Abstract
In Drosophila, two chromosome-wide compensatory systems have been characterized: the dosage compensation system that acts on the male X chromosome and the chromosome-specific regulation of genes located on the heterochromatic fourth chromosome. Dosage compensation in Drosophila is accomplished by hypertranscription of the single male X chromosome mediated by the male-specific lethal (MSL) complex. The mechanism of this compensation is suggested to involve enhanced transcriptional elongation mediated by the MSL complex, while the mechanism of compensation mediated by the painting of fourth (POF) protein on the fourth chromosome has remained elusive. Here, we show that POF binds to nascent RNA, and this binding is associated with increased transcription output from chromosome 4. We also show that genes located in heterochromatic regions spend less time in transition from the site of transcription to the nuclear envelope. These results provide useful insights into the means by which genes in heterochromatic regions can overcome the repressive influence of their hostile environment.
INTRODUCTION
Aneuploidy of entire chromosomes and chromosome segments is an important evolutionary driving force that increases variation but is accompanied by problems associated with changes in gene dosage and genomic instability. The evolution of buffering systems that compensate for dosage differences must therefore allow for a balance between allowing genomic variability and avoiding genomic instability (66). Buffering systems of this kind have been described in Drosophila; in haploid conditions, they cause the transcription output to increase by a factor of approximately 1.4 (22, 46, 67, 82). However, the evolution of heteromorphic sex chromosomes, such as the X and Y chromosome pair in flies and mammals, is accompanied by an expression problem that requires more extensive compensation. Since most genes on the X chromosome should be expressed at the same levels in males and females, dosage compensation mechanisms coevolve as the X-Y chromosome pair is formed (34, 44, 77). Notably, while the ancient homology between the mammalian X and Y is clear, the evolutionary origin of the Drosophila Y is more complicated (8, 77). The evolution of dosage compensation mechanisms is attributable to evolutionary pressures that act at all levels of expression to compensate for the losses of functional gene copies. Two dosage compensation systems have been studied in Drosophila: the male-specific lethal (MSL) complex, which targets and upregulates the male X chromosome (18, 23, 56), and the painting of fourth (POF) protein, which stimulates the expression of the fourth chromosome in Drosophila melanogaster but is believed to have originated from a dosage-compensating mechanism (26, 33–35). The MSL complex and POF coexist; they are on different chromosomes in, e.g., D. melanogaster, but are colocalized on the same chromosome in, e.g., Drosophila ananassae. Their coexistence suggests that they probably act on different levels of gene regulation (66).
The MSL complex is a ribonucleoprotein complex consisting of five male-specific lethal proteins (MSL1, MSL2, MSL3, MLE, and MOF) and two noncoding RNAs, roX1 and roX2. Because expression of MSL2 is sex restricted, the complex is formed only in males and specifically targets the male X chromosome (41). A model of its action has been proposed: MOF (a histone acetyltransferase) acetylates H4K16, and this modification leads to decompaction of the chromatin and hypertranscription of the male X chromosome genes (18, 56). The prevailing idea is that the MSL complex stimulates transcriptional elongation (40). This idea is supported by genome-wide mappings showing that expression of the MSL complex and the associated H4K16 acetylation are both enhanced within gene bodies with a bias to their 3′ end (2, 3, 19, 30, 57). A recent study confirmed that the MSL complex enhances transcription by facilitating the progression of RNP2 across the active X chromosomal genes (32).
Less is known about the regulatory level at which POF acts. POF is a 55-kDa protein containing an RNA recognition motif (RRM). Like the MSL complex, POF binds within the bodies of expressed genes. However, in D. melanogaster, POF specifically targets the fourth chromosome in both males and females (27, 33). The targeting of POF to the fourth chromosome is associated with a chromosome-specific increase in transcript levels that primarily affects differentially expressed genes (26, 67). Flies can survive without POF or missing one copy of the fourth. However, haplo-fourth animals die if they also lack POF. Importantly, the expression of nonubiquitously expressed genes on the fourth chromosome has been shown to be compensated by POF in haplo-4 flies; suppressing or eliminating this compensation causes haplo-fourth lethality (67).
The fourth chromosome of D. melanogaster has several unique characteristics. It is the smallest chromosome in the Drosophila genome, with an approximate size of 5 Mb. Of these 5 Mb, 3 to 4 Mb consists exclusively of simple AT-rich satellite repeats and does not contain any known genes. The sequenced part of chromosome 4 is only 1.3 Mb and represents the banded, polytenized, and gene-rich portion corresponding to cytological sections 101E to 102F (4, 37). In principle, the entire fourth chromosome can be considered heterochromatic; more specifically, it consists of the HP1 enriched “green chromatin” as defined by van Steensel and coworkers (17). This means that the fourth chromosome is enriched in the heterochromatin protein HP1 and in specific histone modification markers of heterochromatin, e.g., methylated H3K9 (13, 15, 60, 61). In keeping with its heterochromatic nature, the polytenized part of chromosome 4 contains large blocks of repeated sequences and transposable elements that are interspersed with the genes (29, 36, 38, 48, 55, 68). Transgenes inserted on the fourth chromosome often show partially silenced, variegated expression (70, 71, 79, 80). In fact, the structure and sequence composition of the fourth chromosome, with its scattered repetitive elements, is more reminiscent of the organization of mammalian chromosomes than that of the other D. melanogaster autosomes. It appears as though the genes located on the fourth chromosome have adapted to function in this repressive milieu.
To study the fundamental compensatory processes acting on the fourth chromosome, we have performed RNA immunoprecipitation (RIP) experiments followed by tiling array analysis (RIP-chip) and transcript profiling experiments. We show here that POF associates with the newly transcribed RNA produced from the fourth chromosome. Our data indicate that POF binds to the spliced form of the transcript and that this binding is associated with an increase in the amount of chromosome 4 transcripts. We also show that transcripts encoded by the fourth chromosome or pericentromeric heterochromatin have a shorter transition time from the site of transcription to the nuclear envelope.
MATERIALS AND METHODS
Immunostaining of polytene chromosomes.
For heat shock, vials were incubated for 45 min in a 38°C water bath and allowed to recover at room temperature for the time indicated. Polytene chromosomes from the salivary glands of 3rd-instar larvae were prepared and stained essentially as previously described (81). Salivary glands were fixed in 2% formaldehyde in phosphate-buffered saline (PBS), 0.1% Triton X-100, and 0.2% NP-40 for 40 s followed by 2 to 3 min in 50% acetic acid containing 1% formaldehyde. Polytene chromosomes were squashed as described previously (81). The slides were washed for 30 min in 1× PBS and 0.1% Triton X-100, transferred to blocking solution (0.1 M maleic acid, 0.15 M NaCl, 1% Boehringer blocking reagent), and incubated for 30 min at room temperature. They were then incubated overnight at 4°C with primary antibodies raised against POF (1:400 dilution) and RNP2 (ab5408; 1:200; Abcam). The slides were washed two times for 10 min each in a solution containing 0.1 M maleic acid, 0.15 M NaCl, and 0.3% Tween 20 and then blocked for 30 min. Goat anti-rabbit and goat anti-mouse antibodies conjugated with Alexa Fluor 555 or Alexa Fluor 488 (Molecular Probes; diluted 1:300) were used as secondary antibodies and incubated at room temperature for 2 h. The squashes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) and washed two times for 10 min each before being mounted with Vectashield (Vector). For DRB (5,6-dichloro-1-β-d-ribofuranoxylbenzimidazole) (Sigma) treatment, salivary glands were dissected in Drosophila serum-free medium (SFM) (Express Five; Invitrogen) and incubated for 60 or 90 min in SFM supplemented with 250 μM DRB and 5% dimethyl sulfoxide (DMSO) or 5% DMSO only (control). Fixation and immunostainings were done as described above. For RNase treatment, salivary glands were dissected in Drosophila SFM (Invitrogen) and pretreated for 2 min in PBS and 0.2% Triton X-100. The glands were then incubated for 12 min in PBS alone (control) or PBS and RNase (45U/ml RNase A, 1,800 U/ml RNase T1; Ambion). For MLE/POF double stainings, primary antibodies against POF (chicken, 1:100) and MLE (rabbit, 1:1,000) were used, and for MSL2/POF, primary antibodies against POF (rabbit, 1:400) and MSL2 (goat, catalog no. sc32459, 1:100) were used. Goat anti-rabbit and goat anti-chicken or donkey anti-goat and donkey anti-rabbit antibodies conjugated with Alexa Fluor 555 or Alexa Fluor 488 (Molecular Probes; diluted 1:300) were used as secondary antibodies. Preparations were analyzed using a Zeiss Axiophot microscope equipped with a KAPPA DX20C charge-coupled-device camera. Images were assembled and merged electronically using Adobe Photoshop. For quantitative comparisons of stains at different time points or with different treatments, preparations and staining were run in parallel. Nuclei with clear cytology were chosen on the basis of DAPI staining and photographed. All settings were identical for each specific antibody. At least 20 nuclei for each time point or treatment were used in these comparisons, and at least four slides for each time point or treatment were analyzed.
RNA immunoprecipitation.
For each sample treatment condition, we used 300 ml of D. melanogaster cells from Schneider's line 2 (ATCC CRL-1963), grown as a suspension culture at 25°C in Erlenmeyer flasks at a density of 1 × 107 cells/ml in Drosophila SFM (Invitrogen) supplemented with 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, and 2 mM l-glutamine. The cells were pelleted, washed twice in 100 ml of a solution containing 10 mM HEPES (pH 7.4) and 140 mM NaCl, and then resuspended in 50 ml of lysis buffer (20 mM HEPES [pH 7.4], 3 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 10 U/ml RNasin, and 0.5× protease inhibitor cocktail; Roche). The cells were allowed to swell on ice for 10 min and then homogenized on ice with 30 strokes of a Dounce homogenizer. The nuclei were pelleted at 2,000 × g for 5 min and used either in the native state (i.e., non-cross-linked) or after formaldehyde cross-linking. For the cross-linked sample (FA), the pelleted nuclei were resuspended in 50 ml lysis buffer and cross-linked by adding formaldehyde to a final concentration of 0.5% and incubating for 10 min at room temperature. The cross-linking was stopped by adding glycine (final concentration of 0.125 M), after which the nuclei were washed once in lysis buffer and resuspended in 2 ml of sonication buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.1 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM PMSF, 10 U/ml RNasin, and 0.5× protease inhibitor cocktail) and sonicated using a Bioruptor (Diagenode) for 4 min, on the high setting (30 s on, 30 s off). For the native samples (N1, N3, and N6), the pelleted nuclei were resuspended in 2 ml sonication buffer and sonicated for 1, 3, or 6 min (N1, N3, and N6, respectively). Cellular debris were removed by centrifugation for 25 min, 4°C at 14,000 × g, and the supernatants were used for immunoprecipitations.
For the whole-cell sample (WCFA), we used 40 ml of D. melanogaster cells from Schneider's line 2, grown as described above. The cells were cross-linked by adding formaldehyde to a final concentration of 0.5% and incubated for 10 min at room temperature. The cross-linking was stopped by adding glycine (final concentration of 0.125 M) and incubating for 5 min on ice. The fixed cells were pelleted, resuspended in 5 ml PBS, washed for 10 min in wash A solution (10 mM HEPES [pH 7.6], 10 mM EDTA [pH 8.0], 0.25% Triton X-100) for 10 min at 4°C, and then washed once with wash B solution (10 mM HEPES [pH 7.6], 100 mM NaCl, 1 mM EDTA [pH 8.0], 0.5 mM EGTA [pH 8.0], 0.01% Triton X-100) for 10 min. The cells were resuspended in 5 ml TEN140 (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 140 mM NaCl), PMSF was added to a final concentration of 1 mM, and the samples were sonicated using a Bioruptor (Diagenode), 4 min, on the high setting (30 s on, 30 s off). To the resuspended cell extract, 50 μl 10% SDS, 500 μl 1% sodium deoxycholate, 500 μl 10% Triton X-100, and 140 μl 5 M NaCl were added sequentially. The extract was incubated for 10 min at 4°C and then centrifuged for 5 min at 4°C and 14,000 × g, after which the supernatants were used for immunoprecipitation.
For immunoprecipitation, 1 to 2 mg nuclear or whole-cell extract was incubated with 8 μl anti-POF, 5 μl anti-MSL2, or 5 μl anti-MOF antibody for 45 min at 4°C with agitation. The antibody complexes were precipitated by incubation with 75 μl of Dynabeads conjugated to protein A (Invitrogen) for 30 min at 4°C with agitation. The supernatant was removed, and the beads were washed twice with PBS (150 mM NaCl), 0.1% Triton X-100, 32 U/ml RNasin, and 0.5× protease inhibitor cocktail and twice in PBS (300 mM NaCl), 0.1% Triton X-100, 32 U/ml RNasin, and 0.5× protease inhibitor cocktail. The cross-links in the FA and WCFA samples were reversed by adding 200 μl 0.45 M LiCl to the beads and incubating 3 to 4 h at 65°C. RNA was isolated using TRI reagent (Ambion) followed by a purification using the RNeasy kit (Qiagen) according to the instructions by the suppliers. The RNA samples were concentrated and reverse transcribed into cDNA using random primers with the ImPromII first-strand synthesis kit (Promega). The single-stranded cDNA was purified with the QIAquick PCR purification kit (Qiagen). The purified cDNA samples were amplified using the WGA2 GenomePlex complete whole-genome amplification kit (Sigma) according to the recommendations by the supplier.
Microarray analysis.
For the RIP tiling array analysis, amplified DNA samples were fragmented, labeled, and hybridized to an Affymetrix Drosophila Genome 2.0 array. The signal intensity data were analyzed with the Affymetrix tiling analysis software (version 1.1.02), using a 90-bp bandwidth and perfect match only. When determining absolute amounts (i.e., to obtain transcript profiles), the bandwidth was set to zero and RNA enrichment ratios for all genes were calculated as the average enrichment ratio value of all probes located within the exons of each gene. The total nuclear amounts of each gene transcript were calculated from the input sample as the average value of all probes located within exons of each gene. Only genes with at least 10 probes within exons were included. To compare the POF-RIP profile to the chromatin immunoprecipitation with microarray technology (ChIP-chip) POF profile and transcriptome profile, we used the data previously obtained from the same cell line (27). The transcriptome profile data from S2 cells was converted to D. melanogaster genomic release 5 using the coordinate converter tool at FlyBase (74).
Whole-cell/nuclear transcript ratios.
To compare the relative amount of transcripts in the whole cell to the amount of transcripts in the nucleus from all chromosome arms, we did the following. (i) Gene transcript values were calculated as the average of all probes located within exons of each gene. This was done both for the whole-cell (WCFA) and the nuclear (FA) extracts. (ii) For each gene, we calculated the ratio between the whole-cell and the nuclear transcript values. (iii) For each chromosome arm, the mean ratio and confidence interval was calculated. (iv) The genomic average ratio (i.e., all genes) was set to zero. The pericentromeric region was compiled based on the breakpoints in S2 cells defined in reference 60, including 92 genes; region 2L:31 was defined as the 77-gene region starting at Pros35 and ending with CG5198.
RNP2 pausing index and elongation density index.
To analyze the density of engaged RNP2 for all chromosome arms, we used the previously calculated pausing index (PI) and elongation density index (EdI) provided as supplementary data in reference 32. The PI is the ratio of the GRO-seq signal at the 5′ peak (first 500 bp downstream of the transcription start point) to the average signal over gene bodies. The EdI is the signal density ratio of the 3′ region of each gene compared to its 5′ region (excluding the first 500 bp) (32).
ChIP of DRB-treated S2 cells.
Schneider's Drosophila line 2 cells (ATCC CRL-1963) were grown in suspension at 25°C in Erlenmeyer flasks to a density of 1 × 107 cells/ml in Drosophila SFM (Invitrogen) supplemented with 100 U/ml of penicillin G, 100 μg/ml of streptomycin sulfate, and 2 mM l-glutamine. DRB treatment was performed using two biological replicates. For each replicate, the culture was divided into two 10-ml fractions. To one fraction of each replicate, DRB was added to a final concentration of 100 μM in 1% DMSO, and the other fraction was used as the control. The cells were then incubated for 60 min prior to cross-linking, sonication, and immunoprecipitation against POF as previously described (27). DNA from the ChIP reactions was quantified by real-time PCR as described previously (26).
Transcriptome profile.
Total RNA was prepared from three biological replicates for both wild type and PofΔ119. For each replicate, RNA was isolated from 20 adult female flies using TRI reagent (Ambion) followed by a purification using the RNeasy kit (Qiagen) according to the instructions of the suppliers. The six labeled cDNA probes were hybridized to the Affymetrix Drosophila Genome 2.0 tiling array. The signal intensity data were analyzed with the Affymetrix tiling analysis software (version 1.1.02). Gene transcript values were calculated as the average enrichment ratio value of all probes located within exons of each gene. Metagene profiles were made by including all annotated exons between the first annotated transcription start site and the last annotated transcription stop site of each gene. The genes were then rescaled to the same relative length, the transcripts were divided into 10 bins, and the average intensity for each bin was calculated. For the metagene profiles, we included only expressed genes. We used strict criteria for expression (average gene values were greater than the average plus 2 standard deviations based on all intergenic probes on the array), resulting in 27 expressed genes on the fourth chromosome and 380 expressed genes on chromosome 2L.
Microarray data accession no.
The microarray data reported in this paper have been deposited at http://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE28519).
RESULTS
POF binding depends on active transcription.
We have previously shown that POF binds to active genes on the fourth chromosome with a preference for exons and that the levels of POF binding to genes correlate with transcript levels (27). We therefore wanted to determine whether there is a relationship between the binding of POF and that of RNA polymerase II (RNP2). To address this question, we heat shocked Drosophila larvae for 45 min at 38°C and monitored the reassociation of POF and RNP2 to chromatin. Upon heat shock, most transcription stops and RNP2 is released from most genes and redistributed to heat shock-induced genes (65). As was the case with RNP2, POF was also released from its normal chromatin target sites after the heat shock, but it was not redistributed to heat shock-induced genes. Both POF and the RNP2 were restored to their normal distributions on the chromosomes over a period of approximately 2 h (Fig. 1). The temporal profiles of the release and the subsequent recovery of POF and RNP2 binding after heat shock were very similar, suggesting that there is a relationship between the binding of the one and that of the other. To test if POF binding depends on active transcription, we monitored POF association with chromosomes following DRB treatment in S2 cells and salivary glands. DRB inhibits phosphorylation of Ser2 in the C-terminal domain (CTD) of RNP2, causing its failure to advance from the initiation to the elongation phase of transcription (9, 10, 45, 58). After DRB treatment of S2 cells, POF was released from target genes on the fourth chromosome (Fig. 2A). DRB treatment of salivary glands showed a corresponding decrease in POF binding to the fourth chromosome. Notably, upon DRB treatment, RNP2 redistributes from a diffuse binding pattern to a more specific banded pattern, consistent with its release from the main part of the gene bodies (Fig. 2B). We conclude that the binding of POF to the fourth chromosome correlates with active transcription and that the release of POF upon DRB treatment suggests that recruitment of POF to specific chromatin domains occurs through nascent RNA.
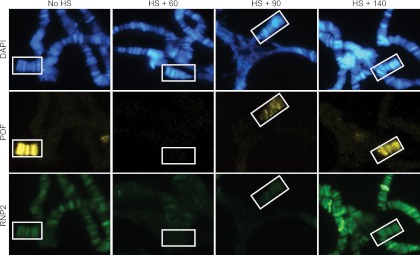
Fig 1.
The time course for the release and reassociation of POF from its chromosomal targets closely resembles that for RNP2. POF (yellow) and RNP2 (green) are released from chromosomes after heat shock. Third-instar Drosophila larvae were heat shocked for 45 min and allowed to recover for 60, 90, or 140 min prior to dissection. After a recovery time of approximately 140 min, the distributions of both POF and RNP2 on the fourth chromosome (indicated by boxes) have been restored to their original patterns. DNA is stained with DAPI (blue).
Fig 2.
POF binding to chromosome 4 genes depends on active transcription. Binding of POF on chromosome 4 genes is decreased upon blocking transcription by DRB treatment. (A) The binding of POF to three chromosome 4 genes (CG31998, Zyx102EF, and MED26) was analyzed by ChIP using antibodies against POF. The y axis indicates enrichment plotted as percentage of input. Results are shown for two biological replicates. DRB treatment causes release of POF from the main part of the gene body. Note that the genes are expressed from right to left, and the primers used are indicated according to reference 26. CG4016 represents a control gene located on chromosome 2R, i.e., not targeted by POF. (B) POF is reduced on the fourth chromosome after a 60- or 90-min DRB treatment of salivary glands. Note that in DRB-treated nuclei, RNP2 redistributes from a diffuse binding pattern to a more specific banded pattern, consistent with its release from the main part of the gene bodies. Arrowheads indicate distal ends of chromosome 4.
POF associates to transcripts from the fourth chromosome.
To delineate the relationship between POF and transcription, we performed RNA immunoprecipitation (RIP) experiments on nuclear extracts from S2 cells with and without cross-linking followed by tiling array analysis (RIP-chip). We analyzed three different sonication conditions; N1, N3, and N6 denote protocols in which the native nuclear extracts were sonicated for 1, 3, and 6 min, respectively (Fig. 3A). In order to assess the extent (if any) to which POF dissociated from its normal targets during the preparation of the native extracts, we prepared, in parallel to the preparation of the native extracts, extracts in which the nuclei were cross-linked with formaldehyde (FA) and also cross-linked whole-cell extracts (WCFA) (Fig. 3A). The higher enrichment ratios on non-fourth chromosomes in native samples (Fig. 3B) compared to cross-linked samples (Fig. 3E) demonstrates that POF has a general RNA affinity and can be dissociated from its normal targets during sample preparation and reassociated to any mRNA.
Fig 3.
POF binds preferentially to chromosome 4 transcripts rather than transcripts originating from the other chromosome arms. (A) Schematic outline of the RIP method. The light gray indicates the period in the sample preparation where POF can be released from its target and reassociate with free RNA. Dark gray indicates the point at which the RNA molecules bound by POF are fixed. N1, N3, and N6 denote native nuclear extracts sonicated for 1, 3, or 6 min, respectively. FA denotes formaldehyde cross-linking. (B, C, D) Mean ratios for native samples of POF-RIP/Input (B), MSL2-RIP/Input (C) and MOF-RIP/Input (D) calculated for all probes within exons for each chromosome arm (log2 scale). Squares indicate the mean value, and whiskers indicate the 95% confidence interval. Sample N1 is indicated in red, N3 in green, and N6 in blue. (E, F, G) Mean ratios for cross-linked samples of POF-RIP/Input (E), MSL2-RIP/Input (F), and MOF-RIP/Input (G) calculated for all probes within exons for each chromosome arm (log2 scale). Squares indicate the mean value, and whiskers indicate the 95% confidence interval. FA is indicated in red and WCFA in green.
Independent of the conditions used to prepare the extract, the results showed that POF has a general affinity for RNA and a significantly higher preference for RNA transcribed from the fourth chromosome than from other chromosomes (Fig. 3B and E; P ≪ 0.001, two-tailed t test). This suggests that POF associates directly with nascent RNA but does not exclude the possibility that chromosome 4 RNAs are enriched indirectly by association of POF with chromatin and that RNA molecules linked to chromatin by active transcription are pulled down indirectly. To test this possibility, we included two chromatin-bound proteins from the MSL complex (MSL2 and MOF) as controls. MSL2 and MOF are involved in the chromosome-specific targeting of the male X chromosome. The chromosome specificity of the MSL complex has several evolutionary and mechanistic similarities to POF (27, 34, 35, 66, 67). In the native extracts (Fig. 3B to D), we observed only minor enrichment of X chromosome transcripts in RIPs using MSL2 and MOF antibodies. Conversely, strong enrichment of chromosome 4 was observed in the POF-RIP. This suggests that whereas MSL2 and MOF bind directly to the chromatin of the X chromosome, POF binds to the nascent RNA transcribed from the chromosome 4 genes. In the cross-linked samples, strong significant enrichments of the chromosome 4 transcripts were observed in the POF-RIP (Fig. 3E; P ≪ 0.001, two-tailed t test). Similarly, using extracts fixed with formaldehyde, MSL2-RIP and MOF-RIP experiments showed enriched levels of chromosome X transcripts compared to autosomes (Fig. 3F and G; P ≪ 0.001, two-tailed t test). These results indicate that after formaldehyde cross-linking, RNA molecules linked to chromatin by transcription are immunoprecipitated by chromatin-associated proteins. The enrichments of X-linked transcripts seen in MSL2 and MOF cross-linked samples are much lower than the observed enrichments of chromosome 4 transcripts in the POF samples. This indicates that POF, in contrast to the MSL complex, is directly linked to RNA. Notably, chromosome 4 transcripts are slightly reduced in MSL2 and MOF RIPs (Fig. 3C, D, F, and G). We also detected an increase in the levels of non-chromosome 4 transcripts in the POF-RIPs, confirming that POF has a general affinity for RNA. This general affinity is most pronounced in the native samples, as expected, since some of the POF proteins are likely to be released from their normal target sites during sample preparation and are then free to associate with any available RNA. The complete results of the MSL2-RIPs and the MOF-RIPs are analyzed and discussed in detail elsewhere (25).
POF binds to nascent processed RNA.
We have previously shown that POF is tightly and very specifically linked to chromosome 4 (26, 27, 33). The RIP results showed that POF associates with RNA transcribed from the fourth chromosome. Thus, the most likely explanation is that POF associates with the fourth chromosome by binding to nascent RNA transcribed from the fourth chromosome. However, we could not exclude the possibility that POF both binds directly to chromatin and has an additional affinity for soluble nuclear chromosome 4-generated RNA. To determine whether POF associates selectively with soluble nuclear transcripts or if the observed association is with nascent RNA that is still linked to chromatin by RNP2, we compared the native RIP profile to a ChIP-chip profile previously generated using the same cell line (Fig. 4) (27). The enrichment profile from the ChIP-chip experiment (chromatin IP) very closely resembled that from the RIP. The high similarity of these two profiles supports a model in which POF binds cotranscriptionally to the nascent RNA. We next asked whether POF binding to the fourth chromosome is RNase sensitive. It has previously been shown that associations of MLE and MOF with the X chromosome are RNase sensitive while MSL1 and MSL2 associations are RNase resistant (1, 59). The RNase sentivity of MLE suggests that it may bind the remaining MSL complex through an RNA molecule, presumably the roX RNAs (11, 59). The result shows that at the RNase treatment condition that causes consistent complete release of MLE and none or very low release of MSL2, POF is not affected (Fig. 5). We conclude that the binding of POF to chromatin is RNase resistant.
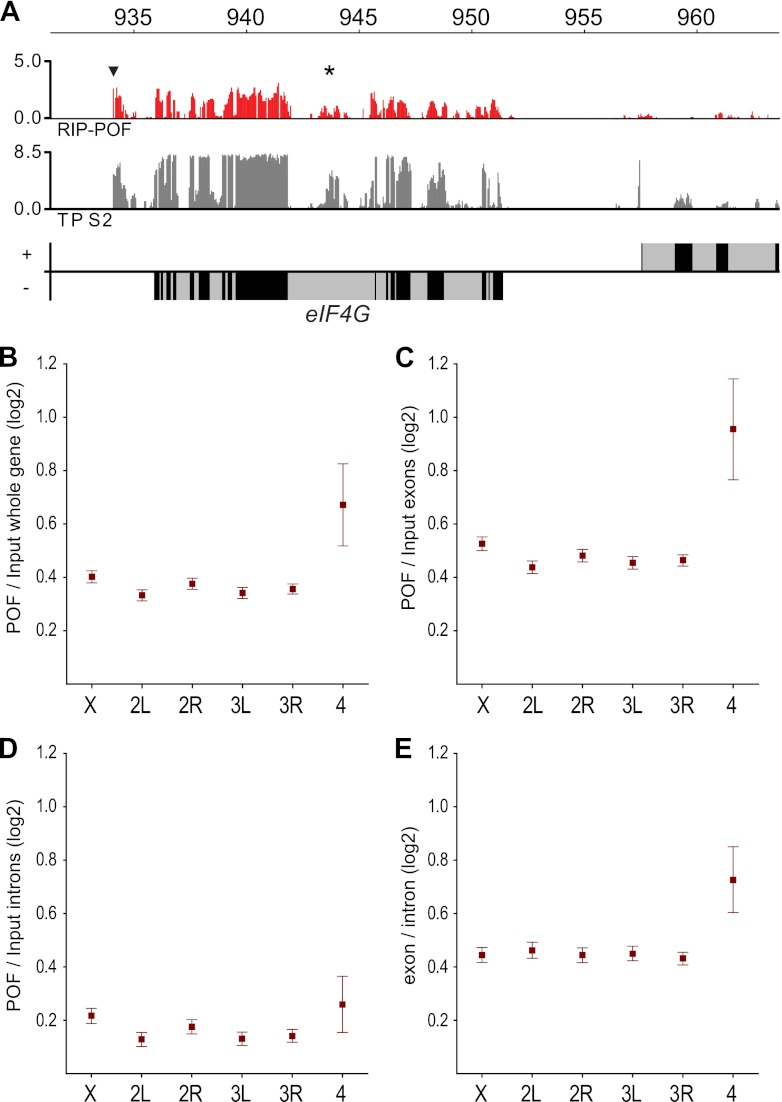
Fig 4.
POF associates with chromatin via RNP2. The figure shows the ChIP-chip POF and RIP-chip POF binding profiles and the transcriptome profile. (A) POF binding profile for the entire fourth chromosome. The chromatin IP (ChIP-chip) profile is shown in orange. The RNA IP (RIP-chip) profiles are shown for the three independent replicates N1, N3, and N6 in red, and the transcriptome profile (TP) is shown in gray. Genes expressed from left to right are represented by rectangles above the horizontal line; genes expressed in the opposite direction are shown below the line. Exons are indicated in black and introns in gray. POF binding profiles and transcriptome profile at a 100-kb region (B) and at the MED26 locus (C). Regions that are repeat masked and therefore not represented on the arrays are shaded. Numbers on the x axis denote chromosomal positions along the fourth chromosome in kilobases.
Fig 5.
POF binding to chromosome 4 is RNase resistant. Isolated salivary glands were treated with PBS ± RNase and double stained with MLE-POF or MSL2-POF. MLE (red) is completely released from the X chromosome after RNase treatment, while POF (green) remains unaffected. MSL2 (red; lower panel) is not affected by the RNase treatment. The rightmost column shows enlargements of the chromocenters and chromosome 4 regions.
Our ChIP-chip data show that POF associates preferentially with exons. By visualizing the POF-RIP enrichment at the gene level, it was found that POF has a strong preference for binding to the exons rather than the introns of enriched RNA sequences (Fig. 6A). We plotted the mean ratio between the POF-RIP and the input RNA for every probe within genes and also for probes exclusively within exons for each chromosome arm (Fig. 6B and C). The enrichment of chromosome 4 RNAs was higher when introns were excluded, indicating that POF associates preferentially with spliced RNAs.
Fig 6.
POF binds to chromosome 4 transcripts with an exon bias. (A) POF-RIP enrichment at the eIF-4G locus. Genes expressed from left to right are represented by rectangles above the horizontal line, and genes expressed in the opposite direction are shown below the line. Exons are indicated in black and introns in gray. The POF-RIP profile is smoothed with a 90-bp bandwidth. Note that the previously detected unannotated exon and the novel gene are enriched (indicated by star and arrowhead, respectively) (27). The mean ratios of POF-RIP/Input (B) for all probes within genes, all probes within exons only (C), and all probes within introns only (D), sorted by chromosome arms. (E) Exon versus intron ratios for all chromosome arms. Squares indicate the mean values, and whiskers indicate the 95% confidence intervals.
Chromosome 4 genes show decreased RNP2 pausing and elongation.
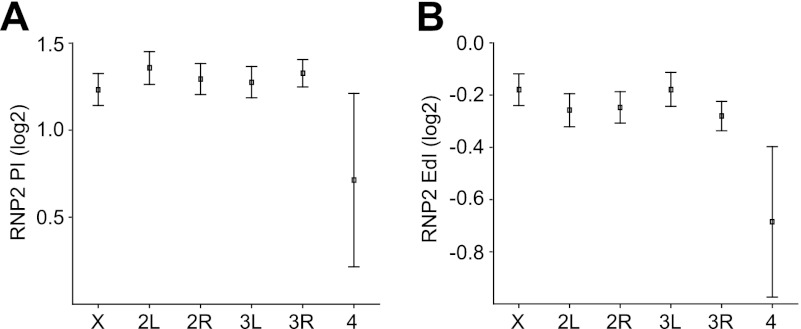
We have shown that POF stimulates the expression of genes on the fourth chromosome (26, 67). The RIP-chip data indicated that POF binds to the nascent RNA of chromosome 4 genes. This binding could be linked to an enhanced transcription or to stimulation of any posttranscriptional process. Dosage compensation of the X chromosome in Drosophila is accomplished by an increased transcription output from the single male X chromosome. The prevailing model is that the MSL complex stimulates transcriptional elongation (40). This model recently gained experimental support when it was shown by global run-on sequencing (GRO-seq) that the male X chromosome has an enhanced transcriptional elongation measured as a higher elongation density index (EdI) (32). We therefore sought to determine whether there was any difference in the transcriptionally engaged RNP2 distribution on the fourth chromosome compared to the other autosomes. Using the previously published genome-wide data of RNP2 pausing index (PI) and RNP2 elongation density index (EdI) (32), we found that RNP2 pausing is significantly decreased on the fourth chromosome compared to on all other autosomes (Fig. 7A; P = 0.0196, two-tailed t test; n = 48 and 5,946 genes). In addition, there is a significant decrease of the RNP2 elongation density index compared to the other autosomes (Fig. 7B; P = 0.0035, two-tailed t test; n = 51 and 6,039 genes). The decreased PI is consistent with an increased transcriptional output, while the decreased EdI is consistent with a decreased transcriptional output. We conclude that the fourth chromosome displays an altered RNP2 profile over active gene bodies compared to the other autosomes.
Fig 7.
The fourth chromosome shows a decrease in RNP2 pausing and a more pronounced decrease of RNP2 density over the gene body compared to the other autosomes. (A) The average RNP2 pausing index (PI) for genes grouped by chromosome arms; (B) the average RNP2 elongation density index (EdI) for genes grouped by chromosome arms. Squares indicate the mean values, and whiskers indicate the 95% confidence intervals. The PI and EdI values are from reference 32.
Loss of Pof causes a general decrease in the number of chromosome 4 transcripts.
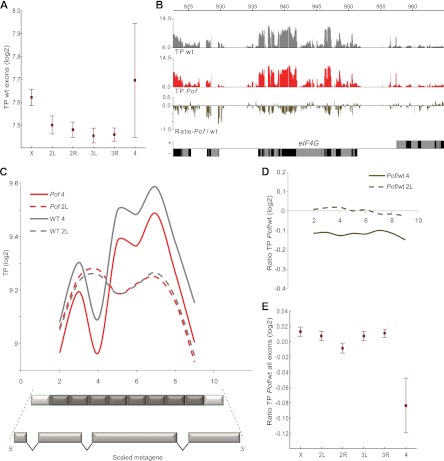
In addition to the possibility that transcription is enhanced per se, at least three posttranscriptional models are compatible with POF-mediated stimulation of expression output from chromosome 4. First, POF may be involved in the splicing machinery; a decrease in the efficiency of splicing could cause a decrease in expression output. Second, the binding of POF to transcripts from the fourth chromosome may protect them from degradation. Third, the binding of POF to nascent RNA may facilitate the export of chromosome 4 transcripts from the nucleus. To further explore the function of and mechanism underpinning POF-mediated stimulation of expression, we performed transcriptome mapping experiments comparing wild-type flies and Pof mutant flies. We prepared total RNA from three biological replicates for both wild-type and Pof mutant adult female flies and hybridized the total RNA to tiling arrays. By averaging the levels of all exon probes for each chromosome arm, we found a slight overexpression of the female X chromosomes relative to autosomes in the wild type (Fig. 8A). This is in line with what has been previously reported (22, 69, 83). We also found that the fourth chromosome is slightly overexpressed relative to all other autosomes (Fig. 8A). The two transcriptome profiles, wild type and Pof, are exceptionally similar, and we detected no difference between the two transcriptome profiles in terms of exon usage or intron boundaries (Fig. 8B), indicating that loss of POF causes no obvious splicing defects or splicing alterations. Next, we rescaled all genes into a metagene profile, including all annotated exons between the first annotated transcription start site and the last annotated transcription stop site. The genes were rescaled to the same relative length, and the transcripts were divided into 10 bins. A plot of the decrease against gene length did not reveal any bias, i.e., the decrease in the levels of the chromosome 4 gene products is most compatible with a decrease in the number of complete transcripts (Fig. 8C). The gene expression profiles for chromosome 4 are based on an average of 27 expressed genes; the ups and downs in the profile represent the exon usage of a few individual genes. Note that the profile is perfectly reproduced in Pof mutants, demonstrating the stability of the method even when the average intensity of the transcript signal was reduced along the whole gene body (Fig. 8C and D). No difference between the wild type and Pof mutants was found for genes on the control chromosome. These results are less consistent, with a difference in RNA stability between the wild type and Pof mutants. In keeping with previous results obtained using expression arrays (26, 67), the levels of chromosome 4 transcripts in the Pof mutants were found to be significantly lower than those of all other chromosomes in the Pof mutants but not in the wild type (Fig. 8E; P = 4.3 × 10−6, two-tailed t test). We conclude that loss of Pof causes a general decrease in the number of chromosome 4 transcripts but no obvious alteration in the structure of individual transcripts.
Fig 8.
POF does not affect splicing nor does it protect RNA from degradation. (A) Mean expression of all probes within exons for each chromosome arm. The y axis shows the absolute levels of exon transcripts (log2 scale) in wild-type flies. Note the increased expression on chromosome 4 and chromosome X. (B) Transcriptome profile for wild-type (gray) and PofD119 mutant (red) adult female flies at the eIF-4G locus. The lower panel shows the ratio between the Pof mutant and wt. Genes expressed from left to right are represented by rectangles above the horizontal line, and genes expressed in the opposite direction are shown below the line. Exons are indicated in black and introns in gray. The transcriptome profiles are smoothed with a bandwidth of 90 bp. (C) Metagene transcriptome profiles for expressed genes are shown for chromosome 4 in the wild type (solid gray) and on chromosome 2L in the wild type (dashed gray) and compared with chromosome 4 in PofD119 mutants (solid red) and chromosome 2L in PofD119 (dashed red). To construct a metagene (schematically illustrated below the graph), all exons between the first annotated transcriptions start site and the last transcription stop site for each gene were fused. The genes were rescaled to the same relative length, and the transcripts were divided into 10 bins. The first and last bins were subjected to annotation artifacts and were excluded from the plot. (D) Ratios of PofD119/wild-type metagene profiles for chromosome 4 (solid line) and chromosome 2L (dashed line). Note that the decrease in transcript signals for chromosome 4 genes is uniform along the gene body. (E) Mean ratio between transcript profile for PofD119 and the wild type for each chromosome arm. Squares indicate the mean value and whiskers indicate 95% confidence intervals.
Transcripts from chromosome 4 and pericentromeric heterochromatin are more efficiently exported.
To test the role of POF in nuclear transport, we calculated the amount of nuclear transcripts for each gene and the corresponding amount of whole-cell transcripts. Next, we calculated whole cell/nuclear ratios for all genes and then average ratios for all chromosome arms. Importantly, this will not give an estimate on how much of a transcript resides in the nuclei but only on the relative ratio compared to all other genes. Intriguingly, we observed a clear increase in the whole cell/nuclear ratio for the chromosome 4 transcripts relative to the other chromosome arms (Fig. 9A; P = 6.7 × 10−18, two-tailed t test). The whole-cell levels of RNA from the fourth chromosome genes were comparable to those of the other chromosome arms, but the amount of nuclear chromosome 4 RNA was reduced. This indicates that chromosome 4 mRNA spends less time in the nucleus than do transcripts from other chromosomes. Chromosome 4 is highly heterochromatic, as demonstrated by its binding of HP1. We therefore sought to determine whether such increases in the whole-cell RNA/nuclear RNA ratio was typical for genes in other heterochromatic regions defined by HP1 binding. We identified two additional regions that are enriched in HP1: cytological region 31 on chromosome arm 2L (12, 16, 24) and the pericentromeric regions of chromosome arms 2L, 2R, and 3L as defined in reference 60. We then calculated the whole cell/nuclear ratio of the transcripts from these HP1-rich regions (Fig. 9B). The results showed a decreased whole cell/nuclear ratio for RNAs from the region 2L:31 but a clear increase in the whole-cell RNA/nuclear RNA ratio for the pericentromeric region (Fig. 9B). We conclude that mRNA from expressed genes located in heterochromatic regions such as the fourth chromosome or pericentromeric heterochromatin spend less time in the nuclei compared to mRNA from euchromatic regions. Taken together, these results suggest that the increased export rate of chromosome 4 transcripts is not dependent on POF (since it is also seen for pericentromeric regions) but rather reflects a characteristic of genes located on the fourth or in the pericentromeric region.
Fig 9.
Transcripts encoded from chromosome 4 and pericentric heterochromatin are more efficiently exported relative to transcripts from other chromosomes. Mean relative ratio in log2 scale of RNA from whole-cell extract (WCFA) compared to RNA from nuclear extract (FA) for all genes on every chromosome arm (A) and for chromosome 2L and 4 compared to the heterochromatin-rich regions 2L:31 and pericentric heterochromatin from chromosome arms 2L, 2R, and 3L (B). Squares indicate the mean values, and whiskers indicate the 95% confidence intervals.
We conclude that POF is enriched on nascent RNAs transcribed from genes on the fourth chromosome and that it stimulates expression output by increasing the number of chromosome 4 transcripts. Furthermore, expressed genes in some HP1-rich regions, such as the pericentromeric regions and the fourth chromosome, exhibit increased transition rates, i.e., the RNA encoded from genes in these regions spends less time in the nuclei than does that from genes in typical euchromatic regions.
DISCUSSION
In Drosophila melanogaster, two chromosome-wide compensatory systems have been characterized: the MSL complex, which targets and stimulates the expression of the X chromosome in males, and POF, which targets and stimulates the expression of the fourth chromosome (34, 66). It has been hypothesized that the MSL complex stimulates expression by facilitating transcriptional elongation, and this hypothesis was recently confirmed experimentally (32). Here, we have used RNA immunoprecipitation and transcriptome profiling techniques to further clarify the mechanism by which POF stimulates gene expression.
POF associates with transcripts from the fourth chromosome.
We have previously shown that POF binds to active genes on the fourth chromosome (27). Since our previously reported ChIP-chip results were obtained using cross-linked extracts, they could not be used to determine whether POF associates directly with the chromatin or binds via interactions with other components and was frozen in place by the cross-linking. In the work reported herein, we observed a relationship between the binding of the POF protein to the chromosome and that of the RNA polymerase. This connection with transcription, together with the fact that POF possesses an RNA binding domain, suggested that POF binds to RNA rather than directly to chromatin. To test this hypothesis, a genome-wide POF RIP-chip experiment was performed. The POF-RIPs verified the association of POF with chromosome 4 transcripts.
Two components of the MSL complex (MSL2 and MOF) were also investigated, both as controls and to determine whether the MSL complex also binds to transcripts from the chromosome it regulates, i.e., the male X chromosome. In contrast to the observed association of POF with transcripts from the fourth chromosome, no unambiguous evidence was found to support any binding of MSL2 or MOF to X-linked transcripts. However, at this point, we cannot exclude the possibility that other components of the MSL complex, especially the more loosely bound MLE, may have a general affinity for transcripts of the X chromosome. We also noticed a slight reduction in the number of transcripts from the fourth chromosome in both the MSL2-RIPs and the MOF-RIPs, suggesting that the general RNA affinity of the MSL complex is less pronounced for the fourth chromosome than for the other chromosomes. This relative reduction in the number of chromosome 4 transcripts associated with the MSL complex might reflect the binding of POF to these transcripts, which could block their association with other RNA binding proteins.
POF binds to nascent chromosome 4 RNAs.
Our POF-RIP results, together with the previously reported strong link between POF and the fourth chromosome, suggested that POF binds to nascent RNAs, while these RNAs are still bound to the RNP2. The most parsimonious explanation of the high similarity between the ChIP-chip and RIP-chip profiles for POF is that POF associates with nascent RNA and the ChIP profile is indicative of chromatin linked via RNP2. However, at this point we cannot exclude the possibility that POF in addition to binding nascent RNAs also associates with fully processed mature mRNAs en route to the cytoplasm. It also remains possible that POF, in addition to binding nascent RNA, also associates with chromatin via interactions with other proteins (for instance, HP1). No evidence of physical association between POF and HP1 has so far been reported, but POF and Setdb1 (the HKMT responsible for H3K9me on the fourth chromosome) have been shown to interact in vitro (75). Thus, POF may be an element of an adaptor system linking histone marks to nascent RNA via HP1 and Setdb1, in a fashion similar to MRG15 and PTB (42, 43). The fact that POF binding to chromatin is RNase resistant may be explained by a stabilization of POF via interaction with a chromatin-associated factor like HP1 or Setdb1. The RNase resistance may also be caused by inaccessibility, i.e., the nascent RNA associated with POF is not accessible to the RNase.
We also report that POF was observed to bind to RNA from other chromosomes, indicating that it possesses a general affinity for RNA. This association with transcripts other than those from the fourth chromosome is more pronounced in the native samples, suggesting that it occurs during sample preparation as an equilibrium reaction rather than accurately reflecting in vivo associations. We speculate that during the preparation of the native samples, some POF molecules are released from their normal target sites and become free to associate with any transcript in the nucleoplasm. In contrast, in the cross-linked samples, the in vivo POF binding is “frozen” before any sample treatment steps are performed. Consequently, POF will only be observed to bind to chromosome 4 transcripts, and the apparent enrichment of RNAs encoded from other chromosomes is lost.
POF shows a preference for exons.
Splicing is commonly regarded as a process that takes place after transcription. However, cotranscriptional splicing was visualized in Drosophila more than 25 years ago (51). More recent studies have revealed that cotranscriptional splicing is more common than was previously believed (52) and also that splicing can begin as a cotranscriptional event and continue posttranscriptionally (6, 50). Therefore, the strong exon bias reported here is compatible with POF binding to nascent RNAs. Cotranscriptional splicing is believed to depend on cooperation between exon recognition and the speed of transcription by RNP2. Further, it has been shown that the density of nucleosomes is higher within exons; they may thus function as “speed bumps” to slow down the RNP2 elongation rate (62, 63, 73). Since POF is connected to nascent RNAs, this reduction in the speed of transcription over exons would explain the exon bias observed at the chromatin level in ChIP-chip experiments (27).
POF stimulates expression output of chromosome 4 mRNAs.
Given that POF presumably binds to nascent RNA via its RRM1 domain, it is interesting to consider hypothetical mechanisms by which POF might regulate the expression of genes on chromosome 4. The binding of POF to nascent RNA may directly or indirectly (i.e., via chromatin structure modifications) stimulate transcription. There are several examples of interactions between chromatin-associated proteins recognizing histone modification marks and proteins that bind nascent RNA (20, 21, 39, 43, 54, 64).
To explore the potential difference in engaged RNP2 distribution on chromosome 4 compared to other autosomes, we used the previously published GRO-seq data for Drosophila S2 cells (32). The cited study shows that the male X chromosome has a higher elongation density index (EdI) than the autosomes, which is interpreted as an enhanced transcription elongation (32). In contrast, comparing the fourth chromosome to all other autosomes, we find a significantly decreased EdI, which is consistent with a less efficient transcription elongation. The fourth chromosome also shows a decreased pausing index (PI). This is, on the other hand, in line with an increased transcription output from chromosome 4 genes. It is tempting to speculate that the heterochromatic nature of the fourth chromosome, with HP1 enriched over gene bodies of active genes, causes the decreased EdI. Considering that the fourth chromosome is expressed at levels equal to (or slightly higher than) those of the other chromosomes, this elongation disadvantage may be counteracted by a decreased RNP2 pausing. Since POF is bound to in principle all active chromosome 4 genes, it remains elusive whether POF is connected to the observed decrease in chromosome 4 PI.
It is also possible that the binding of POF to nascent RNA has posttranscriptional effects. There are at least three possible posttranscriptional scenarios we must consider: splicing, protection, and transport. If splicing were the main function of POF targeting, we would expect a difference in the transcriptome profiles between Pof mutants and the wild type. However, the transcriptome profiles of Pof mutants are very similar to those of the wild type, and there is no evidence of an increased rate of incorrect splicing or more frequent use of introns in the Pof mutant. The only striking difference between the transcriptome profile of the Pof mutant and that of the wild type is the reduction in the amount of processed transcripts from chromosome 4. The same reduction is observed whether we look at the 5′ end, 3′end, or middle part of the genes and is thus less consistent with the hypothesis that chromosome 4 transcripts are more prone to degradation in Pof mutants. This demonstrates that POF has a positive effect on the amount of chromosome 4 transcripts which is not caused by improved splicing efficiency.
Improved export of transcripts from chromosome 4 and pericentromeric heterochromatin.
Analysis of the ratio of input RNA levels from WCFA (whole cells) to input RNA levels from FA (nuclei) revealed that the relative amounts of chromosome 4 transcripts and transcripts from genes in the pericentromeric region are higher in the cytoplasm than in the nucleoplasm, in relation to transcripts from the other chromosomes. Notably, although we have not measured export rates per se, our results are consistent with pericentromeric and chromosome 4 transcripts being more efficiently exported. Chromatin is highly organized within the nucleus: euchromatic blocks are preferentially located in the center, while heterochromatic regions, such as the fourth chromosome and pericentromeric heterochromatin, tend to localize closer to the nuclear rim (31, 53, 72). Whether the nuclear periphery is a repressive or permissive environment for gene expression has been debated. The transition time for the export of transcribed mRNAs from the site of synthesis to the nuclear pore would be minimized for chromosomal regions close to the nuclear pore. The gene-gating hypothesis postulates that nucleoporins associate with active genes and facilitate the export of the corresponding mRNAs (5). Components of the nuclear pore complex (nucleoporins) have been reported to interact with transcriptionally active genes (7, 28) and the MSL complex (47, 76). It has been shown in a mammalian system that transport of an mRNA from the site of transcription to the nuclear pore occurs within a time frame of 5 to 40 min. In the same study, no pileup of mRNAs at the nuclear pore was found, and export through the pore was rapid (0.5 s) (49). Thus, a closer proximity to the nuclear pore may increase transcription output, especially in rapidly dividing cells, since reducing the transition time would allow a more rapid initiation of protein synthesis after cell division. We observed that the whole cell/nuclei ratio for transcripts produced from the fourth chromosome and the pericentromeric heterochromatin was increased compared to the transcript ratio of the entire genome. It should be stressed that although these genes are located in seemingly repressive environments, both the number of expressed genes and gene expression (as shown here) are comparable to euchromatic chromosome regions (60). It may be that genes located in these heterochromatic regions benefit from their relative proximity to the nuclear pore, which would facilitate the export of transcribed mRNA. This may in fact be one reason why genes located in pericentric heterochromatin, such as light and rolled, are repressed by rearrangements that move them into euchromatic surroundings (14, 78).
It is tempting to speculate that the evolution of more efficient logistics for the export of transcripts from the fourth chromosome was driven by the need to facilitate the expression of its genes. We have also shown that the quantity of chromosome 4 transcripts is reduced in Pof mutants, and our data indicate that this decrease is not caused by splicing defects or increased degradation. It is not yet known whether the binding of POF to nascent RNAs increases the efficiency of transcription or whether it facilitates their efficient export. However, it should be stressed that transcription levels are probably influenced by a number of stimulatory and repressive influences and that during the course of evolution these factors become increasingly interdependent.
ACKNOWLEDGMENTS
We thank M. Kuroda for MSL antibodies and Tracy Nissan for comments on the manuscript.
This work was supported by grants from the Carl Tryggers and Erik Philip-Sörensens Foundations (P.S.), the Kempe Foundation (A.-M.J. and A.A.), Nilsson-Ehle Foundation (A.A.), and the Swedish Research Council and Magnus Bergvalls Foundation to J.L.
Footnotes
Published ahead of print 2 April 2012
REFERENCES
- 1. Akhtar A, Zink D, Becker PB. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405–409 [DOI] [PubMed] [Google Scholar]
- 2. Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20:848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alekseyenko AA, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ashburner M, Golic KG, Hawley RS. 2005. Drosophila: a laboratory handbook. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 5. Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. U. S. A. 82:8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brody Y, et al. 2011. The in vivo kinetics of RNA polymerase II elongation during cotranscriptional splicing. PLoS Biol. 9:e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capelson M, et al. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140:372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carvalho A, Koerich L, Clark A. 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng B, Price DH. 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282:21901–21912 [DOI] [PubMed] [Google Scholar]
- 10. Chodosh LA, Fire A, Samuels M, Sharp PA. 1989. 5,6-Dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264:2250–2257 [PubMed] [Google Scholar]
- 11. Copps K, et al. 1998. Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J. 17:5409–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cryderman DE, et al. 2005. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 232:767–774 [DOI] [PubMed] [Google Scholar]
- 13. Czermin B, et al. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185–196 [DOI] [PubMed] [Google Scholar]
- 14. Eberl DF, Duyf BJ, Hilliker AJ. 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics 134:277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eissenberg JC, Morris GD, Reuter G, Hartnett T. 1992. The heterochromatin-associated protein HP1 is an essential protein in Drosophila with dosage-dependent effects on position effect variegation. Genetics 131:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fanti L, Berloco M, Piacentini L, Pimpinelli S. 2003. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica 117:135–147 [DOI] [PubMed] [Google Scholar]
- 17. Filion GJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143:212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gelbart ME, Kuroda MI. 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136:1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilfillan GD, et al. 2006. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 20:858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunderson FQ, Johnson TL. 2009. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in cotranscriptional spliceosome assembly. PLoS Genet. 5:e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gunderson FQ, Merkhofer EC, Johnson TL. 2011. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc. Natl. Acad. Sci. U. S. A. 108:2004–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta V, et al. 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hallacli E, Akhtar A. 2009. X chromosomal regulation in flies: when less is more. Chromosome Res. 17:603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. James TC, et al. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50:170–180 [PubMed] [Google Scholar]
- 25. Johansson A-M, Allgardsson A, Stenberg P, Larsson J. 2011. msl2 mRNA is bound by free nuclear MSL complex in Drosophila melanogaster. Nucleic Acids Res. 39:6428–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johansson AM, Stenberg P, Bernhardsson C, Larsson J. 2007. Painting of fourth and chromosome-wide regulation of the fourth chromosome in Drosophila melanogaster. EMBO J. 26:2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johansson AM, Stenberg P, Pettersson F, Larsson J. 2007. POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet. 3:e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140:360–371 [DOI] [PubMed] [Google Scholar]
- 29. Kaminker JS, et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3:RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kind J, et al. 2008. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133:813–828 [DOI] [PubMed] [Google Scholar]
- 31. Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8:104–115 [DOI] [PubMed] [Google Scholar]
- 32. Larschan E, et al. 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471:115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsson J, Chen JD, Rasheva V, Rasmuson Lestander A, Pirrotta V. 2001. Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 98:6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larsson J, Meller VH. 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14:417–431 [DOI] [PubMed] [Google Scholar]
- 35. Larsson J, Svensson MJ, Stenberg P, Mäkitalo M. 2004. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl. Acad. Sci. U. S. A. 101:9728–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Locke J, Howard TL, Aippersbach N, Podemski L, Hodgetts BR. 1999. The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108:356–366 [DOI] [PubMed] [Google Scholar]
- 37. Locke J, McDermid H. 1993. Analysis of Drosophila chromosome four by pulse field electrophoresis. Chromosoma 102:718–723 [DOI] [PubMed] [Google Scholar]
- 38. Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R. 1999. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 9:137–149 [PMC free article] [PubMed] [Google Scholar]
- 39. Loomis RJ, et al. 2009. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33:450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lucchesi JC. 1998. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr. Opin. Genet. Dev. 8:179–184 [DOI] [PubMed] [Google Scholar]
- 41. Lucchesi JC, Kelly WG, Panning B. 2005. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 39:615–651 [DOI] [PubMed] [Google Scholar]
- 42. Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144:16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luco RF, et al. 2010. Regulation of alternative splicing by histone modifications. Science 327:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mank JE. 2009. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet. 25:226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marshall NF, Price DH. 1992. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 12:2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McAnally AA, Yampolsky LY. 2010. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol. Evol. 2:44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mendjan S, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21:811–823 [DOI] [PubMed] [Google Scholar]
- 48. Miklos GLG, Yamamoto MT, Davies J, Pirrotta V. 1988. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome four and the β-heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 85:2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mor A, et al. 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12:543–552 [DOI] [PubMed] [Google Scholar]
- 50. Neugebauer KM. 2002. On the importance of being cotranscriptional. J. Cell Sci. 115:3865–3871 [DOI] [PubMed] [Google Scholar]
- 51. Osheim YN, Miller OL, Beyer AL. 1985. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell 43:143–151 [DOI] [PubMed] [Google Scholar]
- 52. Pandya-Jones A, Black DL. 2009. Cotranscriptional splicing of constitutive and alternative exons. RNA 15:1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parada L, Misteli T. 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12:425–432 [DOI] [PubMed] [Google Scholar]
- 54. Piacentini L, et al. 2009. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 5:e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pimpinelli S, et al. 1995. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. U. S. A. 92:3804–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prestel M, Feller C, Becker PB. 2010. Dosage compensation and the global rebalancing of aneuploid genomes. Genome Biol. 11:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prestel M, Feller C, Straub T, Mitlöhner H, Becker PB. 2010. The activation potential of MOF is constrained for dosage compensation. Mol. Cell 38:815–826 [DOI] [PubMed] [Google Scholar]
- 58. Price DH. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Richter L, Bone JR, Kuroda MI. 1996. RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells 1:325–336 [DOI] [PubMed] [Google Scholar]
- 60. Riddle NC, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21:147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schotta G, et al. 2002. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schwartz S, Ast G. 2010. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 29:1629–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schwartz S, Meshorer E, Ast G. 2009. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 16:990–995 [DOI] [PubMed] [Google Scholar]
- 64. Sims RJ, et al. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28:665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Spradling A, Penman S, Pardue ML. 1975. Analysis of Drosophila mRNA by in situ hybridization: sequences transcribed in normal and heat shocked cultured cells. Cell 4:395–404 [DOI] [PubMed] [Google Scholar]
- 66. Stenberg P, Larsson J. 2011. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120:213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stenberg P, et al. 2009. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 5:e100302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stenberg P, Pettersson F, Saura AO, Berglund A, Larsson J. 2005. Sequence analysis of chromosome identity in three Drosophila species. BMC Bioinformatics 6:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sturgill D, Zhang Y, Parisi M, Oliver B. 2007. Demasculinization of X chromosomes in the Drosophila genus. Nature 450:238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sun FL, et al. 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. U. S. A. 97:5340–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sun FL, et al. 2004. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24:8210–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tanabe H, et al. 2002. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. U. S. A. 99:4424–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tilgner H, et al. 2009. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 16:996–1001 [DOI] [PubMed] [Google Scholar]
- 74. Tweedie S, et al. 2009. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37:D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tzeng TY, Lee CH, Chan LW, Shen CK. 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Natl. Acad. Sci. U. S. A. 104:12691–12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vaquerizas JM, et al. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6:e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vicoso B, Bachtrog D. 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 17:585–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wakimoto BT, Hearn MG. 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125:141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wallrath LL, Elgin SC. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9:1263–1277 [DOI] [PubMed] [Google Scholar]
- 80. Wallrath LL, Guntur VP, Rosman LE, Elgin SC. 1996. DNA representation of variegating heterochromatic P-element inserts in diploid and polytene tissues of Drosophila melanogaster. Chromosoma 104:519–527 [DOI] [PubMed] [Google Scholar]
- 81. White RAH. 1998. Immunolabelling of Drosophila, p 215–240. In Roberts DB. (ed), Drosophila, a practical approach. IRL Press, Oxford, United Kingdom [Google Scholar]
- 82. Zhang Y, et al. 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8:e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang Y, Oliver B. 2010. An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]