Abstract
Infection of the chestnut blight fungus Cryphonectria parasitica with Cryphonectria hypovirus 1 (CHV1) causes disruption of virulence, pigmentation, and sporulation. Transcriptional downregulation of key developmentally regulated fungal genes occurs during infection, but vegetative growth is unaffected. Previous studies showed that CHV1 utilizes trans-Golgi network (TGN) secretory vesicles for replication. In this study, the fungal cell surface hydrophobin cryparin was chosen as a marker to follow secretion in virally infected and noninfected strains. Subcellular fractionation, cryparin-green fluorescent protein (GFP) fusion, and Western blot studies confirmed that vesicles containing cryparin copurify with the same fractions previously shown to contain elements of the viral replication complex and the TGN resident endoprotease Kex2. This vesicle fraction accumulated to a much greater concentration in the CHV1-infected strains than in noninfected strains. Pulse-chase analysis showed that the rates and amount of cryparin being secreted by the CHV1 containing strains was much lower than in noninfected strains, and the dwell time of cryparin within the cell after labeling was significantly greater in the CHV1-infected strains than in the noninfected ones. These results suggest that the virus perturbs a specific late TGN secretory pathway resulting in buildup of a key protein important for fungal development.
INTRODUCTION
The filamentous ascomycete Cryphonectria parasitica (Murrill) M. E. Barr is one of the most destructive tree pathogens known. It is the causal agent of chestnut blight, a disease that devastated both the European and American chestnut populations in the first part of the 20th century (5). This fungus is itself parasitized by a mycovirus, Cryphonectria hypovirus 1 (CHV1). Unlike many other viral infections, CHV1 does not cause severe cytopathic effects. Specific traits of mature development in fungi such as pigmentation, sporulation, female fertility, and virulence toward the host are repressed by the presence of the virus, while growth is not affected (28). Lack of development is not lethal but it prevents dissemination of the fungus via spore dispersal, the most common method of infecting new hosts (24). These effects of the virus have led to its use as a biological control agent in Europe (30).
The cytoplasm of virus-infected strains shows little evidence of adverse effects caused by the virus; all organelles are intact and the only indication of infection is that virus-infected strains contain increased numbers of membrane-enclosed vesicles (33). Virions of CHV1 are not found because it lacks a protein coat. Typically, RNA viruses replicate in close association with host membranes (42), and CHV1 has been found to utilize host vesicles for replication (12). Vesicles from noninfected strains lack viral RNA and are present in smaller numbers but otherwise have properties and composition similar to those that can be isolated from infected strains (17).
Viral double-stranded RNA (dsRNA), now believed to be the replicative form of the virus (20), and RNA-dependent RNA polymerase activity are copurified with vesicles from the virus-infected strains (13). Subcellular fractionation has shown that these same virus-containing vesicles copurify with markers for the late trans-Golgi network (TGN) and include the serine protease Kex2 (23). One of the genes that is downregulated by CHV1, the cryparin gene, corresponds to an abundantly expressed cell surface hydrophobin necessary for eruption of the fruiting bodies of the fungus through the bark of the host (24). Cryparin contains the Lys-Arg consensus sequence and has been shown to be processed by Kex2 in vivo (22). Without the signal peptide for secretion recognition, sequence analysis has shown cryparin to be 9,050 Da (52). However, prior to secretion the protein could be found in a 36-kDa glycosylated form along with a 24-kDa unglycosylated form in a fraction enriched for putative secretory vesicles (29). The same study showed that radioactively labeled cryparin exits the cell within 10 min of labeling and is rapidly rebound to the cell wall (29). During log-phase growth, approximately 25% of the total mRNA produced by the fungus is cryparin mRNA (52). CHV1 infection reduces levels of cryparin expression and secretion by up to 70% (6, 52). This along with the observation that viral elements copurify with fungal Kex2 (23) led us to hypothesize that the replication of CHV1 may interfere with the secretion of developmentally important proteins such as cryparin.
Extracellular enzymes potentially involved in virulence have been studied from virus-infected and noninfected strains of the fungus (14, 19, 48). These studies have identified differences between virulent and hypovirulent strains but have not successfully led to an understanding of the basis of C. parasitica virulence. Many enzymes and other secreted compounds probably act in concert to cause pathogenicity; thus, disruption of a regulatory mechanism that controls their expression is another way that the virus can affect virulence. Protein transport and secretion pathways contribute directly to the overall pathogenic potential of fungi in general (45), and Kex2 specifically has been shown to be essential for full virulence in C. parasitica (22). This study demonstrates that virally infected cells accumulate more vesicle material than noninfected cells. In infected cells, cryparin was found to cofractionate with Kex2. Noninfected cells showed a similar distribution of Kex2, but cryparin could not be detected using standard methods. The buildup of cryparin in the infected strains was confirmed by pulse-chase studies that showed that infected cells secrete cryparin at a much lower rate, and as a consequence the protein accumulates to very high levels compared to noninfected strains.
MATERIALS AND METHODS
Strains and growth conditions.
The following Cryphonectria parasitica strains were used: strain EP67 (ATCC 38753) and its isogenic CHV1-containing strain EP802 (ATCC 52574); strain EP155 (ATCC 38751) and its isogenic CHV1-containing strain UEP1 (38); and cryparin deletion strain Δ119 and rescue strain WT6 (24). Inoculum for liquid culture was grown at 25°C on PDAmb plates (39). Plates were grown for 7 days, homogenized in EP complete liquid medium (39) for 1 min at full speed in a Waring blender (New Hartford, CT), and used to inoculate Fernbach flasks containing 1 liter of EP complete (39). The cultures were grown on an orbital shaker at 136 rpm at room temperature under ambient light for 3 days.
Plasmid constructs and generation of GFP-expressing strains.
The recombinant plasmid pCrpGfp was constructed to express green fluorescent protein (GFP) within the coding region of cryparin. This was done using the genomic clone of cryparin (52), plasmid pCT74 containing a modified version of the green fluorescent protein gene, sgfp, kindly provided by Lynda Ciuffetti (27), and plasmid pSV50, containing the benomyl resistance gene (49). Plasmid pCT74 was used as a template for PCR, and using the oligonucleotides AAAGGATCCGCTTTACTTGTACAGE and AAAGGATCCATGGTGAGCAAGGC (forward and reverse primers with a BamHI site underlined), we obtained a 700-bp fragment with the gfp gene flanked by in-frame BamHI sites, which was then cloned into the newly created BamHI site at nucleotide position 710 of the coding region of the cryparin gene as previously described (16). A-2.4 kb SalI band from pSV50 comprising the benomyl resistance gene tub-2 (49) was inserted at the 3′ end of the cryparin gene in pCRP (Fig. 1). This vector was used to transform the cryparin deletion strain, Δ119 (24), using transformation protocols described previously (9). The transformants were plated on PDAmb containing 0.5 μg/ml benomyl (ChemService, West Chester, PA), and resistant fluorescent transformants were selected using a Leica MZ FLIII stereoscope equipped with a GFP2 excitation filter with a band pass of 460 to 500 nm and an emission filter of 510 nm (Leica Microsystems, Wetzlar, Germany). Single-spore isolates were selected from approximately 10 individual transformants. Insertion of the pCrpGfp vector into the transformants was confirmed by Southern hybridization analysis (43). Transfer of the viral RNA into recipient strains was confirmed by isolation of viral nucleic acid as described previously (12). CHV1 containing strain UEP1 (38) was paired with 3 independent transformants and the virus transferred to them via hyphal anastomosis. A positive control for constitutive expression of GFP was created by transforming ΕP155 with a linearized copy of pCT74 (27). As a negative control, the rescued strain of Δ119,WT6 (24), was included as appropriate.
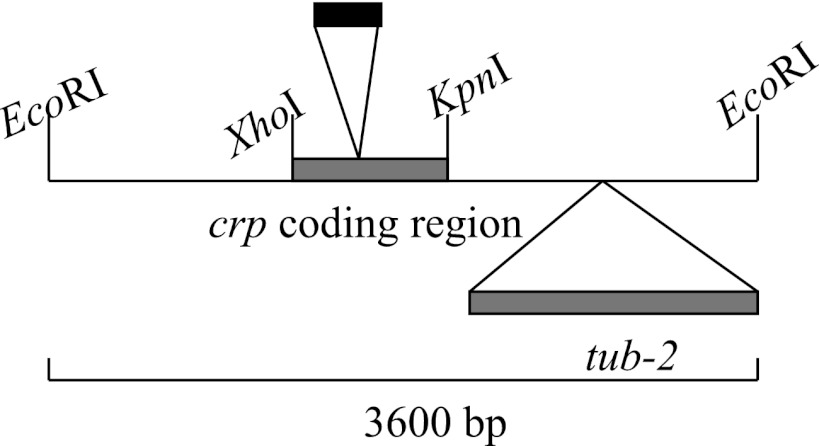
Fig 1.
Recombinant plasmid containing the GFP-coding region under the control of cryparin regulatory elements. Modification of a 3,600-bp genomic clone of cryparin was done by inserting a 700-bp fragment coding for the GFP protein near the end of the coding region of the cryparin gene. A 2,400-bp fragment containing the gene coding for benomyl resistance was inserted at the 3′ end of the genomic clone, outside the coding region for cryparin. This plasmid was used to transform strain Δ119, a cryparin-null mutant.
Subcellular fractionation of vesicle accumulation.
Subcellular fractionation was performed essentially as described previously (23) with the following modifications. A 1.0-ml aliquot of the microsomal fraction harvested as previously described was loaded onto 10-ml linear gradients of Ficoll-2H2O. Each 1.0-ml aliquot was adjusted with buffer B (0.1 M MES [morpholineethanesulfonic acid]-NAOH, pH 6.5, 1 mM EDTA, 0.5 M MgCl2, 1 mM dithiothreitol, 0.5 M phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin) to contain the dry weight equivalent of 1.5 g mycelia harvested. This was done by taking 10% of the fresh weight used for microsomal extraction, lyophilizing this portion, and extrapolating back to the dry weight of the remaining portion of the mycelia pad. Centrifugation was carried out in an L8-70 ultracentrifuge (Beckman Coulter, Palo Alto, CA) at 90,000 × g for 16 h in a swinging SW41 Ti rotor, and 1.0-ml fractions were collected, diluted with 8 ml buffer B, and centrifuged for 2 h at 110,000 × g. Pellets were resuspended in 200 μl of buffer B and stored at −80°C until further processing. Insoluble material at the bottom of the gradients was not processed.
Gel electrophoresis and Western blotting.
Proteins from subcellular fractions were analyzed by 12.5% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis and Western blotting as described previously (23). Blots were incubated with cryparin antibody (24) at a dilution of 1/200 or Kex2 antibody (22) at a dilution of 1/8,000 in phosphate-buffered saline (PBS), 0.3% Tween 20, and 7% nonfat dried milk overnight at 4°C. Secondary anti-rabbit horseradish peroxidase conjugate (Sigma, St. Louis, MO) was then used at a 1/2,000 for detection of cryparin antibodies and 1/5,000 for detection of Kex2 antibodies in the buffer described above. Incubation with secondary antibody was done for 2 h at room temperature. Detection was done using SuperSignal chemiluminescent subtrates (Thermo Scientific, Rockford, IL) according to the manufacturer's directions.
Proteins obtained from culture filtrates and mycelia of strains expressing CRP::GFP were analyzed as described for the above samples, with the exception of the primary antibody being anti-GFP (Clontech, Palo Alto, CA) used at a 1/200 dilution.
Microscopy.
Mycelia harvested as described above were placed as a suspension in 50 μl of H2O on the surface of a microscope slide and covered with a coverslip. Emission of green and red fluorescence was examined with a Nikon Microphot SA fluorescence microscope (Nikon, Melville, NY) equipped with a 100× oil objective, numerical aperture (NA) 1.4, and a 41018 Endow filter with an excitation band pass of 450 to 490 nm and a 500-nm long-pass emitter from Chroma Technologies (Brattleboro, VT). The images were captured using a Spot 2.2 digital camera using Spot 4.1 software (Diagnostic Instruments, Inc., Sterling Heights, MI). This software was then used to measure approximate vesicle size.
This filter allowed simultaneous viewing of GFP and FM4-64 fluorescence. The plasma membrane was identified by adding 50 μl of a freshly diluted 16 mM stock of solution of FM4-64 (Molecular Probes, Eugene, OR) in 100% dimethyl sulfoxide (DMSO). The dilution was made in water to a molarity of 4 μM and was added to one side of the coverslip. The solution was drawn through to the other side by placing a Kim-Wipe (Kimberly-Clark, Roswell, GA) adjacent to the coverslip. Staining for cell walls and septa was done using calcofluor white (Molecular Probes, Eugene, OR) at a concentration of 10 μg/ml. A small amount of mycelia was placed into 1.0 ml of the calcofluor solution and incubated for 5 min at room temperature (18). Cells were spun down for 1 min in at 15,000 × g to pellet the mycelia, which were then resuspended in 500 μl of water, spun again, and resuspended in 100 μl of water for observation. The cells were placed on a glass slide and observed by fluorescence microscopy using a Nikon UV filter cube uv2A DM400, excitation band pass of 330 to 380 nm and an emission filter at 420 nm.
Images were captured using a Spot 2.2 digital camera using Spot 4.1 software (Diagnostic Instruments, Inc., Sterling Heights, MI).
Assay of GFP expression and visualization of cellular accumulation.
Mycelia were grown as described, harvested by centrifugation at 6,000 x g. The resulting pellet was rinsed 3 times with double-distilled water. Fifty milligrams (wet weight) of this material was suspended in 1.0 ml of buffer B (23) and homogenized in a Mini-Bead-Beater (Biospec Products, Bartlesville, OK) for 30 s. Cell-free supernatants were obtained by centrifugation at 20,000 × g for 10 min. One hundred and fifty microliters of the supernatant was removed for protein analysis using Coomassie Plus protein reagent kit (Pierce, Rockford, IL). Triton X-100 was added to the remaining sample to a final concentration of 1%. The sample was processed again using the bead beater for 30 s and centrifuged once more at 20,000 × g. Two hundred microliters of the supernatant was added to a 96-well black microtiter plate with a clear bottom (Corning Costar, Cambridge, MA). GFP fluorescence was measured at an excitation wavelength of 494 nm with an emission cutoff of 520 nm using a plate-reading fluorometer, Spectramax M2 (Molecular Devices, Sunnyvale, CA). Results were expressed as mean GFP fluorescence normalized to total protein, experiments were repeated 3 times, and the standard error of the mean (SEM) was calculated. Three independent transformation isolates were analyzed and all produced similar data.
Protein isolation from culture filtrates and mycelia.
Culture filtrate and mycelia were collected as described previously (29). Fifty-milliliter aliquots of culture filtrates were lyophilized and resuspended in 5 ml of 60% ethanol. Fifty milligrams of lyophilized mycelia was suspended in 1 ml of 60% ethanol and homogenized in a Mini-Bead-Beater (Biospec Products, Bartlesville, OK) for 30 s and spun at 20,000 × g for 10 min. Proteins from both preparations were aliquoted and frozen at −80°C until further analysis.
Pulse-chase analysis.
Pulse-chase analysis was performed as described previously (29). Briefly, mycelia grown on PDAmb for 7 days were used as the inoculum. One-sixth of a plate (90 mm diameter) was homogenized in a Virtis handheld homogenizer (The Virtis Company, Gardiner, NY) and inoculated into 100 ml of EP complete liquid medium in 250-ml baffled flasks. The culture was grown for 3 days with shaking, at which time 50 μCi (1,000 Ci/mol) [35S]cysteine (Amersham International, Arlington Heights, IL) was added to the medium. Radiolabeled cysteine was chosen because the mature form of cyparin contains a minimum of 14 cysteine residues (52). Cells were transferred to fresh medium 30 min after the addition of [35S]cysteine, and at given times following transfer into fresh medium, 10 ml of the mixture of cells and culture medium was removed and filtered through a Büchner funnel and cells were collected on Miracloth (Calbiochem-Novabiochem, La Jolla, CA). Samples from time points up to 8 h were collected in this manner. The collected cells were lyophilized and cryparin was extracted in 60% ethanol, which was previously shown to remove all contaminating molecules except pigments (6). This extraction technique removed both cryparin from the cell wall and cytoplasmic cryparin. As reported in previous work, this fraction was considered representative of cell wall, since cytoplasmic cryparin was negligible compared to that accumulated on the cell wall (29). The proteins were separated on 12.5% SDS-PAGE gels. The gel was dried and exposed for 24 h at −80°C to a phosphorimager bioimaging screen (Fuji Medical Systems USA, Inc., Stanford CT). Confirmation of the labeled protein as cryparin was based on its size and a specific reaction with cryparin antibody and a comparison carried out with a cryparin deletion strain control (24, 29). Cryparin is one of the few C. parasitica proteins soluble in 60% ethanol (6).
To monitor the cytosplasmic accumulation of cryparin, 20 ml of the growing culture was removed at each sampling time. Following filtration and washing steps, cells were immediately added to a tube containing 0.5 g of glass beads (0.5 mm diameter; Biospec Products, Bartlesville, OK) and 1 ml of TMD buffer (50 mM Tris [pH 8.0], 10 mM MgCl2, 5 mM dithiothreitol) on ice. Cells were broken in a Mini-Bead-Beater (Biospec Products, Bartlesville, OK), and the cell debris were removed by centrifugation for 10 min at 12,800 × g. The supernatant was lyophilized overnight, and cryparin was extracted in 60% ethanol and analyzed as described above. However, exposure times to the phosphorimager screens ranged from 5 to 11 days due to the paucity of labeled cryparin present in the cytoplasm.
In pulse-chase analysis for both cell wall and cytoplasmic accumulation of cryparin, the density of the protein per gram of mycelia was calculated with the quantification program on the phosphorimager. Each experiment was repeated a minimum of three times, and the data shown are representative of multiple experiments. As in previous studies on the secretion of cryparin, SEM could not be calculated because quantitative counts for each experiment varied widely. However, the pattern of labeling between noninfected and infected was similar in all experiments (29).
RESULTS
A CRP::GFP fusion protein was constructed using the sgfp variant of GFP (27) placed within the coding region of the cryparin gene (52) (Fig. 1). Three independent transformants of a previously described cryparin deletion strain, Δ119 (24), were chosen for further analysis on the basis of GFP expression, and CHV1 was transferred into these strains via hyphal anastomosis. These strains were named GFP0-3 and GFP0.8, respectively.
Vesicle accumulation and subcellular localization of cryparin.
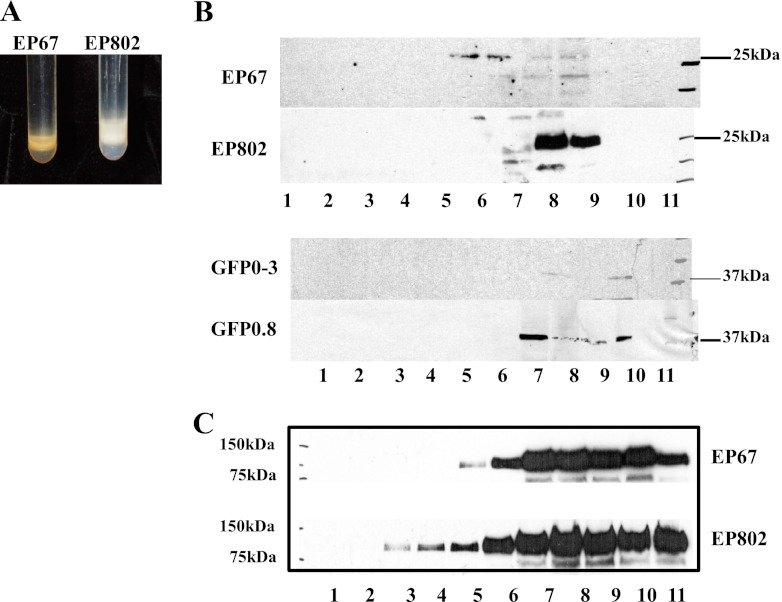
Subcellular fractionation and immunoblotting were used to detect vesicle proliferation and cryparin accumulation, respectively. Previous work (12, 17) noted that vesicles accumulated to greater abundance in CHV1-infected strains, but these studies did not correct for possible differences in amounts of mycelium used in the study, as freshly harvested nonlyophilized material was used in these protocols. Figure 2A compares the gradients of EP67 and EP802. Equal dry weight equivalents of microsomal material were applied to the gradients. EP802 showed accumulation of a much thicker band of opalescent material. EP67 material was in contrast pigmented and present in a smaller portion of the gradient. Western blots of the gradient fractions using cryparin antibodies, shown in Fig. 2A, revealed an accumulation of cryparin in fractions 8 and 9 in the virus-infected strain EP802. In contrast, negligible amounts of cryparin were detected in the collected fractions of wild-type EP67 (Fig. 2B, top). Similarly, cryparin-GFP fusion protein was detected at the predicted molecular mass of 36 kDa and accumulated in fractions 7 to 10 in virally infected GFP0.8 strains. No fusion protein was detected in the same fractions in noninfected GFP0-3 strains (Fig. 2B, bottom). Distribution of Kex2 in the gradient fractions of EP67 and EP802 is shown in Fig. 2C.
Fig 2.
Ficoll-2H2O gradients and Western blot analysis of microsomal fractions. (A) Dry weight equivalents of microsomal fractions extracted from mycelia were loaded onto Ficoll gradients, centrifuged, and photographed. (B) Western blot analysis showing gradient fractions for EP67, EP802, GFP0-3, and GFP0.8 using cryparin antibodies. (C) Western blot analysis of gradient fractions from EP67 and EP802 using antibodies to Kex2.
Fluorometric quantification and cellular localization of CRP::GFP.
To monitor the effects of CHV1 in vivo, expression of the CRP::GFP fusion protein was examined microscopically (Fig. 3A). Hyphae transformed with pCT74-59 expressed GFP under the control of the toxA promoter from Pyrenophora tritici-repentis (27) and resulted in a constitutively expressed protein located in the cytoplasm (Fig. 3A, left [pCT74-59]). Figure 3A, middle (GFP0-3), shows hyphae expressing the cryparin-GFP fusion protein. At 3 days postinoculation, the protein could be seen localizing intracellularly to discrete vesicles approximately 100 nm in width. In contrast, the CHV1-infected transformant GFP0.8 cryparin-GFP fusion protein within vesicles appeared to merge with the plasma membrane. Colocalization done with the styryl dye FM4-64, specific for the plasma membrane, confirmed this observation. After a 1-min pulse of FM4-64, red fluorescence began to almost immediately displace the areas of green fluorescence in the virus-containing strain GFP0.8 (Fig. 3B). GFP0-3 and the control strain pCT74-59 show red fluorescence in the same location; however, green fluorescence is limited to vesicles in the case of GFP0-3 and to the cytoplasm in the case of pCT74-59 (Fig. 3B). Two other independent transformants showed similar patterns of GFP expression and FM4-64 dye accumulation. Septal regions and cell walls were confirmed with visualization using calcofluor white, a fluorescent dye specific for chitin (data not shown). GFP accumulation detected by fluorescence measurement from cell extracts harvested from 3-day shake culture is shown graphically in Fig. 3C. The CHV1-infected strain GFP0.8 expressed approximately twice as much fluorescence as the noninfected strain GFP0-3. The WT6 cryparin deletion mutant rescue strain (24) was used as a background control. Two other transformants showed similar patterns of fluorescence accumulation.
Fig 3.
Localization and quantification of GFP. (A) Epifluorescence microscopy of strains pCT74-59, GFP0-3, and GFP0.8. (B) Epifluorescence microscopy of these same strains stained with FM4-64. Bars, 10 μm. (C) GFP expression values obtained from fluorometric analysis of cell-free lysates of these strains including the non-GFP-expressing strain WT6 as a background control. Data were normalized to total protein and represent the averages of replicates from 3 independent experiments. The error bars represent the SEM.
Western analysis of cell-free lysates using anti-GFP (Fig. 4) revealed production of the intact fusion protein coinciding with the calculated molecular mass of 36 kDa, comprising the GFP portion of the protein (27-kDa) and 9.0 kDa for cryparin. The actual molecular mass of cryparin is 9,050 kDa, although purified preparations of cyparin run at 24 to 30 kDa (52). The vector-only transformant pCT74-59, expressing only the GFP protein, contained a band at the expected molecular mass of 27 kDa. Some of the fusion protein appears to be processed in both GFP0-3 and GFP0.8 and appears as the 27-kDa band in the Western blot. WT6 is the rescued strain derived from the cryparin-null mutant, Δ119, and as expected does not show expression of the fusion protein and was included as a negative control (Fig. 4A).
Fig 4.
Western blot analysis of cryparin-GFP fusion protein using GFP antibodies. (A) Cryparin-GFP fusion protein extracted from mycelia. WT6, the cryparin rescue strain, was included as a negative control, and pCT74-59 expressing GFP was included as a positive control. GFP0-3 and GFP0.8, Δ119 and CHV1-infected Δ119, respectively, expressing GFP under the control of cryparin regulatory elements. (B) Secreted cryparin-GFP fusion protein extracted from culture filtrates.
Culture filtrates from these strains were also analyzed for detection of secreted CRP::GFP fusion protein. Since gfp was inserted into a created BamHI site at position 710 in the 978-nucleotide coding region of cryparin (Fig. 1), the fusion protein retained both the signal sequence for secretion as well as the Kex2 processing site. However, only negligible amounts of extracellular fusion protein could be detected from the culture filtrates of GFP0-3 or GFP0.8, and only when blots had been extremely exposed. In addition, small amounts of processed fusion protein were detected in the culture filtrates of GFP0-3 but never appeared in filtrates of GFP0.8 (Fig. 4B).
Cryparin secretion is slowed in CHV1-infected cells compared to noninfected cells.
Pulse-chase analysis was used to measure and compare the relative amounts of cryparin accumulating on cell walls versus that remaining in the cytoplasm of infected and noninfected cells. As reported in previous work, the relative amount of cryparin in the cytoplasm at all time points was insignificant compared to that on the cell wall, so all experiments reporting the cryparin accumulation on cell walls used lyophilized whole cells for the analysis (29). Figure 5 shows phosphorimager scans of radiolabeled cryparin accumulation on the cell wall of EP67 for up to 300 min in agreement with previous work (29). EP802 in contrast shows less labeled cryparin bound to the cell wall during the same chase period. Confirmation of the bands obtained by phosphorimager analysis as cryparin was based on size and a specific reaction with cryparin antibody (29). The data suggested that less labeled cryparin was being produced in EP802 and were in agreement with previous work that showed that CHV1-infected strains transcribed 50% less cryparin than noninfected strains (25).
Fig 5.
Cell wall accumulation of 35S-labeled cryparin. A 20-min pulse of [35S]cysteine was added to a 3-day old-liquid culture of the fungus. Samples were taken at the indicated time points during the chase period. Radiolabeled cryparin was extracted from lyophilized mycelia using 60% ethanol and resolved on SDS-PAGE. The gel was dried and exposed to a phosphorimage screen for 24 h. Density was measured from the phosphorimager screens. Units are arbitrary phosphorimager units. The data shown are representative of multiple experiments.
To examine amounts of cryparin remaining within cells, pulse-chase experiments and harvesting of samples were identical to that for the cell wall accumulation experiments presented above. However, instead of lyophilizing collected material and extracting cryparin directly in 60% ethanol, samples were processed using TMD buffer. Cell walls were removed by centrifugation and the supernatants were lyophilized. This method revealed radiolabeled cryparin within the cells, free from contaminating cell wall cryparin. The chase in these experiments extended up to 8 h. Phosphorimager scans of cryparin density within cells revealed a very rapid exit of the labeled hydrophobin, within 30 min from the CHV1-free strain EP67 (Fig. 6). In contrast, high levels of labeled cryparin could still be detected in the infected strains for up to 4 h during the chase period, and only after 8 h had all labeled cryparin disappeared within cells, suggesting eventual turnover of the protein. These results illustrate that the presence of CHV1 lowers the rate of cryparin secretion by more than 3-fold and support the results shown in Fig. 2B.
Fig 6.
Cytoplasmic accumulation of cryparin. A 20-min pulse of [35S]cysteine was added to a 3-day-old liquid culture of the fungus. Samples were taken at the indicated time points during the chase period. Radiolabeled cryparin was extracted from the cytoplasm and analyzed as described for Fig. 5 with an extension of the sampling period to 8 h. Density was measured from the phosphorimager screens exposed for a minimum of 5 days. Units are arbitrary phosphorimager units. The data shown are representative of multiple experiments.
DISCUSSION
In this study, we have shown biochemical and cytological evidence that CHV1 causes vesicle accumulation, lowers the rate of secretion of a key developmental protein, and increases the retention of host vesicles that copurify with the TGN endoprotease Kex2. Previous assays of solubilized vesicles revealed RNA-dependent RNA polymerase activity in these fractions which contain virus (13). Further characterization of these fractions showed that they contained markers for the TGN as well as the viral polymerase, helicase, and the nonstructural viral protein p29 (23). CHV1 is the type viral species of the Hypoviridae; it has sequence similarities to plant potyviruses and falls within the picorna-like virus superfamily (26). CHV1 replication occurs, as in all positive-strand RNA viruses, in association with cellular membranes (1). Our current findings are in general agreement with these results. Many viral and cellular components of both plant and animal viruses that contribute to these replication complexes have been characterized (42). We show here that CHV1 causes an enrichment of the membrane fraction compared to virus-free isolates. However, CHV1 does not cause the same cytopathic effects induced by poliovirus, another member of the picorna-like virus superfamily (2).
CHV1 significantly reduces the expression of many host genes (3, 6, 8, 24, 40, 51). A number of these genes contain consensus sequences for secretion and further processing by Kex2 (8, 51, 52). Cryparin is a highly expressed cell surface hydrophobin that was coded for by one of the Kex2-processed and CHV1-downregulated genes. Because of its abundant expression in liquid culture, it provided a convenient marker to follow the effects of the virus on secretion. We report here the accumulation of cryparin in the same subcellular fractions as those containing markers for the TGN, including Kex2, the viral genome, polymerase, helicase, and the nonstructural viral protein p29 (23).
Previous work with noninfected cells revealed that cryparin secretion occurred rapidly through the cell wall without being retained in it. Over time, the amount of labeled cryparin in the culture medium decreased as the amount on the cell wall increased (29). Our work here is in agreement with those results. However, in contrast to the rapid secretion of cryparin from CHV1-free cells, infected cells retain pulse-labeled cryparin for up to 4 h. As a consequence of this retention within cells, less cryparin is secreted into the medium for association on the cell wall, and therefore less cryparin can be isolated from the cell walls of CHV1-infected strains. Previous work showed that cryparin was downregulated by CHV1 at the level of transcription (25). We now believe that this buildup of cryparin within the cell may actually be responsible for its transcriptional downregulation, as this has been shown to be the case in Trichoderma reesei (35) and Aspergillus niger (4).
It is not unusual for a virus to slow or disrupt host secretion. Members of the families Potyviridae and Picornaviridae, to which CHV1 is related, have been shown to affect secretory events. Tobacco etch virus, one of the best-characterized potyviruses, produces a 6-kDa viral protein that induces the formation of the endoplasmic reticulum (ER)-derived vesicles, and the virus has been found in association with these vesicles (44). Recent work has shown that secretion of a soluble marker was inhibited by overexpression of this protein (50). The most well-known member of the Picornaviridae, poliovirus, causes a disruption of secretion most likely caused by disruption of the Golgi apparatus (10). Unlike infection with the viruses mentioned here, CHV1 infection does not disrupt cellular organelles, cause major cytopathic effects, or reduce vegetative growth. The fact that CHV1 downregulates a subset of proteins that are secreted without affecting growth suggests that the pathway for secretion of these proteins is not used for secretion of proteins necessary for hyphal growth. Our results also suggest that not all secretory pathways are affected by the virus, since growth is not reduced. Evidence for multiple secretion systems in fungi has been described in the yeast Yarrowia lipolytica (46) and in the filamentous fungus T. reesei (47).
We have shown here that CHV1 not only slows secretion of a key developmental protein but also changes the location of expression of a cryparin-GFP-tagged fusion protein. When examined microscopically in early-log-phase cultures expressing cryparin-GFP fusion protein in infected as well as noninfected strains, fluorescence localizes to spherical vesicles within the hyphae. They ranged in size from 60 to 100 nm in diameter and were distributed most frequently in subapical regions. Golgi-to-plasma membrane transport vesicles described in yeast are similar in size and shape (31). In several filamentous fungi, secreted proteins fused to GFP also revealed fluorescence localizing to vesicles similar in size and appearance (37). Many reports of Golgi and secretory vesicles in fungi show evidence of their presence in the apical region of growing hypha, near the Spitzenkorper (21). However, studies done on cryparin secretion suggest that the older hyphae secrete the most cryparin (29). Indeed antibodies revealed cryparin to be specific to aerial hyphae which form just behind the margin of the colony. It was most abundantly present in sexual and asexual fruiting bodies (6). Recently, two proteins of the secretory pathway, a v-snare and two t-snares of T. reesei, were imaged by colocalization studies using fluorescence lifetime imaging microscopy. One of the interacting pairs was found to be localized to older subapical regions of fungal colonies, suggesting that not all protein secretion in filamentous fungi occurs at hyphal tips (47).
When late-log-phase cultures of CHV1-infected cryparin-GFP tagged strains were examined microscopically, fluorescent vesicles were observed in very close association with the plasma membrane and in some cases appeared to be merging with it. The cryparin-GFP fusion protein in these strains also accumulated near septal regions. Accumulation of fluorescence in septal regions was reported in A. niger when using a glucoamylase-GFP tagged protein to study secretion (15). This group postulated that the fluorescent septa may be due to accumulation of secretory vesicles in this region. Why CHV1 would change accumulation patterns of cryparin tagged-GFP in this way is unknown. Data from these time points confirmed that the fusion protein accumulated in the same fractions as reported previously. Several well-studied plant and animal viruses have been shown to relocate host proteins by altering signals on endomembranes (7, 11, 32, 34, 36). It is possible that CHV1 factors present in the vesicles alter their original destination.
Only negligible amounts of cryparin-GFP tagged fusion protein could be detected in culture filtrates of either CHV1-infected or virus-free strains. We could detect the 36-kDa intact fusion protein as well as the 27-kDa GFP protein in whole-cell lysates. However, culture filtrates contained only trace amounts of the fusion protein in the infected strains, while the noninfected strains revealed trace amounts of both the fusion protein and cryparin, suggesting that only the noninfected strains secreted the fully processed fusion protein. This is in general agreement with previous reports from A. niger in which gene fusions of a secreted protein glucoamylase and GFP were difficult to detect in all culture filtrates. They postulated that the presence of fungal proteases produced in the culture fluid, as well as the acidic pH, were responsible for the lack of detectable heterologous proteins in the culture filtrate (15).
This work continues the characterization of the effect of CHV1 on C. parasitica. Our hypothesis that the virus hijacks a subset of host secretory vesicles which are used for viral replication, transcription, and movement is supported by the results presented here. Using 2 different virulent North American strains, EP67 and EP155, as well as their virus-infected counterparts, we have shown that CHV1 causes a proliferation of membrane vesicles of TGN origin (23) in which the secreted sporulation protein, cryparin (24), is retained and as a result less is secreted to be reassociated with the cell wall.
ACKNOWLEDGMENTS
We thank Tzion Fahima for his helpful review of the manuscript, Matthew Pye for his help with phophoimager data, and Linda Ciuffetti for the plasmid pCT74.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Ahlquist P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldabe R, Carrasco L. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64–76 [DOI] [PubMed] [Google Scholar]
- 3. Allen TD, Dawe AL, Nuss DL. 2003. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonectria parasitica to infection by virulence-attenuating hypoviruses. Eukaryot. Cell 2:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Sheikh H, et al. 2004. Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol. Microbiol. 53:1731–1742 [DOI] [PubMed] [Google Scholar]
- 5. Anagnostakis SL. 1987. Chestnut blight - the classical problem of an indroduced pathogen. Mycologia 79:23–37 [Google Scholar]
- 6. Carpenter CE, et al. 1992. Effect of a virus on accumulation on a tissue-specific cells-surface protein of the fungus Cryphonectria (Endothia) parasitica. Mol. Plant Microb. Interact. 5:55–61 [DOI] [PubMed] [Google Scholar]
- 7. Chen YL, et al. 2009. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virol. J. 6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi GH, Larson TG, Nuss DL. 1992. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol. Plant Microb. Interact. 5:119–128 [DOI] [PubMed] [Google Scholar]
- 9. Churchill ACL, Ciuffetti LM, Hansen DR, Van Etten HD, Van Alfen NK. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Cur. Genet. 17:25–32 [Google Scholar]
- 10. Cornell CT, Kiosses WB, Harkins S, Whitton JL. 2006. Inhibition of protein trafficking by coxsackievirus B3: multiple viral proteins target a single organelle. J. Virol. 80:6637–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Culver JN, Padmanabhan MS. 2007. Virus-induced disease: altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 45:221–243 [DOI] [PubMed] [Google Scholar]
- 12. Fahima T, Kazmierczak P, Hansen DR, Pfeiffer P, Van Alfen NK. 1993. Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 195:81–89 [DOI] [PubMed] [Google Scholar]
- 13. Fahima T, Wu Y, Zhang L, Van Alfen NK. 1994. Identification of the putative RNA polymerase of Cryphonectria hypovirus in a solubilized replication complex. J. Virol. 68:6116–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao S, Choi GH, Shain L, Nuss DL. 1996. Cloning and targeted disruption of enpg-1, encoding the major in vitro extracellular endopolygalacturonase of the chestnut blight fungus, Cryphonectria parasitica. Appl. Environ. Microbiol. 62:1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gordon CL, et al. 2000. A glucoamylase :: Gfp gene fusion to study protein secretion by individual hyphae of Aspergillus niger. J. Microbiol. Meth. 42:39–48 [DOI] [PubMed] [Google Scholar]
- 16. Gullusci M, Turina M. 2007. Silencing of cryparin, a cell wall hydrophobin, in Cryphonectria parasitica. J. Plant Pathol. 89:141–147 [Google Scholar]
- 17. Hansen D, Van Alfen NK, Gilles K, Powell WA. 1985. Naked dsRNA associated with hypovirulence of Endothia parasitica is packaged in fungal vesicles. J. Gen. Virol. 66:2605–2614 [Google Scholar]
- 18. Harris SD, Morrell JL, Hamer JE. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Havir EA, Anagnostakis SL. 1985. Oxaloacetate acetylhydrolase activity in virulent and hypovirulent strains of (Endothia) Cryphonectria parasitica. Physiol. Plant Pathol. 26:1–10 [Google Scholar]
- 20. Hillman BI, Suzuki N. 2004. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 63:423–472 [DOI] [PubMed] [Google Scholar]
- 21. Hubbard MA, Kaminskyj SGW. 2008. Rapid tip-directed movement of golgi equivalents in growing Aspergillus nidulans hyphae suggests a mechanism for delivery of growth-related materials. Microbiol. 154:1544–1553 [DOI] [PubMed] [Google Scholar]
- 22. Jacob-Wilk D, Turina M, Kazmierczak P, Van Alfen NK. 2009. Silencing of kex2 significantly diminishes the virulence of Cryphonectria parasitica. Mol. Plant Microbe Interact. 22:211–221 [DOI] [PubMed] [Google Scholar]
- 23. Jacob-Wilk D, Turina M, Van Alfen NK. 2006. Mycovirus Cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica. J. Virol. 80:6588–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kazmierczak P, Kim DH, Turina M, Van Alfen NK. 2005. A hydrophobin of the chestnut blight fungus, Cryphonectria parasitica, is required for stromal pustule eruption. Eukaryot. Cell 4:931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kazmierczak P, Pfeiffer P, Zhang L, Van Alfen NK. 1996. Transcriptional repression of specific host genes by the mycovirus Cryphonectria hypovirus 1. J. Virol. 70:1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koonin E, Choi G, Nuss D, Shapira R, Carrington J. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U. S. A. 88:10647–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lorang JM, et al. 2001. Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 67:1987–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCabe PM, Van Alfen NK. 2002. Molecular basis of symptom expression by the Cryphonectria hypovirus, p 125–143 In Stellos MT. (ed), dsRNA genetic elements:concepts and applications in agriculture, forestry, and medicine. CRC Press, Boca Raton, FL [Google Scholar]
- 29. McCabe PM, Van Alfen NK. 1999. Secretion of cryparin, a fungal hydrophobin. Appl. Environ. Microbiol. 65:5431–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milgroom MG, Cortesi P. 2004. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42:311–338 [DOI] [PubMed] [Google Scholar]
- 31. Morin-Ganet M-N, Rambourg A, Deitz SB, Franzusoff A, Képès F. 2000. Morphogenesis and dynamics of the yeast Golgi apparatus. Traffic 1:56–68 [DOI] [PubMed] [Google Scholar]
- 32. Murray JL, et al. 2005. Rab9 gtpase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J. Virol. 79:11742–11751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newhouse JR, Hoch HC, Macdonald WL. 1983. The ultrastructure of Endothia parasitica. Can. J. Botany 61:389–399 [Google Scholar]
- 34. Padmanabhan MS, Shiferaw H, Culver JN. 2006. The tobacco mosaic virus replicase protein disrupts the localization and function of interacting AUX/IAA proteins. Mol. Plant Microbe Interact. 19:864–873 [DOI] [PubMed] [Google Scholar]
- 35. Pakula TM, et al. 2003. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei - evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 278:45011–45020 [DOI] [PubMed] [Google Scholar]
- 36. Pathak KB, Sasvari Z, Nagy PD. 2008. The host pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379:294–305 [DOI] [PubMed] [Google Scholar]
- 37. Poggeler S, Masloff S, Hoff B, Mayrhofer S, Kuck U. 2003. Versatile eGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54–61 [DOI] [PubMed] [Google Scholar]
- 38. Powell WA, Alfen NKV. 1987. Differential accumulation of poly(A)+ RNA between virulent and double-stranded RNA-induced hypovirulent strains of Cryphonectria (endothia) parasitica. Mol. Cell. Biol. 7:3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Puhalla JE, Anagnostakis S. 1971. Genetic and nutritional requirements of endothia parasitica. Phytopathology 61:169–173 [Google Scholar]
- 40. Rigling D, Van Alfen NK. 1993. Extra- and intracellular laccases of the chestnut blight fungus, Cryphonectria parasitica. Appl. Environ. Microbiol. 59:3634–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reference deleted.
- 42. Salonen A, Ahola T, Kaariainen L. 2005. Viral RNA replication in association with cellular membranes, p 139–173 In Marsh M. (ed), Membrane trafficking in viral replication. Springer-Verlag, Heidelberg, Germany: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, New York [Google Scholar]
- 44. Schaad MC, Jensen PE, Carrington JC. 1997. Formation of plant virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siriputthaiwan P, Jauneau A, Herbert C, Garcin D, Dumas B. 2005. Functional analysis of clpt1, a rab/gtpase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 118:323–329 [DOI] [PubMed] [Google Scholar]
- 46. Swennen D, Beckerich J-M. 2007. Yarrowia lipolytica vesicle-mediated protein transport pathways. Evol. Biol. 7:219–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valkonen M, et al. 2007. Spatially segregated snare protein interactions in living fungal cells. J. Biol. Chem. 282:22775–22785 [DOI] [PubMed] [Google Scholar]
- 48. Varley DA, Podila GK, Hiremath ST. 1992. Cutinase in Cryphonectria parasitica, the chestnut blight fungus: suppression of cutinase gene expression in isogenic hypovirulent strains containing double-stranded RNAs. Mol. Cell. Biol. 12:4539–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vollmer SJ, Yanofsky C. 1986. Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 83:4869–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei T, Wang A. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82:12252–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L, Baasiri RA, Van Alfen NK. 1998. Viral repression of fungal pheromone precursor gene expression. Mol. Cell. Biol. 18:953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang L, Villalon D, Sun Y, Kazmierczak P, Van Alfen NK. 1994. Virus-associated down-regulation of the gene encoding cryparin, an abundant cell surface protein from the chestnut blight fungus, Cryphonectria parasitica. Gene 139:59–64 [DOI] [PubMed] [Google Scholar]