Abstract
Summary: The yeast two-hybrid system pioneered the field of in vivo protein-protein interaction methods and undisputedly gave rise to a palette of ingenious techniques that are constantly pushing further the limits of the original method. Sensitivity and selectivity have improved because of various technical tricks and experimental designs. Here we present an exhaustive overview of the genetic approaches available to study in vivo binary protein interactions, based on two-hybrid and protein fragment complementation assays. These methods have been engineered and employed successfully in microorganisms such as Saccharomyces cerevisiae and Escherichia coli, but also in higher eukaryotes. From single binary pairwise interactions to whole-genome interactome mapping, the self-reassembly concept has been employed widely. Innovative studies report the use of proteins such as ubiquitin, dihydrofolate reductase, and adenylate cyclase as reconstituted reporters. Protein fragment complementation assays have extended the possibilities in protein-protein interaction studies, with technologies that enable spatial and temporal analyses of protein complexes. In addition, one-hybrid and three-hybrid systems have broadened the types of interactions that can be studied and the findings that can be obtained. Applications of these technologies are discussed, together with the advantages and limitations of the available assays.
INTRODUCTION
The creativity and technical diversity that reside in the tools developed to investigate the binding of one protein to another show how eagerly scientists have been anticipating the characterization of protein-protein interactions (PPIs), from a small-scale atomic level to a large-scale interactomics level. Many genetic, biochemical, biophysical, and computational technologies are now developed that contribute to the knowledge on which proteins interact with each other, taking advantage of specific phenomena that occur during an interaction. These include isothermal titration calorimetry (514), where emission of heat during a protein association is analyzed, and fluorescence anisotropy (417), in which the reduced speed of rotational movement of a protein is detected after it binds another protein. Other biophysical methods include dual polarization interferometry (111), surface plasmon resonance (567), static light scattering (18), and circular dichroism (218) methods. Examples of biochemical interaction technologies are the proximity ligation assay (606), cross-linking (661), the pulldown assay (67), coimmunoprecipitation (316), and tandem affinity purification (TAP) (524). Genetic approaches comprise phage display (78), the yeast two-hybrid system (178), protein fragment complementation assays (PCAs) (312), and protein microarrays (327).
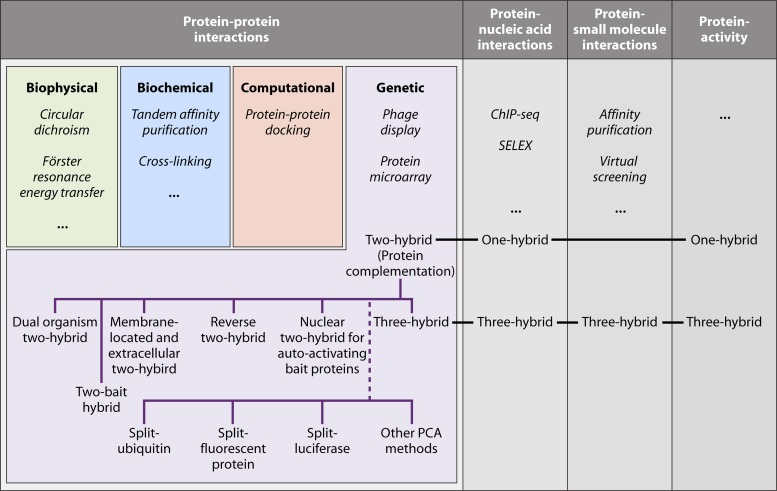
The broad spectrum of available technologies (Fig. 1) is explained by the complementary output that each of them provides. While biophysical methods such as isothermal titration calorimetry have the advantage of giving details on the kinetics of an interaction, several biochemical and genetic techniques can be used to screen for the identification of undiscovered binding partners. Different techniques are also complementary in the identity of PPIs that can be investigated. Affinity purification is the method of preference for the characterization of stable multiprotein complexes, in contrast to the yeast two-hybrid system, which is more suitable for identification of transient and binary PPIs.
Fig 1.
Overview of protein-protein interaction technologies and alternative applications for two-hybrid assay-derived methods. ChIP-seq, chromatin immunoprecipitation followed by sequencing; SELEX, systematic evolution by exponential enrichment; PCA, protein fragment complementation assay. Technologies in italics are not discussed in this review.
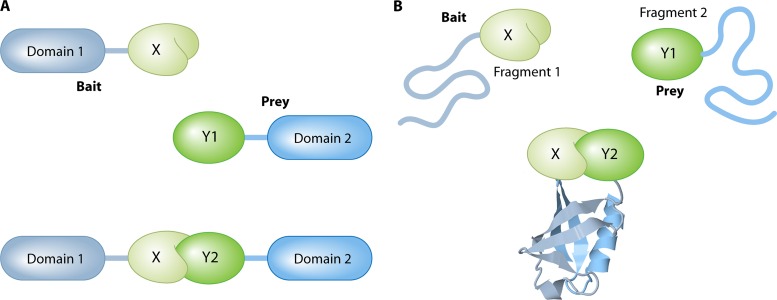
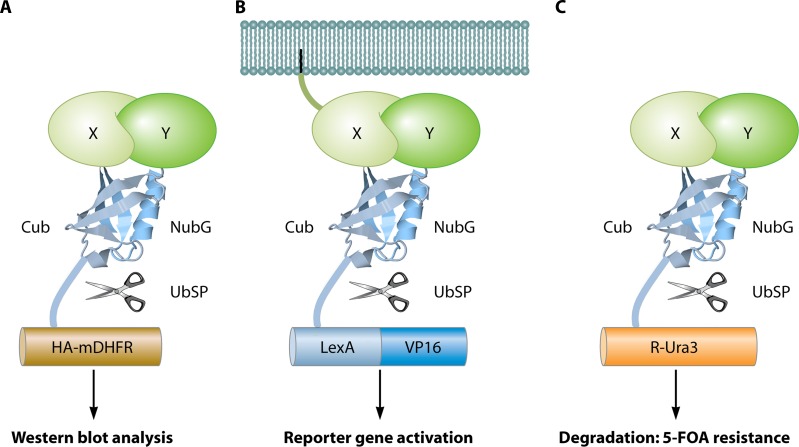
In this review, we focus on genetic in vivo methods for PPI studies. The technologies described can be divided into two main categories: two-hybrid systems and PCAs. The clear distinction between these groups lies in the fact that PCAs depend on the PPI-induced refolding of two protein fragments to reconstitute a functional reporter (Fig. 2A). On the other hand, two-hybrid systems do not depend on PPI-induced refolding of protein fragments but rather on the colocalization of two protein domains (Fig. 2B). These two definitions need to be taken with some practical flexibility. For example, some two-hybrid systems use only one hybrid protein (e.g., G protein fusion systems), and with some PCAs, the refolded protein is not the final reporter by itself but initiates a process that results in the appearance of the actual reporter (e.g., split-ubiquitin system). Nevertheless, the distinction between refolding of protein fragments (PCA) and colocalization of a protein domain(s) (two-hybrid assay) remains true in all cases. There are many limitations and advantages of PCAs in comparison to two-hybrid systems. In general, two-hybrid assays take place artificially in a specific compartment of the cell, which prevents analysis of the genuine subcellular locations of PPIs and can result in false-positive interactions between proteins that normally are found in separate cellular compartments. PCAs usually do not require specific localization and therefore more closely reflect the native environment of the proteins under study. In most cases, two-hybrid systems have reporter gene activation as an output, which is an important factor of signal amplification to increase the sensitivity of the method, but with the cost of lowered selectivity. This balance between sensitivity and selectivity is also seen in PCAs, where the output of the method (e.g., transcription activation, enzymatic activity, or fluorescence), the efficiency of protein fragment refolding, and the stability of the refolded reporter complex define how likely it is that false-negative or false-positive results will be detected. Due to the requirement of the two reporter fragments to refold, PCAs tend to be more sensitive to steric hindrance than two-hybrid systems. PCA selectivity is also affected by the spontaneous reassembly of the reporter independent of a PPI, an issue that concerns mainly PCA methods in which the reconstituted reporter cannot reverse back to the unfolded fragments. A clear advantage of PCAs over two-hybrid systems lies in the fact that some PCAs have the ability to detect PPIs with a high temporal resolution (e.g., the split-luciferase method). Finally, many PCA technologies can very easily be transferred to other organisms, while two-hybrid systems often contain many components (reporter genes and DNA-binding domain [DBD] and activation domain [AD] constructs) that need to be adapted specifically for application in a new organism. Therefore, it must be emphasized that any PCA method described in this review can be applied to any organism of interest that can be transformed or transfected with a vector.
Fig 2.
Two-hybrid systems versus PCAs. (A) Colocalization in two-hybrid systems. Two proteins of interest (X and Y) are each fused to a fixed protein domain, forming the bait and the prey, respectively. In the absence of an interaction (upper part with Y1), the domains remain distant, preventing a detectable output. If the two proteins do interact (lower part with Y2), the bait recruits the prey to a specific cellular location (e.g., reporter gene or plasma membrane), where it can stimulate a detectable output (e.g., gene activation or signal transduction). The domains do not need to be in physical contact. (B) Protein refolding in PCAs. Two proteins of interest (X and Y) are each fused to a fixed protein fragment. If there is no interaction between X and Y (upper part with Y1), the fragments remain unstructured and lack any functional abilities. Upon interaction between the bait and prey (lower part with Y2), the two fragments refold into a fully functional reporter protein (e.g., ubiquitin in the figure). In most cases, the interaction does not need to take place in a specific cellular location, but a minimal time frame of physical contact between the two fragments is necessary to establish complete refolding. The image of the ubiquitin protein is based on the Protein Data Bank (PDB) structure under accession number 1UBQ (671).
For reasons of consistency, we always use the general term “PCA” to address the category of reporter folding technologies, but it should be noted that they are also known as “split-protein sensors.” Specific PCA techniques are named split-“X” methods, such as the split-ubiquitin and split-luciferase methods, because this is the most commonly used way to address them.
Fluorescence resonance energy transfer (FRET) and bioluminescence resonance energy transfer (BRET) are similar to two-hybrid and PCA methods. They are not discussed here, but several reviews can be found that elucidate the uses of FRET and BRET for PPI research (117, 118, 394, 413, 517, 646, 677). Likewise, in vitro PCA applications are not mentioned, but there are public reports on the use of these techniques for discovery of PPI-inhibiting compounds (243).
This review provides insights into two-hybrid systems and PCAs, highlighting their applications, advantages, and limitations. The first part describes the evolution of the original yeast two-hybrid system, from the original design to high-throughput genomewide screens. The second part explains alternative two-hybrid systems in Saccharomyces cerevisiae for the study of PPIs but also for other purposes, such as the detection of PPI inhibitors and the examination of associations between proteins and RNA, DNA, or small molecules. The third part deals with the current applications of PCAs in S. cerevisiae. Importantly, this section also contains a general introduction to three of the most commonly applied PCAs (the split-mouse dihydrofolate reductase [split-mDHFR], split-luciferase, and split-fluorescent protein [split-FP] methods), with considerations that apply to all organisms. The last part covers the development and applications of two-hybrid systems and PCAs in different organisms ranging from bacteria to mammalian cells. Table 1 gives an overview of validated applications for the currently available systems.
Table 1.
Validated applications for genetic protein-protein interaction technologies
| Purpose | Technology [reference(s)]a |
|---|---|
| Confirmation of PPIsb | All endogenous PCAs |
| All endogenous two-hybrid systems | |
| Large-scale screening for PPIsc | Two-hybrid systems (yeast, bacteria) (689, 721) |
| Split-ubiquitin system (yeast) (447) | |
| Split-DHFR system (yeast) (633) | |
| Mammalian two-hybrid system (mammalian cells) (535) | |
| Mammalian protein-protein interaction trap (mammalian cells) (395) | |
| Split-FP system (mammalian cells) (376) | |
| Small-scale screening for PPIsd | Sos recruitment system (yeast) (455) |
| Ras recruitment system (yeast) (331) | |
| Repressed transactivator system (yeast) (569) | |
| Split-FP system (mammalian cells) (544) | |
| RNA Pol III system (yeast) (597) | |
| One-hybrid system for PPIs (yeast) (229) | |
| Association and dissociation of PPIs (temporal dynamics) | Split-luciferase system (yeast, mammalian cells) (429, 615) |
| Localization of PPIs | Split-FP system (yeast, bacteria, fungi, plants, animal cell cultures) (17, 262, 602) |
| Split-DHFR system with fluorescein-conjugated substrate (plants, mammalian cells) (545, 625) | |
| Discovery of PPI inhibitors | Reverse two-hybrid system (yeast, bacteria) (115, 710) |
| Forward two-hybrid system (yeast, mammalian cells) (199, 594) | |
| Repressed transactivator system (yeast) (313) | |
| Split-FP system (bacteria) (457) | |
| Split-CyaA system (bacteria) (495) | |
| Discovery of amino acids perturbing a PPI | Reverse two-hybrid system (yeast, bacteria) (239, 510) |
| Forward two-hybrid system (yeast) (717) | |
| Two-bait hybrid systems (yeast) (539) | |
| Split-ubiquitin system (yeast) (74) | |
| Split-yCD system (yeast) (156) |
Many other technologies exist, for these and also for alternative applications. These are discussed in the text.
Preference goes to methods which mimic the appropriate cellular environment and native expression as closely as possible.
Includes technologies with unbiased screening applications with a significant number of bait proteins in parallel. Prey libraries can originate from a rational (open reading frames) or random (cDNA or genomic DNA libraries) source.
Includes technologies with applications in library screening for PPIs limited to one or a few bait proteins in parallel. Several methods showed promise in prototype experiments with controlled libraries (e.g., see references 190 and 355). Some techniques have been used for module-scale experiments, with multiple bait and prey proteins but without a library (e.g., see reference 161). These are all not included in the table but discussed in the text.
THE YEAST TWO-HYBRID SYSTEM
Development of the Yeast Two-Hybrid System
Fields and Song.
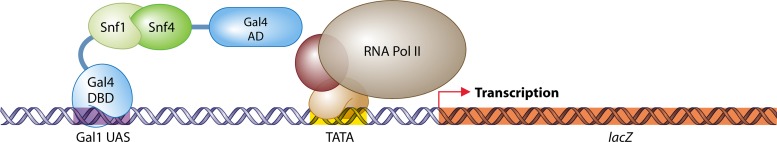
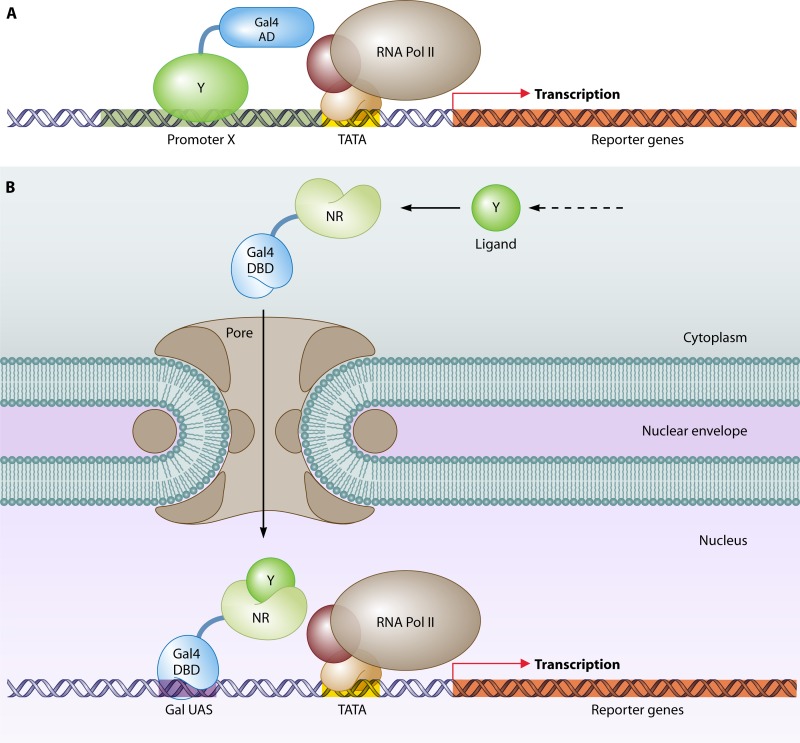
A milestone in the field of PPI studies came with the publication of “A Novel Genetic System To Detect Protein-Protein Interactions” by Stanley Fields and Ok-Kyu Song in 1989 (178). They took advantage of previous studies on the modular arrangement of transcription factors, including S. cerevisiae Gal4 (63, 330, 419, 420, 618). The N-terminal domain of Gal4 (amino acids [aa] 1 to 147) binds to the upstream activating sequence of GAL1, and the C-terminal part of Gal4 (aa 768 to 881) serves as the activation domain, which stimulates gene expression. Both domains carry out their functions independently, but only when they are brought together is a DNA-binding and gene expression-activating protein formed. The principle of their system lies in the fusion of each domain with a protein of interest. Binding of the two proteins of interest results in the reassembly of the transcription factor Gal4, which in turn induces expression of one or more reporter genes. In the study of Fields and Song, the physical association of two S. cerevisiae proteins, Snf1 and Snf4, was confirmed by showing that a strain that expresses the hybrid genes GAL4(1-147)-SNF1 and GAL4(768-881)-SNF4 is capable of inducing expression of an Escherichia coli lacZ reporter gene controlled by the GAL1 promoter (Fig. 3). The DNA-binding domain fusion was named the “bait” that is used to “capture” the so-called “prey” activation domain fusion.
Fig 3.
The first two-hybrid experiment (178). For the study of the interaction of two proteins of interest (in this case, Snf1 and Snf4), one protein is fused to the DNA-binding domain (DBD) of Gal4 (the bait) and the other protein is fused with the activation domain (AD) of Gal4 (the prey). The bait fusion binds upstream activating sequences (UAS) of the reporter gene lacZ. Association of Snf1 with Snf4 brings the Gal4 AD to lacZ, followed by recruitment of the basal transcriptional machinery, which establishes lacZ transcription, detected by chromogenic analysis.
Sensitivity and selectivity.
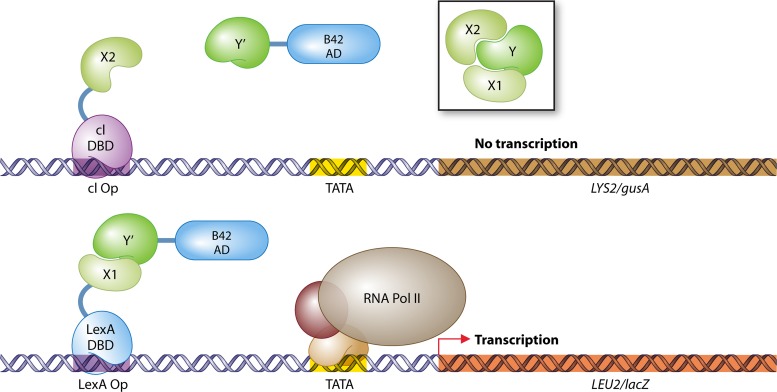
Since 1989, the two-hybrid system has been the subject of many improvements of all its fundamental components, e.g., the reporter genes, the AD, and the DBD. Table 2 gives an overview of the currently available possibilities for each factor. Besides chromogenic reporters such as E. coli LacZ, prototrophic reporter genes were introduced to single out colonies that include interacting bait and prey proteins on prototrophy-selective medium (e.g., see references 234 and 673). This step greatly facilitated the use of prey plasmid libraries to identify the proteins that interact with the bait of interest in a large collection of noninteracting proteins, a significant advantage of the two-hybrid system over many other technologies. Most two-hybrid strains contain multiple reporter genes, with a different promoter region for each reporter, to enable a wider spectrum of sensitivity and selectivity. As an example, strain PJ69-4A (299) contains HIS3, ADE2, and lacZ, controlled by the GAL1, GAL2, and GAL7 promoters, respectively. The HIS3 reporter provides the highest sensitivity, as the Gal4 DBD binds the GAL1 promoter very efficiently. In contrast, PPI assays based on the GAL2p-ADE2 module are very stringent and can be used to exclude dubious results. Finally, the reporter gene lacZ can be applied for ultimate confirmation of the interaction in a semiquantitative (169) galactosidase assay. A more recent selection approach takes advantage of the yeast enhanced green fluorescent protein (yEGFP) as a reporter to screen for interacting pairs by fluorescence-associated cell sorting (FACS) (87–89).
Table 2.
Reporter genes, activation domains, and DNA-binding domains used in yeast two-hybrid experiments
| Componenta | Description (reference) |
|---|---|
| Reporter genes | |
| E. coli lacZ* | β-Galactosidase chromogenic reporter (178) |
| S. cerevisiae MEL1 | Secretory α-galactosidase chromogenic reporter (5) |
| E. coli gusA | β-Glucuronidase chromogenic reporter (580) |
| Aspergillus oryzae lacA3 | Engineered secretory β-galactosidase chromogenic reporter (318) |
| S. cerevisiae HIS3* | Prototrophic reporter for histidine biosynthesis (673) |
| S. cerevisiae LEU2* | Prototrophic reporter for leucine biosynthesis (234) |
| S. cerevisiae URA3 | Prototrophic reporter for uracil biosynthesis (374) |
| S. cerevisiae ADE2* | Prototrophic reporter for adenine biosynthesis (299) |
| S. cerevisiae LYS2 | Prototrophic reporter for lysine biosynthesis (580) |
| Aequorea victoria GFPuv | Fluorescent reporter (107) |
| EGFP | Fluorescent reporter (613) |
| Yeast EGFP | Fluorescent reporter for flow cytometry screens (88) |
| Aureobasidium pullulans AUR1-C | Aureobasidin A resistance reporter (167) |
| Prey activation domains | |
| S. cerevisiae Gal4 AD | Gal4 activating region II (aa 768 to 881), moderate strength (178) |
| Herpes simplex virus VP16 AD | VP16 activating region (aa 413 to 490), high strength (673) |
| E. coli B42 AD | Bacterial polypeptide, weak strength (234) |
| Bait DNA-binding domains | |
| S. cerevisiae Gal4 DBD* | Binds GAL1, GAL2, and GAL7 upstream activating sequences (178) |
| E. coli repressor LexA DBD* | Binds LexA operator sequences (234) |
| H. sapiens estrogen receptor DBD | Binds estrogen receptor elements (374) |
| Bacteriophage λ repressor cI | Binds cI operator sequences (580) |
| Tet repressor | Binds Tet operator sequences (716) |
*, most popular options.
Apart from the reporter promoter region, other factors influence the balance between sensitivity and selectivity. The copy numbers of the episomal bait and prey plasmids, together with the bait and prey promoters, affect expression levels, which in turn have an impact on the likelihood of detecting an interaction (379). CEN-based plasmids and truncated ADH1 promoters lower bait and prey expression levels for high selectivity, while 2μm-based vectors and GAL1 or full-length ADH1 promoters contribute to an increased sensitivity of the system. Libraries of prey plasmids cannot be integrated due to the low transformation efficiency linked with genomic integration. However, bait integration in combination with a strong promoter, to reduce the variability of bait expression, provides a very attractive alternative to the traditional episomal approach. It has proven to reduce both false-positive and false-negative results in library screening experiments (453) and also in alternative two-hybrid systems (22, 451, 502).
In a recent study, the impact of bait and prey vector identity on the positive output of interaction assays was studied with Treponema pallidum and Escherichia coli motility-related proteins (531). Remarkably, different combinations of bait and prey vectors led to differences in the number of positive results, the subset of detected PPIs, and the reliability of the outcome. The authors suggested not only that the expression levels of bait and prey affect output but also that a potential role may be present for less obvious factors, such as the presence or absence of a stop codon in the backbone vectors or the size of the linker sequences within the fusion constructs. Similar conclusions could be drawn using a standard positive and negative reference set (91). In these analyses, many of the positive results acquired by only one bait-prey vector combination were part of the positive reference set, which suggests that the small overlap seen in large-scale studies (e.g., see reference 294 versus reference 650) does not necessarily point to a large number of false-positive results but rather to a variation in experimental procedures. Another possible underappreciated influence on two-hybrid results is the exact composition of the medium (410).
In addition, the identities of the DNA-binding domain, the activation domain, and the reporter genes likely further influence the number of positive (true and false) results. For example, most reporter genes are integrated into the genome, but in some experimental designs, the lacZ gene is retained on an episomal plasmid for an increased response (169, 693). Moreover, some activation domains are strong expression inducers, particularly VP16, while the B42 AD is known to be a weaker activator (62, 557). Finally, Uetz and colleagues recently illustrated the impact of steric hindrance in a study on the interactome of varicella-zoster virus (616). All 70 proteins of this virus were cloned as both N- and C-terminal fusions with the DBD and AD. Interestingly, Uetz et al. discovered three times more interactions than would have been found using only C-terminal fusions, due to increased accessibility of N-terminal interaction domains in the N-terminal fusion constructs. This result shows the intrinsic capacity of the system to find a substantially larger number of PPIs by partially overcoming the steric hindrance problem. In conclusion, the combined use of different setups should extensively enlarge the interactome subspace detectable by two-hybrid methods.
Screening procedures.
In parallel with the technical evolution of the system, several strategies have been developed and optimized for screening experiments (Table 3). In accordance with the specific desire and availability, genomic DNA (gDNA), cDNA, normalized cDNA, full-length cDNA, or open reading frame (ORF) libraries may form the option of choice. Originally, a screening experiment involved the sequential or simultaneous transformation of a two-hybrid strain with the bait plasmid and a prey library. An alternative strategy consists of the construction of mating type a and α two-hybrid strains with the bait and prey plasmids, respectively (37, 180, 197). The screening step is performed by mating of both strains on medium selective for an interaction. The possibility to recycle these mating type-specific bait and prey strains provides a strong advantage over the classic transformation approach, especially for high-throughput screening experiments. Finally, prey plasmids can be pooled in a library or tested individually for one-to-one interactions; the latter is called the array or matrix approach. By separating prey ORFs, the finding of positive clones automatically leads to the identification of the interacting protein without the need for sequencing. In addition, prey constructs that activate reporter genes independently from the nature of the bait are easily discarded when screenings with several baits are performed. While it is difficult to estimate the coverage of an experiment using libraries, the coverage of the matrix approach is much more controlled. Several studies assayed both ORF library and ORF matrix screens and showed that the proportional output of interactions is higher and the number of false-positive results is lower with the matrix strategy (120, 650, 682). However, one-to-one characterization of PPIs in genome-scale two-hybrid screenings is cumbersome, and therefore small prey pool strategies are still the common way to screen in these cases (e.g., see references 294, 494, and 721), or one-to-one experiments are completed after a first selection round with pools (734). In only one study, for which a PCA technology was used, 15 million mating experiments were performed with individual bait and prey strains (633). In an alternative approach, prey plasmids are grouped in unique combinations of pools (smart pools) that allow for the fast identification of the interacting prey protein (310, 311). This technique takes advantage of an inventive pooling-deconvolution procedure. 2N preys are grouped in 2 × N unique pools, with each prey protein represented N times. For example, with 8 prey proteins, A to H (N = 3), we have 6 (2 × 3) unique pools (pool 1, A/B/C/D; pool 2, E/F/G/H; pool 3, A/B/E/F; pool 4, C/D/G/H; pool 5, A/C/E/G; and pool 6, B/D/F/H), with each protein present 3 times. If pools 1, 3, and 5 lead to growth on selective medium, the distinctive conclusion is that prey A interacts with the bait. This strategy relies on the small number of positive clones to be expected. The simple deconvolution step to fish out the protein responsible for the interaction simplifies the postscreening protocol. The relative number of pools to be screened decreases exponentially with the number of prey proteins, from 4 pools for 4 proteins to 10 pools for 32 proteins and 14 pools for 128 proteins. Moreover, with increasing size, each protein is represented more frequently. In conclusion, smart-pool arrays offer an elegant method to screen for PPIs. Finally, bait libraries can also be pooled after removal of autoactivating bait constructs (e.g., see references 30 and 294).
Table 3.
Strategies to screen for PPIs in the yeast two-hybrid system
| Library type | Features, with advantages (+) and disadvantages (−) |
|---|---|
| Genomic DNA | For organisms with low intron occupancy and small intergenic regions; genomic DNA is cut with ClaI-compatible restriction enzymes (95, 299); +, cheap, incomplete fragments may facilitate positive outcome (e.g., with membrane proteins or incorrectly annotated protein-encoding genes); −, small fraction of in-frame protein coding fragments, introns are present |
| cDNA | For organisms with high intron occupancy and large intergenic regions (614); +, cheap, exclusion of noncoding fragments and introns, correct orientation; −, only partial fraction of in-frame protein coding fragments, strong difference in abundance between different cDNA fragments |
| Normalized cDNA | Normalizes the amount of cDNA fragments for each gene (683); +, exclusion of noncoding fragments and introns, correct orientation, better representation of each cDNA fragment; −, relatively expensive, only partial fraction of in-frame protein coding fragments |
| Full-length cDNA | Full-length cDNA fragments created with gene-specific forward primers (627); +, exclusion of noncoding fragments and introns, correct frame and orientation, balanced representation of each gene; −, expensive, complete cDNA fragments reduce the positive outcome rate for specific types of interactions (e.g., with membrane proteins) |
| Open reading frame DNA | Each open reading frame is individually cloned into the prey library by in vivo (gap repair) or in vitro (Gateway from Invitrogen) recombinational cloning (294, 650); +, exclusion of noncoding fragments and introns, correct frame and orientation, balanced representation of each gene; −, expensive, introns are present, complete ORF fragments reduce the positive outcome rate for specific types of interactions (e.g., with membrane proteins) |
Protocols.
Several reviews and method papers report in detail the available strains and plasmids (91, 298, 656) and provide experimental procedures (124, 198, 364, 403, 422, 423, 530, 577). Companies that provide kits for two-hybrid experiments include Dualsystems Biotech (DUALhybrid kit), Invitrogen (ProQuest), Clontech (Matchmaker Gold), Agilent Technologies (HybriZAP), and Promega (CheckMate). Two-hybrid services are available from Hybrigenics and Dualsystems Biotech. For large-scale (multiple-bait) screening, a new strategy was proposed for the identification of bait-prey pairs by en masse next-generation sequencing after PCR stitching (fusing) of the associated bait and prey genes, for significant cost reduction of the analysis step (723).
Application of the Yeast Two-Hybrid System on a Small Scale
The yeast two-hybrid system has established a prominent position in cell biological research and led to the confirmation and discovery of thousands of PPIs. The method complements affinity purification, particularly tandem affinity purification, for the unbiased detection of new protein associations. It has played a crucial role as the starting point of very diverse studies, such as the analysis of light responses and abscisic acid (ABA) signaling in plants (407, 491), the mechanism of protein degradation in the endoplasmic reticulum (654), the molecular basis of limb regeneration in adult vertebrates (366), the interactome topology between herpesviral and human proteins (649), the impact of a bacterial scaffold on human endomembrane trafficking (572), and the regulation of eye development (125). In these examples, a defined bait protein was screened for interactions with a complementary or genomic DNA prey library, leading to the discovery of new PPIs, followed by functional analysis of the newly identified proteins. This second step is crucial as part of the validation process to increase the reliability of two-hybrid data.
Interaction dynamics.
One major future challenge in the study of PPIs is the characterization of their dynamic features. The two-hybrid system in most cases is appropriate to answer only the question of whether two proteins can associate, but it does not provide details on when or in which circumstances the interaction happens. A yeast two-hybrid experiment brings the proteins of interest into a rather unnatural situation, because they are directed to the nucleus (forced colocalization), their corresponding genes are not expressed under the control of their own promoters (forced coexpression), and, with nonyeast proteins, the whole cellular environment differs from the native context. As a result, external influences caused by gene deletions, nutrient sources, or stress conditions may not affect the two-hybrid interaction status due to the absence of mediating factors (e.g., signaling molecules). Therefore, context-dependent interaction studies are ideally performed with the organism from which the proteins of interest originate, in the subcellular compartment in which the proteins of interest naturally reside, and with expression of the proteins under the control of their own promoters. Following this rule, only yeast nuclear protein associations can be investigated for context-specific interaction dynamics by use of the yeast two-hybrid system. For nonnuclear yeast proteins, alternative genetic PPI methods are available (see Alternative Yeast Two-Hybrid Systems and Protein Fragment Complementation Assays in Yeast), and for nonyeast proteins, a large group of interaction technologies has been developed in other host organisms (see Genetic Protein-Protein Interaction Methods in Other Organisms).
Several examples exist where the two-hybrid system was applied for context-dependent PPI studies. Interaction between two catalytic and two regulatory subunits of protein kinase A in S. cerevisiae is stimulated by the kelch repeat proteins Krh1 and Krh2. In the absence of Krh1 and Krh2, the formation of the tetrameric protein kinase A complex is partially inhibited, as shown by a yeast two-hybrid experiment (506). Nutrient sources also affect many protein associations in S. cerevisiae. Formation of the Snf1 protein kinase complex is repressed by glucose, as illustrated in a two-hybrid assay where interactions between subunits of the complex were detected only in media containing alternative carbon sources (305). Taking advantage of the two-hybrid system, it was demonstrated that the transcription factor Rgt1 binds hexokinase 2 (Hxk2) only at high glucose concentrations and binds Med8, a subunit of the RNA polymerase (RNAP) II mediator complex, only at low glucose concentrations (487). An example of stress-dependent interactions is the association of the Hsp90 chaperone with the mitogen-activated protein (MAP) kinase Slt2 under high-temperature conditions or after addition of caffeine (449).
Genetic modifications.
In some PPI studies, the two-hybrid system required specific modifications to generate accurate data. For example, the essential S. cerevisiae F-box protein Cdc4 binds Sic1, an inhibitor of cell cycle proteins, and stimulates its degradation (175). However, direct association in a wild-type strain was difficult to show because of the fast degradation of Sic1 induced by Cdc4. Therefore, a temperature-sensitive cdc4-1 yeast two-hybrid strain was constructed to prove the interaction between Sic1 and catalytically inactive Cdc4 (348). The two-hybrid system is generally considered to detect binary and direct interactions, a notion which may be true for many nonnuclear and nonyeast PPIs. However, to establish a clear direct interaction network of autophagy-related (Atg) proteins in S. cerevisiae, a two-hybrid strain with deletions of 24 ATG genes was created, and multiple Atg PPIs were confirmed, showing a direct physical interaction independent of the presence of other Atg proteins (75). Finally, deletion of endogenous genes can reduce competitive binding for bait or prey proteins (670).
Application of the Yeast Two-Hybrid System on a Large Scale
Although PPI technologies such as surface plasmon resonance, FRET, phage display, and protein microarrays have been applied to some extent for large-scale experiments (71, 174, 327, 522, 719, 738), affinity purification followed by mass spectrometry (MS) (14, 61, 69, 170, 200, 201, 261, 365), PCAs (447, 633), and the two-hybrid system are most commonly used, to date, for high-throughput interactome analysis.
Interactome studies.
The first S. cerevisiae two-hybrid screening on a large scale was carried out by Fields, Rothberg, and colleagues in 2000 (650). They conducted both matrix and library high-throughput experiments. In the matrix approach, 192 individual bait strains were mated each time with 1 of 6,000 ORF prey strains, identifying 281 protein associations that were found in two parallel experiments (20% of the total PPIs discovered). Alternatively, 5,300 ORF bait strains were screened using a pooled prey library, leading to the identification of 691 interactions. A second large-scale assay (294) was performed by making approximately 6,000 ORF bait and prey strains. Four hundred mating reactions were carried out, each time with 96 bait strains against 96 prey strains, revealing 4,549 PPIs. Positive results that were found three times were grouped into a core collection of 841 interactions. Surprisingly, of this core set, only 141 PPIs had been identified in the first study (650). In the most exhaustive screening to date, by Vidal and colleagues (721), 3,917 nonautoactivating bait strains were individually mated with 5,246 prey strains (merged in 94 pools), uncovering 1,809 PPIs, of which 274 interactions were found in the two previous high-throughput experiments. The low level of overlap between the three data sets can be explained by false-positive records (i.e., the precision of the method) and by false-negative results, with the latter dependent on the screening completeness (the fraction of the total number of ORF pairs tested to the total possible number of ORF pairs of the organism under study), the assay sensitivity (the fraction of interactions that can possibly be identified by the assay), and the sampling sensitivity (the fraction of all identifiable interactions in a single trial of an assay) (493, 662, 663, 721). The yeast two-hybrid system has also been applied in large-scale research to investigate intraviral (19, 30, 182, 184, 231, 246, 437, 488, 513, 530, 555, 616, 641, 728) and pathogen-host (120, 155, 187, 339, 717) interactions. Moreover, it has also been employed for interactome mapping of Campylobacter jejuni (494), Helicobacter pylori (529), Synechocystis sp. PCC6803 (562), Bacillus subtilis (431, 472), Plasmodium falciparum (369), Arabidopsis thaliana (59, 235), Drosophila melanogaster (183, 208, 612), Caenorhabditis elegans (55, 116, 389, 538, 601, 681, 682), and Homo sapiens (7, 105, 210, 381, 397, 428, 467, 551, 617, 723).
Analysis of high-throughput data.
Cost reduction and technological improvements allowed for high-throughput two-hybrid screenings but shifted the limiting step toward the confirmation and validation of interaction data. Small-scale two-hybrid results can be verified by alternative interaction techniques, such as glutathione S-transferase (GST) pulldown assay or coimmunoprecipitation, or by the proof of a functional correlation. Although full experimental validation of a medium-scale two-hybrid assay has been reported (349), this type of verification for genomewide PPI studies remains limited to a subset of positive results (389, 721). On the other hand, many computational studies have evaluated the false-positive and false-negative rates of two-hybrid results by using random and positive reference sets, respectively. Specifically, for two-hybrid studies, a positive reference set of binary interactions has been proposed to accurately validate high-throughput data (721). Estimates go from 24% to 51% for the false discovery rate (119, 278) and from 45% to 96% for the false-negative rate (159). In general, two proteins are more likely to be true interactors if they tend to share common features, including coexpression (119, 223, 301, 333, 640), colocalization (301), functional correlation (425, 497), and shared interaction partners (11, 212, 346), and have homologous proteins that bind each other (119, 186, 405, 436, 533, 722). However, coexpression and colocalization analyses for interaction validation, supporting cocomplex analysis and biased small-scale experiments, have been criticized because many true interactors show an anticorrelation with expression (463, 663). Comparison of interaction data for validation across different species has been criticized as well (340, 445, 586). Even a functional correlation is not a necessary prerequisite, as many interactions may truly appear in the cell without a functional context (384). At present, there is no clear consensus on the strategies to accurately validate interaction results, but the ever-increasing availability of PPI data by a variety of experimental tools will support the accuracy of computational validation, which in turn will allow for precise predictions of true interactions.
Computational analysis of PPI data further revealed several biases toward different protein properties. Nuclear, conserved, essential, weakly autoactivating, and structurally disordered proteins are overrepresented in two-hybrid data, but no biases toward protein function were found (44, 93, 676, 724). For high-throughput affinity purification assays, detection of PPIs is skewed toward highly abundant proteins due to the use of native promoters and of proteins associated with specific cellular functions involving protein complexes, such as transcription and protein synthesis (44, 93, 295, 676, 724).
High-throughput PPI data have assisted in the functional characterization of proteins (4, 23, 260, 333, 383, 540, 583), the analysis of interaction network topologies (8, 121, 185, 303, 399, 496, 527, 680, 685, 743; reviewed in references 24, 532, and 568), and the computational prediction of interactions and interactomes (66, 154, 164, 226, 249, 280, 332, 377, 404, 438, 469, 571, 604, 688, 703, 733), protein localization (591), interacting domains (227, 409, 609) and interactome sizes (224, 241, 301, 559, 622, 663, 676, 721).
Limitations of the Yeast Two-Hybrid System
The two-hybrid system suffers from three major drawbacks: (i) it produces a significant number of false-positive results, (ii) it can detect only a subset of the complete interactome, and (iii) it can provide only very limited information on the kinetics or dynamics of a PPI.
False-positive results.
In screening for interacting partners by use of a library, a relatively large number of false-positive results is frequently observed. Sometimes, interacting proteins are detected that are not present in the same subcellular location or time under natural conditions. These proteins might indeed be able to interact, but the interactions have no biological relevance (biological artifacts). Other examples of false-positive results are proteins that overcome nutritional selection, proteins that bind and activate the reporter gene directly, “sticky” or incorrectly folded proteins that nonspecifically bind many baits, plasmid rearrangements or copy number changes that generate autoactivators, or alterations at one of the reporter genes that result in constitutive expression (technical artifacts). However, several approaches exist that deal with these spurious results during screening (251, 579, 669), including the application of two-bait systems (see “Two-Bait Hybrid Systems”). Although initial analyses of high-throughput two-hybrid data suggested large proportions of false-positive results (223, 300, 463), recent examinations indicate an overestimation of the false discovery rates due to misevaluation of the data (663, 721). It is clear now that different technologies, such as the two-hybrid system and tandem affinity purification, lead to detection of different types of PPIs (112, 213, 302) and therefore that each data set should be analyzed by specific appropriate validation methods (663, 721). Nevertheless, spurious two-hybrid results remain a considerable drawback, and only more experimental in-depth analysis can provide conclusive confirmation of a true biologically relevant interaction. Alternative two-hybrid and PCA technologies can play a central role in this data verification.
A particular case of a false-positive result is autoactivation by the bait protein, i.e., the bait by itself can induce expression of the reporter genes independently of the prey. This issue can be solved by removing the domain that induces autoactivation or by increasing 3-aminotriazole (3-AT) levels when using HIS3 as a selection marker. 3-AT is a competitive inhibitor of the His3 enzyme, and addition of this compound elevates the required His3 levels for survival on histidine-deficient medium. Alternative two-hybrid technologies can also be used to address this problem (see “Nuclear Two-Hybrid Systems for Autoactivating Bait Proteins”).
Detection of only a subset of the interactome.
Due to the necessity of the bait and prey proteins to enter the nucleus, a number of interacting protein pairs are not able to induce reporter gene expression. Extracellular proteins, membrane proteins, and generally all proteins with a strong localization signal will often not move to the nucleus, despite their fusion with a nuclear localization sequence (NLS). Moreover, in most cases, membrane proteins need the phospholipid bilayer to fold in the right conformation, and therefore an interaction may not be observed. To overcome the problem of mislocalization, it can be necessary to use a truncated version of the protein. As an example, the S. cerevisiae G-protein-coupled receptor (GPCR) Gpr1 is a membrane protein, but the cytoplasmic C-terminal region was shown to interact with the Gα protein Gpa2 in a yeast two-hybrid experiment (363, 725). Alternatively, PCAs or modified two-hybrid systems may be used to detect membrane PPIs (see “Membrane-Localized and Secretory Pathway Two-Hybrid Systems” and Protein Fragment Complementation Assays in Yeast). Occasionally, proteins might be present in the nuclear environment but unable to interact, such as proteins of the secretory compartments that require oxidative conditions or glycosylation for proper folding (354). Another cause of false-negative interactions results from bait or prey proteins that are toxic to the cell when overexpressed. This problem can be circumvented by the use of inducible promoters, such as in the LexA-based system, where prey genes are expressed under the control of the inducible GAL1 promoter. The use of chimeras has also been criticized because the addition of fusion constructs to the protein of interest could obstruct the interaction. However, the addition of flexible glycine linkers to stimulate independent folding of the different components of the fusion protein partly deals with this problem. As mentioned above, N-terminal fusion of the protein of interest to the AD or DBD can also improve the outcome (e.g., see references 521 and 616). Several PPIs, particularly those of higher eukaryotes, might not be detected in S. cerevisiae when the machinery for a specific posttranslational modification is lacking in yeast cells. Coexpression of the modifying enzyme in the heterologous host system can solve this issue, or the interaction analysis can be performed in vivo in other organisms (see “Three-Hybrid Systems” and Genetic Protein-Protein Interaction Methods in Other Organisms, respectively).
Limited information on the kinetics or dynamics of a PPI.
The artificial environment of a two-hybrid experiment, with forced coexpression and nuclear localization of chimeric proteins, limits the application of the system to semiquantification of binding affinities (169). Furthermore, the length of an experiment, on a multiple-day scale for prototrophic reporter activation, does not allow the detection of fast changes in interaction affinity induced by external factors. Finally, the native location of the protein association under study cannot be analyzed by two-hybrid experiments. PCAs such as the split-luciferase method and the split-FP method have been shown to be highly versatile systems for research on dynamic PPIs (see Protein Fragment Complementation Assays in Yeast and Genetic Protein-Protein Interaction Methods in Other Organisms).
Despite these drawbacks, the track record of the two-hybrid system proves how efficient this method has been for discovering many new PPIs from a large number of organisms due to a number of advantageous properties. The yeast two-hybrid system provides a technique to investigate interactions in the environment of a eukaryotic cell and with the easy handling characteristics of S. cerevisiae. In contrast to the case for affinity purification methods, transient and weak associations can be detected due to signal amplification provided by reporter gene expression. The lack of a cumbersome purification step also adds to the efficiency of the system. The method allows the identification of new binding partners of a protein of interest, which plays an important role in the functional analysis of uncharacterized proteins. The convenience of working with S. cerevisiae makes it possible to screen for drug compounds that disrupt interactions or to screen for mutated versions of a protein that lose the ability to associate with binding partners (see “Reverse Two-Hybrid Systems”). Finally, the concept of the method permits the development of many alternative technologies.
ALTERNATIVE YEAST TWO-HYBRID SYSTEMS
The basic concept of functional reconstitution of a transcription factor has been used as the blueprint for several alternative technologies, many of which deal with the limitations of the original system. These methods are discussed here.
Nuclear Two-Hybrid Systems for Autoactivating Bait Proteins
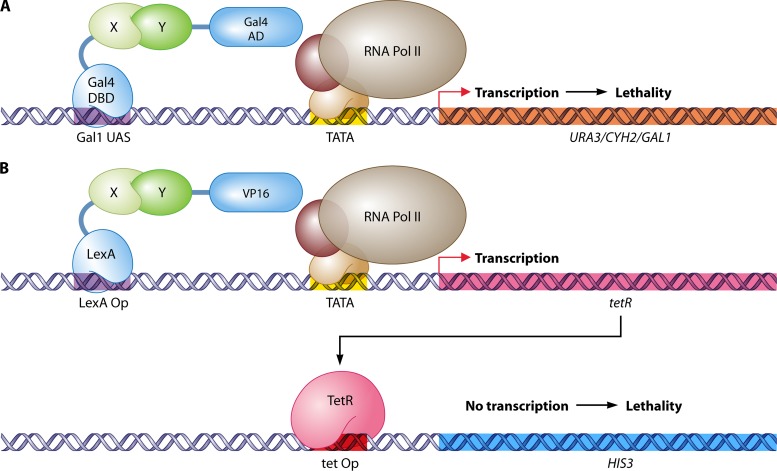
RNA Pol III system.
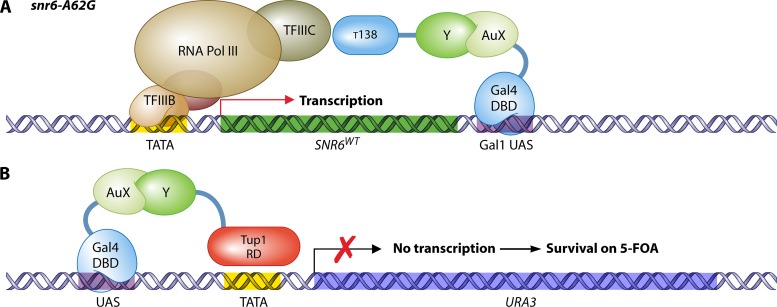
If the bait protein is able to autoactivate transcription by recruiting the traditional RNA polymerase II, an alternative option is to use the RNA polymerase III (RNA Pol III) system (433). RNA Pol III transcribes genes that encode untranslated RNA molecules such as rRNA, tRNA, and other small RNAs. Most of the genes controlled by RNA Pol III do not contain upstream activating sequences or a TATA box but rather have an intragenic regulation. However, the essential gene SNR6, which encodes U6 snRNA, is an exception to the rule and is positioned between a TATA box and a downstream sequence to which subunit τ138 of the Pol III-specific transcription factor TFIIIC (τ) binds. In the RNA Pol III system, this downstream sequence is replaced by Gal4 activating sequences and SNR6 expression depends on the binding of a bait protein, in fusion with the Gal4 DBD, with the prey protein, attached to the τ138 subunit (Fig. 4A). The possibility for selection is acquired by using a temperature-sensitive mutant of SNR6 under the control of its own promoter, with a wild-type version as a reporter gene (511). This strategy was later used to discover the interaction between the A. thaliana transcriptional regulators FIL and NZZ (597).
Fig 4.
Nuclear two-hybrid systems for autoactivating bait proteins. (A) RNA Pol III system (511). Through its fusion with the Gal4 DBD, an autoactivating bait (AuX) protein is tethered to the Gal1 upstream activating sequence, located downstream of the reporter gene SNR6. Interaction of the AuX bait with a prey protein (Y) fused to the τ138 subunit of transcription factor III C (TFIIIC) brings the RNA polymerase III holoenzyme to SNR6. Transcription of SNR6 leads to survival under temperature-restrictive (inactive snR6-A62G) conditions. (B) Repressed transactivator system (259). In the repressed transactivator (RTA) system, association of an autoactivating AuX bait with the prey protein Y, attached to the repressor Tup1, inhibits transcription of the reporter gene URA3. This results in survival of the cell on 5-FOA, a substrate for the production of the toxic compound 5-fluorouracil by the gene product of URA3. Pol, polymerase; TF, transcription factor; Y, prey; AuX, autoactivating bait; AD, activation domain; DBD, DNA-binding domain; RD, repressor domain; UAS, upstream activating sequence.
RTA system.
Another two-hybrid system for autoactivating bait proteins is the repressed transactivator (RTA) system (259). In this method, the transcriptional repressor domain of Tup1 replaces the AD in the prey (Fig. 4B). Upon interaction of the autoactivating bait with the prey, the reporter genes URA3 and HIS3 are repressed, leading to survival on medium containing 5-fluoroorotic acid (5-FOA), which is toxic in the presence of Ura3-mediated uracil synthesis, and to a growth deficiency on medium without histidine. The RTA method has been applied to screen for interactors of the transcriptional activators VP16 (259), androgen receptor (536, 636, 679), c-Myc (259, 276, 516), and microphthalmia-associated transcription factor Mitf (569). Alternatively, the setup of the RTA method allows for positive selection of interaction disruption by growth on selective medium without histidine or uracil. Using an adaptation of the RTA system, specific and nonspecific PPI inhibitors of four well-established protein pairs were identified by screening a compound library (313). This screening experiment required fine-tuning of the procedure with 3-AT and introduction of Leu3 binding sites for moderate basal expression of HIS3.
Alternative strategies.
An autoactivating bait protein can also be attached to the AD instead of the DBD, followed by screening with a library in fusion with the DBD (145). The reliance of the DBD fusion protein (prey) on the presence of the bait for reporter activation is examined by using the counterselectable marker CYH2 on the bait plasmid (152). Colonies sensitive to cycloheximide on selective medium, resulting from CYH2 expression, contain prey proteins that are interacting with the bait, without autoactivation. Despite the creative approaches to establish specific two-hybrid systems that deal with trans-activating bait proteins, the most prevalent strategy remains the removal of the region within the bait protein that initiates transcription in order to use the classic two-hybrid method (e.g., see references 287, 519, and 552) or a one-hybrid system with the promoter target of the transcription factor of interest in front of the reporter genes (229, 598–600). Finally, protein complementation assays (see Protein Fragment Complementation Assays in Yeast) and membrane-localized assays (see “Membrane-Localized and Secretory Pathway Two-Hybrid Systems”) can be applied for the detection of interactions involving autoactivating proteins.
Membrane-Localized and Secretory Pathway Two-Hybrid Systems
Small-G-protein-based methods.
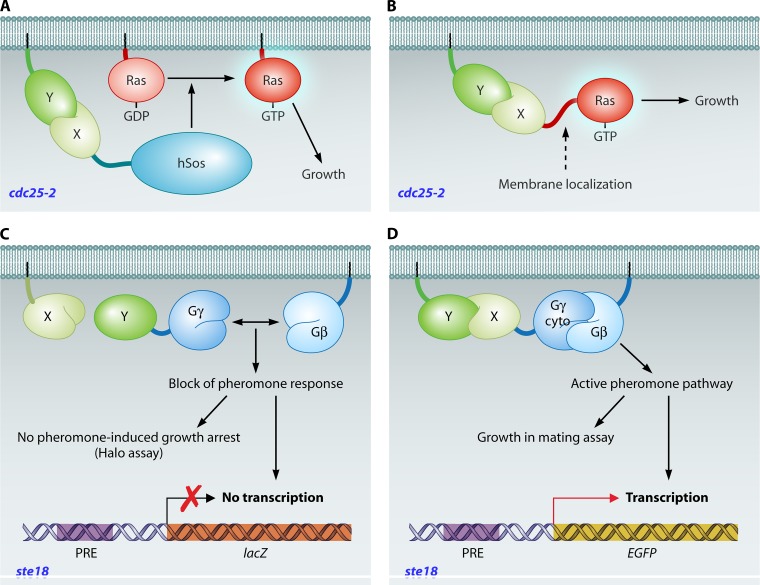
S. cerevisiae needs a functional Ras signaling pathway in order to survive and proliferate. Thus, deletion of Cdc25, the GTP-GDP exchange factor and activator of the small membrane-bound GTPases Ras2 and Ras1, makes the cells inviable (65). Based upon this notion, a strain was constructed with a temperature-sensitive mutant of Cdc25, cdc25-2 (16). In the Sos recruitment system, the human Cdc25 homologue Sos (hSos) is fused with the bait protein of interest and the prey protein is fused with a membrane localization signal. When the bait and prey interact within a cdc25-2 background strain, hSos is recruited to the membrane, where it can activate the Ras proteins, resulting in cell survival. The method was further improved by introduction of a mammalian GTPase activating protein to lower the background activity of false-positive Ras prey proteins, specifically when using mammalian cDNA libraries (15). This Sos recruitment system (Fig. 5A) provides an interesting assay for interactions when proteins are unable to enter the nucleus or when posttranslational modifications in the cytoplasmic milieu are required. In addition, the method has been applied for the discovery of inhibitors of HIV-1 Gag dimerization (652), the analysis of protein membrane localization, and the determination of numbers of transmembrane domains (297, 565). An improved version was created by using mammalian activated Ras (mRas) instead of Sos in the cdc25-2 strain (64). In this Ras recruitment system (Fig. 5B), mRas is used in the bait construct after removal of the CAAX C-terminal peptide responsible for membrane attachment. When a membrane-bound prey protein binds the bait, the constitutively active mRas protein is tethered to the membrane, where it can activate adenylate cyclase, the first target of the essential Ras signaling pathway. The small size of mRas reduces the steric hindrance problem observed with the large hSos protein. Moreover, false-positive Ras proteins are not selected by this method. Both Sos and Ras recruitment systems allow the detection of interactions between soluble bait proteins and soluble or membrane-bound prey proteins. To enable the use of membrane proteins as bait, the reverse Ras recruitment system was introduced, in which mRas is fused to the prey rather than the bait (283). This method typically gives a very large number of false-positive results, as any membrane-bound prey protein brings mRas in close proximity to adenylate cyclase. To circumvent this problem, the bait gene is put under the control of an inducible promoter to investigate the bait dependence of Ras activity and to discard all noninteracting membrane proteins in a prescreening step (283). Small-G-protein-based methods suffer from technical constraints, such as the occurrence of temperature revertants (102), growth at suboptimal temperatures, and the obligatory replica plating step (281), but they nevertheless remain popular and very successful alternatives to the original two-hybrid system (e.g., see references 147, 157, 256, 331, 455, 647, and 692). The Ras recruitment system was further developed for use in mammalian cells (432).
Fig 5.
G-protein-based two-hybrid systems. (A) Sos recruitment system (16). A chimeric protein of bait X with hSos is recruited to the plasma membrane upon interaction of X with a membrane protein (Y). Subsequent GDP-GTP exchange of Ras by membrane-localized hSos enables cell growth by virtue of Ras activity in a temperature-sensitive cdc25-2 background strain. (B) Ras recruitment system (64). Similar to the case in the Sos recruitment system, a membrane-bound prey protein Y that interacts with bait X, fused to mammalian Ras (mRas), will bring mRas to the membrane, where it can activate its downstream target adenylate cyclase to establish cell growth at a restrictive temperature (36°C) in a cdc25-2 background strain. (C) G protein fusion system (162). Association of the γ (Ste18) and β pheromone G protein subunits is required for a response to the addition of pheromones. Strong interaction of a membrane bait protein (X) with the prey protein Y, in fusion with Ste18 (Gγ), pulls Gγ away from Gβ. This dissociation hinders the pheromone response in a ste18 strain, which can be detected by growth in the presence of α pheromone (halo assay) or by the lack of gene expression of reporters under the control of pheromone-responsive elements (PREs). (D) Gγ recruitment system (190). Interaction between a membrane-bound prey protein (Y) and a bait protein (X), fused to Gγ lacking a membrane attachment sequence, unites the G protein β and γ subunits to induce the pheromone pathway after addition of α pheromone. Readouts are reporter genes regulated by PREs or the observation of growth in a mating assay with a ste18 strain.
Trimeric-G-protein-based methods.
Pheromone treatment in yeast cells activates a trimeric αβγ G protein complex, and association of the β (Ste4) and γ (Ste18) subunits is required for signal transduction. In the G protein fusion system, the bait is a membrane protein and the prey is fused to Ste18 (162) (Fig. 5C). When the prey strongly associates with the bait, Ste18 loses its interaction with Ste4, thereby blocking the pheromone signaling pathway in an ste18Δ strain. In case of an interaction, cells in the pheromone-dependent growth inhibition assay (halo assay) retain growth in the presence of pheromone and display reduced expression of a pheromone-controlled lacZ gene. The fact that only one of the two proteins is a hybrid has been suggested to be a major advantage of this technique, but its limitation is found in the large background signals it produces when screening for interactions. A recent, improved interaction tool combines the advantages of the G protein fusion system (growth at 30°C) with the benefits of the Sos recruitment system (higher sensitivity) (190) (Fig. 5D). This Gγ recruitment system makes use of a cytosolic variant of the Gγ subunit (Ste18cyto) fused to a soluble bait protein of interest. The prey protein is attached to the membrane, intrinsically or artificially by addition of a lipidation site. Interaction of bait with prey leads to membrane localization of Ste18cyto and subsequent activation of the pheromone pathway, which can be detected by fluorescence through expression of EGFP under the control of a pheromone-responsive FIG1 promoter (190) or by a mating assay using selective markers exclusively present in either of the haploid strains (191). Increased selectivity of the system is provided by the introduction of an interaction competitor protein (191), and increased sensitivity is given by the integration of STE18 under the control of the pheromone response, leading to feedback signal amplification (189).
Secretory pathway two-hybrid systems.
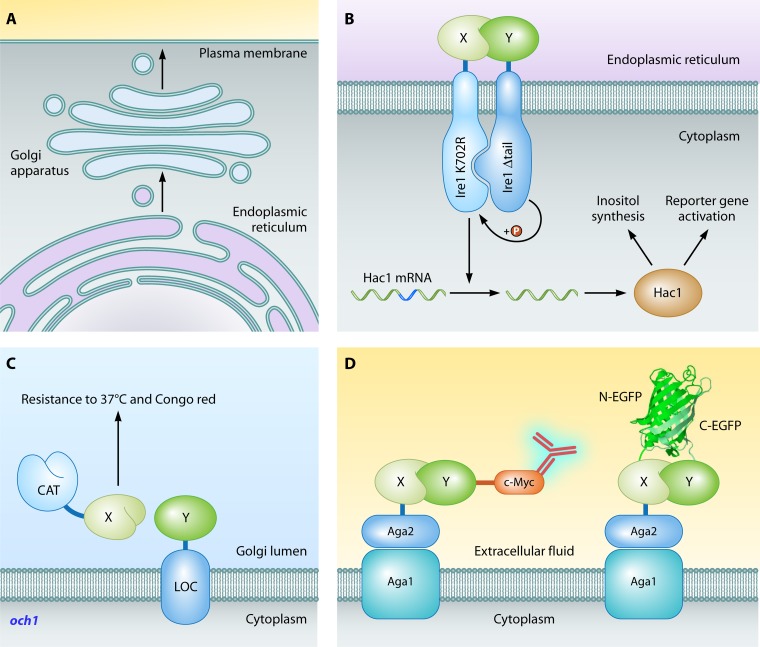
Extracellular proteins or proteins that are naturally found within the lumina of secretory pathway organelles depend for correct folding on distinctive features of these compartments, such as glycosylation, calcium concentration, and oxidizing conditions for disulfide bond formation. To study PPIs among such proteins in their native environment, several two-hybrid systems were developed in different subcompartments of the secretory pathway, ranging from the endoplasmic reticulum to the extracellular space (Fig. 6A).
Fig 6.
Secretory pathway two-hybrid systems. (A) Secretory pathway. Proteins for secretion are synthesized at the endoplasmic reticulum, transported in vesicles to the Golgi apparatus (optionally for further processing), and secreted into the extracellular fluid by fusion of a secretory vesicle with the plasma membrane. Each compartment hosts one of the available two-hybrid systems. (B) Screening for interactions between extracellular proteins (SCINEX-P) (653). Wild-type Ire1 detects the presence of unfolded proteins in the endoplasmic reticulum. Activation of Ire1 leads to homodimerization, trans-phosphorylation, and, finally, splicing of Hac1 mRNA into correctly translatable mRNA. In SCINEX-P, the bait protein X is fused to an Ire1 variant (Ire1 K702R) that can splice Hac1 mRNA only while the prey protein Y is attached to an Ire1 variant (Ire1 Δtail) that can phosphorylate only its associated Ire1 partner. Upon an X-Y interaction within the lumen of the endoplasmic reticulum, Ire1 Δtail phosphorylates Ire1 K702R, which in turn splices Hac1 mRNA, leading to correct translation of Hac1. Hac1 activity can be sensed by detection of inositol synthesis and by activation of reporters regulated by Hac1-dependent promoters. (C) Golgi two-hybrid system (146). The catalytic (CAT) and membrane-attaching (LOC) domains of the mannosyltransferase Och1 are fused to the bait protein X and prey protein Y, respectively. Interaction of the bait with the prey within the Golgi lumen prevents loss (by secretion) of the Och1 catalytic domain, leading to cell survival at 37°C or at 30°C in the presence of Congo red. (D) Yeast surface two-hybrid system (274). The membrane protein Aga1 keeps the Aga2-bait X fusion at the cellular membrane in the extracellular fluid. Binding of the bait with the prey protein Y, tagged with c-Myc, is detected with anti-c-Myc fluorescent antibodies. Alternatively, both bait and prey proteins are linked with fragments of EGFP. The formation of an X-Y heterodimer results in reassembly of the EGFP fragments and in concomitant fluorescence. The EGFP structure image is based on the PDB structure under accession number 2Y0G (550).
The membrane-bound receptor Ire1 senses stress caused by accumulation of unfolded proteins within the endoplasmic reticulum (109). Activation of Ire1 is followed by homodimerization, trans-phosphorylation, and correct splicing of Hac1 mRNA, encoding a transcriptional activator of genes involved in the unfolded protein stress response (328). Two mutant versions of Ire1, Ire1K702R and Ire1Δtail, lack the kinase and Hac1-activating regions of the wild-type protein, respectively (460, 582). Upon dimerization of Ire1K702R with Ire1Δtail, Ire1Δtail can phosphorylate and activate Ire1K702R, which in turn initiates Hac1 mRNA splicing for correct Hac1 translation. In the two-hybrid variant called SCINEX-P (screening for interactions between extracellular proteins) (Fig. 6B), the bait and prey proteins of interest are N-terminally fused to Ire1K702R and Ire1Δtail, respectively (653). Interaction between the bait and prey proteins, located within the endoplasmic reticulum, leads to dimerization of both mutants of Ire1, correct translation of Hac1, and expression of the reporter genes lacZ and HIS3, both under the control of a Hac1-regulated promoter. Deletion of IRE1 and DER1, involved in misfolded protein degradation, causes viability of the two-hybrid strain to depend on Hac1 activity through interaction-induced Ire1 dimerization at elevated temperatures and in the absence of inositol, providing a wide range of selection procedures for increased stringency. In the original study (653), the system was applied to confirm interactions between Gcn4 and anti-Gcn4 antibodies and between the leucine zipper domains of c-Jun and c-Fos.
The recently developed Golgi complex two-hybrid system (Fig. 6C) is based on the complementation of the Golgi complex-resident mannosyltransferase Och1 (146). Like many Golgi complex-based enzymes, Och1 consists of two modular domains: the N-terminal LOC domain for membrane attachment and the C-terminal CAT domain, which performs the mannose transfer reaction within the Golgi complex lumen, an essential reaction for the production of large-chain cell wall mannans. Deletion of OCH1 results in increased cell binding of chitin-binding reagents, such as wheat germ agglutinin, and in strongly reduced growth at a nonpermissive temperature (37°C) or in the presence of the benzidine-type dye Congo red (146). Fusion of the two modular fragments of Och1 to the human transcription factor MyoD and the inhibitor of differentiation protein 2 (Id2) reverses all of the och1 phenotypes through the reassembly of Och1 upon MyoD-Id2 interaction. In addition, an interaction between the transcriptional activator Gal4 and five binding partners was confirmed with the Golgi complex two-hybrid system, suggesting this method to be an alternative tool for the study of interactions involving transcription-activating or extracellular proteins.
The yeast surface two-hybrid system (Fig. 6D), which is based on yeast surface display (49), detects interactions that take place outside the cell. In one version of the method (153), the interaction of two fragments of the 10th type III domain of human fibronectin (FNfn10) was shown by first fusing the N-terminal part of FNfn10 with the cell wall agglutinin protein Aga2, which displays the N-terminal fragment on the yeast surface. Next, the C-terminal part of FNfn10, with a V5 epitope tag, was shown to be attached to the surface by immunofluorescence detection through its interaction with the N-terminal fragment of FNfn10. The method enabled quantitative analysis of interactions between mutated FNfn10 fragments (153). A highly similar system was developed for a study on coiled-coil interactions and antigen-antibody recognition (274). In addition to immunofluorescence detection, the appearance of fluorescence upon interaction-induced green fluorescent protein complementation (also see The Split-FP Method) was used to examine protein-protein binding. Both types of readout are suitable for quantitative interpretation (274). This method enabled the quantitative analysis of antigen-antibody binding after initial selection procedures involving directed evolution with yeast surface display and panning through phage display (275). Finally, an independent group created yet another yeast surface two-hybrid system, based upon the same principle, to study the peptide recognition of major histocompatibility complex (MHC) class II proteins (308).
Three-Hybrid Systems
Three-hybrid systems rely on the intervention of a third component for PPI detection. This third factor can be a protein involved in posttranslational modification of one of the interacting proteins, or it can directly interfere with the PPI. Moreover, three-hybrid systems have been created to study protein–small-molecule and protein-RNA interactions and to screen for enzymes that cleave or bind specific molecular structures.
Posttranslational modifiers.
Many posttranslational modifications present in higher eukaryotes also occur in S. cerevisiae. However, some alterations, such as tyrosine phosphorylation, are absent in yeast. These modifications may be crucial for the structural recognition of two interaction domains. For example, Src homology 2 (SH2) domains bind only proteins with a phosphotyrosine residue (504). Introduction of an exogenous tyrosine kinase into the yeast two-hybrid system enabled the identification of numerous interactions, including the discovery of novel interaction partners for the γ subunit of the IgE receptor FCεRI, the tyrosine phosphatase SHPTP2, the human insulin receptor, the C. elegans adaptor protein CED-2, and the Schistosoma mansoni Tyr kinase TK4 (36, 70, 329, 362, 481, 482). Not surprisingly, many of these interactions involved SH2 domains. In a variation to this theme, the Ras recruitment system (see “Trimeric-G-protein-based two-hybrid methods”) was applied to detect human proteins that interact with the phosphorylated transcription factor c-Jun after the introduction of JNK1 kinase, controlled by the inducible MET3 promoter (3, 468).
Although acetylation and serine/threonine phosphorylation occur commonly in S. cerevisiae, the artificial character of a two-hybrid experiment may prevent posttranslational modifications, for example, if the modifier is not colocalized with its target (bait or prey) protein in the nucleus. The tethered catalysis two-hybrid system deals with this problem by fusion of the posttranslational modifier with the bait protein (230). Fusion of a Gal4 DBD/histone 3 chimera with the histone acetyltransferase Gcn5 resulted in acetylation of histone 3 and the discovery of two binding partners, Rpm2 and Rtm1, that interact with histone 3 only when it is acetylated. Similarly, three tandem repeats of the CTD peptide of the largest RNA polymerase II subunit were fused to the Gal4 DBD and the serine/threonine kinase Kin28, which phosphorylates CTD. Several phosphorylation-dependent interactions were found in a screening assay, mostly with proteins that regulate transcription (230). To identify polyubiquitin-binding proteins, a ternary chimera of Gal4 DBD, the tumor suppressor BRCA1, and the related protein BARD1 was created (711). Autoubiquitination of the BRCA1-BARD1 complex enabled the identification of prey proteins that bind polyubiquitin. The tethered catalysis two-hybrid system has also been adapted to mammalian cells (607).
Trimeric complexes and competitive binding.
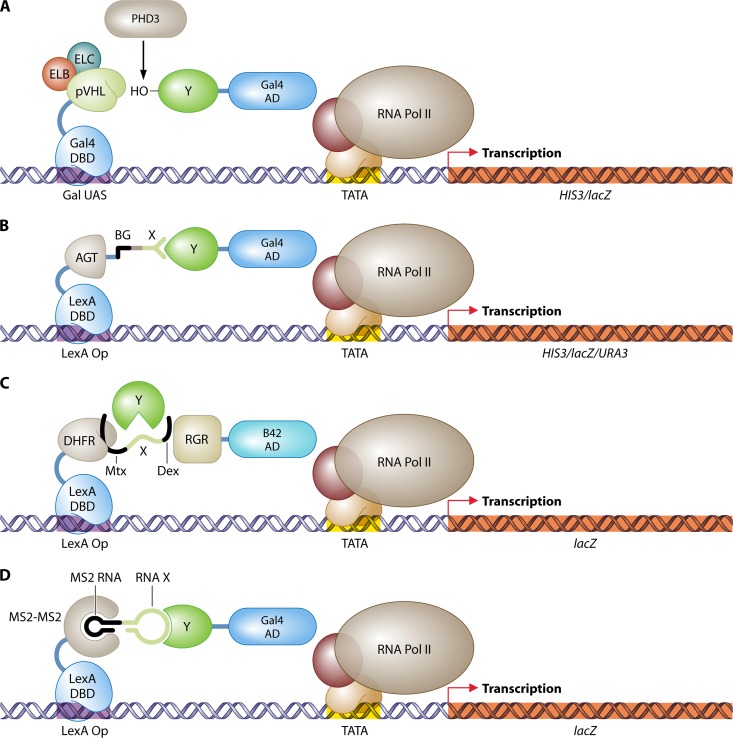
Three-hybrid systems are further used to examine the reliance of a PPI on a third protein as a bridging molecule. The interaction between the epidermal growth factor receptor (EGFR) as bait and the Sos protein as prey was illustrated to depend on the presence of a third adaptor protein, Grb2 (727). The third gene can be put under the control of an inducible promoter, for example, the MET25 promoter, repressed by methionine or cysteine, or the tetracycline-inducible promoter, to investigate how crucial the presence of the bridging protein is for the interaction (461, 639). The three-hybrid system has been used frequently to investigate nonyeast ternary complexes (e.g., see references 192, 408, 560, 563, 576, and 584). In some cases, more than one bridging protein may be required (503, 588). Interaction of the tumor suppressor protein pVHL with the cullin family member CUL-2 depends on the pVHL-stabilizing effect of elongins B and C (503). Analysis of the interacting domains by a four-hybrid analysis revealed a structural resemblance of this complex (CBCVHL) with the E3-like ubiquitin ligase complex SKP1/Cullin/F-box protein and later led to the confirmation of the complex for its involvement in ubiquitin-regulated protein degradation (296). Interaction analysis of this E3 ubiquitin ligase CBCVHL complex and its target proteins has been hampered in mammalian cells due to the fast ubiquitination-induced degradation of the targets. This problem was overcome by application of the yeast three-hybrid system, which involves the introduction of pVHL as a bait protein together with elongins B and C to stabilize the native conformation of pVHL, and by additional inclusion of the prolyl hydroxylase PHD3 for hydroxylation of target prey proteins for pVHL recognition (41) (Fig. 7A). As a result, all components are present to identify prey proteins as targets of the CBCVHL complex, and the absence of CUL-2 prevents unwanted induction of target protein degradation. After confirmation of the binding of pVHL with the known targets HIF1 and -2α, a library approach resulted in the identification of eight novel interactors (41).
Fig 7.
Three-hybrid systems. (A) Identification of target proteins of the E3 ubiquitin ligase complex CBCVHL (41). The pVHL subunit is fused to the LexA DBD, and coexpression of elongins B and C (ELB and ELC) stabilizes the native conformation of pVHL. A human cDNA library in fusion with the Gal4 AD is screened for interactions with pVHL. The human prolyl hydroxylase PHD3 delivers the hydroxyl group to interacting prey proteins, which is essential for recognition by pVHL. (B) Protein–small-molecule interactions (94). O6-Alkylguanine-DNA alkyltransferase (AGT) is fused to the Gal4 DBD and can bind covalently with the small molecule O6-benzylguanine (BG) in vivo. A library of BG small-molecule heterodimers, produced in vitro, can be screened for interactions with a prey protein Y attached to the Gal4 AD. (C) Detection of enzymatic substrate recognition (21). A chimeric protein of LexA and DHFR binds the promoter region of the reporter gene lacZ and interacts with a tripartite small molecule through association of DHFR with Mtx. This small-molecule trimer further consists of a bait linker X and dexamethasone (Dex). Dexamethasone interacts with a fusion of the rat glucocorticoid receptor (RGR) and the B42 AD. The whole complex stimulates expression of lacZ. An enzyme Y that targets and cleaves linker X can be identified by disruption of the transcription activating complex and the loss of lacZ expression. (D) RNA-protein interactions (265). The LexA DNA-binding domain is fused to a head-to-tail dimer of the RNA-binding protein MS2. This hook protein associates with an RNA dimer of bacteriophage MS2 RNA and a bait RNA stretch (X). Interaction of this RNA X with a protein Y fused to the Gal4 AD is detected by stimulation of reporter gene expression. DBD, DNA-binding domain; AD, activation domain; UAS, upstream activating sequence; Op, operator; Pol, polymerase.
Alternatively, the third protein may disrupt an interaction (639). In response to blue light, the A. thaliana cryptochrome 1 blue-light receptor CRY1 competitively interfered with the association of the E3 ubiquitin ligase COP1 with the phytochrome A suppressor SPA1 in a yeast three-hybrid experiment (391, 406). Because the whole experiment was performed in a heterologous organism (S. cerevisiae), both CRY1 activation by blue light and subsequent interaction disruption were shown to be strictly independent of any other A. thaliana protein, a general notion for yeast-based interaction experiments with nonyeast proteins. Other reports exploited the three-hybrid system for studies on the interaction-disrupting abilities of a third protein (e.g., see references 76, 361, 385, and 628).
Protein–small-molecule interactions.
The three-hybrid system has been adapted to investigate associations that go beyond PPIs (Fig. 7B to D). The bridging molecule is not necessarily a protein. A fusion of the LexA DNA-binding protein with the rat glucocorticoid receptor (LexA-RGR; the “hook”) associates with a covalently linked heterodimer of two small molecules (Dex-FK506; the “bait”) by binding of protein RGR with the steroid hormone agonist dexamethasone (Dex) (392). Another hybrid protein, consisting of the human protein FKBP12 and the B42 activation domain (FKBP12-B42; the “prey”), interacts with Dex-FK506 through association of FKBP12 with the immunosuppressant FK506 (566). This ultimately leads to the noncovalent and indirect reassembly of the transcription factor LexA-B42, detected by activation of the reporter gene lacZ (392). Alternative approaches were developed with substitute small-molecule heterodimers, small-molecule-binding proteins, reporter genes, and DNA-binding and activation domains (22, 160, 195, 253, 285, 401). Initially, the sensitivity and stringency of these three-hybrid approaches were shown by recovering known small-molecule-binding proteins from a prey cDNA library (253, 392), by demonstrating the reduction or absence of interaction with specific mutant proteins (2, 122, 253, 285, 392), and by competitive assays with interfering monomeric small molecules (253, 285, 392, 401). However, the full ability of the method was illustrated by the identification of novel targets of small-molecule kinase inhibitors (35). Cyclin-dependent kinase (CDK) inhibitors were found to bind both known and new CDK and CDK-like proteins in a screening assay that utilized HIS3 as a selective reporter gene and methotrexate (Mtx), which binds very tightly to a DNA-bound LexA-dihydrofolate reductase (DHFR) fusion protein (2, 35), as a fixed small molecule in the heterodimer.
Recently, a highly optimized version of the three-hybrid system for small-molecule–protein interactions uncovered numerous novel interaction partners for a variety of drugs (94) (Fig. 7B). The LexA DNA-binding domain was fused with human O6-alkylguanine-DNA alkyltransferase (AGT), which associates covalently with O6-benzylguanine (BG) (334). The covalent linkage between LexA-AGT and BG significantly increases the sensitivity of the assay (382), in contrast to the noncovalent binding partners in previous setups. BG derivatives were created by fusion of BG with a set of drug compounds. To acquire a sensitive three-hybrid strain, three genes (PDR5, SNQ2, and YOR1) encoding broad-spectrum drug transporters were deleted to prevent efflux of the BG derivative. The three-hybrid strain, with the reporter genes HIS3, lacZ, and URA3 and the fusion gene lexA-AGT, was transformed with a human cDNA library fused to the Gal4 AD. To exclude false-positive transformants, which induce reporter genes without the BG derivative, cells were grown in the absence of the BG derivative on medium with 5-FOA. True positive transformants were further selected by growth on medium without histidine and in the presence of the BG derivative. Validation of recovered binding partners of the drug compounds was performed with GST pulldown assays using GST-coupled AGT-BG constructs. With this approach, the confirmation of previously known drug targets was demonstrated, and novel interaction partners were found for the drugs purvalanol B, erlotinib, atorvastatin, and sulfasalazine (94). The experimental approach in this study sets a standard for future protein–small-molecule assays.
Detection of enzymatic activity.
Chemical complementation forms an adaptation of the three-hybrid system for small-molecule–protein interactions and allows for detection of cleavage or a covalent junction of molecules mediated by enzymes (21). The linker, connecting two fixed small molecules, methotrexate and dexamethasone, consists of a molecule of interest that serves as a target of enzymatic catalysis. This tripartite bait connects the DNA-binding hook LexA-DHFR with the transcription activating prey RGR-B42. As a result, any enzyme that cleaves within the molecule of interest and therefore disrupts the Mtx-Dex link can be detected by a loss of reporter gene activity. In the pioneering study (21) (Fig. 7C), the antibiotic cephalosporin, bordered by Mtx and Dex, was hydrolyzed by the β-lactamase cephalosporinase, which resulted in a loss of lacZ reporter expression. Quantitative evaluation of enzymatic activity by chemical complementation was applied to distinguish between β-lactamases with low and high catalytic efficiencies (573), to enhance the activity of glycosynthases and cellulases by directed evolution (400, 509, 632), and to investigate the modes of resistance to cephalosporin induced by β-lactamase mutations (77). Improvements of the original approach included an increase of cell permeability (77), fine-tuning of hook and prey expression (22, 77), and introduction of a sensitive counterselection reporter for detection of bond-forming enzymes (707). Similar methods were developed for application in E. coli (9, 181).
RNA-protein interactions.
Interactions between RNA and proteins play an essential role in many fundamental cellular processes. Associations between mRNA and proteins are crucial for control of mRNA stability, splicing, and translation and for nuclear-cytoplasmic RNA shuttling. Aminoacyl tRNA-synthetases bind tRNA to add the corresponding amino acid, and chromosome ends are maintained by telomerases, complexes of RNA and protein molecules. For the study of interactions between RNA and proteins in three-hybrid systems, the bridging hybrid comes in the form of an RNA heterodimer (622). While one fixed RNA stretch of this dimer binds a hook protein, comprised of a DBD and a fixed RNA-binding protein, the other RNA sequence is tested for interaction with a protein of interest fused to an AD (526, 575). Interaction between the RNA and protein of interest induces expression of the reporter genes HIS3 and lacZ. For one method, the fixed and hook RNA-binding proteins are the Rev-responsive element (RRE) and the RevM10 mutated form of the HIV-1 Rev protein (526), respectively, while in another setup, the binding of two copies of a specific stretch of bacteriophage MS2 RNA to the coat protein of MS2 is taken as a fixed component of the system (575). Both approaches have been used for a number of RNA-protein interaction studies (e.g., see references 290 and 585), but the MS2-based system has seen more applications, mainly because of the high affinity of the MS2 RNA-protein interaction (40) and because of specific improvements. For example, the RNA three-hybrid system is susceptible to revealing a large number of false-positive results, due mainly to direct binding of nonspecific prey proteins to the hook fusion. Introduction of a head-to-tail dimer of a high-affinity mutated version of the MS2 coat protein into the hook reduces these nonspecific associations by steric hindrance and increases the efficiency of hook association with MS2 RNA (265) (Fig. 7D). Other approaches to exclude RNA-independent false-positive results depend on the auxotrophic marker on the RNA plasmid (40, 492) or on an inducible promoter for RNA hybrid gene expression (20). The RNA three-hybrid system has been used to study RNA-protein complexes such as RNase P (268, 307) and telomerase (240) and to investigate interactions between RNA and proteins involved in translation stimulation (585), translation inhibition (250, 480), RNA methylation (712), transport (143, 345), degradation (674), and replication (90). The method enables screening of RNA molecules that bind an RNA-binding protein of interest (574, 712) or the discovery of RNA-binding proteins that interact with a specific artificial (359) or native (583) RNA stretch. Furthermore, RNA-protein binding affinities can be measured based upon three-hybrid experiments (713) when specific considerations are taken into account, such as focused RNA mutagenesis and introduction of flanking RNA regions that allow correct folding of the RNA sequence of interest (714). Recently, two independent groups discovered the Pumilio and FBF homology (PUF) protein repeats that recognize cytosine in RNA, based upon three-hybrid experiments (139, 179). Because the amino acids that recognize adenine, uracil, and guanine were discovered previously, artificial PUF proteins can be created to bind specific mRNA sequences for translation control (179), a method that could become complementary to RNA interference (163). Alternative RNA three-hybrid setups have enabled the identification of trimeric protein-RNA complexes (54, 411) and RNA-RNA interactions (515) and the design of transcription-activating RNA stretches (687). Finally, an ingenious bacterial one-hybrid method is available for detection of heterologous RNA-protein interactions, based on lacZ expression upon relief from antitermination (704).
Reverse Two-Hybrid Systems
The forward two-hybrid system is suitable for identifying the specific interaction domains of two binding proteins by gradual truncation of each protein in the system. However, the identification of single residues that are crucial for the interaction is not straightforward in this approach. Therefore, an adaptation of the traditional method was developed in which a counterselectable marker is used.
Reporter genes.
The counterselectable reporter gene, which is activated following a PPI, expresses a compound that is toxic for the cell. Three commonly used reporters are URA3 (667), CYH2 (373), and GAL1 (228), which render the cells sensitive to 5-FOA, cycloheximide, and galactose in a gal7 background, respectively (Fig. 8A). Repression of a positive selection marker can also be used to investigate dissociation of a PPI. For example (Fig. 8B), a PPI activates expression of the Tet repressor, which then represses the activation of the positive marker HIS3 under the control of the Tet operator (589). The URA3 counterselectable system has been very successful due to the introduction of the basal SPO13 promoter, which tightly regulates URA3 expression in combination with an optimized number of GAL4 binding sites and is strongly repressed under most growth conditions. As a result, mutations in the bait or prey protein of interest that block an interaction lead to an easily detectable resistance of the cells to 5-FOA.
Fig 8.
Reverse two-hybrid systems. (A) Repression of toxic genes (228, 373, 667). Interaction between bait X and prey Y stimulates transcription of URA3, which results in cell toxicity in the presence of 5-fluoroorotic acid; CYH2, which makes the cells sensitive to cycloheximide; or GAL1, which converts galactose into galactose-1-phosphate, a toxic compound that accumulates in the absence of Gal7. Compounds or mutations (in X or Y) that interfere with the association of the bait with the prey are identified by growth on selective medium. (B) Activation of positive selection markers (589). Interaction between bait X and prey Y leads to expression of tetR, encoding the Tet repressor. Next, TetR represses activation of HIS3, hindering cell growth on medium without histidine. Disruption of the interaction between bait and prey is analyzed by the appearance of histidine prototrophy. DBD, DNA-binding domain; AD, activaion domain; UAS, upstream activating sequence; Pol, polymerase; Op, operator.
Selection for missense mutations.
An important concern in screening for residues that are required for an interaction is the presence of nonsense mutations. The original URA3 method provides a second selection step to discover mutations that do not completely block the interaction, as an indication of missense rather than nonsense alterations (667). Alternatively, the wild-type bait or prey protein to be mutagenized can bear a C-terminal linkage with an additional reporter gene, e.g., β-galactosidase (589), GFP (168), or URA3 (393). In this case, missense mutations can be selected by the dissociation of the PPI together with the expression of the C-terminally linked reporter gene. In the one- plus two-hybrid system, the mutated prey protein of interest is present between a Gal4 DBD and the B42 AD. An initial one-hybrid screening for removal of nonsense mutations is performed by selecting for cells that grow on medium lacking histidine, due to the expression of HIS3 with Gal upstream activating sequences stimulated by a complete Gal4-prey-B42 fusion protein (342). In the next step, interaction-defective missense prey constructs are selected for loss of LexA-controlled lacZ reporter gene activation in a two-hybrid assay with a LexA-DBD fused bait protein. Finally, nonsense mutations can also be excluded prior to the two-hybrid screening by cloning the mutated genes N-terminally to a kanamycin resistance marker and preselecting for the right reading frame in E. coli (217). The strength of this strategy lies in the removal of the kanamycin marker by subcloning the library into a new vector by recombinational cloning with the Gateway technique.
Identification of residues that moderate an interaction.
Examples of the use of the reverse two-hybrid system include the isolation of multimerization-defective mutants of human HIV-1 integrase INI1 (114) and the identification of crucial residues for interactions between A. thaliana phytochrome B and its signaling partner, PIF3 (341), between the ArsD metallochaperone and the ArsA ATPase (718), between the proteasome ubiquitin chain receptor Rpn1 and ubiquitin-like domain proteins in S. cerevisiae (214), and between two nonstructural proteins, nsp10 and nsp16, from the severe acute respiratory syndrome (SARS) coronavirus (414). Analysis of allosteric inhibition of an interaction has also been described (510). Mutations in A. thaliana PYR1 that inhibit its pyrabactin-induced association with HAB1 were identified by the reverse two-hybrid system. For 49 mutant versions of PYR1, abscisic acid (ABA) was still able to induce the PYR1-HAB1 interaction, pointing toward mutations that specifically inhibit the binding of one ligand (the ABA agonist pyrabactin) but not the other (ABA). This extension of the two-hybrid method, to mutate regions of the protein outside the protein interaction domain, has also been reported for the forward two-hybrid system (96, 292). The ligand-binding domains (LBDs) of nuclear hormone receptors were modified by directed evolution through site-saturation and random mutagenesis in order to create a receptor that responds to an alternative ligand. A ligand-induced two-hybrid interaction was taken as readout (96, 292). The forward two-hybrid system has been applied successfully in more studies related to mutagenesis, for the selection of mutants that either increase (43) or decrease (282) interaction strength. Another approach consists of mapping the regions of a protein that are not important for protein-protein binding, called the absence-of-interference method, using random mutagenesis and the forward system to select for mutants that do not interfere with the interaction (130). This strategy has the advantages of identifying distant essential interaction regions, in contrast to gradual truncation of the protein, and selecting preferentially for missense mutations over nonsense mutations, in contrast to the reverse two-hybrid method.
Drug discovery.
The reverse two-hybrid system has applications in the discovery of peptides or small molecules that disrupt a PPI. The method shows clear advantages, as cytotoxicity testing is included in the assay, high-throughput screening is possible (279), and there is no need for protein purification. In addition, the permeability of the cell for peptide or small-molecule entrance can be increased by deletion of genes encoding the ergosterol synthesis enzyme Erg6 or the multidrug resistance regulators Pdr1, Pdr3, and Pdr5 or by overexpression of the hexose transporter gene HXT1 or HXT9, which lowers the amount of compound required for the assay (193, 326, 668). Despite initial promising data (279, 720), the use of the reverse two-hybrid system for drug discovery in yeast has been limited (e.g., see reference 710). Other technologies, such as FRET (367), fluorescence anisotropy (525), surface plasmon resonance (315, 570), and virtual screening (726), have more impact on this field, at least in publically available reports. However, other two-hybrid methodologies have also been used for the discovery of PPI inhibitors, including the traditional system (199, 326), the repressed transactivator system (313), and PCAs (see Protein Fragment Complementation Assays in Yeast). In addition, both mammalian and bacterial reverse two-hybrid systems have been developed (see Genetic Protein-Protein Interaction Methods in Other Organisms). Improvements of the current two-hybrid technologies may boost the application of these methods in the field of drug discovery, as seen for the three-hybrid system for protein–small-molecule interactions (94).
Two-Bait Hybrid Systems
A gene deletion disturbs all physical associations of the protein encoded by the removed gene. Removal of such a protein from the interaction network precludes conclusions on the specific influence of each physical association related to this protein. Therefore, edge-specific genetic (edgetic) perturbations, which are mutations that explicitly interrupt only a subset of PPIs of the mutated protein (144, 735), can greatly facilitate the analysis of the distinctive functional properties of a protein in comparison with complete knockout mutations (108). Two-bait hybrid systems provide an interesting platform for discovering crucial amino acid residues that specifically reduce the affinity of a protein with one binding partner but not with another. In these methods, two known interactors of a protein of interest are each fused to a different DNA-binding domain, each of which targets the promoter of a different reporter, while the (mutated) prey protein of interest is attached to an activation domain. A mutation in the prey protein that exclusively activates one reporter and not the other correlates with an edgetic perturbation. To differentiate between mutations in Snf1 that are specific to binding to the activating subunit Snf4 or the kinase domain of Snf1 itself, a double two-hybrid strategy was followed (306). Mutated Snf1 fused to an activation domain was coexpressed with Gal4DBD-Snf4, binding to the GAL1-HIS3 reporter, and LexA-Snf1 kinase domain, binding to the lexA operator (lexA Op)-lacZ. Selection by both a chromogenic (lacZ) and a growth-selective (HIS3) assay resolved all possible influences of mutations (no, specific, or nonspecific interference) in a single screen. A similar method, called the “differential interaction trap,” was applied for identification of missense mutations in the yeast scaffold protein Ste5 that specifically disrupt an interaction with either Ste11 or Ste7, two MAP kinase pathway components (288). Other two-bait techniques are the “two-bait interaction trap” (716) and the “dual-bait system” (580) (Fig. 9). In the latter system, developed by Golemis and coworkers, each bait fusion protein binds the promoters of a chromogenic and a prototrophic reporter gene. The method proved to be efficient in distinguishing interacting partners of two related GTPases, Ras and Krev-1 (580), and discovering edgetic perturbations of the p21-activated kinase Pak1 regarding its interaction partners, the GTPases Cdc42 and Rac (539). Since both GTPases are able to signal to Pak1, identification of Pak1 mutants that were defective in Rac binding shed some light on the respective roles of these small GTPases in mediating the activation of Pak1 by Ras in vivo. An enhanced dual-bait system was configured and addressed the optimization steps of variable expression levels of the baits and sensitivities of the reporters, enrichment for polylinkers for easier cloning, and increased diversity of the selective markers (581). Two-bait systems also have an application in the exclusion of false-positive results. Proteins interacting with the bait protein of interest but not with a control bait can easily be discarded, thereby reducing technical false-positive results (581).
Fig 9.
The dual-bait system (580). Protein Y interacts with two proteins, X1 and X2 (as shown in the box). Bait proteins X2 and X1 are fused with the DNA-binding proteins cI and LexA, respectively, while a library of mutated prey Y proteins is made in fusion with the B42 activation domain. A mutation in protein Y (Y′) that specifically disables the association with X2 but not X1 is detected by reporter activity of LEU2 and lacZ, without expression of LYS2 or gusA. DBD, DNA-binding domain; AD, activation domain; Op, operator; Pol, polymerase.
Other methods for the identification of edgetic perturbations include the split-yeast cytosine deaminase (split-yCD) method (156), traditional two-hybrid experiments combined with site-directed mutagenesis, and reverse two-hybrid screens (82, 144).
One-Hybrid Systems
DNA-protein interactions.
Proteins can bind DNA for transcription, replication, cleavage, ligation, gene regulation, and structural packaging. The detection of DNA-protein interactions is established by several techniques, including SELEX (systematic evolution by exponential enrichment), ChIP-seq (chromatin immunoprecipitation followed by sequencing), and protein microarray analysis (reviewed in reference 715). The one-hybrid system provides an alternative method and is unique in the sense that it can screen for both proteins that bind a specific DNA sequence (686) and DNA sequences recognized by a specific protein (706). In a one-hybrid experiment, the bait DNA is inserted in front of a reporter gene and the prey protein consists of the DNA-binding protein of interest in fusion with an activation domain. The association of the bait DNA with the prey protein results in activation of the reporter gene (Fig. 10A). New developments in the two-hybrid field, such as the application of several reporters (31, 176, 387, 412, 686), Gateway ORF libraries (127), or modified smart-pool assays (666), have been introduced in one-hybrid studies. In general, a heterologous host system is used to prevent interference by endogenous transcriptional activators. The only other available in vivo system, ChIP-seq, requires specific antibodies against a DNA-binding protein or inclusion of epitope tags, which is not straightforward in higher eukaryotes, especially on a large-scale level. Therefore, it is not surprising that the one-hybrid technology is particularly popular for research on plant and animal DNA-protein interactions (79, 128, 254, 483, 665, 737). Derivations of the one-hybrid system were developed to detect DNA-protein dissociations by mutagenesis (34, 667, 706), as well as methylation-dependent interactions (176). Furthermore, other organisms have served as hosts for one-hybrid studies, e.g., mammalian cells (254) and bacteria (440).
Fig 10.
One-hybrid systems. (A) DNA-protein interactions (706). A putative DNA-binding protein Y is in fusion with the Gal4 activation domain. Interaction of protein Y with a promoter X stimulates reporter gene expression. This assay can single out proteins that bind a fixed promoter of interest or DNA sequences that are targeted by a fixed DNA-binding protein of interest. (B) Identification of transcriptional activity (696). A chimeric protein with the Gal4 DNA-binding domain and a transcriptional activator (a nuclear receptor [NR] in this example) stimulates expression of a reporter gene under the control of Gal upstream activating sequences. An application of such a system is the identification of ligands that trigger NR nuclear import and activity. AD, activation domain; DBD, DNA-binding domain; Pol, polymerase; UAS, upstream activating sequence.
Transcriptional activation.
In general, DNA-binding domains can be recognized by in silico analysis. However, activation domains are intrinsically unstructured (32, 648), and only general sequence features of activation domains have been described (207, 557, 645). Therefore, activation domains can be discovered only by experimental analysis. One-hybrid studies can be applied if the native target promoters of a putative transcription factor are not identified. The putative transcriptional activator is fused with a fixed DNA-binding domain that targets upstream activating sequences in the promoter region of a reporter gene. Large-scale analyses of proteins or peptide sequences from E. coli, humans, and S. cerevisiae led to the identification of hundreds of fragments which were able to express the reporter genes (420, 642, 702). The one-hybrid system has been specifically suitable for the discovery of ligands and cofactors of higher eukaryote nuclear receptors, both with yeast as a host organism (264, 380, 696) and in mammalian cells (68, 80, 593) (Fig. 10B). The technique has further seen applications in the pathogenic fungus Candida albicans (554) and the fission yeast Schizosaccharomyces pombe (441).
PROTEIN FRAGMENT COMPLEMENTATION ASSAYS IN YEAST
PCAs exist in many flavors and have many different applications. PCAs provide a wide range of possible applications, depending on the choice of PCA technique. While the two-hybrid system has limited use for studies on the kinetics or spatiotemporal dynamics of PPIs, some PCAs can show the subcellular location of an interacting pair (e.g., the split-FP system) or offer a high resolution in temporal and quantitative analysis of protein-protein binding (e.g., the split-luciferase system).
Similar to the transcriptional readout of the classic two-hybrid system, PCAs need to provide a detectable effect, such as cell survival on selective medium (e.g., the split-mDHFR method), colocalization of the interacting protein as detected by fluorescent antibodies (e.g., the split-lactamase system), or the appearance of fluorescence or luminescence upon interaction of the protein couple (e.g., the split-FP system). Furthermore, the two fragments of the reporter protein should not reassemble spontaneously but only after interaction of the two proteins fused to each fragment. The sensitivity of the assay depends on the presence or absence of signal amplification (e.g., enzymatic activity of the reporter), the signal-to-noise ratio (e.g., the ratio is negatively influenced by autofluorescence), the abundance of bait and prey proteins, and the flexibility of the fragments to reassemble unhindered by the structure or size of the interacting proteins. The development of a PCA requires knowledge of the structure of the candidate reporter to identify possible sites at which to split the protein and to see possibilities for incorporation of specific mutations that either increase or decrease the reassembly efficiency. Potential PCA reporters have gone through many optimizations, by site-directed (312, 633) and random (156) mutagenesis and by selection of different N-terminal and C-terminal fragments from a small (156) or large (629) fragment collection. This ongoing process of reporter optimization, together with advances in optics technologies, promises to make PCAs more and more attractive in the future.
The Split-Ubiquitin System
Introduction of PCAs came with the development of the ubiquitin split-protein sensor (USPS) (or split-ubiquitin) system by Johnsson and Varshavsky (312) (Fig. 11). This method takes advantage of the properties of ubiquitin, a highly conserved 76-amino-acid regulatory protein. Ubiquitin is recognized by ubiquitin-specific proteases that cleave the C-terminal covalent linkage between ubiquitin and the protein to which it is attached (255). When the C-terminal and N-terminal regions of ubiquitin (Cub and Nub) are split and each part is fused to a different protein of interest, functional ubiquitin is formed upon interaction of both fusion proteins. To prevent spontaneous reassociation of ubiquitin, amino acid 13 was converted from isoleucine to glycine (NubG). In the original design, the bait consisted, from the N-terminal to the C-terminal end, of the homodimerization domain of Gcn4 (protein of interest), Cub, mDHFR, and a hemagglutinin (HA) epitope tag. The prey was constructed as a fusion of the homodimerization domain of Gcn4 with NubG. Upon dimerization of Gcn4, ubiquitin was reconstituted and mDHFR-HA was cleaved off by ubiquitin-specific proteases, and this was detected as a shift in a Western blot assay using anti-HA antibodies (Fig. 11A). Later, this rather cumbersome readout was replaced by reporter gene activation (611). The reporter mDHFR was replaced by the hybrid transcription factor LexA-VP16. After interaction of bait and prey, LexA-VP16 is cut off and moves to the nucleus for activation of the reporter genes HIS3 and lacZ (Fig. 11B). This new reporter strategy allows for screening of a library for novel interactors. The bait protein of interest needs to be membrane bound or at least able to exclude the whole fusion construct from entering the nucleus. This makes the technique very complementary with the traditional two-hybrid system. N-terminal relocation of the LexA-VP16 fragment enables the use of membrane-bound bait proteins with cytoplasmic tails on the N-terminal side (209). Also, mating type a and α two-hybrid strains were developed (476) that enable mating of bait and prey transformants for efficient high-throughput studies. Such a large-scale experiment revealed 1,985 S. cerevisiae interactions among 536 integral membrane proteins (447). Employment of the split-ubiquitin system further led to an interactome network of A. thaliana membrane proteins (370). Split-ubiquitin vectors and strains are available at Dualsystems Biotech and MoBiTec.
Fig 11.
The split-ubiquitin system. (A) mDHFR-HA readout (312). Bait protein X and prey protein Y are fused to the C-terminal (Cub) and mutated (I13G) N-terminal (NubG) domains, respectively, of ubiquitin. In addition, a chimera of mDHFR and the HA epitope completes the bait construct. Upon interaction of X with Y, a fully reconstituted ubiquitin is recognized by ubiquitin-specific proteases (UbSP) that cleave HA-mDHFR, resulting in a shift on a Western blot using anti-HA antibodies. (B) LexA-VP16 readout (611). Interaction between a membrane-bound bait X and prey Y leads to cleavage of the artificial transcription factor LexA-VP16. Released LexA-VP16 localizes to the nucleus to activate reporter genes. (C) R-Ura readout (372). Interaction between bait X and prey Y induces the release of Ura3 (with an N-terminal arginine) by proteases. Exposed arginine makes Ura3 highly unstable, resulting in its degradation. Due to the strongly reduced concentration of Ura3, the cells become viable on medium with 5-FOA, a prototoxic substrate of Ura3. The image of the ubiquitin protein is based on the PDB structure under accession number 1UBQ (671).
An alternative version was created using the concept of the N-end rule (Fig. 11C). In Saccharomyces cerevisiae, protein stability depends on the nature of the N-terminal amino acid (660). Amino acids such as glycine, methionine, threonine, alanine, and cysteine stabilize the protein when they are present at its N-terminal end. In contrast, N-terminal basic (e.g., arginine) or bulky hydrophobic amino acids tend to promote protein degradation in a ubiquitin-dependent manner (138). For PPI analysis, the LexA-VP16 construct in the bait is replaced by the reporter protein Ura3, with an arginine residue (R-Ura3) between Ura3 and Cub (708). When two proteins of interest interact, the reassembly of ubiquitin recruits the ubiquitin-specific proteases that cleave off Ura3. As a result, free Ura3 is quickly degraded due to the exposed N-terminal arginine residue. Consequently, the cells become resistant to 5-FOA. While the LexA-VP16 strategy is complementary to the classic two-hybrid system by its application for detection of membrane PPIs, the R-Ura3 method is especially suitable for finding transcription factor partners, both activators and repressors (50, 92, 372). Furthermore, the ambiguity of Ura3 as a reporter protein permits screening for PPIs and PPI inhibition. As an example of the latter, mutations that interrupted the binding of the transcription factor Gal4 with its inhibitor Gal80 were identified by use of the R-Ura3-based method with selective medium without uracil (74). Finally, the N-end rule was employed for the development of mammalian and plant split-ubiquitin systems (528, 548).
As with the original two-hybrid method, the split-ubiquitin system suffers from pulling out a significant number of false-positive results. To cope with this issue, a new strategy was suggested in which the bait protein gene is integrated into the genome and controlled by its native promoter (502). This approach severely decreased the number of false-positive results, as shown by a screening experiment to map the interaction network of the ABC transporter Ycf1 (502). Further adjustments to balance out sensitivity and selectivity are provided with the use of a weak or inducible promoter for controlled bait expression (476, 541) and the availability of low- and high-copy-number bait and prey plasmids (219, 476). For the R-Ura3-based system, careful optimization of 5-FOA levels reduces false discovery rates (133). Protocols for split-ubiquitin experiments can be found elsewhere (149, 605).
The split-ubiquitin method has been adopted for several alternative applications (reviewed in reference 465). Split-ubiquitin three-hybrid techniques were developed for expression of a bridging or competing third protein (220) and for the identification of protein–small-molecule interactions (134). In addition, the unique feature of ubiquitin-induced proteolysis has been exploited to control protein abundance (520), to identify the endoplasmic reticulum pores that transport a specific substrate protein (150), and to eliminate cancer cells in a theoretical design based on conditional maintenance of a toxic protein-encoding vector (659). The steric requirements in a split-ubiquitin experiment further enabled studies on altered protein conformations (148, 534).
In a cytosolic variant (cytoY2H), the S. cerevisiae integral membrane protein Ost4 was added at the N-terminal end of the bait to direct the bait fusion to the membrane. The same strategy as that used for the split-ubiquitin system with the LexA-VP16 reporter was employed, but in this case, the bait protein of interest did not have to be a membrane-bound protein by itself (454). Application of the cytoY2H system revealed several translation-regulating binding partners of Uri1, an uncharacterized yeast protein (454), and a follow-up study confirmed a functional role for Uri1 in translation control (129). Other successful cytoY2H experiments were conducted with the S. cerevisiae ubiquitin ligase Ubr1 (286) and the A. thaliana pentatricopeptide repeat protein PNM1 (237).
The Split-mDHFR Method
General introduction to the split-mDHFR method.
DHFR catalyzes the reduction of dihydrofolate into tetrahydrofolate. Tetrahydrofolate is essential for cell proliferation and growth by acting as a precursor of purine and thymidylate synthesis. This crucial role for DHFR in cell survival can be used for PCA applications with DHFR split into two fragments, the F[1,2] N-terminal and the F[3] C-terminal fragments. In contrast to bacterial and yeast DHFRs, mammalian DHFR enzymes are much less sensitive to the chemical inhibitors methotrexate and trimethoprim. Therefore, murine DHFR (mDHFR) can serve as a reporter protein in bacterial and fungal DHFR systems in which a PPI is detected by survival of the cell in the presence of methotrexate or trimethoprim, with mDHFR taking over the function of the host DHFR protein (508, 587). In mammalian cells, nucleotide-free medium can be used for growth selection (543). Due to the presence of a selection step, the split-mDHFR method can be used to screen for novel PPIs and forms an alternative to transcription-based two-hybrid systems, with the benefit that the proteins under study reside in their natural subcellular compartment. In plant and mammalian cells, reconstituted DHFR has been visualized by addition of fluorescein-conjugated methotrexate (fMTX) (546, 625).
Application of the split-mDHFR method in yeast.
A genome-wide S. cerevisiae PPI screening was performed in which over 4,000 bait proteins were individually examined for interactions with over 4,000 prey proteins in a mating assay (633). After filtering out sticky proteins and benchmarking the results with reference sets, 2,770 high-quality interactions were retained in the final data set. The output was highly complementary with original two-hybrid screens and TAP-MS data. Comparison of interaction results for proteins of the small-subunit (SSU) processome indicated that, at least for this subset of the interactome, the split-mDHFR screening (633) was more complete in identifying true interactions between subunits than high-throughput two-hybrid data (398). The authors suggested that given the poor overlap between different high-throughput two-hybrid experiments (247, 294, 650, 721), the two-hybrid system is not inferior in its ability to detect PPIs, but the screenings were possibly far from reaching saturation. One possible explanation lies in the fact that the split-mDHFR screens were done with individual bait and prey strains, while the classic two-hybrid experiments were performed with pooled prey strains (398).
The Split-yCD Method
A highly versatile PCA is based on S. cerevisiae cytosine deaminase (156). Yeast cytosine deaminase (yCD), encoded by FCY1, is required for the pyrimidine salvage pathway to convert cytosine into uracil. The PCA with this 17-kDa protein was developed by comparing seven combinations of fragments and including three specific mutations that increase thermostability (360). Random mutagenesis of the N- and C-terminal fragments of yCD, followed by fusion to the human GTPase Ras and the Ras-binding domain of c-Raf, respectively, led to the identification of optimized yCD fragment sequences by selection on medium lacking uracil for yCD reassembly in an fcy1 strain. While uracil-deficient medium can be used for positive selection for an interaction, the dissociation of an interaction can be screened for on medium with 5-fluorocytosine, which is converted into toxic 5-fluorouridine triphosphate by a pathway dependent on yCD. This negative and positive selection procedure allows for screening for mutations that disrupt the interaction with one binding partner but not with another (156).
In resemblance to the split-yCD method, the split-Trp system (629) selects for interaction-induced reassembly of Trp1, which enables growth on tryptophan-deficient medium. The technology, developed by creation of randomly circularized permutations of Trp1 (216), was applied to confirm the association of Sec62 and Sec63, two members of the Sec complex.
The Split-Luciferase Method
General introduction to the split-luciferase method.
Luciferases are proteins that bind and catalyze the oxidation of their membrane-permeating substrate luciferin, which ultimately can be observed by the appearance of bioluminescence (705). By separation of luciferase N- and C-terminal fragments and fusion with proteins of interest, a PPI between these proteins can be visualized by the appearance of light. These split-luciferase systems, originally developed in mammalian cells (484), have the very practical characteristic that they provide a high temporal resolution of detection and are reversible, allowing near-real-time association studies. This can be exploited to quantify dynamic changes in protein assemblies (615). Moreover, split-luciferase systems take advantage of the very low cellular background luminescence, leading to a high signal-to-noise ratio. Luciferases applied in PCA technologies originate from the firefly (Photinus pyralis) (484), the sea pansy (Renilla reniformis) (317, 500), the copepod Gaussia princeps (542), and, more recently, the click beetle (258, 343). A particular advantage of the last three luciferases is their much stronger brightness than that of the firefly protein. In addition, click beetle luciferases from Pyrearinus termitilluminans and Pyrophorus plagiophthalamus beetles emit in green and red, respectively, which enables simultaneous investigation of two PPIs (672). A limitation of the method lies in the low photon efflux rates obtained when working with bioluminescence. The chemical reaction required to create excited-state luminescent substrates is a less efficient process than the light absorption-based excitation of fluorescent molecules. As a result, imaging at a subcellular level is difficult, though not impossible (317). Improvements of the luciferase fragments may increase the potential of the method to detect PPIs on a subcellular level. Recently, semirational combinatorial library screening led to the identification of fragments of green click beetle luciferase displaying faster and brighter bioluminescence with a concomitant higher signal-to-noise ratio. Illustrating the advantage of this PCA, time-lapse bioluminescence imaging revealed ligand-induced GPCR–β-arrestin coupling in the submembrane space. In addition, cell lines were generated to enable high-throughput screening for small molecules to disrupt interactions between activated GPCRs and β-arrestin (452). Finally, the split-luciferase system is one of the very few PCAs with applications in living animals.
Application of the split-luciferase method in yeast.
Reports on split-luciferase assays in yeast are rather scarce. However, in a study on the response of the yeast Fus3 MAP kinase pathway to pheromone (429), full advantage was taken of the temporal sensitivity and large dynamic detection range of the split-luciferase system. An ambient threshold concentration of pheromone leads to recruitment of the phosphatase Ptc1 to the scaffold protein Ste5 and to dissociation of the MAP kinase Fus3 from Ste5. Active liberated Fus3 then activates the transcription factor Ste12 for pheromone-responsive gene expression (165). The switch-like response of the pathway to pheromone was shown to depend on competitive binding of Fus3 and Ptc1 to Ste5, the dissonant behavior of Fus3 and Ptc1 in response to the phosphorylation status of four residues on Ste5, and a proposed two-stage binding of both proteins to Ste5 (429). Split-luciferase experiments with the three proteins formed an essential part of this investigation and showed that PCAs enable quantitative and dynamic analysis of PPIs.
The Split-FP Method
General introduction to the split-FP method.
Fluorescent proteins emit light upon excitation by an external light source. The discovery of GFP in Aequorea victoria (590) introduced the concept of fluorescent proteins in biology, and currently, a whole spectrum of natural and genetically optimized proteins is available (reviewed in reference 117). Many of these fluorescent proteins were adapted for interaction analysis by splitting the proteins into two fragments, each attached to a protein of interest (also reviewed in references 335 and 337). This type of PCA, called the split-FP method (or bimolecular fluorescence complementation), was originally developed in E. coli (205) and soon would become the most widespread of all PCA tools. Its easy technology transfer to other organisms led to the application of the split-FP method in many plants, prokaryotes, fungi, and animal cells. The strength of this technique lies in its ability to detect weak interactions at a subcellular resolution, and in contrast to the case for the split-luciferase method, no exogenous agents are required. Variations on the split-FP method have opened up new possibilities in PPI research. Multicolor split-FP assays can be used to monitor multiple PPIs simultaneously, and split FPs can be combined with BRET or FRET fluorescence to study higher-order complexes and with photoswitchable fluorophores to overcome the problems of bleaching and low quantum yield (see the mammalian and plant sections for further details on these variations).
Split-FP systems come with a number of limitations. First, the split-FP method suffers from the same difficulties observed with traditional fluorescence experiments, such as photobleaching, phototoxicity, and autofluorescence. Autofluorescence is not a vast problem in mammalian cells, but plant cells especially are notorious for giving a high background of fluorescence signals (135). Second, results from split-FP assays need to be interpreted with extra caution. Because of the rather low maturation rate of chromophore formation, and hence fluorescence reconstitution, and due to its irreversible nature, split-FP methods do not allow real-time measurements of PPI dynamics. This irreversible chromophore formation, however, offers the advantage of trapping weak (millimolar range) complexes (424). Therefore, split-FP systems, especially the split-Venus (yellow fluorescent protein [YFP] variant) version, are very sensitive, but with the cost of low selectivity. Accordingly, endogenous expression of the constructs is preferred, and positive results from split-FP experiments need to be confirmed by other means or by creation of mutated proteins that colocalize but no longer interact in a split-FP assay. This is important for differentiation between two proteins in close proximity and two proteins that really interact. Finally, slow maturation of split-FP fragments further complicates interpretations of the subcellular location of PPIs. During the time between fluorescence detection and initiation of the PPI, the protein couple can change its position in the cell.
Application of the split-FP method in yeast.
Fluorescent protein complementation of enhanced GFP (EGFP), yellow fluorescent protein (YFP), cyan fluorescent protein (CFP), the YFP variant Venus, and the monomeric Kusabira-Green fluorescent mutant (mKG2) has been employed in yeast (27, 28, 47, 104, 355, 489, 523, 626). Initial results came from studies on the influence of hydrogen peroxide on the location of the interacting couple Rho5 GTPase and Trr1 thioredoxin reductase (602), the dependence of the Rsr1 GTPase self-association on its activator, Bud5 (319), and the reliance for the interaction between Mso1 and the Sm-like protein Sec1 on the Rab GTPase Sec4 and its activator, Sec2 (694). Sec4 was later shown to interact directly with Mso1 in a split-FP assay (695). The use of the split-FP method, in particular that with the highly sensitive Venus fluorescent protein, has been extended further for the discovery of novel interactors in medium-scale experiments (523). Twenty-two lipid droplet proteins used as bait were screened for interactions with 225 mitochondrial and peroxisomal proteins used as prey, resulting in 116 PPIs, indicating a physical interaction of lipid droplets with both mitochondria and peroxisomes. Native promoters were used for bait and prey expression, and the mating approach was conducted to bring bait and prey together within the same strain. It can be expected that several other reports on the use of medium-scale split-FP assays will become public in the near future. A protocol for PCA applications in yeast, including the split-FP method, can be found elsewhere (443).
GENETIC PROTEIN-PROTEIN INTERACTION METHODS IN OTHER ORGANISMS
Genetic Protein-Protein Interaction Methods in Prokaryotes
Although experiments involving eukaryotic PPIs in Escherichia coli are hampered by the lack of an intron splicing machinery and the absence of particular posttranslational modifiers, prokaryotic two-hybrid systems show some clear advantages over the yeast two-hybrid system. First, the use of E. coli as a host organism for two-hybrid experiments enables screening with very large libraries in a very short time, due to the high transformation efficiency and fast growth of this bacterium. Second, two-hybrid screening in bacteria also reduces the chance that the host possesses a eukaryotic homolog that mediates a protein association, which raises the reliability for conclusions on a direct interaction. Furthermore, the absence of endogenous proteins that compete for interactions with the bait or the prey protein increases the sensitivity of the system. Third, the absence of a nuclear envelope avoids the requirement for the fusion proteins to pass a membrane. Finally, proteins that are toxic to yeast at high concentrations may not evoke the same effect in bacteria.
Detection methods for PPIs in bacteria are numerous and are based on fusions to transcriptional repressors and activators, membrane protein dimerization, complementation of biosynthetic enzymes or signaling molecules, and export of folded proteins. Examples are provided below for each of these techniques.
Bacterial two-hybrid systems.
For the development of two-hybrid methods in E. coli, inspiration was found in the dimeric behavior of bacterial repressors. The first bacterial two-hybrid systems were based upon the E. coli λ repressor, which confers immunity to phage infections (158, 273). The N-terminal part of this protein binds DNA, while the C-terminal part is responsible for dimerization, which is necessary for efficient DNA attachment. When the C-terminal region is replaced by a protein of interest, homo-oligomerization of this protein can be evaluated by repression of a lacZ reporter gene. Screening of prey libraries for interactions with this system is hindered by the appearance of homodimerizing prey proteins that repress the reporter independently from the bait. To circumvent this problem, the DNA-binding domains of two allelic variants of the E. coli LexA repressor DBD, i.e., the LexA wild-type DBD and LexA408 DBD, were each fused to a protein of interest (137). Both variants have different binding affinities, depending on the DNA sequence in the promoter. Hetero-oligomerization can then be distinguished from homo-oligomerization by the use of a hybrid sequence bearing an op408/op+ operator sequence in front of lacZ, each of which has a preference to be bound by one of the two LexA variants (Fig. 12A). This technique was recently used to confirm several prokaryotic PPIs (e.g., see references 140 and 664). In order to implement a mating-based strategy such as the one employed in yeast for library screening, an adaptation of this LexA-based bacterial two-hybrid assay was created with bait vectors carrying a mobilization element (103). These vectors can be transferred efficiently by conjugation from an E. coli strain donor expressing all the necessary components for mobilization function to a recipient strain harboring the prey vector.
Fig 12.
Specific bacterial genetic PPI detection methods. (A) Repressor-based two-hybrid system (137). Interaction between two proteins of interest, X and Y, can be monitored by fusion of each with a variant of the LexA repressor (408 or wild type). Heterodimerization of LexA408 and LexA+, induced by X-Y association, is required for efficient repression of the reporter gene lacZ, under the control of the LexA operators op408 and op+. (B) PhaR two-hybrid system (690). The binding of bait X with prey Y, fused to the DNA-binding domain (DBD) of the repressor PhaR and the PHB granule-associated protein PhaP, respectively, results in lacZ expression by recruitment of PhaR to PHB granules. (C) GFP recruitment system (132). Bait X is situated at cell division sites through the action of its chimeric partner, B. subtilis DivIVA. Interaction of protein X with GFP-tagged prey protein Y results in focused fluorescence at the cell division sites. GFP recruitment systems are also available for the pathogenic fungus Candida albicans, for C. elegans, and for mammalian cells. (D) ToxR two-hybrid system (357). The V. cholerae ToxR transcriptional activator requires dimerization of its periplasmic domain for full reporter transcription activation. In the ToxR two-hybrid system, the periplasmic domain is replaced by two proteins of interest, X and Y. An interaction between these two proteins results in efficient ctx promoter binding of the truncated ToxR protein (ToxR′) and subsequent gene expression of lacZ or the chloramphenicol acetyltransferase gene (cat). (E) Tat two-hybrid system (621). The Tat signal sequence (ss) peptide tethers bait protein X to the periplasm. A chimeric fusion of prey Y with the maltose-binding protein without a signal sequence (ΔssMBP) localizes to the periplasm only upon interaction of X with Y. This translocation is required for growth on medium with maltose as the sole carbon source. Alternatively, the prey protein Y is fused to a localization-deficient DsbA enzyme (ΔssDsbA), which catalyzes the formation of active alkaline phosphatase (AP). Active AP converts p-nitrophenyl phosphate (pNPP) to yellow p-nitrophenol (pNP). (F) Bacterial two-hybrid system for DNA-protein interactions (314). To increase sensitivity in the search for zinc finger-DNA associations, the binding of a zinc finger motif X to its target DNA sequence, Y (zinc finger binding motif Zfbm Y), is facilitated by inclusion of two fixed zinc fingers from Zif268 and the target DNA sequence Zif268 bm. The zinc finger fusion further consists of the S. cerevisiae Gal11 interaction domain (Gal11 ID), which binds the S. cerevisiae Gal4 dimerization domain (Gal4 ID). The latter domain is fused to the N-terminal domain of the RNA polymerase α subunit for indirect activation of an operon comprising the S. cerevisiae auxotrophic marker HIS3 and aadA, which confers resistance to spectinomycin. (G) Bacterial reverse two-hybrid system (239). Interaction between chimeric proteins of bait X with the bacteriophage λ cI repressor (which binds the λ cI OR2 operator) and prey Y with the N-terminal domain of the RNA polymerase α subunit results in activation of a gene encoding the C-terminal domain of the bacteriophage 186 cI repressor (186 cI CTD). This truncated protein sequesters and inactivates full-length 186 cI, which normally downregulates cytotoxic 186 prophage genes. Resulting cell death can be circumvented by mutations that block the bait-prey interaction. (H) Intein-mediated split-GFP assay (485). Bait protein X is in fusion with the N-terminal fragments of the intein VDE and GFP, while prey protein Y constructs include their respective C-terminal counterparts. Interaction between X and Y reconstitutes VDE, which splices out and covalently reattaches the GFP fragments to create an isolated GFP monomer, detected by fluorescence. The EGFP structure image is based on the PDB structure under accession number 2Y0G (550).
The implementation of transcriptional activators in prokaryotic two-hybrid systems significantly enhanced the potential for screening experiments by selective growth. Based upon a prototypical experiment involving the bacteriophage λ cI repressor (142), several two-hybrid systems were developed. In one design, the λ cI protein and the α subunit of RNA polymerase are employed as the DBD and AD, respectively (141, 142). The reporter genes are lacZ and the β-lactamase bla gene, which confers resistance to carbenicillin (141). In another system, a triple-zinc-finger motif of murine Zif268 serves as the DBD, and an operon of HIS3 and the spectinomycin resistance gene aadA is applied for efficient screening (314). The methodology was also used to commercially develop the BacterioMatch two-hybrid system and, subsequently, the BacterioMatch II tool, featuring a new HIS3-aadA reporter cassette (Agilent Technologies). In a recent study, the global PPI network of the human pathogen Mycobacterium tuberculosis H37Rv was unraveled using this technique, revealing more than 8,000 interactions among almost 3,000 proteins (689). Alternatively, the ω subunit of RNA polymerase is linked with the prey protein of interest (142). An approach combining the ω subunit and the Zif268 zinc finger domain is particularly suited for studies of PPIs between two monomers (655). The latter system was modified for use with Gateway entry clones, providing a new tool for rapid PPI screening (323). A protocol for bacterial two-hybrid experiments can be found elsewhere (206).
A two-hybrid system in E. coli based on the polyhydroxybutyrate (PHB) synthesis regulatory protein PhaR was recently created (690). This method relies on the fusion of bait and prey proteins carrying the DBD of PhaR and the PHB granule-binding protein PhaP, respectively. The bait fusion protein represses the reporter gene lacZ by binding its promoter. Interaction between bait and prey constructs tethers the bait to the PHB granules, which results in the release of lacZ expression (Fig. 12B). This method displays a reduced technical false-positive rate, resulting from the use of extrinsic components of PHB synthesis, and technology transfer is possible to other bacterial species that can sustain sufficient PHB granule accumulation (690).
Membrane-localized and secretory pathway two-hybrid systems.
There are two main shortcomings of classic two-hybrid tools that prompted researchers to develop alternative methods, namely, the lack of information on the affinity or level of expression of the interacting proteins and the failure to detect interactions within the secretory compartments for proteins that require an oxidizing environment for proper folding. In that respect, the APEx two-hybrid system for anchored periplasmic expression was engineered as a quantitative PPI assay of particular importance for antibody discovery and for selection of high-affinity antibodies (304). The method, which resembles the yeast surface two-hybrid systems, is based on the expression of a soluble epitope-tagged prey protein and a bait protein anchored on the periplasmic side of the inner membrane of E. coli by fusion to a leader peptide and to the first 6 amino acids of the E. coli lipoprotein NlpA. Upon interaction, the prey remains associated with the bait in spheroplasts, which allows quantitative detection by fluorescent anti-epitope-tag antibodies. In such a system, all nonassociated prey proteins are removed in the extracellular fluid upon spheroplast induction. In addition, because the fluorescence signal is a direct function of both the affinity of the interaction and the expression level of the interacting partners, selection for either increased affinity or improved expression is achieved by using multicolor FACS analyses (304).
In a cytology-based screening assay, one protein is fused to DivIVA from B. subtilis or FtsZ from E. coli to target a second protein, fused to GFP, to cell division sites (106, 132) (Fig. 12C). Interactions were observed between soluble proteins, such as the leucine zipper domains of yeast Gcn4, and integral membrane proteins, such as the VirB subunits of the T-DNA transfer system of Agrobacterium tumefaciens. This GFP recruitment system was applied in both E. coli and Agrobacterium tumefaciens (132).
The cholera toxin ToxR regulatory protein of Vibrio cholerae has been exploited as a genetic indicator of PPIs in E. coli in several variations of the two-hybrid approach. ToxR consists of a cytoplasmic gene activating domain linked by a membrane-spanning region to a periplasmic part. ToxR homodimerization at the periplasmic domain is required for proper transcription-inducing activity, and replacement of this domain by proteins of interest allows for PPI experiments (Fig. 12D). Detection of periplasmic PPIs, and of cytoplasmic PPIs after removal of the transmembrane region, is possible in the ToxR-based system (357, 358, 371). In E. coli, ToxR is capable of directly activating transcription at the ctx promoter sequence, which is used as the regulatory element driving a reporter construct such as chromosomal ctx::lacZ (357), plasmid-carried ctx::chloramphenicol acetyltransferase (cat) in the TOXCAT system (553), chromosomal ctx::cat in the POSSYCAT system (positive selection system based on chromosomally integrated cat) to discriminate between interactions of different affinity (233), or the red fluorescent protein variant mCherry for whole-cell detection without an additional substrate (39). The ToxR-based tool has been used as an indicator of folding stability (356), interactions between transmembrane helices (553), heterodimerization in both the periplasm and cytoplasm (252), and sequence motifs required for helix-helix interactions by use of a disabled ToxR fusion as a dominant-negative protein (39). In a variation of the ToxR system, two LexA DBDs (wild-type and 408 DBDs) (137) were coupled to wild-type and mutated glycophorin A transmembrane helices to allow detection in a biological membrane, and lacZ was placed under the regulation of promoter elements, each bound specifically by one LexA repressor domain (564). This system, named GALLEX, can measure both homo- and heterodimerization of membrane proteins, as recently illustrated by the analysis of transmembrane domain interactions between major histocompatibility complex class II proteins (347).
An alternative two-hybrid system in E. coli detects PPIs based on the biological folding quality control mechanism inherent to the twin-arginine transporter pathway (Tat). This mechanism relies on the export of correctly folded proteins by association with a protein carrying a Tat signal peptide (547). In the Tat two-hybrid system, one protein is fused to a Tat signal peptide and the second is fused to a protein reporter that can confer a phenotype only upon export into the periplasmic space (621) (Fig. 12E). In the first attempt, two reporters were used: the maltose-binding protein, whose export permits selection for growth on maltose, and DsbA, which catalyzes the formation of alkaline phosphatase. In an alternative version of the Tat-based system, called FLI-TRAP (functional ligand-binding identification by Tat-based recognition of associating proteins), Waraho and DeLisa (691) exploited the colocalization of the reporter β-lactamase Bla into the periplasm as a semiquantitative and high-throughput readout for interactions. Only those chimeras that were highly expressed and interacted strongly were able to confer β-lactam antibiotic resistance to cells.
Bacterial one-hybrid systems.
Bacterial two-hybrid techniques have been adapted further for protein-DNA studies in one-hybrid assays. The high transformation efficiency of bacteria is especially advantageous for one-hybrid experiments involving randomized DNA or zinc finger motif libraries, enabling screening procedures with 108 transformants. DNA-binding proteins or domains are directly or indirectly fused with the ω (272, 314, 474) or α (151, 439) subunit of RNA polymerase. In addition, weak DNA-protein interactions can be studied by incorporation of a fixed zinc finger-DNA association that facilitates the binding of the DNA and protein of interest (314) (Fig. 12F). Optimized algorithms aid in enhanced predictions of binding motifs from one-hybrid studies (99). Bacterial one-hybrid assays are commonly applied and include studies on selective screening for zinc finger motifs that bind a specified DNA sequence (151, 314) and transcription factors from D. melanogaster (474, 739) and Mycobacterium tuberculosis (232), the latter by application of the commercial BacterioMatch II kit.
Bacterial reverse two-hybrid systems.
The traditional two-hybrid technique can be altered to couple bacterial cell growth to the dissociation of a protein complex, similar to the yeast reverse two-hybrid system. The high cell permeability of bacteria confers a strong advantage regarding experiments that involve addition of small molecules as putative inhibitors of PPIs (204). The first bacterial reverse two-hybrid method was based on reporter gene repression by λ cI, dependent on homodimerization of a protein of interest in fusion with cI (490). Its application led to the discovery of peptides that inhibit HIV-1 protease dimerization. Dissociation of dimerization was observed by derepression of an operon consisting of lacZ and the tetracycline resistance marker tet (490). A similar concept of derepression by interaction inhibition made use of the tricistronic HIS3-Kanr-lacZ operon, originally developed for a forward two-hybrid system (131), as a reporter (267; for the protocol, see reference 266). Coupled with the intracellular synthesis of libraries containing up to 108 cyclic peptides, this system yielded the discovery of inhibitors of an enzymatic dimerization essential to HIV infection (267) or purine synthesis (634). Furthermore, application of this approach led to the detection of peptide inhibitors of the interactions between the tumor suppressor p53 and MDM2 or MDMX (115) and between the HIV Gag protein and human TSG101 (635) and to the elucidation of antiviral defense silencing by the influenza virus NS1 protein (451). To enable stable protein expression independent of the plasmid copy number, as well as to reduce false-positive results due to plasmid loss, chromosomally integrated bait and prey vectors are now available (451). In a different approach, the URA3/5-FOA counterselection system was employed in the bacterial trap system to search for inhibitors of interaction (440). Another recent system exploits a toxic gene as a marker for PPIs (239). This system makes use of the N-terminally truncated version of the bacteriophage 186 cI repressor, lacking the DNA-binding motif, which has a dominant-negative effect on full-length 186 cI, to induce prophage-mediated cell death or to significantly inhibit cell growth upon its expression (Fig. 12G). Based on this reporter concept, disruption of the interaction between a λ cI bait and an RNAPα prey leads to cell growth. The system was applied to the identification of residues important for dimerization of the human transcription factors Arnt and AhR (239).
Bacterial PCA methods.
PPI assays based on the oligomerization-assisted reassembly of split proteins are abundant for use with prokaryotes and include the use of GFP (205), adenylate cyclase from the Gram-negative bacterium Bordetella pertussis (322), and murine DHFR (507), among several others.
(i) Split-FP applications in bacteria.
The split-FP system was originally described for E. coli by Regan and coworkers, in whose study PPI identification was based on fusions to a dissected GFP construct (205). Folding and fluorescence of the split GFP molecule were achieved by bringing into close proximity two fragments of GFP fused to strongly interacting antiparallel leucine zippers. The same research group later provided a set of comaintained plasmids with incorporation of a hexahistidine tag and compatible with E. coli strains expressing the T7 polymerase (424). The study also provided evidence that such a system could be used not only for peptide-peptide interactions but also for identification of interactions between larger proteins. A split-YFP method was used by the group of Ventura to exploit the irreversible behavior of FP fragment folding for detection of transient and weak interactions between individual proteins and between proteins and peptides (for the protocol, see reference 458). With the split-YFP method, weak and strong interactions can be distinguished, suggesting that studies to screen binding affinities could be performed using this technique combined with flow cytometry assays (459). Application of a reverse split-FP strategy is also feasible for identification of competitive inhibitors of a protein association. The interaction of the E. coli heat shock protein DnaK with short hydrophobic segments of proteins was used as a case study to identify pyrrhocoricin-related antibacterial peptides as inhibitors of the interaction (457). Incubation of the cells with the potential inhibitors prior to transcriptional induction of the FP fusion construct is a prerequisite of the method due to the irreversible nature of split-fluorophore assays. Split-FP methods have been applied to other prokaryotes, including A. tumefaciens (17) and B. subtilis (123).
Making use of GFP reconstitution, Umezawa and coworkers worked out an alternative concept based on the intein-mediated protein reconstitution system (PRS) to detect PPIs (485). Inteins are self-splicing proteins that induce the release of reassembled GFP upon interaction of fusion proteins (Fig. 12H) (for the protocol, see reference 320). In the first approach, a variant of the S. cerevisiae Vma1 intein was used as a self-splicing protein element to release GFP following interaction of the fusion proteins (485). To avoid the problem of low splicing efficiency with this intein, the system was improved by integration of the split-intein DnaE from Synechocystis sp., which allowed the formation of GFP after 4 h, instead of the 3 days required in the previous system. A provisional screening experiment with calmodulin and its target peptide, M13, showed that positive transformants could be selected from a negative pool (486).
(ii) The split-CyaA method.
Another PCA technique in E. coli is based on the reconstitution of the Bordetella pertussis adenylate cyclase CyaA (322). The catalytic domain of CyaA can be separated into two complementary fragments, T25 and T18. When each fragment is fused to a protein of interest, a functional adenylate cyclase can be reassembled upon interaction of the two proteins, which is followed by the production of cyclic AMP (cAMP) in an E. coli strain lacking its own adenylate cyclase. Because the activation of genes responsible for the fermentation of maltose and lactose is dependent on cAMP (651), media containing maltose or lactose as the sole carbon source can be used for selection. As one of the most commonly applied and successful bacterial interaction methods, this system has been used widely to discover novel interactors (e.g., see references 85 and 505), to elucidate module-scale interaction networks (e.g., see references 1 and 603), and in particular to establish the network among E. coli membrane proteins (321). Because the confirmation of novel interactions by an independent method is a common practice, bacterial two-hybrid plasmids based on the recombination of adenylate cyclase were modified for easy transfer to vectors for single or double affinity purification (33). The system was also adapted for high-throughput screening for dimerization inhibitors of the type IV secretion protein VirB8 (495). Compounds that reduced VirB8 dimerization were detected by reductions in cAMP reassembly and reduced lacZ expression under the control of an active cAMP pathway. In this assay, the C terminus of the VirB8 protein is positioned in the periplasm, which reflects the natural environment of the protein and therefore is likely more suitable for compound screens than other in vitro or in vivo systems (495).
(iii) Other PCA methods in bacteria.
Some enzymes, such as β-galactosidase and TEM β-lactamase, can be split into two nonfunctional α and ω peptides which lead to proper function only when they are brought into close proximity. A split-galactosidase system was developed in E. coli to confirm known cytoplasmic and membranous PPIs and to validate the association of cytochrome c2 and cytochrome c peroxidase from Rhodobacter capsulatus (51). A split-lactamase system for E. coli enabled the confirmation of the interaction between the human transcription factors Fos and Jun on selective medium with β-lactam antibiotics, and its development highlighted the importance of linker identity (699). Chorismate mutase (CM) from Methanococcus jannaschii is a relatively small enzyme which converts chorismate into prephenate, a crucial step in the biosynthesis of aromatic amino acids. A split-CM system was created for selection on medium lacking aromatic amino acids, with the unique feature that its linker adds strong geometric constraints, which limits its general application but could aid in analyses of the orientation of PPIs (466). As in yeast, the split-DHFR method can be used for E. coli survival selection on medium containing trimethoprim. Bacterial split-DHFR assays are used mostly to optimize peptides for increased binding affinity, for example, for leucine zipper domains (507), and peptides that bind the human transcription factor Jun (110). Other PCA methods include a split-Trp system for E. coli and Mycobacterium smegmatis (479, 556) and a split-adenylate kinase system for Thermus thermophilus (470), both with selective reporters.
Genetic Protein-Protein Interaction Methods in Alternative Fungal Species
Two-hybrid assays are typically carried out in surrogate hosts such as E. coli and S. cerevisiae, which are fast to reproduce, easy to handle, and use the universal genetic code. However, heterologous protein expression of organisms using nonstandard genetic codes is cumbersome in these model host systems. In the fungal kingdom, several Candida species, in particular the human pathogen Candida albicans, have evolved an aberrant codon usage in which the CUG codon encodes a serine instead of a leucine amino acid (561). To circumvent the problem of erroneous translation in heterologous systems, two interaction methods have been developed in C. albicans itself. A classic two-hybrid system was adapted for use in C. albicans (623). Reporter systems, DBDs, and ADs were all compatible for application in C. albicans, as they did not contain interfering CUG codons or those codons were modified. This system identified known and novel PPIs, not previously identified in the yeast system, among signaling pathways involved in virulence of the pathogen. Prey proteins fused to the viral protein VP16 and bait proteins fused to the Staphylococcus aureus repressor LexA were coexpressed from the methionine-regulatable MET3 promoter in order to avoid unwanted overexpression. A second method makes use of Vps32, a protein associated with the cytoplasmic side of endocytic vesicles, as a bait construct to identify interactions by targeted GFP fluorescence in endocytic vesicles (56). This GFP recruitment system, referred to as the vesicle capture interaction (VCI) assay, can yield quantitative data by computational methods of microscopic image analysis. The C. albicans VCI system was employed in a conditional study to illustrate the novel finding that human β-defensins can elicit the interaction between the kinases Pbs2 and Hog1 in this pathogenic fungus (13).
Split-FP assays have been exploited widely in fungi, often as a validation tool for yeast two-hybrid analyses or coimmunoprecipitation assays. In the first split-FP experiment in fungi, nuclear heterodimerization of two transcription factors by a split-EYFP system was shown in the β-lactamase-producing fungus Acremonium chrysogenum (263). All gene fusions were expressed under the control of the Aspergillus nidulans gpdA promoter and trpC terminator to ensure stoichiometric expression of the different constructs. The same split-EYFP vectors were recently applied in the fungus Penicillium chrysogenum (262). Nuclear fluorescence was observed between two components of the Velvet-like protein complex, while this interaction was not identified by yeast two-hybrid analysis. This suggests the requirement of a bridging protein to bring together the two proteins of interest and further highlights the need for interaction identification in homologous systems. The YFP-based approach was further used in Aspergillus nidulans (48), the plant pathogen Magnaporthe grisea (732) (Fig. 13A), the model fungal organism Neurospora crassa (25), and the homothallic ascomycete Sordaria macrospora (166). Alternatively, a Venus-based system similar to the one described for C. elegans (595, 596) was established in the fission yeast Schizosaccharomyces pombe (6). Plasmids were constructed that allow convenient C-terminal tagging of proteins of interest expressed from their endogenous chromosomal locations and under the control of their native promoters. In a case study, the spatial dynamics of the copper transporter Ctr4-Ctr5 complex was shown using Venus complementation in S. pombe. Addition of high concentrations of copper induced internalization of the complex, as the fluorescence signal progressively shifted from the cell surface to the vesicles (289). Finally, a split-EGFP assay using constitutive expression constructs has been described for the plant-symbiotic fungus Epichloë festucae (631).
Fig 13.
Visualization of PPIs by PCAs. (A) Split-YFP assay in the plant pathogen Magnaporthe grisea (732). Pmk1 and Mst7 kinases, which are components of the MAP kinase pathway essential to appressorium formation and plant infection, were fused to the C-terminal and N-terminal parts of YFP, respectively. Interaction between Pmk1 and Mst7 was observed in vivo in appressorium formations only when the putative docking site of Mst7 was intact. A, appressorium; G, germ tube. (Adapted from reference 732 with permission.) (B) Multicolor split-FP assay in tobacco culture cells. Protoplasts were transfected by direct DNA uptake and visualized using laser scanning confocal microscopy. Simultaneous interactions between Agrobacterium tumefaciens VirE2 (VirE2-cCFP) and the Arabidopsis nuclear transport adapter protein importin α-1 (Impa-1-nCerulean) and between VirE2 and importin α-4 (Impa-4-nVenus) were observed in the cytoplasm (cerulean) and nucleus (Venus), respectively. Nuclear localization was confirmed by colocalization of the nuclear marker mCherry-VirD2NLS. Labels below each image indicate the filter set/channel imaged. DIC, differential interference contrast image. (Adapted from reference 375 with permission.) (C) Visualization of odor-evoked calcium release upon formation of functional heteromeric complexes of odorant receptors (ORs) in Drosophila, using a split-YFP assay. (Top) Complementary N-terminal and C-terminal fragments of YFP [YFP(1) and YFP(2)] were fused to the odorant receptor OR83b. (Left) Dimerization of OR83b in neurons lacking native OR83b, visualized by the split-YFP method. (Right) Dimerization of OR83b is still visible in Gr21a neurons, which do not express any native ORs, suggesting a direct PPI. (Bottom) Complementary N-terminal and C-terminal fragments of YFP [YFP(1) and YFP(2)] were fused to the odorant receptor OR43a. YFP complementation is visible in neurons with OR83b but not in neurons lacking OR83b. This implicates that OR43a dimerization depends on the presence of OR83b and may not be a direct PPI. (Adapted from reference 38 with permission.) (D) In vivo imaging of split Renilla luciferase (RLuc) complementation in living mice (344). The strategy to monitor translocation of a particular protein into the nucleus is based on reconstitution of split RLuc by the intein Dna-E (also see Fig. 12H). RLuc-N (N-terminal part) was fused to DnaE-N and a nuclear localization signal. This chimera localizes mainly to the nucleus. RLuc-C (C-terminal part) was fused to DnaE-C and a protein of interest, the nuclear androgen receptor (AR), which localizes to the cytosol. Translocation of AR into the nucleus was visualized upon addition of 5a-dihydrotestosterone (DHT), which binds AR, in COS-7 cells implanted on the backs of mice. DHT-induced translocation of AR results in reconstitution of the DnaE intein and its splicing-reassembly property. Consequently, the spliced and reconstituted RLuc recovers its bioluminescence activity, which is imaged by using a cooled CCD camera and measured as photons per second per cm2. The differential translocation of AR in the presence (+) or absence (−) of DHT could hence be evaluated quantitatively. (Adapted from reference 344 [copyright 2004, National Academy of Sciences].)
Genetic Protein-Protein Interaction Methods in Plants
In vivo protein-protein interaction methods in higher eukaryotes offer the ability to study known or novel PPIs in their native cellular context, and in real time in living cells when visualization is possible. In the last decade, such methods have been established in plants, in particular in the model plant Arabidopsis thaliana (also reviewed in reference 462).
Two-hybrid tools in planta.
The classic two-hybrid method was replicated in A. thaliana protoplasts by use of Gateway-compatible vectors (161). High-copy-number vectors allow expression of DBD- and AD-fused proteins of interest under the control of the strong promoter from cauliflower mosaic virus 35S. Binding of the interacting partners to a GAL4-UAS4::GUS reporter system promotes expression of β-glucuronidase (GUS), used as a semiquantitative readout. Coexpression of a Pro35S/NAN (synthetic neuraminidase gene) vector is employed to normalize GUS measurements for variation in protoplast transfection efficiency. The proof of principle of this method was based on interactions between basic leucine zipper (bZIP) transcription factors and on a direct comparison of the same interacting partners in the yeast two-hybrid system. Novel and weak heterodimerization events that were not detected in the yeast system were identified using the plant two-hybrid approach. An additional two-hybrid method was developed for detection of PPIs involving transcription factors (435). Similar to the repressed transactivation method in yeast, the prey protein is fused to a repressor domain, in this case the ERF-associated amphiphilic repression motif SRDX. The autoactivating bait is linked to the Gal4 DBD, and interaction between bait and prey results in repression of a luciferase reporter gene. Interaction between the human Fos and Jun transcription factors was confirmed with this method, together with the association of two MADS box plant proteins in transgenic Arabidopsis plants (435).
Another two-hybrid method was engineered as an indicator of human estrogenic activities, using transgenic A. thaliana constitutively expressing two effector proteins (643). These consist of a LexA-linked estrogen human receptor and a VP16-fused chimeric human nuclear receptor coactivator. Estrogen-dependent interaction between the two chimeras induces transcriptional activation of the β-glucuronidase reporter. This system illustrates the use of A. thaliana to detect the presence of 17-βa-estradiol at concentrations as low as 50 pM, as well as other estrogenic substrates. This low-cost and sensitive two-hybrid system was further improved by increasing the copy number of the plasmid carrying the prey fusion gene (630). The system was hence rendered five times more sensitive than previously available assays in A. thaliana and other organisms (464, 471, 657).
In a particular study where neither split-FP nor yeast split-ubiquitin assays detected PPIs between two membrane receptors, ERS1 and ETR2, interaction between these proteins was established in a membrane recruitment assay (221). The full-length ethylene receptor ERS1 fused to red fluorescent protein (RFP) served as an anchor to recruit GFP-fused target proteins, which was visualized by colocalization of the fluorescence signals. In this GFP recruitment assay, interaction between ERS1 and the cytoplasmic version of ETR2 was illustrated in plant cells.
Split-FP assays in plants.
Methods of visualization and identification of PPIs in subcellular compartments became available in the context of living plant cells with the development of the split-FP method. The first demonstration of the efficacy of YFP complementation to detect PPIs in plant cells derived from two studies (57, 684). Ohad and his group demonstrated PPIs at the tissue and subcellular levels in Nicotiana benthamiana and Arabidopsis leaves (57). Kudla and coworkers illustrated the dimerization of the Nicotiana tabacum 14-3-3 protein T14-3c in Arabidopsis protoplasts and in Agrobacterium-infiltrated tobacco leaves by using Gateway-compatible split-YFP vectors, which were later used by many other groups (684). Since then, the use of split-FP assays in plants has boomed, and recent reports comprehensively review this PCA method (42, 101, 477, 478). Protocols on how to use the split-FP method as a tool to study PPIs in plant protoplasts are also available (e.g., see references 498 and 701).
The diverse targets in which the assay has been employed range from protoplasts to seedlings, leaves, or epidermal cells in Arabidopsis but also in tobacco, mustard, parsley, leek, and onion plants. Many fluorescent proteins have been reported, including the commonly used YFP (with N- and C-terminal residues YN155 and YC155 or YN173 and YC173), but also Venus, GFP, CFP, SCFP3A (modified CFP), blue fluorescent protein (BFP), cerulean, citrine, and RFP (117). Multicolor split-FP assays for simultaneous or preferential PPI detection have been adapted for plant research, based on the use of the combination of SCFP3A N- and C-terminal fragments with the Venus N-terminal fragment, as well as with fragments from CFP, GFP, YFP, and DsRed-monomer (353, 678) (Fig. 13B). Multicolor expression vectors were also developed in the pSAT series of vectors to facilitate the practice of the method (377). In this study, the differential interaction of the Agrobacterium VirE2 protein with the Arabidopsis importins α-1 and α-4 was illustrated by the cytoplasmic and nuclear localization of the yellow and blue fluorescence signals, respectively. The analysis of interactions between more than two proteins has also been achieved successfully by imaging with combined split-FP and FRET fluorescence (368). Formation of ternary complexes in leaf epidermal cells was visualized as a result of simultaneous interactions between three fluorophore-tagged polypeptides.
The interest in improving fluorophores is evident in the split-FP field, and recent technical efforts on the use of fluorophores in plants have been reported. A truncated version of YFP (lacking two C-terminal amino acids) that greatly eliminated unspecific YFP reconstitution proved to be efficient in split-FP experiments in Arabidopsis protoplasts (388). Although most fluorescence microscopes have the capacity to detect GFP fluorescence, the use of GFP-based PCA has been hindered by the low reconstitution efficiency of split-GFP fragments. However, the combination of the N-terminal region of GFP with either the C-terminal region of CFP or a mutated V163A version of the GFP C-terminal domain showed bright green fluorescence that was 7-fold more efficient than that with the original split-GFP setup (351). An optimized monomeric RFP (mRFP)-based assay was recently described for the investigation of plant-virus interactions in N. benthamiana (740). The new plasmids enable fusion of proteins of interest to either the N- or C-terminal domain of the mRFP fragments, and they possess a linker to improve the flexibility of the chimeric proteins and a c-myc or HA tag to allow immunoblot analysis. Work has also been done in improving the detection sensitivity of PPIs in plant protoplasts by developing a cell sorting procedure (730). Flow cytometry analysis of fluorescence signals in protoplasts isolated from plants with low transformation efficiency can facilitate subsequent PPI identification by confocal microscopy. With the aim of performing high-throughput analyses using the multicolor split-FP assay, Gateway-compatible vectors expressing all possible combinations of SCFP3A and Venus, fused N- or C-terminally, were recently generated (203). In a case study, the vectors were used to show simultaneous interaction between Cnx6 and Cnx7 and between Cnx6 molecules themselves, forming an interacting complex of the molybdopterin synthase.
Despite the advantages offered by the split-FP method, its applicability to whole-interactome mapping has been limited (731). It was employed, however, in parallel experiments with the yeast two-hybrid system to determine the pairwise interactome network of 58 core cell cycle proteins of Arabidopsis (53). GFP fragments were fused C-terminally to target proteins and expressed under the control of a strong 35S promoter. Out of 917 possible interactions, 341 were positively identified with the split-GFP method, while only 77 were established by the yeast-two hybrid approach and only 17% of PPIs were identified by both techniques. For the split-GFP assay, a negative-control set with 40 protein pairs did not show positive results. Interestingly, for 20% of all interactions identified by split-GFP assay, reciprocal expression of the target proteins was necessary for proper GFP refolding. In addition, this method allowed the exclusive detection of 78% of all PPIs, while the yeast two-hybrid assay detected fewer than 5% of PPIs that were not identified by the PCA technique. Overall, these data provide an example of how each technique should be considered and highlight the power of the split-FP strategy and the use of endogenous host cells. In addition, this study resulted in the identification of novel interacting pairs between cyclins of the CDK-CYCD complexes. These binary interactions induced cell division in differentiated tobacco epidermal leaf cells but also in Arabidopsis cells (52). The interaction data were also processed together with gene expression and localization data in a compiling analysis that highlighted distinct protein clusters at each step of the cell cycle. In a comparative analysis between the plant split-FP system, yeast two-hybrid system, tandem affinity purification, and predictive algorithms, the same group revealed platform-specific interactions, a large number of PPIs that were not predicted, and overall limited overlap between the methods (658).
Other PCA methods in plants.
Detection of PPIs based on the reconstitution of reporters other than fluorophores has been attempted in plant cells. Interaction-induced folding of murine DHFR was employed in tobacco protoplasts (625). The reconstituted enzyme binds fMTX, which is retained in cells and can be monitored by spectroscopy, FACS, or fluorescence microscopy. In contrast to the successful implementation of split-DHFR assays, the use of β-galactosidase PCA is poorly suitable for plants due to the high intrinsic level of β-galactosidase activity (637).
As a complement to the split-FP method, split-luciferase assays based on the Renilla reniformis and Photinus pyralis (firefly) luciferase enzymes have been used in protoplasts and whole plants, respectively. PPIs between nuclear histones 2A and 2B and between the membrane proteins SYP51 and SYP61 were demonstrated in the protoplast system (188). This work in protoplasts was applied to construct a series of vectors suitable for high-efficiency transgene expression, to increase the dynamic range of PPI detection levels, and to engineer a large-scale analysis platform of protoplast transfection using 96-well plates (325). This method reliably identified interactions between the membrane-associated SNARE proteins (324). Luminescence was measured within Arabidopsis protoplasts expressing the recombinant proteins at physiological levels. The stringency of the assay was determined by single amino acid substitutions resulting in reduced SNARE-SNARE interaction and by modulating the interactions by use of sodium azide. In plants, the system was adapted to enable both transient expression of fusion proteins and generation of stable transgenic plants (86). Two fragments of the firefly luciferase, i.e., NLuc (aa 2 to 416) and CLuc (aa 398 to 550), were expressed under the control of a strong 35S promoter. Multiple pairs of known interacting proteins were used to validate the system.
Genetic Protein-Protein Interaction Methods in Nonmammalian Animal Models
Interaction methods in invertebrates.
The cellular milieu of the sea hare Aplysia californica, a model organism in neurobiology, is considerably more salty than that of yeast cells. To study interactions between cAMP-dependent transcription factors in their native environment, an Aplysia two-hybrid system was created with the traditional elements from the yeast method, namely, the Gal4 DBD, the Gal4 AD, and lacZ as a reporter gene (97).
The two-hybrid methodology was also adapted in a cultured insect cell model, providing an alternative method for surveying PPIs that cannot be studied in yeast, in particular those affected by posttranslational modifications such as glycosylation, phosphorylation, and acetylation (456). This insect two-hybrid system involves two proteins of interest, fused to the yeast Gal4 DBD and to the AD of mouse nuclear factor kappa B (NF-κB), with firefly luciferase as the reporter. The DBD and AD constructs were placed under the expression of the immediate-early promoter (IE2) from the Orgyia pseudotsugata baculovirus, which is known to allow protein expression in several insects, enabling the use of the method in a wide range of cell lines. The system shows high sensitivity due to low background luciferase activity, with the luciferase gene placed under the control of a minimal HSP70 promoter linked to Gal4 upstream activating sequences. It further permits the elimination of false-positive substrates, such as autoactivators, obtained in the yeast two-hybrid system (456).
In contrast to the extensive use and adaptation of the split-FP method in plants and mammalian systems, PCA technologies to study and visualize PPIs in nonmammalian animal model organisms are sparse. Among invertebrates, the transparent body of the worm Caenorhabditis elegans makes this organism an excellent model for spatiotemporal PPI research involving fluorescence-based applications. A traditional proof-of-principle interaction between leucine zipper polypeptides was detected with split GFP, split CFP, and a combination of GFP and YFP fragments in C. elegans (729). Using a YFP reconstitution assay, temporal and spatial interactions of the stomatin-like protein UNC-1 and the innexin UNC-9 at intercellular junctions were reported (84). A split-Venus system for use in the worm was introduced by Hu and coworkers (257). Direct visualization of the binding of the leucine zipper domains from the C. elegans transcription factors FOS-1 and JUN-1 was demonstrated using an inducible heat shock promoter, with the appearance of fluorescence 30 min after induction. The heat shock promoter was employed to counteract the irreversibility of the split-fluorophore approach and to avoid the potentially detrimental effects of such a system on cellular and developmental behavior. Protocols for this assay are available elsewhere (257, 595). Applications of the split-Venus system in C. elegans include detection of the nucleus-localized interaction between worm BRCA2 and mammalian Rad51, involved in DNA repair (450); oligomerization of DYN-1, essential for endocytosis, at specific membrane regions along the apical surface of intestinal cells (248); nucleus-localized association of the transcriptional regulators MLS-1 and UNC-37 (448); and the PPI between the BK channel subunit SLO-1 and an auxiliary subunit, BKIP-1 (83). The last study did, however, reveal a possible drawback of using the highly sensitive but lowly selective Venus method. While a coimmunoprecipitation experiment clearly showed that the binding of SLO-1 with BKIP-1 depends on the transmembrane and intracellular domains of BKIP-1, split-Venus results remained positive after removal of the intracellular region. This suggests that, in this particular experiment, close proximity rather than actual binding may have caused a positive outcome.
A GFP recruitment assay in C. elegans links interactions to the localization of fluorescence signals to the membrane, in the so-called differential cytolocalization assay (DCLA) (45). A reciprocity test performed by switching the identities of the bait and prey proteins showed that, in most cases, the interactions were retained in this assay. Comparative analyses of the DCLA system with coimmunoprecipitation and yeast two-hybrid data showed only very little overlap in the interaction sets identified, yet controls for false-positive results failed to show interaction. These data support the complementary nature of the different detection methods.
The split-YFP technique has also been applied in the fruit fly model for PPI detection between odorant receptors in olfactory sensory neurons (38) (Fig. 13C). A similar approach was established in Drosophila larvae, based on Venus fragments fused to transcription factors and coexpressed by the heat shock Gal4/UAS system (518). Split-fluorescent constructs, stably expressed under the control of endogenous promoters, were used under physiological conditions in Drosophila embryos to analyze dynamic transcription factor PPIs (284). Protein fusions with Venus, cerulean, and mCherry fluorophore fragments were generated, and PPIs were observed 28 h after embryonic maturation. Finally, Gateway vectors bearing YFP fragments and epitope tags were generated and applied in a study on PPIs between actin nucleation proteins in the wing epithelium and visual system of the host (211).
Interaction methods in vertebrates.
Among vertebrates, the split-FP method has been described for Xenopus laevis (558). A mutated version of Venus was developed to deal with high autofluorescence in Xenopus embryos. The combination of VNm9 and VC155 fragments of Venus gave no fluorescence background in this system, which was also less sensitive to environmental changes such as pH and chloride concentrations. The technique detected an interaction between phosphorylated Smad proteins in vivo and in response to growth factors. Homomers and heteromers of Smad proteins, which are regulatory proteins of cell proliferation and differentiation, were identified at different stages of Xenopus development, and some were translocated to the nucleus after addition of transforming growth factor beta (TGF-β) growth factors, such as activin and nodal-related proteins (236, 558). This Smad2/4 split-FP version provides a direct and quantitative readout for activin-like signaling, with a good signal-to-noise ratio. A similar methodology was followed for zebrafish embryos in a study that revealed the formation of a graded distribution of nodal signaling activity (242).
Mammalian Genetic Protein-Protein Interaction Systems
Although from a strictly technical point of view, two-hybrid and PCA techniques are more demanding to set up in mammalian cells, two main reasons have urged researchers to create such assays.
First, conceptually, PPIs should ideally be studied in their normal physiological context. Many human proteins will not behave properly in nonnative cells, with the main underlying reasons being the different spatiotemporal organization and repertoire of secondary modifications in a mammalian cell compared to those in a unicellular yeast cell. This may not be too problematic for “static” interactions, e.g., PPIs that govern structural elements or molecular machines, but may pose considerable problems in analyzing the dynamics of a protein interaction network. This is particularly true for signaling cascades, from the receptor down to altered PPI complexes at the promoter level, but also applies to various other dynamic processes, including regulated alternative splicing and translation, protein transport mechanisms, vesicle transport, and the plasticity of the actin cytoskeleton and intermediate filaments. Consequently, the effects on a PPI network of altering the cellular milieu, e.g., by external stimuli, can be addressed appropriately only in the native cellular format. Although efforts have been made to introduce some context dependency in yeast cells, e.g., in three-hybrid systems using exogenously added tyrosine kinases, the ever-expanding complexity of posttranslational modifications (N-acetylation, acylation, methylation, glycosylation, various types of ubiquitination, etc.) that control much of the above-mentioned mechanisms strongly argues for the performance of PPI analyses in their proper physiological context. An inherent consequence of this vast heterogeneity is that no single method can be expected to cover the full protein-protein interaction space, especially in mammalian systems.
Second, PPIs are increasingly being recognized as bona fide targets for drug discovery. A growing number of small molecules capable of disrupting designated PPIs are being uncovered, likely through the use of optimized chemical libraries and assay systems (also see examples below). Clearly, potential PPI targets outnumber single-protein targets, such as enzymes, G-protein-coupled receptors (GPCRs), or ion channels, and in fact provide a highly needed alternative, for example, in the case of viral targets where all single-enzyme targets are being exhausted, e.g., for HIV-1. In-cell screening for such PPI modifiers by use of human cell-based assays offers clear advantages. Besides providing the optimal physiological context, compounds are also intrinsically selected for the ability to permeate the membrane of the mammalian phospholipid bilayer. In addition, off-target effects can be detected using three-hybrid systems.
In this review, we divide the mammalian two-hybrid systems into three classes. The first group represents mere adaptations of the yeast two-hybrid assay to the mammalian cellular environment, and the second group comprises various forms of the PCA. The third group encompasses techniques that are based on unique features of mammalian cells, together with strategies that so far have been pioneered only in mammalian cell systems.
Adaptations of the yeast two-hybrid system.
An early example of a conceptual replica of the two-hybrid system was its use to analyze complex formation between leucine zipper transcription factors. The system was based on the Gal4 DBD and VP16 AD, with chloramphenicol acetyltransferase (CAT) as the reporter (113). Later on, similar strategies were used to investigate interactions between SMAD complexes and the transcriptional coactivator CBP (644), as well as protein kinase A (PKA)-dependent changes in the interaction between ERa and SRC1 (744). While such applications relied on transient transfection of bait, prey, and reporter vectors, an optimized version was developed by Isselbacher and colleagues whereby a GFP-based reporter and the bait construct were stably integrated into the cell's genome and prey expression was propagated from a stable, extrachromosomal vector by use of the EBNA-1/ori-P system. In contrast to analytical applications that are limited to selected PPIs, this approach allowed for cDNA library screening using a designated bait (592). More recently, such mammalian two-hybrid assays were optimized to allow medium- and even high-throughput interaction mapping. Pan et al. reported a genomewide SARS coronavirus intraviral PPI map encompassing 40 (often reciprocal) interactions. Notably, several overlapping PPI pairs were found with two previously reported yeast two-hybrid screens, all representing strong signals in the mammalian system. No overlaps were seen between the data sets obtained with yeast as the host (488). The cell array protein-protein interaction assay (CAPPIA) combines a DNA microarray with a two-hybrid readout (177). In brief, bait- and prey-encoding plasmid vectors, together with a reporter directing the expression of an autofluorescent protein, are spotted as transfection mixes on glass slides. Cells are seeded on top and take up the DNAs by reverse transfection. Readout is subsequently performed using a DNA array scanner or by high-throughput microscopy. In the proof-of-concept study, a medium-scale screen using (fragments of) the androgen receptor as bait and a selected set of preys was performed, totaling 160 combinations, demonstrating the cost-effectiveness and efficiency of the procedure. In 2010, extensive mammalian two-hybrid assay-based PPI maps were established using pairwise analyses of 1,988 human and 1,727 mouse transcription factors, revealing a total of 762 and 877 interactions, respectively (535). Interestingly, besides providing novel insights into transcription factor network evolution, evidence that tissue-specific effects were generated was provided by combining broadly expressed with tissue-specific transcription factors, implying tissue-restricted interaction patterns.
Another variation on the two-hybrid theme in mammalian cells is the tetracycline repressor-based system, trM2H (638). In trM2H, hybrids are composed of three functional parts: DNA binding of two tetracycline repressor (TetR) fragments is restored upon bait-prey interaction, leading to activation of transcription by the VP16 AD domain. Bait and prey are thus flanked by the TetR fragments and by the VP16 AD, imposing considerable topological constraints that may in some cases restrict its use. However, the system holds the promise of being highly sensitive, as it can detect the low-affinity (55 μM) sortase A dimerization.
The mammalian two-hybrid system also has applications in drug discovery. Recently, small-compound inhibitors of trimerization of gp41, an HIV-1 protein involved in cell entrance, were discovered by systematic screening for interaction inhibition using 96-well plates and luciferase as a reporter (594). It was suggested that high-throughput compound screening with 384-well plates should be feasible using the mammalian two-hybrid system. In a similar study, more than 3,000 compounds were screened for inhibition of the binding of MDM2, an E3 ubiquitin ligase, to the tumor suppressor p53 by employment of the mammalian two-hybrid method (386). In a Western blot assay, positive hits were found to partially increase the levels of p53 by suppression of MDM2-induced degradation. Commercial mammalian two-hybrid kits are available from Promega (CheckMate system).
Similar to the case for prokaryotes and fungi, FP recruitment systems have been developed for use in mammalian cells. These redistribution assays, based on the colocalization of bait and prey in a particular area of the cell, use fluorescence microscopy as a readout. Baits are typically, but not necessarily, tripartite constructs combining the bait with a tether and an FP that serves as a location control. The prey is also fused to an FP to monitor its position in the cell. These (trans)location biosensors are explored mostly in mammalian cells and were first reported for nuclear targeting (350). In a more sophisticated variant, the bait is linked to the LacI repressor so that bait-prey complexes are targeted to the chromosomal DNA by interacting with a stably integrated array of lac operator sequences (742). Confocal microscopy is then used to visualize DNA-bound prey FPs. The nuclear translocation assay (NTA) uses ligand-induced redistribution of a bait-prey complex whereby the bait is fused to a localization-controllable EGFP construct (136). The translocation cassette is composed of a nuclear export signal (NES), to keep the bait in the cytosol, fused to a nuclear import signal (NLS) and the ligand-binding domain (LBD) of the glucocorticoid receptor. Dexamethasone induces a conformational change leading to exposure of the NLS and subsequent nuclear translocation of the bait. The prey is fused to the dsRed FP, and its translocation to the nucleus is monitored as a parameter for interaction with the bait. Other examples of FP recruitment assays include prey targeting to cell membranes (45, 697), viral intracytoplasmic protein aggregates (446), and P bodies (46).
PCA technologies in mammalian cells.
PCAs have also been adapted very successfully to mammalian cell systems. Reassembly of E. coli β-galactosidase upon bait-prey interaction was pioneered by the Blau group (549) and was initially developed with a colorimetric readout. In a later implementation, this method was used to dissect ligand-dependent interactions between EGFR family members (700). This work provided important new insights into the dynamics of EGFR subunit clustering upon EGF-type ligand addition and into the impact of the anticancer Herceptin antibody thereon. Although PPIs with G-protein-coupled receptors are notoriously difficult to monitor, two adaptations of the β-galactosidase assay were developed based on universal features of the GPCR system: ligand-dependent recruitment of β-arrestin (675) and endocytosis (238). In the latter case, one enzyme moiety was tethered to endosomes, leading to reconstitution of enzymatic activity only after fusion of endocytotic vesicles containing the activated GPCR complex. Besides providing valuable tools for studying GPCR biology, both assays were also adapted to high-throughput compound screening. Intrinsic to this split-β-galactosidase technique is the separate folding of each enzyme fragment and subsequent spontaneous assembly of the whole protein. Although mutations can be introduced to reduce complementation in the absence of a bait-prey interaction, background noise may hinder detection of weak signals.
Such background noise is avoided by using PCAs in which folding depends on fragment proximity that is induced by a bait-prey interaction. Originally explored in yeast cells, such folding-dependent complementation was first reported for murine DHFR in mammalian cells by Michnick and colleagues. In this case, nucleotide-free medium is used for growth selection in DHFR-deficient cells (543). Apart from the survival assay, fluorescein-conjugated methotrexate can be applied as an additional control for interaction. Typical applications of this split-DHFR system in a mammalian context include the demonstration that the erythropoietin receptor exists as a preformed complex that requires ligand-induced conformational changes for activation (546) and the analysis of a signaling network controlling translation initiation (545). Another folding-dependent PPI sensor is the split-lactamase system using fragments of E. coli TEM β-lactamase. Reporter activity can be measured either by in vitro colorimetry in cell lysates or by in vivo fluorescence (194). Taking advantage of the mammalian context, Spotts et al. demonstrated the phosphorylation-dependent CREB-CBP interaction upon elevating cAMP levels by exposure of the cells to forskolin, an activator of adenylate cyclase, or to the cell-permeating cAMP analog CPT-cAMP. Notably, time-lapse microscopic registration of β-lactamase activity could be monitored in single neurons upon cleavage of the CCF-2 fluorophore (608). A more recent application of this assay detected the interaction between the HIV-1-encoded viral infectivity factor (Vif) protein and the human APOBEC 3 cytidine deaminase (72). APOBEC 3G and -F are potent restriction factors that counteract an HIV-1 infection but are themselves targeted by Vif, which marks them for proteasomal degradation. Such straightforward assays to map this interaction can also be used in high-throughput drug screening campaigns to assist in developing novel anti-HIV-1 therapeutics. Protocols for split-DHFR, split-lactamase, and split-FP (see below) applications in mammalian cells can be found elsewhere (544).
Split-luciferase systems have been used frequently in mammalian cell cultures. The reversible character of luciferase reassembly was exploited in the investigation of GPCR-induced deassembly of protein kinase A regulatory and catalytic subunits (615). This study highlighted the use of split-luciferase methods to establish the pharmacological profiles of GPCR-based candidate drugs. Recent examples that demonstrate the flexibility of the split-luciferase approach in mammalian cells include the development of optical probes to monitor fusion of cellular organelles such as mitochondria (277), to detect intraviral PPIs (126), actin polymerization (291), and amyloid-β peptide oligomer formation (244), and to map individual amino acid residues involved in chaperone protein complex formation (309).
Several split-luciferase-based optical sensors have also been used in small animals by use of implanted cells that express the two sensor fragments (344, 416, 501, 542; for a protocol, see reference 672). Imaging is then typically performed using cooled charge-coupled device (CCD) cameras. Examples include ligand-dependent nuclear translocation of the androgen receptor (344) (Fig. 13D) and the intramolecular folding of the estrogen receptor driven by endogenous estradiol levels (499). Such split-luciferase applications are promising tools for studying the pharmacokinetic behavior of pharmaceuticals that target PPIs in experimental animal model systems and were recently suggested to monitor in vivo activation of the EGFR and Her2/neu pathway during (radio)therapy (390, 709). Alternatively, PPIs in animals can be spotted by the split-thymidine kinase (split-TK) system (434). Thymidine kinase from herpes simplex virus 1 phosphorylates nucleoside analogues. Radioactively labeled nucleoside derivatives are retained in the cell upon phosphorylation by TK, which can be observed by positron emission tomography (PET) in living organisms. TK was originally used as a reporter gene in a classic two-hybrid design for living animals (415). However, by a combination of random fragment libraries and rational design, a split-TK method was developed and led to a system which can detect PPIs in deeper tissue (434).
Although the split-FP method was originally reported for E. coli and used GFP fragments (205), Kerppola and coworkers were the first to establish the split-FP method in mammalian cells, based on reconstitution of YFP (270). The excellent spatial resolution in different cellular compartments of mammalian cells by application of the split-FP method is highlighted in numerous reports (reviewed in reference 335; for a protocol, see reference 336). Although the fluorescence intensity of reconstituted FP complexes is estimated to be about 10-fold below that of wild-type GFPs, the low autofluorescence of mammalian cells ensures that signals can still be detected even when the protein pairs are expressed at endogenous levels. The irreversible character of the split-FP system has been exploited in several studies with mammalian cells. As an example, it was suggested for capture of oligomer formation that precedes the protein aggregation that accompanies several neurodegenerative diseases (215).
GFPs exist as a wide range of spectral variants, and accordingly, multicolor split-FP variants were developed to simultaneously capture multiple PPIs in different subcellular locations (271). Multicolor split-FP assays also allow for competition studies between different proteins for a shared partner, which can be especially useful for mapping mutually exclusive interactions with so-called hub proteins (225, 442). Multicolor split-FP assays can also be combined with BRET or FRET readouts, allowing the study of complex formation involving up to four proteins (196, 537). Note that multicolor split-luciferases were also developed recently (258).
In contrast to its widespread use for designated PPIs, high-throughput cDNA library screening applications using the split-FP method are rather limited (544), possibly due to intrinsic topological constraints or to variations of prey expression levels causing strong fluctuations in signal intensity. Very recently, however, an extensive retroviral vector-based human ORFeome screen using a split-Venus configuration identified several novel putative partners of core telomere-associated proteins, holding promise for future large-scale split-FP applications in mammalian cells (376). Direct effects of small molecules on fluorescence intensity are frequently observed and may hamper applications in drug screening. Yet off-target effects of drugs can be monitored using an elaborate panel of FP reporters selected to detect off-target effects on multiple biochemical pathways in human cells (421).
The use of photoswitchable fluorophores may also be an interesting prospect for split-FP applications. In that respect, the group of Miyawaki pioneered the development of a light-induced conversion between the bright and dark states (on-off switch) of a new green fluorescent protein termed Dronpa (10). The unique photochromic property of Dronpa is that it can be excited, erased, and excited again, and this can be used as a tool to monitor the dynamics of molecular processes. Since then, several reversibly switchable fluorescent proteins (RSFPs) have been engineered in the emission spectra of blue-green, green, and red fluorescence (12, 58, 619, 620). In the context of PPI studies, a red RSFP (rsTagRFP) was recently employed to illustrate modulations in both the fluorescence intensity and lifetime of the fluorophore during interaction between a growth factor receptor and a binding protein in live mammalian cells, using photochromic FRET (624). Dronpa was also used successfully in split-FP assays to investigate PPIs in live cells (378). The reversible photoswitching activity was illustrated for full-length and fragmented fluorophores. Photobleaching was induced by irradiation at 488 nm for 1 min, while fluorescence was restored by irradiation at 430 nm for 30 s. These data now open the prospect of the split-RSFP system for PPI studies to overcome the problems of photobleaching and low quantum yield.
Interactions between proteins of the secretory pathway, which make up one-third of the proteome, have typically been difficult to examine. Citrine, a YFP variant with a Q69M mutation, was found to be photostable in various cellular compartments, including the lumen of the secretory pathway (222). A split-FP assay based on this fluorescent protein was able to localize PPIs between the cargo receptor ERGIC-53 and various glycoproteins (475). Recently, interactions among N-glycosyltransferases in the Golgi apparatus were also investigated by this approach (245).
It can be expected that continuously ongoing efforts to prevent spontaneous fragment association, to optimize temperature and pH dependence of chromophore maturation, and to further extend the spectral repertoire will foster even more applications of the split-FP approach (100, 173, 293, 352, 378, 402, 596, 736).
Unique mammalian systems.
The first example of a two-hybrid method whose design is based on intrinsic features of a mammalian cell is the mammalian protein-protein interaction trap (MAPPIT) system (172). MAPPIT relies on the JAK/STAT signaling pathway, which is typically activated via type I cytokine receptors. These receptors lack intrinsic kinase activity but depend on associated cytosolic kinases of the JAK family for signal transduction. Ligands for such receptors include erythropoietin, growth hormone, leptin, and most interleukins and colony-stimulating factors. MAPPIT is a complementation assay whereby the bait is fused to a signaling-deficient receptor that cannot recruit STAT molecules. As the prey is fused to functional STAT recruitment sites, phosphorylation-dependent complementation initiates STAT recruitment and activation, followed by nuclear translocation and a transcriptional response (Fig. 14A). In MAPPIT, the interactor (cytosol) and detector (nucleus) zones are physically separated by taking advantage of the nuclear shuttling of STATs. As a consequence, activation of a reporter gene thus depends on the normal transcriptional machinery, preventing false-positive results at that level. A second characteristic of MAPPIT that reduces the false-positive rate is its ligand dependency, which adds an additional control level: only upon activation of the chimeric bait receptor by cytokine treatment is the system activated. Note that MAPPIT is characterized by a high degree of intrinsic topological flexibility, allowing detection of various PPIs without any structural optimization. This is reminiscent of the yeast two-hybrid system, where the flexibility of DNA allows activation of the RNA polymerase II complex relatively independent of the precise spatial positioning of bait and prey. Likewise, the nonstructured cytokine receptor tails provide flexibility in MAPPIT, allowing prey chimeras to be contacted easily by the JAK kinase. MAPPIT has found wide applications in the study of signal transduction processes and also many other (cytosolic) PPIs (recently reviewed in references 395 and 396a). Because of its favorable sensitivity/specificity ratio, MAPPIT is being used as an orthogonal assay to validate large-scale interactome maps. Examples include the interactomes of S. cerevisiae (721), C. elegans (601), and humans (663). In addition, MAPPIT can be used as a high-throughput assay in both arrayed and cDNA library screening formats (396). MAPPIT is a flexible concept and could be reconfigured to allow for high-throughput drug screening and to study interactions between proteins and small molecules. In reverse MAPPIT, the prey is linked to an inhibitory moiety, e.g., a phosphatase, which inactivates the system upon bait-prey interaction. Disruption of the PPI thus leads to a positive readout that is advantageous in high-throughput campaigns because it discriminates between disruptors and toxic compounds (Fig. 14B) (171). Finally, in the MASPIT three-hybrid format, DHFR is fused to the receptor, allowing display of a chemical dimerizer consisting of methotrexate and a small molecule of interest. Examples include the target recognition of various kinase inhibitors and their use in cDNA screening campaigns (73).
Fig 14.
Unique mammalian genetic PPI methods. (A) Mammalian protein-protein interaction trap (MAPPIT) (172). The bait protein is a fusion with a leptin receptor (LR), which contains three Y-to-F mutations so it is unable to activate STATs spontaneously (one representative phenylalanine [F] is shown). The prey fusion contains a domain of gp130 which can recruit STATs. After interaction of the bait with the prey, Janus kinases (JAKs) phosphorylate gp130, which stimulates binding of gp130 with the STATs. The STATs themselves are phosphorylated by the JAKs, which results in the formation of a STAT complex. The STAT complex binds the rat PAP1 promoter (rPAP1p) and activates luciferase transcription. The leptin receptor is further fused with the extracellular domain of EpoR, a receptor of erythropoietin (Epo), and therefore LR complex formation, which is necessary to make the association with the JAKs, is induced by addition of Epo. (B) Reverse MAPPIT (171). For reverse MAPPIT, a functional LR protein is used with one tyrosine (Y) residue that can be phospharylated upon activation. (Left) The prey Y fusion contains a phosphatase (PaseDom) domain which, upon interaction with bait X, removes the phosphate of JAK2, thereby preventing STAT recruitment. (Right) Inhibition of the bait-prey association by competing proteins (Comp) or compounds (C) reestablishes normal JAK-STAT signaling, which ultimately leads to luciferase reporter gene transcription. (C) Split-TEV method (698). Bait protein X and prey protein Y are fused to the N-terminal and C-terminal domains, respectively, of TEV protease. Association of X with Y initiates the reconstitution of a fully functional TEV protease, which cleaves TEV-specific recognition sequences (rsTEV). A membrane-bound or cytoplasmic protein (MCP), linked by an rsTEV to either luciferase or the artificial transcription factor LexA-VP16, prevents strong bioluminescence or LexA-VP16 nuclear localization, respectively. TEV protease activity releases luciferase, for induction of strong luminescence, or LexA-VP16, for reporter gene activation. The image of TEV protease is based on the PDB structure under accession number 1LVM (512).
A PCA tool that has been developed uniquely for mammalian cells is the split-tobacco etch virus protease (split-TEV protease) system (698) (Fig. 14C). In this system, fragments of the NIa protease of TEV regain activity upon bait-prey interaction. Reporter activity can be either transcription coupled, whereby proteolytic cleavage allows translocation of a transcription factor from the cytosol to the nucleus, or proteolysis only, due to the activation of a luciferase enzyme upon proteolytic release from an inactive complex. In these configurations, a bait-prey interaction does not directly activate the reporter but functions via a reconstituted protease. This approach leads to a stable reporter activity which may affect the capture of dynamic interactions. Yet constitutive GABA-B1aR and GABA-B2R, as well as ligand-dependent ErbB2-ErbB4 receptor heterodimerization, could be monitored. Tango is also a TEV-based system and finds its conceptual origins in the Notch signaling pathway (29). The receptor is fused to a transcription factor via a linker containing a TEV protease cleavage site. Receptor activation leads to the recruitment of a signaling protein fused to TEV protease, thus liberating the transcription factor. Note that the system uses the prokaryotic tTA-driven tetracycline-responsive promoter and thus avoids interference from endogenous signaling pathways. Since, in addition, transient receptor activation is turned into a constitutive signal, the system is highly specific and sensitive. Proof-of-concept experiments demonstrated applications for GPCRs, receptor tyrosine kinases, and steroid hormone receptors. This method is particularly suited to the study of early steps in receptor activation. Applications can include the identification of ligands of orphan receptors and high-throughput screening for signaling modifiers.
Finally, a new method based on trans-SUMOylation enables the observation of interactions by covalent attachment of a SUMO protein followed by shift detection in Western blot analysis (610). The bait chimera contains Ubc9, which adds the small ubiquitin-related modifier SUMO to the prey protein of interest. A number of PPIs were confirmed by application of this method, which forms an in vivo alternative to traditional coimmunoprecipitation.
Dual-Organism Two-Hybrid Systems
To allow direct comparison between interaction data sets obtained from two organisms, dual-organism two-hybrid methods have been developed, relying on the use of a single bait design or compatible vectors. A combined yeast-bacterium two-hybrid system was engineered (578) based on the λ repressor DBD fused to a bait gene and placed under the control of the lpp/lacUV5 and TEF1 promoters in E. coli and S. cerevisiae, respectively. For each organism, a different prey plasmid was used, bearing the B42 AD in yeast and the α subunit of E. coli RNA polymerase (RNAPα) as the AD in E. coli. Reporter genes for S. cerevisiae were gusA and LYS2, and those for E. coli were HIS3 and lacZ, all of which were located in front of λ repressor binding sites. This method allows the sensitivities and specificities of the systems to be compared between the two organisms. In quantifiable assays, S. cerevisiae displayed a larger dynamic range for detecting interactions than the prokaryotic model. However, growth on selective medium was clearly faster for E. coli cells, and moreover, autoactivation did not occur in the bacterial cells. In screens of a human cDNA library against human Ras as bait, different hits were found in E. coli and S. cerevisiae. This result suggests that either the screening was not exhaustive or the specific environment of the screening influenced the outcome. Consequently, it became apparent that screening libraries in different organisms may lead to a broader spectrum of identifiable PPIs.
In an alternative approach, site-specific recombination and reading frame-independent mammalian two-hybrid (M2H) vectors were generated to be fully compatible with the site-specific yeast two-hybrid system (426, 427). This method was developed to address the shortcoming of time-consuming recloning of yeast two-hybrid candidates into mammalian two-hybrid vectors required for retesting of interacting candidates in the endogenous host environment. Vectors expressing bait fusions with the GAL4 DBD and prey fusions with VP16 were made Gateway compatible, fully functional in the mammalian system, and directly compatible with existing yeast two-hybrid vectors. These new vectors did not influence the capacity of the selection method in the mammalian background and did not create autoactivators. This system hence provides a fast way to check interactions in yeast and mammalian systems.
CONCLUSIONS
A bird's-eye view on PPI networks may suggest that nature has evolved a highly inefficient and promiscuous communication system between proteins to perform essential cellular functions. However, this large number of protein-protein associations, directly linked with organism complexity (241), is crucial for cellular robustness and network evolvability (8). The concept of keeping it as simple as possible, seen in man-made systems, does not work for interactomes. In that respect, evolution plays a crucial role because it essentially lacks a sense of overview but requires flexibility in response to genetic and environmental perturbations. Examples of this flexible behavior are seen in the subunits of protein complexes, which can serve as basic building blocks for other present or future complexes, and the presence of parallel and interconnected signaling pathways with multiple PPIs to provide error-tolerant and balanced regulation. The complexity in interaction studies also comes from the variety in the character of PPIs. Distinctions in the lifetime, strength, and obligatory nature of protein associations need to be considered. PPIs differ in being permanent or transient, obligate or nonobligate, and direct or indirect and in having high or low affinity (473). These features correlate with the interaction surfaces, which are generally larger and hydrophobic in stable core complexes and smaller and chemically more versatile for transient PPIs (338). Prediction of these interfaces is not straightforward because proteins often undergo conformational changes during association. Allosteric regulation by covalent modification, third-partner binding, or environmental changes further influences the shape of the interaction domain (473).
The large number and complex nature of PPIs necessitate the development of various technologies. Fortunately, many tools are now available for discovery and characterization of PPIs. Key contributions arose from immense technical improvements in molecular biology tools and joint efforts among research fields as diverse as genetics, live imaging, chemical biology, biophysics, and computational biology. As for genetic PPI methods, microbial two-hybrid systems have already produced a significant number of PPIs based on unbiased large-scale screens and play an important role in functional genomics (e.g., see references 690 and 721). Moreover, recent studies (616) give evidence that these methods are still far from saturation. Reiteration of genomewide two-hybrid studies with different setups could substantially increase the output and further aid in differentiating true from false-positive results. With the increasing availability of ORFeome libraries based on Gateway vectors, large-scale two-hybrid experiments for nonmodel organisms are becoming realistic. Such assays offer additional information on the structure of interaction networks and the evolvability of interactomes. Remarkable developments in two-hybrid assay-based techniques in higher eukaryotes, especially mammalian cells (177, 395), are on the verge of ushering in a breakthrough in high-throughput application and serve as very attractive alternatives to the traditional yeast system. With these techniques together with selection-based PCA technologies, fast progression in interactome mapping may be expected for organisms from viruses to animals to plants. Furthermore, technical improvements in the fields of affinity purification (202), protein microarrays (81), and cross-linking (418) will reveal complementary PPI data essential for full characterization of networks.
Such a blueprint of a static PPI network forms the basis for exploring the dynamics of PPIs. PCAs with high temporal or spatial resolution facilitate studies on the influence of genetic or environmental changes. The easy technology transfer of such methods has made them widespread tools for very diverse applications. With some creativity in experimental setup, profound results can be obtained by these straightforward assays. As an example, the yeast cyclin-dependent kinase Cdc28 phosphorylates and concomitantly inactivates Swi4, a component of the cell cycle-regulating SBF complex. Hence, a positive split-FP assay with these two proteins implicates inactivation of the SBF complex. This readout was employed in a study with 25 deletion strains to uncover a genetic interaction between SBF inactivation and the kinase Elm1 (430). Instead of deletion strains, compound screenings to identify small molecules interfering with an interaction pathway could be performed as well (444). By application of the split-ubiquitin system in a medium-scale screen, an interaction network around the yeast phosphatase Ptc1 and its binding partner Nbp2 was created (269). However, it is by integration of deletion strains and truncated versions of the proteins under study in these split-ubiquitin experiments that extensive conclusions could be drawn on the dynamic control of regulatory circuits by Ptc1 and Nbp2. Not surprisingly, both studies made use of semibiased module-scale screens, which form a nice balance between time-consuming large-scale analyses and prejudiced one-to-one experiments.
The importance of studying PPIs also comes from the increasing awareness that they form valid drug targets. The general view that interaction surfaces are large and unstructured, making them difficult targets for small molecules, is not always true. Many surfaces are covered with pockets and clefts, and smaller hot spots and allosteric sites act as ideal binding regions for drugs (741). Interaction domains can be very specific, as seen, for example, with the docking sites of MAP kinase-binding proteins (26). To date, many small molecules or peptides are validated PPI inhibitors, and some of them have reached the clinical phase (741). Apart from providing the first step in PPI drug discovery, i.e., the identification of a suitable target PPI (e.g., see reference 98), two-hybrid systems have been applied for the discovery of PPI inhibitors, as discussed in this review. The use of these methods for detection of PPIs in pathogenic bacteria, protists, or fungi has been limited, but we are hopeful that in the future, pathogen-specific PPI modulation may become a new therapeutic approach to combat infectious diseases.
With all the tools available, one may ask which would be most suitable for a specific project. The answer lies in the identities of the species and proteins under investigation, the goal of the experiment, and the available equipment. The feasibility of a particular method may be evaluated by the published output. However, each system needs some momentum to become widely used. Even for the yeast two-hybrid system, it took several years before it became a common lab technique. Therefore, it is advisable to try out different methods and evaluate each of them. This not only increases the success rate but also helps to benchmark the existing technologies, as partially done already for two-hybrid systems (91) and a diverse set of in vitro and in vivo interaction methods (60). Careful optimization of available techniques could further increase their usefulness, such as the case for the three-hybrid system for protein–small-molecule interactions (94). With the basic setups already available, improvement of publicly available methods could launch their widespread use, which is especially true for alternative two-hybrid systems. In conclusion, the impact of PPI identification and characterization in cell biology is vast and promises exciting and advanced findings essential to our understanding of biological processes, disease development, host-pathogen interactions, and drug discovery.
ACKNOWLEDGMENTS
We thank Leslie B. Vosshall, Yoshio Umezawa, Jin-Rong Xu, and Stan Gelvin for providing us with pictures of their original work.
B.S. was supported by grants from the Fund for Scientific Research Flanders (G.0242.04) and the Institute for Promotion and Innovation by Science and Technology in Flanders (IWT; SBO project 060839). H.T. was supported by a CREA grant from the Research Council of KU Leuven.
Biographies

Bram Stynen obtained a master's degree in bioengineering at the Katholieke Universiteit of Leuven (KU Leuven) in 2005, with a focus on cell and gene biotechnology. In 2011, he received a Ph.D. in biology at the same university, studying the development of a Candida albicans two-hybrid system, with Patrick Van Dijck as his supervisor. Currently, he is finishing a project on the context-dependent activation of C. albicans transcriptional regulators.

Hélène Tournu received her Ph.D. in molecular microbiology from the University of Aberdeen, Scotland, where she studied the transcriptional regulation of filamentous growth in the model yeast Saccharomyces cerevisiae. She is currently a project leader in the laboratory of Patrick Van Dijck. Her research interests include the study of signaling pathways important to morphogenesis and biofilm formation of the human fungal pathogen Candida albicans.

Jan Tavernier obtained his Ph.D. degree in 1984, by studying the cloning of interferon and interleukin genes. After an extended stay at Biogen and later at Roche, he returned to academia in 1996 at the VIB Department of Medical Protein Research, Ghent University. He founded the Cytokine Receptor Laboratory, which currently consists of over 30 researchers. Based on insights into cytokine receptor activation, he developed the mammalian MAPPIT two-hybrid technology. Detailed information can be found at www.mappit.be. Dr. Tavernier has published more than 200 refereed manuscripts, 20 of which have been cited over 100 times. He also holds 19 patent applications. His main areas of expertise are cytokine receptor activation and signal transduction, also linking to pathways involved in innate immunity, and the analysis of protein-protein interactions, including interactome mapping, pathway walking, and molecular description of interdomain interactions.

Patrick Van Dijck obtained his Ph.D. in 1991, by studying mechanisms of transcriptional activation of androgen- and estrogen-regulated genes. His first postdoctoral work was performed in the Laboratory of Molecular Cell Biology at KU Leuven, with a focus on trehalose metabolism and yeast stress resistance mechanisms. After a second postdoctoral position at Janssen Pharmaceutica (J&J) between 1995 and 1997, he returned to KU Leuven to become a group leader on a VIB-sponsored project. Since 2002, he has been group leader of the VIB Department of Molecular Microbiology, and since 2003, he has been a professor at KU Leuven. There are two research topics in his group. He is investigating the role of plant trehalose metabolism, but the main interest of the group is nutrient-induced signal transduction pathways that affect morphogenesis and virulence in the human fungal pathogen Candida albicans. He has published 100 refereed manuscripts and holds 15 patent applications. He currently leads a group of 20 researchers.
REFERENCES
- 1. Abel S, et al. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abida WM, Carter BT, Althoff EA, Lin H, Cornish VW. 2002. Receptor-dependence of the transcription read-out in a small-molecule three-hybrid system. Chembiochem 3:887–895 [DOI] [PubMed] [Google Scholar]
- 3. Aguilera C, et al. 2011. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature 469:231–235 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed KS, Saloma NH, Kadah YM. 2011. Improving the prediction of yeast protein function using weighted protein-protein interactions. Theor. Biol. Med. Model. 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aho S, Arfman A, Pummi T, Uitto J. 1997. A novel reporter gene MEL1 for the yeast two-hybrid system. Anal. Biochem. 253:270–272 [DOI] [PubMed] [Google Scholar]
- 6. Akman G, MacNeil SA. 2009. MCM-GINS and MCM-MCM interactions in vivo visualised by bimolecular fluorescence complementation in fission yeast. BMC Cell Biol. 10:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albers M, et al. 2005. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol. Cell. Proteomics 4:205–213 [DOI] [PubMed] [Google Scholar]
- 8. Albert R, Jeong H, Barabasi AL. 2000. Error and attack tolerance of complex networks. Nature 406:378–382 [DOI] [PubMed] [Google Scholar]
- 9. Althoff EA, Cornish VW. 2002. A bacterial small-molecule three-hybrid system. Angew. Chem. Int. Ed. Engl. 41:2327–2330 [DOI] [PubMed] [Google Scholar]
- 10. Ando R, Mizuno H, Miyawaki A. 2004. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306:1370–1373 [DOI] [PubMed] [Google Scholar]
- 11. Andreopoulos B, Winter C, Labudde D, Schroeder M. 2009. Triangle network motifs predict complexes by complementing high-error interactomes with structural information. BMC Bioinformatics 10:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andresen M, et al. 2008. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat. Biotechnol. 26:1035–1040 [DOI] [PubMed] [Google Scholar]
- 13. Argimon S, Fanning S, Blankenship JR, Mitchell AP. 2011. Interaction between the Candida albicans high-osmolarity glycerol (HOG) pathway and the response to human β-defensins. Eukaryot. Cell 10:272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arifuzzaman M, et al. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16:686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aronheim A. 1997. Improved efficiency SOS recruitment system: expression of the mammalian GAP reduces isolation of Ras GTPase false positives. Nucleic Acids Res. 25:3373–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Atmakuri K, Ding Z, Christie PJ. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 tranfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Attri AK, Minton AP. 2005. Composition gradient static light scattering: a new technique for rapid detection and quantitative characterization of reversible macromolecular hetero-associations in solution. Anal. Biochem. 346:132–138 [DOI] [PubMed] [Google Scholar]
- 19. Bailer SM, Haas J. 2009. Connecting viral with cellular interactomes. Curr. Opin. Biotechnol. 12:453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bak G, et al. 2010. On-off controllable RNA hybrid expression vector for yeast three-hybrid system. BMB Rep. 43:110–114 [DOI] [PubMed] [Google Scholar]
- 21. Baker K, et al. 2002. Chemical complementation: a reaction-independent genetic assay for enzyme catalysis. Proc. Natl. Acad. Sci. U. S. A. 99:16537–16542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker K, Sengupta DJ, Salazar-Jimenez G, Gornish VW. 2003. An optimized dexamethasone-methotrexate yeast 3-hybrid system for high-throughput screening of small molecule-protein interactions. Anal. Biochem. 315:134–137 [DOI] [PubMed] [Google Scholar]
- 23. Bandyopadhyay S, Sharan R, Ideker T. 2006. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 16:428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barabási AL. 2009. Scale-free networks: a decade and beyond. Science 325:412–413 [DOI] [PubMed] [Google Scholar]
- 25. Bardiya N, et al. 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178:593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bardwell AJ, Frankson E, Bardwell L. 2009. Selectivity of docking sites in MAPK kinases. J. Biol. Chem. 284:13165–13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barnard E, McFerran NV, Trudgett A, Nelson J, Timson DJ. 2008. Detection and localisation of protein-protein interactions in Saccharomyces cerevisiae using a split-GFP method. Fungal Genet. Biol. 45:597–604 [DOI] [PubMed] [Google Scholar]
- 28. Barnard E, McFerran NV, Trudgett A, Nelson J, Timson DJ. 2008. Development and implementation of split-GFP-based bimolecular fluorescence complementation (BiFC) assays in yeast. Biochem. Soc. Trans. 36:479–482 [DOI] [PubMed] [Google Scholar]
- 29. Barnea G, et al. 2008. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. U. S. A. 105:64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bartel PL, Roecklein JA, SenGupta D, Fields S. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72–77 [DOI] [PubMed] [Google Scholar]
- 31. Barz T, Ackermann K, Pyerin W. 2002. A positive control for the green fluorescent protein-based one-hybrid system. Anal. Biochem. 304:117–121 [DOI] [PubMed] [Google Scholar]
- 32. Baskakov IV, et al. 1999. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J. Biol. Chem. 274:10693–10695 [DOI] [PubMed] [Google Scholar]
- 33. Battesti A, Bouveret E. 2010. Improvement of bacterial two-hybrid vectors for detection of fusion proteins and transfer to pBAD-tandem affinity purification, calmodulin binding peptide, or 6-histidine tag vectors. Proteomics 8:4768–4771 [DOI] [PubMed] [Google Scholar]
- 34. Bauer P, et al. 2004. Regulation and a conserved intron sequence of liguleless3/4 knox class-I homeobox genes in grasses. Planta 219:359–368 [DOI] [PubMed] [Google Scholar]
- 35. Becker F, et al. 2004. A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chem. Biol. 11:211–223 [DOI] [PubMed] [Google Scholar]
- 36. Beckmann S, Buro C, Dissous C, Hirzmann J, Grevelding CG. 2010. The Syk kinase SmTK4 of Schistosoma mansoni is involved in the regulation of spermatogenesis and oogenesis. PLoS Pathog. 6:e1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bendixen C, Gangloff S, Rothstein R. 1994. A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 22:1778–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benton R, Sachse S, Michnick SW, Vosshall LB. 2006. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berger BW, et al. 2010. Consensus motif for integrin transmembrane helix association. Proc. Natl. Acad. Sci. U. S. A. 107:703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bernstein DS, Buter N, Stumpf C, Wickens M. 2002. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26:123–141 [DOI] [PubMed] [Google Scholar]
- 41. Bex C, Knauth K, Dambacher S, Buchberger A. 2007. A yeast two-hybrid system reconstituting substrate recognition of the von Hippel-Lindau tumor suppressor protein. Nucleic Acids Res. 35:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhat RA, Lahaye T, Panstruga R. 2006. The visible touch: in planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bichet A, Hannemann F, Rekowski M, Bernhardt R. 2007. A new application of the yeast two-hybrid system in protein engineering. Protein Eng. Des. Sel. 20:117–123 [DOI] [PubMed] [Google Scholar]
- 44. Björklund O, Light S, Hedin L, Elofsson A. 2008. Quantitative assessment of the structural bias in protein-protein interaction assays. Proteomics 8:4657–4667 [DOI] [PubMed] [Google Scholar]
- 45. Blanchard D, Hutter H, Fleenor J, Fire A. 2006. A differential cytolocalization assay for analysis of macromolecular assemblies in the eukaryotic cytoplasm. Mol. Cell. Proteomics 5:2175–2184 [DOI] [PubMed] [Google Scholar]
- 46. Bloch DB, Nobre RA, Bernstein GA, Yang WH. 2011. Identification and characterization of protein interactions in the mammalian mRNA processing body using a novel two-hybrid assay. Exp. Cell Res. 317:2183–2199 [DOI] [PubMed] [Google Scholar]
- 47. Blondel M, et al. 2005. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 24:1440–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blumenstein A, et al. 2005. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15:1833–1838 [DOI] [PubMed] [Google Scholar]
- 49. Boder ET, Wittrup KD. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553–557 [DOI] [PubMed] [Google Scholar]
- 50. Bongards C, Chew BS, Lehming N. 2003. The TATA-binding protein is not an essential target of the transcriptional activators Gal4p and Gcn4p in Saccharomyces cerevisiae. Biochem. J. 370:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borloo J, De Smet L, Vergauwen B, Van Beeumen JJ, Devreese B. 2007. A beta-galactosidase based bacterial two-hybrid system to assess protein-protein interactions in the correct cellular environment. J. Proteome Res. 6:2587–2595 [DOI] [PubMed] [Google Scholar]
- 52. Boruc J, InzÉ D, Russinova E. 2010. A high-throughput bimolecular fluorescence complementation protein-protein interaction screen identifies functional Arabidopsis CDKA/B-CYCD4/5 complexes. Plant Signal. Behav. 5:1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boruc J, et al. 2010. Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22:1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bouffard P, Barbar E, Brière F, Boire G. 2000. Interaction cloning and characterization of RoBPI, a novel protein binding to human Ro ribonucleoproteins. RNA 6:66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boulton SJ, et al. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295:127–131 [DOI] [PubMed] [Google Scholar]
- 56. Boysen JH, Fanning S, Newberg J, Murphy RF, Mitchell AP. 2009. Detection of protein-protein interactions through vesicle targeting. Genetics 182:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bracha-Drori K, et al. 2004. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40:419–427 [DOI] [PubMed] [Google Scholar]
- 58. Brakemann T, et al. 2011. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat. Biotechnol. 29:942–947 [DOI] [PubMed] [Google Scholar]
- 59. Braun P, et al. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333:601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Braun P, et al. 2009. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods 6:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Breitkreutz A, et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328:1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brent R, Finley RL., Jr 1997. Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet. 31:663–704 [DOI] [PubMed] [Google Scholar]
- 63. Brent R, Ptashne M. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43:729–736 [DOI] [PubMed] [Google Scholar]
- 64. Bröder YC, Katz S, Aronheim A. 1998. The ras recruitment system, a novel approach to the study of protein-protein interactions. Curr. Biol. 8:1121–1124 [DOI] [PubMed] [Google Scholar]
- 65. Broek D, et al. 1987. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 48:789–799 [DOI] [PubMed] [Google Scholar]
- 66. Brown KR, Jurisica I. 2005. Online predicted human interaction database. Bioinformatics 21:2076–2082 [DOI] [PubMed] [Google Scholar]
- 67. Brymora A, Valova VA, Robinson PJ. 2004. Protein-protein interactions identified by pull-down experiments and mass spectrometry. Curr. Protoc. Cell Biol. Chapter 17:Unit 17.5 [DOI] [PubMed] [Google Scholar]
- 68. Bubulya A, Wise SC, Shen XQ, Burmeister LA, Shemshedini L. 1996. c-Jun can mediate androgen receptor-induced transactivation. J. Biol. Chem. 271:24583–24589 [DOI] [PubMed] [Google Scholar]
- 69. Butland G, et al. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537 [DOI] [PubMed] [Google Scholar]
- 70. Cabello J, et al. 2010. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 8:e1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Caberoy NB, Zhou Y, Jiang X, Alvarado G, Li W. 2010. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J. Mol. Recognit. 23:74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cadima-Couto I, Saraiva N, Santos AC, Goncalves J. 2011. HIV-1 Vif interaction with APOBEC3 deaminases and its characterization by a new sensitive assay. J. Neuroimmune Pharmacol. 6:296–307 [DOI] [PubMed] [Google Scholar]
- 73. Caligiuri M, et al. 2006. MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem. Biol. 13:711–722 [DOI] [PubMed] [Google Scholar]
- 74. Campbell RN, Westhorpe F, Reece RJ. 2011. Isolation of compensatory inhibitor domain mutants to novel activation domain variants using the split-ubiquitin screen. Yeast 28:569–578 [DOI] [PubMed] [Google Scholar]
- 75. Cao Y, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. 2009. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy 5:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Caro E, Castellano MM, Gutierrez C. 2007. A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447:213–217 [DOI] [PubMed] [Google Scholar]
- 77. Carter BT, et al. 2005. Investigation of the mechanism of resistance to third-generation cephalosporins by class C beta-lactamases by using chemical complementation. Chembiochem 6:2055–2067 [DOI] [PubMed] [Google Scholar]
- 78. Castaño AR, et al. 1995. Peptide binding and presentation by mouse CD1. Science 269:223–226 [DOI] [PubMed] [Google Scholar]
- 79. Castrillo G, et al. 2011. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS One 6:e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chanda D, et al. 2009. Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes. J. Biol. Chem. 284:28510–28521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chandra H, Srivastava S. 2010. Cell-free synthesis-based protein microarrays and their applications. Proteomics 10:717–730 [DOI] [PubMed] [Google Scholar]
- 82. Charloteaux B, et al. 2011. Protein-protein interactions and networks: forward and reverse edgetics. Methods Mol. Biol. 759:197–213 [DOI] [PubMed] [Google Scholar]
- 83. Chen B, et al. 2010. A novel auxiliary subunit critical to BK channel function in Caenorhabditis elegans. J. Neurosci. 30:16651–16661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen B, Liu Q, Ge Q, Xie J, Wang ZW. 2007. UNC-1 regulates gap junctions important to locomotion in C. elegans. Curr. Biol. 17:1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen C, et al. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 107:21755–21760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen H, et al. 2008. Firefly luciferase complementation imaging assays for protein-protein interactions in plants. Plant Physiol. 146:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen J, Carter MB, Edwards BS, Cai H, Sklar LA. 2012. High throughput flow cytometry based yeast two-hybrid array approach or large-scale analysis of protein-protein interactions. Cytometry A 81:90–98 doi:10.1002/cyto.a.21144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen J, et al. 2008. A yEGFP-based reporter system for high-throughput yeast two-hybrid assay by flow cytometry. Cytometry A 73:312–320 [DOI] [PubMed] [Google Scholar]
- 89. Chen J, Zhou J, Sanders CK, Nolan JP, Cai H. 2009. A surface display yeast two-hybrid screening system for high-throughput protein interactome mapping. Anal. Biochem. 390:29–37 [DOI] [PubMed] [Google Scholar]
- 90. Chen SC, Olsthoorn RC. 2010. In vitro and in vivo studies of the RNA conformational switch in alfalfa mosaic virus. J. Virol. 84:1423–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen YC, Rajagopala SV, Stellberger T, Uetz P. 2010. Exhaustive benchmarking of the yeast two-hybrid system. Nat. Methods 7:667–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chew BS, Lehming N. 2007. TFIIB/SUA7(E202G) is an allele-specific suppressor of TBP1 (E186D). Biochem. J. 406:265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chiang T, Scholtens D, Sarkar D, Gentleman R, Huber W. 2007. Coverage and error models of protein-protein interaction data by directed graph analysis. Genome Biol. 8:R186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chidley C, Haruki H, Pedersen MG, Muller E, Johnsson K. 2011. A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis. Nat. Chem. Biol. 7:375–383 [DOI] [PubMed] [Google Scholar]
- 95. Chien CT, Bartel PL, Sternglanz R, Fields S. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. U. S. A. 88:9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. 2005. Directed evolution of specific receptor-ligand pairs for use in the creation of gene switches. Proc. Natl. Acad. Sci. U. S. A. 102:5691–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Choi JH, et al. 2003. Using an Aplysia two-hybrid system to examine the interactions between transcription factors involved in long-term facilitation in the nervous system of Aplysia. Learn. Mem. 10:40–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Christ F, et al. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192–1202 [DOI] [PubMed] [Google Scholar]
- 99. Christensen RG, et al. 2011. A modified bacterial one-hybrid system yields improved quantitative models of transcription factor specificity. Nucleic Acids Res. 39:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chu J, et al. 2009. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens. Bioelectron. 25:234–239 [DOI] [PubMed] [Google Scholar]
- 101. Citovsky V, Gafni Y, Tzfira T. 2008. Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods 45:196–206 [DOI] [PubMed] [Google Scholar]
- 102. Claessens A, Weyn C, Merregaert J. 2008. The cytoplasmic domain of chondrolectin interacts with the beta-subunit of Rab geranylgeranyl transferase. Cell. Mol. Biol. Lett. 13:250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Clarke P, Cuiv PO, O'Connell M. 2005. Novel mobilizable prokaryotic two-hybrid system vectors for high-throughput protein mapping in Escherichia coli by bacterial conjugation. Nucleic Acids Res. 33:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cole KC, McLaughlin HW, Johnson DI. 2007. Use of bimolecular fluorescence complementation to study in vivo interactions between Cdc42p and Rdi1p of Saccharomyces cerevisiae. Eukaryot. Cell 6:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Colland F, et al. 2004. Functional proteomics mapping of a human signaling pathway. Genome Res. 14:1324–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Corbin BD, Geissler B, Sadasivam M, Margolin W. 2004. Z-ring-independent interaction between a subdomain of FtsZ and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cormack RS, Hahlbrock K, Somssich IE. 1998. Isolation of putative plant transcriptional coactivators using a modified two-hybrid system incorporating a GFP reporter gene. Plant J. 14:685–692 [DOI] [PubMed] [Google Scholar]
- 108. Costanzo M, Baryshnikova A, Nislow C, Andrews B, Boone C. 2009. You too can play with an edge. Nat. Methods 6:797–798 [DOI] [PubMed] [Google Scholar]
- 109. Cox JS, Shamu CE, Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206 [DOI] [PubMed] [Google Scholar]
- 110. Crooks RO, Rao T, Mason JM. 2011. Truncation, randomization, and selection: generation of a reduced length c-Jun antagonist that retains high interaction stability. J. Biol. Chem. 286:29470–29479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cross GH, et al. 2003. A new quantitative optical biosensor for protein characterisation. Biosens. Bioelectron. 19:383–390 [DOI] [PubMed] [Google Scholar]
- 112. Cusick ME, Klitgord N, Vidal M, Hill DE. 2005. Interactome: gateway into systems biology. Hum. Mol. Genet. 14(Spec No 2):R171–R181 [DOI] [PubMed] [Google Scholar]
- 113. Dang CV, et al. 1991. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol. Cell. Biol. 11:954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Das S, Cano J, Kalpana GV. 2009. Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance. J. Biol. Chem. 284:19903–19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Datta S, Bucks ME, Koley D, Lim PX, Savinov SN. 2010. Functional profiling of p53-binding sites in Hdm2 and Hdmx using a genetic selection system. Bioorg. Med. Chem. 18:6099–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Davy A, et al. 2001. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Day RN, Davidson MW. 2009. The fluorescent protein palette: tools for cellular imaging. Chem. Soc. Rev. 38:2887–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Day RN, Schaufele F. 2005. Imaging molecular interactions in living cells. Mol. Endocrinol. 19:1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Deane CM, Salwinski L, Xenarios I, Eisenberg D. 2002. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics 1:349–356 [DOI] [PubMed] [Google Scholar]
- 120. de Chassey B, et al. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Deeds EJ, Ashenberg O, Shakhnovich EI. 2006. A simple physical model or scaling in protein-protein interaction networks. Proc. Natl. Acad. Sci. U. S. A. 103:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. de Felipe KS, Carter BT, Althoff EA, Cornish VW. 2004. Correlation between ligand-receptor affinity and the transcription readout in a yeast three-hybrid system. Biochemistry 43:10353–10363 [DOI] [PubMed] [Google Scholar]
- 123. Defeu Soufo HJ, Graumann PL. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol. Microbiol. 62:1340–1356 [DOI] [PubMed] [Google Scholar]
- 124. de Folter S, Immink RG. 2011. Yeast protein-protein interaction assays and screens. Methods Mol. Biol. 754:145–165 [DOI] [PubMed] [Google Scholar]
- 125. Del Bene F, Tessmar-Raible K, Wittbrodt J. 2004. Direct interaction of geminin and Six3 in eye development. Nature 427:745–749 [DOI] [PubMed] [Google Scholar]
- 126. Deng Q, et al. 2011. Application of a split luciferase complementation assay for the detection of viral protein-protein interactions. J. Virol. Methods 176:108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Deplancke B, Dupuy D, Vidal M, Walhout AJ. 2004. A gateway-compatible yeast one-hybrid system. Genome Res. 14:2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Deplancke B, et al. 2006. A gene-centered C. elegans protein-DNA interaction network. Cell 125:1193–1205 [DOI] [PubMed] [Google Scholar]
- 129. Deplazes A, Mockli N, Luke B, Auerbach D, Peter M. 2009. Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J. 28:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dhayalan A, et al. 2008. Mapping of protein-protein interaction sites by the ‘absence of interference’ approach. J. Mol. Biol. 376:1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Di Lallo G, Castagnoli L, Ghelardini P, Paolozzi L. 2001. A two-hybrid system based on chimeric operator recognition for studying protein homo/heterodimerization in Escherichia coli. Microbiology 147:1651–1656 [DOI] [PubMed] [Google Scholar]
- 132. Ding Z, et al. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Dirnberger D, Messerschmid M, Baumeister R. 2008. An optimized split-ubiquitin cDNA-library screening system to identify novel interactors of the human Frizzled 1 receptor. Nucleic Acids Res. 36:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Dirnberger D, Unsin G, Schlenker S, Reichel C. 2006. A small-molecule-protein interaction system with split-ubiquitin as sensor. Chembiochem 7:936–942 [DOI] [PubMed] [Google Scholar]
- 135. Dixit R, Cyr R, Gilroy S. 2006. Using intrinsically fluorescent proteins for plant cell imaging. Plant J. 45:599–615 [DOI] [PubMed] [Google Scholar]
- 136. Dixon AS, Lim CS. 2010. The nuclear translocation assay for intracellular protein-protein interactions and its application to the Bcr coiled-coil domain. Biotechniques 49:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Dmitrova M, et al. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205–212 [DOI] [PubMed] [Google Scholar]
- 138. Dohmen RJ, Madura K, Bartel B, Varshavsky A. 1991. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. U. S. A. 88:7351–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Dong S, et al. 2011. Specific and modular binding code for cytosine recognition in pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 286:26732–26742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Donovan C, Schwaiger A, Krämer R, Bramkamp M. 2010. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 192:3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Dove SL, Hochschild A. 2004. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 261:231–246 [DOI] [PubMed] [Google Scholar]
- 142. Dove SL, Joung JK, Hochschild A. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627–630 [DOI] [PubMed] [Google Scholar]
- 143. Dozhanskaya N, Sung YJ, Conti J, Currie JR, Denman RB. 2003. The fragile X mental retardation protein interacts with U-rich RNAs in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 305:434–441 [DOI] [PubMed] [Google Scholar]
- 144. Dreze M, et al. 2009. ‘Edgetic’ perturbation of a C. elegans BCL2 ortholog. Nat. Methods 6:843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Du W, Vidal M, Xie JE, Dyson N. 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10:1206–1218 [DOI] [PubMed] [Google Scholar]
- 146. Dube DH, et al. 2010. A two-hybrid assay to study protein interactions within the secretory pathway. PLoS One 5:e15648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Dudak A, Kim J, Cheong B, Federoff HJ, Lim ST. 2011. Membrane palmitoylated proteins regulate trafficking and processing of nectins. Eur. J. Cell Biol. 90:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Dues G, Müller S, Johnsson N. 2001. Detection of a conformational change in G gamma upon binding G beta in living cells. FEBS Lett. 505:75–80 [DOI] [PubMed] [Google Scholar]
- 149. Dünkler A, Müller J, Johnsson N. 2012. Detecting protein-protein interactions with the split-ubiquitin sensor. Methods Mol. Biol. 786:115–130 [DOI] [PubMed] [Google Scholar]
- 150. Dünnwald M, Varshavsky A, Johnsson N. 1999. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell 10:329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Durai S, Bosley A, Abulencia AB, Chandrasegaran S, Ostermeier M. 2006. A bacterial one-hybrid selection system for interrogating zinc finger-DNA interactions. Comb. Chem. High Throughput Screen. 9:301–311 [DOI] [PubMed] [Google Scholar]
- 152. Durfee T, et al. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555–569 [DOI] [PubMed] [Google Scholar]
- 153. Dutta S, Koide A, Koide S. 2008. High-throughput analysis of the protein sequence-stability landscape using a quantitative yeast surface two-hybrid system and fragment reconstitution. J. Mol. Biol. 382:721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Dyer MD, Murali TM, Sobral BW. 2007. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics 23:i159–i166 [DOI] [PubMed] [Google Scholar]
- 155. Dyer MD, et al. 2010. The human-bacterial pathogen protein interaction networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS One 5:e12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ear PH, Michnick SW. 2009. A general life-death selection strategy for dissecting protein functions. Nat. Methods 6:813–816 [DOI] [PubMed] [Google Scholar]
- 157. Ecker K, et al. 2009. A RAS recruitment screen identifies ZKSCAN4 as a glucocorticoid receptor-interacting protein. J. Mol. Endocrinol. 42:105–117 [DOI] [PubMed] [Google Scholar]
- 158. Edgerton MD, Jones AM. 1992. Localization of protein-protein interactions between subunits of phytochrome. Plant Cell 4:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Edwards AM, et al. 2002. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 18:529–536 [DOI] [PubMed] [Google Scholar]
- 160. Edwards SR, Wandless TJ. 2007. The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain. J. Biol. Chem. 282:13395–13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ehlert A, et al. 2006. Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46:890–900 [DOI] [PubMed] [Google Scholar]
- 162. Ehrhard KN, Jacoby JJ, Fu XY, Jahn R, Dohlman HG. 2000. Use of G-protein fusions to monitor integral membrane protein-protein interactions in yeast. Nat. Biotechnol. 18:1075–1079 [DOI] [PubMed] [Google Scholar]
- 163. Elbashir SM, et al. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 164. Elefsinioti A, et al. 2011. Large-scale de novo prediction of physical protein-protein association. Mol. Cell. Proteomics 10:M111.010629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Elion EA, Satterberg B, Kranz JE. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell 4:495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Elleuche S, et al. 2010. The small serine-threonine protein SIP2 interacts with STE12 and is involved in ascospore germination in Sordaria macrospora. Eur. J. Cell Biol. 89:873–887 [DOI] [PubMed] [Google Scholar]
- 167. Endo M, Takesako K, Kato I, Yamaguchi H. 1997. Fungicidal action of aureobasidin A, a cyclic depsipeptide antifungal antibiotic, against Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 41:672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Endoh H, Walhout AJ, Vidal M. 2000. A green fluorescent protein-based reverse two-hybrid system: application to the characterization of large numbers of potential protein-protein interactions. Methods Enzymol. 328:74–88 [DOI] [PubMed] [Google Scholar]
- 169. Estojak J, Brent R, Golemis EA. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ewing RM, et al. 2007. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Eyckerman S, et al. 2005. Reverse MAPPIT: screening for protein-protein interaction modifiers in mammalian cells. Nat. Methods 2:427–433 [DOI] [PubMed] [Google Scholar]
- 172. Eyckerman S, et al. 2001. Design and application of a cytokine-receptor-based interaction trap. Nat. Cell Biol. 3:1114–1119 [DOI] [PubMed] [Google Scholar]
- 173. Fan JY, et al. 2008. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem. Biophys. Res. Commun. 367:47–53 [DOI] [PubMed] [Google Scholar]
- 174. Faye C, Chautard E, Olsen BR, Ricard-Blum S. 2009. The first draft of the endostatin interaction network. J. Biol. Chem. 284:22041–22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221–230 [DOI] [PubMed] [Google Scholar]
- 176. Feng SY, Ota K, Ito T. 2010. A yeast one-hybrid system to screen for methylated DNA-binding proteins. Nucleic Acids Res. 38:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fiebitz A, et al. 2008. High-throughput mammalian two-hybrid screening for protein-protein interactions using transfected cell arrays. BMC Genomics 9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Fields S, Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–246 [DOI] [PubMed] [Google Scholar]
- 179. Filipovska A, Razif MF, Nygard KK, Rackham O. 2011. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7:425–427 [DOI] [PubMed] [Google Scholar]
- 180. Finley RL, Jr, Brent R. 1994. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. U. S. A. 91:12980–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Firestine SM, Salinas F, Nixon AE, Baker SJ, Benkovic SJ. 2000. Using an AraC-based three-hybrid system to detect biocatalysts in vivo. Nat. Biotechnol. 18:544–547 [DOI] [PubMed] [Google Scholar]
- 182. Flajolet M, et al. 2000. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242:369–379 [DOI] [PubMed] [Google Scholar]
- 183. Formstecher E, et al. 2005. Protein interaction mapping: a Drosophila case study. Genome Res. 15:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Fossum E, et al. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. 2002. Evolutionary rate in the protein interaction network. Science 296:750–752 [DOI] [PubMed] [Google Scholar]
- 186. Frech C, et al. 2009. Improved homology-driven computational validation of protein-protein interactions motivated by the evolutionary gene duplication and divergence hypothesis. BMC Bioinformatics 10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Friedel CC, Haas J. 2011. Virus-host interactomes and global models of virus-infected cells. Trends Microbiol. 19:501–508 [DOI] [PubMed] [Google Scholar]
- 188. Fujikawa Y, Kato N. 2007. Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52:185–195 [DOI] [PubMed] [Google Scholar]
- 189. Fukuda N, Ishii J, Kondo A. 2011. Gγ recruitment system incorporating a novel signal amplification circuit to screen transient protein-protein interactions. FEBS J. 278:3086–3094 [DOI] [PubMed] [Google Scholar]
- 190. Fukuda N, Ishii J, Tanaka T, Fukuda H, Kondo A. 2009. Construction of a novel detection system for protein-protein interactions using yeast G-protein signaling. FEBS J. 276:2636–2644 [DOI] [PubMed] [Google Scholar]
- 191. Fukuda N, Ishii J, Tanaka T, Kondo A. 2010. The competitor-introduced Ggamma recruitment system, a new approach for screening affinity-enhanced proteins. FEBS J. 277:1704–1712 [DOI] [PubMed] [Google Scholar]
- 192. Fukuura M, et al. 2011. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol. Biol. Cell 22:2620–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9:3447–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Galarneau A, Primeau M, Trudeau LE, Michnick SW. 2002. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 20:619–622 [DOI] [PubMed] [Google Scholar]
- 195. Gallagher SS, Miller LW, Cornish VW. 2007. An orthogonal dexamethasone-trimethoprim yeast three-hybrid system. Anal. Biochem. 363:160–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Gandia J, et al. 2008. Detection of higher-order G protein-coupled receptor oligomers by a combined BRET-BiFC technique. FEBS Lett. 582:2979–2984 [DOI] [PubMed] [Google Scholar]
- 197. Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Garcia-Cuellar MP, Mederer D, Slany RK. 2009. Identification of protein interaction partners by the yeast two-hybrid system. Methods Mol. Biol. 538:347–367 [DOI] [PubMed] [Google Scholar]
- 199. Gardiner L, Coyle BJ, Chan WC, Soultanas P. 2005. Discovery of antagonist peptides against bacterial helicase-primase interaction in B. stearothermophilus by reverse yeast three-hybrid. Chem. Biol. 12:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gavin AC, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636 [DOI] [PubMed] [Google Scholar]
- 201. Gavin AC, et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147 [DOI] [PubMed] [Google Scholar]
- 202. Gavin AC, Maeda K, Kühner S. 2011. Recent advances in charting protein-protein interactions: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22:42–49 [DOI] [PubMed] [Google Scholar]
- 203. Gehl C, Waadt R, Kudia J, Mendel R-R, Hänsch R. 2009. New Gateway vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2:1051–1058 [DOI] [PubMed] [Google Scholar]
- 204. Gerhardt P, Judge JA. 1964. Porosity of isolated cell walls of Saccharomyces cerevisiae and Bacillus megaterium. J. Bacteriol. 87:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Ghosh I, Hamilton AD, Regan L. 2000. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146–157 [DOI] [PubMed] [Google Scholar]
- 206. Giesecke AV, Joung JK. 2007. The bacterial two-hybrid system as a reporter system for analyzing protein-protein interactions. CSH Protoc. 2007:pdb.prot4672. doi:10.1101/pdb.prot4672 [DOI] [PubMed] [Google Scholar]
- 207. Giniger E, Ptashne M. 1987. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature 330:670–672 [DOI] [PubMed] [Google Scholar]
- 208. Giot L, et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302:1727–1736 [DOI] [PubMed] [Google Scholar]
- 209. Gisler SM, et al. 2008. Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system. Mol. Cell. Proteomics 7:1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Goehler H, et al. 2004. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15:853–865 [DOI] [PubMed] [Google Scholar]
- 211. Gohl C, Banovic D, Grevelhörster A, Bogdan S. 2010. WAVE forms hetero- and homo-oligomeric complexes at integrin junctions in Drosophila visualized by bimolecular fluorescence complementation. J. Biol. Chem. 285:40171–40179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Goldberg DS, Roth FP. 2003. Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. U. S. A. 100:4372–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Goll J, Uetz P. 2006. The elusive yeast interactome. Genome Biol. 7:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. 2011. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 9:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Gonçalves SA, Matos JE, Outeiro TF. 2010. Zooming into protein oligomerization in neurogeneration using BiFC. Trends Biochem. Sci. 35:643–651 [DOI] [PubMed] [Google Scholar]
- 216. Graf R, Schachman HK. 1996. Random circular permutation of genes and expressed polypeptide chains: application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U. S. A. 93:11591–11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Gray PN, Busser KJ, Chappell TG. 2007. A novel approach for generating full-length, high coverage allele libraries for the analysis of protein interactions. Mol. Cell. Proteomics 6:514–526 [DOI] [PubMed] [Google Scholar]
- 218. Greenfield NJ. 2004. Circular dichroism analysis for protein-protein interactions. Methods Mol. Biol. 261:55–78 [DOI] [PubMed] [Google Scholar]
- 219. Grefen C, Lalonde S, Obrdlik P. 2007. Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. Chapter 5:Unit 5.27 [DOI] [PubMed] [Google Scholar]
- 220. Grefen C, Obrdlik P, Harter K. 2009. The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 479:217–233 [DOI] [PubMed] [Google Scholar]
- 221. Grefen C, et al. 2008. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1:308–320 [DOI] [PubMed] [Google Scholar]
- 222. Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. 2001. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276:29188–29194 [DOI] [PubMed] [Google Scholar]
- 223. Grigoriev A. 2001. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 29:3513–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Grigoriev A. 2003. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 31:4157–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Grinberg AV, Hu CD, Kerppola TK. 2004. Visualization of Myc/Max/Mad family dimers and the competition or dimerization in living cells. Mol. Cell. Biol. 24:4294–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Gu H, Zhu P, Jiao Y, Meng Y, Chen M. 2011. PRIN: a predicted rice interactome network. BMC Bioinformatics 12:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Guda C, King BR, Pal LR, Guda P. 2009. A top-down approach to infer and compare domain-domain interactions across eight model organisms. PLoS One 4:e5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Gunde T, Tanner S, Auf der Maur A, Petrascheck M, Barberis A. 2004. Quenching accumulation of toxic galactose-1-phosphate as a system to select disruption of protein-protein interactions in vivo. Biotechniques 37:844–852 [DOI] [PubMed] [Google Scholar]
- 229. Guo B, et al. 2006. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol. Cell. Biol. 26:4529–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Guo D, et al. 2004. A tethered catalysis, two-hybrid system to identify protein-protein interactions requiring post-translational modifications. Nat. Biotechnol. 22:888–892 [DOI] [PubMed] [Google Scholar]
- 231. Guo D, Rajamaki ML, Saarma M, Valkonen JP. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935–939 [DOI] [PubMed] [Google Scholar]
- 232. Guo M, et al. 2009. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 19:1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Gurezka R, Langosch D. 2001. In vitro selection of membrane-spanning leucine zipper protein-protein interaction motifs using POSSYCAT. J. Biol. Chem. 276:45580–45587 [DOI] [PubMed] [Google Scholar]
- 234. Gyuris J, Golemis E, Chertkov H, Brent R. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791–803 [DOI] [PubMed] [Google Scholar]
- 235. Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. 2005. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U. S. A. 102:4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Hagemann AI, Xu X, Nentwich O, Hyvonen M, Smith JC. 2009. Rab5-mediated endocytosis of activin is not required for gene activation or long range signalling in Xenopus. Development 136:2803–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Hammani K, et al. 2011. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23:730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Hammer MM, Wehrman TS, Blau HM. 2007. A novel enzyme complementation-based assay for monitoring G-protein-coupled receptor internalization. FASEB J. 21:3827–3834 [DOI] [PubMed] [Google Scholar]
- 239. Hao N, Whitelaw ML, Shearwin KE, Dodd LB, Chapman-Smith A. 2011. Identification of residues in the N-terminal PAS domains important for dimerization of Arnt and AhR. Nucleic Acids Res. 39:3695–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Harrington L, et al. 1997. A mammalian telomerase-associated protein. Science 275:973–977 [DOI] [PubMed] [Google Scholar]
- 241. Hart GT, Ramani AK, Marcotte EM. 2006. How complete are current yeast and human protein-interaction networks? Genome Biol. 7:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Harvey SA, Smith JC. 2009. Visualisation and quantification of morphogen gradient formation in the zebrafish. PLoS Biol. 7:e1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Hashimoto J, et al. 2009. Novel in vitro protein fragment complementation assay applicable to high-throughput screening in a 1536-well format. J. Biomol. Screen. 14:970–979 [DOI] [PubMed] [Google Scholar]
- 244. Hashimoto T, Adams KW, Fan Z, McLean PJ, Hyman BT. 2011. Characterization of oligomer formation of amyloid-beta peptide using a split-luciferase complementation assay. J. Biol. Chem. 286:27081–27091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. 2010. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J. Biol. Chem. 285:17771–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Häuser R, Sabri M, Moineau S, Uetz P. 2011. The proteome and interactome of Streptococcus pneumoniae phage Cp-1. J. Bacteriol. 193:3135–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Hazbun TR, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353–1365 [DOI] [PubMed] [Google Scholar]
- 248. He B, et al. 2010. Live-cell imaging in Caenorhabditis elegans reveals the distinct roles of dynamin self-assembly and guanosine triphosphate hydrolysis in the removal of apoptotic cells. Mol. Biol. Cell 21:610–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. He F, Zhang Y, Chen H, Zhang Z, Peng YL. 2008. The prediction of protein-protein interaction networks in rice blast fungus. BMC Genomics 9:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Henao-Mejia J, et al. 2009. Suppression of HIV-1 Nef translation by Sam68 mutant-induced stress granules and nef mRNA sequestration. Mol. Cell 33:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Hengen PN. 1997. False positives from the yeast two-hybrid system. Trends Biochem. Sci. 22:33–34 [DOI] [PubMed] [Google Scholar]
- 252. Hennecke F, Müller A, Meister R, Strelow A, Behrens S. 2005. A ToxR-based two-hybrid system for the detection of periplasmic and cytoplasmic protein-protein interactions in Escherichia coli: minimal requirements for specific DNA binding and transcriptional activation. Proten Eng. Des. Sel. 18:477–486 [DOI] [PubMed] [Google Scholar]
- 253. Henthorn DC, Jaxa-Chamiec AA, Meldrum E. 2002. A GAL4-based yeast three-hybrid system for the identification of small molecule-target protein interactions. Biochem. Pharmacol. 63:1619–1628 [DOI] [PubMed] [Google Scholar]
- 254. Herrmann F, et al. 2011. p53 gene repair with zinc finger nucleases optimised by yeast 1-hybrid and validated by Solexa sequencing. PLoS One 6:e20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. 1980. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. U. S. A. 77:1783–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Hershkovitz D, et al. 2011. Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J. Invest. Dermatol. 131:662–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Hiatt SM, Shyu YJ, Duren HM, Hu C-D. 2008. Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in Caenorhabditis elegans. Methods 45:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Hida N, et al. 2009. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One 4:e5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Hirst M, et al. 2001. A two-hybrid system for transactivator bait proteins. Proc. Natl. Acad. Sci. U. S. A. 98:8726–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Hishigaki H, Nakai K, Ono T, Tanigami A, Takagi T. 2001. Assessment of prediction accuracy of protein function from protein-protein interaction data. Yeast 18:523–531 [DOI] [PubMed] [Google Scholar]
- 261. Ho Y, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183 [DOI] [PubMed] [Google Scholar]
- 262. Hoff B, et al. 2010. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot. Cell 9:1236–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Hoff B, Kück U. 2005. Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Curr. Genet. 47:132–138 [DOI] [PubMed] [Google Scholar]
- 264. Hollenberg SM, Evans RM. 1988. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell 55:899–906 [DOI] [PubMed] [Google Scholar]
- 265. Hook B, Bernstein D, Zhang B, Wickens M. 2005. RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11:227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Horswill AR, Benkovic SJ. 2006. Identifying small-molecule modulators of protein-protein interactions. Curr. Protoc. Protein Sci. 46:19.15.1–19.15.19 [DOI] [PubMed] [Google Scholar]
- 267. Horswill AR, Savinov SN, Benkovic SJ. 2004. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 101:15591–15596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Houser-Scott F, et al. 2002. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl. Acad. Sci. U. S. A. 99:2684–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Hruby A, et al. 2011. A constraint network of interactions: protein-protein interaction analysis of the yeast type II phosphatase Ptc1p and its adaptor protein Nbp2p. J. Cell Sci. 124:35–46 [DOI] [PubMed] [Google Scholar]
- 270. Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 271. Hu CD, Kerppola TK. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Hu JC, Kornacker MG, Hochschild A. 2000. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods 20:80–94 [DOI] [PubMed] [Google Scholar]
- 273. Hu JC, O'Shea EK, Kim PS, Sauer RT. 1990. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science 250:1400–1403 [DOI] [PubMed] [Google Scholar]
- 274. Hu X, Kang S, Chen X, Shoemaker CB, Jin MM. 2009. Yeast surface two-hybrid for quantitative in vivo detection of protein-protein interactions via the secretory pathway. J. Biol. Chem. 284:16369–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Hu X, Kang S, Lefort C, Kim M, Jin MM. 2010. Combinatorial libraries against libraries for selecting neoepitope activation-specific antibodies. Proc. Natl. Acad. Sci. U. S. A. 107:6252–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Huang A, et al. 2005. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 65:5607–5619 [DOI] [PubMed] [Google Scholar]
- 277. Huang H, Choi SY, Frohman MA. 2010. A quantitative assay for mitochondrial fusion using Renilla luciferase complementation. Mitochondrion 10:559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Huang H, Jedynak BM, Bader JS. 2007. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Huang J, Schreiber SL. 1997. A yeast genetic system for selecting small molecule inhibitors of protein-protein interactions in nanodroplets. Proc. Natl. Acad. Sci. U. S. A. 94:13396–13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Huang TW, Lin CY, Kao CY. 2007. Reconstruction of human protein interolog network using evolutionary conserved network. BMC Bioinformatics 8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Huang W, Wang SL, Lozano G, de Crombrugghe B. 2001. cDNA library screening using the SOS recruitment system. Biotechniques 30:94–98 [DOI] [PubMed] [Google Scholar]
- 282. Huang YC, Tseng SF, Tsai HJ, Lenzmeier BA, Teng SC. 2010. Direct interaction between Utp8p and Utp9p contributes to rRNA processing in budding yeast. Biochem. Biophys. Res. Commun. 393:297–302 [DOI] [PubMed] [Google Scholar]
- 283. Hubsman M, Yudkovsky G, Aronheim A. 2001. A novel approach for the identification of protein-protein interaction with integral membrane proteins. Nucleic Acids Res. 29:E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Hudry B, Viala S, Graba Y, Merabet S. 2011. Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay. BMC Biol. 9:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Hussey SL, Muddana SS, Peterson BR. 2003. Synthesis of a beta-estradiol-biotin chimera that potently heterodimerizes estrogen receptor and streptavidin proteins in a three-hybrid system. J. Am. Chem. Soc. 125:3692–3693 [DOI] [PubMed] [Google Scholar]
- 286. Hwang CS, Shemorry A, Auerbach D, Varshavsky A. 2010. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 12:1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Immink RG, Gadella TWJ, Ferrario S, Busscher M, Angenent GC. 2002. Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. U. S. A. 99:2416–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Inouye C, Dhillon N, Durfee T, Zambryski PC, Thorner J. 1997. Mutational analysis of STE5 in the yeast Saccharomyces cerevisiae: application of a differential interaction trap assay for examining protein-protein interactions. Genetics 147:479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Ioannoni R, Beaudoin J, Mercier A, Labbé S. 2010. Copper-dependent trafficking of the Ctr4-Ctr5 copper transporting complex. PLoS One 5:e11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Irion U, St Johnston D. 2007. Bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445:554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Ishimoto T, Ozawa T, Mori H. 2011. Real-time monitoring of actin polymerization in living cells using split luciferase. Bioconjug. Chem. 22:1136–1144 [DOI] [PubMed] [Google Scholar]
- 292. Islam KM, et al. 2009. Directed evolution of estrogen receptor proteins with altered ligand-binding specificities. Protein Eng. Des. Sel. 22:45–52 [DOI] [PubMed] [Google Scholar]
- 293. Isogai M, et al. 2011. Structure and characteristics of reassembled fluorescent protein, a new insight into the reassembly mechanisms. Bioorg. Med. Chem. Lett. 21:3021–3024 [DOI] [PubMed] [Google Scholar]
- 294. Ito T, et al. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Ivanic J, Yu X, Wallqvist A, Reifman J. 2009. Influence of protein abundance on high-throughput protein-protein interaction detection. PLoS One 4:e5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Iwai K, et al. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. U. S. A. 96:12436–12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Jaaro H, Levy Z, Fainzilber M. 2005. A genome wide screening approach for membrane-targeted proteins. Mol. Cell. Proteomics 4:328–333 [DOI] [PubMed] [Google Scholar]
- 298. James P. 2001. Yeast two-hybrid vectors and strains. Methods Mol. Biol. 177:41–84 [DOI] [PubMed] [Google Scholar]
- 299. James P, Halladay J, Craig EA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Jansen R, Greenbaum D, Gerstein M. 2002. Relating whole-genome expression data with protein-protein interactions. Genome Res. 12:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Jansen R, et al. 2003. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science 302:449–453 [DOI] [PubMed] [Google Scholar]
- 302. Jensen LJ, Bork P. 2008. Biochemistry not comparable, but complementary. Science 322:56–57 [DOI] [PubMed] [Google Scholar]
- 303. Jeong H, Mason SP, Barabasi AL, Oltvai ZN. 2001. Lethality and centrality in protein networks. Nature 411:41–42 [DOI] [PubMed] [Google Scholar]
- 304. Jeong KJ, Seo MJ, Iverson BL, Georgiou G. 2007. APEx 2-hybrid, a quantitative protein-protein interaction assay for antibody discovery and engineering. Proc. Natl. Acad. Sci. U. S. A. 104:8247–8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Jiang R, Carlson M. 1996. Glucose regulates protein inetractions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105–3115 [DOI] [PubMed] [Google Scholar]
- 306. Jiang R, Carlson M. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Jiang T, Guerrier-Takada C, Altman S. 2001. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA 7:937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Jiang W, Boder ET. 2010. High-throughput engineering and analysis of peptide binding to class II MHC. Proc. Natl. Acad. Sci. U. S. A. 107:13258–13263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Jiang Y, et al. 2010. Split Renilla luciferase protein fragment-assisted complementation (SRL-PFAC) to characterize Hsp90-Cdc37 complex and identify critical residues in protein/protein interactions. J. Biol. Chem. 285:21023–21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Jin F, Avramova L, Huang J, Hazbun T. 2007. A yeast two-hybrid smart-pool-array system for protein-interaction mapping. Nat. Methods 4:405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Jin F, et al. 2006. A pooling-deconvolution strategy for biological network elucidation. Nat. Methods 3:183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Johnsson N, Varshavsky A. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Joshi PB, et al. 2007. Identification of protein interaction antagonists using the repressed transactivator two-hybrid system. Biotechniques 42:635–644 [DOI] [PubMed] [Google Scholar]
- 314. Joung JK, Ramm EI, Pabo CO. 2000. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 97:7382–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Jung SO, et al. 2005. Surface plasmon resonance imaging-based protein arrays for high-throughput screening of protein-protein interaction inhibitors. Proteomics 5:4427–4431 [DOI] [PubMed] [Google Scholar]
- 316. Kaboord B, Perr M. 2008. Isolation of proteins and protein complexes by immunoprecipitation. Methods Mol. Biol. 424:349–364 [DOI] [PubMed] [Google Scholar]
- 317. Kaihara A, Kawai Y, Sato M, Ozawa T, Umezawa Y. 2003. Locating a protein-protein interaction in living cells via split Renilla luciferase complementation. Anal. Chem. 75:4176–4181 [DOI] [PubMed] [Google Scholar]
- 318. Kamiya T, Ojima T, Sugimoto K, Nakano H, Kawarasaki Y. 2010. Quantitative Y2H screening: cloning and signal peptide engineering of a fungal secretory LacA gene and its application to yeast two-hybrid system as a quantitative reporter. J. Biotechnol. 146:151–159 [DOI] [PubMed] [Google Scholar]
- 319. Kang PJ, Beven L, Hariharan S, Park HO. 2010. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell 21:3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Kanno A, Ozawa T, Umezawa Y. 2011. Detection of protein-protein interactions in bacteria by GFP-fragment reconstitution. Methods Mol. Biol. 705:1–258 [DOI] [PubMed] [Google Scholar]
- 321. Karimova G, Dautin N, Ladant D. 2005. Interactions network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Karna SLR, et al. 2010. A bacterial two-hybrid system that utilizes Gateway cloning for rapid screening of protein-protein interactions. Biotechniques 49:831–833 [DOI] [PubMed] [Google Scholar]
- 324. Kato N, et al. 2010. Luminescence detection of SNARE-SNARE interaction in Arabidopsis protoplasts. Plant Mol. Biol. 72:433–444 [DOI] [PubMed] [Google Scholar]
- 325. Kato N, Jones J. 2010. The split luciferase complementation assay. Methods Mol. Biol. 655:359–376 [DOI] [PubMed] [Google Scholar]
- 326. Kato-Stankiewicz J, et al. 2002. Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells. Proc. Natl. Acad. Sci. U. S. A. 99:14398–14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Kaushansky A, et al. 2010. Quantifying protein-protein interactions in high throughput using protein domain microarrays. Nat. Protoc. 5:773–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Kawahara T, Yanagi H, Yura T, Mori K. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8:1845–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Keegan K, Cooper JA. 1996. Use of the two-hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene 12:1537–1544 [PubMed] [Google Scholar]
- 330. Keegan L, Gill G, Ptashne M. 1986. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231:699–704 [DOI] [PubMed] [Google Scholar]
- 331. Kehat I, Accornero F, Aronow BJ, Molkentin JD. 2011. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol. 193:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Kelley BP, et al. 2004. PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 32:W83–W88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Kemmeren P, et al. 2002. Protein interaction verification and functional annotation by integrated analysis of genome-scale data. Mol. Cell 9:1133–1143 [DOI] [PubMed] [Google Scholar]
- 334. Keppler A, et al. 2003. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21:86–89 [DOI] [PubMed] [Google Scholar]
- 335. Kerppola TK. 2008. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37:465–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Kerppola TK. 2006. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1:1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Kerppola TK. 2009. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 38:2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Keskin O, Gursoy A, Ma B, Nussinov R. 2008. Principles of protein-protein interactions: what are the preferred ways for proteins to interact? Chem. Rev. 108:1225–1244 [DOI] [PubMed] [Google Scholar]
- 339. Khadka S, et al. 2011. A physical interaction network of dengue virus and human proteins. Mol. Cell. Proteomics 10:M111.012187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Kiemer L, Cesareni G. 2007. Comparative interactomics: comparing apples and pears? Trends Biotechnol. 25:448–454 [DOI] [PubMed] [Google Scholar]
- 341. Kikis EA, Oka Y, Hudson ME, Nagatani A, Quail PH. 2009. Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Kim JY, Park OG, Lee JW, Lee YC. 2007. One- plus two-hybrid system, a novel yeast genetic selection for specific missense mutations disrupting protein/protein interactions. Mol. Cell. Proteomics 6:1727–1740 [DOI] [PubMed] [Google Scholar]
- 343. Kim SB, Otani Y, Umezawa Y, Tao H. 2007. Bioluminescent indicator for determining protein-protein interactions using intramolecular complementation of split click beetle luciferase. Anal. Chem. 79:4820–4826 [DOI] [PubMed] [Google Scholar]
- 344. Kim SB, Ozawa T, Watanabe S, Umezawa Y. 2004. High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc. Natl. Acad. Sci. U. S. A. 101:11542–11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Kimura T, Hashimoto I, Nishizawa M, Ito S, Yamada H. 2010. Novel cis-active structures in the coding region mediate CRM1-dependent nuclear export of IFN-α 1 mRNA. Med. Mol. Morphol. 43:145–157 [DOI] [PubMed] [Google Scholar]
- 346. King AD, Przulj N, Jurisica I. 2004. Protein complex prediction via cost-based clustering. Bioinformatics 20:3013–3020 [DOI] [PubMed] [Google Scholar]
- 347. King G, Dixon AM. 2010. Evidence for role of transmembrane helix-helix interactions in the assembly of the class II major histocompatibility complex. Mol. Biosyst. 6:1650–1661 [DOI] [PubMed] [Google Scholar]
- 348. Kishi T, Ikeda A, Koyama N, Fukada J, Nagao R. 2008. A refined two-hybrid system reveals that SCF(Cdc4)-dependent degradation of Swi5 contributes to the regulatory mechanism of S-phase entry. Proc. Natl. Acad. Sci. U. S. A. 105:14497–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Klopffleisch K, et al. 2011. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Knauer SK, Stauber RH. 2005. Development of an autofluorescent translocation biosensor system to investigate protein-protein interactions in living cells. Anal. Chem. 77:4815–4820 [DOI] [PubMed] [Google Scholar]
- 351. Kodama Y. 2011. A bright green-colored bimolecular fluorescence complementation assay in living plant cells. Plant Biotechnol. 28:95–98 [Google Scholar]
- 352. Kodama Y, Hu CD. 2010. An improved bimolecular fluorescence complementation assay with a high signal-to-noise ratio. Biotechniques 49:793–805 [DOI] [PubMed] [Google Scholar]
- 353. Kodama Y, Wada M. 2009. Simultaneous visulaization of two protein complexes in a single plant cell using multicolour fluorescence complementation analysis. Plant Mol. Biol. 70:211–217 [DOI] [PubMed] [Google Scholar]
- 354. Koegl M, Uetz P. 2007. Improving yeast two-hybrid screening systems. Brief. Funct. Genomic. Proteomic. 6:302–312 [DOI] [PubMed] [Google Scholar]
- 355. Kojima T, Karasawa S, Miyawaki A, Tsumuraya T, Fujii I. 2011. Novel screening system for protein-protein interactions by bimolecular fluorescence complementation in Saccharomyces cerevisiae. J. Biosci. Bioeng. 111:397–401 [DOI] [PubMed] [Google Scholar]
- 356. Kolmar H, Frisch C, Götze K, Fritz HJ. 1995. Immunoglobulin mutant library genetically screened for folding stability exploiting bacterial signal transduction. J. Mol. Biol. 251:471–476 [DOI] [PubMed] [Google Scholar]
- 357. Kolmar H, Frisch C, Kleemann G, Kgötze Stevens FJ, Fritz HJ. 1994. Dimerization of Bence Jones proteins: linking the rate of transcription from an Escherichia coli promoter to the association constant of REIV. Biol. Chem. Hoppe Seyler 375:61–70 [DOI] [PubMed] [Google Scholar]
- 358. Kolmar H, et al. 1995. Membrane insertion of the bacterial signal transduction protein ToxR and requirements of transcription activation studied by modular replacement of different protein substructures. EMBO J. 14:3895–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. König J, et al. 2007. Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers. RNA 13:614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Korkegian A, Black ME, Baker D, Stoddard BL. 2005. Computational thermostabilization of an enzyme. Science 308:857–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Korolik V. 2010. Aspartate chemosensory receptor signalling in Campylobacter jejuni. Virulence 1:414–417 [DOI] [PubMed] [Google Scholar]
- 362. Kotani K, Wilden P, Pillay TS. 1998. SH2-Balpha is an insulin-receptor adapter protein and substrate that interacts with the activation loop of the insulin-receptor kinase. Biochem. J. 335:103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Kraakman L, et al. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002–1012 [DOI] [PubMed] [Google Scholar]
- 364. Kraichely DM, MacDonald PN. 2001. Confirming yeast two-hybrid protein interactions using in vitro glutathione-S-transferase pulldowns. Methods Mol. Biol. 177:135–150 [DOI] [PubMed] [Google Scholar]
- 365. Krogan NJ, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637–643 [DOI] [PubMed] [Google Scholar]
- 366. Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. 2007. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318:772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Kumar S, et al. 2011. FLIM FRET technology for drug discovery: automated multiwell-plate high-content analysis, multiplexed readouts and application in situ. Chemphyschem 12:609–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R. 2010. Combined bimolecular fluorescence complementation and Förster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol. 152:1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. LaCount DJ, et al. 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438:103–107 [DOI] [PubMed] [Google Scholar]
- 370. Lalonde S, et al. 2010. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. Langosch D, Brosig B, Kolmar H, Fritz HJ. 1996. Dimerization of the glycophorin A transmembrane segment in membranes probed with the ToxR transcription activator. J. Mol. Biol. 263:525–530 [DOI] [PubMed] [Google Scholar]
- 372. Laser H, et al. 2000. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl. Acad. Sci. U. S. A. 97:13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Leanna CA, Hannink M. 1996. The reverse two-hybrid system: a genetic scheme for selection against specific protein/protein interactions. Nucleic Acids Res. 24:3341–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Le Douarin B, Pierrat B, Vom Baur E, Chambon P, Losson R. 1995. A new version of the two-hybrid assay for detection of protein-protein interactions. Nucleic Acids Res. 23:876–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Lee LY, Fang MJ, Kuang LY, Gelvin SB. 2008. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Lee OH, et al. 2011. Genome-wide YFP fluorescence complementation screen identifies new regulators or telomere signaling in human cells. Mol. Cell. Proteomics 10:M110.001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Lee SA, et al. 2008. Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC Bioinformatics 9(Suppl 12):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Lee YR, et al. 2010. Development of bimolecular fluorescence complementation using Dronpa for visualization of protein-protein interactions in cells. Mol. Imaging Biol. 12:468–578 [DOI] [PubMed] [Google Scholar]
- 379. Legrain P, Dokhelar MC, Transy C. 1994. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 22:3241–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Le Guével R, et al. 2009. Identification of small molecule regulators of the nuclear receptor HNF4alpha based on naphthofuran scaffolds. Bioorg. Med. Chem. 17:7021–7030 [DOI] [PubMed] [Google Scholar]
- 381. Lehner B, Sanderson CM. 2004. A protein interaction framework for human mRNA degradation. Genome Res. 14:1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Lemercier G, Gendreizig S, Kindermann M, Johnsson K. 2007. Inducing and sensing protein-protein interactions in living cells by selective cross-linking. Angew. Chem. Int. Ed. Engl. 46:4281–4284 [DOI] [PubMed] [Google Scholar]
- 383. Letovsky S, Kasif S. 2003. Predicting protein function from protein/protein interaction data: a probabilistic approach. Bioinformatics 19:i197–i204 [DOI] [PubMed] [Google Scholar]
- 384. Levy ED, Landry CR, Michnick SW. 2009. How perfect can protein interactomes be? Sci. Signal. 2:pe11. [DOI] [PubMed] [Google Scholar]
- 385. Li C, Distelfeld A, Comis A, Dubcovsky J. 2011. Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with nuclear factor-Y complexes. Plant J. 67:763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Li J, Zhang S, Gao L, Chen Y, Xie X. 2011. A cell-based high-throughput assay for the screening of small-molecule inhibitors of p53-MDM2 interaction. J. Biomol. Screen. 16:450–456 [DOI] [PubMed] [Google Scholar]
- 387. Li JJ, Herskowitz I. 1993. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262:1870–1874 [DOI] [PubMed] [Google Scholar]
- 388. Li M, et al. 2010. Detection of in vivo interactions between Arabidopsis class A-HSFs, using a novel BiFC fragment, and identification of novel class B-HSF interacting proteins. Eur. J. Cell Biol. 89:126–132 [DOI] [PubMed] [Google Scholar]
- 389. Li S, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303:540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Li W, et al. 2008. Noninvasive imaging and quantification of epidermal growth factor receptor kinase activation in vivo. Cancer Res. 68:4990–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Lian HL, et al. 2011. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Licitra EJ, Liu JO. 1996. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci. U. S. A. 93:12817–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Lickfeld M, Schmitz HP. 2011. Selection of STOP-free sequences from random mutagenesis for ‘loss of interaction’ two-hybrid studies. Yeast 28:535–545 [DOI] [PubMed] [Google Scholar]
- 394. Lidke DS, Wilson BS. 2009. Caught in the act: quantifying protein behaviour in living cells. Trends Cell Biol. 19:566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Lievens S, Lemmens I, Tavernier J. 2009. Mammalian two-hybrids come of age. Trends Biochem. Sci. 34:579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Lievens S, et al. 2009. Array MAPPIT: high-throughput interactome analysis in mammalian cells. J. Proteome Res. 8:877–886 [DOI] [PubMed] [Google Scholar]
- 396a. Lievens S, De Bosscher K, Lemmens I, Peelman F, Tavernier J. 2011. MAPPIT: a protein interaction toolbox built on insights in cytokine receptor signalling. Cytokine Growth Factor Rev. 22:321–329 [DOI] [PubMed] [Google Scholar]
- 397. Lim J, et al. 2006. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125:801–814 [DOI] [PubMed] [Google Scholar]
- 398. Lim YH, Charette JM, Baserga SJ. 2011. Assembling a protein-protein interaction map of the SSU processome from existing datasets. PLoS One 6:e17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Lima-Mendez G, Van Helden J. 2009. The powerful law of the power law and other myths in network biology. Mol. Biosyst. 5:1482–1493 [DOI] [PubMed] [Google Scholar]
- 400. Lin H, Tao H, Cornish VW. 2004. Directed evolution of a glycosynthase via chemical complementation. J. Am. Chem. Soc. 126:15051–15059 [DOI] [PubMed] [Google Scholar]
- 401. Lin HN, Abida WM, Sauer RT, Cornish VW. 2000. Dexamethasone-methotrexate: an efficient chemical inducer of protein dimerisation in vivo. J. Am. Chem. Soc. 122:4247–4248 [Google Scholar]
- 402. Lin HP, Vincenz C, Eliceiri KW, Kerppola TK, Ogle BM. 2010. Bimolecular fluorescence complementation analysis of eukaryotic fusion products. Biol. Cell 102:525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Lin HY, Lin SE, Chien SF, Chern MK. 2011. Electroporation for three commonly used yeast strains for two-hybrid screening experiments. Anal. Biochem. 416:117–119 [DOI] [PubMed] [Google Scholar]
- 404. Lin M, Shen X, Chen X. 2011. PAIR: the predicted Arabidopsis interactome resource. Nucleic Acids Res. 39:D1134–D1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Lin X, Liu M, Chen XW. 2009. Assessing reliability of protein-protein interactions by integrative analysis of data in model organisms. BMC Bioinformatics 10(Suppl 4):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Liu B, Zuo Z, Liu H, Liu X, Lin C. 2011. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25:1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Liu H, et al. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–1539 [DOI] [PubMed] [Google Scholar]
- 408. Liu JX, Howell SH. 2010. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22:782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Liu M, Chen XW, Jothi R. 2009. Knowledge-guided inference of domain-domain interactions from incomplete protein-protein interaction networks. Bioinformatics 25:2492–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Liu Y, et al. 2011. Media composition influences yeast one- and two-hybrid results. Biol. Proced. Online 13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. 2000. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19:6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Lopato S, et al. 2006. Isolation of plant transcription factors using a modified yeast one-hybrid system. Plant Methods 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Loving GS, Sainlos M, Imperiali B. 2010. Monitoring protein interactions and dynamics with solvatochronic fluorophores. Trends Biotechnol. 28:73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Lugari A, et al. 2010. Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J. Biol. Chem. 285:33230–33241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415. Luker GD, et al. 2002. Noninvasive imaging of protein-protein interactions in living animals. Proc. Natl. Acad. Sci. U. S. A. 99:6961–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Luker KE, et al. 2004. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U. S. A. 101:12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Lundblad JR, Laurance M, Goodman RH. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol. Endocrinol. 10:607–612 [DOI] [PubMed] [Google Scholar]
- 418. Luo J, Fishburn J, Hahn S, Ranish J. 2012. An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol. Cell. Proteomics 11:M111.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419. Ma J, Ptashne M. 1988. Converting a eukaryotic transcriptional inhibitor into an activator. Cell 55:443–446 [DOI] [PubMed] [Google Scholar]
- 420. Ma J, Ptashne M. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847–853 [DOI] [PubMed] [Google Scholar]
- 421. MacDonald ML, et al. 2006. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2:329–337 [DOI] [PubMed] [Google Scholar]
- 422. MacDonald PN. 2001. Two-hybrid systems: methods and protocols. In Methods in molecular biology, vol 177 Humana Press Inc., Totowa, NJ: [PubMed] [Google Scholar]
- 423. MacFarlane SA, Uhrig JF. 2008. Yeast two-hybrid assay to identify host-virus interactions. Methods Mol. Biol. 451:649–672 [DOI] [PubMed] [Google Scholar]
- 424. Magliery TJ, et al. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146–157 [DOI] [PubMed] [Google Scholar]
- 425. Mahdavi MA, Lin YH. 2007. False positive reduction in protein-protein interaction predictions using gene ontology annotations. BMC Bioinformatics 8:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Maier CJ, Maier RH, Hintner H, Bauer JW, Önder K. 2010. Coupled yeast 2-hybrid-mammalian 2-hybrid reading-frame-independent and site-specific recombinational cloning vector system. Assay Drug Dev. Technol. 8:625–629 [DOI] [PubMed] [Google Scholar]
- 427. Maier R, Brandner C, Hintner H, Bauer J, Önder K. 2008. Construction of a reading frame-inedependent yeast two-hybrid vector system for site-specific recombinational cloning and protein interaction screening. Biotechniques 45:235–244 [DOI] [PubMed] [Google Scholar]
- 428. Makhnevych T, et al. 2009. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33:124–135 [DOI] [PubMed] [Google Scholar]
- 429. Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. 2010. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature 465:101–105 [DOI] [PubMed] [Google Scholar]
- 430. Manderson EN, Malleshaiah MK, Michnick SW. 2008. A novel genetic screen implicates Elm1 in the inactivation of the yeast transcription factor SBF. PLoS One 3:e1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431. Marchadier E, et al. 2011. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: exploration by an integrative approach. Proteomics 11:2981–2991 [DOI] [PubMed] [Google Scholar]
- 432. Maroun M, Aronheim A. 1999. A novel in vivo assay for the analysis of protein-protein interaction. Nucleic Acids Res. 27:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433. Marsolier MC, Prioleau MN, Sentenac A. 1997. A RNA polymerase III-based two-hybrid system to study RNA polymerase II transcriptional regulators. J. Mol. Biol. 268:243–249 [DOI] [PubMed] [Google Scholar]
- 434. Massoud TF, Paulmurugan R, Gambhir SS. 2010. A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography. Nat. Med. 16:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Matsui K, Ohme-Takagi M. 2010. Detection of protein-protein interactions in plants using the transrepressive activity of the EAR motif repression domain. Plant J. 61:570–578 [DOI] [PubMed] [Google Scholar]
- 436. Matthews LR, et al. 2001. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs.” Genome Res. 11:2120–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. McCraith S, Holtzman T, Moss B, Fields S. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 97:4879–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. McDowall MD, Scott MS, Barton GJ. 2009. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 37:D651–D656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Meng X, Brodsky MH, Wolfe SA. 2005. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23:988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Meng X, Smith RM, Giesecke AV, Joung JK, Wolfe SA. 2006. Counter-selectable marker for bacterial-based interaction trap systems. Biotechniques 40:179–184 [DOI] [PubMed] [Google Scholar]
- 441. Mercier A, Watt S, Bähler J, Labbé S. 2008. Key function of the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7:493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442. Mervine SM, Yost EA, Sabo JL, Hynes TR, Berlot CH. 2006. Analysis of G protein betagamma dimer formation in live cells using multicolor bimolecular fluorescence complementation demonstrates preferences of beta1 or particular gamma subunits. Mol. Pharmacol. 70:194–205 [DOI] [PubMed] [Google Scholar]
- 443. Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. 2011. Protein-fragment complementation assays for large-scale analysis, functional dissection and dynamic studies of protein-protein interactions in living cells. Methods Mol. Biol. 756:395–425 [DOI] [PubMed] [Google Scholar]
- 444. Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. 2007. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 6:569–582 [DOI] [PubMed] [Google Scholar]
- 445. Mika S, Rost B. 2006. Protein-protein interactions more conserved within species than across species. PLoS Comput. Biol. 2:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446. Miller CL, et al. 2007. Virus-derived platforms for visualizing protein associations inside cells. Mol. Cell. Proteomics 6:1027–1038 [DOI] [PubMed] [Google Scholar]
- 447. Miller JP, et al. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. U. S. A. 102:12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Miller RR, Okkema PG. 2011. The Caenorhabditis elegans T-box factor MLS-1 requires Groucho co-repressor interaction for uterine muscle specification. PLoS Genet. 7:e1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Millson SH, et al. 2005. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450. Min J, Park PG, Ko E, Choi E, Lee H. 2007. Identification of Rad51 regulation by BRCA2 using Caenorhabditis elegans BRCA2 and bimolecular fluorescence complementation analysis. Biochem. Biophys. Res. Commun. 362:958–964 [DOI] [PubMed] [Google Scholar]
- 451. Miranda E, Forafonov F, Tavassoli A. 2011. Deciphering interactions used by the influenza virus NS1 protein to silence the host antiviral sensor protein RIG-1 using a bacterial reverse two-hybrid system. Mol. Biosyst. 7:1042–1045 [DOI] [PubMed] [Google Scholar]
- 452. Misawa N, et al. 2010. Rapid and high-sensitivity cell-based assays of protein-protein interactions using split click beetle luciferase complementation: an approach to the study of G-protein-coupled receptors. Anal. Chem. 82:2552–2560 [DOI] [PubMed] [Google Scholar]
- 453. Miyashita S, Shirako Y. 2008. Chromosomal integration of a binding domain:bait gene into yeast enhances detection in the two-hybrid system. J. Microbiol. Methods 73:179–184 [DOI] [PubMed] [Google Scholar]
- 454. Möckli N, et al. 2007. Yeast split-ubiquitin-based cytosolic screening system to detect interactions between transcriptionally active proteins. Biotechniques 42:725–730 [DOI] [PubMed] [Google Scholar]
- 455. Moerdyk-Schauwecker M, Destephanis D, Hastie E, Grdzelishvili VZ. 2011. Detecting protein-protein interactions in vesicular stomatitis virus using a cytoplasmic yeast two hybrid system. J. Virol. Methods 173:203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456. Mon H, et al. 2009. Analysis of protein interactions with two-hybrid system in cultured insect cells. Anal. Biochem. 392:180–182 [DOI] [PubMed] [Google Scholar]
- 457. Morell M, et al. 2008. Monitoring the interference of protein-protein interactions in vivo by bimolecular fluorescence complementation: the DnaK case. Proteomics 8:3433–3442 [DOI] [PubMed] [Google Scholar]
- 458. Morell M, Espargaro A, Aviles FX, Ventura S. 2007. Detection of transient protein-protein interactions by bimolecular fluorescence complementation: the Abl-SH3 case. Proteomics 7:1023–1036 [DOI] [PubMed] [Google Scholar]
- 459. Morell M, Espargaro A, Aviles FX, Ventura S. 2008. Study and selection of in vivo protein interactions by coupling bimolecular fluorescence complementation and flow cytometry. Nat. Protoc. 3:22–33 [DOI] [PubMed] [Google Scholar]
- 460. Mori K, Ma W, Gething MJ, Sambrook J. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:743–756 [DOI] [PubMed] [Google Scholar]
- 461. Moriyoshi K. 2009. pBT, a novel vector for tetracycline-regulated yeast three-hybrid assay. Nucleic Acids Res. 37:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Morsy M, et al. 2008. Charting plant interactomes: possibilities and challenges. Trends Plant Sci. 13:183–191 [DOI] [PubMed] [Google Scholar]
- 463. Mrowka R, Patzak A, Herzel H. 2001. Is there a bias in proteome research? Genome Res. 11:1971–1973 [DOI] [PubMed] [Google Scholar]
- 464. Mueller SO. 2004. Xenoestrogens: mechanisms of action and detection methods. Anal. Bioanal. Chem. 378:582–587 [DOI] [PubMed] [Google Scholar]
- 465. Müller J, Johnsson N. 2008. Split-ubiquitin and the split-protein sensors: chessman for the endgame. Chembiochem 9:2029–2038 [DOI] [PubMed] [Google Scholar]
- 466. Müller MM, Kries H, Csuhai E, Kast P, Hilvert D. 2010. Design, selection, and characterization of a split chorismate mutase. Protein Sci. 19:1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Nakayama M, Kikuno R, Ohara O. 2002. Protein-protein interactions between large proteins: two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res. 12:1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468. Nateri AS, Riera-Sans L, Da Costa C, Behrens A. 2004. The ubiquitin ligase SCFbw7 antagonizes apoptotic JNK signaling. Science 303:1374–1378 [DOI] [PubMed] [Google Scholar]
- 469. Navlakha S, Kingsford C. 2011. Network archaeology: uncovering ancient networks from present-day interactions. PLoS Comput. Biol. 7:e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Nguyen PQ, Silberg JJ. 2010. A selection that reports on protein-protein interactions within a thermophilic bacterium. Protein Eng. Des. Sel. 23:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471. Nishihara T, Nishikawa J. 2001. Bioassay for endocrine disruptors by using yeast two-hybrid system. Nippon Yakurigaku Zasshi 118:203–210 [DOI] [PubMed] [Google Scholar]
- 472. Noirot-Gros MF, et al. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473. Nooren IM, Thornton JM. 2003. Diversity of protein-protein interactions. EMBO J. 22:3486–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474. Noyes MB, et al. 2008. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 36:2547–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475. Nyfeler B, Michnick SW, Hauri HP. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. U. S. A. 102:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476. Obrdlik P, et al. 2004. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. U. S. A. 101:12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. Ohad N, Shichur K, Yalovsky S. 2007. The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol. 145:1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. Ohad N, Yalovsky S. 2010. Utilizing bimolecular fluorescence complementation (BiFC) to assay protein-protein interactions in plants. Methods Mol. Biol. 655:347–358 [DOI] [PubMed] [Google Scholar]
- 479. O'Hare H, Juillerat A, Dianiskova P, Johnsson K. 2008. A split-protein sensor for studying protein-protein interaction in mycobacteria. J. Microbiol. Methods 73:79–84 [DOI] [PubMed] [Google Scholar]
- 480. Ortega AD, Willers IM, Sala S, Cuezva JM. 2010. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 123:2685–2696 [DOI] [PubMed] [Google Scholar]
- 481. Osborne MA, Dalton S, Kochan JP. 1995. The yeast tribrid system—genetic detection of trans-phosphorylated ITAM-SH2-interactions. Biotechnology (New York) 13:1474–1478 [DOI] [PubMed] [Google Scholar]
- 482. Osborne MA, et al. 1996. The inositol 5′-phosphatase SHIP binds to immunoreceptor signaling motifs and responds to high affinity IgE receptor aggregation. J. Biol. Chem. 271:29271–29278 [DOI] [PubMed] [Google Scholar]
- 483. Ou B, et al. 2011. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4:546–555 [DOI] [PubMed] [Google Scholar]
- 484. Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. 2001. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal. Chem. 73:2516–2521 [DOI] [PubMed] [Google Scholar]
- 485. Ozawa T, Nogami S, Sato M, Ohya Y, Umezawa Y. 2000. A fluorescent indicator for detecting protein-protein interactions in vivo based on protein splicing. Anal. Chem. 72:5151–5157 [DOI] [PubMed] [Google Scholar]
- 486. Ozawa T, Takeuchi TM, Kaihara A, Sato M, Umezawa Y. 2001. Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein-protein interactions in bacteria: improved sensitivity and reduced screening time. Anal. Chem. 73:5866–5874 [DOI] [PubMed] [Google Scholar]
- 487. Palomino A, Herrero P, Moreno F. 2006. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 34:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488. Pan J, et al. 2008. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3:e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Park K, et al. 2007. A split enhanced green fluorescent protein-based reporter in yeast two-hybrid system. Protein J. 26:107–116 [DOI] [PubMed] [Google Scholar]
- 490. Park SH, Raines RT. 2000. Genetic selection for dissociative inhibitors of designated protein-protein interactions. Nat. Biotechnol. 18:847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491. Park SY, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492. Park YW, Wilusz J, Katze MG. 1999. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. U. S. A. 96:6694–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493. Parrish, Gulyas KD, Finley RL., Jr 2006. Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 17:387–393 [DOI] [PubMed] [Google Scholar]
- 494. Parrish JR, et al. 2007. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495. Paschos A, et al. 2011. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun. 79:1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496. Pastor-Satorras R, Smith E, SolÉ RV. 2003. Evolving protein interaction networks through gene duplication. J. Theor. Biol. 222:199–210 [DOI] [PubMed] [Google Scholar]
- 497. Patil A, Nakamura H. 2005. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics 6:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498. Pattanaik S, Werkman JR, Yuan L. 2011. Bimolecular fluorescence complementation as a tool to study interactions of regulatory proteins in plant protoplasts. Methods Mol. Biol. 754:185–193 [DOI] [PubMed] [Google Scholar]
- 499. Paulmurugan R, Gambhir SS. 2007. Combinatorial library screening or developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal. Chem. 79:2346–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500. Paulmurugan R, Gambhir SS. 2003. Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 75:1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501. Paulmurugan R, Umezawa Y, Gambhir SS. 2002. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U. S. A. 99:15608–15613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Paumi CM, et al. 2007. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26:15–25 [DOI] [PubMed] [Google Scholar]
- 503. Pause A, Peterson B, Schaffar G, Stearman R, Klausner RD. 1999. Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc. Natl. Acad. Sci. U. S. A. 96:9533–9538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504. Pawson T, Gish GD, Nash P. 2001. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11:504–511 [DOI] [PubMed] [Google Scholar]
- 505. Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. 2008. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322:1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506. Peeters T, et al. 2006. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. U. S. A. 103:13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507. Pelletier JN, Arndt KM, Plückthun A, Michnick SW. 1999. An in vivo library-versus-library selection of optimized protein-protein interactions. Nat. Biotechnol. 17:683–690 [DOI] [PubMed] [Google Scholar]
- 508. Pelletier JN, Campbell-Valois FX, Michnick SW. 1998. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. U. S. A. 95:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509. Peralta-Yahya P, Carter BT, Lin H, Tao H, Cornish VW. 2008. High-throughput selection for cellulase catalysts using chemical complementation. J. Am. Chem. Soc. 130:17446–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510. Peterson FC, et al. 2010. Structural basis for selective activation of ABA receptors. Nat. Struct. Mol. Biol. 17:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511. Petrascheck M, Castagna F, Barberis A. 2001. Two-hybrid selection assay to identify proteins interacting with polymerase II transcription factors and regulators. Biotechniques 30:296–298, 300,, 302 [DOI] [PubMed] [Google Scholar]
- 512. Phan J, et al. 2002. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277:50564–50572 [DOI] [PubMed] [Google Scholar]
- 513. Phillips SL, Bresnahan WA. 2011. Identification of binary interactions between human cytomegalovirus virion proteins. J. Virol. 85:440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Pierce MM, Raman CS, Nall BT. 1999. Isothermal titration calorimetry of protein-protein interactions. Methods 19:213–221 [DOI] [PubMed] [Google Scholar]
- 515. Piganeau N, Schauer UE, Schroeder R. 2006. A yeast RNA-hybrid system for the detection of RNA-RNA interactions in vivo. RNA 12:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516. Pineda-Lucena A, et al. 2005. A structure-based model of the c-Myc/Bin1 protein interaction shows alternative splicing of Bin1 and c-Myc phosphorylation are key binding determinants. J. Mol. Biol. 351:182–194 [DOI] [PubMed] [Google Scholar]
- 517. Piston DW, Kremers GJ. 2007. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 32:407–414 [DOI] [PubMed] [Google Scholar]
- 518. Plaza S, et al. 2008. Cross-regulatory protein-protein interactions between Hox and Pax transcription factors. Proc. Natl. Acad. Sci. U. S. A. 105:13439–13444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Polte TR, Hanks SK. 1995. Interaction between focal adhesion kinase and Crk-associated tyrosine kinase substrate p130Cas. Proc. Natl. Acad. Sci. U. S. A. 92:10678–10682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520. Pratt MR, Schwartz EC, Muir TW. 2007. Small-molecule-mediated rescue of protein function by an inducible proteolytic shunt. Proc. Natl. Acad. Sci. U. S. A. 104:11209–11214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. Printen JA, Sprague GF., Jr 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522. Ptacek J, et al. 2005. Global analysis of protein phosphorylation in yeast. Nature 438:679–684 [DOI] [PubMed] [Google Scholar]
- 523. Pu J, et al. 2011. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell 2:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Puig O, et al. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229 [DOI] [PubMed] [Google Scholar]
- 525. Puliyappadamba VT, et al. 2011. Antagonists of anaphase-promoting complex (APC)-2-cell cycle and apoptosis regulatory protein (CARP)-1 interaction are novel regulators of cell growth and apoptosis. J. Biol. Chem. 286:38000–38017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526. Putz U, Skehel P, Kuhl D. 1996. A tri-hybrid system for the analysis and detection of RNA-protein interactions. Nucleic Acids Res. 24:4838–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M. 2001. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J. Mol. Biol. 314:1053–1066 [DOI] [PubMed] [Google Scholar]
- 528. Rahim G, Bischof S, Kessler F, Agne B. 2009. In vivo interaction between atToc33 and atToc159 GTP-binding domains demonstrated in a plant split-ubiquitin system. J. Exp. Bot. 60:257–267 [DOI] [PubMed] [Google Scholar]
- 529. Rain JC, et al. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211–215 [DOI] [PubMed] [Google Scholar]
- 530. Rajagopala SV, Casjens S, Uetz P. 2011. The protein interaction map of bacteriophage lambda. BMC Microbiol. 11:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Rajagopala SV, Hughes KT, Uetz P. 2009. Benchmarking yeast two-hybrid systems using the interactions of bacterial motility proteins. Proteomics 9:5296–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532. Raman K. 2010. Construction and analysis of protein-protein interaction networks. Autom. Exp. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533. Ramani AK, Marcotte EM. 2003. Exploiting the co-evolution of interacting proteins to discover interaction specificity. J. Mol. Biol. 327:273–284 [DOI] [PubMed] [Google Scholar]
- 534. Raquet X, Eckert JH, Müller S, Johnsson N. 2001. Detection of altered protein conformations in living cells. J. Mol. Biol. 305:927–938 [DOI] [PubMed] [Google Scholar]
- 535. Ravasi T, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536. Ray MR, et al. 2006. Cyclin G-associated kinase: a novel androgen receptor-interacting transcriptional coactivator that is overexpressed in hormone refractory prostate cancer. Int. J. Cancer 118:1108–1119 [DOI] [PubMed] [Google Scholar]
- 537. Rebois RV, et al. 2008. Combining protein complementation assays with resonance energy transfer to detect multipartner protein complexes in living cells. Methods 45:214–218 [DOI] [PubMed] [Google Scholar]
- 538. Reboul J, et al. 2003. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34:35–41 [DOI] [PubMed] [Google Scholar]
- 539. Reeder MK, Serebriiskii IG, Golemis EA, Chernoff J. 2001. Analysis of small GTPase signalling pathways using p21-activated kinase mutants that selectively couple to Cdc42. J. Biol. Chem. 276:40606–40613 [DOI] [PubMed] [Google Scholar]
- 540. Reguly T, et al. 2006. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Reinders A, et al. 2002. Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 10:763–772 [DOI] [PubMed] [Google Scholar]
- 542. Remy I, Michnick SW. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3:977–979 [DOI] [PubMed] [Google Scholar]
- 543. Remy I, Michnick SW. 1999. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. U. S. A. 96:5394–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544. Remy I, Michnick SW. 2004. Mapping biochemical networks with protein-fragment complementation assays. Methods Mol. Biol. 261:411–426 [DOI] [PubMed] [Google Scholar]
- 545. Remy I, Michnick SW. 2001. Visualisation of biochemical networks in living cells. Proc. Natl. Acad. Sci. U. S. A. 98:7678–7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546. Remy I, Wilson IA, Michnick SW. 1999. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283:990–993 [DOI] [PubMed] [Google Scholar]
- 547. Rodrigue A, Chanal A, Beck K, Muller M, Wu LF. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274:13223–13228 [DOI] [PubMed] [Google Scholar]
- 548. Rojo-Niersbach E, Morley D, Heck S, Lehming N. 2000. A new method for the selection of protein interacions in mammalian cells. Biochem. J. 348:585–590 [PMC free article] [PubMed] [Google Scholar]
- 549. Rossi F, Charlton CA, Blau HM. 1997. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc. Natl. Acad. Sci. U. S. A. 94:8405–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550. Royant A, Noirclerc-Savoye M. 2011. Stabilizing role of glutamic acid 222 in the structure of enhanced green fluorescent protein. J. Struct. Biol. 174:385–390 [DOI] [PubMed] [Google Scholar]
- 551. Rual JF, et al. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173–1178 [DOI] [PubMed] [Google Scholar]
- 552. Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH. 2010. Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol. 10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553. Russ WP, Engelman DM. 1999. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc. Natl. Acad. Sci. U. S. A. 96:863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554. Russell CL, Brown AJ. 2005. Expression of one-hybrid fusions with Staphylococcus aureus lexA in Candida albicans confirms that Nrg1 is a transcriptional repressor and that Gcn4 is a transcriptional activator. Fungal Genet. Biol. 42:676–683 [DOI] [PubMed] [Google Scholar]
- 555. Sabri M, et al. 2011. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent phage Dp-1. J. Bacteriol. 193:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556. Sacco E, et al. 2007. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 104:14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557. Sadowski I, Ma J, Triezenberg S, Ptashne M. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335:563–564 [DOI] [PubMed] [Google Scholar]
- 558. Saka Y, Hagemann A, Piepenburg O, Smith JC. 2007. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development 134:4209–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559. Sambourg L, Thierry-Mieg N. 2011. New insights into protein-protein interaction data lead to increased estimates of the S. cerevisiae interactome size. BMC Bioinformatics 11:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560. Sandrock K, Bartsch I, Busse A, Busse E, Zieger B. 2011. Characterization of human septin interactions. Biol. Chem. 392:751–761 [DOI] [PubMed] [Google Scholar]
- 561. Santos MA, Cheesman C, Costa V, Moradas-Ferreira P, Tuite MF. 1999. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 31:937–947 [DOI] [PubMed] [Google Scholar]
- 562. Sato S, et al. 2007. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 14:207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563. Schittenhelm RB, Althoff F, Heidmann S, Lehner CF. 2010. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromere is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 123:3768–3779 [DOI] [PubMed] [Google Scholar]
- 564. Schneider D, Engelman DM. 2003. GALLEX, a measurement of heterologous association of transmembrane helices in a biological membrane. J. Biol. Chem. 278:3105–3111 [DOI] [PubMed] [Google Scholar]
- 565. Schönhofer-Merl S, Torres-Ruiz RA. 2010. The Sos-recruitment system as a tool to analyze cellular localization of plant proteins: membrane localization of Arabidopsis thaliana PEPINO/PATICCINO2. Mol. Genet. Genomics 283:439–449 [DOI] [PubMed] [Google Scholar]
- 566. Schreiber SL. 1991. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251:283–287 [DOI] [PubMed] [Google Scholar]
- 567. Schuck P. 1997. Reliable determination of binding affinity and kinetics using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 8:498–502 [DOI] [PubMed] [Google Scholar]
- 568. Schüler A, Bornberg-Bauer E. 2011. The evolution of protein interaction networks. Methods Mol. Biol. 696:273–289 [DOI] [PubMed] [Google Scholar]
- 569. Schwarz T, Sohn C, Kaiser B, Jensen ED, Mansky KC. 2010. The 19S proteosomal lid subunit POH1 enhances the transcriptional activation by Mitf in osteoclasts. J. Cell. Biochem. 109:967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570. Scognamiglio PL, et al. 2011. Discovery of small peptide antagonists of PED/PEA15-D4α interaction from simplified combinatorial libraries. Chem. Biol. Drug Des. 77:319–327 [DOI] [PubMed] [Google Scholar]
- 571. Scott MS, Barton GJ. 2007. Probabilistic prediction and ranking of human protein-protein interactions. BMC Bioinformatics 8:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572. Selyunin AS, et al. 2011. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469:107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573. SenGupta DJ, Lin H, Goldberg SD, Mahal JJ, Cornish VW. 2004. Correlation between catalytic efficiency and the transcription read-out in chemical complementation: a general assay for enzyme catalysis. Biochemistry 43:3570–3581 [DOI] [PubMed] [Google Scholar]
- 574. SenGupta DJ, Wickens M, Fields S. 1999. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA 5:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575. SenGupta DJ, et al. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Seok HY, et al. 2010. Rice ternary MADS protein complexes containing class B MADS heterodimer. Biochem. Biophys. Res. Commun. 401:598–604 [DOI] [PubMed] [Google Scholar]
- 577. Serebriiskii IG. 2010. Yeast two-hybrid system for studying protein-protein interactions—stage 1: construction and characterization of a bait protein. Cold Spring Harb. Protoc. 2010:pdb.prot5429 [DOI] [PubMed] [Google Scholar]
- 578. Serebriiskii IG, et al. 2005. A combined yeast/bacteria two-hybrid system: development and evaluation. Mol. Cell. Proteomics 4:819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579. Serebriiskii IG, Golemis EA. 2001. Two-hybrid system and false positives. Approaches to detection and elimination. Methods Mol. Biol. 177:123–134 [DOI] [PubMed] [Google Scholar]
- 580. Serebriiskii IG, Khazak V, Golemis EA. 1999. A two-hybrid dual bait system to discriminate specificity of protein interactions. J. Biol. Chem. 274:17080–17087 [DOI] [PubMed] [Google Scholar]
- 581. Serebriiskii IG, et al. 2002. Detection of peptides, proteins, and drugs that selectively interact with protein targets. Genome Res. 12:1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582. Shamu CE, Walter P. 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028–3039 [PMC free article] [PubMed] [Google Scholar]
- 583. Sharan R, et al. 2005. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. U. S. A. 102:1974–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584. Sheerin DJ, et al. 2011. Inter- and intra-molecular interactions of Arabidopsis thaliana DELLA protein RGL1. Biochem. J. 435:629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585. Shi Y, et al. 2008. IL-6-induced stimulation of c-myc translation in multiple myeloma cells is mediated by myc internal ribosome entry site function and the RNA-binding protein hnRNP A1. Cancer Res. 68:10215–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586. Shi YY, Miller GA, Qian H, Bomsztyk K. 2006. Free-energy distribution of binary protein-protein binding suggests cross-species interactome differences. Proc. Natl. Acad. Sci. U. S. A. 103:11527–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587. Shibasaki S, Sakata K, Ishii J, Kondo A, Ueda M. 2008. Development of a yeast protein fragment complementation assay (PCA) system using dihydrofolate reductase (DHFR) with specific activities. Appl. Microbiol. Biotechnol. 80:735–745 [DOI] [PubMed] [Google Scholar]
- 588. Shih AM, Shin OH. 2011. Interactions among the SNARE proteins and complexin analyzed by a yeast four-hybrid assay. Anal. Biochem. 416:107–111 [DOI] [PubMed] [Google Scholar]
- 589. Shih HM, et al. 1996. A positive genetic selection for disrupting protein-protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc. Natl. Acad. Sci. U. S. A. 93:13896–13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590. Shimomura O, Johnsson F, Saiga Y. 1962. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 59:223–239 [DOI] [PubMed] [Google Scholar]
- 591. Shin CJ, Wong S, Davis MJ, Ragan MA. 2009. Protein-protein interaction as a predictor of subcellular location. BMC Syst. Biol. 3:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592. Shioda T, Andriole S, Yahata T, Isselbacher KJ. 2000. A green fluorescent protein-reporter mammalian two-hybrid system with extrachromosomal maintenance of a prey expression plasmid: application to interaction screening. Proc. Natl. Acad. Sci. U. S. A. 97:5220–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Shrivastav S, et al. 2008. Human immunodeficiency virus (HIV)-1 viral protein R suppresses transcriptional activity of peroxisome proliferator-activated receptor gamma and inhibits adipocyte differentiation: implications for HIV-associated lipodystrophy. Mol. Endocrinol. 22:234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594. Shui X, et al. 2011. A mammalian two-hybrid system-based assay for small-molecular HIV fusion inhibitors targeting gp41. Antiviral Res. 90:54–63 [DOI] [PubMed] [Google Scholar]
- 595. Shyu YJ, et al. 2008. Visualization of protein interactions in living Caenorhabditis elegans using bimolecular fluorescence complementation analysis. Nat. Protoc. 3:588–596 [DOI] [PubMed] [Google Scholar]
- 596. Shyu YJ, Liu H, Deng X, Hu C-D. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 40:61–66 [DOI] [PubMed] [Google Scholar]
- 597. Sieber P, Petrascheck M, Barberis A, Schneitz K. 2004. Organ polarity in Arabidopsis, NOZZLE physically interacts with members of the YABBY family. Plant Physiol. 135:2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598. Siep M, et al. 2004. Basic helix-loop-helix transcription factor Tcfl5 interacts with the calmegin gene promoter in mouse spermatogenesis. Nucleic Acids Res. 32:6425–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599. Sieweke MH. 2000. Detection of transcription factor partners with a yeast one hybrid screen. Methods Mol. Biol. 130:59–77 [DOI] [PubMed] [Google Scholar]
- 600. Sieweke MH, Tekotte H, Frampton J, Graf T. 1996. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85:49–60 [DOI] [PubMed] [Google Scholar]
- 601. Simonis N, et al. 2009. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods 6:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602. Singh K, Kang PJ, Park HO. 2008. The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 105:1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603. Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. 2011. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol. Microbiol. 79:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604. Skrabanek L, Saini HK, Bader GD, Enright AJ. 2008. Computational prediction of protein-protein interactions. Mol. Biotechnol. 38:1–17 [DOI] [PubMed] [Google Scholar]
- 605. Snider J, et al. 2010. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat. Protoc. 5:1281–1293 [DOI] [PubMed] [Google Scholar]
- 606. Söderberg O, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3:995–1000 [DOI] [PubMed] [Google Scholar]
- 607. Spektor TM, Rice JC. 2009. Identification and characterization of posttranslational modification-specific binding proteins in vivo by mammalian tethered catalysis. Proc. Natl. Acad. Sci. U. S. A. 106:14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608. Spotts JM, Dolmetsch RE, Greenberg ME. 2002. Time-lapse imaging of a dynamic phosphorylation-dependent protein-protein interaction in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 99:15142–15147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609. Sprinzak E, Margalit H. 2001. Correlated sequence-signatures as markers of protein-protein interaction. J. Mol. Biol. 311:681–692 [DOI] [PubMed] [Google Scholar]
- 610. Srivastav RK, et al. 2011. Monitoring protein-protein interactions in mammalian cells by trans-SUMOylation. Biochem. J. 438:495–503 [DOI] [PubMed] [Google Scholar]
- 611. Stagljar I, Korostensky C, Johnsson N, te Heesen S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 95:5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612. Stanyon CA, et al. 2004. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 5:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613. Starling AL, et al. 2003. Evaluation of alternative reporter genes for the yeast two-hybrid system. Genet. Mol. Res. 2:124–135 [PubMed] [Google Scholar]
- 614. Staudinger J, Perry M, Elledge SJ, Olson EN. 1993. Interactions among vertebrate helix-loop-helix proteins in yeast using the two-hybrid system. J. Biol. Chem. 268:4608–4611 [PubMed] [Google Scholar]
- 615. Stefan E, et al. 2007. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. U. S. A. 104:16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616. Stellberger T, et al. 2010. Improving the yeast two-hybrid system with permutated fusions proteins: the varicella zoster virus interactome. Proteome Sci. 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617. Stelzl U, et al. 2005. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122:957–968 [DOI] [PubMed] [Google Scholar]
- 618. Stern S, Tanaka M, Herr W. 1989. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624–630 [DOI] [PubMed] [Google Scholar]
- 619. Stiel AC, et al. 2008. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescent nanoscopy. Biophys. J. 95:2989–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620. Stiel AC, et al. 2007. 1.8 A bright structure of the reversible switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem. J. 402:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621. Strauch E-M, Georgiou G. 2007. A bacterial two-hybrid system based on the twin-arginine transporter pathway of E. coli. Protein Sci. 16:1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622. Stumpf CR, Opperman L, Wickens M. 2008. Chapter 14. Analysis of RNA-protein interactions using a yeast three-hybrid system. Methods Enzymol. 449:295–315 [DOI] [PubMed] [Google Scholar]
- 623. Stynen B, Van Dijck P, Tournu H. 2010. A CUG codon adapted two-hybrid system for the pathogenic fungus Candida albicans. Nucleic Acids Res. 38:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624. Subach FV, et al. 2010. Red fluorescent protein with reversibly photoswitchable absorbance for photochromic FRET. Chem. Biol. 17:745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625. Subramaniam R, Desveaux D, Spickler C, Michnick SW, Brisson N. 2001. Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19:769–772 [DOI] [PubMed] [Google Scholar]
- 626. Sung MK, Huh WK. 2007. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interactions in Saccharomyces cerevisiae. Yeast 24:767–775 [DOI] [PubMed] [Google Scholar]
- 627. Suzuki H, et al. 2001. Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 11:1758–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628. Suzuki H, et al. 2009. Differential expression and affinities of Arabidopsis gibberellin receptors can explain variation in phenotypes of multiple knock-out mutants. Plant J. 60:48–55 [DOI] [PubMed] [Google Scholar]
- 629. Tafelmeyer P, Johnsson N, Johnsson K. 2004. Transforming a (beta/alpha)8-barrel enzyme into a split-protein sensor through directed evolution. Chem. Biol. 11:681–689 [DOI] [PubMed] [Google Scholar]
- 630. Takahashi Y, Tojo T, Nagahora S, Yamazaki K-I. 2007. Direct determination of estrogenic and antiestrogenic activities using an enhanced plant two-hybrid system. J. Agric. Food Chem. 55:2923–2929 [DOI] [PubMed] [Google Scholar]
- 631. Takemoto D, et al. 2011. Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc. Natl. Acad. Sci. U. S. A. 108:2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632. Tao H, Peralta-Yahya P, Lin H, Cornish VW. 2006. Optimized design and synthesis of chemical dimerizer substrates for detection of glycosynthase activity via chemical complementation. Bioorg. Med. Chem. 14:6940–6953 [DOI] [PubMed] [Google Scholar]
- 633. Tarassov K, et al. 2008. An in vivo map of the yeast protein interactome. Science 320:1465–1470 [DOI] [PubMed] [Google Scholar]
- 634. Tavassoli A, Benkovic SJ. 2005. Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew. Chem. Int. Ed. Engl. 44:2760–2763 [DOI] [PubMed] [Google Scholar]
- 635. Tavassoli A, et al. 2008. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem. Biol. 3:757–764 [DOI] [PubMed] [Google Scholar]
- 636. Tavassoli P, et al. 2010. TAF1 differentially enhances androgen receptor transcriptional activity via its N-terminal kinase and ubiquitin-activating and -conjugating domains. Mol. Endocrinol. 24:696–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637. Teeri TH, et al. 1989. Gene fusions to lacZ reveal new expression patterns of chimeric genes in transgenic plants. EMBO J. 8:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638. Thibodeaux GN, Cowmeadow R, Umeda A, Zhang Z. 2009. A tetracycline repressor-based mammalian two-hybrid system to detect protein-protein interactions in vivo. Anal. Biochem. 386:129–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639. Tirode F, et al. 1997. A conditionally expressed third partner stabilizes or prevents the formation of a transcriptional activator in a three-hybrid system. J. Biol. Chem. 272:22995–22999 [DOI] [PubMed] [Google Scholar]
- 640. Tirosh I, Barkai N. 2005. Computational verification of protein-protein interactions by orthologous co-expression. BMC Bioinformatics 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641. Titz B, et al. 2008. The binary protein interactome of Treponema pallidum—the syphilis spirochete. PLoS One 3:e2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642. Titz B, et al. 2006. Transcriptional activators in yeast. Nucleic Acids Res. 34:955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643. Tojo T, Tsuda K, Wada TS, Yamazaki K. 2006. A simple and extremely sensitive system for detecting estrogenic activity using transgenic Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 64:106–114 [DOI] [PubMed] [Google Scholar]
- 644. Topper JN, et al. 1998. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 95:9506–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645. Triezenberg SJ. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5:190–196 [DOI] [PubMed] [Google Scholar]
- 646. Truong K, Ikura M. 2001. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 11:573–578 [DOI] [PubMed] [Google Scholar]
- 647. Ueno A, et al. 2011. Toxoplasma gondii: a bradyzoite-specific DnaK-tetratricopeptide repeat (DNAK-TPR) protein interacts with p23 co-chaperone protein. Exp. Parasitol. 127:795–803 [DOI] [PubMed] [Google Scholar]
- 648. Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. 1997. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science 277:1310–1313 [DOI] [PubMed] [Google Scholar]
- 649. Uetz P, et al. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242 [DOI] [PubMed] [Google Scholar]
- 650. Uetz P, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627 [DOI] [PubMed] [Google Scholar]
- 651. Ullmann A, Danchin A. 1983. Advances in cyclic nucleotide research, p 1–53 Raven Press, New York, NY [Google Scholar]
- 652. Urano E, et al. 2011. Novel postentry inhibitor of human immunodeficiency virus type 1 replication screened by yeast membrane-associated two-hybrid system. Antimicrob. Agents Chemother. 55:4251–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653. Urech DM, Lichtlen P, Barberis A. 2003. Cell growth selection system to detect extracellular and transmembrane protein interactions. Biochim. Biophys. Acta 1622:117–127 [DOI] [PubMed] [Google Scholar]
- 654. Ushioda R, et al. 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321:569–572 [DOI] [PubMed] [Google Scholar]
- 655. Vallet-Gely I, Donovan KE, Fang R, Joung JK, Dove SL. 2005. Repression of phase-variable cup gene expression by H-NS-like proteins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 102:11082–11087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656. Van Criekinge W, Beyaert R. 1999. Yeast two-hybrid: state of the art. Biol. Proced. Online 2:1–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657. Van del Belt K, Verheyen R, Witters H. 2003. Comparison of vitellogenin responses in zebrafish and rainbow trout following exposure to environmental estrogens. Ecotoxicol. Environ. Saf. 56:271–281 [DOI] [PubMed] [Google Scholar]
- 658. Van Leene J, Boruc J, De Jaeger G, Russinova E, De Veylder L. 2011. A kaleidoscopic view of the Arabidopsis core cell cycle interactomes. Trends Plant Sci. 16:141–150 [DOI] [PubMed] [Google Scholar]
- 659. Varshavsky A. 2007. Targeting the absence: homozygous DNA deletions as immutable signposts for cancer therapy. Proc. Natl. Acad. Sci. U. S. A. 104:14935–14940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660. Varshavsky A. 1996. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U. S. A. 93:12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661. Vasilescu J, Guo X, Kost J. 2004. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics 4:3845–3854 [DOI] [PubMed] [Google Scholar]
- 662. Vazquez A, Rual JF, Venkatesan K. 2011. Quality control methodology for high-throughput protein-protein interaction screening. Methods Mol. Biol. 781:279–294 [DOI] [PubMed] [Google Scholar]
- 663. Venkatesan K, et al. 2009. An empirical framework for binary interactome mapping. Nat. Methods 6:83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664. Venkatesh GR, et al. 2010. BglJ-RcsB heterodimers relieve repression of the Escherichia coli Bgl operon by H-NS. J. Bacteriol. 192:6456–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665. Vermeirssen V, et al. 2007. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 17:1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666. Vermeirssen V, et al. 2007. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat. Methods 4:659–664 [DOI] [PubMed] [Google Scholar]
- 667. Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 93:10315–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668. Vidal M, Endoh H. 1999. Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol. 17:374–381 [DOI] [PubMed] [Google Scholar]
- 669. Vidalain PO, Boxem M, Ge H, Li S, Vidal M. 2004. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32:363–370 [DOI] [PubMed] [Google Scholar]
- 670. Vignols F, Bréhélin C, Surdin-Kerjan Y, Thomas D, Meyer Y. 2005. A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins. Proc. Natl. Acad. Sci. U. S. A. 102:16729–16734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 671. Vijay-Kumar S, Bugg CE, Cook WJ. 1987. Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194:531–544 [DOI] [PubMed] [Google Scholar]
- 672. Villalobos V, Naik S, Piwnica-Worms D. 2008. Detection of protein-protein interactions in live cells and animals with split firefly luciferase protein fragment complementation. Methods Mol. Biol. 439:339–352 [DOI] [PubMed] [Google Scholar]
- 673. Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205–214 [DOI] [PubMed] [Google Scholar]
- 674. Vollmeister E, et al. 2009. Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA 15:2206–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675. von Degenfeld G, Wehrman TS, Hammer MM, Blau HM. 2007. A universal technology for monitoring G-protein-coupled receptor activation in vitro and noninvasively in live animals. FASEB J. 21:3819–3826 [DOI] [PubMed] [Google Scholar]
- 676. von Mering C, et al. 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417:399–403 [DOI] [PubMed] [Google Scholar]
- 677. Voss TC, Demarco IA, Day RN. 2005. Quantitative imaging of protein interactions in the cell nucleus. Biotechniques 38:413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678. Waadt R, et al. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56:505–516 [DOI] [PubMed] [Google Scholar]
- 679. Wafa LA, et al. 2003. Isolation and identification of l-dopa decarboxylase as a protein that binds to and enhances transcriptional activity of the androgen receptor using the repressed transactivator yeast two-hybrid system. Biochem. J. 375:373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680. Wagner A. 2001. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol. Biol. Evol. 18:1283–1292 [DOI] [PubMed] [Google Scholar]
- 681. Walhout AJ, et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12:1952–1958 [DOI] [PubMed] [Google Scholar]
- 682. Walhout AJ, et al. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287:116–122 [DOI] [PubMed] [Google Scholar]
- 683. Walhout AJ, Vidal M. 1999. A genetic strategy to eliminate self-activator baits prior to high-throughput yeast two-hybrid screens. Genome Res. 9:1128–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684. Walter M, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40:428–438 [DOI] [PubMed] [Google Scholar]
- 685. Wang H, et al. 2009. A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol. Cell. Proteomics 8:1361–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686. Wang MM, Reed RR. 1993. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364:121–126 [DOI] [PubMed] [Google Scholar]
- 687. Wang S, Shepard JR, Shi H. 2010. An RNA-based transcription activator derived from an inhibitory aptamer. Nucleic Acids Res. 38:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688. Wang TY, He F, Hu QW, Zhang Z. 2011. A predicted protein-protein interaction network of the filamentous fungus Neurospora crassa. Mol. Biosyst. 7:2278–2285 [DOI] [PubMed] [Google Scholar]
- 689. Wang Y, et al. 2010. Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome Res. 9:6665–6677 [DOI] [PubMed] [Google Scholar]
- 690. Wang Z-H, et al. 2011. A transferable heterogeneous two-hybrid system in Escherichia coli based on polyhydroxyalkanoates synthesis regulatory protein PhaR. Microbiol. Cell Fact. 10:21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691. Waraho D, DeLisa MP. 2009. Versatile selection technology for intracellular protein-protein interactions mediated by a unique bacterial hitchhiker transport mechanism. Proc. Natl. Acad. Sci. U. S. A. 106:3692–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692. Wasserman T, et al. 2010. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21:117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693. Watson MA, Buckholz R, Weiner MP. 1996. Vectors encoding alternative antibiotic resistance for use in the yeast two-hybrid system. Biotechniques 21:255–259 [DOI] [PubMed] [Google Scholar]
- 694. Weber M, et al. 2010. Mso1p regulates membrane fusion through interactions with the putative N-peptide-binding area in Sec1p domain 1. Mol. Biol. Cell 21:1362–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695. Weber-Boyvat M, Aro N, Chernov KG, Nyman T, Jäntti J. 2011. Sec1p and Mso1p C-terminal tails cooperate with the SNAREs and Sec4p in polarized exocytosis. Mol. Biol. Cell 22:230–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696. Webster NJ, Green S, Jin JR, Chambon P. 1988. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell 54:199–207 [DOI] [PubMed] [Google Scholar]
- 697. Weghuber J, et al. 2010. Detection of protein-protein interactions in the live cell plasma membrane by quantifying prey redistribution upon bait micropatterning. Methods Enzymol. 472:133–151 [DOI] [PubMed] [Google Scholar]
- 698. Wehr MC, et al. 2006. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods 3:985–993 [DOI] [PubMed] [Google Scholar]
- 699. Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. 2002. Protein-protein interactions monitored in mammalian cells via complementation of beta-lactamase enzyme fragments. Proc. Natl. Acad. Sci. U. S. A. 99:3469–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700. Wehrman TS, et al. 2006. A system for quantifying dynamic protein interactions defines a role for Herceptin in modulating ErbB2 interactions. Proc. Natl. Acad. Sci. U. S. A. 103:19063–19068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701. Weinthal D, Tzfira T. 2009. Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 14:59–63 [DOI] [PubMed] [Google Scholar]
- 702. Wiesner C, Hoeth M, Binder BR, de Martin R. 2002. A functional screening assay for the isolation of transcription factors. Nucleic Acids Res. 30:e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703. Wiles AM, et al. 2010. Building and analyzing protein interactome networks by cross-species comparisons. BMC Syst. Biol. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704. Wilhelm JE, Vale RD. 1996. A one-hybrid system for detecting RNA-protein interactions. Genes Cells 1:317–323 [DOI] [PubMed] [Google Scholar]
- 705. Wilson T, Hastings JW. 1998. Bioluminescence. Annu. Rev. Cell Dev. Biol. 14:197–230 [DOI] [PubMed] [Google Scholar]
- 706. Wilson TE, Fahrner TJ, Johnston M, Milbrandt J. 1991. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252:1296–1300 [DOI] [PubMed] [Google Scholar]
- 707. Wingler LM, Cornish VW. 2011. A library approach for the discovery of customized yeast three-hybrid counter selections. Chembiochem 12:715–717 [DOI] [PubMed] [Google Scholar]
- 708. Wittke S, Lewke N, Müller S, Johnsson N. 1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10:2519–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709. Wolf F, Li W, Li F, Li CY. 2011. Novel luciferase-based reporter system to monitor activation of ErbB2/Her2/neu pathway noninvasively during radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 79:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710. Wu G, et al. 2008. Small molecule targeting the Hec1/nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 68:8393–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711. Wu-Baer F, Ludwig T, Baer R. 2010. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol. Cell. Biol. 30:2787–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712. Wurm JP, et al. 2010. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 38:2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 713. Wurster SE, Bida JP, Her YF, Maher L. 2009. Characterization of anti-NF-kappaB RNA aptamer-binding specificity in vitro and in the yeast three-hybrid system. Nucleic Acids Res. 37:6214–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714. Wurster SE, Maher L. 2010. Selections that optimize RNA display in the yeast three-hybrid system. RNA 16:253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715. Xie Z, Hu S, Qian J, Blackshaw S, Zhu H. 2011. Systematic characterization of protein-DNA interactions. Cell. Mol. Life Sci. 68:1657–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716. Xu CW, Mendelsohn AR, Brent R. 1997. Cells that register logical relationships among proteins. Proc. Natl. Acad. Sci. U. S. A. 94:12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717. Yang H, et al. 2011. Insight into bacterial virulence mechanisms against host immune response via the Yersinia pestis-human protein-protein interaction network. Infect. Immun. 79:4413–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718. Yang J, Salam AA, Rosen BP. 2011. Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Mol. Microbiol. 79:872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719. You X, et al. 2006. Intracellular protein interaction mapping with FRET hybrids. Proc. Natl. Acad. Sci. U. S. A. 103:18458–18463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720. Young K, et al. 1998. Identication of a calcium channel modulator using a high throughput yeast two-hybrid screen. Nat. Biotechnol. 16:946–950 [DOI] [PubMed] [Google Scholar]
- 721. Yu H, et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722. Yu H, et al. 2004. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 14:1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 723. Yu H, et al. 2011. Next-generation sequencing to generate interactome datasets. Nat. Methods 8:478–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724. Yu X, Ivanic J, Memisevic V, Wallqvist A, Reifman J. 2011. Categorizing biases in high-confidence high-throughput protein-protein interaction data sets. Mol. Cell. Proteomics 10:M111.012500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725. Yun CW, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. 1997. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240:287–292 [DOI] [PubMed] [Google Scholar]
- 726. Zhang B, et al. 2011. An in silico approach for the discovery of CDK5/p25 interaction inhibitors. Biotechnol. J. 6:871–881 [DOI] [PubMed] [Google Scholar]
- 727. Zhang J, Lautar S. 1996. A yeast three-hybrid method to clone ternary protein complex components. Anal. Biochem. 242:68–72 [DOI] [PubMed] [Google Scholar]
- 728. Zhang L, et al. 2009. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J. Proteome Res. 8:4311–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729. Zhang S, Ma C, Chalfie M. 2004. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell 119:137–144 [DOI] [PubMed] [Google Scholar]
- 730. Zhang X, Wong SM. 2011. Development of a cell sorting procedure to increase the sensitivity of detection of protein-protein interactions in plant protoplasts. J. Virol. Methods 173:347–352 [DOI] [PubMed] [Google Scholar]
- 731. Zhang Y, Gao P, Yuan JS. 2010. Plant protein-protein interaction network and interactome. Curr. Genomics 11:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732. Zhao X, Xu JR. 2007. A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63:881–894 [DOI] [PubMed] [Google Scholar]
- 733. Zhao XM, Zhang XW, Tang WH, Chen L. 2009. FPPI: Fusarium graminearum protein-protein interaction database. J. Proteome Res. 8:4714–4721 [DOI] [PubMed] [Google Scholar]
- 734. Zhong J, Zhang H, Stanyon CA, Tromp G, Finley RLJ. 2003. A strategy for constructing large protein interaction maps using the yeast two-hybrid system: regulated expression arrays and two-phase mating. Genome Res. 13:2691–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735. Zhong Q, et al. 2009. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 5:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736. Zhou J, Lin J, Zhou C, Deng X, Xia B. 2011. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta Biochim. Biophys. Sin. 43:239–244 [DOI] [PubMed] [Google Scholar]
- 737. Zhou J, et al. 2011. Krüppel-like factor 15 activates hepatitis B virus gene expression and replication. Hepatology 54:109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738. Zhu H, et al. 2001. Global analysis of protein activities using proteome chips. Science 293:2101–2105 [DOI] [PubMed] [Google Scholar]
- 739. Zhu LJ, et al. 2011. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39:D111–D117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740. Zilian E, Maiss E. 2011. An optimized mRP-based bimolecular fluorescence complementation system for the detection of protein-protein interactions in planta. J. Virol. Methods 174:158–165 [DOI] [PubMed] [Google Scholar]
- 741. Zinzalla G, Thurston DE. 2009. Targeting protein-protein interactions or therapeutic intervention: a challenge for the future. Future Med. Chem. 1:65–93 [DOI] [PubMed] [Google Scholar]
- 742. Zolghadr K, et al. 2008. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol. Cell. Proteomics 7:2279–2287 [DOI] [PubMed] [Google Scholar]
- 743. Zotenko E, Mestre J, O'Leary DP, Przytycka TM. 2008. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 4:e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744. Zwart W, et al. 2007. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J. 26:3534–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]