Abstract
We report the molecular epidemiology of 27 clinical multidrug-resistant Staphylococcus epidermidis (MDRSE) isolates collected between 2003 and 2007 in an Australian teaching hospital. The dominant genotype (sequence type 2 [ST2]) accounted for 85% of the isolates tested and was indistinguishable from an MDRSE genotype identified in European hospitals, which may indicate that highly adaptable health care-associated genotypes of S. epidermidis have emerged and disseminated worldwide in the health care setting.
TEXT
Staphylococcus epidermidis is an important pathogen involved in health care-associated bloodstream infections and infections related to vascular catheters and prosthetic devices (17). Several investigations have demonstrated that certain multidrug-resistant S. epidermidis (MDRSE) genotypes become established as opportunistic pathogens in the health care setting as a novel ecological niche (8, 10, 12). In addition, recent studies identified several worldwide epidemic clonal lineages (9, 12, 13, 18, 21). Currently, little information is available on the molecular epidemiology of S. epidermidis in the health care setting in Australia (16, 18).
We have previously documented the occurrence and potential dissemination of two genotypes of MDRSE in 11 hospitals in northern Europe between 2001 and 2008. The aims of this study were to examine the molecular epidemiology of clinical isolates of MDRSE collected in a teaching hospital in Australia and determine the possible presence of previously described health care-associated MDRSE clones.
Twenty-seven MDRSE isolates were collected between 2003 and 2007 from patients admitted to Royal Perth Hospital, Western Australia. One isolate per patient was included in the study.
The S. epidermidis isolates were identified using a phenotypic disc method (2) or bioMérieux Vitek2 Compact GP identification card (1). In addition, one isolate from each pulsed-field gel electrophoresis (PFGE) type was identified as S. epidermidis using rpoB sequencing (11). Antimicrobial susceptibility testing was performed by disc diffusion on Mueller-Hinton agar (BBL; Becton Dickinson, Cockeysville, MD) using Oxoid discs according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (4). The antimicrobials tested included cefoxitin (10 μg), erythromycin (15 μg), ciprofloxacin (5 μg), fusidic acid (10 μg), gentamicin (10 μg), rifampin (5 μg), tetracycline (30 μg), and mupirocin (5 μg). CLSI susceptibility interpretive criteria (5) were utilized for all antimicrobials with the exception of fusidic acid (3) and mupirocin (7).
Retrospective medical chart review of the 27 patients whose isolates were included in the study showed that 23 were treated for hospital-acquired MDRSE infections (Table 1). The median age among these 9 women and 14 men was 53 years (range, 20 to 88 years). Eight patients with hematological malignancy, all on hematology wards, had positive blood cultures and presumed line-related sepsis between February 2005 and June 2007. Six of these were treated with either vancomycin or linezolid, one with meropenem and ciprofloxacin, and in one case the treatment details are not available. Four patients with intracranial hemorrhage and shunt insertion were diagnosed with ventriculitis or ventriculoperitoneal shunt infection between January 2004 and February 2006. These patients were treated with IV vancomycin with or without intraventricular vancomycin. Four patients with prosthetic-joint infections were all treated with vancomycin with or without other antibiotics between April 2006 and April 2007. In addition, six patients were diagnosed with other health care-associated MDRSE infections and treated with vancomycin: one pacemaker infection, one postoperative discitis following laminectomy, one postoperative endophthalmitis, one postoperative infection following surgery for trauma, one postoperative thigh collection, and one meliodosis complicated by line-related sepsis. Lastly, one patient with line-related sepsis was treated by line removal and other antibiotics (isolate 11). The time span for these positive cultures was June 2003 to June 2007. The median length of stay (LOS) for patients with significant infection (available data on 21 patients) was 36 days (range, 10 to 282 days).
Table 1.
Epidemiological and clinical data for the 27 methicillin-resistant Staphylococcus epidermidis isolates included in the study in order of appearance in Fig. 1c
| No. | Isolate |
Patient |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PFGE type | ST typeb | SCCmec type | Antibiogram result | Source | Gender and age (yr) | Underlying condition | Diagnosis | Admission LOS (no. of days/yr) | Antibiotic treatment |
||
| Preculture | Postculture | ||||||||||
| 1a | I | 5 | IV (2B) | CIP ERY GEN MUP | Blood | NA | NA | Presumed line-related sepsis | NA/2006 | NA | NA |
| 2 | I | III (3A&5) | CIP ERY GEN MUP | Knee tissue | F 58 | Osteoarthritis | Septic total knee replacement | 27/2006 | CEF | VAN | |
| 3 | D | IV (2B) | CIP ERY FUS GEN | Blood | F 63 | Burkitt's lymphoma | Line-related sepsis | 39/2006 | TZP AZM | MEM CIP | |
| 4 | D | IV (2B) | CIP ERY FUS GEN | Blood | M 48 | AML | Line-related sepsis | 23/2006 | TZP | VAN | |
| 5 | D | IV (2B) | CIP ERY FUS GEN MUP | Blood | M 29 | Heart transplant, idiopathic myopathy | Septic thrombophlebitis second to pacemaker insertion | 16/2006 | SXT TIM | VAN | |
| 6 | D | IV (2B) | CIP ERY FUS GEN MUP | Blood | F 72 | AML | Line-related sepsis | 24/2006 | None | LZD | |
| 7 | D | IV (2B) | CIP ERY FUS GEN | Blood | M 56 | Multiple myeloma, allogeneic stem cell transplant, GVHD | Line-related sepsis | 37/2007 | NA | FEP VAN | |
| 8 | D | IV (2B) | CIP ERY FUS GEN MUP RIF | Blood | M 19 | ALL, bone marrow transplant | Line-related sepsis | 47/2007 | NA | VAN | |
| 9 | D | IV (2B) | CIP ERY FUS GEN | Blood | M 67 | AML | Line-related sepsis | 36/2007 | TZP | FEP LZD VAN | |
| 10 | D | IV (2B) | CIP ERY FUS GEN MUP | Hip tissue | F 88 | Total hip replacement | Septic total hip replacement | 63/2007 | CFZ CIP TIM TMP | LZD TZP VAN | |
| 11 | D | 2 | IV (2B) | CIP ERY FUS GEN | Blood | M 37 | Metastatic carcinoma, Staphylococcus aureus sepsis | Continuing fever, IV line in situ | 18/2007 | FCX | GEN MEM TIM line removed |
| 12 | D | IV (2B) | CIP ERY FUS GEN MUP | Hip liquid | M 63 | Metastatic renal cell carcinoma | Thigh collection postsurgery to femur for metastic lesion | 12/2006 | None | VAN | |
| 13a | D | IV (2B) | CIP FUS MUP | Bone | M 85 | Diabetes mellitus foot infection | Necrotic bone from 3rd metatarsal head | O/P/2005 | NA | CIP DOX | |
| 14 | D | IV (2B) | CIP ERY FUS GEN MUP | Hip tissue | F 54 | Hemiarthroplasty | Septic hemiarthroplasty | 70/2006 | AMC CRO TIM VAN | MEM TIM VAN | |
| 15 | D | IV (2B) | CIP ERY FUS GEN MUP | Disc tissue | M 42 | Laminectomy | Discitis postsurgery | 24/2006 | CFZ LEX | VAN | |
| 16a | D | IV (2B) | CIP ERY FUS GEN MUP | Bone | F 51 | ALL, bone marrow transplant | GVHD, manibular mucormycosis, surgical debridement on date of positive culture; no other obvious source | 106/2007 | AMC CIP CRO FEP LZD MEM TZP VRC | AMB, no anti-staphylococcal therapy added | |
| 17 | D | IV (2B) | CIP ERY FUS MUP TET | Blood | M 20 | AML, stem cell transplant, severe cutaneous GVHD | Febrile illness | NA/2005 | NA | NA | |
| 18 | D | IV (2B) | CIP ERY FUS GEN MUP | Blood | M 22 | ALL, blood stem cell transplant | Line-related sepsis | 282/2007 | CIP MEM TZP | LZD | |
| 19 | H | 2 | III (3A&5) | CIP ERY FUS GEN RIF | Blood | F 39 | Meliodosis | Line-related sepsis | 25/2007 | MEM | VAN |
| 20 | G | III (3A) Variant SCCmercury absent | ERY FUS GEN MUP | Vitreous fluid | F 77 | NA | Postoperative endophthalmitis | 10/2003 | NA | OFX CAZ Intravitreal VAN | |
| 21a | G | 2 | III (3A&5) | CIP ERY GEN TET | Sclera | M 73 | Lung transplant scedosporium eye infection | Enucleation of eye | 1/2005 | TRB VRC | TRB VRC |
| 22 | F | III (3A&5) | CIP ERY FUS GEN MUP RIF | Hip tissue | F 55 | Rheumatoid arthritis | Infected total hip replacement | 108/2007 | FUS RIF | VAN | |
| 23 | F | 2 | III (3A&5&4) Extra ccrA4B4 | CIP ERY FUS GEN RIF | CSF | M 83 | Subarachnoid hemorrhage | Ventriculitis | NA/2004 | LZD MEM VAN | FEP VAN |
| 24 | F | III (3A&5&4) Extra ccrA4B4 | CIP ERY FUS GEN RIF | CSF | M 49 | Subarachnoid hemorrhage | VP shunt infection | 32/2004 | MEM VAN | VAN | |
| 25 | F | Isolate not available | CIP ERY FUS GEN RIF | Leg wound | M 46 | Trauma, splenic rupture | Extensive soft tissue infection requiring amputation of leg | 61/2007 | CIP CLI | CIP MEM VAN | |
| 26 | J | 23 | IV (2B) | CIP ERY MUP RIF TET | CSF | M 67 | Cerebral hemorrhage | Ventriculitis | 105/2006 | None | VAN (IV and intraventricular), MEM LZD |
| 27 | J | IV (2B) | CIP GEN MUP RIF | CSF | F 58 | Subarachnoid hemorrhage | Ventriculitis | 164/2004 | MEM TZP VAN | VAN (IV and intraventricular) | |
Isolate of uncertain significance.
Performed according to Thomas et al. Allelic profile of the seven housekeeping loci (arcC, aroE, gtr, mutS, pyrR, tpi, yqiL) (18a).
AMC, amoxicillin-clavulanic acid; AZM, azithromycin; CFZ, cefazolin; FCX, flucloxacillin; FEP, cefepime; CAZ, ceftazidime; CRO, ceftriaxone; LEX, cephalexin; CEF, cephalothin; CIP, ciprofloxacin; CLI, clindamycin; DOX, doxycycline; ERY, erythromycin; GEN, gentamicin: LZD, linezolid; MEM, meropenem; MUP, mupirocin; OFX, ofloxacin; TZP, piperacillin-tazobactam; RIF, rifampin; TET, tetracycline; TIM, ticarcillin-clavulanic acid; TMP, trimethoprim; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin; AMB, Amphotericin B; TRB, terbinafine; VRC, voriconazole; CSF, cerebrospinal fluid; GVHD, graft versus host disease; NA, none available; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; VP, ventriculoperitoneal; LOS, length of stay; F, female; M. male.
After initial identification and antimicrobial susceptibility testing, the isolates were stored at −70°C until further analysis. Isolates resistant to cefoxitin and at least three of the other antibiotics tested were defined as multidrug resistant (MDR). PFGE, staphylococcal cassette chromosome mec (SCCmec) typing, and multilocus sequence typing (MLST) were performed on the 27 isolates as previously described (6, 19). One isolate from each PFGE type was analyzed using MLST. One isolate was not available for SCCmec typing. In addition, the PFGE patterns of the 27 strains were compared with a previously described compilation of 277 MDRSE isolates collected between 2001 and 2008 at 11 hospitals in northern Europe (14, 19, 20).
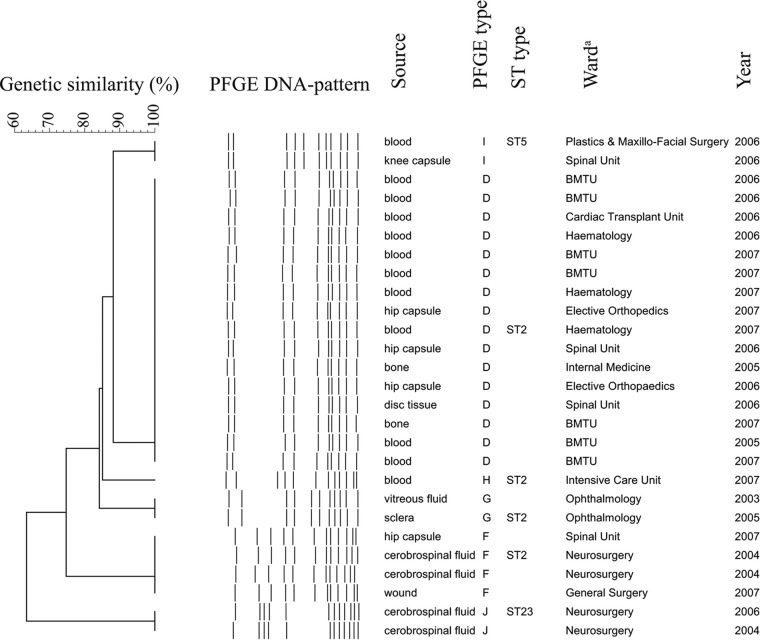
DNA macrorestriction analysis of the 27 MDRSE isolates revealed six PFGE types (Fig. 1). The predominant PFGE type detected, PFGE type D, consisted of 16 isolates (57%). Four PFGE types (G, H, I, J) contained one or two isolates, and PFGE type F consisted of four isolates (Fig. 1). PFGE type D isolates were from patients in six different wards between 2005 and 2007 and accounted for 11 of the 12 blood culture isolates (92%) (Fig. 1). Multilocus sequence type 2 (ST2) included four different PFGE types (D, F, G, and H), representing 85% of isolates (Fig. 1). PFGE type D was indistinguishable from an MDRSE genotype previously described in two Swedish hospitals (19). SCCmec characterization, performed on 26 isolates, identified two SCCmec types, type III (27%) and type IV (73%). ST2 MDRSE isolates harbored both SCCmec types. Type IV SCCmec was found in all 16 PFGE type D isolates.
Fig 1.
Cluster analysis of the genetic similarity of 27 isolates of multidrug-resistant Staphylococcus epidermidis using pulsed-field gel electrophoresis (PFGE). The horizontal upper bar represents genetic similarity (percent). The dotted lines in the center represent digitalized transformation of the PFGE-DNA pattern. Source of culture, PFGE type, sequence type (ST), ward, and year of isolation are described in the columns to the right. a BMTU, bone marrow transplant unit.
The results document the occurrence and possible endemicity of one PFGE type which represented the majority of the examined S. epidermidis isolates. The prevailing genotype in the current study, PFGE type D, was identified among patients treated in six different wards over a 3-year period. Interestingly, all PFGE type D isolates harbored type IV SCCmec, which presently is found in the majority of methicillin-resistant Staphylococcus aureus (MRSA) strains of community origin (15). Furthermore, MLST results showed that four of the six identified genotypes (PFGE types D, F, G, and H) belonged to ST2. This genotype has been detected in strains isolated in as many as 25 different countries across the world (18) (http://sepidermidis.mlst.net). A limitation of this study is that the isolates were a convenience sample; hence, it is not possible to determine the burden of disease. A prospective study characterizing all S.epidermidis isolates causing health care-associated infections would be necessary to determine the prevalence of this clone.
In summary, this report demonstrates the occurrence, persistence, and potential spread of an MDR genotype of S. epidermidis causing health care-associated infections in an Australian teaching hospital. This genotype (ST2) accounted for 85% of isolates tested, was indistinguishable from an MDRSE genotype identified in European hospitals, and has been reported as the most widely disseminated health care-associated ST type. More studies are needed to increase our understanding of the mechanisms that contribute to the evolutionary success of this extremely versatile microorganism.
ACKNOWLEDGMENTS
The authors declare that they have no conflict of interest.
This work was supported by a grant from the Research and Development Unit, Jämtland County Council, Sweden, and the Faculty of Medicine, Umeå University, Umeå, Sweden.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. bioMérieux 2004. VITEK 2 compact instrument user manual, vol 510772-1EN1 (REV 03/2004). bioMérieux, Durham, NC [Google Scholar]
- 2. Casals JB. 2007. User's guide, DIATABS™ diagnostic tablets for bacterial identification, 7th ed Rosco Diagnostica, Taastrup, Denmark [Google Scholar]
- 3. CA-SFM 1996. Report of the Comité de l'Antibiogramme de la Société Française de Microbiologie. Clin. Microbiol. Infect. 2:S48. [DOI] [PubMed] [Google Scholar]
- 4. CLSI 2009. Performance standards for antimicrobial disk susceptibility tests, 7th ed Approved standard M02-A10. CLSI, Wayne, PA [Google Scholar]
- 5. CLSI 2009. Performance standards for antimicrobial susceptibility testing, 19th informational supplement. M100-S18. CLSI, Wayne, PA [Google Scholar]
- 6. Coombs GW, et al. 2011. Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiol. 11:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finlay JE, Miller LA, Poupard JA. 1997. Interpretive criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob. Agents Chemother. 41:1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin. Microbiol. Infect. 14:1020–1027 [DOI] [PubMed] [Google Scholar]
- 9. Kozitskaya S, et al. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43:4751–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li M, Wang X, Gao Q, Lu Y. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58:456–461 [DOI] [PubMed] [Google Scholar]
- 11. Mellmann A, et al. 2006. Sequencing and staphylococci identification. Emerg. Infect. Dis. 12:333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Monk AB, Archer GL. 2007. Use of outer surface protein repeat regions for improved genotyping of Staphylococcus epidermidis. J. Clin. Microbiol. 45:730–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monsen T, Karlsson C, Wistrom J. 2005. Spread of clones of multidrug-resistant, coagulase-negative staphylococci within a university hospital. Infect. Control Hosp. Epidemiol. 26:76–80 [DOI] [PubMed] [Google Scholar]
- 15. Nimmo GR, Coombs GW. 2008. Community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Australia. Int. J. Antimicrob. Agents 31:401–410 [DOI] [PubMed] [Google Scholar]
- 16. Raimundo O, et al. 2002. Molecular epidemiology of coagulase-negative staphylococcal bacteraemia in a newborn intensive care unit. J. Hosp. Infect. 51:33–42 [DOI] [PubMed] [Google Scholar]
- 17. Rogers KL, Fey PD, Rupp ME. 2009. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23:73–98 [DOI] [PubMed] [Google Scholar]
- 18. Schoenfelder SM, et al. 2010. Success through diversity—how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300:380–386 [DOI] [PubMed] [Google Scholar]
- 18a. Thomas JC, et al. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45:616–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Widerstrom M, et al. 2009. Clonality among multidrug-resistant hospital-associated Staphylococcus epidermidis in northern Europe. Scand. J. Infect. Dis. 41:642–649 [DOI] [PubMed] [Google Scholar]
- 20. Widerstrom M, Monsen T, Karlsson C, Wistrom J. 2006. Molecular epidemiology of meticillin-resistant coagulase-negative staphylococci in a Swedish county hospital: evidence of intra- and interhospital clonal spread. J. Hosp. Infect. 64:177–183 [DOI] [PubMed] [Google Scholar]
- 21. Widerstrom M, Wistrom J, Sjostedt A, Monsen T. 2011. Coagulase-negative staphylococci: update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur. J. Clin. Microbiol. Infect. Dis. 31:7–20 [DOI] [PubMed] [Google Scholar]