Abstract
We previously constructed a recombinant monoclonal antibody (rec-MAb 63P4) that detects immediate-early protein IE63 encoded by varicella-zoster virus (VZV) in the cytoplasm of productively infected cells. Here, we used ORF63 truncation mutants to map the rec-MAb 63P4 binding epitope to amino acids 141 to 150 of VZV IE63, a region not shared with other widely used anti-IE63 antibodies, and found that the recombinant antibody does not bind to the simian IE63 counterpart.
TEXT
Varicella-zoster virus (VZV) causes childhood varicella (chickenpox), after which the virus becomes latent in multiple ganglia along the entire neuraxis. Decades later, as cell-mediated immunity to VZV declines, VZV can reactivate to cause zoster (shingles) (13). During latency, a number of VZV open reading frames (ORFs) are transcribed (6, 8, 16, 26). Of the VZV ORF transcripts analyzed quantitatively in latently infected human ganglia, ORF63 transcripts are the most prevalent and abundant (7). VZV ORF63 encodes immediate-early 63 (IE63), a 278-amino-acid protein expressed early after VZV infection in cell culture (10). Phosphorylated IE63 is found predominantly in the nuclei of productively infected cells (4, 9, 23, 24, 27) but exclusively in the cytoplasms of latently infected human ganglia, where its function in maintaining latency and its posttranslational modification are unknown (21).
Characterization of VZV IE63 cellular localization and protein interactions is critically dependent on the availability of well-defined antibodies. Previously, four anti-IE63 antibodies have been described: mouse monoclonal antibody (MAb) 9A12 (17), rabbit polyclonal anti-IE63 (RαIE63) (10), recombinant MAb 63E4 (rec-MAb 63E4) (24), and rec-Ab 63P4 (22). Cellular localization of IE63 has been determined using both RαIE63 and mouse MAb 9A12; however, concerns have recently arisen about detection of IE63 by immunohistochemistry, since intraneuronal deposition of lipofuscin and neuromelanin confounds analysis of IE63 in human trigeminal ganglia using rabbit anti-IE63 polyclonal antibodies (28). Moreover, random lots of commercially obtained ascites fluid-derived mouse anti-VZV antibodies contain mouse ascites Golgi-reactive antibodies that cross-react with blood type A1 determinants localized in the cytoplasm of human trigeminal ganglionic neurons (12, 18, 29). The large discrepancy in the frequency of immunohistochemical detection of cytoplasmic IE63 in latently infected neurons, ranging from often (19) to rarely (21, 28), further underscores the need for well-characterized anti-IE63 antibodies.
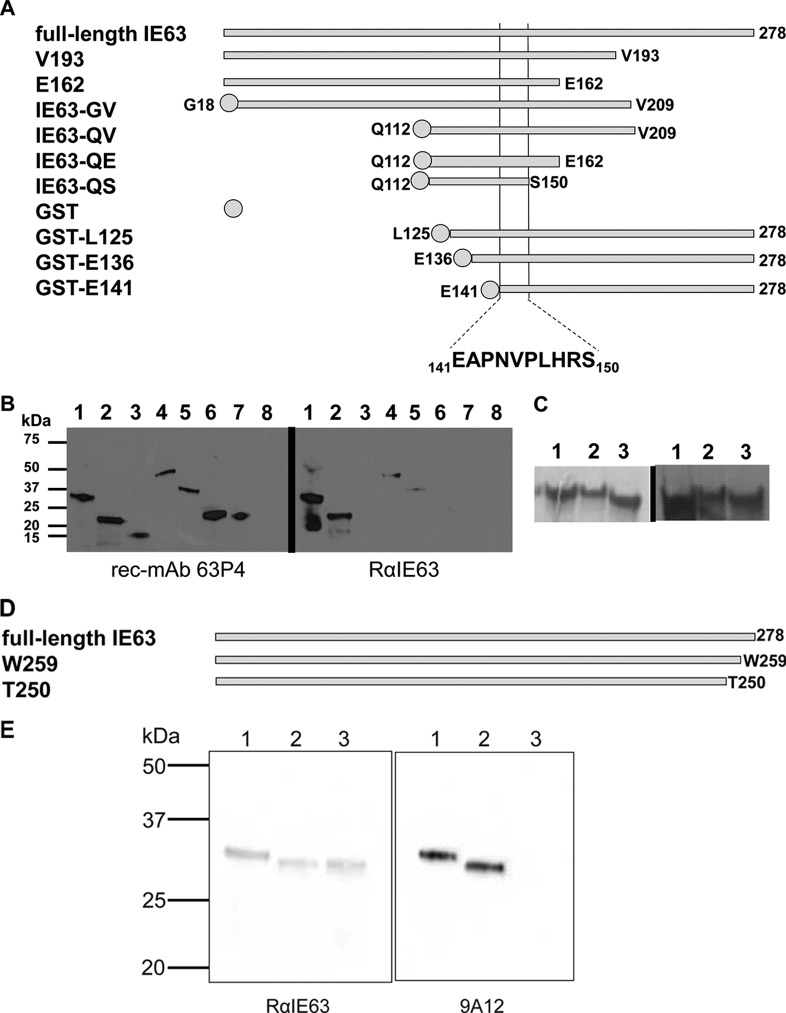
Here, we identified the binding epitope of rec-MAb 63P4, which detects the cytoplasmic form of IE63 (22), and compared it to the IE63 epitopes recognized by the widely used anti-VZV IE63 antibodies RαIE63 and mouse MAb 9A12, all of which were raised against bacterially expressed IE63. To locate the epitopes on IE63 recognized by rec-MAb 63P4 and RαIE63 antibodies, six IE63 truncation mutants were constructed by PCR-based cloning using oligonucleotide primers 1 to 7 (Table 1). Plasmid constructions were verified by DNA sequence analysis. Each recombinant protein was expressed, purified, and analyzed by Western blotting (22). Recombinant proteins included the full-length 278-amino-acid IE63, IE63 with C-terminal deletions at valine 193 (V193) and glutamic acid 162 (E162), internal segments of IE63 from glycine 18 to glutamic acid 209 (IE63-SE), glutamine 112 to glutamic acid 209 (IE63-AE), and glutamine 112 to glutamic acid 162 (IE63-QE), or IE63 with N-terminal deletions, stabilized with N-terminal glutathione S-transferase (GST) coding sequences, from leucine 125 (GST-L125), glutamic acid 136 (GST-E136), or glutamic acid 141 (GST-E141) (Fig. 1A). Western blot analysis of these proteins (Fig. 1B) showed that rec-MAb 63P4 detected full-length IE63 (Fig. 1B, lane 1), C-terminal truncations at V193 (lane 2), E162 (lane 3), and S150 (lane 7), and GST fusion proteins with IE63 N-terminal deletions at G18 (lane 4) and Q112 (lanes 5 to 7), but not the GST tag alone (lane 8), whereas RαIE63 detected full-length IE63 (lane 1) and only IE63 containing a C-terminal deletion at V193 (lane 2) and internal IE63 segments IE63-GV (lane 4) and IE63-QV (lane 5). Thus, the rec-MAb 63P4 epitope consists of 39 amino acids within IE63 between amino acids 112 and 150, whereas the epitope for RαIE63 is located between E162 and V193. The slanted protein bands in Fig. 1B result from imperfections in the 12% polyacrylamide gel, while the smaller bands (Fig. 1B, right, lanes 1 and 2) reflect IE63 degradation products, similar to those seen previously in Western blots using RαIE63 antibody to detect the protein (9). An additional three IE63 truncation mutants containing N-terminal GST tags were constructed (Table 1, primers 8 to 11), sequence verified, expressed, and purified, and they showed single bands with the appropriate mass in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1C, left). Western blot analysis showed that rec-MAb 63P4 detected N-terminal IE63 truncations at amino acids L125 (Fig. 1C, right, lane 1), E136 (lane 2), and E141 (lane 3). Together, these data reveal that rec-MAb 63P4 recognizes an IE63 epitope with the sequence 141EAPNVPLHRS150.
Table 1.
Oligonucleotide primers
| Primer | Sequencea |
|---|---|
| 1 | GGGACATGTTTTGCACCTCACCGG |
| 2 | TTTAAGCTTCACGCCATGGGGGGG |
| 3 | TTTTCTAGACTAATGATGATGATGATGATGAAGCTTCACTATAAAGTCTTC |
| 4 | TTTTCTAGACTAATGATGATGATGATGATGAAGCTTTTCACCACCATCATC |
| 5 | TTTGGATCCTCCAGCTTCAACCCACC |
| 6 | CCGGAATTCTTAGTCTTCACCACCATC |
| 7 | GGCGAATTCTTAGCTCCTATGCAAAGG |
| 8 | TCTGGATCCTAATGGAAGTGTCCC |
| 9 | TTTGGATCCAGGAAGACGGGTTCATTG |
| 10 | TTTGGATCCTTGAGGCGCCGAATGTTC |
| 11 | AAAGAATTCCTACACGCCATGGGG |
| 12 | TTTTCTAGACTAATGATGATGATGATGATGAAGCTTCCAATCTACAC |
| 13 | TTTCTAGACTAATGATGATGATGATGATGAAGCTTGGTGAGCGCTTTCGC |
| 14 | GAAGACGGGTTCATTGCGGCGCCGAATGTTCC |
| 15 | GGAACATTCGGCGCCGCAATGAACCCGTCTTC |
| 16 | GACGGGTTCATTGAGCCGCCGAATGTTCCTTTG |
| 17 | CAAAGGAACATTCGGCGGCTCAATGAACCCGTC |
| 18 | GAAGACGGGTTCATTGCGCCGCCGAATGTTCC |
| 19 | GGAACATTCGGCGGCGCAATGAACCCGTCTTC |
| 20 | GCCGAATGTTCCTTTGGCTGCGAGCGCACTGGAATG |
| 21 | CATTCCAGTGCGCTCGCAGCCAAAGGAACATTCGGC |
| 22 | GGAAGACGGGTTCATTGCTGCCGCAGCGGCTCCTTTGCATAGG |
| 23 | CCTATGCAAAGGAGCCGCTGCGGCAGCAATGAACCCGTCTTCC |
| 24 | GAGGCGCCGAATGTTGCCGCAGCGGCTGCCGCACTGGAATGTGACG |
| 25 | CGTCACATTCCAGTGCGGCAGCCGCTGCGGCAACATTCGGCGCCTC |
| 26 | CTTACTGACATCCACTTTGCC |
| 27 | CAAATAAAGCAATAGCATCACAAATTTCAC |
Restriction endonuclease sites used for cloning are underlined.
Fig 1.
Epitope mapping of rec-MAb 63P4, RαIE63, and mouse MAb 9A12 on VZV IE63. (A) Schematic representation of IE63 proteins used to map the epitopes of rec-MAb 63P4 and rabbit polyclonal anti-IE63 includes full-length IE63 protein and IE63 truncation mutants V193, E162, IE63-GV, IE63-QV, IE63-QE, IE63-QS, GST-L125, GST-E136, and GST-E141. (B) Western blot analysis using rec-MAb 63P4 (left), which detected metal affinity-purified full-length IE63 (lane 1), V193 (lane 2), E162 (lane 3), IE63-GV (lane 4), IE63-QV (lane 5), IE63-QE (lane 6), and IE63-QS (lane 7) but not the GST control (lane 8). Rabbit anti-IE63 antibodies (right) detected full-length IE63 (lane 1), V193 (lane 2), IE63-GV (lane 4), and IE63-QV (lane 5) but not E162 (lane 3), IE63-QE (lane 6), IE63-QS (lane 7), or the GST control (lane 8). (C) SDS-PAGE of Coomassie brilliant blue-stained, affinity-purified recombinant N-terminal truncation IE63 mutants (left) L125 (lane 1), E136 (lane 2), and E141 (lane 3) and detection with rec-MAb 63P4 on Western blots (right). (D) Schematic representation of IE63 proteins used to map the epitope of mouse anti-IE63 MAb 9A12 includes full-length 278-amino-acid IE63 and deletion mutants W259 and T250. (E) Western blot analysis of affinity-purified full-length IE63 (lane 1), W259 (lane 2), and T250 (lane 3) probed with rabbit polyclonal anti-IE63 (left) or mouse MAb 9A12 (right).
To map the IE63 mouse MAb 9A12 epitope, two additional C-terminal-deletion mutants of IE63 were constructed (Table 1, primers 1, 12, and 13), W259 (19 amino acids deleted) and T250 (28 amino acids deleted) (Fig. 1D). RαIE63 detected full-length IE63 and both deletion mutants (Fig. 1E, left), while mouse MAb 9A12 detected full-length IE63 (Fig. 1E, right, lane 1) and W259 (lane 2) but not T250 (lane 3). These findings localized the mouse MAb 9A12 IE63 epitope to a 10-amino-acid segment in IE63 at the C terminus between residues 250 and 259.
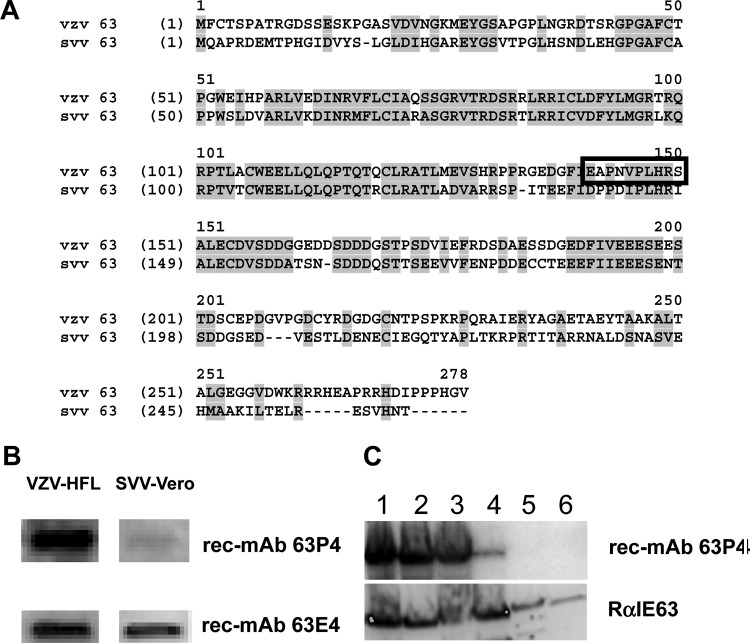
Like VZV, simian varicella virus (SVV) is a neurotropic alphaherpesvirus in the genus Varicellovirus. VZV and SVV share significant biological and molecular characteristics and express homologous IE63 proteins (14). Sequence alignment of VZV and SVV IE63 (Fig. 2A) indicates 52% amino acid identity. When the 10-amino-acid rec-MAb 63P4 VZV IE63 epitope is mapped on SVV IE63, five amino acids are identical and three are semiconserved (E141, N144, and V145); the remaining two VZV amino acids, A142 and S150, correspond to SVV P141 and I149, respectively. Substitution of alanine for S150 did not affect rec-MAb 63P4 binding (data not shown), indicating that the only notable difference between VZV and SVV IE63 within the VZV IE63 rec-MAb 63P4 epitope is A142 (VZV) versus P141 (SVV). Western blot analysis of VZV-infected human lung fibroblasts and SVV-infected Vero cells showed that both rec-MAbs 63P4 and 63E4 detected VZV IE63, while only rec-MAb 63E4 detected SVV IE63 (Fig. 2B). The epitope for rec-MAb 63E4 maps to a site 36 amino acids C-terminal to the rec-MAb 63P4 epitope, and antibody binding to IE63 requires phosphorylation at S186 (24). These findings indicate that the VZV IE63 epitope recognized by rec-MAb 63P4 is blocked by a proline at 142 in SVV. Mapping of the rec-MAb 63P4 binding site on IE63 in Fig. 1 was based on deletion mutations of IE63. It is conceivable that deletion mutations produce local protein folding changes that alter antibody binding, even under the denaturing conditions used in Western blot analysis. To eliminate this concern, specific mutations within the rec-MAb 63P4 binding site of IE63 (141EAPNVPLHRS150) were introduced into the full-length protein (Table 1, primers 14 to 27). The plasmid constructions were verified by DNA sequence analysis, and the recombinant protein was expressed and analyzed on Western blots (Fig. 2C). In the context of full-length IE63, mutations of 141E to 141A (Fig. 2C, lane 1), 142A to 142P (lane 2), 142EA to 142AP (lane 3), and 148HR to 148AA (lane 4) did not eliminate binding of rec-MAb 63P4. However, changing 141EAPNV145 to 141AAAAA145 (Fig. 2C, lane 5) or 146DLHRS150 to 146AAAAA150 (lane 6) inhibited rec-MAb 63P4 binding. All substitutions were detected with the polyclonal rabbit antibody (RαIE63) whose epitope mapped between E162 and V193. In summary, rec-Ab 63P4 recognizes 10 amino acids in IE63 (141EAPNVPLHRS150), in which minor substitutions are permitted but major substitutions abrogate antibody binding.
Fig 2.
Fine-structure mapping rec-MAb 63P4 binding site on IE63. (A) Amino acid alignment of VZV IE63 and SVV IE63, with conserved residues between VZV and SVV IE63 shaded and the rec-MAb 63P4 epitope boxed. (B) Western blot analysis of VZV-infected HFL cells and SVV-infected Vero cells, indicating that rec-MAb 63P4 detected VZV IE63 but not SVV IE63, whereas control antibody rec-MAb 63E4 detected both VZV and SVV IE63. (C) Western blot analysis of full-length VZV IE63 with substitutions in the rec-MAb 63P4 epitope. The substitution of E141 with alanine (lane 1) or A142 with proline (lane 2) or the double substitution EA to AP (lane 3) did not affect rec-MAb 63P4 binding. Also, substitution of H148 and R149 with two alanines (lane 4) did not eliminate rec-MAb 63P4 binding. Alanine substitution of either the first 5 amino acids (E141 to V145) (lane 5) or the last 5 amino acids (P146 to S150) (lane 6) eliminated rec-MAb 63P4 binding.
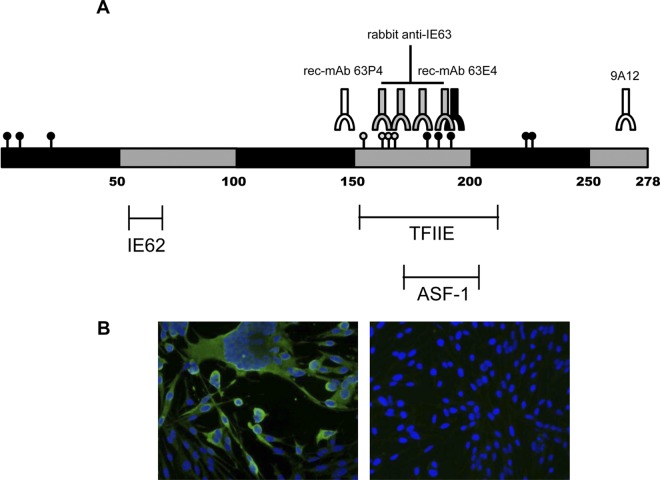
Figure 3A shows the phosphorylation sites and regions required for antibody binding to VZV IE63. Note that unlike rec-MAb 63E4 and RαIE63, the epitope for rec-MAb 63P4 binding is located toward the N terminus of the protein. While the central region contains multiple phosphorylation sites, and rec-MAb 63E4 binding is dependent on phosphorylation of S186, rec-MAb 63P4 binding is independent of posttranslational modifications (24). Also, rec-MAb 63P4 detects a cytoplasmic form of IE63 (Fig. 3B), indicating that the epitope is blocked by either structural rearrangement (epitope sequestering), posttranslational modification, or competing protein interactions within the nucleus. Alternatively, nuclear IE63 may be in the form of a dimer, while cytoplasmic IE63 may exist in a monomeric form. Since the rec-MAb 63P4 epitope (EAPNVPLHRS) does not contain any predicted posttranslational modification sites, the lack of nuclear IE63 staining most likely reflects an IE63 structural rearrangement or binding to RNA polymerase II, ASF-1, TFIIE, VZV IE62, or an as-yet-unidentified nuclear protein (1–3, 11, 20).
Fig 3.
(A) Location of protein and antibody binding sites on phosphorylated VZV IE63. Black and gray circles indicate confirmed and suspected IE63 phosphorylated amino acid residues, respectively. The binding sites for RαIE63, mouse MAb 9A12, and two rec-MAbs are indicated, as well as locations of cellular and viral protein binding sites on IE63. IE62, VZV immediate-early protein 62; TFIIE, transcription factor IIE; ASF-1, antisilencing factor 1. (B) Probing of VZV-infected MeWo (left panel) and control MeWo (right panel) cells with rec-MAb 63P4 (green) shows predominantly cytoplasmic staining. Nuclei were identified by 4′,6′-diamidino-2-phenylindole (DAPI) (blue) staining. Magnification, ×40.
The cytoplasmic localization of IE63 has been identified both in productively infected cells in culture and in latently infected neurons, suggesting functions in addition to those in the nucleus (5, 15, 19, 21). Since rec-MAb 63P4 recognizes IE63 in the cytoplasm, this antibody may facilitate identification of cellular proteins involved in IE63 function(s) by coimmunoprecipitation, as well as providing a well-defined antibody for further in situ histochemistry studies.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants NS32623 (R.J.C., R.M., and D.H.G.), AG032958 (R.J.C., R.M., and D.H.G.), and AG006127 (D.H.G.) from the National Institutes of Health.
We thank Nancy J. Tyson (Regis University) for expert technical assistance, Sebastien Bontems (University of Liège; Liège, Belgium) for the 9A12 antibody, Marina Hoffman for editorial review, and Lori DePriest for manuscript preparation.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Ambagala AP, Cohen JI. 2007. Varicella-zoster virus IE63, a major viral latency protein, is required to inhibit the alpha interferon-induced antiviral response. J. Virol. 81:7844–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambagala AP, et al. 2009. Varicella-zoster virus immediate-early 63 protein interacts with human antisilencing function 1 protein and alters its ability to bind histones H3.1 and H3.3. J. Virol. 83:200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baiker A, et al. 2004. The immediate-early 63 protein of varicella-zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo. J. Virol. 78:1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bontems S, et al. 2002. Phosphorylation of varicella-zoster virus IE63 protein by casein kinases influences its cellular localization and gene regulation activity. J. Biol. Chem. 277:21050–21060 [DOI] [PubMed] [Google Scholar]
- 5. Chen JJ, Zhu Z, Gershon AA, Gershon MD. 2003. Latent and lytic infection of isolated guinea pig enteric ganglia by varicella-zoster virus. J. Med. Virol. 70:S71–S78 [DOI] [PubMed] [Google Scholar]
- 6. Cohrs RJ, Barbour M, Gilden DH. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70:2789–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohrs RJ, Gilden DH. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J. Virol. 81:2950–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohrs RJ, Gilden DH, Kinchington PR, Grinfeld E, Kennedy PG. 2003. Varicella-zoster virus gene 66 transcription and translation in latently infected human ganglia. J. Virol. 77:6660–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohrs RJ, Wischer J, Essman C, Gilden DH. 2002. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 76:7228–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Debrus S, Sadzot-Delvaux C, Nikkels AF, Piette J, Rentier B. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 69:3240–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Valentin E, et al. 2005. Varicella-zoster virus IE63 protein represses the basal transcription machinery by disorganizing the pre-initiation complex. Biol. Chem. 386:255–267 [DOI] [PubMed] [Google Scholar]
- 12. Finstad CL, et al. 1991. Some monoclonal antibody reagents (C219 and JSB-1) to P-glycoprotein contain antibodies to blood group A carbohydrate determinants: a problem of quality control for immunohistochemical analysis. J. Histochem. Cytochem. 39:1603–1610 [DOI] [PubMed] [Google Scholar]
- 13. Gilden DH, Cohrs RJ, Mahalingam R. 2003. Clinical and molecular pathogenesis of varicella virus infection. Viral Immunol. 16:243–258 [DOI] [PubMed] [Google Scholar]
- 14. Gray WL, Starnes B, White MW, Mahalingam R. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123–130 [DOI] [PubMed] [Google Scholar]
- 15. Hood C, et al. 2006. Varicella-zoster virus ORF63 inhibits apoptosis of primary human neurons. J. Virol. 80:1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kennedy PG, Grinfeld E, Bell JE. 2000. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 74:11893–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy PG, Grinfeld E, Bontems S, Sadzot-Delvaux C. 2001. Varicella-zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289:218–223 [DOI] [PubMed] [Google Scholar]
- 18. Kliman HJ, et al. 1995. A mucin-like glycoprotein identified by MAG (mouse ascites golgi) antibodies. Menstrual cycle-dependent localization in human endometrium. Am. J. Pathol. 146:166–181 [PMC free article] [PubMed] [Google Scholar]
- 19. Lungu O, Panagiotidis CA, Annunziato PW, Gershon AA, Silverstein SJ. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. U. S. A. 95:7080–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lynch JM, Kenyon TK, Grose C, Hay J, Ruyechan WT. 2002. Physical and functional interaction between the varicella-zoster virus IE63 and IE62 proteins. Virology 302:71–82 [DOI] [PubMed] [Google Scholar]
- 21. Mahalingam R, et al. 1996. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. U. S. A. 93:2122–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller NH, et al. 2008. Construction of recombinant mouse IgG1 antibody directed against varicella zoster virus immediate early protein 63. Hybridoma 27:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mueller NH, Graf LL, Orlicky D, Gilden D, Cohrs RJ. 2009. Phosphorylation of the nuclear form of varicella zoster virus immediate-early protein 63 by casein kinase II at serine 186. J. Virol. 83:12094–12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mueller NH, et al. 2010. Identification of phosphorylated residues on varicella zoster virus immediate-early protein ORF63. J. Gen. Virol. 91:1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reference deleted.
- 26. Nagel MA, Choe A, Traktinskiy I, Cordery-Cotter R, Gilden D, Cohrs RJ. 2011. Varicella-zoster virus transcriptome in latently infected human ganglia. J. Virol. 85:2276–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ng TI, Keenan L, Kinchington PR, Grose C. 1994. Phosphorylation of the varicella-zoster virus open reading frame (ORF) 62 regulatory product by viral ORF47-associated protein kinase. J. Virol. 68:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zerboni L, et al. 2010. Expression of varicella-zoster virus immediate-early protein IE63 in neurons of latently infected human sensory ganglia. J. Virol. 84:3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zerboni L, et al. 2012. Apparent expression of varicella-zoster virus proteins in latency resulting from reactivity of murine and rabbit antibodies with human blood group A determinants in sensory neurons. J. Virol. 86:578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]