Abstract
The impact of Structured Treatment Interruption (STI) in peripheral blood mononuclear cell (PBMC) proviral reservoirs in 41 highly active antiretroviral therapy (HAART)-treated viremic individuals at baseline and 12 weeks after STI was determined using quantitative PCR (qPCR). Viral load increased 0.7 log10 and CD4 decreased 97.5 cells/mm3 after 12 weeks. A total of 28 of the 41 individuals showed an increased proviral load, 19 with a statistically significant increase above 10%. An increase in active viral replication is an important factor in the replenishment of the proviral reservoir even for short time periods.
TEXT
The persistent presence of proviral DNA associated with the generation of replication-competent viruses in various cellular compartments has been a major barrier to virologic control of the infection (1, 2). Latent viral reservoirs are established very early during primary infection, and latently infected CD4+ T lymphocytes can serve as a long-term stable reservoir for the virus, with a nearly 44-year half-life (3–5, 8). Structured Treatment Interruption (STI) among individuals harboring replicating resistant HIV strains leads to reemergence of wild-type viruses, with a consequent increase in viral load (VL) (5, 7). We developed a quantitative PCR (qPCR) assay targeting proviral HIV-1 DNA and attempted to determine the impact of STI in the proviral reservoir. A total of 41 samples from patients failing highly active antiretroviral therapy (HAART) were subjected to a 12-week STI. Patients were sampled between 1999 and 2000 at baseline (before STI) and after 12 weeks of STI. Informed written consent was approved by the Institutional Review Board of the Federal University of São Paulo (UNIFESP; 0189/05). Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) (QIAamp blood kit; Qiagen, Valencia, CA). VL was measured using a Cobas Amplicor HIV-1 Monitor, version 1.5 (Roche Molecular Systems, Inc., Branchburg, NJ), and CD4 levels were determined using a Becton, Dickinson FACScan Flow Cytometer (Foster City, CA). The qPCR primers and probe amplified a 202-bp fragment of the integrase gene (bp 4309 to 4511 in HXB2). The albumin gene was amplified according to the method of Désiré et al. (6). The standard curve was generated by a 10-fold serial dilution from 1 × 108 copies of plasmid DNA down to one starting molecule. Amplification was performed using an iCycler (Bio-Rad Laboratories, Hercules, CA). For relative quantifications, the normalized value of the HIV-1 proviral load (HIV PL) was calculated as [HIV copy number/(albumin copy number × 2 × 20)]. The values of HIV-1 PL are expressed as HIV copy number/albumin genomic equivalents. We used a plasmid as a positive control at a concentration of 1,000 copies/ml (in duplicate). The intra-assay reproducibility was calculated by testing the recombinant plasmid dilutions at from 107 to 102 copies/ml seven times in the same experiment. To estimate the interassay reproducibility, the later dilutions were analyzed independently on different days by different technicians. We initially aimed to validate the qPCR before application to clinical samples. As shown in Fig. 1, amplification of the recombinant plasmid at various concentrations showed linearity of 8 orders of magnitude. The slope of the standard curve was −3.6 (R2 = 0.99), and PCR efficiency was 88.9%. For specificity analysis, HIV PL in culture supernatants was determined for subtypes B (5 log10/albumin), F (4 log10/albumin), and C (5 log10/albumin). The reproducibility was determined by calculating percent coefficient of variation (CV) from 10 copies and 107 (1.63% and 3.74%, respectively). CVs for interassay determinations were 3.29% and 6.07% (107 and 102, respectively). Standard curve analysis for albumin revealed 88% amplification efficiency (R2 = 0.98; slope = −3.62; intercept = 41).
Fig 1.
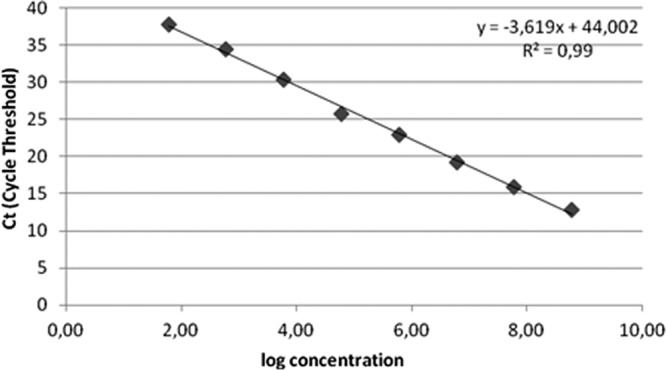
HIV real-time PCR plasmid integrase standard curve, showing linearity over a range of 8 orders of magnitude. The least-squares regression was calculated from plots of values measured for cycle threshold (Ct) (y axis) versus input plasmid DNA over a range of 101 to 108/ml (x axis) tested in duplicate PCR wells per dilution. The correlation coefficient was 0.99, and the slope of the line was −3.6.
All individuals presented genotypic resistance to all three antiretroviral classes at baseline, followed by the reemergence of wild-type virus at week 12 (data not shown). Of the 41 study patients, 83% were males; the median age was 46 years (range, 39 to 53), and the median HIV-1 PL was 0.61 log10. The averages of HIV-1 PL at baseline and week 12 were 0.61 log10 and 0.63 log10, respectively (t test = 1.64; P ≥ 0.11). At 12 weeks, the average plasma VL was significantly higher than the baseline (t test = 6.08; P ≤ 0.01). Fifty percent of patients showed a decrease of 50% or higher in CD4+ T cell counts after 12 weeks of STI (Table 1). Comparisons showed that individual rates of change of HIV-1 PL at week 12 tended to increase in 28 patients (70%), among whom19 (47.5%) showed an increase higher than 10%. For those 19 patients, the median HIV-1 PL was 0.68 log10/albumin genomic equivalents (t test = 6.74; P ≤ 0.01). Fifteen patients (37.5%) showed an increase in VL of ≥1 log10 after 12 weeks, and 10 of those (75%) also showed an increase higher than 10% in the HIV-1 PL at week 12. Conversely, among the 19 patients who showed a VL increase higher than 10% at week 12, 10 had an increase in VL of ≥1 log10, whereas 5 of 21 patients (23.8%) who presented with an increase in HIV-1 PL below 10% had an increase in VL of ≥1 log10 (Fisher's exact test; P = 0.06). There was no correlation between HIV-1 PL and CD4+ counts. HIV-1 PL quantitation is potentially a useful research tool for (i) understanding HIV-1 dynamics and compartmentalization, (ii) refining the monitoring of antiretroviral treatment, or (iii) use in studies aiming at HIV-1 eradication. We therefore developed two systems for absolute quantification that target the HIV-1 integrase gene and the human albumin gene. We developed a methodology that enables us to retrospectively analyze either stored samples that lack previous cell quantification or samples of nonviable cells accurately quantifying distinct HIV clades. We attempted to analyze variations in the HIV-1 PL in samples collected from viremic individuals before and after 12 weeks of STI. Interestingly, 28 of 41 individuals tended toward an increase in the HIV-1 PL after 12 weeks. Of these, 19 had a statistically significant increase of HIV-1 PL greater than 10%, which was accompanied by a notable increase in plasma VL. This suggests that, even for short time periods, active viral replication is an important factor in the replenishment of the proviral reservoir. Almost all individuals showed an important drop in CD4+ T cell counts after STI; nevertheless, the relative numbers of infected cells did tend to increase. STI among individuals failing HAART leads to the reemergence of wild-type HIV, an increase in VLs, and a decrease of CD4+ T cell counts, and it can be a limiting factor in the increase of CD4 counts after antiretroviral treatment is resumed (9, 10). In addition, we think it is highly significant that there is an increase in the pool of HIV-1-infected cells after STI, even among previously viremic individuals. Additional studies quantifying HIV provirus in both different compartments and different cell subpopulations vis-à-vis antiretroviral exposure or interruption, or in the presence of HIV strains with different replicative capacities, may be fundamental for fine-tuning the understanding of HIV dynamics among infected individuals.
Table 1.
Characteristics of STI patients
| Patient characteristic | Median (range) resulta |
|
|---|---|---|
| Baseline | Wk 12 | |
| CD4+ count (cells/mm3) | 218 (10–760) | 97.5 (1–550) |
| Plasma HIV RNA (log10 copies/ml) | 4.90 (1.72–6.23) | 5.60 (4.2–6.89) |
| HIV DNA (log10 copies/genomic equivalent) | 0.61 (0.23–0.92) | 0.62 (0.53–1.04) |
Statistical analyses were performed using Minitab software, version 15.
ACKNOWLEDGMENTS
We thank Sandra Mara Sampaio Andreo and Dercy José de Sá-Filho for technical assistance and comments, Rafael Gonçalves de Azevedo, Michelle Zanoni (Federal University of São Paulo), and Gisele Braz (Lusíada Foundation/Santos) for text revision, and Antonio Charlys da Costa for administrative assistance and secretarial help. We are also grateful to the patients enrolled in this study.
We have declared that no competing interests exist.
This work was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (grant 03/11781-1 to R.S.D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 14 March 2012
REFERENCES
- 1. Christopherson C, et al. 2000. PCR-based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chun TW, et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 3. Chun TW, et al. 2002. Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4+ T cells and CD4+:CD8+ T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 185:1672–1676 [DOI] [PubMed] [Google Scholar]
- 4. Chun TW, et al. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Invest. 115:3250–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deeks SG, et al. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472–480 [DOI] [PubMed] [Google Scholar]
- 6. Désiré N, et al. 2001. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J. Clin. Microbiol. 39:1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller V, et al. 2000. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS 14:2857–2867 [DOI] [PubMed] [Google Scholar]
- 8. Nottet HS, et al. 2009. HIV-1 can persist in aged memory CD4+ T lymphocytes with minimal signs of evolution after 8.3 years of effective highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 50:345–353 [DOI] [PubMed] [Google Scholar]
- 9. Yerly S, et al. 2004. Proviral HIV-DNA predicts viral rebound and viral setpoint after structured treatment interruptions. AIDS 18:1951–1953 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, et al. 1998. Human immunodeficiency virus type 1 in the semen of men receiving highly active antiretroviral therapy. N. Engl. J. Med. 339:1803–1809 [DOI] [PubMed] [Google Scholar]


