Abstract
Distal intraexon (iE) regulatory elements in 4.1R pre-mRNA govern 3′ splice site choice at exon 2 (E2) via nested splicing events, ultimately modulating expression of N-terminal isoforms of cytoskeletal 4.1R protein. Here we explored intrasplicing in other normal and disease gene contexts and found conservation of intrasplicing through vertebrate evolution. In the paralogous 4.1B gene, we identified ∼120 kb upstream of E2 an ultradistal intraexon, iEB, that mediates intrasplicing by promoting two intricately coupled splicing events that ensure selection of a weak distal acceptor at E2 (E2dis) by prior excision of the competing proximal acceptor (E2prox). Mutating iEB in minigene splicing reporters abrogated intrasplicing, as did blocking endogenous iEB function with antisense morpholinos in live mouse and zebrafish animal models. In a human elliptocytosis patient with a mutant 4.1R gene lacking E2 through E4, we showed that aberrant splicing is consistent with iER-mediated intrasplicing at the first available exons downstream of iER, namely, alternative E5 and constitutive E6. Finally, analysis of heterologous acceptor contexts revealed a strong preference for nested 3′ splice events at consecutive pairs of AG dinucleotides. Distal regulatory elements may control intrasplicing at a subset of alternative 3′ splice sites in vertebrate pre-mRNAs to generate proteins with functional diversity.
INTRODUCTION
Eukaryotic genes utilize a versatile array of RNA processing mechanisms to generate an enormous proteome from a modest number of genes (22). The majority of human genes exhibit alternative splicing of one or more internal exons, many of which are subject to tissue-specific or developmental-stage-specific regulation that contributes to functional differentiation. Moreover, it is becoming apparent that many if not most genes also possess alternative promoters associated with distinct first-exon sequences that can be distributed over wide regions at the 5′ ends or can be internal to the gene (2, 18).
Genes in the protein 4.1 family of cytoskeletal adaptor proteins employ alternative transcription initiation at multiple promoters/first exons, as well as alternative pre-mRNA splicing at internal cassette exons, to encode a complex complement of tissue-specific polypeptides (3, 7, 28). Many of the protein 4.1 isoforms encoded by these genes are expressed in tissue- or differentiation-specific patterns, where they exhibit different subcellular localization and fulfill a variety of cytoskeletal linking functions. Proteins encoded by the 4.1B gene are important for heart development via an integrin αvβ8-dependent mechanism (16, 20); they also recruit neurotransmitter receptors to the synapse (14), play a role in paranodal and juxtaparanodal adhesion complexes (13), and function in maintaining the structure of the Golgi (17). 4.1B is also a reported tumor suppressor (6, 12, 27, 30).
We are studying the molecular mechanisms by which pre-mRNA splicing pathways regulate synthesis of 4.1B protein isoforms. Of particular interest is the 4.1B gene's capability of encoding two size classes of protein by utilization of alternative translation initiation sites AUG1 and AUG2 in exons 2 and 4, respectively. Interestingly, expression of AUG1 in mature mRNA is regulated by a poorly understood mechanism that involves coupling between transcription at mutually exclusive alternative promoters and splicing at alternative 3′ splice acceptor sites far downstream in exon 2 (28). All transcripts initiated at exon 1B (E1B) splice directly to the first (proximal) 3′ splice site at exon 2 (E2), designated E2prox (28). This is consistent with previous studies showing in general that proximal acceptors are favored over distal sites (9) and in particular that E2prox is predicted to be a stronger acceptor than E2dis (28). Paradoxically, however, exon 1A (E1A) splices almost exclusively to the weaker distal 3′ acceptor, E2dis. This unusual splice site choice has a major impact on protein structure and function, because the translation initiation codon AUG1 is located between E2prox and E2dis. The E1A-to-E2dis splice deletes AUG1 and produces an alternative mRNA isoform that encodes shorter proteins lacking the N-terminal headpiece of ∼200 amino acids. Physiologically, the presence or absence of the headpiece likely has a substantial effect on protein 4.1B binding, since a similar phenomenon has already been reported with regard to binding of the paralogous 4.1R system to components of the erythroid membrane skeleton (23).
Our study explored the splicing mechanism by which 4.1B E1A bypasses the strong E2prox acceptor to splice selectively at the weaker E2dis acceptor. We tested the hypothesis that E1A splicing is determined by an intron-regulatory element(s) uniquely present in E1A-initiated transcripts, not transcripts originating at E1B, that is, by elements mapping between E1A and E1B. The prototype for promoter position-dependent regulation of this type is the protein 4.1R gene (15, 25, 28). This study utilized minigene splicing reporters in cultured cells and antisense vivo-morpholinos in animal experiments to demonstrate that an unannotated, ultradeep intron element in 4.1B pre-mRNA is required for proper E1A-E2dis splicing. Additional experiments revealed specific alternative 3′ splice site architectural features that are required for accurate intraexon-mediated regulation, opening the possibility that deep intron elements might have a wider role in 3′ splice site choice for a subset of heterologous exons.
MATERIALS AND METHODS
Splicing reporter construction.
Minigene −iEB consisted of two regions from the 5′ region of the human 4.1B gene: the E1A promoter region together with E1A exon sequences and a short region of downstream intron sequence (in the February 2009 version of the human genome assembly, chr18:5,630,171-5,632,114) and E2 with a short upstream intronic region (chr18:5,489,000-5,489,451). Splicing reporter +iEB contained an additional 0.5-kb fragment spanning the 4.1B intraexon (chr18:5,609,935-5,610,458) inserted between E1A and E2, while the size-matched control −iEBsc lacked the intraexon but contained an equal length of additional intron sequence downstream of E1A. Finally, reporter +iER contained the 4.1R gene intraexon substituted for the 4.1B intraexon. All genomic fragments were cloned into the pcDNA3 expression vector from which the cytomegalovirus (CMV) promoter region had been removed by deleting a 740-nucleotide (nt) region between NruI and EcoRV sites, so that transcription was dependent on the E1A promoter. The intermediate splicing reporter (see Fig. 5D) was similar to construct +iEB, except that intron sequences between the intraexon and E2 were removed, leaving iE immediately adjacent to E2.
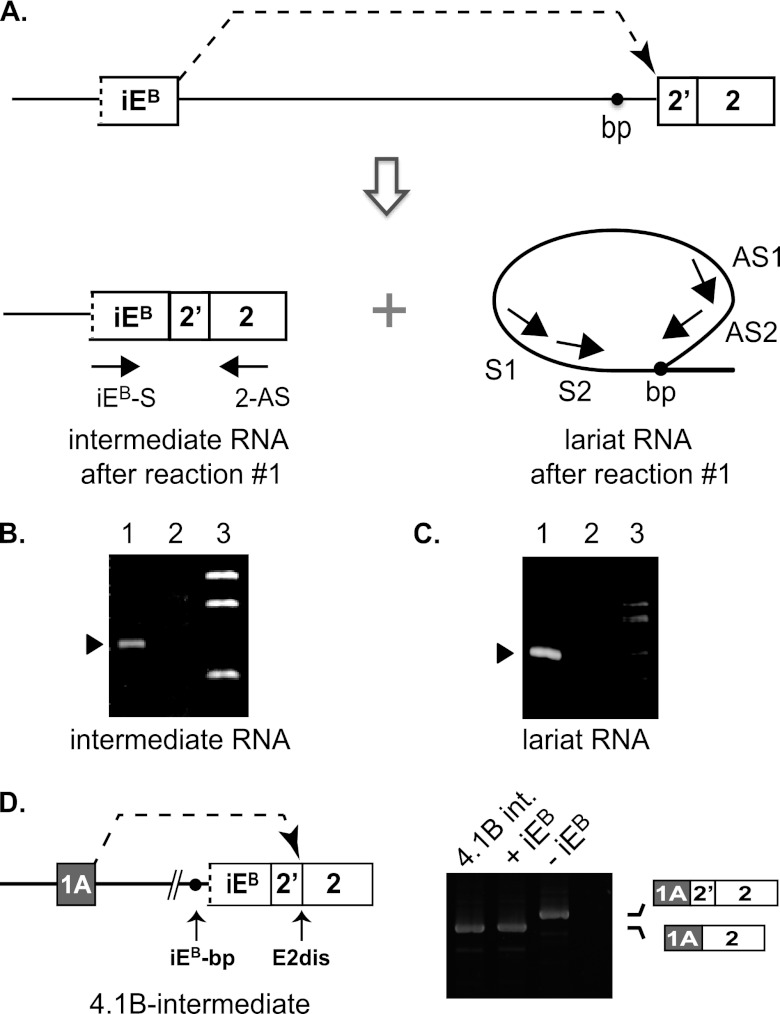
Fig 5.
Analysis of intermediate products generated in the first intrasplicing reaction in 4.1B pre-mRNA. (A) Model showing the expected products of the first intrasplicing event: iEB spliced to E2prox, and a lariat intron released in this reaction. Amplification of the branch point region of the lariat RNA was performed via nested PCRs involving antisense primers (AS1 and AS2) located downstream of the iEB 5′ splice site that loops back to the branch point and two sense primers (S1 and S2) located upstream of the branch point. (B) Gel analysis of the predicted 4.1B intermediate RNA in mouse kidney by RT-PCR analysis using primers in iE and E2. Lane 1, detection of intermediate RNA from normal kidney; lane 2, absence of intermediate RNA from kidney exposed to vMO against the 4.1B iEB-5′ss; lane 3, molecular size standards. (C) Gel analysis of the predicted lariat RNA branch point formed in the first intrasplicing reaction, using the nested primers diagrammed in panel A to detect small amounts of this RNA. Lane 1, product obtained after both nested PCRs; lane 2, no product observed after the first PCR with primers AS1 + S1; lane 3, molecular size standards. (D) (Left) Diagram of a reporter construct used to test whether the predicted splicing intermediate is a bona fide precursor of the fully processed mature E1A-E2dis mRNA. (Right) Analysis of spliced products generated in HEK293 cells transfected with the following reporters: intermediate RNA (4.1B-int), +iEB reporter, shown in Fig. 2, and −iEB reporter, also from Fig. 2. The last lane is a negative RT-PCR control.
Splicing analysis.
Splicing reporter minigenes in most experiments were transfected into mouse Swiss 3T3 cells using the Fugene transfection reagent (Roche) as described previously (25). Splicing analysis of the 4.1B intrasplicing intermediate (see Fig. 5) was analyzed in human HEK293 cells. Total RNA was extracted from cells with Qiagen's RNeasy minikit and then reverse transcribed into cDNA using the Superscript III first-strand synthesis system (Invitrogen) with a reverse primer specifically targeting the pcDNA3 vector (5′-TACAAGGCACAGTCGAGG-3′). The identity of all PCR products was confirmed by sequence analysis. The following primers were used in subsequent PCR assays designed to distinguish between correct and incorrect splicing at the alternative splice acceptors at E2: E1A forward primer, 5′-CTGTGAGCAGCCCTACCTCTCTCT-3′; E2 reverse primer, 5′-AGCGGCGGCGAACTGCTCCAG-3′. All experiments were performed at least three times with results similar to those presented in the figures.
Branch point mapping.
Amplification of lariat intermediates was performed as described previously (29). Nested PCRs were performed using forward primers upstream of the branch point region and reverse primers downstream of the intraexon: first reaction, forward primer, 5′-TCCAGCGTGTGATGTAGGACTG-3′, and reverse primer, 5′-ATTGGCTGGTTTCTGTTTCTCG-3′; second reaction, forward primer, 5′-TAGGACTGACTGACTGTGCTTGGC-3′, and reverse primer, 5′GGGTCACATAAACTCCATACAGAC-3′. DNA sequence analysis of the product confirmed joining of the intraexon 5′ splice site to a consensus branch point upstream of exon 2. As reported earlier (29), the branch point A nucleotide was read as a T in the amplified product.
Vivo-morpholinos.
All vivo-morpholino experiments were repeated at least twice. Antisense morpholinos were 25 nt in length and were obtained from Gene Tools, LLC (Philomath, OR). For mouse experiments, we used vivo-morpholinos containing a covalently linked octaguanidine dendrimer as a delivery moiety to facilitate entry into cells in vivo. Mice were injected into the tail vein at 15 mg/kg on two consecutive days, and then RNA was purified from selected tissues on the third day. Tissues were rinsed in 1× phosphate-buffered saline (PBS), snap-frozen in liquid nitrogen, and stored at −80°C until RNA was isolated by using Qiagen's RNeasy minikit, per the manufacturer's instructions. For zebrafish, the morpholino blocking the intraexon 5′ splice site was diluted to a final concentration of 2 ng/nl in 0.2 M KCl and 0.1% phenol red and injected into the yolks of 1-cell-stage embryos. Doses ranging from 2 to 10 ng of morpholinos (MO) were tested, with no toxicity observed. Embryos were incubated at 28.5°C for 24 h, and then RNA was purified from whole embryos by solubilization and extraction in TRIzol (Invitrogen) according to the manufacturer's instructions, followed by further purification using a Macherey-Nagel NucleoSpin RNA II kit. Vivo-morpholino sequences were as follows: mouse 4.1B iE-5′ss, 5′-ATATTAACTCCAGGACTCACCATGT-3′; mouse 4.1B iE-bp, 5′-CCATGTTCCCGGAGGCAGTGAGACA-3′ (bold and underlined nucleotide indicates the branch point position); mouse 4.1R iE-5′ss, 5′-TACATCAAAGAAGTACTCACCCAGA-3′. The morpholino sequence used in zebrafish to block the intraexon 5′ splice site was 5′-CTTTTGGAAATCAACTCACCAGCGC-3′.
RESULTS
Splicing of 4.1B exon 1A is almost exclusively coupled to the distal acceptor in E2.
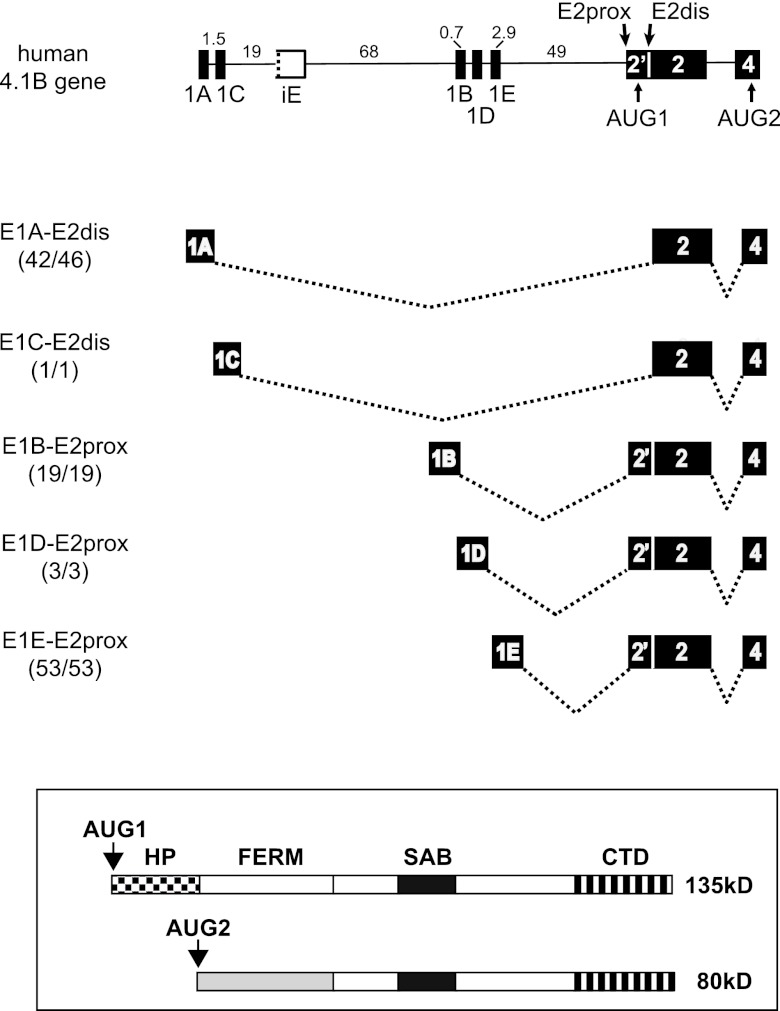
Analysis of protein 4.1B transcripts displayed at the UCSC genome browser revealed a strong coupling between specific alternative first exons and alternative 3′ splice site selection at E2. As shown in Fig. 1, E1A splices mostly to E2dis (42/46 transcripts; three of the nonconforming clones derive from “synovial membrane from rheumatoid arthritis” and could represent aberrant splicing in diseased tissue). A minor subclass of 4.1B transcripts initiated 2 kb downstream at newly designated E1C also splices to E2dis. The splicing pattern exhibited by E1A and E1C leads to skipping of the translation start site AUG1 and to synthesis of smaller 4.1B protein isoforms from an alternative start site in exon 4 (Fig. 1, bottom). In contrast, all E1B- (19/19), E1D- (3/3), and E1E-initiated transcripts (53/53) splice to E2prox. These results confirm and greatly extend our previous observations (28). Among 4.1B cDNAs in the mouse, the same correlation was observed: all E1A transcripts (5/5) splice to E2dis, while E1B transcripts (12/12) select E2prox. Splicing to E2prox impacts 4.1B protein expression by including AUG1 so that the resulting mRNAs encode isoforms with an extended N-terminal domain, the headpiece (Fig. 1, bottom).
Fig 1.
Promoter location relative to the iE regulator determines alternative splicing decisions that determine the N-terminal structure of protein 4.1B isoforms. (Top) Exon-intron arrangement at the 5′ end of the 4.1B gene. Numbers indicate intron lengths (in kb) for the human gene. Alternative first exons are indicated E1A-E1E; E2prox and E2dis represent alternative 3′ splice sites in E2; AUG1 and AUG2 represent alternative translation initiation sites. The putative intraexon, iE, is drawn with a broken line on one side to indicate the lack of a functional 3′ splice site. Depicted below the gene model are pre-mRNA splice patterns derived from databases showing that alternative first exons 1A and E1C splice to E2dis, while E1B, 1D, and 1E splice to E2prox. Numbers in parentheses indicate the fraction of all database transcripts initiated at a given promoter that splice to E2 as shown. (Bottom) Larger protein isoforms encoded by transcripts that splice to E2prox and include AUG1, and smaller proteins encoded by transcripts that splice to E2dis so as to skip AUG1 and initiate translation downstream at AUG2. Domains of 4.1R protein: HP, headpiece at N terminus; FERM, 4.1/ezrin/radixin/moesin homology domain; SAB, spectrin-actin binding domain; CTD, C-terminal domain.
Together these results support the hypothesis that E1A splicing to E2dis is regulated by a mechanism that allows bypass of the stronger proximal acceptor site (E2prox) in favor of the weaker distal site at E2dis (28). A clue to the mechanism lies in the observation that promoter location is strongly correlated with splice site selection: transcripts initiated at the set of upstream promoters splice differently from those initiated at downstream promoters. Similar findings with the paralogous 4.1R gene were explained by the discovery of a deep intron regulatory element, the intraexon (iER), which mapped between upstream and downstream promoters and determined splicing behavior (25). We therefore proposed that an iER-like element might regulate splicing at alternative acceptors in the 4.1B gene.
A deep intron splicing regulator acts at a distance of >100 kb to determine E2dis splice site choice in 4.1B pre-mRNA.
The prototypical iER in the protein 4.1R gene has a unique structure, resembling an exon in size and possessing strong branch point and 5′ splice site sequences but including no AG dinucleotides and lacking a functional 3′ splice site (25). To determine whether an intraexon regulatory element was present in the 4.1B gene, we analyzed 142 kb of genomic DNA between E1A and E2 in the human 4.1B gene and 134 kb of the orthologous intron in the mouse 4.1B gene. For both human and mouse introns, a single candidate intraexon meeting these criteria was identified. These predicted intraexons mapped to ultradeep intron regions, ∼21 kb downstream of E1A and 121 kb upstream of E2 in the human gene (Fig. 1), with analogous distances of 21 kb and 111 kb in the mouse gene. This gene structure would allow the intraexons to be included in, and to regulate splicing of, only the upstream-initiated transcripts.
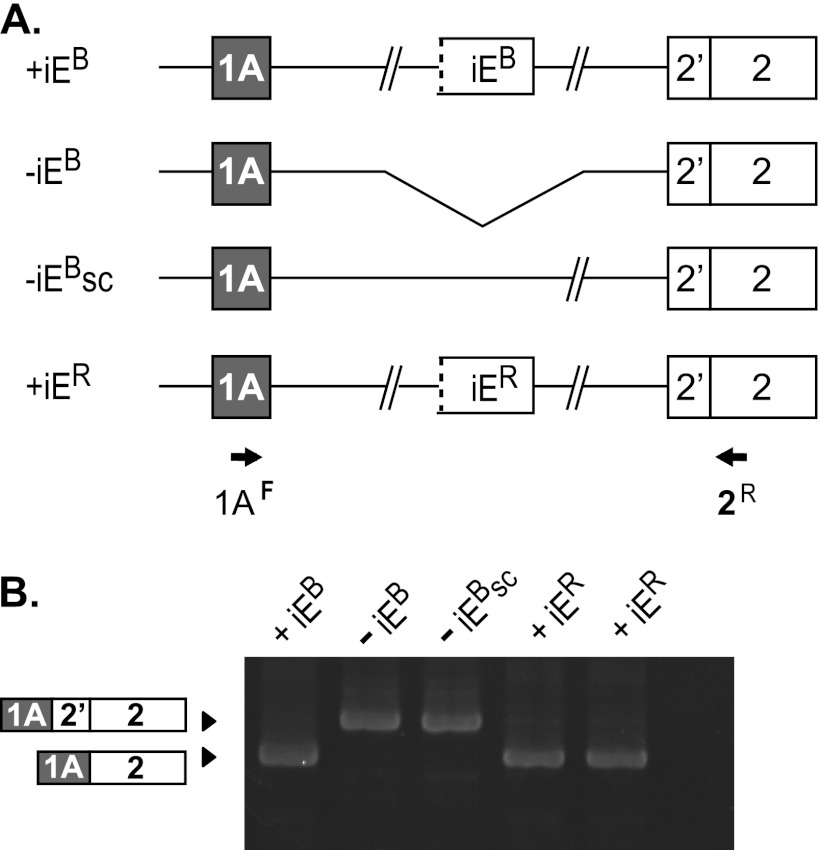
The splicing-regulatory activity of the 4.1B intraexon was tested in the context of human 4.1B minigene splicing reporters that either lacked or contained this element (Fig. 2A). Construct −iEB consisted of two blocks of sequence: E1A with its natural transcriptional promoter region, and E2 with its alternative 3′ splice sites. Both exons retained modest flanking intron sequences, but most of the very long intervening sequence between these elements in endogenous pre-mRNA was not included. Construct +iEB was made by insertion of a 0.5-kb fragment containing the putative iEB with some flanking intron sequence. When these reporters were transfected into mouse 3T3 cells, construct +iEB exhibited correct splicing of E1A to E2dis (Fig. 2B, lane 1). In marked contrast, the shorter construct −iEB yielded splicing of E1A only to the (inappropriate) E2prox acceptor site (Fig. 2B, lane 2). A size control generated by adding 0.5 kb of intron sequence to −iEB (−iEBsc) also failed to splice E1A correctly to E2dis (Fig. 2B, lane 3). Finally, iER could be substituted for iEB with no loss of functionality (Fig. 2B, lane 4), demonstrating that these two intraexons have similar regulatory activity. Together these results suggest that the iEB is a powerful regulatory element that can function at a remarkable distance deep in the intron to enable E1A to bypass the default proximal acceptor in favor of the distal site.
Fig 2.
The 4.1B intraexon is a powerful splicing regulator required for E1A splicing to E2dis. (A) Splicing reporter constructs either lacking (−iEB) or containing inserts of 0.5 kb encompassing the 4.1B intraexon element (+iEB), additional intron sequence downstream of E1A as a size control (−iEsc), or the 4.1R intraexon (+iER). (B) Splicing analysis of the reporters after transfection into HEK293 cells, using the indicated PCR primers. Structure of the PCR products was confirmed by sequence analysis. Lanes are labeled with names of the constructs.
Intraexon function is essential for proper E1A-E2dis splicing in natural full-length 4.1B pre-mRNA in vivo.
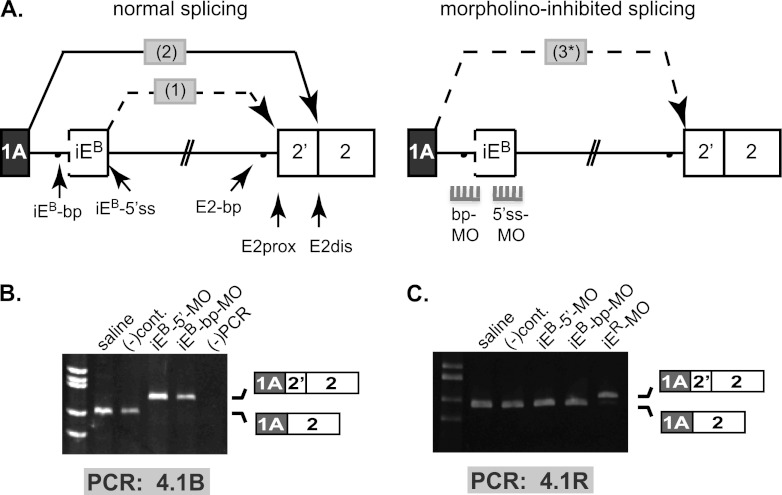
We next investigated iEB function in the context of natural full-length pre-mRNA, where it is quite remote (in the linear sequence) from the regulated splice site. Our strategy was to use antisense vivo-morpholinos (vMOs) that can be introduced systemically into mice via tail vein injection, after which they can cross cell membranes and hybridize to complementary cis-regulatory signals in pre-mRNA so as to induce splicing changes in vivo (21, 26). According to the model (Fig. 3A, left), the iEB 5′ splice site should splice to E2prox (splicing event 1) to generate an intermediate product; then, E1A should splice to the composite E2dis acceptor generated by juxtaposition of the iEB branch point with the AG dinucleotide at E2dis (splicing event 2). To test this model, we used antisense vMOs complementary to either the iEB 5′ splice site (iEB-5′ss) or the iEB branch point (iEB-bp) to block function in vivo. The predicted consequence of blocking iEB function is that E1A would splice in a single step directly to the proximal 3′ splice site at E2 (splicing event 3) (Fig. 3A, right).
Fig 3.
Splice-blocking vivo-morpholinos validate intraexon regulatory function in full-length endogenous 4.1B pre-mRNA in vivo. (A) (Top) The two nested splicing events (1 and 2) proposed for joining E1A to E2dis (left). Splice-blocking morpholinos targeting the iEB 5′ splice site (5′ss-MO) and branch point (bp-MO) were predicted to abolish iEB function, leading to aberrant one-step splicing (3*) of E1A to E2prox (right). (B) RT-PCR analysis of endogenous mouse kidney 4.1B transcripts with primers located in E1A and E2, using RNA from animals treated with sterile saline, a negative-control vMO [(-)cont], or iEB-specific vMOs against iEB-5′ss and iEB-bp. The first lane shows size standards, and the last lane is an RT-PCR negative control. (C) RT-PCR analysis using 4.1R-specific primers to show that splicing of endogenous mouse kidney 4.1R transcripts was not altered by vMOs against the paralogous 4.1B pre-mRNA. Sources of RNA (indicated above each lane) were isolated from animals injected with sterile saline, with negative-control vMO, with 4.1B-specific vMOs against iEB-5′ss or iEB-bp, or with a 4.1R-specific vMO against iER-5′ss. The first lane shows size standards, and the last lane is an RT-PCR negative control. For both panels B and C, correct splicing of E1A to E2dis was observed in mice treated with sterile saline or control vMOs. Gene-specific vMOs induced aberrant splicing of E1A to E2prox only in the cognate 4.1R or 4.1B pre-mRNA; no cross-regulation of the paralogous pre-mRNA was observed.
As shown in Fig. 3B, the normal physiological E1A→E2dis splicing pattern was observed in kidney RNA from saline- or control vMO-injected mice (first two lanes). In contrast, treatment with the gene-specific 4.1B iEB-5′ss vMO induced a virtually complete switch of E1A splicing to E2prox in kidney 4.1B transcripts. Similarly, a second independent vMO targeting the putative 4.1B intraexon branch point also induced a strong switch in E1A splicing from E2dis to E2prox (Fig. 3B, lane iEB-bp-MO). DNA sequence analysis confirmed the identity of all splice junctions in these products. As additional controls for vMO specificity, we showed that neither the 4.1B vMOs nor the negative-control vMO altered splicing of the paralogous 4.1R transcript (Fig. 3C) under conditions in which a 4.1R iER-directed vMO did switch splicing in 4.1R pre-mRNA. vMO directed against the paralogous 4.1R−iER did not alter splicing in 4.1B (data not shown).
RNA intermediate and lariat structures predicted by the 4.1B intrasplicing model are present in normal mouse kidney.
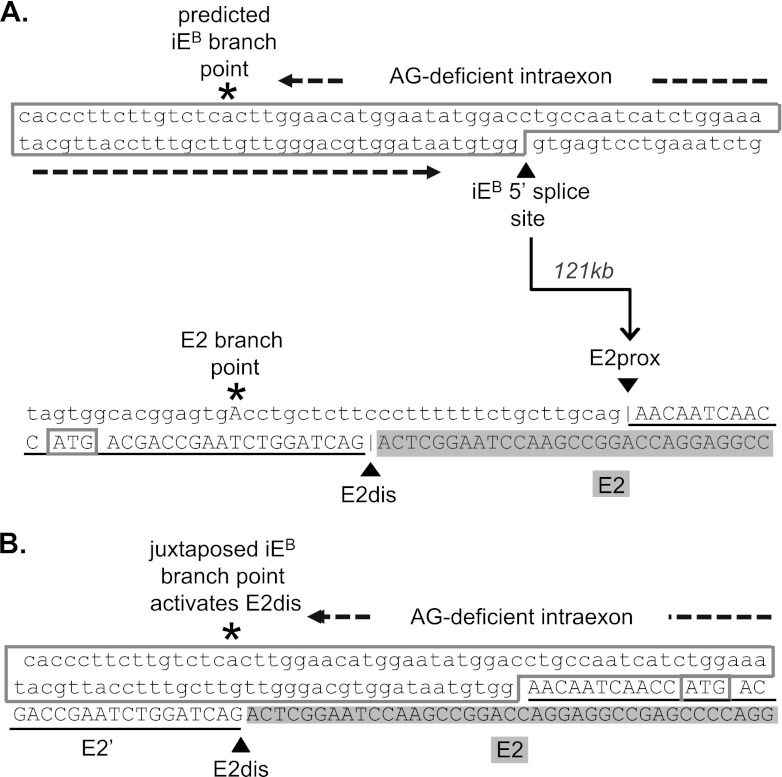
The original splice junction sequences of iEB and E2prox, and the predicted composite 3′ splice site formed by juxtaposition of the iEB branch point to the AG-dinucleotide at E2dis after splicing event 1, are shown in Fig. 4A and B, respectively. The indicated AG-deficient region in iEB is essential to prevent activation of any cryptic 3′ splice sites that might otherwise compete with E2dis.
Fig 4.
Sequences of the 4.1B intraexon, the alternative 3′ splice sites in exon 2, and the composite 3′ splice site formed by juxtaposition of iEB-bp and E2′. (A) The top sequence shows the predicted iEB-branch point and 5′ splice site, with an AG-deficient sequence of 85 nt separating these motifs. The bottom sequence represents the E2 branch point and the two alternative 3′ splice sites at the AG dinucleotides labeled E2prox and E2dis. The translation start site ATG1 is boxed. The arrow indicates the first splicing reaction in which the iEB-5′ss splices, across 121 kb of intron, to acceptor site E2dis. (B) Sequence of the composite 3′ splice site region predicted in the intermediate RNA after splicing of iEB to E2prox.
The same splicing intermediate RNA, along with the expected lariat intron structure, is depicted in Fig. 5A. Consistent with the model, we were able to successfully amplify and confirm by DNA sequence analysis the predicted intermediate from endogenous mouse kidney RNA using primers iEB-S and E2-AS (Fig. 5B, lane 1). As expected, little or none of this intermediate was detected in kidney RNA from mice treated with the splice-blocking iEB-5′ss vMO (lane 2). Further confirmation of the iEB-E2prox splicing event was obtained by branch point mapping (Fig. 5A). Successful amplification across the branch point, followed by sequence analysis of the product, confirmed that the iEB 5′ splice site can loop across ∼111 kb to the branch point sequence, TAGTGA*C, 30 nt upstream of E2prox (Fig. 5C), to generate a lariat intermediate. The TAGTGA*C sequence, in which A* represents the branch site, is a good match for the consensus mammalian branch point (10).
To test whether the intermediate RNA is a functional precursor of mature E1A transcripts, an intermediate splicing reporter was engineered to contain the structure E1AB-intron−iEB-E2 in which iEB is already spliced to E2prox (Fig. 5D, left). After transfection of this construct into HEK293 cells, mature E1A-E2dis product was recovered (Fig. 5D, lane1), consistent with the second step of intrasplicing between E1A and E2dis. This E1A-E2dis RNA structure was identical to that produced by the +iEB splicing reporter (lane 2) and distinct from the aberrant RNA produced from the −iEB reporter lacking the intraexon (lane 3). Splicing of E1A to the composite E2dis acceptor site is thus consistent with the proposed intrasplicing mechanism and corresponds to the second nested splicing event of this process. Together these results indicate that iEB plays an essential role in formation of mature 4.1B mRNAs, even though iEB is not present in the mature mRNA and has never been annotated in the genome browsers.
Intrasplicing of 4.1B transcripts is conserved through vertebrate evolution.
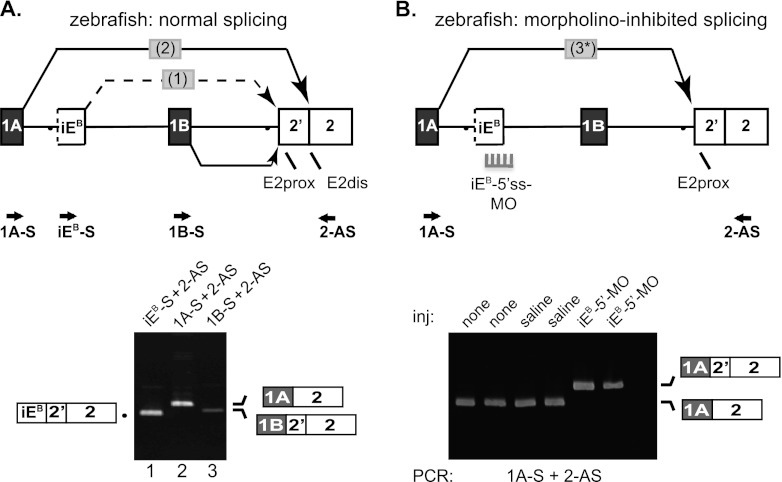
Our previous study reported that the branch point and 5′ splice site motifs of the 4.1R iER were highly conserved in mammals and were in every case separated by an AG-deficient region of 84 to 92 nt (25). Comparative genomic analysis revealed that nearly identical motifs are highly conserved among vertebrate 4.1B genes, consistently located deep within otherwise divergent intron regions (data not shown). To test evolutionary conservation of the intrasplicing mechanism, we examined the orthologous zebrafish 4.1B gene. Like its mammalian counterpart, the zebrafish 4.1B gene couples transcription and splicing to generate E1A-E2dis and E1B-E2prox mRNAs. This correlation was observed previously among transcripts in the genetic sequence databases (28) and then confirmed experimentally by reverse transcription-PCR (RT-PCR) analysis of zebrafish 4.1B transcripts (Fig. 6).
Fig 6.
Analysis of intrasplicing in the zebrafish 4.1B gene. (A) (Top) 5′ structure of the zebrafish 4.1B gene, including the putative intraexon and two nested splicing reactions predicted to be essential for E1A to E2dis splicing. Locations of PCR primers are indicated. Bottom: RT-PCR analysis of normal zebrafish 4.1B transcripts. Lane 1, intermediate RNA product obtained using primers iEB-S and 2-AS; lane 2, E1A-E2dis splicing revealed using primers 1A-S and 2-AS; lane 3, E1B-E2prox splicing demonstrated using primers 1BF and 2R. B. Top: Model for switch in splice acceptor site usage induced in vivo by inhibition of intraexon function. Splice-blocking MO directed against the intraexon 5′ splice site (5′ss-MO) inhibited splicing events 1 and 2, leading to direct splicing of E1A to E2prox in a single aberrant splicing event 3. Bottom: RT-PCR analysis of duplicate experiments using primers 1A-S and 2-AS to amplify zebrafish RNA prepared from uninjected embryos (none); embryos injected with 0.2 M KCl buffer alone (saline); or injected with 2 or 10 ng of 5′ss-MO against the 4.1B intraexon (iEB-5′-MO).
Based on these results, we hypothesized that a functionally orthologous iE likely lies in the region between alternative first exons E1A and E1B in the zebrafish gene. Analysis of genomic sequences revealed a single candidate iE located downstream of E1A and upstream of E1B that contained three essential features: an AG-deficient sequence (109 nt) flanked by a candidate branch point (CTCTCA*C) and a 5′ splice site (CTG/gtgagt, where the boundary between the iE sequence in uppercase and the intron sequence in lowercase is indicated by a slash). These features in the zebrafish gene are nearly identical to the mammalian counterparts. As predicted by the intrasplicing model, this putative zebrafish iEB splices directly to E2prox to form an intermediate RNA structure, while E1A splices to E2dis (Fig. 6A, bottom, lanes 1 and 2). In contrast, transcripts initiated at the apparent downstream promoter associated with E1B do not include iEB and splice directly to E2prox (Fig. 6A, bottom, lane 3). When formation of the intermediate in E1A transcripts was blocked by an antisense MO that masks the iEB 5′ splice site, E1A splicing in vivo robustly switched from E2dis to E2prox (Fig. 6B, last two lanes). Control experiments showed that uninjected zebrafish embryos and those injected with saline solution alone were able to execute the normal E1A→E2dis splice pattern. These experiments demonstrate that the zebrafish 4.1B gene exhibits intrasplicing analogous to that of mammalian 4.1R and 4.1B and that it is mediated by the first known zebrafish iE located deep in the upstream intron and comprised of key regulatory elements similar to those in the mammalian iEB and iER.
Intrasplicing selectivity at heterologous alternative splice acceptor sequences.
Intrasplicing-regulated 3′ splice site selection in 4.1R (25) and 4.1B (this study) occurs in the context of two closely related genes sharing primary sequence similarities in the region between E2prox and E2dis (28) that could play a mechanistic role in splice site choice (15). If specific sequences at or near the alternative acceptor sites are functionally important, intrasplicing might regulate splicing outcomes in only a limited number of transcripts. Alternatively, intrasplicing might employ the nested splicing strategy in a relatively sequence-independent manner to promote use of distal 3′ splice sites in other exons. Here, additional splicing reporters were designed to test the flexibility of intrasplicing to regulate heterologous splice acceptors.
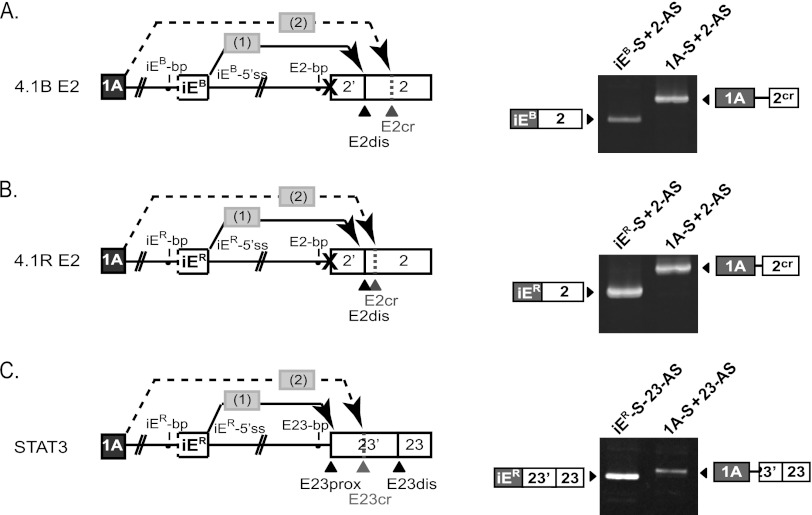
First, we modified the model 4.1B splicing reporter by inactivating E2prox but leaving intact E2dis and all other sequence features. Figure 7A shows that mutating E2prox caused E2dis to become the default acceptor for the first step of intrasplicing, as indicated by iEB→E2dis splicing (first lane). Interestingly, E1A then spliced to a newly activated cryptic site (E2cr) at the next downstream AG (Fig. 7A, second lane). E1A→E2cr splicing was iEB dependent, since splicing reporters lacking iEB yielded E1A→E2dis splicing exclusively (data not shown). This result suggested that iEB-mediated splice site choice is independent of specific primary sequences flanking either of the acceptor sites used sequentially in the nested splicing reactions. Further testing of this hypothesis was pursued in the context of a 4.1R splicing reporter. As with 4.1B, a comparable AG shift occurred upon mutation of E2prox, so that iER→E2dis and E1A→E2cr splicing were observed (Fig. 7B). In this minigene, E2cr splicing did not occur at the first downstream AG, which was immediately adjacent to E2dis and might be sterically unavailable, but E2cr splicing did occur at the next AG just a few nucleotides downstream. Thus, in both 4.1B and 4.1R we observed that a sequence normally selected in the second nested splicing event (E2dis) was entirely capable of functioning in the first step, with a concomitant shift of the second step to the next available downstream AG dinucleotide. The suggestion that intraexon-mediated splicing sequentially activates the second acceptor based primarily on its proximity to the first has important implications. First, the apparent primary sequence independence of intrasplicing implies that an alternative model to explain E2 splice site choice, based on competitive sequence-specific binding of splice-promoting and splice-inhibiting factors (15), is less likely to be a viable regulatory mechanism under these conditions. Second, only alternative 3′ splice site pairs in which the proximal site is dominant would be good candidates for regulation by intrasplicing, because selection of the distal site in the first step would lead to aberrant cryptic splicing at the next downstream AG in the second step.
Fig 7.
Intrasplicing splice site selection in heterologous sequence contexts. (A) Mutation of the E2prox AG dinucleotide shifts nested splicing events 1 and 2 from E2prox and E2dis to E2dis and E2cr. The gel shows results of PCR analysis of the nested splicing events using the indicated primers. (B) Mutation of E2prox induces an AG shift in the nested splicing reactions, leading to activation of a cryptic acceptor in E2. As in panel A, the diagram and RT-PCR analysis show the intermediate product from step 1 and final spliced product from step 2. (C) Nested splicing at E23 of the STAT3 gene shows that step 2 of intrasplicing selects the first downstream AG at E23cr rather than the authentic distal acceptor site E23dis.
If the iE-mediated nested splicing mechanism sequentially selects consecutive AG dinucleotides as alternative acceptor sites, then exons having 3′ss pairs with one or more AG dinucleotides located between the authentic sites would not be good candidates for regulation by intrasplicing. One such exon is exon 23 of the STAT3 gene, alternative splicing of which leads to synthesis of alternative C-terminal domains with differential transcriptional activity and antitumorigenic potential (31). In E23, the E23prox and E23dis sites are separated by 50 nt, with an additional AG (E23cr) only 12 nt downstream of E23prox. We tested the ability of the well-characterized 4.1R iER to regulate STAT3 isoform expression via intrasplicing using the splicing reporter shown in Fig. 7C. RT-PCR analysis revealed predominant iER→E23prox splicing (Fig. 7C, first lane), as expected. However, this was followed by exclusively E1A→E23cr splicing (Fig. 7C, second lane), with no correct E1A→E23dis splicing being observed. These findings suggest that intrasplicing can regulate 3′ splice site choice only for a subset of exons in which alternative acceptors map to consecutive AG dinucleotides in the context where the dominant proximal site is selected in step 1 and the weaker second site is utilized in step 2.
Misdirected intrasplicing can explain aberrant 3′ splice site choice in mutant 4.1R transcripts from a human patient with hereditary elliptocytosis and severe hemolytic anemia.
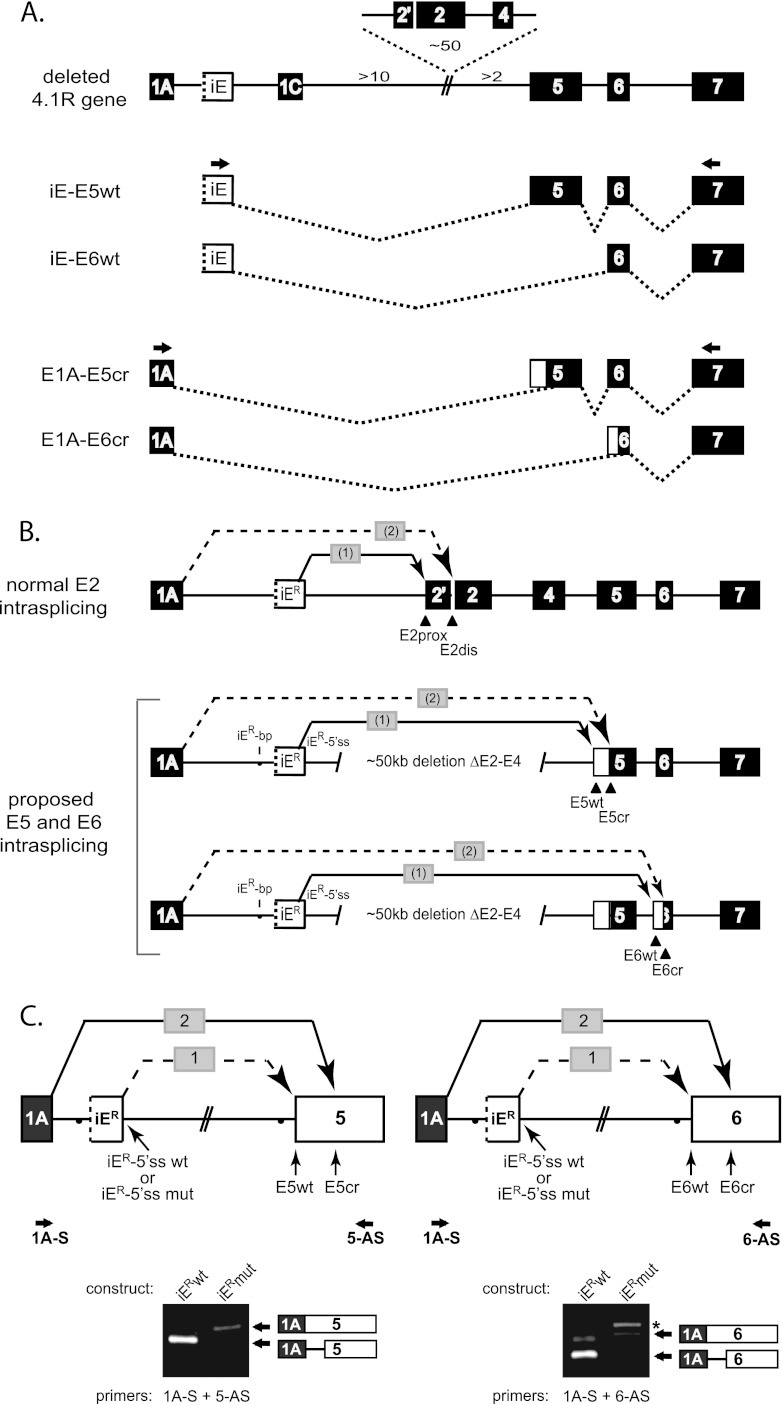
Mechanically unstable red blood cell membranes were reported in a human patient with hemolytic anemia due to a partial 4.1R gene deletion of exons 2 to 4 (E2-4). In conjunction with the deletion, aberrant splicing of unknown etiology was observed at the 3′ splice sites of alternative exon E5 and constitutive exon E6 (4). The mechanism for aberrant splicing was not clear, since the gene deletion boundary was at least 2 kb upstream of E5 and >16 kb upstream of E6. Such a mutation should have spared any splicing-regulatory information in the proximal upstream introns, and this result suggested that aberrant splicing was due to more distal effects. A clue to the mechanism was inferred by comparing these aberrant splicing events (redrawn in Fig. 8A from data in reference 4) to the splicing patterns observed at E2 in normal individuals (Fig. 8B). That is, the differential coupling of alternative splice site pairs in the patient's transcripts (iER→E5wt and E1A→E5cr; iER→E6wt and E1A→E6cr) strongly resembles the nested intrasplicing events observed at E2 (iER→E2prox and E1A→E2dis) in the wild-type 4.1R gene. Notably, the splicing of iER to E5wt and E6wt indicates that these splice acceptor sites remain perfectly functional. We therefore hypothesized that, in the absence of deleted E2, selection of cryptic sites in E5 and E6 is due to iER-mediated nested intrasplicing that has shifted its action to new targets E5 and E6 in the patient.
Fig 8.
Misdirected intrasplicing activates cryptic splice sites in a patient with deletion of 4.1R exons 2 to 4. (A) Structure of a reported 4.1R gene deletion and major aberrant splicing events observed at E5 and E6 (4), except that exon 1B in reference 4 is designated here as iER due to its unique properties mediating intrasplicing. Whereas iER in the patient spliced to the normal wild type 3′ splice sites (E5wt and E6wt), E1A spliced exclusively to aberrant cryptic splice sites (E5cr and E6cr). (B) Proposed intrasplicing model to explain aberrant splicing at E5 and E6. Splicing data are consistent with nested splicing events precisely analogous to those reported originally at E2 (top) (25), except that the deletion of E2-E4 shifts iER-mediated intrasplicing to alternative exon E5 and constitutive exon E6. (C) Intrasplicing analysis in minigene splicing reporters containing a functional (iER-5′ss) or inactivated (iER-5′ss mut) intraexon (top) upstream of either E5 or E6. RT-PCR analysis revealed that E1A splices to the normal acceptors at E5 and E6 in the absence of functional iE (lanes iERwt). In contrast, iER-mediated intrasplicing resulted smaller products representing aberrant splicing at cryptic sites, exactly as observed in the patient (lanes iERmut).
This proposed mechanism was tested in minigene splicing reporters designed to evaluate E1A splicing to either E5 or E6 in the presence or absence of functional iER sequences (Fig. 8C). For both minigenes, the presence of intact iER resulted in E1A splicing predominantly to cryptic splice acceptors (intrasplicing step 2; lanes iERwt); the expected intermediate products generated by intrasplicing step 1 (iER→E5wt and iER→E6wt) were also detected (results not shown). Both cryptic sites were located at the first AG dinucleotide downstream of the corresponding normal acceptor sites, and both corresponded exactly to the major sites detected in endogenous transcripts from erythroid and lymphoid cells of the patient (4). Moreover, selection of these cryptic sites was iER-dependent, since mutation of the iER-5′ss blocked intrasplicing step 1 and resulted in robust splicing of E1A directly to the wild type sites at E5wt and E6wt (Fig. 8C, lanes iERmut). These results further support the model that two iER-mediated nested splicing events recursively select the most proximal AG followed by the next downstream AG dinucleotide, with no apparent requirement for other specific sequences at the distal acceptor.
DISCUSSION
This study documents the ability of an ultradeep intron regulatory element to promote a specific splicing outcome between flanking exons, without itself being represented in the mature spliced product. The functional importance of the 4.1B intraexon was demonstrated not only in minigene splicing reporter assays but also in the context of full-length endogenous pre-mRNA in mouse and zebrafish models in vivo. The remarkable ability of the 4.1B intraexon to regulate downstream splicing from a distance of ∼121 kb, and of the 4.1R intraexon to regulate splicing at E2 located ∼94 kb downstream (25, 26), suggests that other deep intron splicing control elements and multistep splicing mechanisms of biological and/or medical importance have yet to be discovered.
In the protein 4.1R and 4.1B genes, intrasplicing provides a novel mechanism by which transcription and splicing decisions may be coupled, distinct from the kinetic coupling and recruitment models (1, 19) that depend ultimately on sequence properties of the promoter rather than promoter location. The physiological importance of coupling in the protein 4.1 genes lies in its regulation of N-terminal protein structure and function. In both genes, first exons downstream of the intraexon splice directly to E2prox, allowing retention of translation initiation site AUG1 and leading to synthesis of larger protein isoforms with N-terminal extensions of ∼200 amino acids (the “headpiece”). In contrast, upstream first exons execute iER- or iEB-mediated intrasplicing to E2dis, excising AUG1 and resulting in production of smaller isoforms from the downstream AUG2 in exon 4. The presence or absence of the headpiece region modulates the affinity of protein 4.1 interactions with other cytoskeletal and membrane proteins (23). Thus, membrane functional properties can be regulated via a pathway that begins with differential transcriptional activation at alternative promoters, coupled to alternative splicing events that ultimately control alternative translation initiation sites responsible for synthesis of the different 4.1 protein isoforms. Although less is known about the functional differences between large and small isoforms of 4.1B protein, differential tissue-specific utilization of alternative first exons can explain the abundance of larger 4.1B isoforms in selected tissues (e.g., brain) but not in others (kidney) (11).
At the level of pre-mRNA splicing, the nested splicing reactions provide a mechanism to explain selection of E2dis, a 3′ splice site that consistently scores as a very weak splice acceptor in all vertebrate species for which the sequence is available (28). The nested mechanism can physically excise the E2prox 3′ splice site in the first step, and the AG-deficient nature of the intraexon sequence allows splicing in the second step to occur at the AG dinucleotide at E2dis. Activation of splicing at consecutive AG dinucleotides normally occurs in E2 via two nested splicing reactions, but surprisingly, this intrasplicing mechanism appears completely transferable to E5 and E6 in transcripts from an elliptocytosis patient deleted for E2-E4. Our studies suggest that a 4.1R-type intraexon could potentially regulate alternative 3′ splice site choice in other exons due to its powerful activation of splicing at even “weak” AG dinucleotides in the second nested reaction, if they meet two specific criteria: (i) the proximal AG should be the default acceptor selected when intraexon activity is missing or silenced, and (ii) the distal site should reside at the next downstream AG dinucleotide so as to be uniquely selected in the second step of intrasplicing. Figure 9 illustrates the latter point, showing that the next available AG was selected in 6 of 7 cases tested, the only exception being when the next AG was directly adjacent to 3′prox.
Fig 9.
The second step of intrasplicing preferentially activates the next downstream AG dinucleotide. The primary sequence of 3′ splice site choice in the first and second steps of intrasplicing for several exons is shown. The second step exhibits a strong preference for activating the next available AG based on position rather than splice site strength.
Intrasplicing as described here may represent a special case of a more general phenomenon proposed on theoretical grounds to facilitate removal of long introns via nested splicing reactions (24). This specialized version of intrasplicing bears some resemblance to, but ultimately is distinct from, recursive splicing (5), which excises long introns via multiple smaller steps involving splicing events at a series of 3′ splice sites across the intron. Both intrasplicing and recursive splicing are multistep resplicing processes promoted by (mostly unannotated) deep intron motifs that have properties of both introns and exons but ultimately are excised from the mature mRNA product. Both mechanisms also utilize the juxtaposition of splicing motifs in one splicing event to activate a subsequent splicing event. However, the molecular details of intrasplicing as currently understood appear to be different from those of recursive splicing. In various splicing reporters, normal 4.1 pre-mRNAs, and aberrant patient transcripts, the nested intrasplicing step consistently occurred at a composite 3′ splice site generated by juxtaposition of the iE branch point with the next AG dinucleotide downstream of the first-step acceptor. We speculate that the second acceptor is selected by a scanning mechanism that would not require de novo exon definition of the ligated sequences in the first step (8). In contrast, each successive step of recursive splicing involves a new composite 5′ splice donor site formed when an exon is spliced to a downstream “zero-length exon” containing adjacent 3′ and 5′ splice sites. The biological functions of these two resplicing mechanisms may also be different, since the primary purpose of intrasplicing events thus far known is to provide a control mechanism for selection of a distal 3′ splice site, while recursive splicing has been proposed primarily as a mechanism for excision of long introns.
We speculate that intrasplicing plays a broader role than is currently appreciated in regulating gene expression. For example, intraexon function may not be limited to 5′ regions in conjunction with alternative first exons but could also be located within genes to regulate additional classes of 3′ splice sites. Moreover, intraexon recognition could be regulated by splicing enhancers and silencers that would allow functional control of the pathway. We therefore anticipate two classes of intraexons: “constitutive intraexons,” of the type found in 4.1R and 4.1B, and “alternative intraexons,” subject to temporal or spatial regulation. Finally, it is not difficult to imagine that a variation on the mechanism could allow regulation of alternative 5′ splice sites within an exon. Future studies will be required to devise strategies for global identification of such elements in order to understand their full contribution to gene expression in the human genome.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants DK32094 (to M.N.) and GM061952 (to S.L.A.) and by the Director, Office of Biological and Environmental Research, U.S. Department of Energy, under contract DE-AC02-05CH11231.
J.G.C., M.P., and T.L.G. designed the experiments. M.P. performed the minigene and vivo-MO experiments, and T.L.G. carried out the zebrafish experiments. J.G.C., N.M., T.L.G., and S.L.A. wrote the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1. Auboeuf D, Batsche E, Dutertre M, Muchardt C, O'Malley BW. 2007. Coregulators: transducing signal from transcription to alternative splicing. Trends Endocrinol. Metab. 18:122–129 [DOI] [PubMed] [Google Scholar]
- 2. Baek D, Davis C, Ewing B, Gordon D, Green P. 2007. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 17:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baklouti F, et al. 1997. Organization of the human protein 4.1 genomic locus: new insights into the tissue-specific alternative splicing of the pre-mRNA. Genomics 39:289–302 [DOI] [PubMed] [Google Scholar]
- 4. Baklouti F, et al. 2011. Homozygous deletion of EPB41 genuine AUG-containing exons results in mRNA splicing defects, NMD activation and protein 4.1R complete deficiency in hereditary elliptocytosis. Blood Cells Mol. Dis. 47:158–165 [DOI] [PubMed] [Google Scholar]
- 5. Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. 2005. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavanna T, Pokorna E, Vesely P, Gray C, Zicha D. 2007. Evidence for protein 4.1B acting as a metastasis suppressor. J. Cell Sci. 120:606–616 [DOI] [PubMed] [Google Scholar]
- 7. Conboy J. 1999. The role of alternative pre-mRNA splicing in regulating the structure and function of skeletal protein 4.1. Proc. Soc. Exp. Biol. Med. 220:73–78 [DOI] [PubMed] [Google Scholar]
- 8. Crabb TL, Lam BJ, Hertel KJ. 2010. Retention of spliceosomal components along ligated exons ensures efficient removal of multiple introns. RNA 16:1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu XD, Mayeda A, Maniatis T, Krainer AR. 1992. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl. Acad. Sci. U. S. A. 89:11224–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao K, Masuda A, Matsuura T, Ohno K. 2008. Human branch point consensus sequence is yUnAy. Nucleic Acids Res. 36:2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gascard P, et al. 2004. Putative tumor suppressor protein 4.1B is differentially expressed in kidney and brain via alternative promoters and 5′ alternative splicing. Biochim. Biophys. Acta 1680:71–82 [DOI] [PubMed] [Google Scholar]
- 12. Gerber MA, Bahr SM, Gutmann DH. 2006. Protein 4.1B/differentially expressed in adenocarcinoma of the lung-1 functions as a growth suppressor in meningioma cells by activating Rac1-dependent c-Jun-NH(2)-kinase signaling. Cancer Res. 66:5295–5303 [DOI] [PubMed] [Google Scholar]
- 13. Horresh I, Bar V, Kissil JL, Peles E. 2010. Organization of myelinated axons by Caspr and Caspr2 requires the cytoskeletal adapter protein 4.1B. J. Neurosci. 30:2480–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoy JL, Constable JR, Vicini S, Fu Z, Washbourne P. 2009. SynCAM1 recruits NMDA receptors via protein 4.1B. Mol. Cell. Neurosci. 42:466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang SC, et al. 2009. Coupled transcription-splicing regulation of mutually exclusive splicing events at the 5′ exons of protein 4.1R gene. Blood 114:4233–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jung Y, Kissil JL, McCarty JH. 2011. β8 integrin and band 4.1B cooperatively regulate morphogenesis of the embryonic heart. Dev. Dyn. 240:271–277 [DOI] [PubMed] [Google Scholar]
- 17. Kang Q, Wang T, Zhang H, Mohandas N, An X. 2009. A Golgi-associated protein 4.1B variant is required for assimilation of proteins in the membrane. J. Cell Sci. 122:1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura K, et al. 2006. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16:55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kornblihtt AR. 2005. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 17:262–268 [DOI] [PubMed] [Google Scholar]
- 20. McCarty JH, Cook AA, Hynes RO. 2005. An interaction between αvβ8 integrin and band 4.1B via a highly conserved region of the band 4.1 C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 102:13479–13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morcos PA, Li Y, Jiang S. 2008. Vivo-morpholinos: a non-peptide transporter delivers morpholinos into a wide array of mouse tissues. Biotechniques 45:613–623 [DOI] [PubMed] [Google Scholar]
- 22. Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nunomura W, et al. 2009. Marked difference in membrane-protein-binding properties of the two isoforms of protein 4.1R expressed at early and late stages of erythroid differentiation. Biochem. J. 417:141–148 [DOI] [PubMed] [Google Scholar]
- 24. Ott S, Tamada Y, Bannai H, Nakai K, Miyano S. 2003. Intrasplicing—analysis of long intron sequences. Pac. Symp. Biocomput. 2003:339–350 [DOI] [PubMed] [Google Scholar]
- 25. Parra M, Tan JS, Mohandas N, Conboy JG. 2008. Intrasplicing coordinates alternative first exons with alternative splicing in the protein 4.1R gene. EMBO J. 27:122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parra MK, Gee S, Mohandas N, Conboy JG. 2011. Efficient in vivo manipulation of alternative pre-mRNA splicing events using antisense morpholinos in mice. J. Biol. Chem. 286:6033–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robb VA, Gerber MA, Hart-Mahon EK, Gutmann DH. 2005. Membrane localization of the U2 domain of protein 4.1B is necessary and sufficient for meningioma growth suppression. Oncogene 24:1946–1957 [DOI] [PubMed] [Google Scholar]
- 28. Tan JS, Mohandas N, Conboy JG. 2005. Evolutionarily conserved coupling of transcription and alternative splicing in the EPB41 (protein 4.1R) and EPB41L3 (protein 4.1B) genes. Genomics 86:701–707 [DOI] [PubMed] [Google Scholar]
- 29. Vogel J, Hess WR, Borner T. 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 25:2030–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong SY, et al. 2007. Protein 4.1B suppresses prostate cancer progression and metastasis. Proc. Natl. Acad. Sci. U. S. A. 104:12783–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zammarchi F, et al. 2011. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc. Natl. Acad. Sci. U. S. A. 108:17779–17784 [DOI] [PMC free article] [PubMed] [Google Scholar]