Abstract
Summary: Proteolytic cleavage of proteins that are permanently or transiently associated with the cytoplasmic membrane is crucially important for a wide range of essential processes in bacteria. This applies in particular to the secretion of proteins and to membrane protein quality control. Major progress has been made in elucidating the structure-function relationships of many of the responsible membrane proteases, including signal peptidases, signal peptide hydrolases, FtsH, the rhomboid protease GlpG, and the site 1 protease DegS. These enzymes employ very different mechanisms to cleave substrates at the cytoplasmic and extracytoplasmic membrane surfaces or within the plane of the membrane. This review highlights the different ways that bacterial membrane proteases degrade their substrates, with special emphasis on catalytic mechanisms and substrate delivery to the respective active sites.
INTRODUCTION
Bacterial membrane-associated proteases have important functions in the processing, quality control, and regulated turnover of proteins that are transported to the plasma membrane or to extracytoplasmic compartments such as the periplasm. In the general protein secretion pathway of bacteria, the membrane proteases signal peptidase (SP) and signal peptide hydrolase play critical roles. Signal peptidases function to remove the N-terminal targeting peptides from secretory preproteins. These proteases cleave juxtamembrane peptide bonds of the substrate during or shortly after their translocation across the membrane. Signal peptides are typically further degraded by signal peptide hydrolases to release them from the membrane or to generate cleaved products that serve in signaling to the cell (113). Quality control and regulated turnover of membrane proteins are necessary not only for the removal of malfolded or damaged proteins in the membrane but also to respond appropriately to stressful environmental conditions. This ensures the fidelity of processes in the membrane that are critical for bacterial growth, division, and survival. For example, in response to an external stress such as heat, the sigma E pathway of the Gram-negative bacterium Escherichia coli is switched on. This leads to the synthesis of chaperones and proteases for repair or destruction of damaged proteins. To activate this pathway, the periplasmic site 1 protease DegS cleaves RseA, a transmembrane protein that normally sequesters sigma E. RseA is then cleaved further by the site 2 membrane protease (S2P) RseP, which liberates the cytoplasmic domain of RseA bound to sigma E. It is further degraded by cytoplasmic ATP-dependent proteases, which results in the release of sigma E into the cytoplasm and in sigma E-dependent gene expression (5, 69). RseP and certain signal peptide hydrolases are highly intriguing enzymes because they catalyze hydrolytic reactions within the membrane plane. Moreover, in the Gram-positive bacterium Bacillus subtilis, the S2P RasP (YluC) plays an important role in the regulation of cell division (19, 199). RasP is responsible for the rapid turnover of FtsL, a small bitopic membrane protein, which is an essential part of the cell division machinery. Finally, the membrane protease FtsH plays a key quality control role in degrading misassembled and damaged membrane proteins in bacteria (65). FtsH, which is an essential protein, degrades its substrates in an ATP-dependent manner.
This review focuses on signal peptidases that cleave secretory preproteins, signal peptide hydrolases that degrade signal peptides, site 1 and site 2 proteases that are involved in the regulated turnover of membrane proteins, and the quality control protease FtsH, involved in the degradation of malfolded membrane proteins. These proteases highlight the problem of cleavage of substrates in juxtamembrane regions proximal to the membrane surface or within the membrane plane, as well as the degradation of membrane proteins following extraction of the substrate from the membrane.
GENERAL CONSIDERATIONS
Before we discuss the different membrane proteases involved in the protein secretion and quality control pathways, it is instructive to focus attention on two major differences in the known proteases that cleave membrane-associated substrates. One group of membrane proteases is represented by extramembrane enzymes that have active sites outside the membrane and cleave their membrane-associated substrates at the aqueous membrane boundary. A key question here is what allows them to cleave right at the membrane boundary with exquisite accuracy and not at other sites far removed from the membrane surface, which might have disastrous consequences for the cell. Other extramembrane proteases cleave their substrates only after these have been released or extracted from the membrane. Some of the latter membrane proteases use ATP hydrolysis to dislocate membrane protein substrates from the lipid bilayer. The second group of membrane proteases is represented by intramembrane proteases that have membrane-embedded active sites. A fascinating question concerning these intramembrane-cleaving proteases is where the hydrolytic water comes from in a non-water-accessible hydrophobic environment. Another challenge is to find out how the transmembrane region of a substrate that is cleaved by the protease enters the substrate binding region of the protease from the lipid phase of the membrane to gain access to the active site region of the protease. These and many other relevant questions related to the function of membrane proteases are addressed in this review, using the available structural information on these enzymes for guidance.
SIGNAL PEPTIDASES CLEAVE AT THE MEMBRANE SURFACE
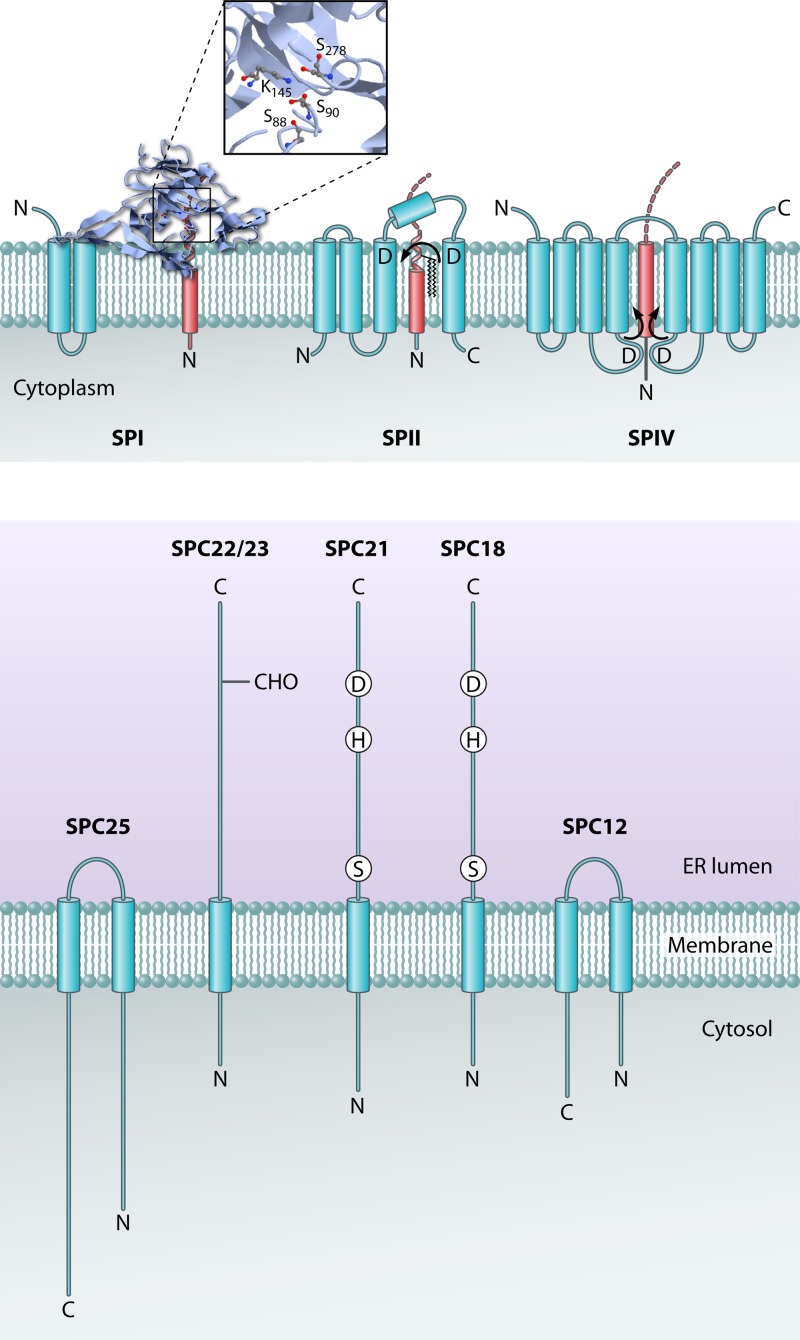
There are several types of SPs in the bacterial cell (Fig. 1) (113). SPs cleave the signal peptides of exported proteins after they have served their purpose in targeting these proteins to the machinery for protein translocation across the membrane. In bacteria, SPI is the general signal peptidase that cleaves the majority of preproteins. The bacterial SPI is homologous to the catalytic subunit(s) of the signal peptidase complex (SPC) in the endoplasmic reticulum (ER) of eukaryotes, which is responsible for processing of preproteins that are translocated into the ER lumen (150, 168). SPII (lipoprotein signal peptidase) is responsible for cleaving lipoprotein precursor proteins. The prepilin signal peptidase (SPIV) is responsible for processing signal peptides of type IV pilins, as well as a variety of pseudopilins involved in the secretion of proteins across the outer membranes of Gram-negative bacteria (type II secretion) or in DNA uptake by Gram-positive bacteria (140).
Fig 1.
Signal peptide cleavage of precursor proteins by signal peptidases. Signal peptidase I (SPI) employs a Ser-Lys catalytic dyad for signal peptide cleavage from secretory precursor proteins at the extracytoplasmic surface of the membrane. The Protein Data Bank (PDB) structure of the catalytic domain (accession number 1T7D) and the program JMol were used to generate the three-dimensional (3D) structure image of SPI. Signal peptidase II (SPII) is an aspartic acid protease that cleaves signal peptides from bacterial lipoprotein precursors just beneath the extracytoplasmic membrane surface. The lipoprotein precursor protein is diacylglyceride modified prior to SPII cleavage. Signal peptidase IV (SPIV) is an aspartic protease that cleaves signal peptides from prepilins and pseudopilins at the cytoplasmic surface of bacterial membranes. The eukaryotic ER signal peptidase complex (SPC) is composed of five subunits, of which SPC18 and SPC21 are catalytic. Transmembrane helices of signal peptidases are depicted as blue barrels, and substrate helices are depicted as red barrels. A zoomed-in view of the active site residues of SPI is shown. The locations of the N and C termini of the signal peptidases and their substrates are indicated.
Signal Peptidase I
SPI serves a crucial role in the liberation of translocated secretory precursor proteins from the cytoplasmic membrane through the removal of the signal peptide (171). This cleavage is essential for protein release into the periplasmic space, transport of proteins to the outer membrane in Gram-negative bacteria, and secretion of proteins into the extracellular medium. Notably, SPI-catalyzed membrane release of translocated proteins applies not only to proteins transported by the general protein secretion (Sec) pathway but also to those transported by the twin-arginine translocation (Tat) pathway, which facilitates the export of fully folded proteins (67, 96). Additionally, SPI has been shown to cleave internal signal peptides in a few polytopic membrane proteins (8, 12, 70, 118, 147). E. coli SPI is the best-studied signal peptidase in this family. It spans the cytoplasmic membrane twice, with a large C-terminal domain containing the active site protruding into the periplasmic space (132). The SPI family of proteases is unusual in that it is not inhibited by standard serine protease inhibitors, most likely because they do not bind with high enough affinity (200). However, these enzymes are inhibited by β-lactams (16, 49), lipopeptides (97, 112, 126), and lipoglycopeptides (83). SPI is an unconventional serine protease containing an active site Ser-Lys dyad configuration instead of the canonical Ser-His-Asp triad architecture (36). SPI of E. coli requires serine 90 and lysine 145 for activity (15, 114, 142, 160). Why a Ser-Lys dyad is used is not entirely clear. It might be just by chance, or the alternate active site configuration may allow for activity in a different cellular environment, as the pH optimum is typically higher for Ser-Lys proteases than for Ser-His-Asp proteases. However, it should be noted that the extracytoplasmic side of the bacterial cytoplasmic membrane has a relatively low pH (<6) due to the transmembrane proton gradient. At such low pH values, SPI enzymes are barely active, which suggests that they might be pH regulated. This would minimize the potentially deleterious proteolysis of membrane proteins by SPI until this enzyme is somehow activated for preprotein cleavage. Alternatively, the pKa of the active site lysine residue of SPI could be lowered by hydrophobic interactions with membrane constituents such as phospholipids (171).
A significant breakthrough in the signal peptidase field was the 1.9-Å X-ray crystal structure determination of the periplasmic domain (Δ2-75) (111) (Fig. 1). The Δ2-75 domain is catalytically active (159) and cleaves substrates at the normal cleavage site, despite lacking its two transmembrane anchors and cytoplasmic domain (22). The structure of the Δ2-75 domain revealed that this protease not only employs a Ser-Lys dyad catalytic mechanism but also contains an exposed hydrophobic surface for membrane association (111). In the structure, serine 90 (the nucleophile) is covalently attached to the cleaved 5S penem inhibitor and within H-bonding distance of lysine 145 (the general base).
In contrast to Gram-negative bacteria, many Gram-positive bacteria contain multiple SPI enzymes. This became evident from studies on the SPI enzymes of B. subtilis, a soil bacterium that is widely used for the biotechnological production of secreted enzymes (198). The biological function of the SPI enzymes in B. subtilis is that they cleave secretory preproteins for release of the mature product from the membrane. This release is needed to target proteins to the thick Gram-positive bacterial cell wall and the extracellular milieu. However, the SPI enzymes of B. subtilis also seem to prevent jamming of the Sec machinery with secretory preproteins (150). The SPI SipS of B. subtilis 168 was the first characterized signal peptidase from a Gram-positive bacterium (168). Subsequently, it was found that B. subtilis contains four other chromosomally encoded type I signal peptidases, denoted SipT, SipU, SipV, and SipW (150, 151). In addition, some strains of B. subtilis were shown to contain endogenous plasmids (pTA1015 or pTA1040) specifying an SPI denoted SipP (105, 153). Multiple related SPI enzymes were subsequently identified in a range of Gram-positive bacteria, including major pathogens such as Bacillus anthracis and Staphylococcus aureus as well as biotechnologically relevant bacteria such as streptomycetes (171). Importantly, the amino acid sequence of SipS allowed the identification of conserved domains in SPI enzymes from bacteria, archaea, the mitochondrial inner membrane, the chloroplast thylakoidal membrane, and the endoplasmic reticular membrane (150, 168). These conserved domains focused attention on the critical roles of the conserved Ser and Lys residues in the prokaryotic and organellar SPI enzymes that were subsequently shown to be responsible for catalysis (111, 169).
Mitochondria and chloroplasts, which probably evolved from bacterial endosymbionts, contain related SPI enzymes in the inner membrane and the thylakoidal membrane, respectively. The mitochondrial Imp1 and Imp2 enzymes remove signal peptides from proteins that are targeted to the inner membrane, and the chloroplast's TPP removes signal peptides from proteins that are transported into the thylakoid luminal space. Like the homologous SPI enzymes of bacteria, Imp1, Imp2, and TPP contain active site Ser-Lys residues that carry out the catalytic reaction (23, 26). The eukaryotic SPC is responsible for the removal of signal peptides from proteins that are translocated into the ER. In contrast to the case for prokaryotic and organellar SPI enzymes, the catalytic subunits of the ER SPC seem to employ conserved Ser, His, and Asp residues for catalysis (Fig. 1) (150, 152, 168, 175). It is unclear why the ER SPC catalytic subunits use the standard Ser-His-Asp active site residues instead of the Ser-Lys dyad configuration.
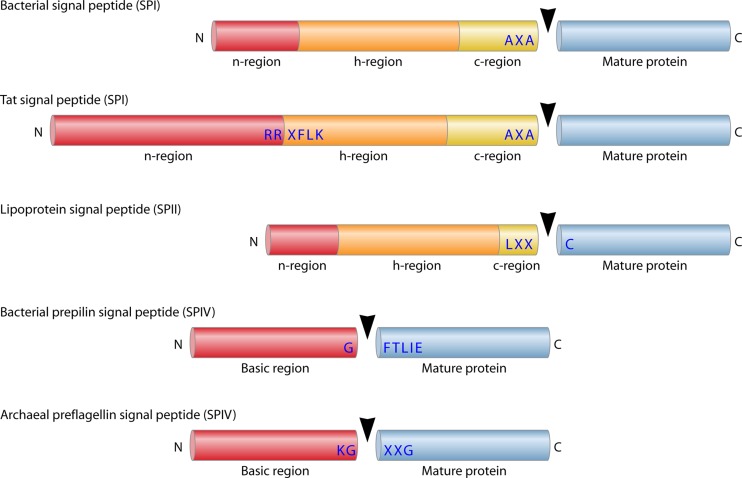
All SPI substrates contain a signal peptide that has three conserved domains: the positively charged N region, the central hydrophobic H region, and the C region, which contains the substrate specificity determinants for signal peptidase cleavage (Fig. 2). In B. subtilis, secretory signal peptides have been analyzed at the proteome level, and the lengths of the signal peptide domains have been determined for 58 proteins (148). Cleavage is predicted to be at or near the extracytoplasmic membrane surface, because the length of the H regions of secretory signal peptides, up to the Ala-X-Ala signal peptidase recognition motif (positions −3 to −1), is typically 19 amino acids. If one assumes ∼3.6 amino acid residues per turn of the helix, a rise of ∼1.5 Å per residue along the helix axis, and a membrane thickness of ∼30 Å, then the cleavage site at the end of the C region will be presented at the membrane surface. The signal peptides of Gram-negative bacteria, and also those of eukaryotes, are substantially shorter than those of B. subtilis and other Gram-positive bacteria (13). This implies that the smallest functional signal peptides, with total lengths of ∼15 residues from the N to the C region, will not completely span a membrane of 30 Å. In this case, the end of the C region will be presented to the SPI below the extracytoplasmic membrane surface.
Fig 2.
Signal peptide substrates of different classes of signal peptidases. Schematic representations are shown for bacterial (Sec-type) signal peptides cleaved by SPI, twin-arginine (Tat) signal peptides cleaved by SPI, lipoprotein signal peptides cleaved by SPII, bacterial prepilin signal peptides cleaved by SPIV, and archaeal preflagellin signal peptides cleaved by the SPIV homologue FlaK. The N, H, C, and basic regions of the respective signal peptides, mature protein parts, and conserved SP recognition sites are indicated. The SP cleavage site is marked with a black arrowhead. N, N terminus; C, C terminus.
How does SPI cleave substrates at or below the membrane surface? A clue came from the X-ray structure of the inhibitor-SPI catalytic domain complex, which revealed the substrate binding groove (112). Structural analyses showed that SPI forms main chain hydrogen bond interactions with residues −1 to −7 of the signal peptide C region. This implies that for SPI binding, the signal peptide has to undergo a conformational change within the membrane and switch from an α-helical conformation to an extended structure. One hypothesis is that this conformational change is triggered by the binding of SPI to this region. In this model, SPI most likely interacts reversibly with the membrane via its hydrophobic surface-exposed membrane association domain. The catalytic domain could thus swing in and out of the membrane. Upon penetrating the lipid phase of the membrane, the catalytic domain would gain access to the signal peptide region up to the residue at position −7 (Fig. 1). Importantly, this mechanism explains the cleavage of short signal peptides that cannot completely span the membrane. Indeed, the Δ2-75 derivative of E. coli SPI has been shown to bind to lipid vesicles and to interact with phospholipid monolayers (170). Moreover, detergents or phospholipids are also required for optimal activity of the Δ2-75 domain (159). Alternatively, the signal peptide region at positions −1 to −7 may have a tendency to undergo the conformational change spontaneously, before SPI can bind. This would move the signal peptide region at positions −1 to −7 toward the aqueous extracytoplasmic environment, where it would be recognized and cleaved by SPI.
The Ala-X-Ala substrate specificity of SPI is determined by the S1 and S3 pockets (111, 112). The E. coli S1 pocket residues Met91, Ile144, Leu95, and Ile86 make direct van der Waals contact with the P1 residue. Residues forming the S3 pocket are Phe84, Ile144, Val132, and Ile86. These S1 and S3 pocket residues coming into contact with the P1 and P3 substrate residues are highly conserved in the SPI family. Interestingly, Ile144 plays a profound role in cleavage fidelity, as mutation of Ile144 to Cys results in “sloppy cleavage” at multiple sites in a preprotein substrate (71). One possible explanation for the lack of fidelity is that the cysteine mutation in the binding site results in sliding of the substrate within the active site such that alternative peptide bonds can be hydrolyzed. Moreover, Ile144 and Ile86 control substrate specificity, because mutation of these residues results in substrate cleavage even with an arginine at the −3 position (35).
The SPI enzymes of Gram-positive bacteria such as B. subtilis are much smaller than SPI of E. coli. Nevertheless, they contain the 4 conserved regions, denoted boxes B, C, D, and E, which are found within all SPI enzymes. Mutagenesis studies showed that these enzymes also employ a Ser-Lys catalytic dyad (169). As demonstrated for the SPI SipS of B. subtilis, the Ser43 and Lys83 residues form the catalytic dyad. The Leu74 and Tyr81 residues of SipS contribute to catalysis, most likely by lowering the pKa of the active site Lys83 residue to such an extent that it can function as a general base (17, 169). Furthermore, Asp146 and Asp153 are also important for the activity of SipS, but these residues appear to be critical conformational determinants. Notably, domain swapping studies on the SPI enzymes of B. subtilis have shown that the N-terminal regions, which comprise the unique transmembrane anchors of these enzymes, are important determinants of their substrate specificity (173). However, these specificity-determining N-terminal regions of the Bacillus SPI enzymes include neither the S1 and S3 pockets nor the active sites (172). These findings therefore suggest that the main role of the N-terminal regions with respect to substrate specificity is to correctly position the active site for substrate binding at or below the membrane surface. This is in line with a model that suggests that membrane penetration of the active site of SPI is necessary to bind particular signal peptides, especially those that are too short to fully span the membrane. The membrane penetration may vary somewhat between the multiple SPI enzymes (i.e., SipS, SipT, SipU, SipV, and SipW) in B. subtilis, which might explain why the different SPI proteases cannot fully substitute for one another. Some of these enzymes have different substrate preferences despite recognizing the same Ala-X-Ala motif within signal peptide substrates.
While the Ser-Lys catalytic dyad is conserved in the vast majority of SPI enzymes from prokaryotes, several bacterial species belonging to the Actinobacteria (e.g., Arthrobacter, Rhodococcus, and Xylanimonas), Firmicutes (Bacillus, Clostridium, Desulfitobacterium, Eubacterium, and Ruminococcus), and Mollicutes (Sphaerobacter) possess SPI enzymes that more closely resemble the catalytic subunits of the ER SPC and archaeal SPI enzymes. SipW of B. subtilis was the first identified representative of these atypical SPI enzymes in bacteria (150). Site-directed mutagenesis of residues of SipW that are conserved in all known SPI enzymes showed that Ser47, His87, and Asp106 are indispensable for activity. Thus, SipW and other closely related SPI enzymes most likely employ a conventional Ser-His-Asp catalytic triad or a Ser-His catalytic dyad similar to that of the ER SPC (152, 175). Very recently, it was reported that the 20 C-terminal residues of SipW, which localize in the cytoplasm, serve a potentially nonenzymatic function in the regulation of biofilm formation on solid surfaces (145). Site-directed mutagenesis data show that the signal peptidase activity of SipW is dispensable for the formation of this type of biofilms. Conversely, a SipW mutant protein that lacks the 20 C-terminal residues is still active as a signal peptidase but does not facilitate biofilm formation on solid surfaces. Thus, SipW seems to be a bifunctional enzyme. How the second activity of SipW works needs to be investigated further. In any case, the available data suggest that the C terminus of SipW is needed to activate the eps genes for the formation of a biofilm matrix when cells are on a solid surface. Most likely, this involves direct or indirect interactions with the SinR protein, which is a repressor of the eps genes and the tapA-sipW-tasA genes. Thus, it seems that SipW is a bifunctional SPI. Judged by the conservation of the C terminus, it seems that SipW proteins of other bacilli may have similar functions (166), but this remains to be demonstrated.
Signal Peptidase II
SPII plays a crucial role in the subcellular localization and export of lipid-modified bacterial proteins. These proteins are lipid modified by the diacylglyceryl transferase Lgt prior to processing by SPII. In Gram-negative bacteria, the resulting mature lipoproteins are retained in either the cytoplasmic membrane or the outer membrane via their diacylglyceryl moiety. Alternatively, they are secreted into the growth medium, where they form micelle-like structures (120, 155). In Gram-positive bacteria, the mature lipoproteins are retained predominantly in the cytoplasmic membrane via their diacylglyceryl moiety, but they can also be released into the growth medium upon alternative processing by as yet unidentified proteases (8, 154). Like the secreted proteins that are processed by SPI, lipid-modified precursor proteins can be delivered to SPII via either the Sec pathway or the Tat pathway for protein export (146, 155, 188). However, to date, Tat-dependent export of lipoproteins has been shown only in streptomycetes, whereas this does not seem to occur in other bacteria, such as E. coli and B. subtilis (66, 146). Why streptomycetes export a substantial number of lipoproteins (presumably in a folded state) by the Tat pathway is not known, but it may relate to the ecological niche in the soil that is occupied by these organisms. If so, this niche would impose strong selective pressure for folding of secretory proteins in the cytoplasm and their subsequent Tat-dependent export, as seems to be evidenced by the large numbers of Tat-dependently exported proteins encountered in streptomycetes.
Notably, the SPII recognition site in the C region of lipoprotein signal peptides overlaps with the recognition site for the diacylglyceryl transferase Lgt. This region is generally known as the “lipobox.” The lipobox in lipoprotein signal peptides from E. coli and B. subtilis has the consensus sequence L-A/S-A/G-C (154) (Fig. 2). The invariable cysteine residue of the lipobox is the target for lipid modification and the first residue of the mature lipoprotein after cleavage by SPII (155). Based on proteomic analyses, the lipobox of lipoproteins from B. subtilis has been defined more precisely as having the sequence L/I/T/A/G/M/V-A/S/G/T/I/M/V/F-A/G-C-S/G/E/N/T/A/Q/R at positions −3 to +2 around the SPII cleavage site (154).
The bacterial SPII enzyme employs a variation of the SPI cleavage mechanism for catalysis. SPII is an aspartic acid protease with its catalytic residues positioned at the extracytoplasmic membrane surface (156). In B. subtilis SPII, both active site Asp residues 102 and 129 are positioned at the ends of transmembrane segments (Fig. 1). In contrast to the case for SPI, where the protease interacts reversibly with the membrane, the catalytic residues of SPII appear to be fixed just beneath the membrane surface for signal peptide cleavage from lipoprotein precursors (156). Therefore, the signal peptide cleavage site of these precursors needs to be more exposed to the lipid phase of the membrane, which is probably facilitated by the relatively short H region of lipoprotein signal peptides. The H region has an average length of 12 residues in B. subtilis (7 amino acids shorter than the H region of secretory signal peptides) (149), and it has an average length of 10 residues in E. coli (155). Additionally, the diacylglyceryl modification of lipoprotein precursors may serve to correctly present their cleavage site to SPII (32). To allow SPII to bind and cleave its preprotein substrate, the signal peptide α-helix would have to be disrupted, like the case in SPI-mediated catalysis. Indeed, many lipoprotein signal peptides have helix-breaking residues within the region of positions −4 to −6 that may facilitate substrate recognition and binding of the substrate specificity residues located in the lipobox (37, 177). Further detailed mechanistic insights into substrate binding and cleavage by SPII enzymes await the elucidation of an SPII structure.
Signal Peptidase IV and Preflagellin Signal Peptidase
The precursors of type IV pilins and related pseudopilins are specifically processed by prepilin signal peptidases (SPIV) (31, 141). Unlike SPI or SPII signal peptides, the consensus SPIV recognition sequence, Gly-Phe-Thr-Leu-Ile-Glu, where cleavage occurs between the Gly and Phe residues, is located between the N and H regions (Fig. 2) (95). In addition to signal peptide cleavage, SPIV is also responsible for N-methylation of the phenylalanine at position +1 relative to the cleavage site (116). Pseudopilin signal peptides show clear structural similarities to other types of bacterial signal peptides, and the pseudopilin precursors are most likely targeted to the membrane by signal recognition particle (SRP) and inserted by Sec pathways (9, 43). The subsequent assembly of the pseudopilins into a type IV pilus is facilitated by a dedicated pilin assembly pathway. Interestingly, recent studies by Saller et al. have implicated the YidC homologue YqjG of B. subtilis in the biogenesis of type IV pili required for DNA uptake (131), as yqjG is critical in the development of genetic competence. This suggests that YidC, a well-conserved membrane protein insertase and assembly catalyst, could be involved in the membrane insertion and/or assembly of type IV pilus subunits.
The SPIV enzymes are aspartic acid proteases, like SPII (84), belonging to the GXGD type of intramembrane-cleaving proteases. Consistent with this view, they are sensitive to the specific aspartic acid protease inhibitor combination 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride plus glycinamide (84). The SPIV enzymes span the membrane eight times (140) and have their active site Asp residues positioned close to the membrane boundaries, within short cytoplasmic loops (Fig. 1). In contrast to other intramembrane-cleaving proteases of the GXGD family, which cleave within the membrane, SPIV cleaves its substrates within the cytoplasm, just proximal to the membrane surface (140). This is consistent with the location of the SPIV recognition motif between the N and H regions of the signal peptide (Fig. 2) (149).
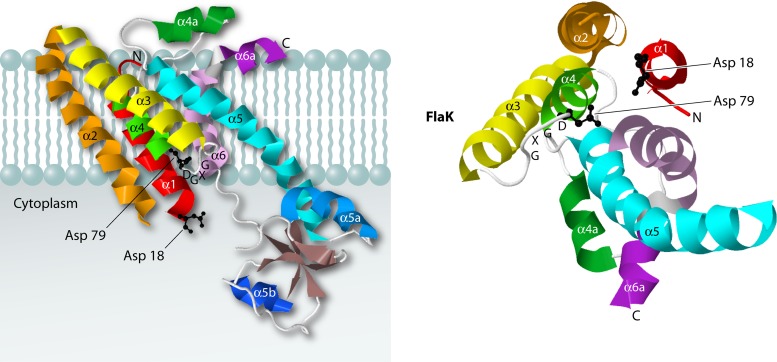
Recently, the crystal structure of the preflagellin signal peptidase FlaK from the archaeon Methanococcus maripaludis was solved at 3.6-Å resolution (Fig. 3) (59). FlaK is a GXGD-type membrane protease with six transmembrane segments. It is related to SPIV as well as to the intramembrane-cleaving proteases presenilin and signal peptide peptidase (SPP) (see the following sections). The structure revealed that FlaK is composed of a mostly α-helical membrane-embedded domain and a soluble cytoplasmic domain with four antiparallel β-strands (Fig. 3). The catalytic Asp residues of FlaK, like those of SPIV, are located near the ends of transmembrane segments (i.e., transmembrane segments 1 and 4) that face the cytoplasmic side of the membrane. The particular arrangement of transmembrane helices 1 and 4 with transmembrane helix 6 around the catalytic site is conserved in FlaK and presenilin. Intriguingly, it seems that conformational changes are needed to bring the GXGD motif and the catalytic Asp residue in the first membrane-spanning helix together for catalysis. In the crystal structure, the catalytic Asp18 and Asp79 residues lie 12 Å apart, which suggests that the structure represents an inactive conformation of FlaK. Indeed, the results of cross-linking studies support the view that FlaK can adopt an inactive conformation in the absence of a substrate (59). It is presently not known how the conformational switch to an active state is brought about upon substrate binding. The nonactive conformation of FlaK in the absence of substrate may be advantageous, because it would ensure that proteolysis does not occur until it is needed for substrate cleavage. This could help to avoid the potentially deleterious cleavage of other proteins.
Fig 3.
Structure of the preflagellin signal peptidase FlaK of Methanococcus maripaludis. The structure on the left shows a side view of FlaK, and the structure on the right represents a view from the cytoplasmic side, with the cytoplasmic domain removed. Each of the six α helices is colored differently. The two catalytic Asp residues are shown as ball-and-stick models, in black. The crystal structure data were obtained using PDB accession number 3S0X, and JMol was used to generate the 3D structure images.
SIGNAL PEPTIDE HYDROLASES DEGRADE SIGNAL PEPTIDES WITHIN OR OUTSIDE THE PLANE OF THE MEMBRANE
After cleavage by signal peptidase, signal peptides are typically further degraded by signal peptide hydrolases, often also referred to as signal peptide peptidases (SPPs). This degradation is important, because signal peptides may be harmful to the cell, as they can interfere with membrane integrity and block protein translocation via the Sec machinery (25, 46, 165). In some cases, the cleaved fragments of signal peptides are released from the membrane and function in signal transduction pathways, both in eukaryotes (87, 101) and in bacteria (6, 41). Notably, multiple enzymes, such as the bacterial RseP and signal peptide peptidase A (SPPA) enzymes, appear to be involved in signal peptide degradation in species ranging from bacteria to humans. This functional redundancy makes it difficult to pinpoint one particular group of enzymes as “the SPP” of a particular organism. Accordingly, only a limited number of endogenous substrates have been identified for particular SPPs. Recently, in-depth proteomic studies by Ravipaty and Reilly identified signal peptide fragments of 18 secreted S. aureus proteins (122). Specifically, these were C-terminal signal peptide portions that were generated through consecutive cleavage by SPI at the signal peptidase cleavage site and by an unidentified protease within the H region. Presumably, one or more SPPs were responsible for the observed cleavage of the H region. Interestingly, the signal peptide fragments were identified in the growth medium, indicating that they were released from the extracytoplasmic side of the membrane. To date, it is not known whether the release of C-terminal signal peptide fragments into the extracellular milieu is specific for S. aureus or whether this process also occurs in other bacteria. Furthermore, the fates of the respective N-terminal signal peptide fragments remain unknown. Notably, the studies by Ravipaty and Reilly also showed that, in certain cases, signal peptide hydrolysis within or at the membrane of S. aureus may not be fully effective, as five full-length signal peptides were also identified in the growth medium (122). These excreted signal peptides belonged to the secreted proteins Sle1, SACOL0723, SceD, IsaA, and SACOL2295. It is presently not clear why these five signal peptides were not further degraded or whether they might serve signaling functions after their excretion.
SPPA
For bacteria, relatively little is known about what happens to the signal peptide after it is cleaved from a preprotein. Almost 30 years ago, it was shown that SPPA (also known as protease IV) can cleave the Braun's major lipoprotein signal peptide in E. coli (61). Like SPI, SPPA is a Ser-Lys dyad protease (76, 180) that spans the membrane once, with a large C-terminal domain localized to the periplasmic space (180). However, it is different from SPI in that SPPA is inhibited by common serine protease inhibitors (63). This suggests that these inhibitors bind with higher affinities to SPPA than to SPI.
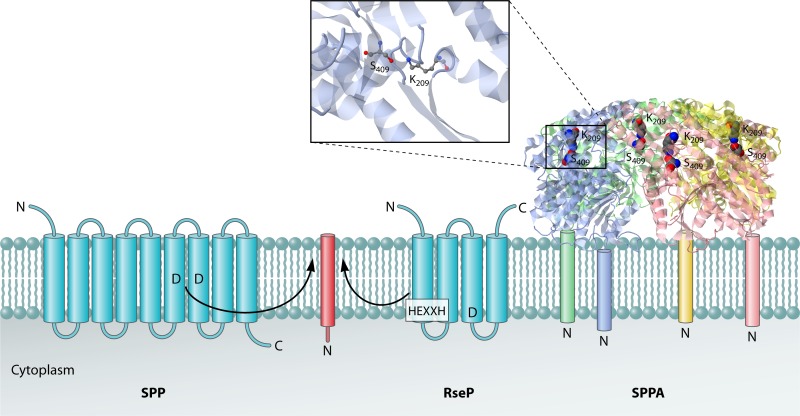
In contrast to the eukaryotic SPP, SPPA does not cleave the signal peptide within the membrane plane (Fig. 4) (76, 180). The 2.8-Å crystal structure of the periplasmic domain of SppA (Δ2-46) revealed that the active site serine and lysine residues are positioned approximately 80 Å from the membrane surface (76), conclusively showing that SPPA can cleave only signal peptides that are either released or extracted from the membrane. The structure of the periplasmic domain of SPPA showed that it forms a tetrameric bowl-like structure, with a large opening at the base facing the membrane and a smaller opening at the top, with diameters of 96 Å and 22 Å, respectively. The bowl opening at the membrane surface is positioned such that it can capture the signal peptide after it is released from the membrane. Once the signal peptide is captured, it makes its way to the active site, where cleavage takes place. Notably, SPPA belongs to the same clan of proteases as the Ser-His-Asp ClpP protease (123). Although the protein folds of SPPA and ClpP are similar, the oligomeric nature of these proteases is different, as ClpP forms a two-stacked 7-fold assembly oriented in a back-to-back fashion, with axial openings at the ends (179). Furthermore, while SPPA is an ATP-independent protease, ClpP associates with an ATPase subunit (either ClpA or ClpX in E. coli) and uses ATP hydrolysis to unfold and translocate proteins into the proteolytic chamber for degradation.
Fig 4.
Cleavage of signal peptides by signal peptide hydrolases. The eukaryotic signal peptide hydrolase/peptidase SPP is an aspartic acid protease with catalytic Asp residues located within the plane of the membrane. The RseP protease, which seems to function as a general bacterial signal peptide hydrolase, is a metalloprotease with an intramembrane catalytic site facing the cytoplasmic side of the membrane. The HEXXH motif that binds the catalytic Zn2+ ion is indicated. The bacterial signal peptide peptidase SPPA is a homotetramer with catalytic Ser-Lys dyads in domains that are juxtapositioned to the extracytoplasmic membrane surface. Transmembrane helices of SPP and RseP are depicted as blue barrels, and the substrate helix is depicted as a red barrel. Furthermore, the four subunits of the SPPA complex are depicted in blue, green, red, and yellow. A zoomed-in view of the active site residues of SPPA is shown. The PDB structure 3BF0 and the program JMol were used to generate the 3D structure image of SPPA. The proposed position of the SPPA N-terminal transmembrane segment for each monomer (which was missing from the construct used to the solve the structure) is shown schematically. The locations of the N and C termini of the signal peptide hydrolases and their substrates are indicated.
RseP
Recently, it was shown that the S2P protease RseP (originally annotated as YaeL [2, 128]) is involved in cleavage of signal peptides (128). RseP is a homologue of human S2P, a protease involved in proteolytic activation of the sterol regulatory element-binding protein (SREBP). S2Ps are the “founding members” of proteases that carry out regulatory intramembrane proteolysis (RIP) (20, 129). RIP is conserved from bacteria to humans and regulates many signal transduction pathways (21). Proteases in these pathways cleave a membrane-spanning regulatory protein and employ active site residues positioned within the transmembrane segments of the proteins.
RseP is a metalloprotease with its active site membrane embedded in a partially exposed aqueous milieu (78) (Fig. 4). RseP spans the membrane four times, with an Nout-Cout topology, and contains an HEXXH zinc-binding motif near the C-terminal end of the first transmembrane segment (68). The essential glutamic acid residue within the HEXXH motif functions either as a general base to activate a water molecule (68) or as a proton shuttle, as found in the enzyme deformylase (see reference 121). Furthermore, RseP has two periplasmic PDZ domains, one of which has a critical role in binding the newly exposed C terminus of its substrate (e.g., RseA) upon site 1 cleavage by proteases such as DegS (34, 64, 91).
Judging by recent observations reported by Saito et al., it seems likely that RseP is a general signal peptide hydrolase in prokaryotes such as E. coli and B. subtilis (128). In addition, RseP also cleaves other substrates, such as RseA. In its role as a signal peptide hydrolase, RseP cleaves the signal peptide after the preprotein is first processed by SPI or SPII. RseP was furthermore shown to cleave a β-lactamase signal peptide fused to the maltose binding protein in E. coli (2) and also the signal peptides of several prelipoproteins in Enterococcus faecalis and S. aureus, to generate sex pheromones (6, 41). An SPP function of the S2P RasP would be consistent with the secretion defects described for a rasP mutant of B. subtilis (54), as signal peptides have a known inhibitory effect on preprotein translocation across the membrane (25, 46, 165). Processing of substrates by the prokaryotic S2P RseP and eukaryotic S2Ps is facilitated by helix destabilization at the site of cleavage (2, 195). In addition, RseP requires a C-terminal hydrophobic amino acid in its substrate to allow cleavage (91). This explains why RseP, like all S2P proteases, can cleave substrates only after they have been cleaved by a separate site 1 protease (5, 69). Once the substrate's C terminus has been liberated, the N-terminal PDZ domain of RseP binds to the C-terminal 3 to 5 residues of the substrate. This then activates the protease, allowing it to cleave the substrate (64). It thus seems that SPI and SPII enzymes fulfill the role of site 1 proteases in delivering cleaved signal peptides as substrates to the S2P protease RseP.
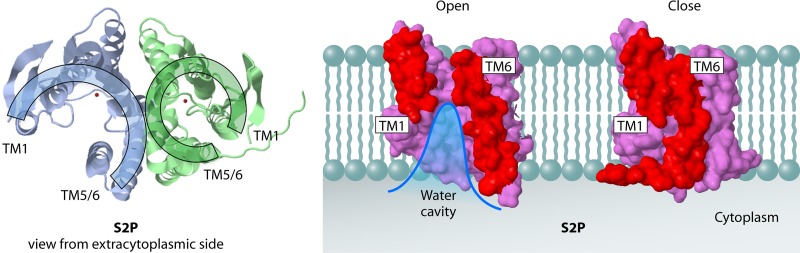
Recently, the structure of the S2P from the archaeon Methanocaldococcus jannaschii was solved at 3.3-Å resolution (40). The archaeal S2P is a metalloprotease with six membrane-spanning helices and an active site within the membrane plane. The active site Zn2+ (Fig. 5), shown in red in the left structure, and Glu55, which activates the nucleophilic water molecule of the M. jannaschii S2P, are localized within the membrane, 14 Å from the cytoplasmic membrane surface. There is also an aqueous channel within the protease, in the plane of the membrane, that is continuous with the cytoplasmic region (Fig. 5). Presently, it is not understood how a substrate accesses the active site region of the M. jannaschii S2P from the lipid bilayer, although it has been hypothesized that this protease contains a lateral gate, comprised of transmembrane helix 1 and transmembrane helices 5 and 6, that can open and close (40) (Fig. 5). This idea was based on the structure of the protease, as two conformations of the protease—the open and closed forms—were found in the crystal (40). The caveats with this hypothesis are that one-third of the protein was deleted and the resulting fragment was crystallized in detergent, not in membrane lipid. It also remains to be determined how substrates access the E. coli S2P RseP, since it spans the membrane only four times and lacks the region that was proposed to be involved in substrate gating in the M. jannaschii S2P.
Fig 5.
Structure of Methanocaldococcus jannaschii S2P. (Left) The top view of S2P (PDB accession number 3B4R) shows the presumed lateral gate formed by transmembrane helix 1 (TM1) on one side and TM5 and TM6 on the other side. S2P with an opened lateral gate is shown in blue, and S2P with a closed lateral gate is shown in green. The catalytic Zn2+ ion is depicted as a red ball. (Based on reference 40.) (Right) Side views of the molecular surface of S2P in the open and closed states. To illustrate the relative distance, TM1 and TM6 are shown in red. The rest of the molecule is shown in purple.
SPP
The biochemically best-characterized SPPs are those of eukaryotes which function in signaling and protein trafficking, as the cleaved signal peptide fragments have downstream functions in these pathways (98). These SPPs are members of a family of aspartic acid proteases that can cleave substrates within the membrane plane (183). They contain two invariant Asp residues that are essential for catalysis (183) and can be inhibited by standard inhibitors against the aspartic acid protease presenilin/γ-secretase (184). Eukaryotic SPPs have nine predicted transmembrane segments, with their N termini located in the ER lumen and their C termini located in the cytoplasm (Fig. 4) (183). The catalytic Asp residues are located within membrane-spanning regions 6 and 7 (Fig. 4) (185). The human SPP was first identified and purified in 2002 (183). Members of the SPP family are found in a wide variety of organisms, including Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, Mus musculus, and Homo sapiens. Importantly, the SPPs are similar to the presenilin family of proteases implicated in Alzheimer's disease, which also catalyze intramembrane proteolysis (117, 183). Both protease groups belong to the aforementioned GXGD-type aspartyl protease family and have nine transmembrane segments (see reference 42).
In addition to cleaving amino-terminal signal peptides, SPP processes the hepatitis C virus (HCV) polyprotein during viral infection by cleaving an internal segment of the viral polypeptide (104). For processing of this internal signal peptide by SPP, prior cleavage of the preprotein by the SPC must occur (88). A helix-breaking residue in the substrate's transmembrane region and flanking sequences can be important for cleavage (88). Interestingly, recent studies by Schrul et al. addressed SPP interactions with signal peptides and other membrane proteins by coimmunoprecipitation (134). This revealed that SPP can interact specifically with a range of signal peptides and newly synthesized preproteins as well as with membrane proteins. Preproteins and misfolded membrane proteins were also shown to interact with SPP without being degraded. Taken together, these results indicate that SPP not only hydrolyzes certain signal peptides but also collects preproteins and misfolded membrane proteins destined for disposal in large oligomeric membrane protein aggregates. Such aggregates may ultimately be degraded in the cytoplasm, by the proteasome, for example.
An unanswered key question is how SPP binds its substrates in the membrane environment. The structure of SPP with or without bound substrate will be needed to determine how this is accomplished. Such structure-function studies will also elucidate where the hydrolytic water molecule originates. Whether SPPs function primarily in signal peptide hydrolysis or membrane protein quality control is unclear.
MEMBRANE PROTEASES THAT DEGRADE MISFOLDED AND MISASSEMBLED PROTEIN SUBSTRATES
Proteins embedded in the bacterial cytoplasmic membrane serve a plethora of essential functions, not only in protein transport processes but also in the uptake of ions and nutrients, the excretion of waste products, and cell signaling. It is therefore of crucial importance that the cell is able to control the quality of essential membrane proteins and to remove mistranslated, damaged, misfolded, or aggregated membrane proteins (1, 57).
The AAA Protease FtsH
FtsH (also called HflB) is a key protease involved in the quality control of membrane protein folding in bacteria. It is a widely conserved protein found in bacteria, mitochondria, and chloroplasts (65). Expression of FtsH is induced at high temperature, indicating that it is a heat shock protein. FtsH is an essential protein for E. coli and functions to degrade misfolded membrane protein substrates or damaged membrane proteins, as well as misassembled membrane protein subunits (65). For example, FtsH degrades the SecY subunit of the SecYEG protein-conducting channel when it is overexpressed in the absence of SecE (75), and it also degrades SecY when the SecYEG channel is jammed by a LacZ fusion protein (174). Similar results were obtained when subunit a of the Fo sector of ATP synthase was overexpressed in the absence of subunits b and c, as this led to the degradation of subunit a by FtsH (3).
FtsH is a zinc metalloprotease with two N-terminal transmembrane segments in the inner membrane followed by the widely conserved AAA (ATPase associated with diverse cellular activities) domain and a protease domain, both localized in the cytoplasm. Unlike the proteases described so far, FtsH extracts its membrane protein from the membrane and then degrades the protein. It uses ATP hydrolysis by the AAA domain to accomplish this dislocation task. FtsH degrades the membrane protein substrate in a processive manner by using its ATPase activity to also unfold the substrate, allowing it to enter the protease chamber for degradation. Notably, degradation of the membrane protein polypeptide by FtsH cannot occur if it contains a domain that is tightly folded, because FtsH does not possess a strong unfolding activity (56). This is consistent with the role of FtsH in the quality control of membrane proteins, where it degrades loosely or improperly folded proteins (74).
The activity of FtsH can be regulated by the prohibitin homologues HflK and HflC (72). HflK and HflC are single-pass inner membrane proteins that can form a complex with FtsH and strongly inhibit FtsH activity for degradation of membrane protein substrates but not soluble substrates (72). HflKC may inhibit degradation of membrane substrates simply by blocking their entry into the proteolytic complex (73). In addition, it has been proposed that prohibitins may function as membrane chaperones and stabilize proteins, based on studies in mitochondria (108). Interestingly, FtsH, HflK, and HflC are copurified as a large complex with YidC and other proteins (164). YidC functions as a chaperone involved in the insertion and folding of membrane proteins (29, 82). Accordingly, it was proposed that YidC and FtsH-HflKC function early in the biosynthesis of nascent membrane proteins and participate in a quality control process. In this process, YidC facilitates the folding of newly inserted membrane proteins and FtsH degrades membrane proteins if they are not properly integrated and folded. Determining the precise roles of YidC and FtsH-HflKC in the complex will require further investigation.
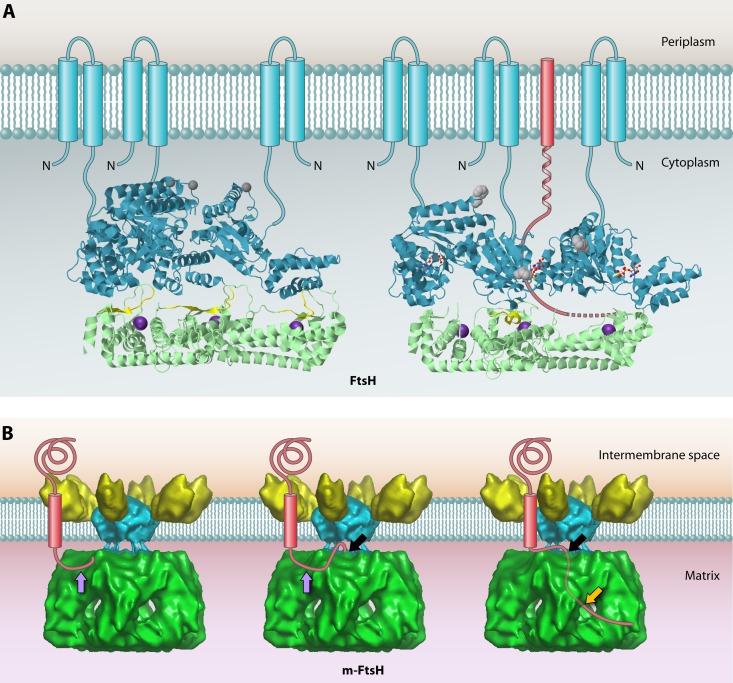
Recently, the structure of the cytoplasmic domain of FtsH, including the AAA ATPase domain and the protease domain, was solved (143) (Fig. 6A). The protease domain has a 6-fold symmetry, whereas the six AAA domains—each with bound ADP—alternate in an open and a closed form (Fig. 6A; note that only 3 subunits forming half of the holoenzyme are shown). The catalytic metal ion (indicated in purple in the figure) in the protease domain is coordinated by His422, His418, and Glu496. Interestingly, when the AAA domain is in the open form, there is a channel from the exterior region via the adjacent closed AAA subunit that may allow substrate polypeptides to enter the protease active site region. A model of how the ATPase cycle is used to allow FtsH to perform processive degradation of protein substrates is presented in Fig. 6A (structure on the right).
Fig 6.
Structures of bacterial FtsH and the homologous m-AAA protease in the inner mitochondrial membrane. (A) Structures of the cytoplasmic domains of apo-FtsH (left) and ADP-FtsH (right). The AAA domains are shown in cyan, and protease domains are shown in green. To give a clear inside view of the hexamer chamber, only 3 subunits, forming half of the holoenzyme, are shown. Amino acids 450 to 460 are proposed to form an active site switch (highlighted in yellow). These residues change from a β-sheet conformation in the apoprotein to an α-helical conformation in the ADP-bound form, which closes the proteolytic site of the corresponding subunit. The inward movement of this AAA domain opens the substrate tunnel, and the substrate polypeptide chain (red) is pulled through the chamber toward the open proteolytic site of the adjacent subunit. Phe234 at the substrate binding pore is shown in gray. The proposed positions of the N-terminal transmembrane segments of each monomer (which were missing from the construct used to the solve the structure) are shown schematically. Bound ADP is shown as a ball-and-stick model. The Zn2+ ion at the proteolytic site is shown as an enlarged purple sphere. The apo-FtsH structure was obtained from PDB accession number 3KDS, and that of ADP-FtsH was obtained from PDB accession number 2CEA. JMol was used to generate the 3D structure images. (B) Proposed mode of action of the mitochondrial m-AAA protease based on the cryo-electron microscopy (cryo-EM) structure. The structure data were obtained from EMDataBank (accession number 1712), and the Astex Viewer was used to generate the image. From left to right, the model depicts subsequent stages in substrate degradation. First, an unfolded terminal peptide of the substrate binds the surface of the AAA domain at the initial contact site (purple arrow). Subsequently, the unfolded peptide is transferred to the secondary binding site (black arrow), where it enters the AAA ring through the center pore. Lastly, the substrate is degraded in the chamber of the protease domain, and the resulting degradation products are released from the side pores (orange arrow).
Similar to bacterial FtsH, the homologous AAA proteases in mitochondria play vital quality control roles and are critical for mitochondrial biogenesis (79). In fact, yeast mitochondria contain two inner membrane AAA proteases: one with the protease domain located in the matrix compartment (i.e., the m-AAA protease) and the other with the protease domain in the intramembrane space (i.e., the i-AAA protease). Both proteases degrade proteins that are not assembled properly in the inner membrane (125, 144). Since the mitochondrial matrix is equivalent to the bacterial cytoplasm, the mitochondrial m-AAA protease is the functional equivalent of the bacterial FtsH protease. Interestingly, a 12-Å-resolution structure has been determined for the intact hetero-oligomeric yeast m-AAA protease that is composed of the homologous Yta10 and Yta12 subunits and includes the transmembrane domain (86) (Fig. 6B). This structure revealed that the catalytic matrix domain and the transmembrane domain are separated by 13 Å, which provides enough space for the protease to bind an unfolded polypeptide in this region (Fig. 6B; a black arrow indicates where a substrate enters the AAA ring). The authors hypothesized that upon initial contact with the surface of the AAA domain, unfolded polypeptides are passed through the AAA chamber in an ATP-dependent manner and then transferred into the protease chamber, where they are degraded. Intriguingly, the distance between the proposed substrate binding site of the m-AAA protease and the central pore of the AAA domain matches the length of an unfolded peptide of 20 residues. This explains why substrates of the m-AAA protease and FtsH need to have a presumably unfolded tail of at least 20 residues in order to initiate the processive degradation of protein substrates (27, 90).
The role of FtsH and the m-AAA protease in dislocation of membrane proteins from the bacterial membrane is reminiscent of the degradation of membrane proteins in the ER, where the AAA ATPase p97 plays a key role (106). In the case of the eukaryotic system, however, the extracted proteins are degraded by the cytosolic proteasome (80, 158).
HtpX
The heat shock-inducible protease HtpX is also implicated in the quality control of membrane protein folding and assembly (130). HtpX is a metalloprotease, like FtsH, and has two amino-terminal transmembrane segments, with the protease domain facing the cytoplasm (136). However, unlike FtsH, the HtpX protein lacks an ATPase domain. Interestingly, in an ftsH knockout strain containing a suppressor mutation in the fabZ gene (110), the heat shock protease HtpX is essential for growth. This suggests that HtpX and FtsH have one or more overlapping functions (136). Indeed, it was shown that in the absence of FtsH, HtpX can cleave SecY when SecY is overexpressed (130). To date, unfortunately, there are no structures available for HtpX.
The Rhomboid Protease GlpG
As observed for the S2Ps, the “founding members” of the intramembrane proteases (124), the rhomboid proteases cleave their substrates within a membrane environment by utilizing catalytic residues in an aqueous channel within the membrane plane (Fig. 7). Rhomboid proteases have been implicated in many mitochondrial functions, growth factor signaling, and the activation of the twin-arginine translocase (Tat) apparatus in some Gram-negative bacteria (44). The E. coli rhomboid protease GlpG is encoded by the glpEGR operon (194), where glpE encodes a sulfur transferase and glpR encodes a repressor regulating genes of the glp regulon under glycerol deprivation conditions. However, the chromosomal context of glpG is different in various bacteria. Therefore, the chromosomal context probably does not indicate a possible physiological function of GlpG or point to potential substrates of GlpG in bacteria. In E. coli, GlpG has been shown to cleave the LacY and MdfA proteins (10, 39, 99), suggesting that this rhomboid protease may play a quality control role by degrading misfolded or improperly assembled membrane proteins. In the case of the eukaryotic rhomboid proteases, known substrates include Spitz, Gurken, and Keren, which are membrane-bound precursor forms of ligands that bind to the epidermal growth factor receptor after being released due to cleavage (162, 163). The substrate specificity of the rhomboid proteases is conserved, as E. coli GlpG can also cleave the heterologous Spitz and Gurken proteins. GlpG has a preference for a small side chain at the P1 position for substrate cleavage, while a helix-destabilizing residue is important in the hydrophobic region downstream of the cleavage site (4). More recently, a universal consensus sequence was determined for the rhomboid protease substrates, in which residues at P1, P4, and P2′ are important for substrate binding and cleavage (139).
Fig 7.
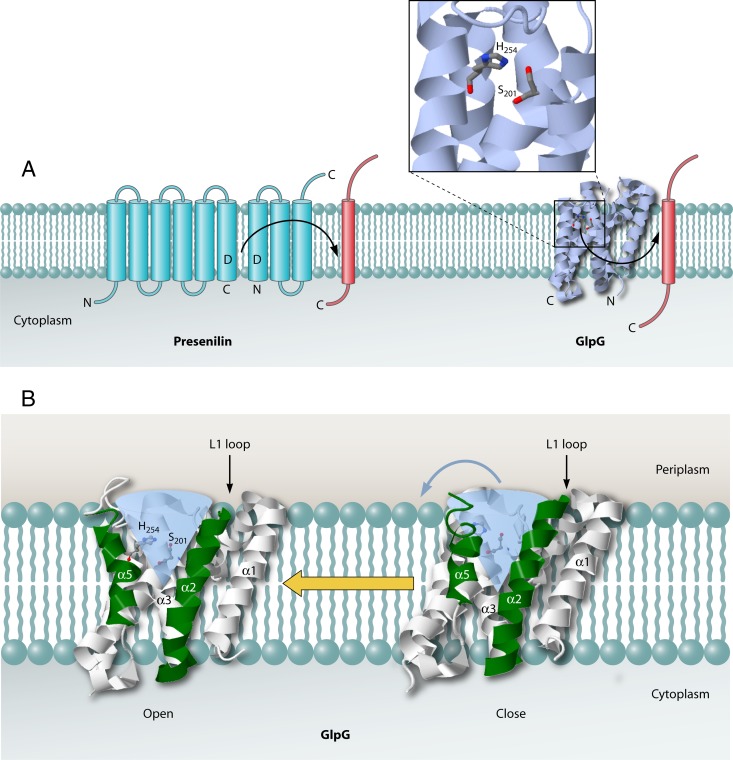
Secondary structure of presenilin and 3D structure of GlpG. (A) The aspartic acid protease presenilin is synthesized as a membrane protein with nine transmembrane helices. It is cleaved upon activation, which results in an N-terminal moiety with six transmembrane regions and a C-terminal moiety with three transmembrane regions. Each of these moieties contains one catalytic aspartic acid residue. GlpG (PDB accession number 2IC8) employs active site Ser and His residues in intramembrane proteolysis. Transmembrane helices are depicted as blue barrels for presenilin and as a 3D ribbon structure for GlpG. Substrate helices are depicted as red barrels. The locations of the N and C termini of presenilin, GlpG, and their substrates are indicated. (B) Side views of the rhomboid protease GlpG (PDB accession number 2NRF) show the lateral gate formed by transmembrane helices TM2 (α2) and TM5 (α5). α2 and α5 are highlighted in green. The water channel is marked as a transparent blue cone. JMol was used to generate the 3D structure images.
Major breakthroughs in understanding how intramembrane-cleaving proteases work were achieved in 2007 with the elucidation of the structure of GlpG by X-ray crystallography (Fig. 7) (14, 89, 182, 193). This first structure of an intramembrane-cleaving protease solved a big problem in cell biology, because up to this point, it was not clear how these proteases bury their active site residues within the membrane and cleave their substrates in an environment that excludes water. Also, the GlpG structure provided clues to how substrates integrated into the lipid bilayer could gain access to an intramembrane protease.
The GlpG crystal structure reveals an aqueous channel below the membrane surface that is continuous with the active site (Fig. 7, bottom panel; the water channel is shown as a blue cone). The active site contains catalytic Ser-His residues that function as the nucleophile and general base, respectively (Fig. 7, top panel). Several models have been proposed for substrate passage from the lipid phase into the active site. The first model proposes that there is a lateral gate comprised of helices 2 and 5 (Fig. 7, bottom panel) (10, 14, 161, 181). Movement of transmembrane helix 5 away from the protease core would allow substrate access to the catalytic serine residue within transmembrane helix 4 (193). Evidence for this model comes from studies which showed that mutations within transmembrane helix 5 cause displacement of this helix from the core region and activate the protease (10). In addition, the positions of transmembrane 5 segments within the different X-ray structures of the protein are quite variable (14, 93). The second model proposes that access of the substrate to the active site is controlled by transmembrane helices 1 and 3 and by an L1 loop region (182). Movement of the L1 loop from the periplasmic region would allow access of a substrate's transmembrane region to the active site helices 2 and 5, in which the periplasmic region is plugged by the L1 loop. A drawback of this model is that the L1 loop seems not optimally positioned for a gating function at helices 2 and 5. Although a lateral gating model for rhomboid proteases appears attractive, it should be noted that there is currently no direct biochemical evidence for this mechanism in these enzymes.
SHEDDASES
The cleavage of membrane proteins by site 1 proteases is also known as ectodomain shedding, because this process results in the release of the membrane protein's ectodomain into the extracellular milieu. Accordingly, site 1 proteases are also referred to as sheddases. The bacterial sheddases typically cleave their substrates on the extracytoplasmic side of the lipid bilayer, at a juxtamembrane position. As already mentioned above, site 1 proteolysis precedes further intramembrane cleavage of transmembrane segments by site 2 proteases such as S2P. Substrate cleavage by sheddases is often regulated such that it occurs only under special conditions, for example, upon heat stress, membrane perturbation by antimicrobial peptides, or protein secretion stress.
DegS
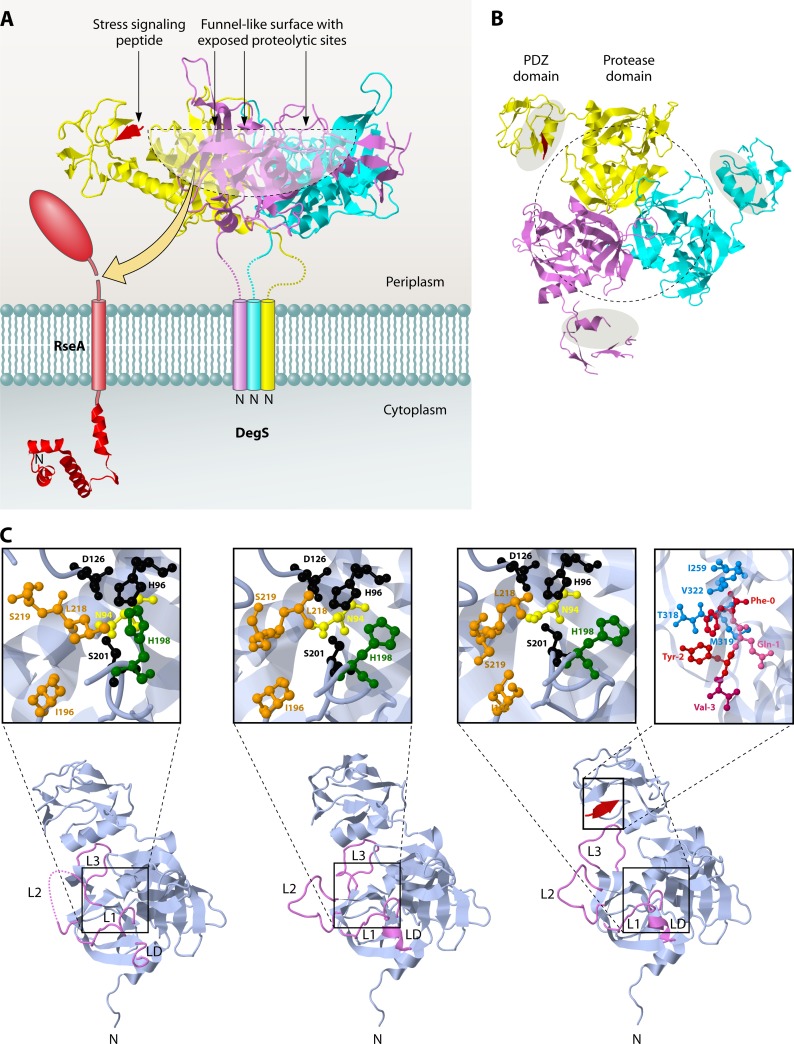
The currently best-studied bacterial sheddase is DegS of E. coli. This protease belongs to the HtrA (high temperature requirement A) class of serine proteases, which have general roles in gene regulation and protein quality control in extracytoplasmic compartments (28, 81). Unlike its paralogues DegP and DegQ, which are released into the periplasm of E. coli upon signal peptide cleavage, DegS remains attached to the inner membrane through an N-terminal membrane anchor (Fig. 8A) (94, 178). DegS has been shown to catalyze the initial (site 1) cleavage in the anti-sigma factor RseA that sequesters sigma E (Fig. 8A) (5, 69). Biochemical and structural analyses have shown that DegS is functional as a trimer in which each subunit contains a protease domain and a PDZ domain (Fig. 8A) (47, 189, 196). With its PDZ domain, DegS can sense the presence of C-terminal peptides from mislocalized or misfolded outer membrane proteins, resulting in the allosteric activation of the protease domain and the stabilization of the active protease (50, 51). In fact, small hydrophobic tripeptides are sufficient to activate the DegS protease (52). Recent structural studies indicate that the binding of a C-terminal peptide from an outer membrane protein to the PDZ domain relieves inhibitory contacts between the PDZ and protease domains, thereby activating the protease domain (138). Interestingly, this allosteric activation seems to be an intrinsic property of the protease domain, as protein substrates must bind tightly and specifically to this domain to facilitate their own degradation (51, 137).
Fig 8.
Trimeric structure of the sheddase DegS. (A) Side view of DegS. (B) View from the periplasmic side. The protease domains in the center surround a funnel-like exposed surface where the proteolytic sites are located (outlined by a black dotted line). The PDZ domains that contain the peptide-binding grooves (gray shaded areas in panel B) are located on the outside of the DegS trimer. They are connected to the protease domains at their C termini. A peptide that is bound to the PDZ domain in one of the subunits is colored red. The structure data were obtained from PDB accession number 1SOZ, and JMol was used to generate the images. (C) Monomer structures of DegS in the peptide-free inactive (left), intermediate (middle), and peptide-bound active (right) states. The L1, L2, L3, and LD loops are highlighted in violet. In the intermediate form, without a substrate, L3 retreats from the substrate docking position (active form) to the inactive conformation, while L1, L2, and LD, located at the active site center, still adopt the active conformation. The active sites are shown in enlarged insets for all structures. The catalytic residues Ser201, His96, and Asp126 are shown in black. Residues forming the S1 pocket are shown in orange. These residues are reorganized upon substrate binding (compare the inactive and active forms). N94 (yellow) and H198 (green), which interfere with the catalytic triad and destabilize the oxyanion hole, as shown in the inactive form, move away in the active form. The stress signaling peptide binding site in the PDZ domain for DegS in the proteolytic active state is also shown in an enlarged inset. The peptide is colored red, and residues forming the hydrophobic pocket surrounding the Phe0 position are colored blue. Structure data were obtained from PDB, using accession numbers 1SOT (inactive state), 1VCW (intermediate state), and 1SOZ (active state). JMol was used to construct the images.
The structure of DegS has been solved in various states, including peptide-free and intermediate states that are inactive (Fig. 8C, bottom left and central structures) and a peptide-bound state that is active (Fig. 8C, bottom right structure). In the peptide-free and intermediate states, the Asn and His residues (shown in yellow and green, respectively) interfere with the catalytic Ser-His-Asp residues (shown in black). Strikingly, the structure of DegS with an activating peptide bound to the PDZ domain (Fig. 8C, bottom right structure, with the peptide bound to the PDZ domain shown in red) results in movement of the Asn94 (shown in yellow in the inset) and His198 (shown in green in the inset) residues from the Ser-His-Asp catalytic residues, leading to an active proteolytic machinery. This explains the activation of this protease by binding to the C-terminal peptide of a misfolded protein.
Homologues of E. coli DegS are present in many bacteria, but their roles in RIP have not yet been studied in much detail. For Salmonella enterica serovar Typhimurium, a causative agent of salmonellosis, DegS was shown to be required for survival in murine macrophages and a degS mutant was highly attenuated in mouse infection models (127). In the enteric pathogen Vibrio cholerae, the causative agent of cholera, DegS is implicated in sigma E-mediated resistance to membrane-acting, host gut-derived antimicrobial peptides (103). In B. subtilis, the DegS homologue proteases HtrA and HtrB are both membrane bound by an N-terminal membrane anchor like E. coli DegS (7, 19, 62, 198, 199). Both enzymes have been implicated in the cellular response to the stresses caused by high-level production of secretory proteins or heat, but whether this involves a sheddase activity of HtrA or HtrB is not known (30, 62, 187). Interestingly, the cellular levels of HtrA and HtrB seem to be controlled by RasP, the RseP homologue of B. subtilis, suggesting that they are subject to intramembrane cleavage (19, 199). This would explain why fragments of HtrA and HtrB are detectable in the growth medium of B. subtilis.
PrsW
In B. subtilis, regulated intramembrane proteolysis of the RsiW anti-sigma factor is controlled by the sheddase PrsW (38, 53, 55). The membrane protein RsiW sequesters the sigma factor SigW, which is needed for the cellular responses to cell envelope damage caused by antimicrobial peptides and alkaline shock. Upon initial cleavage of RsiW by PrsW and subsequent cleavage by the site 2 protease RasP, SigW is released into the cytoplasm, resulting in the activation of about 60 SigW-controlled genes. PrsW is predicted to have five transmembrane domains (167), and it belongs to a family of membrane proteases with representatives in Gram-positive and Gram-negative bacteria as well as in archaea and eukaryotes (38). One of the few characterized proteases of this family is the CaaX protease Rce1p of S. cerevisiae, which removes the C-terminal AAX residues from a prenylated protein following the initial prenylation of the cysteine residue in the CAAX motif (18, 33, 115, 133). To date, neither the catalytic mechanism of PrsW nor that of Rce1p has been investigated, but based on sequence similarities, it has been proposed that these enzymes may be metalloproteases (18, 33, 38, 53, 55, 115, 133).
EUKARYOTIC PROTEASES THAT CATALYZE INTRA- AND JUXTAMEMBRANE PROTEOLYSIS
For a broader understanding of all mechanisms of proteolysis that potentially occur within the bacterial cytoplasmic membrane or at its surface, it is instructive to compare the mechanisms of substrate cleavage by signal peptidases and signal peptide hydrolases with those of the eukaryotic presenilin protease and sheddases, all of which also cleave within the membrane or at the membrane boundaries. Please refer to references 24 and 45 for detailed recent reviews and discussions on the eukaryotic S2P and rhomboid proteases.
Presenilin Protease
The presenilins function in Notch signaling and cell adhesion and have also been implicated in Alzheimer's disease. These proteases belong to the same family of aspartic acid proteases as the eukaryotic SPPs (135, 191). β-Secretase and presenilin 1, the catalytic component of the multisubunit enzyme γ-secretase, cleave the β-amyloid precursor protein to excise Aβ peptides of various lengths that can form neuritic plaque. Similar to the SPP proteases, presenilin 1 cleaves within the transmembrane region of substrates such as the β-amyloid precursor protein. Presenilins are synthesized with nine transmembrane segments (85), but unlike SPP family members, they undergo cleavage during activation that results in an N-terminal fragment with six transmembrane segments and a C-terminal fragment with three transmembrane segments (Fig. 7, top panel). Notably, the transmembrane domains containing the catalytic Asp residues in presenilin have the opposite orientation to the transmembrane domains containing the catalytic Asp residues in SPP (Fig. 4) (109, 183). The different orientations correlate with the opposite orientations of the transmembrane substrates of these proteases for cleavage. The SPP protease cleaves single-pass membrane substrates with a type II orientation (i.e., with the N terminus in the cytoplasm), while presenilin cleaves substrates with type I membrane topology (i.e., with the C terminus in the cytoplasm) (100). Another difference is that presenilin is not active unless the other subunits, i.e., nicastrin, Aph-1, and Pen-2, are present, while SPP appears to be active by itself (77, 183). The Pen-2 subunit within γ-secretase is required to stabilize the presenilin fragment heterodimer for presenilin cleavage (119). Nicastrin is important for both the stability and trafficking of the other components of the presenilin 1–γ-secretase complex (197). In terms of activity, mutations of the invariant Asp residues within presenilin 1 completely eliminate γ-secretase activity in eukaryotic cells, supporting the view that presenilin 1 is the catalytic component. Although many substrates have a valine at the P1′ position, which can be important for processing, presenilin is quite tolerant to mutation of cleavage site residues (11).
Sheddases
Eukaryotic sheddases typically cleave their substrates at the juxtamembrane region outside the lipid bilayer. These enzymes function in the liberation (shedding) of ectodomains from membrane proteins. By analogy, signal peptidases of the SPI type could thus be regarded as a special class of sheddases dedicated to protein secretion. Substrate cleavage by sheddases is often regulated such that it occurs only at certain times or at particular locations in the cell. The eukaryotic sheddases, which are extramembrane proteases tethered to the membrane, use active site residues found in the “standard” proteases, such as the aspartic acid proteases or metalloproteases.
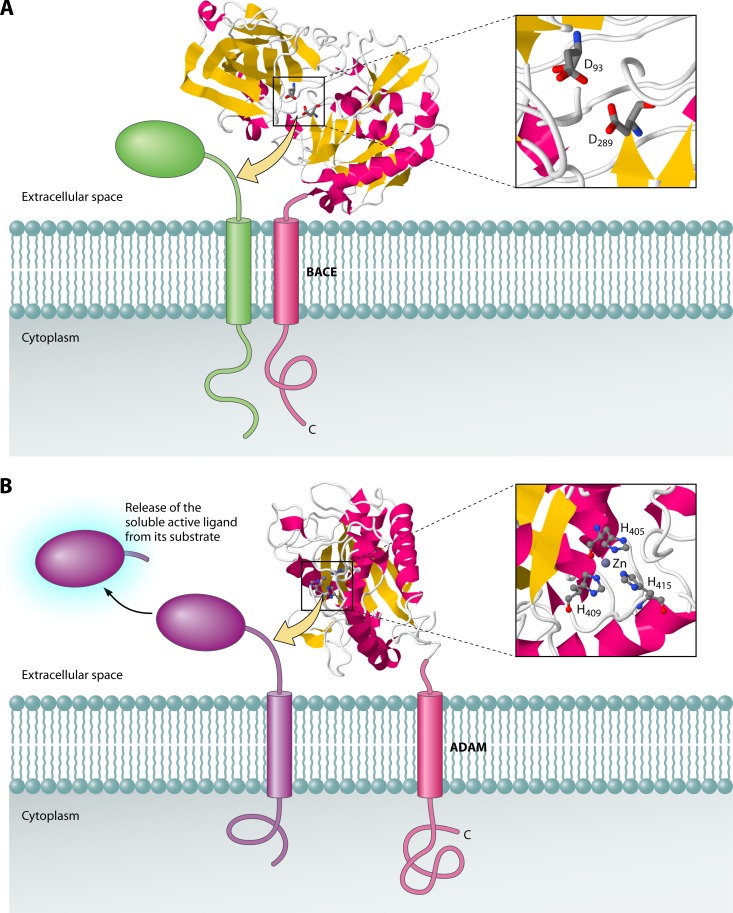
BACE.
The BACE (β-secretase) family of proteases (48) forms a prominent class of sheddases. BACE proteases are membrane-anchored aspartic acid proteases that have been implicated in Alzheimer's disease (176). BACE1 cleaves the amyloid protein precursor 27 residues from the membrane surface (157) and is also involved in shedding of the type III neuregulin 1 protein that is important in nerve myelination (60, 190). BACE is composed of one transmembrane domain and a protease domain facing the extracellular space (Fig. 9A). The structure of the protease domain of human BACE has been solved at 2.0-Å resolution (58). While the exact positioning of the protease domain with respect to the membrane is not known, it is believed that BACE can cleave substrates directly at the extracellular membrane surface, where the substrate cleavage site is accessible.
Fig 9.
The sheddases BACE and ADAM17 cleave substrates at the extracellular membrane surface. Sheddases belonging to the BACE (A) and ADAM (B) families cleave membrane proteins at aqueous juxtamembrane positions. This results in the release of ectodomains from the substrate proteins into the extracellular space. The catalytic domains of both BACE (PDB accession number 1W50) and ADAM (PDB accession number 1BKC) are shown with zoomed-in views of the active site regions. JMol was used to generate the 3D structure images.
ADAM.
Another important class of sheddases is the ADAM (a disintegrin and metalloprotease) group of proteases (192). These sheddases are metalloproteases that cleave a number of membrane-anchored substrates for release into the extracellular space (Fig. 9B). ADAM8 and ADAM10 have been implicated in inflammation, cell adhesion, and neurodegeneration (92). These proteases can cleave CD23, myelin basic protein, and tumor necrosis factor alpha (107, 186). Similarly, ADAM17 (also known as TACE or tumor necrosis factor alpha-converting enzyme) is capable of processing interleukin-1 receptor type II, Kit ligand, L-selectin, transforming growth factor alpha, tumor necrosis factor alpha, and tumor necrosis factor receptors 1 and 2 (107). The structure of the protease domain of ADAM17 has been solved by X-ray crystallography (102). Exactly how ADAM proteases cleave at the membrane surface is currently not understood. One possibility is that these enzymes have a preference for cleavage in their substrates' unstructured regions proximal to the membrane surface.
CONCLUSIONS
After decades of research on membrane proteases in the bacterial protein secretion/quality control area, it has become very clear that no single mechanism is used to cleave substrates at the intra- and juxtamembrane positions. SPI cleaves a preprotein substrate during or after membrane translocation, and its catalytic domain penetrates the lipid phase of the membrane. This allows it to bind the substrate and then cleave it in a water-exposed environment at the extracytoplasmic surface of the membrane. Signal peptide degradation by SPP or RseP is thought to occur within the plane of the membrane. In the cases where the structures of intramembrane proteases are known, it is clear that the active sites employ standard catalytic residues that are embedded within the membrane and contain an aqueous channel where the hydrolytic reaction occurs. In contrast, the membrane protease FtsH extracts its substrates from the membrane and degrades them in the aqueous milieu of the cytoplasm. Similarly, SPPA and DegS degrade their substrates in the aqueous extracytoplasmic milieu. In all cases, whether proteolysis occurs outside or within the membrane, the active sites of the proteases use standard catalytic residues and, where needed, aqueous channels.
While much progress has been made in understanding bacterial proteases that cleave within the cytoplasmic membrane and at the membrane surface, there are still many questions that remain to be investigated. How substrates precisely access the active sites of intramembrane proteases is not understood. Also, it is still somewhat controversial whether there is a lateral gating mechanism that controls access of the substrate to the active site of intramembrane proteases from the lipid bilayer. What are the structural features of the substrate that allow it to be cleaved by one specific intramembrane protease but not by another protease? What are the characteristics of membrane substrates that allow for cleavage only at the proximal surface, such as by signal peptidases of the SPI and SPII types? To answer such questions, it will be necessary to perform further in-depth biochemical studies and to solve the structures of membrane proteases with their bound substrates. Lastly, studies in B. subtilis suggest that membrane proteases can set a limit to the overproduction of membrane proteins needed for structural analyses or biomedical and biotechnological applications (19, 199). Given the very important functions of membrane proteases in bacterial growth and cell viability, further research is needed to determine how the bacterial network of membrane proteases can best be modified to achieve maximal production of valuable membrane proteins with minimal detrimental side effects on the overproducing host cells.
ACKNOWLEDGMENTS
This work was supported by a national science grant (MCB-0316670) (to R.E.D.), by EU grants LSHM-CT-2006-019064 and PITN-GA-2008-215524, by the transnational SysMO Initiative (through projects BACELL SysMO1 and -2), and by grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (to J.M.V.D.).
Biographies

Ross E. Dalbey received his B.S. degree in chemistry from the University of Washington (1978) and his Ph.D. in biochemistry from Washington State University (1983), where he studied the muscle protein myosin in the laboratory of Ralph Yount. He then did his postdoctoral research with William Wickner at UCLA, where he studied the function and membrane insertion of leader peptidase. In 1987, he became an Assistant Professor in the Department of Chemistry at The Ohio State University and was promoted to Professor in 1997. His research focuses on how proteins are inserted and assembled into the inner membranes of bacteria. In addition, he has maintained strong and continuing interests in examining proteases such as signal peptidases in the bacterial protein export and secretion pathway.

Peng Wang received his B.S. degree in biochemistry from Nanjing University (2002) and his Ph.D. degree in biochemistry at The Ohio State University (2009), with Ross Dalbey. His graduate research focused on signal peptide peptidase A and the role of YidC, a membrane insertase, in E. coli membrane protein biogenesis. His current postdoctoral work is with Xin Li at The Ohio State Veterinary School Center and focuses on the DPS protein of the Lyme disease agent Borrelia burgdorferi.

Jan Maarten van Dijl is a Professor in the Department of Medical Microbiology of the University of Groningen and the University Medical Center Groningen, the Netherlands. He earned his M.Sc. in biology in 1985 and his Ph.D. in mathematics and natural sciences in 1990, both at the University of Groningen. Subsequently, he was a postdoctoral research fellow in the laboratories of Gerard Venema at the University of Groningen and Gottfried Schatz at the Biozentrum of the University of Basel, Switzerland. In 1998, he was appointed Assistant Professor in Pharmaceutical Biology, and since 2004, he has been a Professor of Molecular Bacteriology. Dr. van Dijl's research is focused on fundamental and applied aspects of protein transport and sorting in Gram-positive bacteria such as Bacillus subtilis and Staphylococcus aureus. His important achievements include the first characterization of type I and type II signal peptidases of Gram-positive bacteria and the first bioinformatic and proteomic descriptions of the secretomes of B. subtilis and S. aureus. Dr. van Dijl is a coordinator and active participant in national and European research programs on bacterial protein secretion, gene function analysis, genome engineering, systems biology, and synthetic biology.
REFERENCES
- 1. Akiyama Y. 2009. Quality control of cytoplasmic membrane proteins in Escherichia coli. J. Biochem. 146:449–454 [DOI] [PubMed] [Google Scholar]
- 2. Akiyama Y, Kanehara K, Ito K. 2004. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 23:4434–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akiyama Y, Kihara A, Ito K. 1996. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 399:26–28 [DOI] [PubMed] [Google Scholar]
- 4. Akiyama Y, Maegawa S. 2007. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol. Microbiol. 64:1028–1037 [DOI] [PubMed] [Google Scholar]
- 5. Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 16:2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. An FY, Sulavik MC, Clewell DB. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antelmann H, et al. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143–156 [DOI] [PubMed] [Google Scholar]
- 8. Antelmann H, et al. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484–1502 [DOI] [PubMed] [Google Scholar]
- 9. Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189:2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker RP, Young K, Feng L, Shi Y, Urban S. 2007. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc. Natl. Acad. Sci. U. S. A. 104:8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beel AJ, Sanders CR. 2008. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 65:1311–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beltzer JP, Wessels HP, Spiess M. 1989. Signal peptidase can cleave inside a polytopic membrane protein. FEBS Lett. 253:93–98 [DOI] [PubMed] [Google Scholar]
- 13. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 14. Ben-Shem A, Fass D, Bibi E. 2007. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc. Natl. Acad. Sci. U. S. A. 104:462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Black MT. 1993. Evidence that the catalytic activity of prokaryote leader peptidase depends upon the operation of a serine-lysine catalytic dyad. J. Bacteriol. 175:4957–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Black MT, Bruton G. 1998. Inhibitors of bacterial signal peptidases. Curr. Pharm. Des. 4:133–154 [PubMed] [Google Scholar]
- 17. Bolhuis A, et al. 1999. Different mechanisms for thermal inactivation of Bacillus subtilis signal peptidase mutants. J. Biol. Chem. 274:15865–15868 [DOI] [PubMed] [Google Scholar]
- 18. Boyartchuk VL, Ashby MN, Rine J. 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275:1796–1800 [DOI] [PubMed] [Google Scholar]
- 19. Bramkamp M, Weston L, Daniel RA, Errington J. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62:580–591 [DOI] [PubMed] [Google Scholar]
- 20. Brown MS, Goldstein JL. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed] [Google Scholar]
- 21. Brown MS, Ye J, Rawson RB, Goldstein JL. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391–398 [DOI] [PubMed] [Google Scholar]
- 22. Carlos JL, et al. 2000. The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J. Biol. Chem. 275:38813–38822 [DOI] [PubMed] [Google Scholar]
- 23. Chaal BK, Mould RM, Barbrook AC, Gray JC, Howe CJ. 1998. Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana. Implications for the origin and catalytic mechanism of the enzyme. J. Biol. Chem. 273:689–692 [DOI] [PubMed] [Google Scholar]
- 24. Chen G, Zhang X. 2010. New insights into S2P signaling cascades: regulation, variation, and conservation. Protein Sci. 19:2015–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen L, Tai PC, Briggs MS, Gierasch LM. 1987. Protein translocation into Escherichia coli membrane vesicles is inhibited by functional synthetic signal peptides. J. Biol. Chem. 262:1427–1429 [PubMed] [Google Scholar]
- 26. Chen X, Van Valkenburgh C, Fang H, Green N. 1999. Signal peptides having standard and nonstandard cleavage sites can be processed by Imp1p of the mitochondrial inner membrane protease. J. Biol. Chem. 274:37750–37754 [DOI] [PubMed] [Google Scholar]
- 27. Chiba S, Akiyama Y, Mori H, Matsuo E, Ito K. 2000. Length recognition at the N-terminal tail for the initiation of FtsH-mediated proteolysis. EMBO Rep. 1:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell. Biol. 12:152–162 [DOI] [PubMed] [Google Scholar]
- 29. Dalbey RE, Kuhn A. 2004. YidC family members are involved in the membrane insertion, lateral integration, folding, and assembly of membrane proteins. J. Cell Biol. 166:769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darmon E, et al. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Bentzmann S, Aurouze M, Ball G, Filloux A. 2006. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188:4851–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dev IK, Ray PH. 1984. Rapid assay and purification of a unique signal peptidase that processes the prolipoprotein from Escherichia coli B. J. Biol. Chem. 259:11114–11120 [PubMed] [Google Scholar]
- 33. Dolence EK, Dolence JM, Poulter CD. 2001. Solid-phase synthesis of a radiolabeled, biotinylated, and farnesylated Ca(1)a(2)X peptide substrate for Ras- and a-mating factor converting enzyme. Bioconjug. Chem. 12:35–43 [DOI] [PubMed] [Google Scholar]
- 34. Dries DR, Yu G. 2009. Rip exposed: how ectodomain shedding regulates the proteolytic processing of transmembrane substrates. Proc. Natl. Acad. Sci. U. S. A. 106:14737–14738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ekici OD, et al. 2007. Altered −3 substrate specificity of Escherichia coli signal peptidase 1 mutants as revealed by screening a combinatorial peptide library. J. Biol. Chem. 282:417–425 [DOI] [PubMed] [Google Scholar]
- 36. Ekici OD, Paetzel M, Dalbey RE. 2008. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 17:2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ekici OD, et al. 2009. Profiling the substrate specificity of viral protease VP4 by a FRET-based peptide library approach. Biochemistry 48:5753–5759 [DOI] [PubMed] [Google Scholar]
- 38. Ellermeier CD, Losick R. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Erez E, Bibi E. 2009. Cleavage of a multispanning membrane protein by an intramembrane serine protease. Biochemistry 48:12314–12322 [DOI] [PubMed] [Google Scholar]
- 40. Feng L, et al. 2007. Structure of a site-2 protease family intramembrane metalloprotease. Science 318:1608–1612 [DOI] [PubMed] [Google Scholar]
- 41. Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803–817 [DOI] [PubMed] [Google Scholar]
- 42. Fluhrer R, Steiner H, Haass C. 2009. Intramembrane proteolysis by signal peptide peptidases: a comparative discussion of GXGD-type aspartyl proteases. J. Biol. Chem. 284:13975–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189:1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freeman M. 2008. Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42:191–210 [DOI] [PubMed] [Google Scholar]
- 45. Freeman M. 2009. Rhomboids: 7 years of a new protease family. Semin. Cell Dev. Biol. 20:231–239 [DOI] [PubMed] [Google Scholar]
- 46. Gouridis G, Karamanou S, Gelis I, Kalodimos CG, Economou A. 2009. Signal peptides are allosteric activators of the protein translocase. Nature 462:363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grininger M, Ravelli RB, Heider U, Zeth K. 2004. Expression, crystallization and crystallographic analysis of DegS, a stress sensor of the bacterial periplasm. Acta Crystallogr. D Biol. Crystallogr. 60:1429–1431 [DOI] [PubMed] [Google Scholar]
- 48. Haniu M, et al. 2000. Characterization of Alzheimer's beta-secretase protein BACE. A pepsin family member with unusual properties. J. Biol. Chem. 275:21099–21106 [DOI] [PubMed] [Google Scholar]
- 49. Harris DA, Powers ME, Romesberg FE. 2009. Synthesis and biological evaluation of penem inhibitors of bacterial signal peptidase. Bioorg. Med. Chem. Lett. 19:3787–3790 [DOI] [PubMed] [Google Scholar]
- 50. Hasenbein S, et al. 2010. Conversion of a regulatory into a degradative protease. J. Mol. Biol. 397:957–966 [DOI] [PubMed] [Google Scholar]
- 51. Hasselblatt H, et al. 2007. Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress. Genes Dev. 21:2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hauske P, et al. 2009. Peptidic small molecule activators of the stress sensor DegS. Mol. Biosyst. 5:980–985 [DOI] [PubMed] [Google Scholar]
- 53. Heinrich J, Hein K, Wiegert T. 2009. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 74:1412–1426 [DOI] [PubMed] [Google Scholar]
- 54. Heinrich J, Lunden T, Kontinen VP, Wiegert T. 2008. The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology 154:1989–1997 [DOI] [PubMed] [Google Scholar]
- 55. Heinrich J, Wiegert T. 2006. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol. Microbiol. 62:566–579 [DOI] [PubMed] [Google Scholar]
- 56. Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. 2003. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11:659–669 [DOI] [PubMed] [Google Scholar]
- 57. Hinz A, Lee S, Jacoby K, Manoil C. 2011. Membrane proteases and aminoglycoside antibiotic resistance. J. Bacteriol. 193:4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hong L, et al. 2000. Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor. Science 290:150–153 [DOI] [PubMed] [Google Scholar]
- 59. Hu J, Xue Y, Lee S, Ha Y. 2011. The crystal structure of GXGD membrane protease FlaK. Nature 475:528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu X, et al. 2006. Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 9:1520–1525 [DOI] [PubMed] [Google Scholar]
- 61. Hussain M, Ichihara S, Mizushima S. 1982. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257:5177–5182 [PubMed] [Google Scholar]
- 62. Hyyrylainen HL, et al. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159–1172 [DOI] [PubMed] [Google Scholar]
- 63. Ichihara S, Beppu N, Mizushima S. 1984. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J. Biol. Chem. 259:9853–9857 [PubMed] [Google Scholar]
- 64. Inaba K, et al. 2008. A pair of circularly permutated PDZ domains control RseP, the S2P family intramembrane protease of Escherichia coli. J. Biol. Chem. 283:35042–35052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ito K, Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211–231 [DOI] [PubMed] [Google Scholar]
- 66. Jongbloed JD, et al. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068–44078 [DOI] [PubMed] [Google Scholar]
- 67. Jongbloed JD, et al. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54:1319–1325 [DOI] [PubMed] [Google Scholar]
- 68. Kanehara K, Akiyama Y, Ito K. 2001. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene 281:71–79 [DOI] [PubMed] [Google Scholar]
- 69. Kanehara K, Ito K, Akiyama Y. 2002. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 16:2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Karatsa-Dodgson M, Wormann ME, Grundling A. 2010. In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids. J. Bacteriol. 192:5341–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Karla A, Lively MO, Paetzel M, Dalbey R. 2005. The identification of residues that control signal peptidase cleavage fidelity and substrate specificity. J. Biol. Chem. 280:6731–6741 [DOI] [PubMed] [Google Scholar]
- 72. Kihara A, Akiyama Y, Ito K. 1996. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 15:6122–6131 [PMC free article] [PubMed] [Google Scholar]
- 73. Kihara A, Akiyama Y, Ito K. 1998. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol. 279:175–188 [DOI] [PubMed] [Google Scholar]
- 74. Kihara A, Akiyama Y, Ito K. 1999. Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 18:2970–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kihara A, Akiyama Y, Ito K. 1995. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. U. S. A. 92:4532–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim AC, Oliver DC, Paetzel M. 2008. Crystal structure of a bacterial signal peptide peptidase. J. Mol. Biol. 376:352–366 [DOI] [PubMed] [Google Scholar]
- 77. Kimberly WT, et al. 2003. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl. Acad. Sci. U. S. A. 100:6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Koide K, Maegawa S, Ito K, Akiyama Y. 2007. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J. Biol. Chem. 282:4553–4560 [DOI] [PubMed] [Google Scholar]
- 79. Koppen M, Langer T. 2007. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 42:221–242 [DOI] [PubMed] [Google Scholar]
- 80. Kostova Z, Wolf DH. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Krojer T, Sawa J, Huber R, Clausen T. 2010. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat. Struct. Mol. Biol. 17:844–852 [DOI] [PubMed] [Google Scholar]
- 82. Kuhn A, Stuart R, Henry R, Dalbey RE. 2003. The Alb3/OxaI/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13:510–516 [DOI] [PubMed] [Google Scholar]
- 83. Kulanthaivel P, et al. 2004. Novel lipoglycopeptides as inhibitors of bacterial signal peptidase I. J. Biol. Chem. 279:36250–36258 [DOI] [PubMed] [Google Scholar]
- 84. LaPointe CF, Taylor RK. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502–1510 [DOI] [PubMed] [Google Scholar]
- 85. Laudon H, et al. 2005. A nine-transmembrane domain topology for presenilin 1. J. Biol. Chem. 280:35352–35360 [DOI] [PubMed] [Google Scholar]
- 86. Lee S, et al. 2011. Electron cryomicroscopy structure of a membrane-anchored mitochondrial AAA protease. J. Biol. Chem. 286:4404–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. 2001. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167:6441–6446 [DOI] [PubMed] [Google Scholar]
- 88. Lemberg MK, Martoglio B. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735–744 [DOI] [PubMed] [Google Scholar]
- 89. Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. 2007. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc. Natl. Acad. Sci. U. S. A. 104:750–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leonhard K, et al. 2000. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol. Cell 5:629–638 [DOI] [PubMed] [Google Scholar]
- 91. Li X, et al. 2009. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. U. S. A. 106:14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lichtenthaler SF, Steiner H. 2007. Sheddases and intramembrane-cleaving proteases: RIPpers of the membrane. Symposium on regulated intramembrane proteolysis. EMBO Rep. 8:537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lieberman RL, Wolfe MS. 2007. Membrane-embedded protease poses for photoshoot. Proc. Natl. Acad. Sci. U. S. A. 104:401–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lipinska B, Zylicz M, Georgopoulos C. 1990. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 172:1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lory S. 1994. Leader peptidases of type IV prepilins and related proteins, p 31–48 In von Heijne G. (ed), Signal peptidases. RG Landes Company, Austin, TX [Google Scholar]
- 96. Luke I, Handford JI, Palmer T, Sargent F. 2009. Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch. Microbiol. 191:919–925 [DOI] [PubMed] [Google Scholar]
- 97. Luo C, Roussel P, Dreier J, Page MG, Paetzel M. 2009. Crystallographic analysis of bacterial signal peptidase in ternary complex with arylomycin A2 and a beta-sultam inhibitor. Biochemistry 48:8976–8984 [DOI] [PubMed] [Google Scholar]
- 98. Lyko F, Martoglio B, Jungnickel B, Rapoport TA, Dobberstein B. 1995. Signal sequence processing in rough microsomes. J. Biol. Chem. 270:19873–19878 [DOI] [PubMed] [Google Scholar]
- 99. Maegawa S, Ito K, Akiyama Y. 2005. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry 44:13543–13552 [DOI] [PubMed] [Google Scholar]
- 100. Martoglio B, Golde TE. 2003. Intramembrane-cleaving aspartic proteases and disease: presenilins, signal peptide peptidase and their homologs. Hum. Mol. Genet. 12(Spec No 2):R201–R206 [DOI] [PubMed] [Google Scholar]
- 101. Martoglio B, Graf R, Dobberstein B. 1997. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 16:6636–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Maskos K, et al. 1998. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc. Natl. Acad. Sci. U. S. A. 95:3408–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mathur J, Davis BM, Waldor MK. 2007. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 63:848–858 [DOI] [PubMed] [Google Scholar]
- 104. McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Meijer WJ, et al. 1995. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol. Microbiol. 17:621–631 [DOI] [PubMed] [Google Scholar]
- 106. Nakatsukasa K, Brodsky JL. 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Naus S, et al. 2006. Identification of candidate substrates for ectodomain shedding by the metalloprotease-disintegrin ADAM8. Biol. Chem. 387:337–346 [DOI] [PubMed] [Google Scholar]
- 108. Nijtmans LG, et al. 2000. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19:2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nyborg AC, Jansen K, Ladd TB, Fauq A, Golde TE. 2004. A signal peptide peptidase (SPP) reporter activity assay based on the cleavage of type II membrane protein substrates provides further evidence for an inverted orientation of the SPP active site relative to presenilin. J. Biol. Chem. 279:43148–43156 [DOI] [PubMed] [Google Scholar]
- 110. Ogura T, et al. 1999. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31:833–844 [DOI] [PubMed] [Google Scholar]
- 111. Paetzel M, Dalbey RE, Strynadka NC. 1998. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature 396:186–190 (Erratum, 396:707.) [DOI] [PubMed] [Google Scholar]
- 112. Paetzel M, Goodall JJ, Kania M, Dalbey RE, Page MG. 2004. Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based inhibitor. J. Biol. Chem. 279:30781–30790 [DOI] [PubMed] [Google Scholar]
- 113. Paetzel M, Karla A, Strynadka NC, Dalbey RE. 2002. Signal peptidases. Chem. Rev. 102:4549–4580 [DOI] [PubMed] [Google Scholar]
- 114. Paetzel M, et al. 1997. Use of site-directed chemical modification to study an essential lysine in Escherichia coli leader peptidase. J. Biol. Chem. 272:9994–10003 [DOI] [PubMed] [Google Scholar]
- 115. Pei J, Grishin NV. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275–277 [DOI] [PubMed] [Google Scholar]
- 116. Pepe JC, Lory S. 1998. Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa. Effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J. Biol. Chem. 273:19120–19129 [DOI] [PubMed] [Google Scholar]
- 117. Ponting CP, et al. 2002. Identification of a novel family of presenilin homologues. Hum. Mol. Genet. 11:1037–1044 [DOI] [PubMed] [Google Scholar]
- 118. Powers ME, et al. 2011. Type I signal peptidase and protein secretion in Staphylococcus epidermidis. J. Bacteriol. 193:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Prokop S, Shirotani K, Edbauer D, Haass C, Steiner H. 2004. Requirement of PEN-2 for stabilization of the presenilin N-/C-terminal fragment heterodimer within the gamma-secretase complex. J. Biol. Chem. 279:23255–23261 [DOI] [PubMed] [Google Scholar]
- 120. Pugsley AP. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rajagopalan PT, Grimme S, Pei D. 2000. Characterization of cobalt(II)-substituted peptide deformylase: function of the metal ion and the catalytic residue Glu-133. Biochemistry 39:779–790 [DOI] [PubMed] [Google Scholar]
- 122. Ravipaty S, Reilly JP. 2010. Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 9:1898–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rawlings ND, Barrett AJ. 1999. MEROPS: the peptidase database. Nucleic Acids Res. 27:325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rawson RB, et al. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1:47–57 [DOI] [PubMed] [Google Scholar]
- 125. Rep M, Grivell LA. 1996. The role of protein degradation in mitochondrial function and biogenesis. Curr. Genet. 30:367–380 [DOI] [PubMed] [Google Scholar]
- 126. Roberts TC, Schallenberger MA, Liu J, Smith PA, Romesberg FE. 2011. Initial efforts toward the optimization of arylomycins for antibiotic activity. J. Med. Chem. 54:4954–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rowley G, Stevenson A, Kormanec J, Roberts M. 2005. Effect of inactivation of degS on Salmonella enterica serovar Typhimurium in vitro and in vivo. Infect. Immun. 73:459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Saito A, et al. 2011. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc. Natl. Acad. Sci. U. S. A. 108:13740–13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sakai J, et al. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell 85:1037–1046 [DOI] [PubMed] [Google Scholar]
- 130. Sakoh M, Ito K, Akiyama Y. 2005. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J. Biol. Chem. 280:33305–33310 [DOI] [PubMed] [Google Scholar]
- 131. Saller MJ, et al. 2011. Bacillus subtilis YqjG is required for genetic competence development. Proteomics 11:270–282 [DOI] [PubMed] [Google Scholar]
- 132. San Millan JL, Boyd D, Dalbey R, Wickner W, Beckwith J. 1989. Use of phoA fusions to study the topology of the Escherichia coli inner membrane protein leader peptidase. J. Bacteriol. 171:5536–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. 1998. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. U. S. A. 95:11175–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Schrul B, Kapp K, Sinning I, Dobberstein B. 2010. Signal peptide peptidase (SPP) assembles with substrates and misfolded membrane proteins into distinct oligomeric complexes. Biochem. J. 427:523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Shearman MS, et al. 2000. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry 39:8698–8704 [DOI] [PubMed] [Google Scholar]
- 136. Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662 [DOI] [PubMed] [Google Scholar]
- 137. Sohn J, Grant RA, Sauer RT. 2010. Allostery is an intrinsic property of the protease domain of DegS: implications for enzyme function and evolution. J. Biol. Chem. 285:34039–34047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Sohn J, Grant RA, Sauer RT. 2009. OMP peptides activate the DegS stress-sensor protease by a relief of inhibition mechanism. Structure 17:1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Strisovsky K, Sharpe HJ, Freeman M. 2009. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36:1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Strom MS, Bergman P, Lory S. 1993. Identification of active-site cysteines in the conserved domain of PilD, the bifunctional type IV pilin leader peptidase/N-methyltransferase of Pseudomonas aeruginosa. J. Biol. Chem. 268:15788–15794 [PubMed] [Google Scholar]
- 141. Strom MS, Lory S. 2001. Type IV prepilin leader peptidases, p 127–159 In Dalbey RE, Sigman DS. (ed), The enzymes, 3rd ed. Vol XXII. Co- and posttranslational proteolysis of proteins. Academic Press, London, United Kingdom [Google Scholar]
- 142. Sung M, Dalbey RE. 1992. Identification of potential active-site residues in the Escherichia coli leader peptidase. J. Biol. Chem. 267:13154–13159 [PubMed] [Google Scholar]
- 143. Suno R, et al. 2006. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol. Cell 22:575–585 [DOI] [PubMed] [Google Scholar]
- 144. Tatsuta T, Langer T. 2009. AAA proteases in mitochondria: diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 160:711–717 [DOI] [PubMed] [Google Scholar]
- 145. Terra R, Stanley-Wall NR, Cao G, Lazazzera BA. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. J. Bacteriol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Thompson BJ, et al. 2010. Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol. Microbiol. 77:943–957 [DOI] [PubMed] [Google Scholar]
- 147. Thornton J, Blakey D, Scanlon E, Merrick M. 2006. The ammonia channel protein AmtB from Escherichia coli is a polytopic membrane protein with a cleavable signal peptide. FEMS Microbiol. Lett. 258:114–120 [DOI] [PubMed] [Google Scholar]
- 148. Tjalsma H, et al. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Tjalsma H, et al. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12:2318–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Tjalsma H, et al. 1997. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities. Constitutive and temporally controlled expression of different sip genes. J. Biol. Chem. 272:25983–25992 [DOI] [PubMed] [Google Scholar]
- 152. Tjalsma H, et al. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102–25108 [DOI] [PubMed] [Google Scholar]
- 153. Tjalsma H, et al. 1999. The plasmid-encoded signal peptidase SipP can functionally replace the major signal peptidases SipS and SipT of Bacillus subtilis. J. Bacteriol. 181:2448–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Tjalsma H, van Dijl JM. 2005. Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics 5:4472–4482 [DOI] [PubMed] [Google Scholar]
- 155. Tjalsma H, Zanen G, Bron S, van Dijl JM. 2000. The eubacterial lipoprotein-specific (type II) signal peptidases, p 3–26 In Dalbey RE, Sigman DS. (ed), The enzymes, 3rd ed. Vol XXII. Co- and posttranslational proteolysis of proteins. Academic Press, London, United Kingdom [Google Scholar]
- 156. Tjalsma H, Zanen G, Venema G, Bron S, van Dijl JM. 1999. The potential active site of the lipoprotein-specific (type II) signal peptidase of Bacillus subtilis. J. Biol. Chem. 274:28191–28197 [DOI] [PubMed] [Google Scholar]
- 157. Tomasselli AG, et al. 2003. Employing a superior BACE1 cleavage sequence to probe cellular APP processing. J. Neurochem. 84:1006–1017 [DOI] [PubMed] [Google Scholar]
- 158. Tsai B, Ye Y, Rapoport TA. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3:246–255 [DOI] [PubMed] [Google Scholar]
- 159. Tschantz WR, et al. 1995. Characterization of a soluble, catalytically active form of Escherichia coli leader peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34:3935–3941 [DOI] [PubMed] [Google Scholar]
- 160. Tschantz WR, Sung M, Delgado-Partin VM, Dalbey RE. 1993. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J. Biol. Chem. 268:27349–27354 [PubMed] [Google Scholar]
- 161. Urban S, Baker RP. 2008. In vivo analysis reveals substrate-gating mutants of a rhomboid intramembrane protease display increased activity in living cells. Biol. Chem. 389:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Urban S, Freeman M. 2003. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11:1425–1434 [DOI] [PubMed] [Google Scholar]
- 163. Urban S, Schlieper D, Freeman M. 2002. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr. Biol. 12:1507–1512 [DOI] [PubMed] [Google Scholar]
- 164. van Bloois E, et al. 2008. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 582:1419–1424 [DOI] [PubMed] [Google Scholar]
- 165. van der Wal FJ, et al. 1992. The stable BRP signal peptide causes lethality but is unable to provoke the translocation of cloacin DF13 across the cytoplasmic membrane of Escherichia coli. Mol. Microbiol. 6:2309–2318 [DOI] [PubMed] [Google Scholar]
- 166. van Dijl JM. Biofilm research uncovers a novel non-enzymatic signal peptidase function in Bacillus. J. Bacteriol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. van Dijl JM, et al. 2007. In's and out's of the Bacillus subtilis membrane proteome, p 287–330 In Graumann P. (ed), Bacillus: cellular and molecular biology. Caister Academic Press, Wymondham, United Kingdom [Google Scholar]
- 168. van Dijl JM, de Jong A, Vehmaanpera J, Venema G, Bron S. 1992. Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 11:2819–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. van Dijl JM, de Jong A, Venema G, Bron S. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. Structural and functional similarities with LexA-like proteases. J. Biol. Chem. 270:3611–3618 [DOI] [PubMed] [Google Scholar]
- 170. van Klompenburg W, et al. 1998. Phosphatidylethanolamine mediates insertion of the catalytic domain of leader peptidase in membranes. FEBS Lett. 431:75–79 [DOI] [PubMed] [Google Scholar]
- 171. van Roosmalen ML, et al. 2004. Type I signal peptidases of Gram-positive bacteria. Biochim. Biophys. Acta 1694:279–297 [DOI] [PubMed] [Google Scholar]
- 172. van Roosmalen ML, et al. 2001. Detergent-independent in vitro activity of a truncated Bacillus signal peptidase. Microbiology 147:909–917 [DOI] [PubMed] [Google Scholar]
- 173. van Roosmalen ML, et al. 2001. Distinction between major and minor Bacillus signal peptidases based on phylogenetic and structural criteria. J. Biol. Chem. 276:25230–25235 [DOI] [PubMed] [Google Scholar]
- 174. van Stelten J, Silva F, Belin D, Silhavy TJ. 2009. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325:753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. VanValkenburgh C, Chen X, Mullins C, Fang H, Green N. 1999. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J. Biol. Chem. 274:11519–11525 [DOI] [PubMed] [Google Scholar]
- 176. Vassar R, et al. 1999. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286:735–741 [DOI] [PubMed] [Google Scholar]
- 177. von Heijne G. 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133:17–21 [DOI] [PubMed] [Google Scholar]
- 178. Waller PR, Sauer RT. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178:1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wang J, Hartling JA, Flanagan JM. 1997. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447–456 [DOI] [PubMed] [Google Scholar]
- 180. Wang P, et al. 2008. Escherichia coli signal peptide peptidase A is a serine-lysine protease with a lysine recruited to the nonconserved amino-terminal domain in the S49 protease family. Biochemistry 47:6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang Y, Ha Y. 2007. Open-cap conformation of intramembrane protease GlpG. Proc. Natl. Acad. Sci. U. S. A. 104:2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Wang Y, Zhang Y, Ha Y. 2006. Crystal structure of a rhomboid family intramembrane protease. Nature 444:179–180 [DOI] [PubMed] [Google Scholar]
- 183. Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215–2218 [DOI] [PubMed] [Google Scholar]
- 184. Weihofen A, et al. 2003. Targeting presenilin-type aspartic protease signal peptide peptidase with gamma-secretase inhibitors. J. Biol. Chem. 278:16528–16533 [DOI] [PubMed] [Google Scholar]
- 185. Weihofen A, Martoglio B. 2003. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 13:71–78 [DOI] [PubMed] [Google Scholar]
- 186. Weskamp G, et al. 2006. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat. Immunol. 7:1293–1298 [DOI] [PubMed] [Google Scholar]
- 187. Westers H, et al. 2006. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 273:3816–3827 [DOI] [PubMed] [Google Scholar]
- 188. Widdick DA, et al. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol. Microbiol. 80:1395–1412 [DOI] [PubMed] [Google Scholar]
- 189. Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T. 2004. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483–494 [DOI] [PubMed] [Google Scholar]
- 190. Willem M, et al. 2006. Control of peripheral nerve myelination by the beta-secretase BACE1. Science 314:664–666 [DOI] [PubMed] [Google Scholar]
- 191. Wolfe MS, et al. 1999. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 398:513–517 [DOI] [PubMed] [Google Scholar]
- 192. Wolfsberg TG, et al. 1995. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev. Biol. 169:378–383 [DOI] [PubMed] [Google Scholar]
- 193. Wu Z, et al. 2006. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat. Struct. Mol. Biol. 13:1084–1091 [DOI] [PubMed] [Google Scholar]
- 194. Yang B, Larson TJ. 1998. Multiple promoters are responsible for transcription of the glpEGR operon of Escherichia coli K-12. Biochim. Biophys. Acta 1396:114–126 [DOI] [PubMed] [Google Scholar]
- 195. Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. U. S. A. 97:5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Zeth K. 2004. Structural analysis of DegS, a stress sensor of the bacterial periplasm. FEBS Lett. 569:351–358 [DOI] [PubMed] [Google Scholar]
- 197. Zhang YW, et al. 2005. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J. Biol. Chem. 280:17020–17026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Zweers JC, et al. 2008. Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb. Cell Fact. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zweers JC, Wiegert T, van Dijl JM. 2009. Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl. Environ. Microbiol. 75:7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Zwizinski C, Date T, Wickner W. 1981. Leader peptidase is found in both the inner and outer membranes of Escherichia coli. J. Biol. Chem. 256:3593–3597 [PubMed] [Google Scholar]