b). Bootstrap percentages from 2,000 replicates are shown in each node. Scale bar indicates the number of differences. Reference sequences were retrieved from GenBank under the accession numbers shown in parentheses.
Antifungal susceptibility tests were carried out by determining the MIC according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) E.Def 7.1 reference document (11). All isolates were tested against nine common antifungal drugs (Table 2). According to EUCAST version 3.0 data points, all isolates were resistant to fluconazole. Although there are no breakpoints for the other antifungal agents, given the breakpoints determined for other species, all isolates presented high MIC values against anidulafungin and low MICs values to all other drugs (Table 2).
Table 2.
MICs values of strains isolated from patients
| Strain | MIC (mg/liter)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMB | 5FC | FCZ | ITZ | VCZ | KTZ | ANID | CAS | POS | |
| 103837 | 0.5 | 0.25 | 8 | 0.06 | 0.06 | 0.06 | 0.25 | 2 | 0.03 |
| 103838 | 0.25 | 0.5 | 8 | 0.06 | 0.06 | 0.03 | 0.5 | 1 | 0.03 |
| 103839 | 0.25 | 0.25 | 16 | 0.06 | 0.06 | 0.06 | 0.5 | 1 | 0.03 |
| 103840 | 0.5 | 0.25 | 8 | 0.06 | 0.06 | 0.06 | 0.5 | 1 | 0.03 |
| 103841 | 0.25 | 0.25 | 8 | 0.03 | 0.06 | 0.06 | 0.5 | 2 | 0.06 |
| 103842 | 0.25 | 0.25 | 8 | 0.06 | 0.06 | 0.03 | 0.25 | 1 | 0.06 |
Tests were carried out according to the EUCAST E.Def 7.1 document. AMB, amphotericin B; 5FC, 5-flucytosine; FCZ, fluconazole; ITZ, itraconazole; VCZ, voriconazole; KTZ, ketoconazole; ANID, anidulafungin; CAS, caspofungin; POS, posaconazole.
DMic strain no. 103839 was sent to the Fungal Biodiversity Center, Centraalbureau voor Schimmelcultures (CBS), in The Netherlands for identity confirmation. The strain was identified as a new Candida species based on the sequences of the large subunit (LSU) and the internal transcript spacer (ITS) 1 and 2 regions. The most closely related species was Candida pseudorugosa F. Y. Bai and Juan Li, with a 99% similarity with LSU and a 94% similarity with ITS. The strain was deposited at the CBS culture collection as CBS 12267.
Bloodstream infection (BSI) is the most important infection caused by yeast due to its high mortality rate (7, 8, 10, 33, 41). All over the world, different surveys have reported a rising number of yeast-related BSIs in the last decade (2, 10, 30, 41). Furthermore, Candida species are the fourth most common cause of hospital-acquired bloodstream infection in the United States (10, 27, 30, 41). Although Candida albicans has shown to be the most frequent species isolated from blood cultures (41), an increasing number of BSIs caused by yeast species other than C. albicans have been reported (27, 38, 41). Moreover, an emergence has been observed of invasive candidiasis by less common Candida species, some of which stand out due to their decreased susceptibility to azoles and other antifungal agents (29). Some of these species appear to be more prominent in some geographic regions than in others. For example, Candida guilliermondii and C. rugosa seem to be isolated mainly in Latin America (29); a significant number of C. rugosa cases have also been reported from India (34), and Candida inconspicua and Candida norvegensis seem to be isolated mainly in Europe (29). In Argentina, two national surveillance studies on yeast-related BSIs have shown that more than 50% of these infections were produced by species other than C. albicans (8, 33). Early and accurate identification of yeast species is essential to implement an appropriate treatment and to avoid unnecessary drug exposure in patients having, in general, other comorbidities. Accurate yeast identification is also essential to understand the epidemiology of emerging fungal pathogens.
Candida pseudorugosa was described and reported by Li et al. in 2006 (22). These authors isolated two strains from the sputum of an intensive care unit patient with acute pneumonia. The two isolates described were not susceptible to several commonly used antifungal agents.
All the isolates described in the present report were misidentified when using commercial methods available for clinical laboratories. On CHROMagar Candida medium, the first subculture of the isolates produced pink colonies and blue colonies; all these colonies were green in the subsequent subcultures and turned blue after 48 h (Table 1). This feature of color change during the incubation period has also been observed in the yeast species Kodamaea ohmeri (21). These results may lead to the misidentification of the isolates as Candida albicans/Candida dubliniensis (both green on CHROMagar) or Candida tropicalis (blue on CHROMagar) in this medium. In the same way, different colonial morphologies resembling Candida krusei or C. albicans have been described for C. rugosa on CHROMagar (13, 14). The isolates were also misidentified as C. rugosa by the ID32C commercial method and by the phenotypic standard method. C. pseudorugosa had also been misidentified as C. rugosa (22) by this method, and this new species is not included in the databases of the phenotypic methods.
The assimilation profile of the isolates presented some slight differences with the C. pseudorugosa type strain profile. However, various molecular studies have shown that phenotypic characteristic may vary among strains of a single species and that different species may share phenotypic profiles (16, 35). In contrast, DNA-based identification methods, in particular gene sequencing, offer a “constant” upon which species identification may be based (16). In this sense, sequencing of ribosomal DNA has demonstrated to be capable of identifying almost all pathogenic yeasts (4, 5, 6, 12, 17). Moreover, new species cannot be described without sequence analysis since this analysis provides a more objective separation of genera and species than the standard phenotypic method (9, 31, 36, 37, 39).
In order to accurately identify the isolates, we sequenced the ITS regions and the 26S D1/D2 domain, which are the most widely used regions for Candida species identification. In our comparative analysis, the isolates showed 3 nucleotide differences with C. pseudorugosa type strain XH1164 and zero or one gap difference with the C. pseudorugosa-like strains MZKI K-259 and MZKI K-269. In general, strains of a species show no more than 0 to 3 nucleotide differences (0% to 0.5%) in this domain, and strains showing 6 or more noncontiguous substitutions (1%) are considered separate species (15). However, there are exceptions, and species have been documented which have a >1% intraspecific variability (15, 19). For the ITS regions, our comparative analysis showed a 94% similarity (24 nucleotide differences) with the C. pseudorugosa type strain. Unfortunately, there were no ITS1-5.8S-ITS2 region sequences of the strains MZKI K-259 and MZKI K-269 available in public databases. In the case of ITS regions, Nilsson et al. (24) calculated the weighted intraspecific ITS variability in the fungus kingdom as 2.51% (standard deviation [SD], 4.57%) and the weighted intraspecific ITS variability in the Ascomycota phylum as 1.96% (SD, 3.73%). The wide spread in intraspecific variability that these authors observed testifies to the apparent futility of trying to find a single fungal-wide cutoff value to demarcate intraspecific from interspecific variability (24).
Our results and the CBS identification agree and might indicate that the isolates may belong to a new Candida species. Nevertheless, given that the close relationship observed with the C. pseudorugosa type strain and that there is no more information available about this species for further comparison, we thought that more studies and the analysis of other genes are necessary to resolve whether the isolates belong to a new species or a subspecies.
Phylogenetic trees based on the 26S D1/D2 domain and the ITS regions show similar results (Fig. 1). In both trees, the isolates clustered with the Candida pseudorugosa type strain and show their close relationship to C. rugosa. In addition, the 26S D1/D2 domain tree shows the isolates forming a subgroup with the C. pseudorugosa-like strain MZKI K-259. In the same way, the C. rugosa cluster shows different subgroups. These topologies may indicate that further analyses are necessary to define if those subgroups represent variants of the same species or different species.
Candida rugosa has been cited as a potential emerging fungal pathogen (25) that may be most common in Latin America (28, 29). Pfaller et al. (28) utilized the database of the ARTEMIS DISK Antifungal Surveillance Program to describe 452 isolates of this species. Their findings suggest that this species appears to be developing increased resistance to azole antifungal agents, especially in certain geographic regions, and it most often causes BSI and urinary tract infections in patients hospitalized in the medical and surgical inpatient services. As the authors highlight, one limitation of the survey was that most laboratories employed commercial identification methods to identify the isolates, and these methods may misidentify some species (29).
All the isolates present in this report were resistant to fluconazole; were susceptible to amphotericin B, voriconazole, itraconazole, and posaconazole; and exhibited high MIC values to anidulafungin and caspofungin. Similar in vitro behavior against echinocandins has been reported in some Candida species, such as Candida parapsilosis and Candida guilliermondii (26). For these species with elevated MICs to echinocandins, the clinical significance is unclear and remains to be determined. Although they may respond clinically to treatment, there might be no response under conditions of decreased drug penetration. Thus, the more conservative approach would be to consider these isolates as nonsusceptible to the echinocandins tested since there is insufficient evidence to categorize them as resistant (1).
The C. pseudorugosa isolates described by Li et al. (22) seems to be resistant to amphotericin B, caspofungin, itraconazole, and nystatin; dose dependent to fluconazole; and susceptible to flucytosine and voriconazole. The authors proposed the idea of an intrinsic antifungal-insusceptible property of this species to multiple antifungal agents, since the isolates were recovered from patients without previous antifungal treatment. This could be also the case for our isolates, given that the patient did not receive previous antifungal treatment. It is worthy of note that the other close relative species, C. rugosa, may exhibit acquired resistance to one or more of these antifungal agents commonly used in treatment of the yeast BSIs (28).
We reported the first case of a bloodstream infection in humans caused by a Candida sp. closely related to C. pseudorugosa. This paper provides evidence indicating this organism as a potential human pathogen that may have decreased susceptibility to azoles and other antifungal agents.
Footnotes
Published ahead of print 29 March 2012
REFERENCES
- 1. Arendrup MC, et al. 2011. EUCAST technical note on anidulafungin. Clin. Microbiol. Infect. 17:E18–E20 [DOI] [PubMed] [Google Scholar]
- 2. Beck-Sague C, Jarvis WR. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J. Infect. Dis. 167:1247–1251 [DOI] [PubMed] [Google Scholar]
- 3. Butinar L, Strmole T, Gunde-Cimerman N. 2011. Relative incidence of ascomycetous yeasts in arctic coastal environments. Microb. Ecol. 61:832–843 [DOI] [PubMed] [Google Scholar]
- 4. Chen YC, et al. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen YC, et al. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciardo DE, Schar G, Bottger EC, Altwegg M, Bosshard PP. 2006. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J. Clin. Microbiol. 44:77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colombo AL, et al. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cordoba S, et al. 2011. Species distribution and susceptibility profile of yeast isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev. Argent. Microbiol. 43:176–185 [DOI] [PubMed] [Google Scholar]
- 9. Desnos-Ollivier M, et al. 2008. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 46:3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edmond MB, et al. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244 [DOI] [PubMed] [Google Scholar]
- 11. EUCAST 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405 [DOI] [PubMed] [Google Scholar]
- 12. Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50(Pt 3):1351–1371 [DOI] [PubMed] [Google Scholar]
- 13. Horvath LL, Hospenthal DR, Murray CK, Dooley DP. 2003. Direct isolation of Candida spp. from blood cultures on the chromogenic medium CHROMagar Candida. J. Clin. Microbiol. 41:2629–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hospenthal DR, Murray CK, Beckius ML, Green JA, Dooley DP. 2002. Persistence of pigment production by yeast isolates grown on CHROMagar Candida medium. J. Clin. Microbiol. 40:4768–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurtzman CP. 2006. Yeast species recognition from gene sequence analyses and other molecular methods. Mycoscience 47:65–71 [Google Scholar]
- 16. Kurtzman CP, Piskur J. 2006. Taxonomy and phylogenetic diversity among the yeasts, p 29–46 In Sunnderhagen P, Piskur J. (ed), Comparative genomics: using fungi as models, vol 15 Springer-Verlag, Berlin, Germany [Google Scholar]
- 17. Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371 [DOI] [PubMed] [Google Scholar]
- 18. Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lachance MA, et al. 2003. The D1/D2 domain of the large-subunit rDNA of the yeast species Clavispora lusitaniae is unusually polymorphic. FEMS Yeast Res. 4:253–258 [DOI] [PubMed] [Google Scholar]
- 20. Leaw SN, et al. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee JS, et al. 2007. Kodamaea ohmeri isolates from patients in a university hospital: identification, antifungal susceptibility, and pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 45:1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J, Xu YC, Bai FY. 2006. Candida pseudorugosa sp. nov., a novel yeast species from sputum. J. Clin. Microbiol. 44:4486–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lott TJ, Burns BM, Zancope-Oliveira R, Elie CM, Reiss E. 1998. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr. Microbiol. 36:63–69 [DOI] [PubMed] [Google Scholar]
- 24. Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. 2008. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. Online 4:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nucci M, Marr KA. 2005. Emerging fungal diseases. Clin. Infect. Dis. 41:521–526 [DOI] [PubMed] [Google Scholar]
- 26. Pfaller MA, et al. 2008. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J. Clin. Microbiol. 46:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfaller MA, Diekema DJ. 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10(Suppl. 1):11–23 [DOI] [PubMed] [Google Scholar]
- 28. Pfaller MA, et al. 2006. Candida rugosa, an emerging fungal pathogen with resistance to azoles: geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J. Clin. Microbiol. 44:3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfaller MA, et al. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaller MA, et al. 1998. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. The SENTRY Participant Group. J. Clin. Microbiol. 36:1886–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pincus DH, Orenga S, Chatellier S. 2007. Yeast identification—past, present, and future methods. Med. Mycol. 45:97–121 [DOI] [PubMed] [Google Scholar]
- 32. Pryce TM, Palladino S, Kay ID, Coombs GW. 2003. Rapid identification of fungi by sequencing the ITS1 and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41:369–381 [DOI] [PubMed] [Google Scholar]
- 33. Rodero L, et al. 2005. Multicenter study of fungemia due to yeasts in Argentina. Rev. Argent Microbiol. 37:189–195 [PubMed] [Google Scholar]
- 34. Singh RI, et al. 2011. Epidemiology of candidaemia in critically ill trauma patients: experiences of a level I trauma centre in North India. J. Med. Microbiol. 60:342–348 [DOI] [PubMed] [Google Scholar]
- 35. Suh SO, Blackwell M, Kurtzman CP, Lachance MA. 2006. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1006–1017 [DOI] [PubMed] [Google Scholar]
- 36. Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141(Pt 7):1507–1521 [DOI] [PubMed] [Google Scholar]
- 37. Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trick WE, Fridkin SK, Edwards JR, Hajjeh RA, Gaynes RP. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989–1999. Clin. Infect. Dis. 35:627–630 [DOI] [PubMed] [Google Scholar]
- 39. Vaughan-Martini A, Kurtzman CP, Meyer SA, O'Neill EB. 2005. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5:463–469 [DOI] [PubMed] [Google Scholar]
- 40. White TJ, Bruns T, Lee S, Taylor JW. 1990. PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY [Google Scholar]
- 41. Wisplinghoff H, et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 42. Yarrow D. 1998. Methods for the isolation, maintenance and identification of yeasts, p 77–100 In Kurtzman CP, Fell JW. (ed), The yeasts, a taxonomic study, 4th edition Elsevier, Amsterdam, The Netherlands [Google Scholar]