Abstract
Summary: One form of immune evasion is a developmental state called “persistence” whereby chlamydial pathogens respond to the host-mediated withdrawal of l-tryptophan (Trp). A sophisticated survival mode of reversible quiescence is implemented. A mechanism has evolved which suppresses gene products necessary for rapid pathogen proliferation but allows expression of gene products that underlie the morphological and developmental characteristics of persistence. This switch from one translational profile to an alternative translational profile of newly synthesized proteins is proposed to be accomplished by maximizing the Trp content of some proteins needed for rapid proliferation (e.g., ADP/ATP translocase, hexose-phosphate transporter, phosphoenolpyruvate [PEP] carboxykinase, the Trp transporter, the Pmp protein superfamily for cell adhesion and antigenic variation, and components of the cell division pathway) while minimizing the Trp content of other proteins supporting the state of persistence. The Trp starvation mechanism is best understood in the human-Chlamydia trachomatis relationship, but the similarity of up-Trp and down-Trp proteomic profiles in all of the pathogenic Chlamydiaceae suggests that Trp availability is an underlying cue relied upon by this family of pathogens to trigger developmental transitions. The biochemically expensive pathogen strategy of selectively increased Trp usage to guide the translational profile can be leveraged significantly with minimal overall Trp usage by (i) regional concentration of Trp residue placements, (ii) amplified Trp content of a single protein that is required for expression or maturation of multiple proteins with low Trp content, and (iii) Achilles'-heel vulnerabilities of complex pathways to high Trp content of one or a few enzymes.
INTRODUCTION
Life Cycle of the Chlamydiaceae Family
The Chlamydiaceae family includes intracellular pathogens of great importance for human medicine (112). Under conditions favoring the rapid cell proliferation that occurs during the acute disease process, progression through a two-stage developmental cycle occurs. Elementary bodies (EBs), released from lysed host cells, are extracellular infectious entities that gain entry to new host cells and progress to an intracellular developmental stage of replicative capability. These reticulate bodies (RBs) initiate a stage of high metabolic activity, and cell populations increase until the host cell resources are depleted. Transformation back to the EB stage then occurs prior to host cell lysis. (Abbreviations, definitions, and a URL for supplemental files are given in Table 1.)
Table 1.
Abbreviations, definitions, and key URL link to supplemental filesa
| Abbreviation or term | Definition |
|---|---|
| EBs | Infectious elementary body developmental form of chlamydiae |
| RBs | Proliferative reticulate body developmental form of chlamydiae |
| Persistence | Quiescent aberrant body developmental form of chlamydiae |
| LGT | Lateral gene transfer |
| Trp | l-Tryptophan |
| PEP | Phosphoenolpyruvate |
| PRPP | Phosphoribosyl pyrophosphate |
| IDO | Indoleamine dioxygenase |
| IFN-γ | Gamma interferon |
| KDO | 2-Keto-3-deoxyoctulosonic acid |
| LPS | Lipopolysaccharide |
| PMPs | Polymorphic membrane proteins |
| Trp content | Percentage of Trp residues in a given protein |
| p/P Trp ratio | Ratio of the Trp content of a given protein to that of the proteome |
| p/bdh-P Trp ratio | Ratio of the Trp content of a given protein to that fraction of the proteome that shares a best-hit homolog with another proteome to which it is being compared |
| Trp burden | Absolute number of Trp residues per given protein. Where an entire pathway is under consideration, the Trp burdens of individual proteins contribute to a cumulative Trp burden with respect to that pathway. |
| Trp hurdle | A particular step(s) in a multistep pathway that is conspicuous for having a high Trp content |
| Bidirectional best-hit pairs | Protein homologs from two different genomes where each is mutually the best BLAST hit returned when one is used as the query against the proteome of the other |
| Up-Trp selection | Evolutionary selection for increased Trp content in a given protein |
| Down-Trp selection | Evolutionary selection for decreased Trp content in a given protein |
| Achilles heel step | An enzyme in a multistep pathway whose Trp content renders the entire pathway vulnerable to Trp limitation |
| Master-slave mechanisms | Indirect effect of Trp content upon the expression of “slave” proteins that are dependent upon a “master” protein of high Trp content |
Supplemental material for this article may be found at http://semiglobe.lanl.gov/supplement.php.
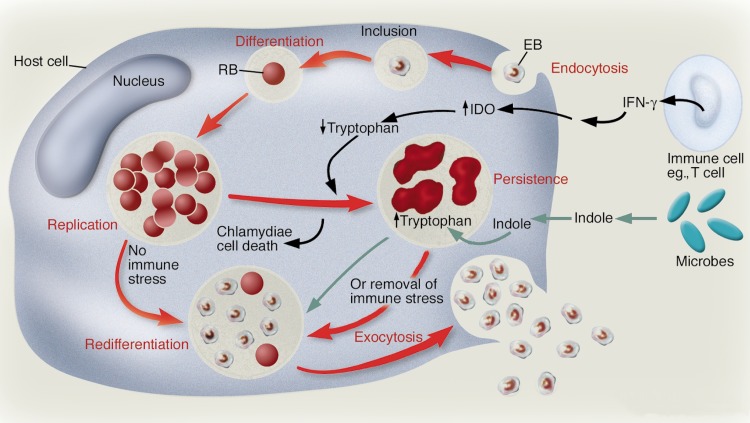
Whenever the innate host immune response is triggered, members of the Chlamydiaceae can respond to the host defense mechanism via progression to an alternative developmental state called “persistence” (8, 54). This amounts to a chronic state of pathogenesis and is a sophisticated survival mode. It can be of prolonged duration, but upon removal of the stimulus imparted by the immune response, the persistent state is competent for return to the rapid proliferation state that is associated with acute episodes of disease. The developmental stages in the life cycle of these pathogens are portrayed in Fig. 1. Also see the recent overview by Wyrick (138), which summarizes the contemporary contrast between the well-defined persistence-inducing conditions for Chlamydia trachomatis in vitro and the ill-defined nature of the persistent state in vivo.
Fig 1.
Overview of the developmental stages in a chlamydial infection. At the upper right, an infectious EB attaches to the host cell, with subsequent differentiation to an RB, which replicates within an inclusion. RBs may complete the cycle by differentiation back to EBs, which are released in conjunction with host cell lysis. Alternatively, RBs may respond to Trp starvation (caused by induction of IDO triggered by IFN-γ) by entering a quiescent stage of persistence. Genital strains of C. trachomatis can utilize environmental indole as a precursor of Trp. (Reprinted from reference 93 with permission of the American Society for Clinical Investigation.)
Role of Trp in One Mechanism Utilized by the Human Host as an Immune Response
Protective responses to chlamydial infections include the production of the proinflammatory cytokine gamma interferon (IFN-γ) by T cells. Antichlamydial effects of IFN-γ in humans include the production of nitric oxide from host arginine via induction of nitric oxide synthase, as well as the triggering of a mechanism for iron deprivation (58). Third, and especially prominent in the human host, is the IFN-γ-mediated induction of indoleamine 2,3-dioxygenase (IDO), a host enzyme which converts Trp to l-formylkynurenine. This was originally discovered by Byrne and coworkers (21, 22). l-Formylkynurenine has multiple potential fates in the host, such as being an NAD(P)+ precursor or progressing to acetyl-coenzyme A (acetyl-CoA) in the mitochondrial organelles. The action of IDO thus creates a state of near-starvation for Trp, which is an essential nutrient for the pathogen because all chlamydial species lack an intact pathway of Trp biosynthesis. Furthermore, IFN-γ strongly induces synthesis of host tryptophanyl-tRNA synthetase, whose elevated level not only favors host access but also tends to sequester the already diminished Trp pool away from parasitic metabolism (36). It appears that the most successful state of persistence from the pathogen's perspective can be equated not with absolute starvation for Trp but rather with conditions of substantial limitation for Trp. Thus, Leonhardt et al. (71) reported that when Trp availability is curtailed to drastically low levels, persistence is largely blocked and reactivation of surviving cells is minimal. Hence, provisioning of Trp at trickle levels could be an all-important feature needed to help sustain persistent cells indefinitely.
It is interesting that the foregoing triad of IFN-γ effects are not necessarily independent and might work cooperatively. Thus, a variety of evidence is advanced in this paper to show that iron deprivation is likely to be exacerbated further by Trp limitation. In addition, Xie et al. (139) have discussed and illustrated the interesting cross-pathway interactions reported in the literature between the IFN-γ-induced IDO and nitric oxide synthase, whereby each tends to counterbalance the effect of the other. Nitric oxide, the product of nitric oxide synthase, inhibits the activity of IDO. On the other hand, 3-hydroxyanthranilate, a downstream metabolite of Trp catabolism initiated by IDO, inhibits the activity of nitric oxide synthase.
The Special Nature of the Trp Codon and Its Gene Product
Among the 20 amino acids, Trp is unique in being specified by only one codon (except in a few very rare cases [61]). Proteins have a generally low content of Trp, which is biochemically the most expensive of the amino acids to synthesize in vivo. It is not uncommon for proteins (especially small proteins) to lack Trp residues altogether. Low-GC-content organisms (which include the pathogenic Chlamydiaceae) exhibit an amino acid preference that selects against GC-rich coding sequences (GARPW) and favors AT-rich codons (FYMINK) (117). The Trp codon is AGG and therefore is generally selected against in Chlamydiaceae. Thus, in comparison with the well-studied bacterium Escherichia coli, which has a proteomic Trp content of 1.50%, C. trachomatis D/UW-3/CX, Chlamydiae muridarum Nigg, Chlamydophila pneumoniae AR39, Chlamydophila caviae GPIC, Chlamydophila abortus S26/3, and Chlamydophila felis Fe/C have proteomic Trp contents of 0.95%, 0.97%, 1.00%, 0.95%, 0.96%, and 0.95%, respectively. “Candidatus Protochlamydia amoebophila” UWE25, a close relative of the pathogenic Chlamydiaceae that has also been selected for comparative analysis in this review, is a low-GC-content organism that has a low proteomic Trp content of 1.13%.
Against this background, one might reasonably expect that in low-GC organisms such as the Chlamydiaceae, retained Trp residues in proteins will tend to be those crucial for catalytic activity or for some other functional role (e.g., exploitation of its physical properties, along with those of tyrosine, as a membrane anchor [41]). Indeed, among all of the amino acids, Trp residues confer unique structural capabilities to some proteins. In orthologous families of globular proteins, Trp has been asserted to be more resistant to replacement than any other amino acid. Since it is hydrophobic, it is usually found at an interior position of a given protein. Trp has the largest rigid side of the 20 amino acids, making it hard to replace without perturbing the folded state of the active three-dimensional (3D) structure. Since Trp residues represent an expensive biosynthetic investment yet have unique potential to provide important functional properties, very strong selective pressures to explain the sets of up-Trp proteins (proteins with selection for more Trp residues than expected) and down-Trp proteins (proteins with selection for fewer Trp residues than expected) existing in the Chlamydiaceae are implicated.
Premise of the Review
Guiding concept.
In the face of a host defense mechanism that creates a nutritional state of limitation for Trp, individual Chlamydiaceae genes may have undergone selection for increased or decreased Trp content of their encoded proteins. This would depend upon whether a given protein is expressed during rapid parasite propagation (when Trp is available in the host cytosol) or during the chronic stage of persistence (when Trp is depleted). Did decreased availability of host-derived Trp actually come to be utilized by Chlamydiaceae as an exploitable cue for the impending onset of an immune response initiative by the host? Evolved changes accomplishing an increase or decrease in the Trp content of protein subsets in the pathogen could directly orchestrate a new developmental profile of translational expression. At one extreme, proteins having a high content of Trp are likely to be expressed poorly during Trp starvation, even if transcription is enhanced. At the other extreme, the expression of proteins that lack Trp residues altogether will not be restricted by starvation for Trp. It is noteworthy that ribosomal proteins and components of the overall translational machinery are generally made up of zero-Trp or low-Trp proteins and are thus available for protein synthesis under conditions of either Trp limitation or Trp sufficiency. Under conditions of minimal expression, residual enzyme activity will depend to a great extent upon enzyme stability in vivo. It is pertinent in this context that chlamydial proteins have been reported to be exceedingly stable (99).
Members of the Chlamydiaceae that persist in a state of continued starvation for Trp exhibit a distinctly altered profile of gene products, as one would expect from the morphological and metabolic changes associated with persistence. Beatty et al. (5) first suggested (in 1994) that the mechanism for the aberrant morphology and gene product profile of chlamydial growth in response to IFN-γ might be a “direct result of IDO depletion of the Trp available for translation of Chlamydia proteins.” These authors in the Byrne group noted that the decline in the major outer membrane protein (MOMP) during persistence correlates with the high Trp content of MOMP, as well as observing the contrasting high expression of cHSP60 (a zero-Trp protein) during persistence (5, 7). Against this background, it was interesting to see if this precocious concept could be expanded and further supported by exploitation of the ever-enlarging body of current databases and genome resources. Accordingly, we screened the identities of high-Trp proteins and low-Trp proteins in the current set of genomes for the pathogenic chlamydiae.
We initially considered the impact of Trp availability upon individual key proteins, such as those needed to mobilize carbon, energy, or Trp itself. More complex mechanisms that go beyond the simplistic consideration of the differential translation potentials of individual proteins having high or low overall Trp content were also pursued. Thus, we looked for (i) special cases where translational malfunction might occur as a result of the regional concentration of Trp residues (Trp hot spots) in a protein; (ii) “master-slave” relationships where a single master protein of high Trp content might affect expression of multiple slave proteins, regardless of their individual Trp contents; (iii) “Achilles heel” vulnerabilities of entire biochemical pathways that might be triggered by individual lynchpin proteins of particularly high Trp content; and (iv) evidence of functionally equivalent analog proteins that might have replaced their nonhomologous counterparts of different Trp content.
Rationale underlying the selection of two genomes outside the Chlamydiaceae for comparative analysis.
One of the genomes (“Ca. Protochlamydia amoebophila”) selected for comparative analysis diverged from the Chlamydiaceae at the phylogenetically near taxon level of family, whereas the other (E. coli) diverged at the phylogenetically distant taxon level of phylum. “Ca. Protochlamydia amoebophila” belongs to the family Parachlamydiae, which belongs to the order Chlamydiales, in common with the family Chlamydiaceae. It is an intracellular parasite of protozoan organisms. As such, it shares many lifestyle characteristics with the Chlamydiaceae, such as the aforementioned two-stage developmental cycle, a reflection of their recent common ancestry. This includes dependence upon the host for various nutrients, including Trp. “Ca. Protochlamydia amoebophila” represents a sister lineage of “environmental chlamydiae” that have only recently become appreciated as an ancient and diverse grouping (13, 56). At a much more distant level, a comparison is made with the well-known E. coli genome. E. coli is a free-living organism with no auxotrophies, and its genome has a high GC content.
The intracellular parasitism exhibited by the Chlamydiales is a relatively ancient capability, as reflected by the host affinities of the current assemblage of five taxon families, other than the Chlamydiaceae, in the order (13). The intracellular parasitism of mammals is a recent innovation, since the mammalian host emerged as only a recent divergence on the geological time scale. Accordingly, the Chlamydiaceae appear to be a correspondingly recent evolutionary divergence within the Chlamydiales. The common ancestor of the Chlamydiaceae must have already possessed well-developed features of reductive evolution and host-interactive machinery to sustain an intracellular lifestyle. Basic capabilities such as ATP and glucose-6-phosphate (glucose-6-P) import from the host were probably already in place, in contrast with many other changes needed for tuning to the new host environment. Although the IFN-γ/Trp depletion mechanisms of mammalian systems are of course absent in hosts such as protozoans, the simple presence or absence of Trp as a nutrient might have been an ancient developmental cue that remains in protozoans. This may then have become a more elaborate and sophisticated developmental cue that coevolved in the pathogenic chlamydiae and their mammalian hosts.
For comparative purposes, E. coli was selected as an organism for which any events of up-Trp or down-Trp selection would be irrelevant to a lifestyle of intracellular parasitism. In contrast, “Ca. Protochlamydia amoebophila” may share with the Chlamydiaceae a limited repertoire of up-Trp or down-Trp events that occurred in a common ancestor. Thus, we asked the following. In contrast to E. coli, to what extent might the common ancestor of “Ca. Protochlamydia amoebophila” and the Chlamydiaceae have experienced up-Trp selection or down-Trp selection? And what incidents of up-Trp and down-Trp selection are unique to the Chlamydiaceae?
Normalization of the Trp Contents of Individual Proteins to That of the Proteome
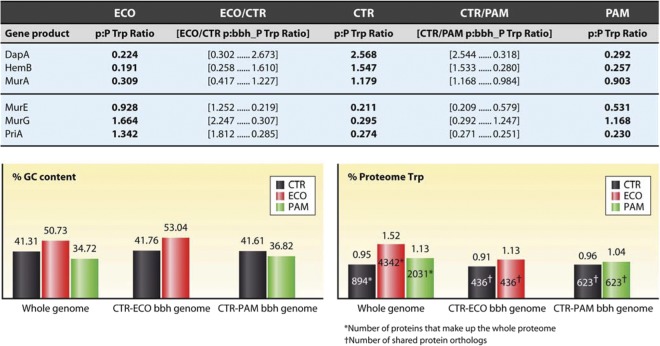
C. trachomatis, E. coli, and “Ca. Protochlamydia amoebophila” (41, 51, and 35% GC, respectively) have proteome Trp contents of 0.95, 1.52, and 1.13%, respectively (Fig. 2). Compared to E. coli, C. trachomatis is under much more selective pressure to avoid Trp residues in its proteome, because C. trachomatis is a low-GC organism, as explained above. Some examples of up-Trp selection (top three) and down-Trp selection (bottom three) for C. trachomatis are illustrated in Fig. 2. Data shown for C. trachomatis are also typical of the other species of Chlamydiaceae. DapA, a protein of about 300 amino acids, is characteristically low in Trp content. It has only one Trp residue in E. coli and “Ca. Protochlamydia amoebophila,” in contrast to DapA of C. trachomatis, which has seven Trp residues. Calculation of p/P Trp ratios (ratios of the Trp content of DapA to the Trp content of the proteome) revealed an enrichment of Trp content in C. trachomatis such that the value of the C. trachomatis ratio is greater by about an order of magnitude than those of “Ca. Protochlamydia amoebophila” and E. coli. HemB represents a similar example of up-Trp selection that occurred specifically in the pathogenic Chlamydiaceae. MurA, another enzyme of generally low Trp content, illustrates a case where up-Trp selection occurred in the common ancestor of C. trachomatis and “Ca. Protochlamydia amoebophila.” MurE illustrates a case of down-Trp selection in C. trachomatis relative to “Ca. Protochlamydia amoebophila” and E. coli. The difference is even more pronounced in other species of Chlamydiaceae, where MurE is a zero-Trp protein. MurG exemplifies another down-Trp selection that is specific to the Chlamydiaceae. On the other hand, down-Trp selection for PriA occurred in the common ancestor of C. trachomatis and “Ca. Protochlamydia amoebophila.” In this case, in a large protein of about 750 residues, E. coli invests 15 Trp residues, compared to only 2 for both C. trachomatis and “Ca. Protochlamydia amoebophila.”
Fig 2.
Normalization of the Trp contents of individual proteins to that of the proteome. ECO, E. coli; CTR, C. trachomatis; PAM, “Ca. Protochlamydia amoebophila.”
Shared-Ortholog Combinations: C. trachomatis-“Ca. Protochlamydia amoebophila” and C. trachomatis-E. coli
At some level, it may be of interest to compare the fractional proteomes of ortholog sets that are common to the C. trachomatis-“Ca. Protochlamydia amoebophila” combination, on the one hand, or the C. trachomatis-E. coli combination, on the other hand. In Fig. 2, these are referred to as the CTR-ECO bbh proteome and the CTR-PAM bbh proteome (bbh stands for “best bidirectional hit”). Of the 894 proteins in the C. trachomatis proteome, only 436 have best-hit matches among the 4,342 E. coli proteins. From its assemblage of 2,031 proteins, the more closely related “Ca. Protochlamydia amoebophila” proteome returns 623 best-hit matches to C. trachomatis queries. Figure 2 (lower left panel) shows that the GC contents of the partial genomes defined by bbh genome combinations are fairly close to those of the complete genomes. However, the partial proteome in E. coli defined by the 436 proteins that return best-hit bidirectional matches to C. trachomatis queries exhibits only a 1.13% Trp content, compared to the 1.52% Trp content of the complete proteome. This may be due in part to the fact that the highly conserved proteins of the translational apparatus inevitably have low Trp content, often being zero-Trp proteins.
One could choose to normalize the individual proteins of any proteome being compared to C. trachomatis to the fraction of the proteome which returns the best bidirectional hits to BLAST queries. Figure 2 shows a comparison of p/P Trp ratios computed using complete proteome data and p/bbh_P Trp ratios computed using the fractional proteomes for the E. coli-C. trachomatis combination or the C. trachomatis-“Ca. Protochlamydia amoebophila” combination. There is very little difference in the p/P Trp ratio and the p/bbh_P Trp ratio for the C. trachomatis-“Ca. Protochlamydia amoebophila” combination. For the C. trachomatis-E. coli combination, use of the p/bbh_P Trp ratio diminishes the magnitude of apparent up-Trp selection (top three entries in Fig. 2) and accentuates the magnitude of apparent down-Trp selection (bottom three entries in Fig. 2). Complete tables of p/bbh_P Trp ratios for the C. trachomatis-“Ca. Protochlamydia amoebophila” and C. trachomatis-E. coli proteome combinations have been generated and are available as supplementary files S9 and S10 at http://semiglobe.lanl.gov/supplement.php.
In contrast to p/P Trp ratios, the values of p/bbh_P Trp ratios of a given proteome will vary in accordance with the proteome to which it is being compared. In an evolutionary context, the use of p/P Trp ratios should be more generally useful, because the placement of Trp residues evolves within complete proteomes and is not restricted to arbitrary partial proteomes. Each organism faces pressure to maintain the number of Trp residues within boundaries dictated by the GC content of the genome. If C. trachomatis has experienced down-Trp selection for given proteins, it has the luxury of allocating Trp residues to other proteins in its proteome, regardless of whether orthologs are present in organisms such as E. coli. Likewise, if E. coli has experienced down-Trp selection for given proteins, it has the luxury of allocating Trp residues among a very large assemblage of proteins for which C. trachomatis has no homologs.
Dilemma Posed by Extreme Differences in Host Responses of Humans and Mice
IFN-γ induction of GTPases in the mouse host.
This article strongly asserts that all of the pathogenic chlamydiae must share an underlying sensitivity to Trp availability as a fundamental developmental cue that senses the onset of the host immune response. This is based upon the finding that the overwhelming majority of chlamydial protein orthologs share a profile of proteins exhibiting selection for high Trp content or low Trp content (up-Trp or down-Trp proteins). This Trp cue thesis fits well with the above-described induction of human IDO mediated by IFN-γ in response to C. trachomatis infection. However, a comparable IDO response is absent in the mouse host, in which the major IFN-γ-mediated host response is the induction of the large subset of p47 GTPases that exist within the GTPase superfamily. These inducible p47 GTPases have been lost in the primate genealogy of vertebrates (9). Individual p47 GTPase members have differing properties (52, 91), most of which have not yet been studied, but they generally bind cardiolipin, phosphoinositides, NSF attachment protein receptor adaptor proteins, and other p47 GTPases in order to mobilize their membrane regulatory capabilities against compartmented pathogens (see reference 65 and references therein). Another subset of GTPases, the p65 guanylate-binding proteins, are strongly induced by IFN-γ in both human and mouse hosts (65). In contrast to the p47 GTPases, this subset has been suggested to actually sensitize chlamydia to Trp starvation, possibly as a result of GTP depletion (128). This would be consistent with the effect of Trp limitation upon nucleotide import from the host, as discussed later.
Chlamydial production of cytotoxin.
GTPases have generally been described as targets of toxins with respect to the pathogenesis of infectious disease (17). Chlamydial organisms are known to produce cytotoxins that are homologous (at least in regional sections) to large clostridial toxins (LCTs) and to enteric LifA cytotoxin (lymphostatin). Cytotoxin genes are located within a unique chlamydial region of variability called the plasticity zone (see reference 82 and references therein). The cytotoxin gene of C. trachomatis D/UW-3 has undergone extensive degradation and is mostly nonfunctional, although one fragment (CT166) retains some function (23). Even at the serovar level, cytotoxin variability is evident. Recently, a comprehensive study of 15 serovars of C. trachomatis demonstrated a distinct relationship between different tissue tropisms and three different cytotoxin deletions (23). Hence, noninvasive oculotropic and urogenitotropic serovars and invasive serovars can be distinguished by their possession of one of three cytotoxin polymorphisms. C. trachomatis, with its degraded cytotoxin, represents one extreme of evolutionary de-emphasis, and C. muridarum, having three paralogous cytotoxin copies, represents the other extreme. In addition to C. muridarum, another mouse pathogen, Citrobacter rodentium, relies on a well-studied cytotoxin homolog (lymphostatin) as a virulence factor (3). Chlamydophila caviae and Cp. felis each possess a single cytotoxin gene encoding one large fully functional cytotoxin of >3,000 amino acids. The cytotoxin molecule has been shown to be present preformed in chlamydial EBs and transferred to infected host cells very early (10). Inactivation of host GTPases by the cytotoxin presumably occurs near the sites of EB entry (82). Subsequently, the cytotoxin molecules are degraded quite rapidly.
The cytotoxin fragment of C. trachomatis D/UW-3 does not impose a particularly great Trp burden, having only four Trp residues, and the p/P Trp ratio is only 0.65. On the other hand, the cytotoxins of C. muridarum, Cp. caviae, and Cp. felis impose very substantial Trp burdens. The full Trp burden information for the cytotoxins is shown in Table 2, in which a detailed comparison between the chlamydial cytotoxins and the enteric cytotoxins is given. The ratio of cytotoxin Trp to proteome Trp (p/P Trp ratio) is much elevated in chlamydial organisms compared to that in enteric organisms, indicating that there has been up-Trp selection in the chlamydial cytotoxins. Among the chlamydial organisms, the absolute number of Trp residues utilized (Trp burden) for cytotoxin input by C. muridarum is truly amazing, since the three adjacent paralogs require a total of 124 Trp residues. This enormous demand for Trp input might be amplified even further, to the extent that the cytotoxin might be multimeric, as has been reported for the large clostridial cytotoxin of Clostridium sordellii (131).
Table 2.
Trp burdens of cytotoxins generated in chlamydial organisms
| Organisma | Locus tag | No. of amino acids in encoded protein | No. of Trp residues | % Trp residues | Proteome Trp % | p/P Trp ratio |
|---|---|---|---|---|---|---|
| Chlamydia muridarum Nigg | TC0437 | 3,255 | 41 | 1.25 | 0.97 | 1.289 |
| TC0438 | 3,335 | 43 | 1.28 | 0.97 | 1.320 | |
| TC0439 | 3,235 | 40 | 1.24 | 0.97 | 1.278 | |
| Chlamydophila caviae GPIC | CCA0058 | 3,346 | 42 | 1.25 | 0.95 | 1.316 |
| Chlamydophila felis Fe/C-56 | CF0442 | 3,298 | 48 | 1.45 | 0.95 | 1.526 |
| Citrobacter rodentium ICC168 | ROD_26401 | 3,208 | 33 | 1.02 | 1.52 | 0.671 |
| Escherichia coli O127:H6 strain E2348/69 | E2348C_3234 | 3,223 | 36 | 1.11 | 1.53 | 0.725 |
| Escherichia coli H30* | p026VIR_p049 | 3,166 | 29 | 0.91 | 1.78 | 0.511 |
| Escherichia coli O157:H7 strain EC4115* | ECH74115_B0076 | 3,169 | 28 | 0.88 | 1.67 | 0.527 |
| Escherichia coli O157:H7 strain EDL933* | L7095 | 3,169 | 28 | 0.88 | 1.67 | 0.527 |
| Escherichia coli O157:H7 strain Sakai* | pO157p58 | 3,169 | 28 | 0.88 | 1.69 | 0.521 |
| Escherichia coli O157:H7 strain TW14359* | ECSP_6070 | 3,169 | 28 | 0.88 | 1.67 | 0.527 |
*, the protein is encoded in the plasmid.
The IFN-γ/GTPase/cytotoxin/Trp depletion thesis.
Dwelling upon the remarkable Trp content of the C. muridarum mouse pathogen cytotoxins suggests a reasonable answer to the following question: if Trp availability is indeed a fundamental cue with developmental consequences, how might Trp depletion occur in the mouse host in the absence of the IFN-γ/IDO host response seen in humans? The mouse host (unlike the human host) expresses multiple p47 GTPases as a defense measure against pathogens, including C. muridarum. C. muridarum (unlike C. trachomatis) expresses a trio of cytotoxins that target at least some of the p47 GTPases. Thus, the mouse host GTPase assemblage and the mouse pathogen cytotoxin have coevolved, as pointed out in several insightful papers (82, 97). The cytotoxin acts early in the developmental cycle and is quickly degraded. This degradative release of Trp could provide a boost of extra Trp for early pathogen growth. The eventual production of new cytotoxin molecules in RBs could, in contrast, act as a Trp sink, leading to Trp starvation. If this is correct, then IFN-γ may evoke the same outcome of Trp starvation for species-adapted chlamydiae in humans and mice, but by different mechanisms. The key host IFN-γ-induced entities involved in these equivalent outcomes are human IDO and mouse p47 GTPases. Since evolutionary analysis indicates that the p47 GTPases have been lost in the primate branch of the vertebrate lineage (9), the early ancestral mammalian state would be one where the p47 GTPases had been discarded. Thus, in response to an absence of its target, a corresponding loss of cytotoxin, i.e., coevolved loss, must have occurred in the human pathogen C. trachomatis. Accordingly, the emergence of IDO as a strongly induced host-specific IFN-γ response in primates must have been a relatively recent evolutionary event. Thus, replacement of an ancestral IFN-γ/GTPase/cytotoxin/Trp depletion mechanism by a contemporary IFN-γ/IDO/Trp depletion mechanism in humans is proposed.
Access to Dynamic, Sortable Tables of Trp Content Data
The website at http://semiglobe.lanl.gov/supplement.php contains links to supplementary files (at the bottom of the page), including Tables S1 and S2. These tables have been prepared such that every gene product of C. trachomatis D/UW-3 has been used as a query for best bidirectional BLAST hits against eight other genomes (C. muridarum Nigg, Cp. abortus S26/3, Cp. caviae GPIC, Cp. felis Fe/C-56, Cp. pneumoniae CWl029, “Ca. Protochlamydia amoebophila” UWE25, E. coli K-12, and Blochmannia pennsylvanicus BPEN). The most generally useful and compact table (Table S1) contains an initial column in which each C. trachomatis gene is hyperlinked to a gene neighborhood resource at the SEED (100). Not only does this SEED page facilitate navigation to orthologs in other genomes, but it allows direct access to DNA or protein sequences and to tools such as BLAST. Table S1 has columns for protein length, number of Trp residues, % Trp content, and protein/proteome Trp ratios (p/P Trp ratios). Each column is sortable (left click) and can be hidden (uncheck boxes after right click). In this paper, we frequently refer specifically to the C. trachomatis data entries, and unless exceptions are noted, the reader can infer that the corresponding data for other Chlamydiaceae organisms are essentially the same.
The primary working table (Table S2) is more extensive (except for the absence of the SEED gene neighborhood links). It contains additional columns showing the underlying statistical data. Entries in either table highlighted with horizontal green bars show proteins having local concentrations of Trp residues, i.e., locations where at least three amino acid residues in a scanning window of six residues are Trp. Table S3 is a version of the sortable, dynamic table used for the previous analysis of aspartokinase (74), but it is expanded to contain Trp content data. Two additional tables (Table S4 and Table S5) are not specifically referred to in this paper, but they may be a resource of interest for some readers. They use two different pathogen genomes as the source of query proteins for the best bidirectional BLAST hits against the proteomes of four other selected genomes.
Trp CONTENT OF THE PROTEOME
General Variation in Trp Content of the Proteome versus GC Content
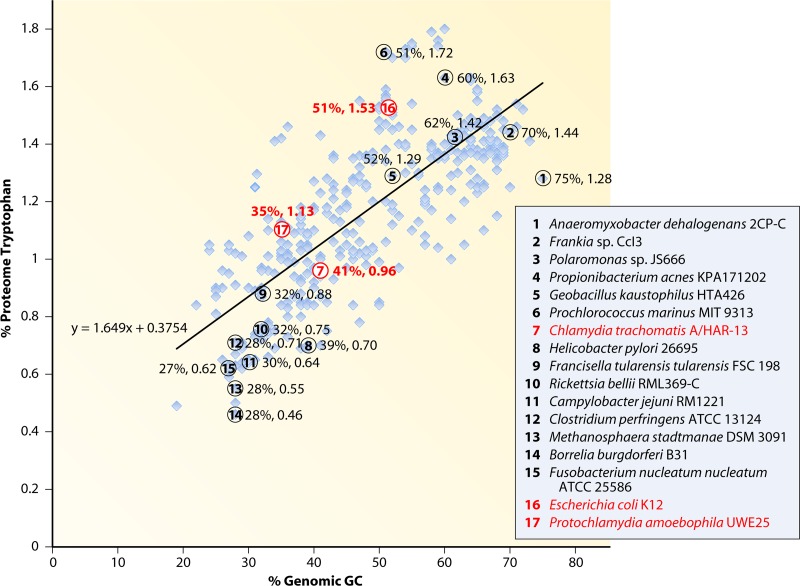
The Trp content of 397 sequenced genomes was calculated as a percentage of the proteome (% proteomic Trp), and this was plotted against the % genomic GC content. Figure 3 shows that the % proteomic Trp increases as a function of % genomic GC content, as expected. Thus, organisms with AT-rich genomes are expected to support a minimal content of Trp in their proteins. While the trend is unmistakable, considerable scatter occurs. For example, Anaeromyxobacter dehalogenans (organism 1 in Fig. 3), with a GC content of 75%, exhibits 1.28% Trp residues in its proteome, in contrast to Prochlorococcus marinus (organism 6), which has 1.72% Trp in its proteome, even though its genomic GC content is lower (51%). C. trachomatis (organism 7), its close phylogenetic neighbor “Ca. Protochlamydia amoebophila” (organism 17), and its distant phylogenetic neighbor E. coli (organism 16) are marked in the figure. The Trp content of C. trachomatis falls somewhat below the trend line predicted by % genomic GC content, whereas both “Ca. Protochlamydia amoebophila” and E. coli have somewhat greater Trp contents than predicted.
Fig 3.
Relationship between genomic %GC and the % Trp content of the proteome. A total of 397 sequenced genomes were accessed from the IMG database in order to obtain the genome and proteome information. Seventeen organisms were selected for specific identification in the figure, and they are numbered at the right and indicated by matching numbers within enclosed circles positioned over the appropriate light blue squares. C. trachomatis (organism 7), E. coli (organism 16), and “Ca. Protochlamydia amoebophila” (organism 17) are specially marked.
Trp Does Not Exhibit Typical Proportionality of Residue Number versus Protein Length
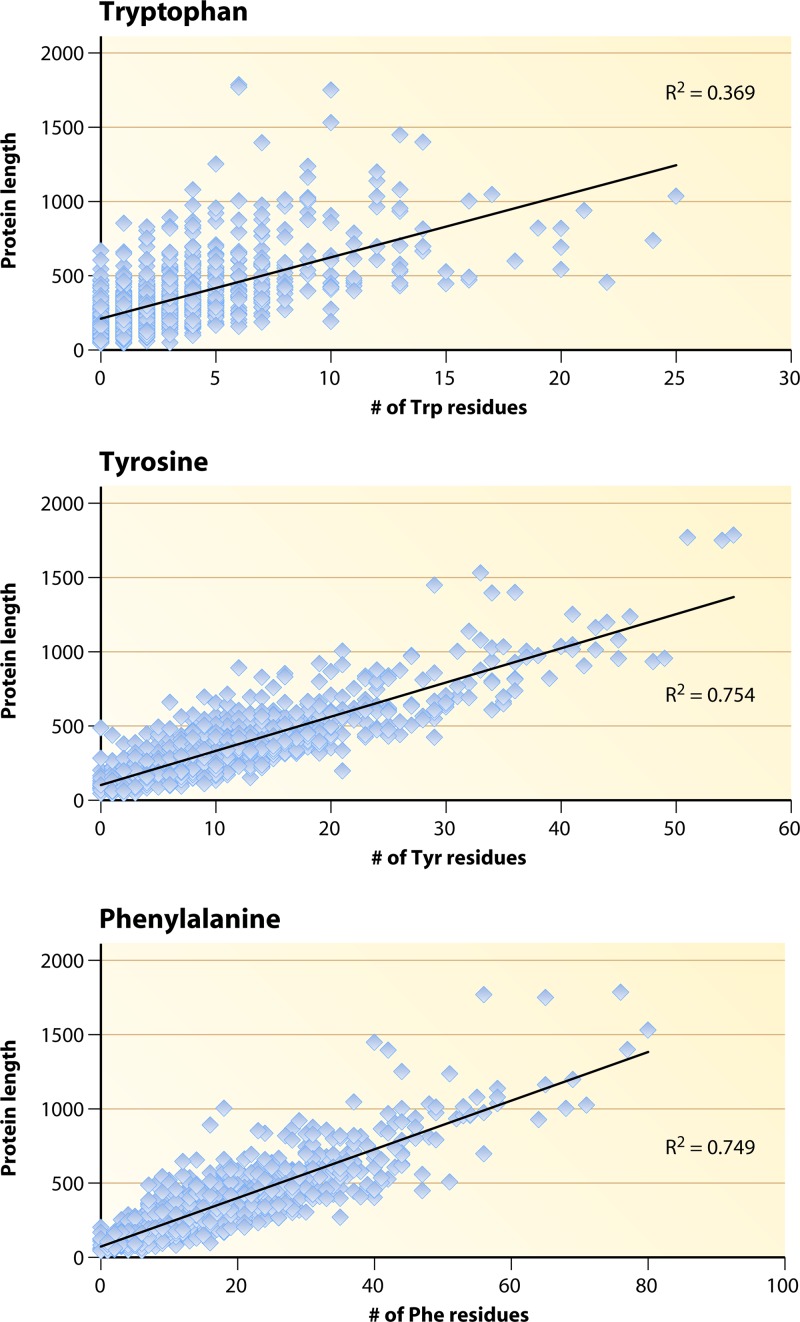
Amino acids are generally distributed with proportionality along the lengths of proteins. Figure 4 illustrates the results obtained when protein length is plotted against residue number for the three aromatic amino acids in C. trachomatis. Values for the coefficient of determination (R2) were calculated, and Trp is exceptional in having a low R2 value (0.369). This means that 37% of the variance in Trp number can be explained by variation in protein length (and vice versa). For comparison, phenylalanine is another hydrophobic amino acid, and it exhibits a high R2 value (0.749). Tyrosine yields a similar high R2 value. The R2 value calculated for the distribution of Trp residues versus protein length has the lowest value among the 20 amino acids (see Table S6 at http://semiglobe.lanl.gov/supplement.php for all 20 plots). The next lowest R2 value is for cysteine (0.455), followed by methionine and histidine (0.620). The deviation of the Trp profile from the proportionality of distribution of Trp residues versus protein length that generally occurs with other amino acids is consistent with up-Trp selection and down-Trp selection in the repertoire of individual C. trachomatis proteins.
Fig 4.
Distribution of aromatic amino acid residues along the lengths of proteins within the proteome of C. trachomatis. The coefficient of determination (R2) is the fraction of the variance that is shared by the two variables.
IMPACT OF Trp LIMITATION ON CARBON SOURCE
The Chlamydiaceae are absolutely dependent upon their host organism for provision of glucose-6-P as a starting point for carbon metabolism. Mammalian glucose-6-P translocase facilitates transfer of cytosolic glucose-6-P to the endoplasmic reticulum, where it is converted to glucose via a tightly coupled glucose-6-phosphatase system (39). Microbial homologs of glucose-6-P translocase exist in various intracellular pathogens, such as Listeria and Escherichia, where they can be viewed as mimics of a normal mammalian process that has been subverted for pathogenic function. The protein responsible for glucose-6-P transport in the Chlamydiaceae is called UhpC/T. It contains both receptor (UhpC) and transport (UhpT) functions that have been documented fully for E. coli (60). These permeases belong to the organophosphate:inorganic phosphate antiporter family of the major facilitator superfamily of permeases (114). It has been shown that the permeases from Chlamydiaceae are closer to the mammalian permeases on a protein tree than they are to other bacterial hexose-P permeases (26), and lateral gene transfer (LGT) in one direction or the other was suggested. Since that time, the genome of “Ca. Protochlamydia amoebophila” has become available, and it (as well as those of members of several other chlamydial families) has a gene encoding UhpC/T. This indicates that intracellular energy parasitism emerged in protozoans and other host organisms as a very ancient event. Because the common ancestor of these chlamydial families must have existed long before the emergence of mammals, it now seems most plausible that a mammalian ancestor acquired the gene encoding its glucose-6-P translocase from a chlamydial donor source.
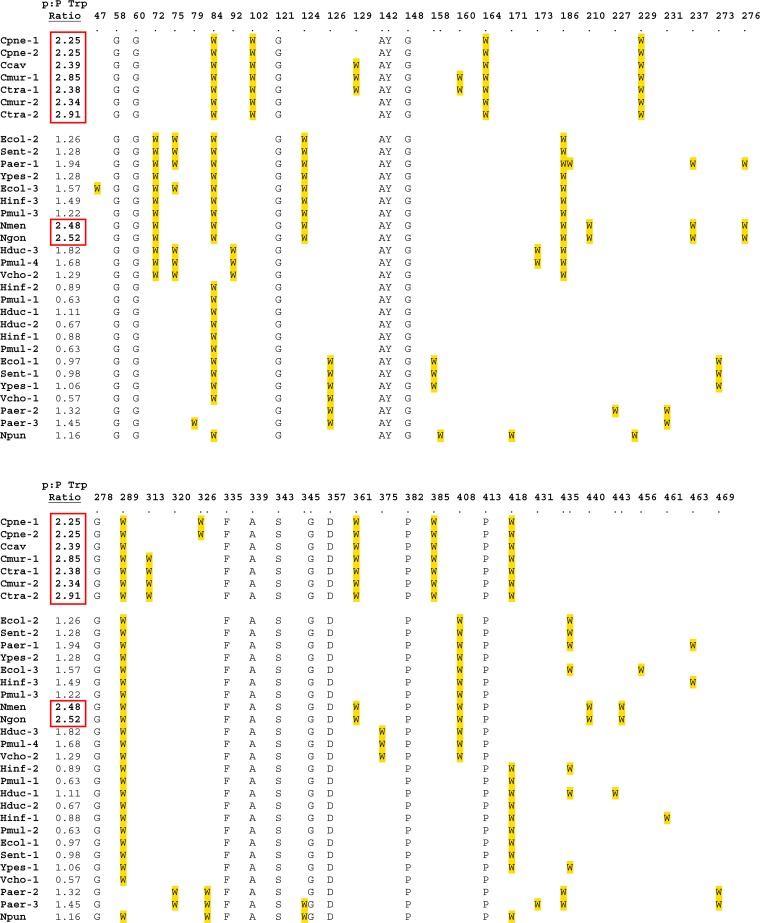
It is quite striking that UhpC/T consists of about 5% Trp (see Table S1 at http://semiglobe.lanl.gov/supplement.php). This is far beyond the average proteomic Trp content of about 1% for the Chlamydiaceae. Furthermore, the large size of this protein (456 amino acid residues) requires an input of 22 or 23 Trp residues per enzyme unit, which is a very high Trp burden. Figure 5 shows the distribution of Trp residues in an amino acid alignment in which only invariant residues and Trp residues are shown. All of the pathogen hexose-phosphate transporters possess a considerable number of Trp residues, five of which are located at invariant positions in the alignment. However, the Chlamydiaceae (middle group; indicated with a red box) exhibit ratios of UhpC/T Trp content to proteomic Trp content (p/P Trp ratio) that average >5.1. Clearly there has been up-Trp selection for additional Trp residues that must not have been otherwise essential. It is noteworthy that the Parachlamydiaceae organism (“Ca. Protochlamydia amoebophila”), which resides in a protozoan host rather than a mammalian host, exhibits a p/P Trp ratio (left column) which does not indicate up-Trp selection. Note that in addition to the overall high Trp content, many Trp residues are distributed within localized hot spots, including a number of tandem Trp residues. This would likely occasion translational pausing. The combination of high Trp content and the existence of Trp hot spots surely must confer extreme sensitivity of glucose-6-P transport to Trp availability.
Fig 5.
Trp contents of the glucose-6-P transporters from Chlamydiaceae family organisms (center group) and from diverse other organisms. Each number in the column under the heading “p:P Trp Ratio” is the ratio of the Trp content of the transporter to the Trp content of the proteome. The genomic abbreviations, from top to bottom, are as follows: Lmon, Listeria monocytogenes 4b F2365; Spro, Serratia proteamaculans 568; Vvul, Vibrio vulnificus YJ016; Ppro, Photobacterium profundum SS9; PSYC, Psychromonas sp. CNPT3; Ecol, Escherichia coli K-12; Ahyd, Aeromonas hydrophila hydrophila ATCC 7966; Pamo, “Candidatus Protochlamydia amoebophila” UWE25; Cpne, Chlamydophila pneumoniae CWL029; Ctra, Chlamydia trachomatis D/UW-3/CX; Cmur, Chlamydia muridarum Nigg; Ccav, Chlamydophila caviae GPIC; Cfel, Chlamydophila felis Fe/C-56; Cdip, Corynebacterium diphtheriae NCTC 13129; Cbot, Clostridium botulinum A ATCC 3502; Saur, Staphylococcus aureus aureus MRSA252; and Bcer, Bacillus cereus ATCC 10987. The occurrences of Trp residues in an alignment are displayed as bold W's highlighted in yellow.
Thus, it follows that under conditions of Trp starvation, one can expect a drastic decline in the ability to translate the enzyme that is needed for the import of host glucose-6-P. This is the prime source of carbon and energy for Chlamydiaceae (60), which lack the ability to import glucose and which further lack hexokinase. It is of interest in this context that persistence can be triggered by manipulation of glucose availability to host cells in a tissue culture system (46).
GLYCOGEN BIOSYNTHESIS AND BREAKDOWN
Glycogen biosynthesis begins with the conversion of glucose-6-P to glucose-1-P by phosphoglucomutase, and this reversible reaction is also the last step of glycogen breakdown. Since Chlamydiaceae have two widely diverged hexose-phosphate mutase paralogs (MrsA), perhaps they are functionally specialized for biosynthesis and catabolism. The remaining steps of glycogen biosynthesis are catalyzed by GlgC, GlgA, and GlgB. The Trp contents of all of these proteins (see Table S1 at http://semiglobe.lanl.gov/supplement.php) are moderately low, except for that of the branching enzyme (GlgB). This enzyme, which produces branched glycogen from glycogen, has a very high Trp content (p/P Trp ratio = 3.42). During Trp starvation, glycogen accumulation would seem to be minimal at the outset in consideration of the drastically diminished import of the beginning substrate (glucose-6-P), as discussed in the preceding section. The additional impact of the high Trp requirement of GlgB should abolish glycogen biosynthesis.
Synthesis of the enzymes needed for glycogen breakdown would be much decreased during Trp limitation, since the initial two degradative enzymes (GlgX [glycogen hydrolase] and GlgP [glycogen phosphorylase]) have very high p/P Trp ratios, of 2.21 and 1.80, respectively. The conclusion that glycogen breakdown is highly attenuated during Trp starvation may seem counterintuitive at first glance. But the attenuation of breakdown may in fact be a crucial and sophisticated mechanism to conserve a potential source of glucose-6-P through an extended duration in which persistent cells are provided with trickle-level amounts of glucose-6-P from glycogen.
HIGH Trp CONTENT OF TWO FUNCTIONALLY DIVERGENT PARALOGS FOR NUCLEOTIDE TRANSPORT
ATP provisioning from the mammalian host is a crucial source of energy for Chlamydiaceae. More broadly, ribonucleoside triphosphates are crucial building blocks to support parasitic nucleic acid biosynthesis, and the Chlamydiaceae are also dependent upon the host as an essential source of these precursor compounds. Specific ATP transport and general ribonucleoside triphosphate transport are accommodated by separate translocase systems. Two paralogs of translocase genes were initially demonstrated in C. trachomatis (129), encoding large integral membrane proteins with high Trp contents. A comprehensive analysis of all known ATP/ADP translocases (116) suggests that an original broad-specificity nucleoside triphosphate transport gene (ttc) in a chlamydial ancestor was duplicated, with one copy (ntt) retaining broad-specificity competence and the other copy (tlc) diverging to encode a transporter with narrow specificity for ATP and ADP. The Tlc divergence was attended by a change in mechanism from H+ symport to antiporter exchange. Although alternative evolutionary conclusions in the literature were considered, the assertions of Schmitz-Esser et al. (116) that an ancestral chlamydial ATP/ADP translocase gene was the donor for LGT transfer to Rickettsiae and to plant plastids seem well supported. Almost half (six) of the total Trp residues are invariant throughout the group of ATP/ADP translocases aligned by Schmitz-Esser et al. (116), and these are also invariant in the homologous set of broad-specificity Ntt transporters. Ntt translocase operates via an H+ symporter mechanism. Ntt (encoded by CT495 in C. trachomatis), similar to its Tlc homolog (encoded by CT065 in C. trachomatis), is a large protein containing well over 500 amino acids and has a high Trp content (2.40%; p/P Trp ratio = 2.53). Likewise, the Tlc protein exerts a strong demand for input of Trp residues (high Trp burden) and has a 2.46% Trp content (p/P Trp ratio = 2.59).
The broad-specificity translocase may be of particular interest with respect to the importance of GTP import because during the elongation stage of translation, GTP is used as an energy source for the binding of a new amino-bound tRNA to the A site of the ribosome. GTP is also used as an energy source for the translocation of the ribosome toward the 3′ end of the mRNA. In addition, GTP in chlamydia is a substrate for the GTP-dependent phosphoenolpyruvate (PEP) carboxykinase, perhaps one of the key enzymes operating in the overall mechanism for developmental sensitivity to the Trp cue. This is discussed in the next section.
PEP CARBOXYKINASE
We consider the impact of Trp's presence or absence upon chlamydial PEP carboxykinase (PckG) to be one of the most pivotal influences upon translational profile transitions. There are two classes of PEP carboxykinase homologs in nature: one of these is ATP dependent (PckA), and the other is GTP dependent (PckG) (55). GTP-dependent homologs are present mainly in higher eukaryotes, most archaea, and some fungi. They are present in only a relatively few bacteria, including chlamydial organisms. PckG of C. trachomatis has a very high p/P Trp ratio (3.16), and its 18 Trp residues represent one of the higher Trp burdens. The individual Trp residues are quite well conserved in other bacteria that possess this homolog type, including “Ca. Protochlamydia amoebophila.” PEP carboxykinase in chlamydial organisms illustrates a case where the evolutionary option to select an analog alternative with high Trp content has been exercised, occurring in this case in an ancient chlamydial ancestor that predated the divergence of the Chlamydiaceae. In comparison, E. coli has the ATP-dependent homolog, with a p/P Trp ratio of only 1.09. The p/P Trp ratio of C. trachomatis (and “Ca. Protochlamydia amoebophila”) PckG is about three times its proteome average, whereas that of E. coli PckA is about the same as the proteome average.
Interlocking metabolic relationships bring to mind several far-reaching aspects of PEP significance. (i) Since the aforementioned ATP/ADP translocase of the Chlamydiaceae is capable of importing ATP from the host in exchange for ADP exported from the pathogen, the Chlamydiaceae were generally regarded as energy parasites (94), but this was later qualified to the extent that ATP can be generated by pyruvate kinase (59). However, the latter capability depends upon how robust carbon metabolism is with respect to the generation of PEP. Under conditions of Trp limitation, the translation of PEP carboxykinase, the main or sole source of PEP, should be quite vulnerable, since it has one of the highest Trp contents in chlamydial proteomes. In addition, enzyme activity should be challenged even further because both substrates, GTP and oxaloacetate, should be at marginal levels due the high Trp contents of both transporters for these, as discussed earlier. Thus, while pyruvate kinase may indeed generate some supplementary ATP under conditions of Trp sufficiency during active growth, it is unlikely to do so during persistence. (ii) PEP limitation for MurA catalysis can represent a crucial Achilles heel vulnerability in a complex pathway such as the cell division/peptidoglycan pathway, as discussed later. (iii) Likewise, PEP limitation for the KdsA step of 2-keto-3-deoxyoctulosonic acid (KDO) biosynthesis in the lipopolysaccharide pathway could be another such roadblock, also discussed later. In view of the Pck relationship to the cell division pathway, it may be meaningful that the chlamydial pckG gene is closely linked to mreB, encoding a “shape-determining protein” of cell division. In fact, mreB and pck appear to constitute an operon, since the genes overlap.
DICARBOXYLATE IMPORTATION
A sodium-dependent 2-oxoglutarate/malate translocator imports 2-oxoglutarate of host origin into chlamydial cells. It also transports oxaloacetate. This enzyme is a closely related homolog of the well-studied plant enzyme which imports 2-oxoglutarate into the plastid compartment (133). CT204 of C. trachomatis encodes a 471-residue 2-oxoglutarate permease that has 16 Trp residues (ranking 11th in Trp burden). It ranks ninth in p/P Trp ratio, with a ratio of 3.57. Importantly, this translocator has locally concentrated placements of Trp residues, including one region that has 6/24 Trp residues (with one run of three consecutive Trp residues). Orthologs in other organisms also have high Trp contents, and many of the Trp residues are highly conserved. The E. coli ortholog also has a high Trp content of 16 residues and has a p/P Trp ratio of 2.2. It thus appears that in Chlamydiaceae, a crucial permease that already had a characteristically high Trp content needed no modification to be responsive to Trp limitation. Surprisingly, this important gene is located on the lagging strand of replication (see below). The “Ca. Protochlamydia amoebophila” proteome seems not to possess an ortholog of this chlamydial transporter, as it did not return a bidirectional best hit to the CT204 query.
SELF-LIMITATION OF Trp IMPORT CAUSED BY Trp STARVATION
The TyrP family of bacterial transport proteins includes enzymes that transport tyrosine or Trp with differing specificities (115). In a phylogenetic tree of these family proteins, E. coli TyrP (tyrosine-specific permease) diverges from clusters centered at nodes for Mtr (high-affinity Trp transport) and TnaB (low-affinity Trp transport). The about-intermediate position of TyrP proteins from Chlamydiaceae in this tree, between the Tyr and Trp groups, is shown in Fig. 9 of the work of Xie et al. (139). The transport specificity of TyrP proteins from Chlamydiaceae has not been elucidated experimentally, but it seems likely that these might be broad-specificity transporters for Trp, tyrosine, and probably phenylalanine.
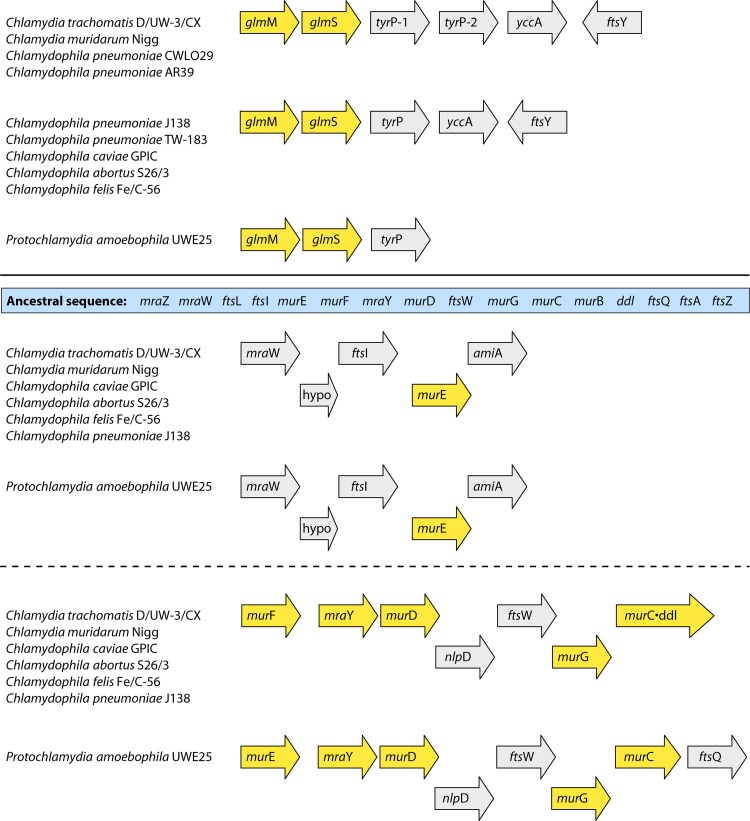
Fig 9.
Intriguing gene cluster arrangements that have functional implications. The top panel illustrates the linkage of glmM and glmS, the first two genes of the lipid II pathway from fructose-6-P (see Fig. 7), with tyrP in species of Chlamydiaceae and “Ca. Protochlamydia amoebophila.” The TyrP gene product is a Trp transporter. In the top grouping, two species of Chlamydia and two strains of Chlamydophila pneumoniae have tandem gene duplicates of tyrP, designated tyrP-1 and tyrP-2. In addition, the Chlamydiaceae organisms possess two linked genes that function in cell division; these genes are not linked to glmM and glmS in “Ca. Protochlamydia amoebophila.” Genes that are depicted as yellow arrows encode enzymes whose functional roles are illustrated in Fig. 7. In the bottom panel, two linkage regions (separated by a dashed line) are shown which have become separated from the extensive putative ancestral sequence of 16 genes given in blue at the top of the panel. Hypo, hypothetical protein gene.
The TyrP transporter for chlamydial import of Trp from the host is itself a high-Trp protein. This is not surprising per se because Trp residues in transmembrane proteins have special properties that can facilitate long-distance functional associations on opposite sides of the membrane. For example, in the staphylococcal multidrug transporter QacA, a number of Trp residues appear to be located at the periphery of transmembrane segments, where they guide the depth of the transmembrane insertion (47). Aromatic stacking interactions accommodated by Trp residues may also assist the stable binding of aromatic substrates. The high counts of both Tyr and Trp residues in the transmembrane domains of integral membrane proteins have also been asserted to have been selected for their antioxidant functions (92). It appears that Tyr is as effective as Trp for exercise of the antioxidant function, and thus selection of Tyr residues for their antioxidant function might generally be the least expensive evolutionary choice. Trp and Tyr have also been shown to have largely equivalent properties at the membrane-water interface region of membrane proteins, especially in their properties as membrane anchors (41).
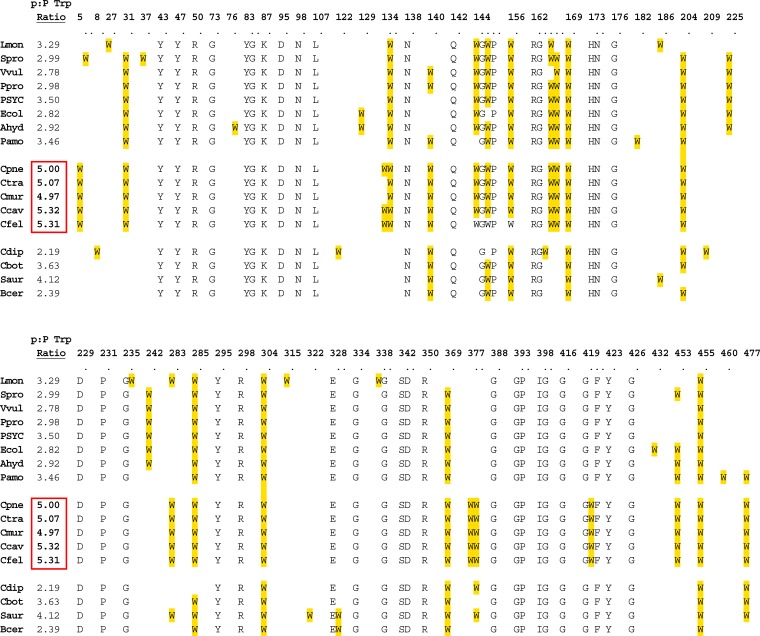
Although TyrP proteins are generally high-Trp proteins, as might be expected for the foregoing reasons, it is striking that TyrP proteins from the Chlamydiaceae have undergone up-Trp selection for an even higher Trp content. The Trp contents of the same set of aromatic permeases displayed in the tree of Xie et al. (139) are shown in Fig. 6. In this figure, the distribution of Trp residues is shown in an alignment of TyrP proteins, and the ratio of the Trp content of TyrP to the Trp content of the proteome (p/P Trp ratio) is given at the left. Only the chlamydial Trp residues at positions 84 and 289 are highly conserved throughout the alignment, and it seems qualitatively obvious that selection has favored the recruitment of far more Trp residues in TyrP proteins of chlamydial organisms than would otherwise be expected. TyrP in “Ca. Protochlamydia amoebophila” has also undergone up-Trp selection, exhibiting a p/P Trp ratio of 2.19 (not shown in Fig. 6). Hence, it appears that an event of up-Trp selection in the TyrP transport protein occurred in the common ancestor of all of the Chlamydiaceae and “Ca. Protochlamydia amoebophila.” This observation, together with other examples described in this review, indicates that Trp availability became a signaling feature between early chlamydial intracellular auxotrophs and their hosts prior to divergence of the Chlamydiaceae.
Fig 6.
Distribution of Trp residues of the TyrP transporters from Chlamydiaceae family organisms (top group) and from other organisms shown in the phylogenetic tree of Xie et al. (139). Each number in the column labeled “p:P Trp Ratio” is the ratio of the Trp content of the TyrP transporter to the Trp content of the proteome. The genomic abbreviations, from top to bottom, are as follows: Cpne, Chlamydophila pneumoniae CWL029; Ccav, Chlamydophila caviae GPIC; Cmur, Chlamydia muridarum Nigg; Ctra, Chlamydia trachomatis D/UW-3/CX; Ecol, Escherichia coli K-12; Sent, Salmonella enterica subsp. enterica serovar Typhi CT18; Paer, Pseudomonas aeruginosa PAO1; Ypes, Yersinia pestis CO92; Hinf, Haemophilus influenzae Rd KW20; Pmul, Pasteurella multocida subsp. multocida Pm70; Nmen, Neisseria meningitidis MC58; Ngon, Neisseria gonorrhoeae FA 1090; Hduc, Haemophilus ducreyi 35000HP; Vcho, Vibrio cholerae O1 bv. eltor N16961; and Npun, Nostoc punctiforme PCC 73102. Ecol-1, Ecol-2, and Ecol-3 have been annotated TyrP (b1907), Mtr (b3161), and TnaB (b3709), respectively. The remaining sets of gene IDs and functional designations for the nonchlamydial organisms are as follows: Hduc-1, HD1875 and TyrP; Hduc-2, HD1499 and TyrP; Hduc-3, HD0648 and TnaB; Hinf-1, HI0528 and TyrP; Hinf-2, HI0477 and TyrP; Hinf-3, HI0287 and Mtr; Ngon, NG02073 and Mtr; Nmen, NMB2031 and Mtr; Npun, Npun_F6003 and TyrP; Pmul-1, PM0732 and TyrP; Pmul-2, PM0810 and TyrP; Pmul-3, PM1192 and Mtr; Pmul-4, Pm1419 and TnaB; Paer-1, PA5434 and Mtr; Paer-2, PA1916 and TyrP; Paer-3, PA3766 and TyrP; Sent-1, STY2145 and TyrP; Sent-2, STY3460 and Mtr; Vcho-1, VCA0772 and TyrP; Vcho-2, VCA0160 and TnaB; Ypes-1, YP01285 and Mtr; and Ypes-2, YP01209 and TyrP.
It seems additionally significant that there is often (but not always) more than one TyrP paralog in chlamydiae, encoded by tandem genes. This is the case for C. trachomatis, C. muridarum, and some strains of Cp. pneumoniae. A phylogenetic tree (data not shown) indicates that a gene duplication occurred in the common ancestor of C. trachomatis and C. muridarum. Gieffers et al. (40) noted that respiratory strains of Cp. pneumoniae (e.g., strains CW and AR) possessed two paralogous copies of tyrP, whereas vascular strains (e.g., strains J and T) had a single tyrP gene. The gene duplication in Cp. pneumoniae strains occurred very recently, independent of the more ancient gene duplication in C. trachomatis and C. muridarum. It was suggested (40) that the vascular tropism and a greater tendency to exist in the persistent state might be tied closely to a lesser capability for Trp import from the host. Recently, Carlson et al. (24) showed that a newly sequenced genome of an oculotropic strain of C. trachomatis (A/HAR-13) possessed a disrupted second copy of the tyrP gene. Thus, it seems that a correspondence of tyrP copy number and tissue tropism may also apply to C. trachomatis, as is the case in Cp. pneumoniae. It is interesting to consider that whereas the expression of two TyrP paralogs should promote increased competence for Trp import during Trp sufficiency (and this has indeed been shown [40]), it also would increase the Trp burden during Trp insufficiency.
CHLAMYDIAL OPERONS ENCODING PARTIAL PATHWAYS OF Trp BIOSYNTHESIS
Chlamydial organisms exhibit an incomplete set of Trp pathway genes that varies in phylogenetic distribution, ranging from having no Trp pathway genes at all (e.g., Cp. pneumoniae) to having a trpREbEa operon (e.g., C. trachomatis) or having a nearly complete Trp pathway which is nevertheless missing the initial two genes, encoding subunits of anthranilate synthase (e.g., Cp. caviae) (139). Some strains of C. trachomatis have been shown (35) to deploy a functional tryptophan synthase (consisting of TrpEb and TrpEa), regulated by a trpR repressor, for scavenging of niche-available indole. This was convincingly asserted to be the basis for the tissue tropism of genital strains. The low Trp content of the TrpR, TrpEb, and TrpEa proteins is consistent with indole scavenging for recycling to Trp under conditions of Trp limitation. Regulation of trpREbEa by trpR via repression and attenuation has been studied (1, 25).
In 2002 (139), we described the unusual 8-member trp operon of Cp. caviae: trpRBDCEbEa-kynU-prsA. At that time, Cp. caviae was classified as Chlamydophila psittaci and kprS was used as an alternative name for prsA. We used the convention of naming the genes in the order of the enzymatic steps encoded, as implemented comprehensively in reference 141. This nomenclature is also used in the annotations at the SEED database, a major resource for this analysis to which each query entry in Table S1 at http://semiglobe.lanl.gov/supplement.php is hyperlinked to the appropriate SEED page. Under this logical nomenclature, the alpha and beta subunits of TrpE (tryptophan synthase) are named TrpEa and TrpEb, respectively. The operon (and the genome) lacks genes encoding the two subunits of anthranilate synthase, and therefore an enzymatic link to Trp from chorismate is missing in Cp. caviae. Unlike the case for any other known trp operon, two genes encoding enzymes outside the linear pathway of Trp biosynthesis have been incorporated. It was suggested that the gene products of this unique operon could allow the circuitous recovery of the equivalent of lost Trp that had been degraded by the host to kynurenine. The key intercepting enzyme is kynureninase (encoded by kynU), which diverts host kynurenine to pathogen anthranilate, the initial precursor needed by the chain of chlamydial enzymes to complete the subsequent steps of Trp biosynthesis. The final gene of the complex operon is prsA, encoding phosphoribosyl pyrophosphate (PRPP) synthase. This enzyme is necessary for the generation of PRPP, used in the reaction catalyzed by TrpD. Both kynU and prsA are not present elsewhere in the Chlamydiaceae. Subsequent experimental work (136) has provided evidence that Trp recycling explains the resistance of Cp. caviae to the effects of IFN-γ in a heterologous system (human host cells). Although the p/P Trp ratios for TrpRBDCEbEa and PrsA are all quite low (with TrpD, TrpEa, and PrsA being zero-Trp proteins), the key recycling enzyme, kynureninase, has a very high p/P Trp ratio (1.74; 7 Trp residues in 425 amino acids). In addition, the PRPP synthase utilizes ATP in a reaction in which two high-energy equivalents are consumed in the production of PRPP and AMP. This suggests that this novel operon may not exist simply as a straightforward mechanism to completely circumvent the Trp depletion strategy of guinea pig host cells. Rather, we suggest the possibility that under conditions where severe Trp limitation has been achieved in vivo, this recycling mechanism may be geared to jump-start metabolic activity only as host Trp becomes newly available to persistent cells. Transition from quiescent persistence to rapid growth will initially be limited by renewal of the chlamydial Trp transporters and the ATP transporter. Translation of kynureninase in concert with increased availability of ATP for PRPP synthase could be the essential features needed to boost overall Trp availability in a concerted way. The transporter for kynurenine is unknown, but the foregoing scenario would be assisted greatly if the kynurenine transporter is a low-Trp protein. Note that the above scenario assumes that the guinea pig host induces IDO in response to IFN-γ, as in humans. If this direct response of Trp levels to IFN-γ is absent in the guinea pig host, as in the case of the mouse host, essentially the same outcome could prevail indirectly due to the IFN-γ/GTPase/cytotoxin/Trp depletion mechanism that was proposed earlier for the mouse host.
DISABLEMENT OF MULTICOMPONENT PATHWAYS AT KEY ACHILLES HEEL STEPS
Iron Transport
Persistence can be induced by at least four different experimental manipulations in cultured human host cells. Although the overall aberrant morphologies elicited are similar, significant differences at the molecular and metabolic levels have been described (67). The true state of in vivo persistence may differ in some respects from each of the model systems used to study persistence. Nevertheless, the manipulated variables that can trigger entry into a persistent state offer important clues to the nature of in vivo persistence. Thus, the particular importance to pathogenic chlamydia of acquisition of iron from host cells, for example, is indicated by the entry of RB populations into a persistent state following the experimental restriction of iron alone (109).
In cases where an entire multistep pathway is functionally prominent during rapid proliferation, the Trp depletion cue that mutes that pathway in concert with the transition to persistence could work by affecting just one (or a few) key component of the system. For example, C. trachomatis possesses a 4-gene operon encoding components of an iron transport system. These genes overlap or are closely adjacent. YtgA (CT067) is a periplasmic iron-binding lipoprotein, and YtgB (CT068) and YtgC (CT069) are transmembrane permeases that probably form the transport pore. YtgD (CT070) is an ATP-binding lipoprotein that energizes transport. YtgA, reported to be highly immunogenic, has been studied experimentally (89). YtgA and YtgB exhibit only slightly elevated p/P Trp ratios, of 1.28 and 1.21, respectively. However, the 451-residue YtgC enzyme possesses 11 Trp residues, yielding a p/P Trp ratio of 2.56. It ranks highly by both the p/P Trp ratio and Trp burden criteria. Most strikingly, YtgC displays one of the most notable Trp hot-spot distribution patterns of the C. trachomatis proteome (WWW and WKTGW). If a translating ribosome is stalled within the ytgC transcript because of a charged-Trp deficiency, this could stall translation of the preceding two genes. With respect to the downstream ytgD gene, the untranslated transcript region could be subject to degradation. Alternatively, the pausing could favor an RNA secondary structure that controls downstream transcription or translation, as exemplified by the variety of Trp pathway mechanisms reviewed recently (86).
It is quite likely that translational pausing within YtgC during Trp starvation might render the multigene transcript vulnerable to degradation. The deployment of additional Trp residues in YtgC is “balanced” by the zero-Trp content of YtgD. Importantly, the 4-gene operon is also present in the control genome of “Ca. Protochlamydia amoebophila.” The up-Trp selection that must have occurred in C. trachomatis YtgC is convincing because the “Ca. Protochlamydia amoebophila” protein has only 4 Trp residues (compared to 11 in the C. trachomatis protein), yielding a p/P Trp ratio of 0.78 (compared to 2.56 in C. trachomatis). On the other hand, the down-Trp selection in YtgD of C. trachomatis is convincing because the “Ca. Protochlamydia amoebophila” protein has 3 Trp residues, giving a p/P ratio of 0.79. Of the four Ytg proteins, E. coli has a bidirectional hit homolog only for YtgB.
It is further compelling that since YtgA and YtgD are lipoproteins, they are additionally vulnerable to Trp limitation because the three genes of lipoprotein biosynthesis that accomplish the crucial posttranslational modifications are conspicuously high in Trp content (see later discussion of the slave-master relationships in lipoprotein biosynthesis).
The Sodium Ion Cycle
Dibrov et al. (32) have discussed the interesting paradox of the chlamydial Na+ cycle and asserted a rationale in support of a crucial role for a sodium gradient in the intracellular life cycle. The primary sodium pump of chlamydial organisms is a Na+-translocating NADH:ubiquinone oxidoreductase (NQR) comprised of six subunits encoded by a four-gene operon plus two isolated genes (all located on the lagging strand of replication [see below]). Other chlamydial elements of the sodium cycle include a sodium-specific A1A0-type ATPase, a Na+-H+ antiporter, and various Na+-dependent transporters.
The Na+- and H+-transporting proteins of C. trachomatis identified by Dibrov et al. (32) are enumerated in Table 3, where Trp content is expressed as p/P Trp ratios. The various C. trachomatis proteins are compared with their bidirectional best-hit orthologs from “Ca. Protochlamydia amoebophila” and E. coli, as retrieved from Table S1 at http://semiglobe.lanl.gov/supplement.php. Phylogenetically, this is a near-neighbor comparison and a distant-neighbor comparison. Table 3 shows that the primary H+ pump, which coexists with the Na+ ion cycle, consists of two subunits which both have very high p/P Trp ratios. These ratios are also rather high in “Ca. Protochlamydia amoebophila” and E. coli. Thus, it appears that proton motive force as a source of energy might be generally sensitive to Trp limitation. The six subunits of the primary NQR sodium pump in both C. trachomatis and “Ca. Protochlamydia amoebophila” exhibit generally high p/P Trp ratios, yielding what would seem to be a distinct cumulative up-Trp selection in the common ancestor of pathogenic chlamydiae and “Ca. Protochlamydia amoebophila” (C. trachomatis has a cumulative 32 Trp residues and “Ca. Protochlamydia amoebophila” has 29 Trp residues in the 6-subunit complex, a heavy Trp burden for a single metabolic entity). E. coli returned only four bidirectional best hits to the six C. trachomatis query proteins, and while one of these (b1630) possessed a high p/P Trp ratio, the remaining three exhibited low values. The four E. coli proteins differ functionally from their chlamydial Na+-translocating NADH:ubiquinone oxidoreductase counterparts in that they are involved in the NADH-quinone reductase for reduction of SoxR. These two redox systems appear to have diverged long ago from a common oxidoreductase. The CT280-b1632 and CT281-b1627 pairs encode proteins of similar size and are homologous throughout. On the other hand, CT278 and CT740 encode proteins that are significantly larger than those encoded by their b1630 and b3844 E. coli counterparts. In this case, homologous alignment is only fractional.
Table 3.
Phylogenetic comparison of Na+ ion cycle membersa
| Chlamydia trachomatis gene | p/P Trp ratio | “Ca. Protochlamydia amoebophila” |
Escherichia coli |
||
|---|---|---|---|---|---|
| Gene | p/P Trp ratio | Gene | p/P Trp ratio | ||
| Primary Na+ pump genes | |||||
| CT278 | 2.08 | PC0301 | 1.73 | b1630 | 2.43 |
| CT279 | 1.66 | PC0300 | 1.14 | ||
| CT280 | 1.47 | PC0299 | 0.83 | b1632 | 0.85 |
| CT281 | 2.15 | PC0298 | 1.79 | b1627 | 0.68 |
| CT634b | 0.22 | PC0095 | 0.19 | ||
| CT740 | 1.95 | PC1533 | 1.33 | b3844 | 0.84 |
| Primary H+ pump genes | |||||
| CT013 | 3.54 | PC1630 | 2.66 | b0978 | 2.30 |
| CT014 | 2.08 | PC1629 | 1.71 | b0979 | 2.95 |
| Na+-specific A1A0-type ATPase genes | |||||
| CT303 | 3.07 | PC1659 | 1.50 | ||
| CT304 | 0 | PC1676 | 0.63 | ||
| CT305 | 0.81 | PC1677 | 1.66 | ||
| CT306 | 0.52 | PC1678 | 0.41 | ||
| CT307 | 0.72 | PC1679 | 0.60 | ||
| CT308 | 1.24 | PC1680 | 0.89 | ||
| CT309 | 2.37 | PC1681 | 1.01 | ||
| CT310 | 1.01 | PC1682 | 0 | ||
| Na+/H+ antiporter gene | |||||
| CT857 | 2.50 | ||||
| Na+-dependent transporter genes | |||||
| CT409 | 1.16 | PC1598 | 1.17 | b0007 | 1.93 |
| CT435 | 1.15 | PC0622 | 1.35 | ||
| CT401 | 0.51 | PC1785 | 0.42 | b4077 | 0.90 |
| CT230 | 0.76 | PC1734 | 0.84 | ||
| CT204 | 3.39 | b0770 | 2.20 | ||
| CT554 | 1.27 | PC0767 | 0.22 | b0401 | 1.20 |
| CT231 | 3.43 | PC1260 | 2.84 | ||
Values in bold indicate high p/P Trp ratios.
The CT634 gene is a target of the iron-responsive regulator encoded by dcrA at the CT296 locus (but see discussion of possible uncertainties in “Regulation by Chlamydial DcrA” in the text). DcrA has a very high Trp content (p/P Trp ratio = 2.72).
The 8-subunit Na+-specific A1A0-type ATPase exhibits a somewhat variable p/P Trp ratio in comparing C. trachomatis and “Ca. Protochlamydia amoebophila,” but the overall cumulative Trp burdens for this complex in the two proteomes are the same (29 Trp residues). Thus, this ATPase complex, like the aforementioned primary sodium pump, appears to have undergone substantial up-Trp selection in an early chlamydial ancestor. Among the Na+-dependent transporters listed at the bottom of Table 3, two are noteworthy for their high Trp content. CT204 encodes an all-important dicarboxylate (2-oxoglutarate and oxaloacetate) transporter of the sodium-sulfate symporter family which was discussed earlier. CT231 encodes an uncharacterized transporter of the neurotransmitter-sodium symporter family.
Type III Secretion
Extensive type III secretion systems comprise numerous and functionally diverse virulence determinants that underlie the pathobiology of chlamydiae (62, 104). In C. trachomatis, a conserved chlamydial gene organization was demonstrated whereby 37 genes are distributed into 10 operons within six genetic regions (48). Most of the gene products possess moderate or very low Trp contents. However, in the CT557-to-CT564 region (a complex operon having an internal promoter), translocation proteins S and T have p/P Trp ratios of 2.23 (CT563) and 2.55 (CT564), respectively. Up-Trp selection that is Chlamydiaceae specific is strongly indicated, since the corresponding “Ca. Protochlamydia amoebophila” bidirectional best hits have values of only 1.13 and 0.63, respectively. The CT559 and CT560 gene products also have rather high p/P Trp ratios, of 1.61 and 1.88, respectively (in comparison with 1.04 and 1.26 in “Ca. Protochlamydia amoebophila”). The remaining four gene products have low Trp contents. Since type III secretion machineries are multicomponent structures requiring various molar ratios of the protein components (48), a requirement facilitated by operon organization, any discontinuities with respect to Trp content could disrupt function during Trp starvation. In one other case, CT091 to CT087 encode five proteins, the first four of which have low p/P Trp ratios (0.28, 0.74, 0, and 0.72). However, the final translation product is a homolog of MalQ, which is, in fact, one of the highest-Trp-content proteins in chlamydia. This does not appear to be the consequence of up-Trp selection, since many of the Trp residues are conserved in multiple alignments. Indeed, the “Ca. Protochlamydia amoebophila” MalQ protein is even more extreme than C. trachomatis MalQ. It has a p/P Trp ratio of 3.65 (ranked first) and has 23 Trp residues (ranked second in Trp burden). The specific role of MalQ in the secretion machinery, if any, would be interesting to know. Perhaps it was recruited as a protein that was already endowed with a high Trp content. If MalQ participates in critical protein-protein interactions with some or all of the other gene products generated by the operon, it could mediate an overall disruption of operon function under conditions of Trp limitation. Five of the six gene regions encoding type III secretion, including the last two, are located on the lagging strand of replication. This suggests that they may be expressed primarily or exclusively at developmental times when DNA replication is minimal.
The type III secretion system is also an example of what we call a master-slave relationship (see the related section below). In such cases, multiple proteins (slave proteins) whose function depends upon a master protein can be influenced negatively by Trp limitation without an expensive requirement for a high Trp content of the slave proteins. In this example, the master protein is actually a key set of proteins, namely, those representing the Achilles heel vulnerabilities of the secretion system. C. trachomatis and other Chlamydiaceae utilize the type III secretion system to deliver many effector proteins directly into the cytosol of host cells. To the extent that the type III secretion system is disabled under conditions of Trp limitation, the entire complement of effector proteins will fail to be exported. One important example is the C. trachomatis protein encoded at the CT456 locus and called Tarp (translocated actin-recruiting protein). Tarp functions as a multivalent phosphorylation-dependent signaling hub that is crucial for success in the early phase of chlamydial infection (27) and was recently shown to interact with the human adaptor protein SHC1 (85). Variations in chlamydial Tarp exhibit interesting correlations with particular variations of clinical phenotype (76). Given the biology of the persistent state, one would not expect Tarp to be competent for export during persistence and the attending Trp limitation. Even though C. trachomatis Tarp has a lower-than-average p/P Trp ratio of 0.62, curtailment of the type III export machinery due to Trp limitation would be an overriding feature to explain the lack of Tarp export. The relatively high Trp burden (6 Trp residues in 1,005 amino acids) might be another factor. Another type III secretion effector protein is encoded by CT621 (53). It also has a low Trp content (p/P Trp ratio = 0.38) but differs from Tarp in being secreted late in the developmental cycle.
MULTIPLE Trp HURDLES IN THE CELL DIVISION PATHWAY
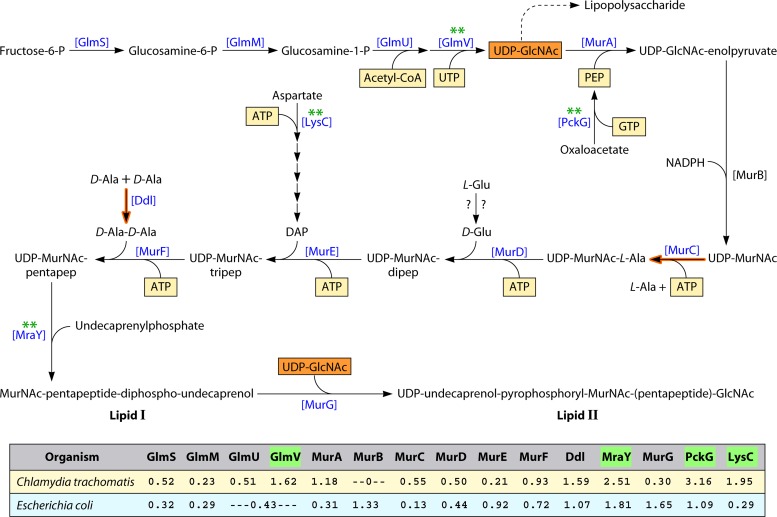
Overview of the Lipid II Pathway
The foregoing are examples of selection of high Trp content within one or more vulnerable links of a multicomponent system that could significantly mute the entire system. Such isolated and seemingly extravagant expenditures of Trp may be offset by selections of low Trp content elsewhere in the system in order to ameliorate the overall proteomic Trp burden. This phenomenon of up-Trp selection events at Achilles heel focal points that are counterbalanced by down-Trp selection events is seen in other complex and even more expansive systems, as illustrated here in the case of the pathway for cell division. It was initially enigmatic that genes encoding enzymes of peptidoglycan biosynthesis are present in chlamydial organisms, since cell walls are not present (121). However, it now seems clear that the main portion of the peptidoglycan pathway, up to the lipid II [UDP-undecaprenol-pyrophosphoryl-MurNAc-(pentapeptide)-GlcNAc] precursor, is a dual-function pathway, with the aforementioned precursors being shared metabolites for the pathways of cell division and peptidoglycan biosynthesis. Hence, in chlamydial organisms, it appears that the lipid II pathway is needed only for cell division. Henrichfreise et al. (51) have discussed the interesting parallels that exist with respect to the lipid II pathway between the pathogenic chlamydiae and Wolbachia (mutualistic endobacteria of many arthropods and filarial nematodes).
Figure 7 illustrates the lipid II pathway as it extends from UDP-GlcNAc. UDP-GlcNAc (shown with orange highlighting) is a key metabolite, being located at a branchpoint which also diverges to lipopolysaccharide biosynthesis. The enzymatic steps required for the origin of UDP-GlcNAc from fructose-6-P, in effect the common trunk of a branched pathway, is shown as well. The overall pathway is enormously demanding of energy input, requiring acetyl-CoA, UTP, GTP, PEP, and multiple molecules of ATP, as emphasized with yellow highlighting in Fig. 7. The table at the bottom of Fig. 7 shows a comparison of enzymic/proteomic (p/P) Trp ratios for the enzymatic steps of the cell division pathway in C. trachomatis and E. coli. The steps which seem to impose a particular hurdle for pathway flow under conditions of Trp limitation are marked with double green asterisks. A single Trp hurdle exists in the common trunk, at the last step of that segment, which is catalyzed by GlmV. In E. coli, the GlmU and GlmV reactions are carried out by a single bifunctional enzyme called GlmU. The GlmV enzyme of C. trachomatis is not homologous with the C-terminal portion of E. coli GlmU. GlmV exemplifies a case where a high-Trp analog has evolved as a functional replacement of a low-Trp analog. A number of such up-Trp (or down-Trp) replacements are enumerated in this article. The extent to which the GlmV Trp hurdle in C. trachomatis decreases the availability of UDP-GlcNAc for the MurA reaction could be a crucially sensitive element of pathway output diminution, since UDP-GlcNAc flow to MurA is not only competed for by the lipopolysaccharide pathway but also utilized a second time in the cell division pathway, at the MurG step, which converts lipid I to lipid II. Figure 7 shows the utilization of d-glutamate and d-alanine as components of the pentapeptide moiety at the MurD and MurF steps, respectively, but it is not certain whether these d-amino acids are actually formed endogenously or acquired from the host. If l-amino acids are perhaps used instead, then the chlamydial pentapeptide composition might differ from that of most bacteria. MraY, which forms lipid I, has a very high p/P Trp ratio (2.51), compared to a ratio of 1.81 in E. coli.
Fig 7.
The lipid II pathway in Chlamydiaceae. The pathway from fructose-6-P to UDP-GlcNAc branches divergently to feed the lipopolysaccharide pathway and the lipid II pathway. The lipid II pathway is synonymous with the peptidoglycan pathway prior to the various cross-linking reactions, and the lipid II pathway is also required for cell division. The genes encoding MurC and Ddl (indicated with red arrows) are fused in the Chlamydiaceae, and the Trp contents of the two domains are shown separately in the lower table for comparison with E. coli, in which the domains are not fused. Four enzymes that exhibit particularly high demand for Trp are marked with double green asterisks in the figure and highlighted green in the lower table. The sequence of reactions catalyzed by MurCDEF are similar ATP-dependent reactions that build the pentapeptide moiety of UDP-MurNAc-pentapeptide. It is presently uncertain if the d-amino acid components of the pentapeptide are possibly replaced by another amino acid, since the genes encoding the racemase enzymes have not been identified in Chlamydiaceae. The three alternative fates of UDP-GlcNAc (two in the lipid II pathway and one in the lipopolysaccharide pathway) are indicated by the use of orange highlighting. Metabolite abbreviations: UDP-GlcNAc, UDP-N-acetylglucosamine; UDP-MurNAc, UDP-N-acetylmuramic acid. In the bottom table, the ratios of the Trp contents of the various enzymes depicted to the Trp content of the C. trachomatis proteome are compared with the corresponding ratios of E. coli. Note that E. coli lacks PckG, instead utilizing a functional analog called PckA. Also, the functions of GlmU and GlmV are carried out by a single protein in E. coli, one which did not yield any bidirectional best hits with GlmU and GlmV queries of C. trachomatis (see the text).
PEP availability seems to be a crucial factor influencing the extent to which certain enzyme steps are Trp hurdles. PEP, which is needed for the MurA step, is produced by GTP-dependent PEP carboxykinase (PckG). PckG ranks 17th in the C. trachomatis genome, with its high p/P Trp ratio of 3.16. Because it is a large protein (599 amino acids), it ranks even higher (9th) on the Trp burden list. In addition to considerations of the effect of Trp limitation upon the translation of PckG, the successful generation of PEP must be impacted heavily by the likely diminution of the oxaloacetate and GTP substrates, which require import by the Trp-rich transporters (dicarboxylate transporter and the broad-specificity nucleoside triphosphate transporter discussed earlier). A second substrate entering the pathway at the MurE step, diaminopimelate (DAP), is likely a significant hurdle in view of the high p/P Trp ratio of the C. trachomatis aspartokinase (LysC). Both LysC and PckG exhibit markedly higher p/P Trp ratios than the corresponding E. coli enzymes (see the bottom of Fig. 7). (Recall from an earlier section that PckG is a Trp-rich analog replacement for PckA, a nonhomologous replacement which occurred in the common ancestor of “Ca. Protochlamydia amoebophila” and the Chlamydiaceae.) The DAP pathway branch is discussed more fully in the next section. Translational discontinuities dictated by the various Trp hurdles can be expected to generate peptidoglycan fragments. This is of considerable interest because such modifications of the structure of peptidoglycan have been discussed widely in the context of pathogen strategies to avoid recognition by the host innate immune system (135).
For clarity of presentation, “Ca. Protochlamydia amoebophila” was not included in the table given at the bottom of Fig. 7. “Ca. Protochlamydia amoebophila” p/P Trp ratios for most of the enzymes listed in the table are generally <1.0. However, several of the C. trachomatis Trp hurdles are also Trp hurdles for “Ca. Protochlamydia amoebophila,” implicating an ancient up-Trp selection for these steps. The following enzymological features of variation that distinguish C. trachomatis and other Chlamydiaceae from E. coli occurred in the common Chlamydiales ancestor. (i) GlmU and GlmV are present as separate proteins in all of the Chlamydiales, including “Ca. Protochlamydia amoebophila” and representatives of two other families (Simkania negevensis and Waddlia chondrophila). (ii) All of the Chlamydiales utilize the GTP-dependent PckG enzyme rather than the ATP-dependent PckA enzyme. The following enzymological features of variation occurred after the divergence of the Chlamydiaceae family from other families of the order. (i) Fusion of the murC and ddl genes exists in all of the Chlamydiaceae. Curiously, this fusion is also present in Parachlamydia ancanthamoebae but not in “Ca. Protochlamydia amoebophila,” even though both are currently classified as genera belonging to the family Parachlamydiaceae. (ii) Except for the Chlamydiaceae, all of the other Chlamydiales families possess an unusual LysC aspartokinase having a catalytic domain but no allosteric domain. A common ancestor of the Chlamydiaceae may have acquired a regulated aspartokinase via LGT (see below). In terms of structure (but not Trp content), LysC of Chlamydiaceae is more like that of E. coli than like those of its sister Chlamydiales members.
How does “Ca. Protochlamydia amoebophila” compare with C. trachomatis, with respect to the four Trp hurdles highlighted by double green asterisks in Fig. 7? GlmV in “Ca. Protochlamydia amoebophila” appears to approach that of C. trachomatis as a Trp hurdle, having a p/P Trp ratio of 1.40. PckG should be a strong Trp hurdle in “Ca. Protochlamydia amoebophila,” exhibiting a p/P Trp ratio of 2.84. Note in Fig. 7 the potential cooperative impact of the fact that GlmV and PckG converge upon the MurA reaction, as both are essential sources of substrate. One could generalize that the ancient chlamydial ancestor already had GlmV and PckG in place as Trp hurdles and that this was followed after divergence of the Chlamydiaceae by some strengthening of these hurdles in this family, as well as by addition of MraY and LysC as further reinforcing Trp hurdles.
Diaminopimelate Biosynthesis
In Fig. 7, the six-step pathway to DAP is shown as the chlamydial pathway instead of the canonical eight-step pathway of E. coli. Until recently, Chlamydiaceae appeared to lack an intact pathway to DAP, since genes encoding orthologs of DapD, DapC, and DapE were absent. However, certain organisms, such as higher plants, cyanobacteria, and the Chlamydiaceae, possess a single aminotransferase (DapL) which carries out the same overall conversion from l-tetrahydrodipicolinate to l,l-diaminopimelate that is accomplished by the DapD-DapC-DapE enzyme trio (83). In Chlamydiaceae, the six-step pathway to DAP is the sole surviving remnant of the reductive evolution process which eliminated the bulk of the multibranched aspartokinase network. The pathway remaining is a simple linear pathway initiated with aspartate plus ATP and ending with the DAP end product. Since DAP decarboxylase is absent, DAP cannot function as a precursor of lysine. Since this simplified Chlamydiaceae pathway arrangement is also seen in “Ca. Protochlamydia amoebophila” (Parachlamydiaceae) and known representatives of other chlamydial families, this simplified pathway appears to be a shared evolutionary feature that extends to the taxon level of order.
Table 4 shows a comparison of the p/P Trp ratios of various chlamydial DAP enzymes with the corresponding DAP enzymes present in E. coli and “Ca. Protochlamydia amoebophila.” LysC in the Chlamydiaceae has a distinctly higher Trp content than those in both E. coli and “Ca. Protochlamydia amoebophila” (indicated in bold in Table 4). The most striking Trp hurdle seems to be the DapA enzyme, which has a very high Trp content in Chlamydiaceae that is almost an order of magnitude more than that seen in both E. coli and “Ca. Protochlamydia amoebophila” (also shown in bold in Table 4). It seems clear that up-Trp selection in DapA of Chlamydiaceae has occurred, since the vast majority of the Trp residues are not conserved for generally important functional roles in catalysis. Since the earlier enzyme, LysC, is also a Trp hurdle, the capability of DapA is likely further compromised due to limitation of substrate.
Table 4.
Trp content of biosynthetic pathway to DAP in Chlamydiaceae species, E. coli, and “Ca. Protochlamydia amoebophila”
| Organism | Ratio of protein Trp content to proteome Trp content (p/P Trp ratio) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| LysC | Asd | DapA | DapB | DapL | DapD | DapC | DapE | DapF | |
| Chlamydia trachomatis D/UW-3/CX | 1.95 | 0.95 | 2.58 | 0 | 0.80 | 1.15 | |||
| Chlamydia muridarum Nigg | 1.42 | 0.93 | 2.52 | 0 | 0.78 | 0.77 | |||
| Chlamydophila felis Fe/C-56 | 0.72 | 1.26 | 2.54 | 0.86 | 0.57 | 1.93 | |||
| Chlamydophila abortus S26/3 | 1.18 | 1.25 | 2.89 | 0.85 | 0.78 | 1.60 | |||
| Chlamydophila caviae GPIC | 1.20 | 1.26 | 2.55 | 0.86 | 0.80 | 1.62 | |||
| Chlamydophila pneumoniae CWL029 | 0.90 | 1.19 | 2.38 | 0.75 | 0.75 | 1.54 | |||
| “Candidatus Protochlamydia amoebophila” | 0.33 | 1.24 | 0.29 | 1.21 | 0.84 | 1.97 | |||
| Escherichia coli K-12 | 0.29 | 1.42 | 0.22 | 0.24 | 0.72 | 0.32 | 1.05 | 0.24 | |
Values in bold indicate low p/P Trp ratios.
Aspartokinase (LysC) merits particular attention. Aspartokinases generally tend to contain relatively few Trp residues, and they exemplify the occasional protein that has low Trp content across all phyla. The average p/P Trp ratio for aspartokinases is only 0.33, and even an aspartokinase with a p/P Trp ratio of 1.0 is conspicuously high (Fig. 8). Zero-Trp aspartokinases are not unusual, and we generated a list of 31 genomes that produce an aspartokinase having zero-Trp content (see Table S7 at http://semiglobe.lanl.gov/supplement.php). These genomes vary from one with a genomic GC% of only 17% (“Candidatus Carsonella ruddii” PV) to one with a genomic GC% of 72% (Streptomyces griseus subsp. griseus NBRC 13350). The values given in the Table 4 LysC column exhibit a fair amount of variability for Chlamydiaceae, but the Trp contents are distinctly greater than that seen in either E. coli or “Ca. Protochlamydia amoebophila.” The aspartokinases (518 sequences from 339 genomes) were recently studied thoroughly by cohesion group analysis (74), a methodology which sorts a given protein into conservative homology groupings. The LysC p/P Trp ratio was calculated for each of the 52 cohesion groups and for each of 31 orphan (insufficient identity to justify membership in a cohesion group) sequences. These data are plotted as a bar graph in Fig. 8, where it can be seen that cohesion group 22, which contains six genomes of the Chlamydiaceae, has the highest Trp content of the entire set of genomes, except for CG-09. It seems clear that up-Trp selection occurred in the Chlamydiaceae. Eight aspartokinases in CG-09 are fused to DAP decarboxylase, the last enzyme of lysine biosynthesis, and have been proposed to have originated from the Bacteroidetes lineage (74). The E. coli enzyme belongs to CG-21, a group which has only a slightly higher Trp content than the average of all the genomes included.
Fig 8.
Elevated Trp content of aspartokinase (Ask) in Chlamydiaceae compared to other organisms. Bars 1 to 52 in the histogram represent 52 cohesion groups, with bars 53 to 84 being individual Ask orphans. This assemblage was obtained from 518 trimmed Ask sequences present in 339 complete genomes. The average ratio of Ask Trp content/proteome Trp content was calculated to be 0.33. A total of six genomes making up the current selection from the Chlamydiaceae phylum each possess a single Ask enzyme, and all of these belong to cohesion group 22. Zero-Trp cohesion groups (e.g., at position 28) or single orphan sequences (e.g., at positions 55 to 57, 62, or 83) are frequent. The complete membership of cohesion groups and all of the organisms indicated by the acronyms can be found in Table S3 in the supplemental material at http://semiglobe.lanl.gov/supplement.php.
In contrast to the aspartokinases from Chlamydiaceae, those from other chlamydial families (such as “Ca. Protochlamydia amoebophila”) are completely different in having a very low Trp content and in lacking the twin ACT domains that aspartokinases typically deploy for allosteric regulation. “Ca. Protochlamydia amoebophila” aspartokinase has only one Trp residue, and the aspartokinases from Waddlia, Simkania, and Parachlamydia organisms similarly have only one or even zero Trp residues. The latter chlamydial enzymes have about 50% identity with the catalytic portion of various aspartokinases from the Actinobacteria phylum, and perhaps they originated via LGT (followed by loss of the allosteric domain). It seems that the common chlamydial ancestor possessed an aspartokinase of low Trp content, as is common in nature, and that it lacked allosteric control. Although the aspartokinases of the Chlamydiaceae do have a relatively high Trp content in the N-terminal catalytic region of the enzyme (three Trp residues for C. trachomatis), the Trp content of the allosteric region (five Trp residues for C. trachomatis) is even greater. The aspartokinases from the Chlamydiaceae family possess twin ACT allosteric domains which lack the signature conserved domain motifs that mark control by lysine, threonine, or methionine. It seems plausible that they might be subject to feedback inhibition by DAP, a pattern of regulation which has not yet been described. This would make sense, since reductive evolution has made DAP the pathway end product in these organisms. One of the most distinctive features attending the divergence of the Chlamydiaceae family within its order appears to be the evolutionary acquisition of an aspartokinase with presumptive allosteric regulation and with high Trp content relative to that of other aspartokinases in nature.
Insight from Clustering of Cell Division Genes
Genes which are clustered together on the chromosome often implicate functional relationships that might not otherwise have been obvious. As shown in the top section of Fig. 9, it was not unexpected (as seen in Fig. 7) that glmM and glmS would be adjacent members of an apparent operon, but it was pleasingly unexpected to find that the Trp transporter encoded by tyrP is within the gene cluster shown. In some cases (the top group of four genomes), two tyrP paralogs are present in tandem. This linkage relationship compels consideration of the possibility that Trp might induce an operon that couples two signal events of pathogen proliferation, namely, entry of host-derived Trp into pathogen cells and acceleration of the process of cell division. YccA is an extremely hydrophobic membrane-associated protein that binds to FtsH (a cell division protein that is unlinked to the genes shown in Fig. 9) (64). It has a very high p/P Trp ratio (2.65).
The two gene clusters shown in the lower panel belong to a conserved cluster of genes that participate in cell division and peptidoglycan biosynthesis in rod-shaped bacteria. Arrows filled with yellow shading indicate genes whose gene products are illustrated in Fig. 7. The 16-gene assemblage given across the top of the middle panel and denoted the “ancestral sequence” has been called the dcw (division and cell wall) cluster (105). The strong conservation of gene order has been suggested to implicate the cotranslational assembly of many dcw gene products (90). The common ancestor of the Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes “superphylum” has been proposed to have had a dcw cluster (mraZ > mraW > ftsL > ftsI > murE > murF > mraY > murD > ftsW > murG > murC > murB > ddl > ftsQ > ftsA > ftsZ) similar to the 16-gene dcw cluster in E. coli (105). If this was so, then the following changes occurred to yield the contemporary gene organization of the pathogenic chlamydiae: (i) mraZ has been lost, (ii) a hypothetical gene has replaced ftsL, (iii) the ancestral operon has been split between murE and murF to yield the two clusters shown in the bottom and middle panels of Fig. 9, (iv) the latter rearrangement was probably accompanied by the insertion of amiA adjacent to murE (see the top grouping in the bottom panel of Fig. 9), (v) nplD (also called lytF) was inserted between murD and ftsW, (vi) murB was transposed out of the cluster (this might have been synonymous with the fusion of murC and ddl in the pathogenic chlamydiae), and (vii) ftsQ, ftsA, and ftsZ were all lost in pathogenic chlamydiae. The nlpD and amiA insertions both encode peptidoglycan-degrading enzymes which are involved in turnover and/or cell separation. “Ca. Protochlamydia amoebophila” underwent the same changes as the pathogenic chlamydiae did, except that both murB and ddl were transposed out of the cluster, leaving ftsQ adjacent to murC. In addition, ddl exists as a fusion of two identical gene duplicates. This “Ca. Protochlamydia amoebophila” fusion may have originated from a higher plant source via LGT (105).
An additional cell division gene, mreB, is unlinked to the two gene groups discussed above, but it resides as the initial gene of an apparent operon with pckG, which encodes the Trp-rich PEP carboxykinase (discussed extensively above as a crucial step in the pathway shown in Fig. 7). Translational pausing of PckG may indirectly affect the expression of MreB.
Another unlinked gene of interest because it has a high Trp content in the pathogenic Chlamydiaceae is located at the CT645 locus on the lagging strand in C. trachomatis and encodes the cell division protein YlmG/Ycf19. It has an extremely high p/P Trp ratio (4.30) and must have been subject to Chlamydiaceae-specific up-Trp selection, since the homolog in “Ca. Protochlamydia amoebophila” has a p/P Trp ratio of 1.09. No bidirectional best hit to the C. trachomatis query was returned from the E. coli genome. Of further interest is the fact that this cell division gene overlaps with CT644, encoding tRNA dihydrouridine synthase. Although the latter enzyme has a moderate p/P Trp ratio (0.94), it is possible that under conditions of Trp limitation, translational pausing of the cell division protein might also affect expression of the tRNA modification enzyme.
LIPOPOLYSACCHARIDE AND ITS KDO COMPONENTS
Lipopolysaccharide (LPS) antigen is known to diminish sharply in the state of persistence (6). LPS biosynthesis can be viewed in the context of metabolic interlock, as it diverges from a common pathway of early steps shared by the cell division pathway, as shown in Fig. 7. It begins with the fatty acylation of UDP-N-acetylglucosamine (UDP-GlcNac) as the first step in a long pathway consisting of about two dozen enzyme reactions (132). Note that UDP-GlcNac is a crucial branchpoint metabolite which diverges to LPS and to lipid II of the cell division pathway (Fig. 7). Availability of UDP-GlcNac under conditions of Trp limitation may be lessened greatly in that it is the product of GlmV, a high-Trp-content enzyme that may additionally be limited for its UTP substrate, a metabolite that is itself dependent upon a nucleotide transporter of high Trp content. Such cumulative effects of Trp limitation are very likely to contribute to an overall effect that could be quite powerful.
A significant step in the assembly of LPS is the incorporation of two KDO molecules at the 6′ position of lipid IVA. This attachment function is carried out by KDO transferase (KdtA), an integral membrane protein which is bifunctional in consideration of the ability to attach two KDO molecules. KdtA is essential for LPS biosynthesis and normal cell growth in E. coli (11). In C. trachomatis (and probably other chlamydia), KdtA is trifunctional in that it is capable of appending an additional, third KDO molecule to position 8 of the outer KDO sugar (12).
Table 5 shows that six Chlamydiaceae genomes (shaded in gray) exhibit KdtA enzymes with distinctly higher p/P Trp ratios than those found in a variety of Gram-negative bacteria. For comparison, the closest relative, “Ca. Protochlamydia amoebophila,” has a KdtA enzyme with roughly half the Trp content of the pathogenic Chlamydiaceae (shown in bold in Table 5). The KdtA enzyme of E. coli (also shown in bold in Table 5) has a Trp content which is even less than the average of its proteome. Thus, chlamydial genomes have experienced a very distinct up-Trp selection for KdtA. The two enzymes of KDO biosynthesis, KdsA and KdsB, are also included in Table 5. These are soluble enzymes which require no essential Trp residues for catalysis and frequently have a zero-Trp content in various proteomes. Although the KdsA p/P Trp ratio in chlamydial species only slightly exceeds the proteome average, its ratio is distinctly higher than that of most of the other organisms in Table 5. Thus, up-Trp selection of chlamydial KdsA proteins has also occurred, in addition to that of KdtA. The functionality of KdsA would also be challenged severely during Trp depletion because it depends upon PEP as a cosubstrate (see the previous discussion of PEP carboxykinase). The Trp content of KdsB, the enzyme that forms the activated CMP derivative of KDO, appears to be roughly similar to that of other genomes.
Table 5.
Genomic comparison of proteins relevant to KDOa
| Organism | Genomic GC% | Proteomic % Trp | KdtA |
KdsA |
KdsB |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Trp residues | Protein length (aa) | p/P Trp ratio | No. of Trp residues | Protein length (aa) | p/P Trp ratio | No. of Trp residues | Protein length (aa) | p/P Trp ratio | |||
| Blochmannia pennsylvanicus BPEN | 29.56 | 1.65 | 6 | 427 | 0.85 | 0 | 284 | 0.00 | 3 | 262 | 0.69 |
| Campylobacter jejuni jejuni NCTC 11168 | 30.55 | 0.65 | 3 | 385 | 1.20 | 0 | 271 | 0.00 | 0 | 239 | 0.00 |
| “Candidatus Protochlamydia amoebophila” UWE25 | 34.72 | 1.13 | 7 | 417 | 1.49 | 1 | 275 | 0.32 | 2 | 259 | 0.68 |
| Bartonella henselae Houston-1 | 38.23 | 1.06 | 7 | 440 | 1.50 | 1 | 279 | 0.34 | 0 | 243 | 0.00 |
| Legionella pneumophila Paris | 38.37 | 1.15 | 6 | 419 | 1.25 | 2 | 274 | 0.63 | 3 | 250 | 1.04 |
| Helicobacter pylori 26695 | 38.87 | 0.7 | 5 | 393 | 1.82 | 1 | 276 | 0.52 | 0 | 243 | 0.00 |
| Chlamydophila caviae GPIC | 39.19 | 0.95 | 10 | 434 | 2.43 | 3 | 269 | 1.17 | 1 | 254 | 0.41 |
| Chlamydophila felis Fe/C-56 | 39.34 | 0.95 | 9 | 434 | 2.18 | 3 | 269 | 1.17 | 1 | 254 | 0.41 |
| Chlamydophila abortus S26/3 | 39.87 | 0.96 | 9 | 411 | 2.28 | 3 | 269 | 1.16 | 1 | 254 | 0.41 |
| Chlamydia muridarum Nigg | 40.3 | 0.97 | 11 | 430 | 2.64 | 3 | 269 | 1.15 | 1 | 275 | 0.37 |
| Chlamydophila pneumoniae CWL029 | 40.58 | 1.01 | 8 | 437 | 1.81 | 3 | 269 | 1.10 | 1 | 254 | 0.39 |
| Chlamydia trachomatis D/UW-3/CX | 41.31 | 0.95 | 11 | 431 | 2.69 | 3 | 269 | 1.17 | 1 | 254 | 0.41 |
| Coxiella burnetii RSA 493 | 42.6 | 1.21 | 5 | 424 | 0.97 | 3 | 280 | 0.89 | 3 | 249 | 1.00 |
| Aquifex aeolicus VF5 | 43.3 | 0.93 | 3 | 353 | 0.91 | 3 | 267 | 1.21 | 1 | 234 | 0.46 |
| Chlorobium chlorochromatii CaD3 | 44.28 | 1.01 | 5 | 436 | 1.14 | 0 | 272 | 0.00 | 2 | 248 | 0.80 |
| Porphyromonas gingivalis W83 | 48.29 | 1.03 | 6 | 412 | 1.41 | 0 | 272 | 0.00 | 2 | 254 | 0.76 |
| Escherichia coli K-12 | 50.73 | 1.52 | 6 | 425 | 0.93 | 0 | 284 | 0.00 | 3 | 248 | 0.80 |
| Neisseria meningitidis Z2491 | 51.81 | 1.19 | 9 | 423 | 1.79 | 0 | 280 | 0.00 | 1 | 253 | 0.33 |
| Brucella suis 1330 | 57.25 | 1.21 | 6 | 446 | 1.11 | 1 | 277 | 0.30 | 0 | 251 | 0.00 |
| Agrobacterium tumefaciens C58 (Cereon) | 59.04 | 1.25 | 7 | 440 | 1.27 | 1 | 282 | 0.28 | 0 | 251 | 0.00 |
| Nitrobacter hamburgensis X14 | 61.62 | 1.35 | 4 | 434 | 0.68 | 1 | 287 | 0.26 | 0 | 258 | 0.00 |
| Rhodopseudomonas palustris BisB18 | 64.96 | 1.23 | 4 | 434 | 0.75 | 1 | 296 | 0.27 | 1 | 245 | 0.33 |
| Pseudomonas aeruginosa UCBPP-PA14 | 66.29 | 1.49 | 9 | 425 | 1.42 | 0 | 281 | 0.00 | 5 | 254 | 1.32 |
Enzyme abbreviations: KdtA, KDO transferase; KdsA, KDOP (2-keto-3-deoxy-d-manno-octulosonate-8-P) synthase; KdsB, CMP-KDO synthetase.
Among other LPS pathway enzymes in Chlamydiaceae, LpxB (a large peripheral membrane enzyme which condenses UDP-2,3-diacyl-GlcN with lipid X to form the 1′-6 linkage in lipid A) appears to impose a Trp hurdle to LPS biosynthesis. Thus, the p/P ratio of C. trachomatis LpxB is 2.074, compared to 0.71 for “Ca. Protochlamydia amoebophila” and 0.68 for E. coli. However, there is complexity in the comparison, because the C. trachomatis protein and its fellow proteins in other Chlamydiaceae possess an N-terminal domain of about 225 amino acids which is absent in “Ca. Protochlamydia amoebophila” or E. coli. Upstream of the “Ca. Protochlamydia amoebophila” lpxB gene is a gene (pc1311) that encodes a lipid A biosynthesis domain-containing protein, denoted LAB_N (pfam07578; COG3952) at NCBI. The COG3952 domain is repeated as symmetrical regions of about 70 amino acids. It is a homolog of the C. trachomatis N-terminal domain of lpxB. Thus, it is clear that a gene fusion occurred in the common ancestor of the Chlamydiaceae to yield the larger protein present in C. trachomatis. Like “Ca. Protochlamydia amoebophila,” all chlamydial organisms other than the Chlamydiaceae possess the free-standing LAB_N gene, as do a number of other bacteria (most notably in the Bacteroidetes phylum). A gene encoding LAB_N is absent in the E. coli genome. LAB_N proteins generally tend to have a high Trp content, with several of the Trp residues being highly conserved. However, the “Ca. Protochlamydia amoebophila” LAB_N protein is remarkable not only for its extremely high Trp content but also for a large Trp hot spot whereby seven Trp residues are deployed within a 38-amino-acid stretch. Furthermore, six of these Trp residues are present in three tandem combinations. The LAB_N protein of “Ca. Protochlamydia amoebophila” consists of 5.72% Trp and exhibits an extremely high p/P Trp ratio (5.72). The LAB_N region fused to LpxB in the Chlamydiaceae, while contributing substantially to the high Trp content of LpxB, has a single Trp residue at each of the three positions where the “Ca. Protochlamydia amoebophila” protein has tandem Trp residues. Thus, it appears that the common chlamydial ancestor possessed adjacent genes encoding the LAB_N protein and LpxB. In “Ca. Protochlamydia amoebophila,” these genes overlap by three base pairs and are probably translationally coupled such that the sensitivity of LAB_N to Trp availability probably impacts the translation of LpxB as well. Fusion of these genes in the common ancestor of Chlamydiaceae produced a single protein of very high Trp content. Therefore, fusion of LAB_N, a protein absent in E. coli, to LpxB has provided a novel mechanism for transforming LpxB into a focal point of sensitivity to Trp limitation.
In striking contrast to the above data, proteins involved in reactions preceding LpxB, such as LpxC and LpxD, are zero-Trp proteins. This suggests that some intermediates might tend to accumulate during the Trp starvation regimen in place during persistence. Indeed, such LPS fragments might reflect the well-known avoidance strategy deployed by Gram-negative bacteria to evade the host immune surveillance system by various modifications of the lipid A component of LPS (28, 107). Note that LpxD is high on the list of zero-Trp proteins sorted according to length, indicating down-Trp selection (Table 6).
Table 6.
Use of a masked-column version of Table S1 (available at http://semiglobe.lanl.gov/supplement.php) with appropriate sorting to evaluate down-Trp selection of C. trachomatis zero-Trp proteins
| C. trachomatis locus | SEED ID | Genea | Productb | Protein length (aa) | E. coli locus | SEED ID | Genea | Product | No. of Trp residues | Protein length (aa) |
|---|---|---|---|---|---|---|---|---|---|---|
| CT586 | fig|272561.1.peg.599 | uvrB | UvrABC system protein B | 668 | b0779 | fig|83333.1.peg.765 | uvrB | UvrABC system protein B | 0 | 673 |
| CT661 | fig|272561.1.peg.678 | gyrB2 | DNA gyrase subunit B | 605 | ||||||
| CT110 | fig|272561.1.peg.111 | mopA | 60-kDa chaperonin | 544 | b4143 | fig|83333.1.peg.4055 | mopA | 60-kDa chaperonin | 0 | 548 |
| CT571 | fig|272561.1.peg.584 | gspE | General secretion protein E | 501 | b3326 | fig|83333.1.peg.3260 | gspE | General secretion protein E | 2 | 493 |
| CT497 | fig|272561.1.peg.510 | dnaB | Replicative DNA helicase | 472 | b4052 | fig|83333.1.peg.3962 | grpA | Replicative DNA helicase | 3 | 471 |
| CT334 | fig|272561.1.peg.339 | dnaX2 | DNA Pol III, gamma subunit | 466 | b0470 | fig|83333.1.peg.466 | dnaZX | DNA Pol III, gamma and tau subunits | 6 | 643 |
| CT491 | fig|272561.1.peg.503 | rho | Transcription termination factor | 464 | b3783 | fig|83333.1.peg.3711 | tsu | Transcription termination factor rho | 1 | 419 |
| CT669 | fig|272561.1.peg.686 | yscN | Yop secretion ATPase | 442 | b1941 | fig|83333.1.peg.1921 | flaC | Flagellum-specific ATP synthase | 3 | 457 |
| CT097 | fig|272561.1.peg.98 | nusA | Transcription antitermination factor | 434 | b3169 | fig|83333.1.peg.3114 | nusA | Transcription elongation protein | 4 | 495 |
| CT089 | fig|272561.1.peg.90 | lcrE | Low calcium response E protein | 421 | ||||||
| CT705 | fig|272561.1.peg.722 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | 419 | b0438 | fig|83333.1.peg.435 | lopC | ATP-dependent Clp protease ATP-binding subunit ClpX | 0 | 424 |
| CT075 | fig|272561.1.peg.76 | dnaN | DNA polymerase III, beta chain | 416 | b3701 | fig|83333.1.peg.3637 | dnaN | DNA polymerase III, beta chain | 1 | 366 |
| CT322 | fig|272561.1.peg.325 | tufB | Elongation factor Tu | 394 | b3339 | fig|83333.1.peg.3273 | tufA | Elongation factor Tu | 1 | 394 |
| CT341 | fig|272561.1.peg.346 | dnaJ | Chaperone protein DnaJ | 392 | b0015 | fig|83333.1.peg.15 | groP | Chaperone protein DnaJ | 0 | 376 |
| CT709 | fig|272561.1.peg.726 | mreB | Rod shape-determining protein MreB | 366 | b3251 | fig|83333.1.peg.3193 | rodY | Rod shape-determining protein MreB | 0 | 367 |
| CT243 | fig|272561.1.peg.246 | lpxD | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | 354 | b0179 | fig|83333.1.peg.179 | omsA | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | 3 | 341 |
| CT773 | fig|272561.1.peg.790 | ldh | Leucine dehydrogenase | 346 | ||||||
| CT391 | fig|272561.1.peg.399 | CT391 | Hypothetical CT391 protein | 335 | ||||||
| CT070 | fig|272561.1.peg.71 | CT070 | Probable metal transport system membrane protein CT070 | 318 | ||||||
| CT547 | fig|272561.1.peg.560 | CT547 | Hypothetical CT547 protein | 318 | ||||||
| CT093 | fig|272561.1.peg.94 | ribF | Riboflavin kinase/flavin adenine dinucleotide synthase | 301 | b0025 | fig|83333.1.peg.25 | ribF | Riboflavin biosynthesis protein | 0 | 313 |
Location on the lagging strand is indicated in bold.
All of these C. trachomatis gene products contained zero Trp residues.
MASTER-SLAVE MECHANISMS FOR Trp SIGNAL AMPLIFICATION
The mobilization of Trp residues to alter whole translational profiles represents quite a challenge to general considerations of cellular economy, as detailed earlier. However, there are mechanisms that have the innovative potential to respond to Trp availability indirectly through the operation of master-slave gene relationships, whereby a given master gene is critical to the functional expression of multiple slave genes. Here the Trp content of a single gene can have meaning for the expression of many other genes, regardless of their individual Trp contents. Two types of master genes come to mind: (i) those that dictate posttranslational modifications and (ii) those that act as regulatory genes. Prominent examples of these are outlined in this section.
Posttranslational Generation of Lipoproteins
Bacterial lipoproteins are a set of proteins having an N-terminal cysteine whose sulfhydryl group is modified with a diacylglyceryl moiety attached via thioether linkages and whose amino group is acylated with a fatty acid (4). The initial unmodified prolipoprotein is converted to a diacylglyceryl prolipoprotein by prolipoprotein diacylglyceryl transferase. Its signal peptide is then cleaved by prolipoprotein signal peptidase to yield apolipoprotein, which is converted to the mature lipoprotein by the action of apolipoprotein transacylase at the cysteine amino group. Because of the relationship of lipoproteins to membrane structure and their recognized roles in morphogenetic transitions, the Trp content of unmodified prolipoproteins, the proteins of the secretory apparatus required for their export, and the three proteins required for posttranslational modifications merit consideration for their ability to influence developmental transitions. C. trachomatis D/UW-3/CX has been reported to possess genes encoding 14 lipoproteins in its genome (79). These are CT067, CT175, CT253, CT381, CT386, CT444, CT541, CT548, CT567, CT600, CT734, CT770, CT775, and CT780. One of these (omp3; CT444), encoding a cysteine-rich outer membrane protein, is a zero-Trp protein. Proteins with high Trp contents and p/P Trp ratios above 2.0 are CT775 (Sn glycerol acyltransferase; ratio of 2.07), CT567 (hypothetical protein; ratio of 2.40), and CT780 (thioredoxin disulfide isomerase ratio of 2.43). Most of the remaining prolipoproteins have low to moderate Trp contents. One case in point is the “macrophage infectivity potentiator” (MIP) of C. trachomatis, one of the best-characterized chlamydial lipoproteins (96). The preprotein (encoded by CT541) has only one Trp among its 243 residues (p/P Trp ratio = 0.43). However, it nevertheless seems likely that posttranslational maturation of MIP would be in jeopardy during Trp limitation.
Although the likely protein components of the secretory machinery (encoded by CT025, CT701, CT448, CT321, and CT510) exhibit a range of low to average p/P Trp ratios, the final posttranslational steps for lipoprotein biosynthesis all have conspicuously high Trp contents. Alipoprotein N-acyltransferase (CT534) has a p/P Trp ratio of 3.88 (ranking sixth by p/P ratio) and has 20 Trp residues (thus ranking similarly highly by Trp burden). Prolipoprotein diacylglyceryl transferase (CT252) has very high p/P Trp ratio (3.10). Finally, the p/P Trp ratio of lipoprotein signal peptidase (CT408) is also quite high (1.88). Thus, together, these three enzymes of posttranslational biosynthesis require a significant cumulative input of Trp. The homologous proteins in E. coli and “Ca. Protochlamydia amoebophila” have similarly high Trp contents, and this set of enzymes appears to be one of consistently high Trp content in nature. The translation of such proteins might be significantly more sensitive to Trp limitation in Trp auxotrophs such as C. trachomatis and “Ca. Protochlamydia amoebophila” than in prototrophs such as E. coli. Thus, the combined Trp burden carried by the posttranslational modification machinery most likely impacts the maturation of all 14 lipoproteins listed in the previous paragraph. It is intriguing to consider the potential significance that under conditions of Trp limitation, some of these preproteins might be made and exported but not matured.
The foregoing section nicely exemplifies a master-slave mechanism of amplification in which the effects of Trp limitation upon master genes can be extended indirectly, with great leverage, to other proteins which do not necessarily have high Trp contents per se.
Regulation by Chlamydial DcrA
Although divalent cation-dependent regulator A (DcrA; encoded by CT296) in C. trachomatis exhibits an unconvincing level of sequence identity with the ferric uptake regulator (Fur) family of iron-responsive regulators, this functionality was indicated by its ability to complement an E. coli fur mutant (137). The DcrA protein of C. trachomatis exhibits a very high p/P Trp ratio (2.72). The potential for far-reaching, amplifying consequences of its Trp content are impressive, in that 28 genes that were asserted to be DcrA targets were identified (108). Interestingly, many of these target genes have been highlighted elsewhere in separate contexts in this article. For example, one asserted target of DcrA is the Trp transporter gene tyrP-1 (CT817), and the binding region was said to be located upstream in the coding region of glmS (108). However, the status of DcrA, although undoubtedly very important and interesting, must be examined further, since a very recent publication (63) disputes the functional assignment of the CT296 protein as a transcriptional repressor that acts as a Fur homolog. Binding of DcrA to the previously reported targets could not be replicated. Instead, the CT296 protein was predicted to utilize iron as a cofactor and probably binds 2-oxoglutarate. Although it is stated (63) to resemble nonheme Fe(II) 2-oxoglutarate-dependent enzymes, key catalytic residues are not conserved, and some unique biochemical role has been suggested. Thus, CT296 encodes a protein having some as yet uncertain relationship with iron metabolism, but it nevertheless seems likely that it represents an intriguing, albeit undefined, link between Trp and iron levels.
Since either Trp starvation or iron starvation is able to trigger a state similar to persistence, it is interesting that both Trp and iron may be integrant signals combined in the trigger mechanism for induction of persistence in vivo. In this respect, it is of interest that iron depletion and induction of IDO by IFN-γ are coupled, in that IFN-γ also downregulates transferrin receptors, resulting in lower levels of host cell iron (19). IFN-γ also activates inducible nitric oxide synthase of host cells, producing nitric oxide, which sequesters iron atoms with high affinity. This leads to inhibition of various iron-dependent enzymes (57). ParB (chromosome partitioning protein) and TrxB (thioredoxin reductase) exemplify proteins whose synthesis was found to be downregulated when persistent infection was induced by iron deficiency (134). These proteins have low p/P Trp ratios (0.37 and 0.59, respectively), so restricted Trp availability does not directly explain the downregulation.
Trip230 Transcriptional Activator Protein
CT647 of C. trachomatis encodes a homolog of the eukaryote-type protein thyroid hormone receptor-interacting protein (Trip). Its function in chlamydial organisms is unstudied, but it presumably acts as a transcriptional activator and may be involved in folate metabolism, as discussed in the next section. The 4-gene operon containing CT647 is specific to the pathogenic chlamydiae and is not present in “Ca. Protochlamydia amoebophila” or E. coli. The Trip230 p/P Trp ratio of 5.47 is the highest in the C. trachomatis proteome, and the Trp burden value of 10 is considerable for a relatively small protein. Thus, this protein has the potential to affect its unknown target genes with great sensitivity in response to Trp availability.
Folate-Dependent Enzymes
C. trachomatis and other chlamydial organisms have been shown to be capable of de novo folate synthesis as well as to transport preformed folates derived from host cells (34). A bioinformatic analysis by de Crecy-Lagard et al. (31) produced an evaluation of a folate operon in C. trachomatis which extends from CT614 to CT610. It consists of genes which encode dihydroneopterin aldolase (FolB), a bifunctional hydroxymethyldihydropterin pyrophosphokinase/dihydropteroate synthase (FolKP), dihydrofolate reductase (FolA), dihydrofolate synthase (FolC replacement), and a PQQ-like protein of unknown function (but shown not to replace FolE). These folate pathway gene products exhibit p/P Trp ratios of 0.84, 2.34, 2.64, 1.30, and 1.36, respectively. Thus, FolA and FolKP constitute two Trp hurdles in the pathway. The folate transporter is unknown, but it seems likely that it has a high Trp content, since membrane transport proteins often have high Trp contents. The regulation of the folate operon is unknown, but a conspicuous possible regulator is the product of CT647, denoted the Trip230 transcriptional activator (see the above section). The gene encoding the latter putative activator is located within a truly intriguing 4-gene operon of tightly linked genes: CT646 encodes a likely inner membrane protein (p/P Trp ratio = 1.83), CT647 encodes the activator protein (p/P Trp ratio = 5.47), CT648 encodes what has been annotated a phosphatidylinositol-4-phosphate 5-kinase (p/P Trp ratio = 1.74), and CT649 encodes formyltetrahydrofolate cycloligase (p/P Trp ratio = 1.18). The annotation of CT648, if correct, is very interesting, but support for this annotation seems questionable. Formyltetrahydrofolate is the most stable of the natural folates and is unique in not serving as a cofactor in one-carbon metabolism. Importantly, formyltetrahydrofolate likely serves as a storage form of folate in chlamydiae, similar to assertions for seeds and fungal spores (see reference 69 and references therein). The cycloligase is the only enzyme known to be able to recycle formyltetrahydrofolate to a usable metabolic form (5,10-methenyltetrahydrofolate), which occurs via an ATP-dependent and irreversible reaction.
To the extent that folate biosynthesis is sensitive to Trp availability, master-slave relationships (with folate biosynthesis being the master entity) may exist with respect to the following folate-dependent slave enzymes in C. trachomatis. Perhaps most noteworthy is methionyl-tRNA formyltransferase, since formylation of initiator tRNA is essential for translation and bacteria are not known to import methionyl-tRNA (31). Other folate-dependent enzymes are formyltetrahydrofolate cycloligase (CT649), methylene tetrahydrofolate dehydrogenase (FolD; encoded by CT078), dihydrofolate reductase (FolA; encoded by CT612), and serine hydroxymethyltransferase (CT432). Except for FolA, which has a very high p/P Trp ratio (2.64), the remaining folate-dependent enzymes have average or low p/P Trp ratios. However, because of the master-slave relationship, it is quite possible that Trp availability may markedly affect the activities of these enzymes (which require folate) even though their Trp contents may be quite low.
Import of SAM for Methylation Reactions
The pathogenic Chlamydiaceae have lost the capacity for synthesis of S-adenosylmethionine (SAM) via SAM synthetase, an enzyme using methionine and ATP as substrates (14). SAM import from host resources is therefore critical for transmethylation reactions, and such import has the additional value of sparing the need for ATP, which would otherwise be needed in order to synthesize SAM. The SAM transporter (SAMHT) has been demonstrated experimentally in C. trachomatis L2, and it has been proposed that import of SAM is coupled with the export of the toxic transmethylation product S-adenosylhomocysteine (14). Such an antiport mechanism is energetically efficient. CT580 in C. trachomatis encodes the SAM transporter, with 7 Trp residues in a 327-amino-acid protein (p/P Trp ratio = 2.25). Two Trp residues (WGW) are nearly in tandem. The “Ca. Protochlamydia amoebophila” homolog has a similarly high Trp content, suggesting up-Trp selection in an ancient chlamydial ancestor. The E. coli genome lacks a gene for this transporter. The high Trp content of SAMHT suggests that the important process of transmethylation is curbed heavily during the Trp limitation that accompanies persistence. Importantly, there are many methyltransferase reactions in which the S-methyl group of SAM is transferred to a large variety of acceptor molecules.
Thus, SAMHT could be regarded as the master element of a master-slave arrangement in which SAM-utilizing enzymes are slave components. A well-documented example of the latter is rRNA adenine dimethyltransferase (KsgA), an enzyme in chlamydial organisms that was used to affirm the functionality of SAM-dependent methylation reactions in Chlamydia (15). Although KsgA has a moderate p/P Trp ratio (1.14) in the pathogenic Chlamydiaceae, it appears to exemplify a distinct incidence of up-Trp selection in these organisms, since the homologs in “Ca. Protochlamydia amoebophila” and E. coli are both zero-Trp proteins. The up-Trp requirement of KsgA in combination with decreased SAM availability for competing methyltransferases during Trp depletion is likely to greatly minimize the KsgA reaction.
REGIONAL Trp HOT SPOTS EXEMPLIFIED BY POLYMORPHIC MEMBRANE PROTEINS
Polymorphic membrane proteins (Pmps) are present in the pathogenic chlamydia in multiple copies which are highly divergent. The nine Pmps of C. trachomatis and the 16 Pmps of Cp. pneumoniae have been estimated to represent 13.6% and 17.5% of the Chlamydia-specific coding capacity, respectively (112). This is quite remarkable considering the extensive reductive evolution in chlamydial organisms, a process which generally discourages paralog expansions. It is noteworthy that the “nearest neighbor” chlamydial organism in this article, the amoeba endosymbiont “Ca. Protochlamydia amoebophila,” possesses only a single candidate gene encoding a Pmp protein (49). It is generally considered likely that Pmp proteins are key virulence factors, probably playing a role in antigenic variation and/or host adhesion. Pmps consist of three domains, which are color coded in Fig. 10: an N-terminal domain which is the surface-exposed portion of the protein, a central domain (Pmp_M), and an autotransporter domain at the C terminus. The autotransporter domain forms a transmembrane barrel through which the N-terminal domain is extruded (50). The exact function of the Pmp_M domain has yet to be established, but Thomson et al. (127), who recently recognized the Pmp_M domain, pointed out that this domain corresponds to a posttranslational cleavage product described for the Cp. pneumoniae PmpD protein (66). It is known that extracellular proteins exhibit a strong compositional bias toward economical amino acids, and generally expensive amino acids such as Trp are distinctly avoided (118). The low content of Trp in the extracellular portion of Pmp proteins is therefore consistent with this general expectation. However, consideration of the fact that the high Trp content of the remainder of Pmp proteins is required in order for their N-terminal sections to achieve an extracellular localization makes the Pmp proteins quite conspicuous in the overall context of pathogenic proteins that interact with host cells.
Fig 10.
Distribution of Trp residues within the various domain regions of Pmp proteins. A total of 63 intact Pmps are displayed and color coded with respect to domain regions as defined across the top. These Pmps are from five genomes: Chlamydophila caviae GPIC (CCA), Chlamydophila abortus S26/3 (CAB), Chlamydophila pneumoniae AR39 (CP), Chlamydia trachomatis D/UW-3/CX (CT), and Chlamydia muridarum Nigg (TC). CAB267 and CCA00275 were eliminated from Fig. 10 because a lack of the Pmp_M domain and extremely short N-terminal domains probably rules out functional competence. Likewise, CP0301 was eliminated from the sequence collection owing to the complete absence of an autotransporter domain. The positions of individual Trp residues are indicated by black vertical lines. Pmp families defined by Grimwood and Stephens (45) are shown with brackets on the left margin. The initial set of cohesion groups obtained using cohesion group analysis (16, 74) is shown by green vertical lines at the left, and orphan sequences are depicted by solid red circles. The membership of the final six cohesion groups obtained is indicated by the four groups and two orphans, as separated by gaps along the right margin.
Figure 10 is drawn to emphasize that the Trp contents of Pmp proteins in five chlamydial organisms exhibit a distinct regional distribution of Trp hot spots. C. trachomatis (CT tags) and C. muridarum (TC tags) each have 9 Pmps, Cp. abortus (CAB tags) has 13 Pmps, and Cp. caviae (CCA tags) and Cp. pneumoniae (CP tags) each have 16 Pmps. These 63 Pmps are drawn in Fig. 10 to display the uneven locations of Trp residues (shown as vertical black lines) distributed within the three domains. The original family designations accompanying the description of the Pmp superfamily (45) are shown along the far left. The considerable variation in the lengths of individual Pmps is almost entirely the consequence of the substantial variability of the N-terminal domain (which is consistent with the putative role of antigenic variation). The Pmps of C. trachomatis and C. muridarum exist as nine high-similarity ortholog pairs. Pmp families A, D, and H are represented by a single ortholog member drawn from each of the five chlamydial organisms. The remaining families exhibit extensive paralog expansion. The C. trachomatis proteins included in Fig. 10 are from a serovar which elicits urogenital disease. However, other serovars cause ocular trachoma or lymphogranuloma venereum. In a study that included 15 serovars representing these three disease groups, it is interesting that the PmpH family produced a tripartite tree that exactly paralleled the disease group into which a given serovar fell (123).
It is qualitatively apparent that the N-terminal domain has a sparse Trp content and that the remaining domains have a high Trp content. The E/F Pmp family at the top of Fig. 10 exhibits the greatest Trp content in the N-terminal domain, but this is still always less than 1%. It is not unusual for an N-terminal domain to lack Trp residues altogether. The average Trp content with respect to domain section is indicated at the bottom of Fig. 10. A region of particularly high Trp concentration is the approximately 80-amino-acid section centered around the linker region. Except for the E/F family in the top grouping, Trp residues are concentrated mostly near the linker region. Frequently, there is a Trp residue positioned at the left end of the autotransporter domain. The average Trp content of the high-density Trp region surrounding the linker is 5.65%. For the CT869 and TC_0261 ortholog pair of the Pmp E/F family, this value is 7.78%.
PHYLOGENETIC RELATIONSHIPS BETWEEN MEMBERS OF THE Pmp SUPERFAMILY
Figure 10 shows the original Pmp family designations, based upon a tree obtained from complete sequences (45). Since the extreme variability of the N terminus seems likely to be a source of phylogenetic noise, we assembled a set of sequences that all begin with the Pmp_M domain in order to construct a tree based upon the much more conserved middle and C-terminal domains. We used the cohesion group approach (16, 74, 140, 141) for the evolutionary analysis. This approach utilizes a process which eliminates bias caused when the sequences available do not exhibit an ideal phylogenetic spacing, i.e., some groups that have very similar sequences will contribute to overrepresentation of some amino acid residues. Cohesion groups are obtained by multiple rounds of tree building. An initial tree is collapsed at any nodes supported by high bootstrap values, and the tree is rebuilt using one arbitrarily chosen sequence to represent the sequences collapsed together into a common cohesion group. This process is repeated until a tree with no branches of high bootstrap value is obtained. In the final tree, the relationships between the cohesion group branches are uncertain by definition, because the bootstrap values are not high. Figure 10 shows the results of an early step in the process. At the left, following the “total Trp burden” column, are vertical lines in green which represent initial cohesion groups. These add up to 10 cohesion groups and 5 orphan sequences (red dots). In this case, families E/F and H were a single cohesion group, families D and B/C were a single cohesion group, and family A was a single cohesion group. The G/I family corresponded to five cohesion groups and the four orphans. At the far right of the figure are the results of the final cohesion group progression, which produced six cohesion groups, two of which are orphans. Nodes of the final cohesion group tree, which suggests the deepest phylogenetic relationships, exhibited bootstraps values of <650.
DOES Trp STARVATION SLOW UTILIZATION OF BRANCHED-CHAIN AMINO ACIDS FOR PROTEIN SYNTHESIS?
Among the amino-acyl tRNA synthetases, those for leucine, isoleucine, and valine are conspicuous for their strikingly high Trp contents (see Table S1 at http://semiglobe.lanl.gov/supplement.php). These are large proteins, which therefore represent a large Trp burden. They exhibit a monophyletic distribution with respect to amino acid specificity and are probably the result of an ancient gene duplication (18). Both a sequence-based phylogeny (18) and a structure-based phylogeny (98) indicate that leucyl-tRNA synthetase diverged from a common ancestor of valyl-tRNA synthetase and isoleucyl-tRNA synthetase. An alignment of isoleucyl-tRNA synthetases shows a distinct separation of two subgroups. The three branched-chain acyl-tRNA synthetases belong to the class I grouping (11 members) and are usually monomeric, in contrast to the distinctly different class II enzymes (11 members), which are usually multimeric (98). Aside from the leucyl-, valyl-, and isoleucyl-tRNA synthetases, none of the remaining tRNA synthetases are remarkable in their Trp content, except for glutamyl-tRNA synthetase. This smaller enzyme has 10 Trp residues in a 506-amino-acid protein, yielding a p/P ratio of 2.074. Methionyl-tRNA synthetase belongs to the same subclass (subclass IA) as the branched-chain acyl-tRNA synthetases on both structural grounds and sequence comparison grounds (18, 98), but it does not exhibit a particularly high Trp content (p/P Trp ratio = 1.526). Within a given specificity grouping of the three branched-chain tRNA synthetases, the Trp residues are mostly highly conserved, but most of these are not conserved across the three specificity groupings. For example, in the group of leucyl-tRNA synthetases, 13 Trp residues are highly conserved, but only 3 of these are conserved across all three specificity groupings. Under conditions of Trp limitation, the synthesis of proteins with a high content of branched-chain amino acids might be curtailed, but this provocative consideration is not within the scope of this review.
RELATIONSHIP OF Trp CONTENT TO GENE LOCATION ON THE LEADING OR LAGGING STRAND OF REPLICATION
G- and U-ending codons are used significantly more often on the leading strand of DNA replication (84). Since the trp codon is UGG, the bias should be for these codons to be located on the leading strand. In C. trachomatis, 1,728 trp codons are on the leading strand and 1,256 trp codons are on the lagging strand (113). It stands to reason that the cumulative influence of multiple trp codons in genes encoding the various high-Trp proteins would favor their location on the leading strand. Location on the leading strand avoids potential head-to-head collisions of DNA polymerase in replication and RNA polymerase in transcription. The still inevitable codirectional collisions of genes on the leading strand are much better tolerated, since initiated transcripts are not truncated and replication may be slowed only transiently (110). Codirectional collisions with arrested elongation complexes can lead to double-strand breaks (33). Codirectional collisions generate replication forks which must be restarted (87). PriA-dependent mechanisms restart forks from stalled sites. PriA, a large enzyme, has a very low p/P Trp ratio in chlamydial organisms, e.g., 0.27 in C. trachomatis (2/753 Trp residues) and 0.23 in “Ca. Protochlamydia amoebophila” (2/745 Trp residues). This is one of the most dramatic examples of down-Trp selection in chlamydial organisms. E. coli PriA, for comparison, has 15/732 residues, giving a p/P Trp ratio of 1.34. It is interesting that the priA gene is located on the lagging strand of replication in both C. trachomatis and E. coli.
For the reasons given above, genes that are collected into operons on the lagging strand of replication should be especially vulnerable to head-on polymerase collisions, owing to the generally large size of the transcript. One such operon is the 4-gene NQR operon, encoding four subunits of the sodium pump. The other two, noncontiguous genes, encoding additional subunits of the primary sodium pump, are also located on the lagging strand. In contrast, the two subunits of the primary H+ pump are located on the leading strand of replication.
In the pathogenic Chlamydiae, there are some very surprising exceptions whereby genes encoding proteins that are absolutely essential and, unlike PriA, have especially high Trp content are on the lagging strand. The most conspicuous example in C. trachomatis is CT544, the gene encoding glucose-6-P translocase. This crucial permease has the second highest p/P Trp ratio and ranks third by Trp burden. Other surprises are CT231, encoding a dicarboxylate transporter (ranking 9th and 11th by p/P Trp ratio and Trp burden, respectively), as well as CT065, encoding the all-important ADP/ATP translocase (ranking 44th and 28th by p/P Trp ratio and Trp burden, respectively). Perhaps this dilemma is lessened if transcripts for such proteins that are so important to the biology of Chlamydiae are unusually stable. The following scenario might explain how such stable transcripts might be available for translation in times of need. During persistence, DNA replication is presumably greatly lessened. Therefore, collision events caused by DNA polymerase should not be as problematic for RNA polymerase producing new transcripts from the lagging strand. Transcripts for key proteins such as glucose-6-P translocase might be stockpiled for translation when Trp becomes available. In short, during transition from persistence to vegetative growth, certain transcripts might already be available to jump-start the rapid propagation process of growth through translation of key proteins. This is consistent with the important observations made recently by Ouellette et al. (99), who showed that transcription and translation are uncoupled in Chlamydia and that transcripts made in the absence of translation can be quite stable. This scenario might be enhanced further if the recent contention that small RNAs promote mRNA stability to activate the synthesis of virulence factors (106) proves to apply to chlamydiae.
INCLUSION MEMBRANE PROTEINS
A number of proteins have been implicated functionally in the morphogenesis that attends the transition to persistence. Among them, inclusion membrane proteins, whose important functions are thought to include nutrient transport and prevention of lysosomal fusion, have been studied extensively since the work of Rockey et al. with IncA in Cp. caviae (111). IncA exhibits species-level variation in that other-species orthologs are absent in Cp. pneumoniae and Cp. psittaci. Following the elucidation of IncB, IncC, IncD, IncE, IncF, and IncG, the list of putative inclusion membrane proteins in C. trachomatis has grown to 50 (see reference 73 and references therein). Six of these appear to be unique to C. trachomatis, based on the criterion that there are no bidirectional best hits to other chlamydial genomes. Thirteen of the Inc proteins (26%) are zero-Trp proteins (CT089, CT116, CT118, CT119, CT164, CT196, CT222, CT226, CT229, CT232, CT345, CT529, and CT618 proteins), indicating the likelihood that expression of these could be important under conditions of Trp limitation. CT089 was discussed above, in “Type III Secretion,” as an interesting member of a five-gene operon. Overall, inclusion membrane proteins tend to have low or moderate Trp contents. Trp content differences of the inclusion membrane proteins may reflect the differential properties of inclusion membranes that accompany different morphological states of the Chlamydiaceae during the life cycle. One protein (CT147 protein) is large (1,449 amino acids), with 13 Trp residues, thus standing out on the criterion of Trp burden even though the p/P Trp ratio is only average (0.94). Six proteins have high Trp contents, with p/P Trp ratios ranging from 1.70 to 2.86. We considered the possibility that localized Trp hot spots might occur at the C termini or N termini of inclusion membrane proteins and might have significance reminiscent of that of such hot spots for the polymorphic membrane proteins discussed earlier, but none were found. However, the CT813 protein, with 6 Trp residues in 264 amino acids, did exhibit a Trp hot spot at a midprotein location, with 3 Trp residues (2 in tandem) located in a 9-amino-acid region. Overall, the 50 genes encoding inclusion membrane proteins are distributed about equally between locations on either the leading strand or the lagging strand. The differential developmental properties of inclusion membrane proteins with respect to Trp content seem worthy of experimental examination.
IDENTIFICATION OF A HIGH-Trp HISTONE METHYLASE SPECIFIC TO CHLAMYDIAL PATHOGENS
CT737 of C. trachomatis encodes a histone methylase which has recently been reported (103) to be translocated to the host cell nucleus via a type III secretion system. This “nuclear effector” (NUE) protein then can methylate the host histones H2B, H3, and H4. It also accelerates its own methylase efficiency through self-methylation. A previous claim that chlamydial histone is a substrate for this enzyme (95) is disputed (103). The Cp. caviae, Cp. abortus, and Cp. felis methylases all exhibit p/P Trp ratios of about 4.3, ranking 2nd by p/P Trp ratio in these organisms. The Cp. pneumoniae methylase (p/P Trp ratio = 3.13) lacks several Trp residues that are conserved in the foregoing methylases, while the C. trachomatis and C. muridarum methylases lack two additional otherwise-conserved Trp residues (p/P Trp ratio is still high, at 2.4). Alignment of NUE with eukaryotic homologs in the SET domain region (130 residues) shows that conserved chlamydial Trp residues are not present in the eukaryotic sequences. This indicates that up-Trp selection for this methylase has occurred within the lineage of pathogenic chlamydiae. Accordingly, the high Trp content of this exported methylase predicts that this virulence effector will not be translated under conditions of Trp limitation, an outcome for which there presumably has been evolutionary selection favoring the persistent state.
Another C. trachomatis protein, encoded by CT373, is potentially of comparable significance. This gene product is secreted by the type III system (124), is presumably translocated to the host, and has a yet unknown target in the host. The CT373 protein has homologs restricted to its pathogenic relatives, and they all have very high Trp contents (p/P Trp ratio is about 2.1).
THE SPECIAL CATEGORY OF ZERO-Trp PROTEINS
Zero-Trp proteins in the pathogenic Chlamydiaceae are of particular interest because of their differential potential to be translated under conditions where Trp availability becomes limited by the host. While many zero-Trp proteins are characteristically low in Trp content across most bacterial phyla, others appear to be the result of down-Trp selection in the pathogenic Chlamydiaceae. Table 6 is a “snapshot” of a greatly condensed portion of Table S1, available at http://semiglobe.lanl.gov/suppl_dwnld/giant_table.html, which is a dynamic, sortable table that has been an essential resource for interpretation of the massive data accessed in this study. Table 6 was obtained by hiding all genome column blocks except for those of C. trachomatis and E. coli. The “Trp%,” “strand,” and “p/P Trp ratio” columns were also masked. We wanted to view the list of zero-Trp proteins in C. trachomatis in descending order from the longest amino acid strings. To do this, two sorting steps were applied. The “protein length” column header was left clicked to sort from the largest number to the smallest number. Next, the “Trp number” column header was left clicked to sort the number of Trp residues from the smallest value to the greatest value. At the top of the column, this yielded the 143-member set of C. trachomatis zero-Trp proteins, in order of size. Only the top 21 members of the set fit into the space allocation for Table 6. Examination of the complete 143-member set online (see Table S1 at the website given above) reveals that fundamental enzymes engaged in transcription and translation processes are conspicuously prevalent. The right side of Table 6 is blank whenever there is no bidirectional best hit between the C. trachomatis query and the E. coli proteome. Interestingly, the C. trachomatis genes shown in Table 6 tend to be located on the lagging strand of replication (these gene names are shown in bold in the table), whereas most of the bidirectional best hits for the corresponding E. coli genes are located on the leading strand of replication.
UvrB (involved in DNA repair) is the largest zero-Trp protein in C. trachomatis (Table 6). Hence, in the zero-Trp set of proteins, UvrB is the member that would be least likely to lack Trp residues by chance. The gene products of uvrB, mopA, clpX, dnaJ, and mreB were shown to be zero-Trp proteins in both C. trachomatis and E. coli, and these exemplify proteins that generally trend to low Trp contents in most proteomes. The “Ca. Protochlamydia amoebophila” UvrB protein has two Trp residues (but still a low p/P ratio [0.26]). UvrB from “Ca. Protochlamydia amoebophila” and other chlamydial families, such as Waddlia, Simkania, and Parachlamydia, does not seem to be as similar to UvrB from the Chlamydiaceae family as one would expect from the overall chlamydial phylogeny.
Given the overall reductive evolution of the Chlamydiaceae, it is unusual that they possess two species of DNA gyrase, each having two subunits. CT189 and CT190 encode subunits A and B, which yield bidirectional best hits to E. coli b2231 and b3699, encoding the single DNA gyrase present in the E. coli genome. The E. coli pair has low p/P Trp ratios (0.22 and 0.49), comparable to the C. trachomatis pair, which has ratios of 0.49 and 0.25. However, the other paralogous pair of DNA gyrase subunits which is present in chlamydial organisms is smaller overall and imposes a much smaller Trp burden: two Trp residues for small subunit A and zero Trp residues for small subunit B (see the CT661 row of Table 6). This compares to a required input of six Trp residues for the larger C. trachomatis subunits encoded by CT189 and CT190. The subunit B encoded by CT661 appears to have evolved by the deletion in one paralog of a region between the cd03366 domain and the pfam00986 domain in the C-terminal half of the original protein. Subunit B of DNA gyrase (CT661) in Table 6 has no bidirectional best hit in E. coli because it is the one of two C. trachomatis paralogs which has the least similarity to the single E. coli DNA gyrase B. One wonders if the smaller, low-Trp pair of DNA gyrase subunits (GyrA_2 and GyrB_2) in C. trachomatis might be a functional backup geared to function under conditions of Trp limitation.
The gene product of CT334 exemplifies a case of down-Trp selection in C. trachomatis (Table 6). This is partly because CT334 is significantly smaller than its E. coli counterpart, which encodes both the gamma and tau subunits of DNA polymerase III. This phenomenon is possible because of ribosomal frameshifting in E. coli that can result in a truncated gene product, which is in fact the gamma subunit. Alternatively, complete translation without a translational frameshift in E. coli produces the longer tau subunit (37). In contrast, CT334 matches sequence expectations for the shorter gamma subunit, and there is no sequence in the C. trachomatis genome that corresponds to the C-terminal region of the E. coli protein (residues 474 to 643). The “Ca. Protochlamydia amoebophila” enzyme (522 amino acids) is essentially like the C. trachomatis enzyme in being a zero-Trp protein and in lacking sequence to form the tau subunit. Newly available sequences from Waddlia (452 amino acids), Simkania (491 amino acids), and Parachlamydia (512 amino acids), representing other chlamydial families, reveal a range of variation of the carboxy end that suggests that degradative trimming of the now irrelevant tau component is still ongoing, since an ancient truncation event in the common chlamydial ancestor. Thus, down-Trp selection for DNA polymerase III of C. trachomatis and “Ca. Protochlamydia amoebophila” seems associated with a simplification of subunit structure.
Using the above-described masked-column version of Table S1 from http://semiglobe.lanl.gov/suppl_dwnld/giant_table.html, the Trp burden column for E. coli was sorted to show zero-Trp proteins at the top. E. coli proved to have 70 zero-Trp proteins which had bidirectional best-hit matches with C. trachomatis proteins. Of these, 41 C. trachomatis proteins were also zero-Trp proteins. It was of interest to see which of the remainder might be suggestive candidates for up-Trp selection in C. trachomatis. CT057 encodes the most striking candidate, IspG (see Fig. 11 and relevant discussion), which has six Trp residues. Other non-Chlamydiaceae chlamydial organisms, including “Ca. Protochlamydia amoebophila,” also have relatively high Trp contents in IspG, thus indicating up-Trp selection in an ancient chlamydial ancestor. These IspG enzymes have been assigned to the TIGR00612 family of the nonmevalonate pathway of isopentenyl pyrophosphate biosynthesis, whose chlamydial members have a long C-terminal insert of about 200 amino acids. Approximately half of the Trp residues are located within the insert. Other up-Trp proteins screened in this way were dimethyladenosine transferase (CT354), deoxyphosphooctonate aldolase (CT655) (discussed elsewhere), and the DNase YcfH (CT594). The C. trachomatis dimethyladenosine transferase appears to be specifically up-Trp selected in the pathogenic chlamydiae, since the “Ca. Protochlamydia amoebophila” homolog is a zero-Trp protein, like the E. coli homolog.
Fig 11.
Methylerythritol phosphate pathway of isoprenoid biosynthesis. The key enzyme (IspE) which dictates MEC accumulation is highlighted in yellow, as are the key high-energy substrates CTP and ATP. The high Trp content of IspE, expressed as the p/P Trp ratio, is also highlighted.
FURTHER EXAMPLES OF UP-Trp SELECTION AND DOWN-Trp SELECTION
When various masked-column versions of Table S1 (http://semiglobe.lanl.gov/supplement.php) were implemented to compare gene products in the C. trachomatis, “Ca. Protochlamydia amoebophila,” and E. coli proteomes, many intriguing cross-genome discrepancies in Trp content, not otherwise presented in this article, could be discerned. Although these point to many exciting possibilities, they have not yet been the subject of specific experimentation by chlamydial researchers. We have selected some proteins that seem destined to be worthy subjects of future research in the context of Trp availability and the developmental state of the chlamydiae.
Primosomal Protein N′
With the online Table S1 sorted as illustrated by the Table 6 snapshot, one can scroll down beyond what is shown in Table 6 to search for low-Trp proteins in the C. trachomatis query that match high-Trp proteins in the E. coli column. Such proteins are candidates for down-Trp selection in C. trachomatis. For example, the CT778 gene of C. trachomatis (encoding primosomal protein N′, a PriA helicase essential for oriC/DnaA-independent DNA replication) has 2 Trp residues in 753 amino acids (p/P ratio = 0.27), compared to 15 Trp residues in 732 amino acids in E. coli (p/P ratio = 1.34). If the Trp number column is sorted in the E. coli section of the table, one can see that E. coli PriA is very high on the Trp burden list. Here, then, is a very clear example of down-Trp selection in C. trachomatis. (An extensive multiple alignment of PriA proteins which illustrates comprehensively that PriA typically exhibits a high Trp burden in nature is available from the authors.) This down-Trp selection must have occurred in the common ancestor of the pathogenic chlamydiae and “Ca. Protochlamydia amoebophila,” since the PriA enzyme of “Ca. Protochlamydia amoebophila” also has an equally low Trp content.
The Seven-Step Pathway of Chorismate Biosynthesis
The chorismate biosynthesis pathway begins with the condensation of erythrose-4-P and PEP and is intact in chlamydial organisms, yet it does not connect with the quantitatively major output branches leading to the three aromatic amino acids because these metabolic branches have been lost via reductive evolution. Although the seven-step pathway is surely functional in chlamydial organisms, flux through it must be very low, since other known connecting pathways are low-flux, vitamin-like pathways. Examples of the latter in nature are steps to the 4-aminobenzoate component of folate or to the 4-hydroxybenzoate precursor for ubiquinone biosynthesis. Neither of these steps are apparent in chlamydial organisms. Perhaps, in one or both of the last two steps, an unknown nonhomolog (analog) carries out the connecting function. In any event, under conditions of Trp limitation, biosynthetic flow to chorismate is likely impeded by a number of the general hurdles discussed previously. Chorismate biosynthesis requires substantial energy input. Both the first enzyme step and the sixth enzyme step require PEP, and the fifth enzyme step requires ATP. Nevertheless, it is possible that this low-flow pathway enjoys a degree of priority for scarce supplies of PEP and ATP that ensue with Trp limitation because of (i) the quantitatively small, vitamin-like demand for an end product(s) and (ii) down-Trp selection of chorismate pathway proteins. Although the cognate bias hypothesis for amino acid biosynthetic pathways (2) engenders a general expectation of low Trp content in chorismate pathway enzymes, down-Trp selection evidently has lowered the Trp content even further in the pathogenic chlamydiae. Thus, the set of chorismate pathway enzymes represents a significantly lower Trp burden in the pathogenic chlamydiae than in “Ca. Protochlamydia amoebophila.” There are a cumulative 12 Trp residues in the combined seven chorismate pathway proteins, which is less than half of the corresponding cumulative total in “Ca. Protochlamydia amoebophila.” It is especially striking that the ATP-dependent shikimate kinase is a zero-Trp protein in all species of Chlamydia and Chlamydophila but the “Ca. Protochlamydia amoebophila” protein possesses six Trp residues.
4-Hydroxybenzoate Octaprenyltransferase
4-Hydroxybenzoate octaprenyltransferase is encoded by ubiA (CT219) on the lagging strand in C. trachomatis and contrasts markedly with its homolog in E. coli, which is encoded on the leading strand (b4040). With 13 Trp residues, the E. coli protein is highly placed by both Trp burden and p/P Trp ratio (2.95), whereas the C. trachomatis enzyme has only two Trp residues. The C. trachomatis ubiA gene is followed by ubiX. The “Ca. Protochlamydia amoebophila” genome did not return bidirectional best hits for UbiA and UbiX (with the C. trachomatis proteome being the query proteome). Ubiquinone is an important functional component of the electron transport chain, but its biosynthetic pathway has not yet been studied fully in chlamydial organisms. The initial enzyme in E. coli, catalyzing the formation of 4-hydroxybenzoate from chorismate, does not appear to be present in chlamydial organisms (or in fact to be common in nature), and presumably an alternative step is utilized. The distinct down-Trp selection of UbiA in the pathogenic Chlamydiaceae suggests that further analysis will indicate the importance of maintaining some ubiquinone biosynthesis under conditions of Trp limitation.
Protease III Precursor
The large protease III precursor protein (over 950 amino acids), encoded by ptr (CT806) on the lagging strand of C. trachomatis, exhibits down-Trp selection, as it has 5 Trp residues compared to the 14 present in E. coli. Although the C. trachomatis p/P Trp ratio is lower (0.95 in E. coli and 0.55 in C. trachomatis), of course, the decrease in Trp burden may be the most significant factor. Protease III has been annotated to function in the RecBCD pathway of DNA repair. A similar case is that of DNA helicase II, encoded by uvrD (CT608 in C. trachomatis) on the lagging strand and having 5 Trp residues, in contrast to 13 Trp residues in the E. coli DNA helicase II, encoded by b3813 on the leading strand. In C. trachomatis, it is interesting that uvrD appears to be in an operon with rpoN, encoding the sigma 54 factor of RNA polymerase.
PgsA
The PgsA enzyme (CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase; also known as phosphatidyl-glycerophosphate synthetase), encoded by b1912, plays a key role in glycerolipid and glycerophospholipid metabolism. The E. coli PgsA enzyme easily has the highest rank in both the p/P Trp ratio column (3.97) and the Trp% column (6.04% Trp) of the E. coli section of Table S1 at http://semiglobe.lanl.gov/supplement.php. (Note that the corresponding homolog of Blochmannia pennsylvanicus has an even more dramatic p/P Trp ratio of 5.06.) The C. trachomatis gene CT797 encodes a gene product that has only two Trp residues (compared to 11 in E. coli), with the result that the C. trachomatis enzyme has a 0.99% Trp content, compared to the 6.04% Trp content of the E. coli enzyme. The placement of Trp residues in E. coli PgsA results in distinctive hot spot regions, most notably a pair of tandem Trp residues and three Trp residues in one nine-residue region. Down-Trp selection of this PgsA species in an ancient Chlamydiales ancestor can be deduced, since other chlamydial matches to the C. trachomatis PgsA enzyme from families other than Chlamydiaceae always have a Trp content as low as or lower than that of C. trachomatis, e.g., “Ca. Protochlamydia amoebophila” PgsA has zero Trp residues and Waddlia PgsA has one Trp residue.
The aforementioned PgsA enzymes from E. coli, C. trachomatis, and “Ca. Protochlamydia amoebophila” are one set of homologs, as defined by retrieval of bidirectional best hits from the CT797 query. All chlamydial organisms (but not E. coli) possess another paralog of PgsA that has a higher Trp content than the preceding set of chlamydial enzymes. The CT496 protein, a somewhat smaller enzyme from C. trachomatis, has a high p/P Trp ratio (3.13), and the pc0242 protein from “Ca. Protochlamydia amoebophila” has a p/P Trp ratio of 1.88. The Trp content of this PgsA paralog varies from five Trp residues in C. trachomatis and C. muridarum to two or three Trp residues in all other chlamydial organisms. Perhaps the low-Trp paralog set (defined by the CT797 gene product) and the high-Trp paralog set (defined by the CT496 gene product) are designed for different functions in the context of glycerolipid and glycerophospholipid metabolism during the life cycle, depending upon Trp availability.
Thio:Disulfide Interchange Protein
The thio:disulfide interchange protein encoded by CT595 is one of the most dramatic examples of up-Trp selection. The C. trachomatis protein has 20 Trp residues, compared to 11 Trp residues in the E. coli protein. The C. trachomatis p/P Trp ratio is 3.04, compared to the E. coli value of 1.28. The “Ca. Protochlamydia amoebophila” homolog also exhibits a high Trp burden and a very high p/P Trp ratio, indicating that up-Trp selection occurred in an ancient Chlamydiales ancestor. The chlamydial enzymes are about 120 amino acids longer than the E. coli enzyme, and this is due almost entirely to an insert that exists at the N terminus in a region denoted the DsbC superfamily (Fig. 12). This region surrounds the smaller DsbC region and scores as a nonspecific hit for COG4233 in the Chlamydiales, but it is absent in E. coli. Figure 12 shows the distribution of Trp residues within the three motif regions of the enzyme, accounting for a total of 14 Trp residues in C. trachomatis and 5 Trp residues in E. coli. Although the COG4233 region (which is absent in E. coli) contributes three Trp residues, the conserved DsbC region contains four Trp residues, compared with only two in E. coli. The DsbD region of C. trachomatis possesses five Trp residues, whereas the E. coli version has only two. The C-terminal domain has two Trp residues in C. trachomatis and only one in E. coli. Thus, aside from the inserted region, C. trachomatis exhibits an increased density of Trp residues throughout the protein relative to that in E. coli. The C. trachomatis and E. coli proteins each have six Trp residues not portrayed in Fig. 12. Each has one Trp residue in the small connecting region between DsbC and DsbD, and each has five Trp residues in the region labeled the “DsbD alpha interface” in Fig. 12. This interface is a localized region of high Trp density in both C. trachomatis and E. coli. While it is unstudied in chlamydial organisms, the enzyme presumably acts as a disulfide isomerase, interacting with incorrectly folded proteins to correct nonnative disulfide bonds. It functions by transferring its disulfide bond to other proteins and is reduced in the process. Under conditions of Trp limitation, this enzyme is likely a Trp hurdle for the general process, which has yet to be elucidated in the chlamydiae.
Fig 12.
Screen shots of the cdd graphics obtained when the thio:disulfide interchange proteins from C. trachomatis (top panel) and E. coli (bottom panel) are used as BLAST queries at the NCBI database. The portions of the sequences corresponding to each of the three motif regions are given within color-coded boxes, with Trp residues highlighted.
CT819 Transport Protein
The CT819 transport protein, a protein of uncertain specific function, might have functional ties to other previously discussed genes. Its gene follows the single or tandem tyrP gene(s) for Trp transport in the pathogenic chlamydiae. It also might participate in cell division, since its homolog in Faecalibacterium prausnitzii L2-6 has been described as an integral membrane protein which interacts with FtsH. In C. trachomatis, the p/P Trp ratio is 2.65, compared to a ratio of 1.12 in the E. coli homolog. The CT819 protein did not return a bidirectional best hit with the “Ca. Protochlamydia amoebophila” proteome. Like the TyrP proteins, the CT819 protein appears to have been subject to up-Trp selection. It is additionally intriguing that this protein exhibits a hot spot region of locally concentrated Trp residues.
Helix-Turn-Helix Transcriptional Regulator (YfgA)
The small helix-turn-helix transcriptional regulator YfgA (about 140 amino acids) is encoded by CT009 of C. trachomatis and exhibits a p/P Trp ratio of 2.94, compared to a ratio of 1.64 in “Ca. Protochlamydia amoebophila.” There is no bidirectional best hit to the E. coli proteome. As discussed elsewhere (i.e., with regard to master-slave relationships), the effect of Trp limitation upon this regulator has the potential to be magnified to the extent that this regulator might have multiple targets. If the regulator were an activator, the negative effect of Trp limitation upon the regulator could even be extended to transcripts for multiple proteins having low or modest Trp contents. In all current genomes of the order Chlamydiales, encoded Trp residues are confined to a 19-amino-acid stretch in the C-terminal region. YfgA regulators from families other than the Chlamydiaceae have two (e.g., from Waddlia or Simkania) or three (e.g., from “Ca. Protochlamydia amoebophila”) Trp residues in this hot spot region. On the other hand, members of the Chlamydiaceae have four (e.g., C. trachomatis) or, more frequently, five (e.g., Cp. pneumoniae) Trp residues in the hot spot region. This comprises 21 to 26% of a 19-amino-acid stretch. The impact of the additional Trp placements is potentiated by the fact that one or two tandem Trp configurations are created within the region. Thus, the difference between the YfgA proteins in pathogenic Chlamydiaceae and their closest relatives involves the addition of only a few Trp residues in an already existing hot spot region. It appears that Chlamydiaceae-specific up-Trp selection has occurred in an example here where an appreciation of the hot spot localization is all-informative.
DOES A MORE SUBTLE Trp LIMITATION REGIMEN IMPACT RB-EB TRANSITIONS?
During acute infection, in the absence of host withdrawal of Trp via IDO, it is interesting to consider whether Trp might nevertheless play a role in the developmental cycle, even when the persistent state is not triggered. With the progression of rapid RB proliferation, the overall resources of infected host cells become depleted and Trp availability might be a sensitive developmental cue. How this might unfold is illustrated by consideration of the unique and absolutely crucial role of histones in the chlamydial developmental cycle. Late-stage RBs express two histones, Hc1 (a zero-Trp protein) and Hc2 (a one-Trp protein), which bind to the chromatin and produce a highly compacted state that effectively curtails metabolic activity. Consequently, the DNA of infectious EB cells exists as condensed, inactive chromatin. The full details of histone regulation may still be incomplete, but a small RNA (IhtA) blocks translation of Hc1 (43) in RBs until a late cycle time. This specific regulation does not affect Hc2 (44). The transition of EB cells to RB cells within newly occupied host cells requires histone release from the chromatin. Grieshaber et al. (42) showed that release of histone from EB DNA, thereby allowing transition to the RB developmental state, is mediated by 2-C-methylerythritol 2,4-cyclodiphosphate (MEC). MEC is an intermediate of the isoprenoid biosynthetic pathway deployed by chlamydial organisms, the nonmevalonate methylerythritol phosphate pathway. This seven-step pathway begins with the initial utilization of pyruvate and glyceraldehyde-3-P, ultimately producing dimethylallyl diphosphate and isopentenyl diphosphate (diagrammed in Fig. 11). The seven-step pathway for isoprenoid biosynthesis (Fig. 11) deploys enzymes that are generally quite low in Trp content, with the sole exception of IspE (p/P Trp ratio = 1.82). Chlamydial IspE has been shown to play a critical role in MEC production that goes beyond its catalytic function, since E. coli IspE could not substitute for it to accomplish histone release (42). Perhaps elevation of IspE levels results in a pathway flow imbalance whereby MEC is produced faster than it is utilized. If so, MEC accumulation, which is important for histone release in the EB-to-RB transition, should be favored by generous Trp availability in newly infected host cells, since translation of IspE demands high Trp input. On the other hand, during the RB-to-EB transition, where Trp availability is low, translation of IspE might be diminished owing to its high Trp content. Furthermore, IspE is part of an ATP-dependent reaction (Fig. 11), and ATP resources ought to largely be exhausted. In addition, the preceding reaction catalyzed by IspD requires CTP (another metabolite likely to be depleted in late-stage RBs) as a cosubstrate. Hence, overall substrate availability to IspE will be greatly reduced.
If the small molecule MEC is sufficient to accomplish histone release, what could be happening to explain the unique requirement that the IspE enzyme be a chlamydial protein? There are many examples of enzymes which have minor catalytic activities that can become operational under conditions where the enzyme level is high, the normal substrate level is low, and the abnormal substrate level is high. Suppose that chlamydial IspE (but not E. coli IspE) can use MEC as an alternative substrate for some unknown reaction and that the product is in fact the real histone release agent. At high IspE levels, the enzyme may not be saturated with its preferred substrate, and it will encounter high levels of the accumulated nonpreferred substrate (MEC). There are quite a few examples of this sort of substrate ambiguity in the literature (88). It can often be the basis for suppressor mutations which elevate the level of an enzyme which can fill in the missing function of another enzyme that was lost due to the primary mutation.
Another element of this puzzle that may have significance is the observation that IspG, the enzyme that normally utilizes MEC, appears to have undergone up-Trp selection. Even though C. trachomatis IspG exhibits what would generally be an unremarkable p/P Trp ratio of 1.04, this enzyme in most proteomes generally has a very low Trp content, e.g., the E. coli ortholog is a zero-Trp protein. In this respect, the chlamydial IspG protein is reminiscent of the chlamydial aspartokinase, where such p/P Trp ratios are also very high (see Fig. 8 and the attending discussion). Up-Trp selection for IspG also appears to have occurred in “Ca. Protochlamydia amoebophila” and in other newly recognized chlamydial families. Since the chlamydial IspG enzymes all have a long C-terminal extension of unknown function, it is interesting to consider that this extension might be a domain that interacts with IspE in the context of the discussion in the preceding paragraph.
DEDUCING THE EVOLUTIONARY HISTORY OF Trp CONTENT SELECTION IN CHLAMYDIAE
The evaluation of the Trp content of the “Ca. Protochlamydia amoebophila” proteome in comparison to that of the pathogenic Chlamydiaceae revealed the unexpected likelihood that availability of Trp from the host was already used to some extent as a primitive cue to facilitate the intracellular lifestyle of ancient chlamydial organisms. The Trp contents of individual proteins have in some cases increased or decreased in both “Ca. Protochlamydia amoebophila” and the Chlamydiaceae, indicating that their selection occurred in a common ancestor. In many other cases, up-Trp or down-Trp selection occurred only after divergence of the Chlamydiaceae. Sometimes a critical protein already had a sufficiently high Trp content to possibly influence the translational profile in response to Trp starvation, e.g., glucose-6-P translocase in “Ca. Protochlamydia amoebophila” has a p/P Trp ratio of 3.46 (Fig. 5). The pathogenic chlamydiae have further enhanced the vulnerability of this enzyme to Trp starvation by up-Trp selection that has resulted in extraordinarily high p/P Trp ratios, in excess of 5.0. The broad-specificity nucleotide translocase present in both “Ca. Protochlamydia amoebophila” and pathogenic chlamydiae is another crucial protein that characteristically has a high Trp content. The pathogenic chlamydiae, but not “Ca. Protochlamydia amoebophila,” have essentially doubled the Trp burden through generation of the ATP/ADP-specific paralog. Both “Ca. Protochlamydia amoebophila” and the pathogenic chlamydiae demonstrate up-Trp selection for the TyrP permease of Trp transport, as well as for the key enzyme that generates GTP-specific PEP carboxykinase. On the other hand, both “Ca. Protochlamydia amoebophila” and the pathogenic chlamydiae exhibit down-Trp selection for primosomal protein N′ and for CDP-diacylglycerol-3-P 3-phosphatidyltransferase. One might suppose that the foregoing proteins that experienced up-Trp or down-Trp selection were key enzymes in an ancient common ancestor that responded to cues of Trp availability from the host. There are many proteins that underwent up-Trp or down-Trp selection in the pathogenic chlamydiae but not in “Ca. Protochlamydia amoebophila,” and these presumably reflect an evolutionary response to the mammalian host. An obvious case is up-Trp selection for iron transport in pathogenic chlamydiae, as iron is particularly important to the physiology of the mammalian host, but not in “Ca. Protochlamydia amoebophila.” Other examples include components of the primary NQR sodium pump (up-Trp selection), type III secretion (up-Trp selection), lipoprotein biosynthesis (up-Trp selection), diaminopimelate biosynthesis (up-Trp selection), and chorismate biosynthesis (down-Trp selection).
Recently, the order Chlamydiales has seen the documentation of genomes for organisms representing four additional families in addition to the Chlamydiaceae and Parachlamydiaceae (which includes “Ca. Protochlamydia amoebophila”): Criblamydiaceae, Rhabdochlamydiaceae, Simkaniaceae, and Waddliaceae (13). The availability of these additional genomes and the many yet to come will make it possible to deduce the detailed evolutionary history of Trp content in proteomes throughout the order Chlamydiales.
EXTENSION OF THE Trp CUE HYPOTHESIS BEYOND THE CHLAMYDIAE
Byrne and Beatty recently reflected upon the relationship of chlamydial persistence and the attendant aspects of pathogenicity to other classic persistent/chronic diseases, such as malaria, toxoplasmosis, syphilis, tuberculosis, and typhoid fever (20). Here we consider the idea that Trp availability might serve as a cue for the response of other mammalian pathogens to enter an alternative developmental stage of chronic resistance.
Other Intracellular Pathogens
Francisella tularensis is a facultative intracellular pathogen whose presence has been shown to induce host IDO within infected mouse lungs (102). Even though the genes for Trp biosynthesis are present in F. tularensis and were indeed shown to be important for survival, unlike the case for chlamydiae, this biosynthetic potential is not completely able to offset the antimicrobial action of host IDO. Since Trp withdrawal by the host is clearly an effective defense mechanism, it would be interesting to evaluate whether there is evidence that selection has decreased the Trp content of at least some crucial proteins of F. tularensis strains. Perhaps Francisella is in the very early stages of evolutionary transition from extreme virulence to the more successful parasitism that would be occasioned by eventual acquisition of an alternative developmental state approaching that of persistence in the pathogenic chlamydiae.
Extracellular Pathogens
The growth of extracellular bacteria can also be vulnerable to IDO-mediated Trp depletion, as has been shown with group B streptococci (Streptococcus agalactiae) in human cord blood macrophages (77), Streptococcus pneumoniae in mouse lungs (102), and Enterococcus faecalis (and close relatives) in human uroepithelial cells (78). It is suggestive that E. faecalis, Streptococcus mutans, Streptococcus pyogenes, Streptococcus equi, Streptococcus gordonii, and S. pneumoniae comprise a phylogenetic clade in which the Trp biosynthetic pathway appears to have been lost independently in some of its members, i.e., E. faecalis, S. pyogenes, and S. equi (141). The pathway of Trp biosynthesis is also absent in S. agalactiae. Perhaps these auxotrophies, which can be inferred to have been recent events of reductive evolution, reflect an emerging strategy for evasion of immune surveillance in response to host-mediated withdrawal of Trp availability. This would be similar to the presumed evolutionary events in the pathogenic chlamydiae.
One physiological state of Streptococcus species that might be enhanced by Trp availability and strongly discouraged by Trp depletion is that of competence for DNA transformation in the pyogenic, bovis, and mutans phylogenetic groups. Competence in Streptococcus is an established virulence determinant (72). Full maturation of the competent state is accomplished by a quorum-sensing system (38). In streptococci, SigX (or ComX) is an alternative sigma factor which acts as a master regulator of the dozens of proteins responsible for induction of the state of competence (70). Two entirely different quorum-sensing proximal circuits connecting with ComX exist in different streptococcal groups. One of these is distributed among members of the pyogenic, bovis, and mutans groups (80). It consists of a small double-tryptophan peptide pheromone which is processed and imported back into the cell via an oligopeptide transporter (multicomponent Opp complex), where it interacts with the ComR regulatory protein. We note here that in S. mutans, SigX has a nearly 1.9% Trp content, with all three Trp residues confined to a Trp hot spot span of 17 residues in the N-terminal region (nearly an 18% localized Trp content). The ComS pheromone peptide has tandem Trp residues near the C terminus. This conspicuous Trp tandem is absolutely conserved within a peptide that is otherwise quite variable. The OppD subunit of oligopeptide transport, an absolutely critical entity for competence (80), has a Trp content of >1.7%, and three Trp residues within a 17-residue span near the C terminus define a Trp hot spot of more than 13% Trp content. We suggest the possibility of the following scenario. A pathogen population reaches the threshold numbers required to satisfy the quorum-sensing mechanism. If the host immune surveillance system has not yet activated IDO (with its consequence of Trp depletion), then sufficient Trp nutrients will exist to support the full expression of the competent state. Cells in the competent state may confer certain temporary growth advantages, such as the ability to utilize DNA as a source of nutrients such as carbon, nitrogen, and phosphorus (e.g., as speculated by van der Ploeg [130]). Yet competence is at the same time a fragile physiological state. Once immune surveillance has triggered the various responses to mount an attack upon vulnerable pathogen cells, it would be to the advantage of newly dividing cells to forego the competent state. This might be accomplished by the following effects of Trp depletion upon components of the ComRS proximal circuit of competence induction: (i) failure to satisfy the tandem Trp requirement during translation of the key pheromone peptide ComS, (ii) a decrease in the import of any successfully processed ComS back into pathogen cells due to the high Trp content (especially at the Trp hot spot) of OppD, and (iii) the demand concentrated at the Trp hot spot (nearly 18%) of SigX. Because of the complex and hierarchical circuitry that controls the competent state, these individual effects can be expected to exert multiplying effects. In light of information linking other processes, such as bacteriocin production and biofilm formation, to the cascade that implements the competent state (130), Trp may be the trigger that dictates expression of many additional genes beyond the dozens required for the competent state. (Note that the mitis and anginosus groups of streptococci possess an entirely different proximal circuit of competence induction that is also quorum sensing in nature and encoded by comCDE. The ComCDE proteins have a strikingly low Trp content, with only one Trp residue in the entire aggregate.)
Role of Trp in General Survival Responses of Bacteria
While the phenomenon of persistence in chlamydiae may seem unique, it may in a larger context reflect a generally evolved survival response of bacteria to nutrient deprivation. For example, nonsporulating bacteria generally persist in a physiological state of nutrient starvation controlled by a genetic program that assists long-term survival (81, 120). The transition to and from a long-term survival state in Sinorhizobium meliloti or Rhodobacter sphaeroides is mediated by the regulatory role of a tryptophan-rich sensory protein (TspO) (30). Although these organisms are quite high in GC content, the Tsp proteins have extraordinarily high Trp contents. R. sphaeroides TspO, for example, has a Trp content of nearly 7%, with one Trp hot spot spanning 13 residues containing 4 Trp residues (2 of them in tandem). TspO homologs also populate the genomes of various pseudomonad and xanthomonad organisms. Thus, Xanthomonas axonopodis TspO has a Trp content of >9% (15 of 166 residues). This high Trp content suggests that Trp availability is probably a specific cue in the regulatory mechanism deployed.
Trp and Viral Infections
Finally, the possible signaling nature of Trp abundance/Trp depletion may even be relevant to viral infections, since IDO is known to mediate antiviral effects against vaccinia virus, measles virus, and a number of herpesviruses, as reviewed recently by Daubener et al. (29). In this context, it is quite suggestive that a Trp-rich carboxy terminus of the small envelope protein of hepatitis B virus is essential for virion assembly (68). Within a span of 63 residues, there are 9 Trp residues (more than 14% Trp content), including one hot spot consisting of 4/11 Trp residues. In fact, this protein is the most striking example that we have found anywhere of multiple Trp residues distributed within a small region.
CONCLUDING PERSPECTIVES
The innate immune response to C. trachomatis, an intracellular pathogen of humans, involves a cascade of events that triggers catabolism of Trp by the host cells, with a consequent starvation of C. trachomatis for Trp. Subsequently, evasion of the immune response can be accomplished by entry of the pathogen into a quiescent but reversible stage of development called persistence. What is the mechanism that allows the pathogen to respond to this host defense mechanism so successfully? Gene products such as ADP/ATP translocase, the nucleoside triphosphate transporter, the hexose-phosphate transporter, PEP carboxykinase, the Trp transporter, the Pmp protein superfamily for cell adhesion and antigenic variation, and components of the cell division pathway (including the aspartokinase pathway to diaminopimelate) exemplify key processes that support the developmental phase of rapid proliferative growth in chlamydial organisms. It is clear that the Trp contents of these proteins, many of which are highly antigenic, are much higher than would be predicted in consideration of the overall proteome Trp content. This is counterbalanced by other gene products, including proteins known to be associated with the state of persistence, that possess low-Trp or even zero-Trp content. For example, primosomal protein N′, a PriA helicase essential for oriC/DnaA-independent DNA replication, is a very conspicuous case of down-Trp selection. Importantly, the pathogen's strategy of selectively increased Trp usage to guide the translational profile can be leveraged further, with minimal overall Trp usage, by (i) regional concentration of Trp residue placements in proteins, (ii) amplified Trp content of a single protein that is required for expression or maturation of multiple proteins, (iii) Achilles heel vulnerabilities of complex pathways to high Trp content of one or a few lynchpin enzymes, and (iv) evolutionary replacement of proteins by same-function analogs with different Trp contents. During persistence, some important genes located on the lagging strand of replication may produce stable transcripts poised for translation upon transition to rapid growth. This is consistent with recent findings that transcription and translation can be uncoupled in Chlamydia. Persistence is a state of immune evasion whereby chlamydial pathogens respond to the host withdrawal of Trp via implementation of a sophisticated survival mode. A mechanism has evolved which suppresses gene products necessary for rapid pathogen proliferation but allows expression of gene products that underlie the morphological and developmental characteristics of persistence. This switch from one translational profile to an alternative translational profile of newly synthesized proteins is asserted to be accomplished by maximizing the Trp content of some proteins needed for rapid proliferation and minimizing the Trp content of other proteins supporting the state of persistence. The similarity of up-Trp and down-Trp proteomic profiles in all of the pathogenic Chlamydiaceae suggests that Trp availability is an underlying cue relied upon by this family of pathogens to trigger developmental transitions. It is suggested that prior to the emergence of Chlamydiaceae and their mammalian hosts, a few key events of up-Trp or down-Trp selection occurred in ancient chlamydial organisms that possessed an intracellular lifestyle.
ACKNOWLEDGMENTS
This project was partially supported by an NIH NIDCR grant (Y1-DE-6006-02 to G.X. and C.-C.L.). Additional support was provided by Taiwan National Science Council grant NSC-97-3112-B-010-019 to C.-C.L.
We dedicate this review to Gerald Byrne, whose early insights sparked our interest in the essential premise of this article, and to Charles Yanofsky, whose lifelong work is fundamental to our current understanding of tryptophan metabolism and regulation.
Biographies

Chien-Chi Lo received a dual B.S. degree in public health and medical technology (2002) from the National Taiwan University and an M.S. degree in bioinformatics (2007) from the National Yang-Ming University, both in Taipei, Taiwan. During this time, he engaged in research projects that required a combination of experimental and computational skills. He developed a strong and continuing interest in how bioinformatics approaches can contribute insights into biological systems. He currently is a research technologist at Los Alamos National Laboratory (Los Alamos, NM) and a graduate student in the Department of Computer Science at the University of New Mexico (Albuquerque, NM). His research interests are in bioinformatics, including sequence analysis, biological database development, computational biology, and programming.

Gary Xie obtained his medical degree (B.M.) from Xianya School of Medicine, Central South University (formerly Hunan Medical University), in China (1992). He received both M.S. (1998) and Ph.D. (2002) degrees from the University of Florida (Gainesville, FL). His Ph.D. studies focused on the comparative genome analysis of aromatic amino acid biosynthesis. He did postdoctoral research on the human chromosome and bacterial genome annotation and analysis at JGI—Los Alamos (2002 to 2004). Subsequently, he was appointed as a Technical Staff Member of the Bioscience Division, Los Alamos National Laboratory. He currently is the principal investigator on a project to annotate and curate all oral pathogen genome sequences for the ORALGEN database. Most recently, he has been (i) working on a metagenome project involving sequenced environmental samples, (ii) developing bioinformatics applications for single-genome/metagenome annotation and analysis, and (iii) conducting comparative studies of human pathogens.

Carol A. Bonner received a B.A. degree in biology at the State University of New York at Binghamton and a Ph.D. degree at the University of Florida (Gainesville, FL). She managed a large program at the University of Florida, where she was in charge of a plant tissue culture laboratory and a microbiology laboratory. She directed laboratory studies focused upon the enzymology and regulation of aromatic amino acid biosynthesis in both higher plants and bacteria. Her present interest lies in bioinformatics analysis of biochemical pathways. She has been working as an annotator for the SEED database and is interested in helping to apply the cohesion-group approaches described in this article to develop a semiautomated program that would facilitate rapid and efficient evaluation of the SEED subsystems.

Roy A. Jensen received an undergraduate degree from Ripon College in Wisconsin and a Ph.D. degree from the University of Texas. Following postdoctoral studies at the University of Washington, he held faculty positions at the State University of New York at Buffalo, Baylor College of Medicine, M. D. Anderson Hospital and Tumor Institute, the State University of New York at Binghamton, and the University of Florida, where he is currently Professor Emeritus. He had a strong interest in evolution before it became fashionable for biochemists and geneticists. This interest, dynamically stimulated by the advent of genome sequencing, has merged nicely in recent times with an enduring fascination with the alternative biochemical pathways that can lead to a given end product, the diverse alternative patterns of regulation that exist to control them, and the complex relationships (metabolic interlock) between what are often considered unrelated pathways.
APPENDIX
Source of Genome Data
Our starting genome resources consisted of 22 bacterial genomes and their protein sequences, obtained from the Integrated Microbial Genomes (IMG) system (version 1.5; http://img.jgi.doe.gov/). The 22 genomes were Chlamydia muridarum Nigg (Cmu), Chlamydia trachomatis D/UW-3/CX, Chlamydophila abortus S26/3 (Cab), Chlamydophila caviae GPIC (Cca), Chlamydophila felis Fe/C-56 (Cfe), Chlamydophila pneumoniae CWL029 (Cpn), “Candidatus Protochlamydia amoebophila” UWE25, Escherichia coli K-12, Blochmannia pennsylvanicus BPEN (Bpe), Bartonella henselae Houston-1 (Bhe), Brucella suis 1330 (Bsu), Nitrobacter hamburgensis X14 (Nha), Rhodopseudomonas palustris BisB18 (Rpa), Anaplasma marginale St. Maries (Ama), Anaplasma phagocytophilum HZ (Aph), Ehrlichia chaffeensis Arkansas (Ech), Neorickettsia sennetsu Miyayama (Nse), Rickettsia bellii RML369-C (Rbe), Wolbachia pipientis wMel (Wpi), Pelagibacter ubique HTCC1062 (Pub), Coxiella burnetii RSA 493 (Cbu), and Legionella pneumophila Paris (Lpn). This 22-genome resource (see Table S8 in the supplementary files at http://semiglobe.lanl.gov/supplement.php) is being maintained as a resource for expanded studies that extend from the current analysis, which was limited to the genomes included in a 9-genome table (see Table S1 in the supplementary files at http://semiglobe.lanl.gov/supplement.php). The nine genomes in Table S1 are those of Chlamydia muridarum, Chlamydia trachomatis, Chlamydophila abortus, Chlamydophila caviae, Chlamydophila felis, Chlamydophila pneumoniae “Candidatus Protochlamydia amoebophila,” Escherichia coli, and Blochmannia pennsylvanicus. Note that while we have used the Chlamydiaceae taxonomy in place at NCBI for practical reasons, a strong case has been made to reunite the members of the Chlamydiaceae into a single genus, Chlamydia (122).
Calculation of Observed and Expected Trp Frequencies
The observed Trp frequencies were calculated using the freqaa.pl script, which was modified from that of Tekaia et al. (126). We derived the observed raw tryptophan counts of the protein sequences from each species and computed the observed percent frequency of amino acids which were Trp residues. The expected Trp frequencies were calculated on the basis of the method suggested by Lobry (75). If the relative frequencies of the four nucleotides in the gene are PA, PC, PG, and PT, then the expected frequency of occurrence of each amino acid in the corresponding protein sequence is the sum of the expected frequencies of occurrence of its codons. Since only one codon (TGG) represents Trp, the expected Trp frequency is expressed as follows: PTrp = PTGG.
PTGG is expressed in terms of the independent probability of the three bases in the codon, normalized to take the three stop codons (TAA, TAG, and TGA) into account, as follows: PTGG = PTPGPG/[1 − (PTPTPA + PTPAPG + PTPGPA)].
To set the values of these parameters, the simplifying assumption that PA = PT and PC = PG was used. The statistical relationships termed parity rule type 2 by Sueoka (125) justify this simplification for the present data set. With this assumption, there is one parameter, G+C content (PC + PG), that can be used to predict the expected Trp frequency. The G+C content corresponding to each species can calculate the expected PTrp. The Trp ratio for each protein of 22 proteomes was calculated using observed frequencies divided by expected frequencies, as follows: Trp ratio = PTrp (observed)/PTrp (expected).
Statistical Inference
We computed the Z values for the deviations between each of the observed frequencies, given their expectations, according to the method of Pe'er et al. (101), using the following equation:
The Z values are positive if observed values are larger than expected values and negative if observed values are smaller than expected values. The null hypothesis is that the observed Trp frequencies are equal to expected Trp frequencies. The probability, or P value, to support the null hypothesis is derived from the Z value under a normal distribution. If the P value is lower than the significance level, such as 0.05, the null hypothesis is rejected, and the test result indicates that there is a significant difference between observed frequencies and expected frequencies. In other words, the Trp frequency of the protein is overexpected or underexpected at that significance level. The software package SPSS 11.5 for Windows was used for these calculations.
Localized Aggregation of Trp Residues (Trp Hot Spots)
The protein sequences of each species were screened using the multi_w.pl script. This script finds patterns of Trp residue concentration in which a window of six scanned residues contains at least three Trp residues. The script reports the positions of these Trp residues in a given sequence of amino acids. Those proteins returning positive hits for the criterion of three or more Trp residues per scan window are depicted with green bars in Table S1, available at http://semiglobe.lanl.gov/supplement.php.
Assignment of Genes to Leading or Lagging Strand of Replication
In order to ascertain whether genes are positioned on the leading or lagging strand of replication, they must be located relative to the origin and terminus of replication. The replication origins and replication termini of C. muridarum, C. trachomatis, Chlamydophila pneumoniae, and E. coli were derived from the study of Song et al. (119). Details of gene assignments are given based on the GenBank ptt file under the “strand” qualifier.
REFERENCES
- 1. Akers JC, Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 188:4236–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alves R, Savageau MA. 2005. Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes. Mol. Microbiol. 56:1017–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Babbin BA, Sasaki M, Gerner-Schmidt KW, Nusrat A, Klapproth JM. 2009. The bacterial virulence factor lymphostatin compromises intestinal epithelial barrier function by modulating rho GTPases. Am. J. Pathol. 174:1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babu MM, et al. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beatty WL, Belanger TA, Desai AA, Morrison RP, Byrne GI. 1994. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62:3705–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beatty WL, Morrison RP, Byrne GI. 1994. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect. Immun. 62:4059–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beatty WL, Morrison RP, Byrne GI. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bekpen C, et al. 2005. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 6:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Belland RJ, et al. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 98:13984–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belunis CJ, Clementz T, Carty SM, Raetz CR. 1995. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 270:27646–27652 [DOI] [PubMed] [Google Scholar]
- 12. Belunis CJ, Mdluli KE, Raetz CR, Nano FE. 1992. A novel 3-deoxy-d-manno-octulosonic acid transferase from Chlamydia trachomatis required for expression of the genus-specific epitope. J. Biol. Chem. 267:18702–18707 [PubMed] [Google Scholar]
- 13. Bertelli C, et al. 2010. The Waddlia genome: a window into chlamydial biology. PLoS One 5:e10890 doi:10.1371/journal.pone.0010890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Binet R, Fernandez RE, Fisher DJ, Maurelli AT. 2011. Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. mBio 2:e00051–11 doi:10.1128/mBio.00051-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Binet R, Maurelli AT. 2009. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 9:279 doi:10.1186/1471-2180-9-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bonner CA, et al. 2008. Cohesion group approach for evolutionary analysis of TyrA, a protein family with wide-ranging substrate specificities. Microbiol. Mol. Biol. Rev. 72:13–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bourne HR, Sanders DA, McCormick F. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125–132 [DOI] [PubMed] [Google Scholar]
- 18. Brown JR, Doolittle WF. 1995. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl. Acad. Sci. U. S. A. 92:2441–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Byrd TF, Horwitz MA. 1989. Interferon gamma-activated human monocytes downregulate transferrin receptors and inhibit the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. J. Clin. Invest. 83:1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Byrne GI, Beatty WL. (ed). 2012. Chlamydial persistence redux. ASM Press, Washington, DC [Google Scholar]
- 21. Byrne GI, et al. 1986. Induction of tryptophan degradation in vitro and in vivo: a gamma-interferon-stimulated activity. J. Interferon Res. 6:389–396 [DOI] [PubMed] [Google Scholar]
- 22. Byrne GI, Lehmann LK, Landry GJ. 1986. Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53:347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carlson JH, et al. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72:7063–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson JH, Porcella SF, McClarty G, Caldwell HD. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carlson JH, Wood H, Roshick C, Caldwell HD, McClarty G. 2006. In vivo and in vitro studies of Chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59:1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chico-Calero I, et al. 2002. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc. Natl. Acad. Sci. U. S. A. 99:431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clifton DR, et al. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Darveau RP. 1998. Lipid A diversity and the innate host response to bacterial infection. Curr. Opin. Microbiol. 1:36–42 [DOI] [PubMed] [Google Scholar]
- 29. Daubener W, Schmidt SK, Heseler K, Spekker KH, MacKenzie CR. 2009. Antimicrobial and immunoregulatory effector mechanisms in human endothelial cells. Indoleamine 2,3-dioxygenase versus inducible nitric oxide synthase. Thromb. Haemost. 102:1110–1116 [DOI] [PubMed] [Google Scholar]
- 30. Davey ME, de Bruijn FJ. 2000. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl. Environ. Microbiol. 66:5353–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Crecy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. 2007. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics 8:245 doi:10.1186/1471-2164-8-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dibrov P, Dibrov E, Pierce GN, Galperin MY. 2004. Salt in the wound: a possible role of Na+ gradient in chlamydial infection. J. Mol. Microbiol. Biotechnol. 8:1–6 [DOI] [PubMed] [Google Scholar]
- 33. Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. 2011. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146:533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fan H, Brunham RC, McClarty G. 1992. Acquisition and synthesis of folates by obligate intracellular bacteria of the genus Chlamydia. J. Clin. Invest. 90:1803–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fehlner-Gardiner C, et al. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893–26903 [DOI] [PubMed] [Google Scholar]
- 36. Fleckner J, Rasmussen HH, Justesen J. 1991. Human interferon gamma potently induces the synthesis of a 55-kDa protein (gamma 2) highly homologous to rabbit peptide chain release factor and bovine tryptophanyl-tRNA synthetase. Proc. Natl. Acad. Sci. U. S. A. 88:11520–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flower AM, McHenry CS. 1990. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Natl. Acad. Sci. U. S. A. 87:3713–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fontaine L, et al. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foster JD, Nordlie RC. 2002. The biochemistry and molecular biology of the glucose-6-phosphatase system. Exp. Biol. Med. (Maywood) 227:601–608 [DOI] [PubMed] [Google Scholar]
- 40. Gieffers J, et al. 2003. Genotypic differences in the Chlamydia pneumoniae tyrP locus related to vascular tropism and pathogenicity. J. Infect. Dis. 188:1085–1093 [DOI] [PubMed] [Google Scholar]
- 41. Granseth E, von Heijne G, Elofsson A. 2005. A study of the membrane-water interface region of membrane proteins. J. Mol. Biol. 346:377–385 [DOI] [PubMed] [Google Scholar]
- 42. Grieshaber NA, Fischer ER, Mead DJ, Dooley CA, Hackstadt T. 2004. Chlamydial histone-DNA interactions are disrupted by a metabolite in the methylerythritol phosphate pathway of isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 101:7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. 2006. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol. Microbiol. 59:541–550 [DOI] [PubMed] [Google Scholar]
- 44. Grieshaber NA, Sager JB, Dooley CA, Hayes SF, Hackstadt T. 2006. Regulation of the Chlamydia trachomatis histone H1-like protein Hc2 is IspE dependent and IhtA independent. J. Bacteriol. 188:5289–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grimwood J, Stephens RS. 1999. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4:187–201 [DOI] [PubMed] [Google Scholar]
- 46. Harper A, Pogson CI, Jones ML, Pearce JH. 2000. Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect. Immun. 68:1457–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hassan KA, Souhani T, Skurray RA, Brown MH. 2008. Analysis of tryptophan residues in the staphylococcal multidrug transporter QacA reveals long-distance functional associations of residues on opposite sides of the membrane. J. Bacteriol. 190:2441–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hefty PS, Stephens RS. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heinz E, et al. 2009. Comprehensive in silico prediction and analysis of chlamydial outer membrane proteins reflects evolution and life style of the Chlamydiae. BMC Genomics 10:634 doi:10.1186/1471-2164-10-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Henderson IR, Lam AC. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 9:573–578 [DOI] [PubMed] [Google Scholar]
- 51. Henrichfreise B, et al. 2009. Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol. Microbiol. 73:913–923 [DOI] [PubMed] [Google Scholar]
- 52. Henry SC, et al. 2009. Balance of Irgm protein activities determines IFN-gamma-induced host defense. J. Leukoc. Biol. 85:877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S. 2009. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holyoak T, Sullivan SM, Nowak T. 2006. Structural insights into the mechanism of PEPCK catalysis. Biochemistry 45:8254–8263 [DOI] [PubMed] [Google Scholar]
- 56. Horn M, et al. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730 [DOI] [PubMed] [Google Scholar]
- 57. Igietseme JU. 1996. Molecular mechanism of T-cell control of Chlamydia in mice: role of nitric oxide in vivo. Immunology 88:1–5 [PMC free article] [PubMed] [Google Scholar]
- 58. Igietseme JU, Ananaba GA, Candal DH, Lyn D, Black CM. 1998. Immune control of chlamydial growth in the human epithelial cell line RT4 involves multiple mechanisms that include nitric oxide induction, tryptophan catabolism and iron deprivation. Microbiol. Immunol. 42:617–625 [DOI] [PubMed] [Google Scholar]
- 59. Iliffe-Lee ER, McClarty G. 2002. Pyruvate kinase from Chlamydia trachomatis is activated by fructose-2,6-bisphosphate. Mol. Microbiol. 44:819–828 [DOI] [PubMed] [Google Scholar]
- 60. Iliffe-Lee ER, McClarty G. 2000. Regulation of carbon metabolism in Chlamydia trachomatis. Mol. Microbiol. 38:20–30 [DOI] [PubMed] [Google Scholar]
- 61. Inagaki Y, Ehara M, Watanabe KI, Hayashi-Ishimaru Y, Ohama T. 1998. Directionally evolving genetic code: the UGA codon from stop to tryptophan in mitochondria. J. Mol. Evol. 47:378–384 [DOI] [PubMed] [Google Scholar]
- 62. Jamison WP, Hackstadt T. 2008. Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb. Pathog. 45:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kemege KE, et al. 2011. Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes. J. Bacteriol. 193:6517–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kihara A, Akiyama Y, Ito K. 1998. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J. Mol. Biol. 279:175–188 [DOI] [PubMed] [Google Scholar]
- 65. Kim BH, et al. 2011. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332:717–721 [DOI] [PubMed] [Google Scholar]
- 66. Kiselev AO, Skinner MC, Lampe MF. 2009. Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PLoS One 4:e5191 doi:10.1371/journal.pone.0005191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Klos A, Thalmann J, Peters J, Gerard HC, Hudson AP. 2009. The transcript profile of persistent Chlamydophila (Chlamydia) pneumoniae in vitro depends on the means by which persistence is induced. FEMS Microbiol. Lett. 291:120–126 [DOI] [PubMed] [Google Scholar]
- 68. Komla-Soukha I, Sureau C. 2006. A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 80:4648–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kruschwitz HL, McDonald D, Cossins EA, Schirch V. 1994. 5-Formyltetrahydropteroylpolyglutamates are the major folate derivatives in Neurospora crassa conidiospores. J. Biol. Chem. 269:28757–28763 [PubMed] [Google Scholar]
- 70. Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leonhardt RM, Lee SJ, Kavathas PB, Cresswell P. 2007. Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of Chlamydia trachomatis. Infect. Immun. 75:5105–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li YH, Tian XL, Layton G, Norgaard C, Sisson G. 2008. Additive attenuation of virulence and cariogenic potential of Streptococcus mutans by simultaneous inactivation of the ComCDE quorum-sensing system and HK/RR11 two-component regulatory system. Microbiology 154:3256–3265 [DOI] [PubMed] [Google Scholar]
- 73. Li Z, et al. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect. Immun. 76:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lo CC, Bonner CA, Xie G, D'Souza M, Jensen RA. 2009. Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol. Mol. Biol. Rev. 73:594–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lobry JR. 1997. Influence of genomic G+C content on average amino-acid composition of proteins from 59 bacterial species. Gene 205:309–316 [DOI] [PubMed] [Google Scholar]
- 76. Lutter EI, et al. 2010. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect. Immun. 78:3678–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. MacKenzie CR, Hadding U, Daubener W. 1998. Interferon-gamma-induced activation of indoleamine 2,3-dioxygenase in cord blood monocyte-derived macrophages inhibits the growth of group B streptococci. J. Infect. Dis. 178:875–878 [DOI] [PubMed] [Google Scholar]
- 78. MacKenzie CR, et al. 1999. Growth inhibition of multiresistant enterococci by interferon-gamma-activated human uro-epithelial cells. J. Med. Microbiol. 48:935–941 [DOI] [PubMed] [Google Scholar]
- 79. Madan Babu M, Sankaran K. 2002. DOLOP—database of bacterial lipoproteins. Bioinformatics 18:641–643 [DOI] [PubMed] [Google Scholar]
- 80. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Matin A, Auger EA, Blum PH, Schultz JE. 1989. Genetic basis of starvation survival in nondifferentiating bacteria. Annu. Rev. Microbiol. 43:293–316 [DOI] [PubMed] [Google Scholar]
- 82. McClarty G, Caldwell HD, Nelson DE. 2007. Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10:47–51 [DOI] [PubMed] [Google Scholar]
- 83. McCoy AJ, et al. 2006. l,l-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. U. S. A. 103:17909–17914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. McInerney JO. 1998. Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 95:10698–10703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mehlitz A, et al. 2010. Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J. Cell Biol. 190:143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Merino E, Jensen RA, Yanofsky C. 2008. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 11:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. 2011. Co-directional replication-transcription conflicts lead to replication restart. Nature 470:554–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miller BG, Raines RT. 2004. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 43:6387–6392 [DOI] [PubMed] [Google Scholar]
- 89. Miller JD, Sal MS, Schell M, Whittimore JD, Raulston JE. 2009. Chlamydia trachomatis YtgA is an iron-binding periplasmic protein induced by iron restriction. Microbiology 155:2884–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mingorance J, Tamames J, Vicente M. 2004. Genomic channeling in bacterial cell division. J. Mol. Recognit. 17:481–487 [DOI] [PubMed] [Google Scholar]
- 91. Miyairi I, et al. 2007. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 179:1814–1824 [DOI] [PubMed] [Google Scholar]
- 92. Moosmann B, Behl C. 2000. Cytoprotective antioxidant function of tyrosine and tryptophan residues in transmembrane proteins. Eur. J. Biochem. 267:5687–5692 [DOI] [PubMed] [Google Scholar]
- 93. Morrison RP. 2003. New insights into a persistent problem—chlamydial infections. J. Clin. Invest. 111:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Murata M, et al. 2007. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology 153:585–592 [DOI] [PubMed] [Google Scholar]
- 96. Neff L, et al. 2007. Molecular characterization and subcellular localization of macrophage infectivity potentiator, a Chlamydia trachomatis lipoprotein. J. Bacteriol. 189:4739–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nelson DE, et al. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658–10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. O'Donoghue P, Luthey-Schulten Z. 2003. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 67:550–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ouellette SP, et al. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNgamma-mediated host cell tryptophan starvation. Mol. Microbiol. 62:1387–1401 [DOI] [PubMed] [Google Scholar]
- 100. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pe'er I, et al. 2004. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins 54:20–40 [DOI] [PubMed] [Google Scholar]
- 102. Peng K, Monack DM. 2010. Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect. Immun. 78:2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 6:e1000995 doi:10.1371/journal.ppat.1000995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Peters J, Wilson DP, Myers G, Timms P, Bavoil PM. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15:241–251 [DOI] [PubMed] [Google Scholar]
- 105. Pilhofer M, et al. 2008. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J. Bacteriol. 190:3192–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Podkaminski D, Vogel J. 2010. Small RNAs promote mRNA stability to activate the synthesis of virulence factors. Mol. Microbiol. 78:1327–1331 [DOI] [PubMed] [Google Scholar]
- 107. Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rau A, Wyllie S, Whittimore J, Raulston JE. 2005. Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Raulston JE. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65:4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rocha EP. 2004. The replication-related organization of bacterial genomes. Microbiology 150:1609–1627 [DOI] [PubMed] [Google Scholar]
- 111. Rockey DD, Heinzen RA, Hackstadt T. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617–626 [DOI] [PubMed] [Google Scholar]
- 112. Rockey DD, Lenart J, Stephens RS. 2000. Genome sequencing and our understanding of chlamydiae. Infect. Immun. 68:5473–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Romero H, Zavala A, Musto H. 2000. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res. 28:2084–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Saier MH, Jr, et al. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257–279 [PubMed] [Google Scholar]
- 115. Sarsero JP, Wookey PJ, Gollnick P, Yanofsky C, Pittard AJ. 1991. A new family of integral membrane proteins involved in transport of aromatic amino acids in Escherichia coli. J. Bacteriol. 173:3231–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Schmitz-Esser S, et al. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J. Bacteriol. 186:683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Singer GA, Hickey DA. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 17:1581–1588 [DOI] [PubMed] [Google Scholar]
- 118. Smith DR, Chapman MR. 2010. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1:00131–10. doi:10.1128/mBio.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Song J, Ware A, Liu SL. 2003. Wavelet to predict bacterial ori and ter: a tendency towards a physical balance. BMC Genomics 4:17 doi:10.1186/1471-2164-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Spector MP. 1998. The starvation-stress response (SSR) of Salmonella. Adv. Microb. Physiol. 40:233–279 [DOI] [PubMed] [Google Scholar]
- 121. Stephens RS, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 122. Stephens RS, Myers G, Eppinger M, Bavoil PM. 2009. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol. Med. Microbiol. 55:115–119 [DOI] [PubMed] [Google Scholar]
- 123. Stothard DR, Toth GA, Batteiger BE. 2003. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 71:1200–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Subtil A, et al. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 56:1636–1647 [DOI] [PubMed] [Google Scholar]
- 125. Sueoka N. 1995. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 40:318–325 [DOI] [PubMed] [Google Scholar]
- 126. Tekaia F, Yeramian E, Dujon B. 2002. Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene 297:51–60 [DOI] [PubMed] [Google Scholar]
- 127. Thomson NR, et al. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tietzel I, El-Haibi C, Carabeo RA. 2009. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One 4:e6499 doi:10.1371/journal.pone.0006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tjaden J, et al. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187:3980–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Voth DE, Qa'Dan M, Hamm EE, Pelfrey JM, Ballard JD. 2004. Clostridium sordellii lethal toxin is maintained in a multimeric protein complex. Infect. Immun. 72:3366–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wang X, Quinn PJ. 2010. Lipopolysaccharide: biosynthetic pathway and structure modification. Prog. Lipid Res. 49:97–107 [DOI] [PubMed] [Google Scholar]
- 133. Weber A, Flugge UI. 2002. Interaction of cytosolic and plastidic nitrogen metabolism in plants. J. Exp. Bot. 53:865–874 [DOI] [PubMed] [Google Scholar]
- 134. Wehrl W, Meyer TF, Jungblut PR, Muller EC, Szczepek AJ. 2004. Action and reaction: Chlamydophila pneumoniae proteome alteration in a persistent infection induced by iron deficiency. Proteomics 4:2969–2981 [DOI] [PubMed] [Google Scholar]
- 135. Wolfert MA, Roychowdhury A, Boons GJ. 2007. Modification of the structure of peptidoglycan is a strategy to avoid detection by nucleotide-binding oligomerization domain protein 1. Infect. Immun. 75:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wood H, Roshick C, McClarty G. 2004. Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol. Microbiol. 52:903–916 [DOI] [PubMed] [Google Scholar]
- 137. Wyllie S, Raulston JE. 2001. Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40:1027–1036 [DOI] [PubMed] [Google Scholar]
- 138. Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201(Suppl 2):S88–S95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Xie G, Bonner CA, Jensen RA. 2002. Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol. 3:research0051–research0051.17. doi:10.1186/gb-2002-3-9-research0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Xie G, Bonner CA, Song J, Keyhani NO, Jensen RA. 2004. Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy. BMC Biol. 2:15 doi:10.1186/1741-7007-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Xie G, Keyhani NO, Bonner CA, Jensen RA. 2003. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303–342 [DOI] [PMC free article] [PubMed] [Google Scholar]