Abstract
The RNA-binding protein hnRNP Q has been implicated in neuronal mRNA metabolism. Here, we show that knockdown of hnRNP Q increased neurite complexity in cultured rat cortical neurons and induced filopodium formation in mouse neuroblastoma cells. Reexpression of hnRNP Q1 in hnRNP Q-depleted cells abrogated the morphological changes of neurites, indicating a specific role for hnRNP Q1 in neuronal morphogenesis. A search for mRNA targets of hnRNP Q1 identified functionally coherent sets of mRNAs encoding factors involved in cellular signaling or cytoskeletal regulation and determined its preferred binding sequences. We demonstrated that hnRNP Q1 bound to a set of identified mRNAs encoding the components of the actin nucleation-promoting Cdc42/N-WASP/Arp2/3 complex and was in part colocalized with Cdc42 mRNA in granules. Using subcellular fractionation and immunofluorescence, we showed that knockdown of hnRNP Q reduced the level of some of those mRNAs in neurites and redistributed their encoded proteins from neurite tips to soma to different extents. Overexpression of dominant negative mutants of Cdc42 or N-WASP compromised hnRNP Q depletion-induced neurite complexity. Together, our results suggest that hnRNP Q1 may participate in localization of mRNAs encoding Cdc42 signaling factors in neurites, and thereby may regulate actin dynamics and control neuronal morphogenesis.
INTRODUCTION
In polarized cells, mRNA targeting coupled to local translation at proper subcellular sites is important for transient and spatially restricted protein expression. In neuronal systems, such a mechanism is involved in both directional growth of axons and synaptic plasticity of dendrites (49). Moreover, translation of localized mRNAs in dendrites can be regulated by neuronal activity. Much evidence has indicated that participation of RNA-binding proteins in local translation of mRNAs is required for neuronal development and function (17). One example is the zip code binding protein 1 (ZBP1), which regulates dendritic arborization and directional axon growth via its involvement in β-actin mRNA transport and translation (10, 43, 53).
Previous reports have indicated that heterogeneous nuclear riboprotein Q (hnRNP Q) has multiple functions in mRNA metabolism, including regulation of precursor mRNA splicing as well as editing, stability control, transport, and translation of mRNA (4, 5, 8, 24, 52). Evidence supporting the role of hnRNP Q in cytoplasmic mRNA trafficking comes from its association with synaptotagmins (membrane-trafficking proteins) and its presence in RNA granules associated with the kinesin family motor protein KIF5 (23, 39). Moreover, hnRNP R, which is related to hnRNP Q, functions together with the survival motor neuron (SMN) protein in transporting β-actin mRNA to axons and growth cones (48). Axonal transport and localized translation of β-actin mRNA are important for axonal growth. Accordingly, depletion of hnRNP R impairs axon growth and elongation of motor neurons, as does SMN deficiency, suggesting a physiological function for hnRNP R in neurons (14). Because hnRNP Q also interacts with SMN (40), it is possible that hnRNP Q also participates in cytoplasmic mRNA trafficking and even localized translation of mRNAs in neurons.
In this study, we demonstrated that knockdown of hnRNP Q in neurons induced dysregulated neurite formation. A search for candidate hnRNP Q targets identified a set of mRNAs encoding factors involved in Cdc42-dependent actin polymerization, suggesting a potential mechanism for hnRNP Q-regulated neuronal morphogenesis. The Cdc42 signaling pathway initially induces a conformational change in the neuronal Wiskott-Aldrich syndrome protein (N-WASP), which subsequently interacts with and activates the Arp2/3 complex (Arp is actin-related protein) (16). The Arp2/3 complex consists of seven subunits, is involved in nucleating actin polymerization, and drives directional cell migration (16, 28). Like β-actin mRNA, the transcripts of all Arp2/3 complex subunits are localized at the leading edge of migrating or elongating cells, which likely allows efficient actin polymerization in cell protrusions (38). However, it is not clear how mRNAs encoding the Arp2/3 components are effectively localized. To address this issue, we went on to characterize the potential role of hnRNP Q in localized expression of the Cdc42/N-WASP/Arp2/3 signaling factors.
MATERIALS AND METHODS
Cell culture and transfection.
Mouse neuroblastoma N2A cells were grown at 37°C in Dulbecco's modified Eagle medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine. Differentiation of N2A cells was induced by serum deprivation (51). Transfection was performed using Lipofectamine 2000 (Invitrogen). For hnRNP Q knockdown, N2A cells cultured in six-well plates were transfected with 80 pmol/well of siQ#1 (sense, GCAAGCAGCAAAGAAUCAAAUGUAU; antisense, AUACAUUUGAUUCUUUGCUGCUUGC) or siQ#2 (sense, GGCUACAGAACAUGUUAAUGGAAAU; antisense, AUUUCCAUUAACAUGUUCUGUAGCC) small interfering RNAs (siRNAs) (Stealth RNAi Syncrip-MSS226121 and MSS226123, respectively; Invitrogen). The control siRNA (siL; sense, GGAUUUCGAGUCGUCUUAAUGUAUA; antisense, UAUACAUUAAGACGACUCGAAAUCC) was complementary to firefly luciferase (Invitrogen). For rescue experiments, siRNA and the expression vector of hnRNP Q1, Q2, Q3, or Q1ΔC (8) were cotransfected into N2A cells. The expression plasmids of all wild-type and mutant Cdc42 and N-WASP were obtained from Jenn-Yah Yu (National Yang-Ming University, Taipei, Taiwan) and Sue Lin-Chao (Academia Sinica, Taipei, Taiwan). After transfection for 24 h, cells were incubated in serum-free medium to allow differentiation for 24 h. Differentiated cells were fixed or harvested for subsequent analysis.
For primary cortical neuron culture, coverslips were precoated with 1 mg/ml poly-d-lysine. Cortical neurons were isolated from rat embryos (day 18 to 19) and seeded at ∼1 × 105 cells/well in 24-well plates with coverslips. Cells were cultured in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), 0.5 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. Transfection was performed using ICAFectin 441R (In Cell Art) per the manufacturer's instructions. Each 100-μl transfection mixture contained 0.5 μg pEGFP-C1, 15 pmol siRNA, and 1.3 μl ICAFectin 441R. After transfection for 3 days, cells were fixed and analyzed for immunofluorescence and morphology.
To induce cell depolarization, 10 mM KCl or 0.5 mM sodium glutamate was used to treat cortical neurons and N2A cells for 5 min or 15 min, respectively.
Antibodies.
Polyclonal antibodies against hnRNP Q1, green fluorescent protein (GFP), Cdc42, and Arpc1a and Arp2 were purchased from AnaSpec, Invitrogen, BioVision, and GeneTex, respectively. Polyclonal TAR DNA-binding protein 43 (TDP-43) and N-WASP antisera were gifts from C.-K. James Shen and Sue Lin-Chao (Academia Sinica, Taipei, Taiwan), respectively. Mouse monoclonal actin, fragile X mental retardation protein (FMRP), MAP2, and Tau1 antibodies were purchased from Millipore. Mouse monoclonal hnRNP Q/R and α-tubulin antibodies were purchased from Sigma and Thermo Scientific, respectively.
Indirect immunofluorescence and image analysis.
Cortical neurons were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde and 4% sucrose for 10 min at room temperature and then permeabilized in PBS containing 0.1% Triton X-100 for 1 min followed by ice-cold methanol for 10 min at 4°C. N2A cells were fixed in PBS containing 4% paraformaldehyde and permeabilized in PBS containing 0.1% Triton X-100 for 5 min. After permeabilization, cells were blocked in PBS containing 10% bovine serum albumin (BSA) for 1 h and then incubated with primary antibodies at room temperature for 2 h. After being washed, cells were incubated with Alexa Fluor 488- or 568-conjugated secondary antibody (Invitrogen) at room temperature for 1 h. F-actin was stained using Alexa Fluor 488-conjugated phalloidin (Invitrogen) during the secondary antibody incubation. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma). For morphology analysis and quantification, images were acquired using an UltraView confocal microscope (PerkinElmer) with a 20×, 40×, or 100× oil objective (Olympus). To reveal hnRNP Q1 granules in neurites, images were acquired at a resolution of 2,048 by 2,048 by using an LSM700 confocal microscope (Carl Zeiss) with a Plan-Apochroma 100× 1.40 NA Oil Ph3 objective lens.
To determine the subcellular distribution of proteins, ImageJ software (National Institutes of Health, Bethesda, MD) was used for analysis and processing of immunofluorescence images. Signals of the soma and neurites were obtained from manually outlined regions according to α-tubulin immunostaining. The nuclear signal was obtained from DAPI-stained regions. To analyze protein distribution along the neurites, linear neurites were selected for signal profile analysis. Each neurite was divided into 450 fragments from the proximal to distal end. Average ratios of signals from each fragment were collected from 50 neurites.
Riboprobe preparation.
The cDNAs corresponding to the full length or each of four fragments of the Cdc42 3′ untranslated region (UTR) (GenBank accession number NM_009861) were cloned into the pGEM-T plasmid (Promega). The resulting plasmids were linearized by restriction enzyme digestion. In vitro transcription was performed using T7 or SP6 RNA polymerase (Promega) in the presence of digoxigenin (DIG; Roche)-, biotin (Roche)-, or 32P-labeled UTP. DIG- and biotin-labeled RNAs were purified by using the RNeasy minikit (Qiagen), and 32P-labeled RNAs were purified by using G-25 resin (GE Healthcare).
Fluorescence in situ hybridization (FISH).
N2A cells were fixed in PBS containing 4% paraformaldehyde for 20 min and permeabilized in PBS containing 0.5% Triton X-100 for 5 min. After being washed with PBS containing 5 mM MgCl2, cells were prehybridized with hybridization buffer containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50 μg/ml heparin, 100 μg/ml tRNA, and 0.1% Tween 20 at 70°C for 1 h in a moisture chamber. Cells were then incubated with hybridization buffer containing 1 ng/μl DIG-labeled RNA probes at 70°C for 3 h. After extensive washing with hybridization buffer and PBS containing 0.1% Tween 20 at 70°C, cells were incubated with PBS containing 5% BSA, 4% FBS, 0.3% Triton X-100, and 2% blocking reagent (Roche) at room temperature for 1 h. Incubation was continued for another 1 h after addition of 1 U/μl peroxidase-conjugated sheep anti-DIG antibody (Roche). After extensive washing with a buffer containing 100 mM Tris (pH 7.5), 150 mM NaCl, and 0.1% Tween 20, cells were incubated with Cy3-tyramide (TSA-PLUS system; PerkinElmer) at a 1:2,000 dilution at room temperature for 15 min. Indirect immunofluorescence of hnRNP Q1 was performed as described above.
Analysis of neuronal morphology.
Axons and dendrites of cortical neurons were indicated by Tau1 and MAP2 immunostaining, respectively. Morphological analysis of neuronal cells was carried out according to five criteria, as follows. (i) A branch was defined as a >10-μm process emerging from a neurite. (ii) The branch order was defined as the number of branch points along the dendrite or axon that had the highest number of branch points on a single neuron. (iii) A dendrite was defined as a >10-μm MAP2-positive process emerging from the soma. (iv) The primary axon length was the longest path from the soma to axonal terminal. (v) The distance between the soma and the first branch point was measured on the primary axon. All of these analyses were performed using MetaMorph (Molecular Devices) and ImageJ software.
N2A filopodia were defined as actin-positive and α-tubulin-negative protrusions greater than 1 μm. For neurite morphological analysis, N2A cell differentiation was induced and cells were immunostained with anti-α-tubulin. Cells with a total neurite length >2-fold higher than the soma diameter were considered to be differentiated. Neurite morphology was analyzed according to the following parameters: (i) a branch was defined as a >5-μm process emerging from a neurite, and (ii) a neurite was defined as an α-tubulin-positive process of >10 μm emerging from the soma.
Immunoprecipitation.
Cell lysates were incubated with antibody-conjugated protein A-Sepharose (GE Healthcare) or anti-FLAG M2 beads (Sigma) at 4°C for 2 h. Beads were washed extensively with NET-2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) containing 0.5% NP-40.
Immunopurification of hnRNP Q1-associated mRNPs.
N2A cells were grown in 10-cm petri dishes to ∼70% confluence and then cultured in serum-free DMEM for 24 h. To induce depolarization, cells were treated with 50 mM KCl for 15 min and then collected in RSB100 buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 2.5 mM MgCl2) containing 40 μg/ml digitonin. Lysates were centrifuged at 1,500 × g for 5 min and subsequently at 4,000 × g for 15 min to remove nuclei and debris. Supernatants were collected as the cytoplasmic fraction. Immunoprecipitation was performed by incubating the cytoplasmic lysates with protein A-Sepharose coated with affinity-purified hnRNP Q1 antibody or control immunoglobulin at 4°C for 2 h. After extensive washing in NET-2 buffer containing 0.5% Triton X-100, 4% of the resin was used for immunoblotting with anti-hnRNP Q1. The remaining resin was suspended in 1 ml TRIzol reagent (Invitrogen) for RNA extraction. To immunopurify overexpressed hnRNP Q1-associated mRNAs, FLAG-hnRNP Q1 was overexpressed in N2A cells. Cells were cultured in serum-free DMEM for 24 h. Immunoprecipitation was performed as described above except that anti-FLAG M2 beads were used.
Microarray and bioinformatics analysis.
RNA was extracted with TRIzol reagent and then purified with the RNeasy minikit (Qiagen). Mock- and FLAG-hnRNP-Q1-coprecipitated RNAs (∼3 μg) were used for microarray hybridization as described previously (26). Affymetrix mouse genome arrays (430_2, comprising 45,101 probe sets corresponding to 22,721 unique genes) were used for hybridization according to the manufacturer's guidelines. Hybridized chips were scanned with an Affymetrix GeneChip 7G scanner. Results were recorded and analyzed using GeneSpring GX 10 software (Silicon Genetics). Probe sets of hnRNP Q1 coprecipitates with a signal that was >300 units and >1.45-fold over the control were scored; these potential hnRNP Q1-associated mRNAs comprised ∼3,000 probe sets corresponding to 2,251 unique genes. Subsequent reverse transcription-PCR (RT-PCR) analysis revealed that hnRNP Q1 could interact with Arp2/Actr2 mRNA (signal ratio, 1.48-fold) but not β-actin, Rac1, or RhoA mRNA (signal ratio, 0.91, 1.18, or 0.68, respectively), indicating the specific selection of hnRNP Q1-associated mRNAs. A nonredundant list of potential hnRNP Q1 target mRNAs was obtained through EGAN software (42) analysis and further subjected to various functional annotations.
To identify potential binding elements of hnRNP Q1, 3′ UTR sequences of the top and bottom 100 candidate genes were retrieved from the NCBI database; the top and bottom represent the pool of transcripts with the highest and lowest signal ratios of hnRNP Q1 to immunoglobulin immunoprecipitation (signal > 300 units), respectively. The 3′ UTR of each gene was then subjected to a computer-assisted pattern search to identify possible sequences ranging from 4 to 7 nucleotides. Unique sequences that appeared over six genes (P < 0.01) in the top set of 100 genes but not in the bottom set were selected for alignment.
RT-PCR.
RT-PCR was performed by using Superscript III reverse transcriptase (Invitrogen) and specific pairs of primers (see Table S1 in the supplemental material).
RNA affinity selection and UV cross-linking.
For RNA affinity selection, streptavidin-agarose (Sigma) was conjugated with 1 μg of DIG-labeled RNA at 4°C for 30 min and then incubated with 400 μg of N2A cell lysates at 4°C for 2 h. After extensive washing with NET-2 buffer containing 0.1% NP-40, the resin was subjected for immunoblotting analysis. For UV cross-linking, 400 μg N2A cell lysates containing overexpressed FLAG-hnRNP Q1 was incubated with 1 × 106 cpm of 32P-labeled RNA in a 100-μl reaction mixture at 30°C for 5 min. The mixtures were irradiated on ice in a UV cross-linker (Stratalinker) for 10 min and then treated with 10 μg RNase A at 37°C for 25 min. The mixtures were then subjected for immunoprecipitation with anti-FLAG M2 beads. Bound proteins were subjected to autoradiography.
Fractionation of N2A neurites and soma.
N2A cells were transfected with control or hnRNP Q siRNAs. After 24 h of transfection, ∼1 × 106 cells were plated on 24-mm transwells with polyethylene tetraphthalate membranes (1-μm pore size; Millipore). After incubation in growth medium for 2 h, cells were incubated in serum-free medium and cultured for another 24 h. For protein and RNA extraction, both the top and bottom membranes were incubated with 50 μl or 150 μl protein denaturing buffer (for protein extraction) or 1 ml TRIzol (RNA extraction).
RESULTS
hnRNP Q1 is localized in neuronal granules.
Previous reports have implied a potential function for hnRNP Q in cytoplasmic mRNA transport (4), but further details have not been explored. Therefore, we first examined whether hnRNP Q is localized in neuronal granules. We used antibodies against hnRNP Q1, the most abundant and cytoplasmic isoform of hnRNP Q (8), for indirect immunofluorescence analysis of rat embryonic cortical neurons that were in vitro cultured for 7 days (DIV7) and differentiated neuroblastoma N2A cells. Immunofluorescence of cortical neurons with anti-hnRNP Q1 revealed that ∼40% of staining signals were located very close to the cell body (Fig. 1A, cortical neurons and bar graph of cortical neurons, region 1) but the rest were distributed along the neuronal processes, albeit with gradually decreasing abundance toward the tips (Fig. 1A, cortical neurons and bar graph, regions 2 to 5). We also examined whether hnRNP Q granules contain any other neuronal RNA-binding proteins. Using antibodies against the fragile X mental retardation protein (FMRP), which is also an RNA granule protein and participates in neuronal mRNA metabolism (13), we observed that the distribution pattern of FMRP was similar to that of hnRNP Q1 and that the granules of hnRNP Q1 and FMRP were partially colocalized in neuronal processes (Fig. 1A). Similar results were observed in N2A cells except that the neurites of N2A cells were generally shorter than those of cortical neurons (Fig. 1A, N2A cells and bar graph of N2A cells). Notably, a fraction of hnRNP Q1, like ZBP1 (43), was detected at branch points of cortical neurons (see Fig. S1 in the supplemental material), suggesting that hnRNP Q1 may have a functional role in neurons, such as neurite growth or arborization control.
Fig 1.
hnRNP Q1 is localized in cytoplasmic granules of neuronal cells. (A) Immunofluorescence was performed in isolated primary rat cortical neurons (upper panels). Magnified images of white-lined rectangles (a to d) in the uppermost panel are shown below. To quantify the distribution of hnRNP Q1 along neuronal processes, a neurite of ∼120 μm in length was evenly divided into five segments from the root (proximal) to the tip (distal). The immunofluorescence signal intensity of hnRNP Q1 of each segment was compared to that of the entire neurite (presented as percentage). The bar graph shows the average percentage of the hnRNP Q1 signal of each segment; the data were obtained from 30 neurites. An analogous experiment was performed in differentiated N2A cells (lower panels). The hnRNP Q1 immunofluorescence signal was quantified as in cortical neurons; the average length of N2A neurites was ∼60 μm. The bar graph shows the average hnRNP Q1 signal of each of three segments from the proximal to the distal region (n = 50). (B) FLAG-hnRNP Q1 was overexpressed in N2A cells and immunoprecipitated from cell lysates. Immunoblotting was performed by using anti-FLAG and anti-FMRP. pEF represents transfection with empty vector. (C) Cortical neurons (upper panels) were mock treated (Con) or treated with 0.5 mM glutamate (Glu) or 10 mM KCl. Immunofluorescence was performed using anti-hnRNP Q (which reacts with both hnRNP Q and R) and anti-TDP-43. A selected region (white-lined rectangle) of neurites is magnified (inset). Arrowhead indicates colocalized hnRNP Q1 and TDP-43. The bar graph shows the relative hnRNP Q signal in the entire neurite (n = 32) of mock-, glutamate-, or KCl-treated cortical neurons. KCl treatment was also performed in differentiated N2A cells (lower panels).
Next, we examined whether FMRP is a component of hnRNP Q-containing RNPs. We transiently expressed FLAG-hnRNP Q1 in N2A cells and then performed immunoprecipitation using anti-FLAG. Immunoblotting revealed that FLAG-hnRNP Q1 coprecipitated with FMRP, but their association was disrupted by RNase treatment (Fig. 1B). Although hnRNP Q1 had no direct interaction with FMRP, these two proteins may still coexist in some mRNPs and even in some neuronal granules (Fig. 1A).
Because neuronal RNA granules are dynamic structures that can respond to changes in neuronal activity (31), we examined whether stimulation or depolarization of cortical neurons has any effect on cellular distribution of hnRNP Q granules. When cortical neurons were treated with glutamate or KCl, hnRNP Q/R granules (detected using anti-hnRNP Q/R; see Materials and Methods) were more evident in neurites, as was another neuronal RNA granule factor, TAR DNA-binding protein 43 (TDP-43) (Fig. 1C). Moreover, cell depolarization also promoted colocalization of hnRNP Q/R and TDP-43 (Fig. 1C, cortical neurons and bar graph). An analogous result was observed with KCl-treated N2A cells (Fig. 1C, N2A cells). These results indicated that cellular localization of hnRNP Q in neuronal protrusions could be modulated in response to neuronal activation. Therefore, hnRNP Q may participate in mRNA regulation in neuronal granules in a dynamic manner.
Knockdown of hnRNP Q affects neuronal morphogenesis.
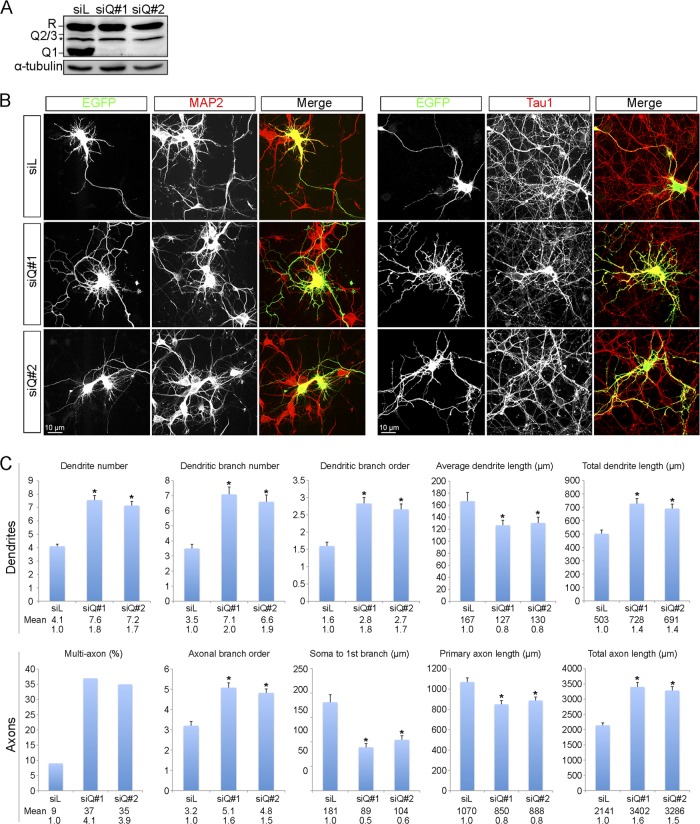
To explore the neuronal functions of hnRNP Q, we knocked down hnRNP Q expression in cortical neurons using small interfering RNAs (siRNAs). We first introduced siQ RNAs (siQ#1 or siQ#2) into N2A cells to evaluate their effectiveness for hnRNP Q knockdown. Immunoblotting showed that both siRNAs almost completely depleted all hnRNP Q isoforms but had no effect on hnRNP R (Fig. 2A). Next, we cotransfected siQ and an expression vector encoding enhanced green fluorescent protein (EGFP) into cortical neurons. Immunofluorescence microscopy was performed with antibodies against MAP2 and Tau1 to distinguish dendrites and axons. Anti-GFP staining indicated positively transfected cells. As shown in Fig. 2B, hnRNP Q knockdown cells exhibited a higher degree of complexity of MAP2-positive and Tau1-positive neurites. The effect of hnRNP Q knockdown on neurite morphological changes was quantified by measuring a variety of neurite parameters such as length and branch order of both types of neurites. For each parameter, we selected 100 GFP-positive neurons for quantification. As indicated by MAP2, depletion of hnRNP Q led to an ∼1.7- to 2-fold increase in the number of dendrites and branches as well as in branch order of DIV3 cortical neurons compared with the control (Fig. 2C), suggesting that hnRNP Q knockdown induced neurite arborization. Moreover, hnRNP Q knockdown increased the total length of dendrites by ∼40% over the control, although the average neurite length decreased (Fig. 2C). We reasoned that the increased total dendrite length was likely due to the increased dendrite number and branching. Transfection of siQ in cortical neurons at DIV7 produced similar phenotypes (see Fig. S2 in the supplemental material). Analysis of dendritic morphology revealed that hnRNP Q depletion resulted in a more complex pattern of neurite morphology.
Fig 2.
Knockdown of hnRNP Q induces neurite arborization in cortical neurons. (A) N2A cells were transiently transfected with siRNA targeting luciferase (siL) or hnRNP Q (siQ#1 or siQ#2). Cell lysates were analyzed by immunoblotting using anti-hnRNP Q/R, which recognizes all hnRNP Q isoforms and hnRNP R; α-tubulin was used as a control. A putative hnRNP R isoform is indicated by an asterisk; the weak signal above it represents hnRNP Q2/3. (B) Cortical neurons were cultured for 3 days after isolation from rat embryos. The EGFP expression vector was cotransfected with siQ RNA for 3 days. Immunostaining with anti-MAP2 and anti-Tau1 identified dendrites and axons, respectively, and anti-GFP staining confirmed positively transfected cells. To highlight neurites, cells are shown in white on a black background; merged images are shown in color. (C) Dendritic (upper panel) and axonal (lower panel) parameters were measured in 100 GFP-positive cells from at least three independent experiments. Bars represent mean values ± standard errors of the means, asterisks indicate a P value of <0.01, and numbers below bars are mean and fold change for each parameter upon hnRNP Q knockdown.
With anti-Tau1, we observed an ∼60% increase in the branch order of axon-like processes of cortical neurons upon hnRNP Q knockdown at DIV3 and a 50% decrease in the distance between the soma and the first branch of these neurites (Fig. 2C). Moreover, ∼35% of hnRNP Q knockdown cells had two or more axons, representing a nearly 4-fold increase compared with the control. Similar to dendrites, total axon length increased ∼1.5-fold, but the length of the primary axon decreased by ∼20%. Because DIV7 neurons displayed a complex projection pattern, we assessed the effect of hnRNP Q knockdown only on axon number and the distance from the soma to the first branch point. The results were similar to those in DIV3 neurons (Fig. 2). Overall, these results indicated that hnRNP Q depletion induced formation of multiple axons and increased neurite branching.
hnRNP Q1 regulates filopodium formation and neurite branching in neuroblastoma cells.
Next, we examined whether the aforementioned effect of hnRNP Q depletion on cortical neurite morphological changes could be reproduced in N2A cells. Filopodia are actin-rich structures that mediate neurite outgrowth in the process of neuronal differentiation (9, 33). Under nondifferentiation conditions, very few neurite processes were observed in control siRNA-transfected N2A cells (Fig. 3A, siL). However, transfection with either siQ RNA induced significant filopodium formation, as evidenced by phalloidin staining (Fig. 3A, siQ#1 and siQ#2). We also examined the effect of hnRNP knockdown in differentiated N2A cells. Compared with the control, siQ transfection caused a 2-fold increase in the number of neurites and an ∼50% increase in their length (Fig. 3B). In general, neurites of N2A cells lack branches (11); however, knockdown of hnRNP Q dramatically increased the number of neurite branches (Fig. 3B). Therefore, as in cortical neurons, depletion of hnRNP Q in N2A cells particularly promotes neurite arborization.
Fig 3.
hnRNP Q1 plays a role in neurite morphogenesis and filopodium formation in N2A cells. (A) N2A cells were transfected as for Fig. 2A and cultured under nondifferentiation conditions, followed by immunostaining with anti-α-tubulin. Actin and cell nuclei were stained with fluorescent phalloidin and DAPI, respectively. (B) N2A cells were transfected as for Fig. 2A and then induced to differentiate. Cells were immunostained with anti-α-tubulin and counterstained with DAPI. (C) N2A cells were cotransfected with siRNA (siL or siQ#1) plus an empty expression vector or a vector expressing hnRNP Q1, Q2, or Q3 and an EGFP expression vector, and then cultured under differentiation conditions. In all three panels, the mean and fold change of each parameter are shown.
To evaluate the specificity of hnRNP Q knockdown, we performed a rescue experiment. As above, transfection of the siQ#1 siRNA increased neurite complexity of N2A cells (Fig. 3C, siQ plus vector). In contrast, overexpression of hnRNP Q1 reproducibly reduced neurite number and length in N2A cells, albeit with minimal effects (Fig. 3C, siL plus Q1). However, siQ-increased neurite branching in N2A cells was largely suppressed by the siQ-resistant hnRNP Q1 but not by either hnRNP Q2 or Q3 (Fig. 3C). This result underscored the specific role of hnRNP Q in neuron morphogenesis and verified that hnRNP Q1, but not two other nuclear isoforms (8), could modulate neurite morphogenesis and particularly branching, which was consistent with the accumulation of hnRNP Q1 at the branch points of neurites.
Putative hnRNP Q1 mRNA targets encode regulators of actin assembly.
The above result indicated that hnRNP Q1 associates with neuronal RNA granules and may help control neurite morphogenesis. Next, we attempted to determine whether hnRNP Q1 exerts its function by regulating mRNA metabolism in neuronal cells. We immunoprecipitated hnRNP Q1 from cytoplasmic lysates of N2A cells and used microarrays to identify coprecipitated mRNAs. Of ∼22,700 gene sets on the chip, ∼2,250 mRNA species (∼10%) showed a >1.45-fold increase in the hnRNP Q1 immunoprecipitates over the control (see Table S2 in the supplemental material).
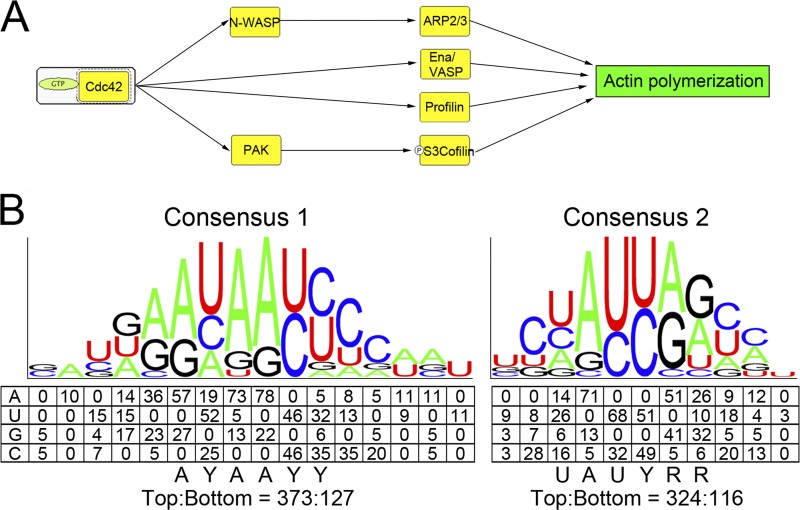
Gene ontology analysis of the ∼2,250 high-scoring candidates revealed that many are involved in protein synthesis, membrane trafficking, ubiquitin or ubiquitin-like protein modification, protein degradation, and signal transduction (see Table S3 in the supplemental material). Further analysis using PANTHER (protein analysis through evolutionary relationships) indicated that several candidates were components of the Rho-GTPase signaling pathway, which regulates the actin cytoskeleton. These candidates included Cdc42, N-WASP, six components of the Arp2/3 complex, p21-activated kinases (PAK), vasodilator-stimulated phosphoprotein (Ena/VASP), profilin, and cofilin (see Fig. 4A for a simplified scheme and Fig. S3 in the supplemental material for a complete scheme). In particular, the Cdc42/N-WASP/Arp2/3 complex has been demonstrated to regulate neuronal morphogenesis (44). Therefore, identification of this set of the candidates could support the morphological changes caused by hnRNP Q depletion.
Fig 4.
Analysis of hnRNP Q1-associated mRNAs reveals actin assembly regulators and consensus motifs in the 3′ UTR. (A) A schematic diagram shows a simplified network of actin assembly regulation by Cdc42. Potential hnRNP Q1 targets are indicated by yellow boxes; among them, Cdc42, N-WASP, and several Arp2/3 components were subsequently verified (see Fig. 5). (B) Two sets of degenerate consensus sequences in the 3′ UTR of hnRNP Q1 targets were identified through a pattern search (see Materials and Methods). Numbers represent the frequency of each nucleotide in the top 100 hnRNP Q1-associated transcripts. The consensus sequence of each motif is shown below the schematic diagram (Y = C/U, R = A/G). The ratio at the bottom represents the frequency of the motif in the 3′ UTR of the top versus bottom 100 transcripts.
Validation of identified hnRNP Q1-associated transcripts.
Next, we attempted to verify whether the identified transcripts are indeed associated with hnRNP Q-containing mRNPs. We immunoprecipitated hnRNP Q1 from N2A cell lysates and performed RT-PCR with gene-specific primers to detect the transcripts of Cdc42, N-WASP, and four components of the Arp2/3 complex (Arp2/Actr2, Arp3/Actr3, Arpc1a/b, and Arpc4) in the hnRNP Q1 precipitates (Fig. 5A). We also examined transcripts of PTEN (a potential target of hnRNP Q identified in this study), β-actin (which is associated with hnRNP R in neuronal transport granules [14]), and two Rho family members, RhoA and Rac1. PTEN but not β-actin, RhoA, or Rac1 mRNA coprecipitated with hnRNP Q (Fig. 5A). Finally, we found that KCl treatment of N2A cells promoted the interaction between hnRNP Q1 and the transcripts of most Cdc42 signaling factors besides N-WASP, but it had no effect on the binding of hnRNP Q1 to PTEN mRNA (Fig. 5A). Together, these results verified a set of identified hnRNP Q1 targets encoding actin assembly factors and further showed that the association of hnRNP Q1 with some of its targets could be modulated by neuronal activity.
Fig 5.
Verification of hnRNP Q-associated mRNAs. (A) Cell lysates of mock-treated (−) or KCl-treated (+) cortical neurons were subjected to immunoprecipitation (IP) using control immunoglobulin (IgG) or anti-hnRNP Q1. The top panel shows immunoblotting of lysates using anti-hnRNP Q1. Coprecipitated RNAs were analyzed by RT-PCR with specific primers; the number of amplification cycles is indicated in parentheses. Input: RT-PCR was performed using mock- and KCl-treated lysates prior to immunoprecipitation. rRNA served as a control. Relative levels of hnRNP Q1-associated RNAs in KCl- versus mock-treated cells are shown; means and standard errors of the means were obtained from three independent experiments. (B) N2A cells were transiently transfected with an empty vector (pEF) or FLAG-hnRNP Q1-expressing vector. The cell lysates were subsequently incubated with 32P-labeled sense and antisense (con) Cdc42 3′ UTR. After UV cross-linking, the reaction mixtures were subjected to immunoprecipitation using anti-FLAG M2 beads. Lanes 1 to 4 show the input.
To examine whether hnRNP Q1 directly binds to its target mRNAs, we performed UV cross-linking using 32P-labeled Cdc42 3′ untranslated region (UTR) as a probe. Although both the sense and antisense (as a control) Cdc42 3′ UTR RNAs were cross-linked to myriad proteins in N2A cell lysates (Fig. 5B, lanes 1 to 4), anti-FLAG immunoprecipitation showed that overexpressed FLAG-hnRNP Q1 bound with higher affinity to the Cdc42 3′ UTR than the control (Fig. 5B, lane 8). This result was further confirmed by the RNA pulldown assay (see below for Fig. 6).
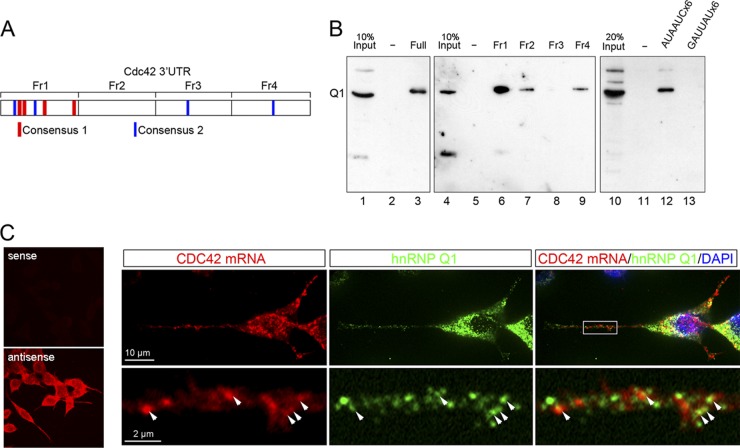
Fig 6.
hnRNP Q1 interacts with the Cdc42 mRNA likely via the identified hnRNP Q1 binding elements and is colocalized with the Cdc42 mRNA in neuritis. (A) Schematic diagram of the Cdc42 3′ UTR and its fragments that were used as probe for RNA affinity selection. Two putative hnRNP Q1 binding elements, of which the core sequence is conserved between humans, mice, and chickens, are indicated by red (consensus 1) and blue (consensus 2) lines. (B) The N2A cell lysate was subjected to RNA affinity selection using biotinylated full length (lane 3) or fragment (lanes 6 to 9) of the Cdc42 3′ UTR or repeated AUAAUC (lane 12) or antisense (lane 13) RNA as a probe. Lanes 1, 4, and 10 are the input. Lanes 2, 5, and 11 are mock affinity selection without RNA probe. (C) FISH of differentiated N2A cells was performed using DIG-labeled sense or antisense probe targeting the 3′ UTR of Cdc42 (left panel). The right panel shows that differentiated N2A cells were subjected to FISH using the antisense Cdc42 probe, indirect immunofluorescence using anti-hnRNP Q1 (as in Fig. 1A), and DAPI staining.
Consensus sequences for RNA binding to hnRNP Q1.
To determine whether hnRNP Q1 interacts directly with its targets via specific cis elements, we first performed a pattern search in the 3′ UTR of the top 100 candidate mRNAs against the bottom 100 transcripts (see Materials and Methods). Alignment of identified cis elements of six or seven residues revealed two sets of motifs with a degenerate consensus sequence, AYAAYY and UAUYRR (Y = C/U, R = A/G) (Fig. 4B). We then searched for the most representative sequences of each motif in the 3′ UTR of the top and bottom 100 transcripts. The results showed that both hexamers appeared nearly 3 times more often in the top 100 transcripts than the bottom group (Fig. 4B), indicating that these motifs are enriched in the highest-potential targets of hnRNP Q1. The compositions of these two motifs agreed with previous reports that hnRNP Q binds preferentially to poly(A) or poly(U)-rich sequences (18, 19). In particular, the (C)UAUYRR motif (Fig. 4B) showed significant sequence homology to the preferred binding site (UCUAUC) of Caenorhabditis elegans HRP-2, the homolog of mammalian hnRNP Q/R (22). The identified consensus sequences also exist in validated hnRNP Q1 targets; for example, the Cdc42 3′ UTR contained four copies of each identified motif that also revealed evolutionary conservation, and the majority of them were located in the first quarter of the 3′ UTR (Fig. 6A).
hnRNP Q1 interacts directly with the 3′ UTR of Cdc42 mRNA.
Next, we performed RNA affinity selection to investigate whether hnRNP Q1 binds the fragments of the Cdc42 3′ UTR that contained the identified motifs. We first used biotinylated full-length 3′ UTR of Cdc42 as probe to pull down proteins from the N2A cell lysate. Immunoblotting demonstrated that hnRNP Q1 was selected (Fig. 6B, lanes 1 to 3). This result also confirmed the binding of hnRNP Q1 to the Cdc42 3′ UTR detected by UV cross-linking (Fig. 5B). We then used four subfragments of the Cdc42 3′ UTR for the RNA affinity selection. The result showed that subfragment 1, which was enriched for the identified motifs, most efficiently pulled down hnRNP Q1 (Fig. 6B, lanes 4 to 9). Finally, we used artificially designed RNA fragments containing either six copies of motif 1 (AUAAUC) or its antisense sequence to pull down hnRNP Q1. As expected, only the sense and not the antisense (GAUUAU) strand could select hnRNP Q1 from the N2A cell lysate (Fig. 6B, lanes 10 to 13), suggesting that hnRNP Q1 may specifically recognize at least one of the identified elements.
We then investigated whether the Cdc42 mRNA exists in hnRNP Q1 granules by indirect immunofluorescence and fluorescence in situ hybridization (FISH) in N2A cells. FISH with a digoxigenin (DIG)-labeled antisense, but not sense, riboprobe directed against the Cdc42 3′ UTR revealed specific signals in N2A cells (Fig. 6C, left panel). Furthermore, we observed that the Cdc42 mRNA puncta were partially colocalized with hnRNP Q1-containing granules in the neuritic processes of N2A cells (Fig. 6C, right panel). Together our data indicated that hnRNP Q1 directly interacted with the Cdc42 mRNA and may play a role in its metabolism in neuronal granules.
Binding of hnRNP Q1 to target mRNAs correlates with enhanced neurite branching.
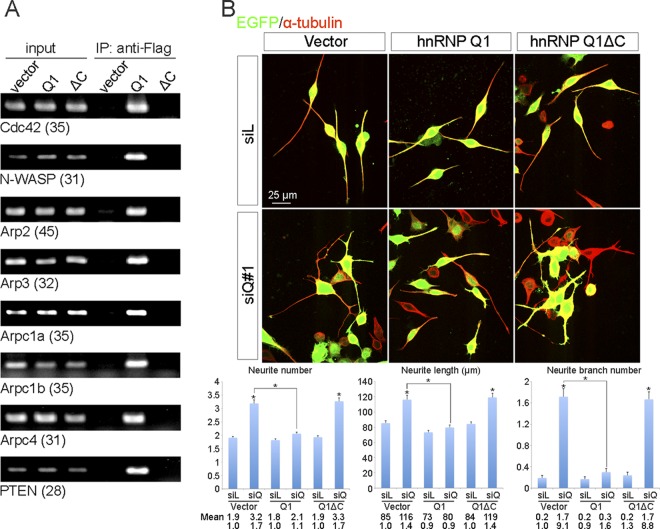
The C-terminal RGG-rich domain of hnRNP Q contributes to RNA binding and is essential for its activity in regulating alternative splicing (8). We thus examined whether this domain is also involved in the binding of hnRNP Q1 to its target mRNAs and in modulating neurite morphogenesis. We transiently expressed FLAG-tagged and C-terminally truncated hnRNP Q1 (FLAG-Q1ΔC) in N2A cells. Using immunoprecipitation coupled with RT-PCR, we found that this ΔC mutant no longer bound to the mRNAs examined (Fig. 7A). Next, we cotransfected the expression vector of FLAG-Q1ΔC with siQ#1 into N2A cells and then induced cell differentiation. Indirect immunofluorescence microscopy using anti-GFP and anti-α-tubulin showed that this mutant failed to rescue the neurite morphological changes caused by hnRNP Q knockdown (Fig. 7B). These data indicated that the C-terminal domain of hnRNP Q1 is required for both target mRNA binding and control of neurite morphogenesis. Moreover, this result suggested that hnRNP Q1 may modulate neuronal morphogenesis by binding to mRNAs encoding multiple components of the actin assembly apparatus.
Fig 7.
The C-terminal domain of hnRNP Q1 is essential for RNA binding and control of neurite morphogenesis. (A) N2A cells were transiently transfected with empty vector or expression vector encoding FLAG-hnRNP Q1 (full-length or ΔC). Cell lysates were subjected to immunoprecipitation (IP) with anti-FLAG followed by RT-PCR using specific primers. (B) Transient transfection of N2A cells with siRNA was performed as for Fig. 2A, except that FLAG-hnRNP Q1ΔC was compared with full-length hnRNP Q1. Indirect immunofluorescence was performed using anti-α-tubulin (red) and anti-GFP (green). Neurite number, total neurite length, and branch number were measured and quantified (n = 100); mean values and standard errors of the means from three independent experiments are shown in the bar graphs.
Depletion of hnRNP Q causes redistribution of Cdc42/N-WASP/Arp in neurites.
To examine how hnRNP Q modulates neurite morphogenesis via its targets, we first examined whether the mRNA and/or protein levels of the Cdc42 signaling factors were altered upon hnRNP Q knockdown. Indeed, neither depletion (data not shown) nor overexpression (Fig. 7A) of hnRNP Q could significantly change the overall level of its mRNA targets. Therefore, we cultured hnRNP Q-depleted N2A cells on Transwell filters to separate the soma and neurites. Using RT-PCR, we found that the levels of all four transcripts examined (Arp2, Arpc1a, Cdc42, and N-WASP) decreased in neurites after hnRNP Q knockdown (Fig. 8A). A similar result was obtained in cortical neurons (data not shown). Using immunoblotting, we also observed that hnRNP Q knockdown considerably reduced all four protein levels in N2A neurites, with the most substantial effect on Cdc42, and also reduced the levels of the three other proteins, albeit with minor effects (Fig. 8B). Therefore, hnRNP Q1 may participate in the transport and/or translation of some of its targets in neurites.
Fig 8.
Knockdown of hnRNP Q modifies the subcellular distribution of its target proteins. (A) N2A cells were transfected with control siL or siQ#1 or siQ#2 siRNA for 24 h and then grown on transwell inserts for 24 h. Soma and growing neurites were collected from the top and bottom compartments, respectively. Total RNAs were isolated from soma and neurites and subjected to RT-PCR analysis as for Fig. 5A. (B) N2A cells were transfected with siL or siQ#1 siRNA as for panel A, and total protein was collected from soma and neurites for immunoblotting using specific antibodies as indicated. In both panels A and B, bar graphs show relative levels of targets in hnRNP Q-knockdown neurites compared to control neurites; target mRNA and protein levels were normalized against actin. (C) N2A cells were transfected with siL, siQ#1, or siQ#2 for 3 days. Immunofluorescence was performed using antibodies against Cdc42, N-WASP, Arpc1a, Arp2, actin, and α-tubulin. Cdc42, N-WASP, Arpc1a, and Arp2 could be detected at the distal neurites of siL-transfected cells (arrowheads) but not those of siQ-transfected cells (arrows). (D) Graphs show relative fluorescence intensity (y axis) of each indicated protein along the neurites (x axis) of control or siQ-transfected cells. Details of measurements are described in Materials and Methods; mean values were obtained from 50 cells. (E) Fluorescence signals of proteins in subcellular fractions (i.e., neurites versus soma) were quantified as described in Materials and Methods. The neurite/soma ratio of each protein is shown for control and knockdown cells; means and standard deviations were obtained from 50 cells.
We then performed immunofluorescence using specific antibodies in siQ-transfected differentiated N2A cells; representative fluorescent images are shown in Fig. 8C and in Fig. S4 in the supplemental material. Moreover, the change in fluorescence intensity of individual proteins was assessed using a series of consecutive optical neurite sections (Fig. 8D). The two Arp2/3 components (Arpc1a and Arp2) were concentrated in the distal tips of neurites in control siRNA-transfected cells, but their levels were reduced in neurite tips upon hnRNP Q knockdown using either siQ#1 or siQ#2 RNA (Fig. 8C and D). A similar result was observed for Cdc42, although its distal distribution was not as prominent as that of the two Arp2/3 proteins (Fig. 8C and D). N-WASP showed a more even distribution along the neurites; however, hnRNP Q knockdown still led to a redistribution of N-WASP from the neurites to the soma (Fig. 8C and D). Localization of α-tubulin and β-actin was not significantly affected by hnRNP Q knockdown (Fig. 8C and data not shown). Moreover, the total fluorescence intensity of individual proteins was quantified separately in neurites and soma. Compared to the control, hnRNP Q knockdown in N2A cells caused a 25 to 40% decrease in the relative abundance of Cdc42, N-WASP, Arp2, and Arpc1a in neurites versus soma, but no significant change was observed in α-tubulin or β-actin distribution (Fig. 8E), consistent with the results obtained from immunoblotting (Fig. 8A and B).
Taken together, hnRNP Q may participate in localized expression of several Cdc42/N-WASP/Arp factors in neurites by binding to their mRNAs. In the absence of hnRNP Q, those factors may be expressed in inappropriate locations, resulting in abnormal neurite arborization (Fig. 2 and 3).
hnRNP Q1 regulates neuronal morphogenesis through the Cdc42/N-WASP/Arp pathway.
To confirm that the hnRNP Q depletion-induced neurite phenotypes involved the Cdc42 signaling pathway, we examined whether blockage of this pathway by using a dominant negative mutant Cdc42 could reverse the effect. Consistent with earlier results (Fig. 3), differentiated N2A cells transfected only with control siRNA generally extended two primary neurites, and hnRNP Q knockdown increased the number of neurites and of branches (Fig. 9A, vector). Overexpression of wild-type Cdc42 in control siRNA-transfected cells promoted neurite overgrowth, more significantly in the branch number (4.5-fold); this effect of Cdc42 was also detected in hnRNP Q-depleted cells, albeit minimally (Fig. 9A, Cdc42). However, overexpression of Cdc42N17, a dominant negative mutant of Cdc42 (55) that binds GDP preferentially over GTP, markedly inhibited the hnRNP Q knockdown-induced neurite phenotype (Fig. 9A, Cdc42N17).
Fig 9.
hnRNP Q1 regulates neuronal morphogenesis through the Cdc42/N-WASP signaling pathway. (A) N2A cells were cotransfected with siRNA (siL or siQ#2) and vectors expressing EGFP and wild-type Cdc42 or dominant negative Cdc42N17. (B) N2A cells were transfected with siRNA (siL or siQ#2) and vectors expressing an EGFP and the C-terminal domain of N-WASP fusion (ECFP-CA) or a cofilin-homologous-domain-lacking N-WASP (N-WASPΔcof). In panels A and B, vector represents transfection with empty expression vector, and indirect immunofluorescence was performed using anti-α-tubulin (red) and anti-GFP (green). Bar graphs show average neurite number, neurite length, and branch point number from 100 GFP-positive cells.
Next, we took advantage of two dominant negative mutants of N-WASP to examine whether N-WASP is involved in hnRNP Q knockdown-induced neurite arborization. The C-terminal domain of N-WASP interacts directly with the Arp2/3 complex, and the cofilin homology region of N-WASP enhances this interaction (25, 47). Previously, it has been shown that overexpression of a fusion protein of enhanced cyan fluorescent protein and the C-terminal domain of N-WASP (ECFP-CA) or of an N-WASP deletion mutant lacking the cofilin homologous domain (N-WASPΔcof) interferes with the interaction of endogenous N-WASP with Arp2/3 (20, 36, 47). In differentiated N2A cells, transient expression of ECFP-CA or N-WASPΔcof had no obvious effect on neurite number or length, but it slightly reduced the number of branch points (Fig. 9B). However, overexpression of ECFP-CA or N-WASPΔcof in hnRNP Q-depleted N2A cells substantially reduced neurite number (by ∼50%) and branch formation (4- to 5-fold) and also decreased neurite length by ∼30% (Fig. 9B).
Taken together, our results showed that the dominant negative mutants of Cdc42 and N-WASP rescued the neuronal morphological phenotypes induced by hnRNP Q knockdown. Therefore, hnRNP Q might play a role in neurite morphogenesis by regulating the Cdc42/N-WASP pathway.
DISCUSSION
In cortical neurons, synaptic connectivity and spine formation require new protein synthesis at proper sites within the dendrites. Numerous RNA-binding proteins have been implicated in coordinating mRNA delivery and local translational control (49). In this study, we found that knockdown of hnRNP Q induced neurite overgrowth in cultured cortical neurons and promoted filopodium formation in N2A cells. Moreover, we identified a set of hnRNP Q1-associated mRNAs that encode factors involved in Cdc42-induced actin polymerization and provided evidence suggesting that hnRNP Q1 may regulate localization of those transcripts, thereby modulating neurite morphogenesis.
hnRNP Q1 binds specific sets of dendritic mRNAs in neuronal cells.
Gene ontology analysis revealed that a set of hnRNP Q1-associated mRNAs encode proteins that have specific functions in neuronal cells, such as neurite outgrowth and motility control (see Table S3 in the supplemental material). Because hnRNP Q knockdown caused prominent neurite overgrowth (Fig. 2), we verified and investigated the target cohort involved in Cdc42-mediated regulation of actin polymerization. In addition to the Cdc42/N-WASP/Arp2/3 signaling axis, potential targets of hnRNP Q1 with known functions in actin dynamics included the small actin regulators cofilin and profilin and the WASP homolog Ena/VASP (Fig. 4). Identification of transcripts encoding the regulatory subunit of protein phosphatase 2A (PP2A) is also interesting because PP2A can dephosphorylate and hence activate the actin depolymerizing factor cofilin (1). These observations reinforce the role of hnRNP Q1 in regulating actin networks. Notably, a group of candidate hnRNP Q targets encode factors involved in protein synthesis (i.e., ribosomal proteins and translation factors) and protein trafficking (i.e., small GTPase Rab and dynein light chain), both of which are important for specific neuronal functions such as neurite outgrowth and morphogenesis. Recent evidence has also indicated that protein degradation mediated by the ubiquitin-proteasome pathway is as important as localized protein synthesis in modulating growth cone dynamics (30). Interestingly, the candidate hnRNP Q1 targets also included transcripts encoding factors with ubiquitin-protein ligase activity. Although this finding remains to be verified, the possibility remains that hnRNP Q1 fine-tunes local protein expression by balancing protein synthesis and degradation.
hnRNP Q1 modulates neuronal morphogenesis by regulating target protein expression in neurites.
Formation of growth cone filopodia requires actin polymerization at the leading edges of neuronal processes (35). The activity of the Cdc42/N-WASP/Arp2/3 complex is crucial for promoting filopodium formation (34). The Arp2/3 complex also plays an important role in branching of actin filaments at the leading edge of motile cells (45). Interestingly, the mRNAs of all seven Arp2/3 complex members are targeted to cellular protrusions, and their transport is likely to be regulated by Rho-GTPase (37, 38); still, the details of how this transport is achieved and regulated remain largely unclear. Our result that hnRNP Q1 directly bound the Cdc42 mRNA and perhaps all other validated targets such as N-WASP and several Arp2/3 members indicates a possible role for hnRNP Q1 in the trafficking, stability control, or translation of these transcripts in cytoplasm (Fig. 4), suggesting that the absence of hnRNP Q may alter the sites of actin assembly in neurites. To determine their subcellular distribution, we fractionated the soma and neurites of N2A cells to measure transcript and protein levels in each compartment, and we performed immunofluorescence microscopy in N2A cells. Coincidentally, we observed that hnRNP Q knockdown reduced the abundance of target transcripts in neurites and relocalized their encoded proteins more proximally to the cell body (Fig. 8). This result suggested that hnRNP Q1 depletion may cause improper deposition of these transcripts and/or uncontrolled translation during mRNA transport. Moreover, inefficient expression of actin polymerization regulatory factors at distal sites of neurites may account for an increased frequency of neurite branching and reduced distance between the soma and the first branch point in cortical neurons (Fig. 2). We also noticed that Cdc42 was located near or at the cell membrane in neuronal cells, but hnRNP Q knockdown reduced this peripheral accumulation (data not shown). Therefore, in the absence of hnRNP Q, Cdc42 mRNA may be inefficiently translocated to sites near the plasma membrane and be translated at perinuclear regions. All our results support the idea that hnRNP Q1 may act to deposit target mRNAs at distal sites of neurites and/or limit their expression during trafficking.
Cytoskeletal dynamics in neuronal cells may be coregulated by hnRNP Q1 and other RNA-binding proteins.
Several RNA-binding proteins have been implicated in regulation of dendritic and axonal outgrowth based on their activity in posttranscriptional regulation of cytoskeletal mRNAs (2, 29, 31, 43). Drosophila FMRP controls actin dynamics by regulating the translation of mRNAs encoding profilin and Rac1 (7, 46). Indeed, the Rac1 signaling pathway also impacts FMRP, resulting in a more complex regulation of neurite outgrowth (27). In addition, FMRP regulates expression of the neuron-specific MAP1b, which is a major microtubule component in neurons (32). FMRP knockout in murine neurons led to excess filopodium formation, consistent with the observation in dfmr1 mutant flies, supporting a role for FMRP in negative regulation of axonal outgrowth and branching (3, 41). In contrast, knockdown of ZBP1 reduces the expression of β-actin in distal dendrites and thereby impairs the growth of dendritic filopodia (10). Importantly, ZBP1 can modulate growth cone dynamics in response to guidance cues, which likely involves a role for ZBP1 in regulating transport and local translation of β-actin and cofilin mRNAs (53). Staufen 1 and 2 proteins can also regulate the dendritic cytoskeleton via β-actin mRNA (15, 50). A more recent report showed that hnRNP K can regulate multiple transcripts encoding various types of cytoskeletal proteins, including neurofilaments and two MAPs, Tau and Arp2 (29). Knockdown of hnRNP K has pleiotropic effects on cytoskeletal organization and impairs axon outgrowth (29). We show here that the effect of hnRNP Q knockdown on neurite arborization could be largely compromised by the presence of the dominant negative GDP-bound Cdc42 mutant (Fig. 9). Similar results were obtained with overexpression of the C-terminal domain of N-WASP that interferes with the binding of endogenous N-WASP to Arp2/3 or a truncated N-WASP that fails to interact with Arp2/3 (Fig. 9). Therefore, hnRNP Q may influence neurite growth primarily by modulating the Cdc42-mediated signaling pathway that dictates actin polymerization. Thus, it appears that neuronal cytoskeleton dynamics can be controlled by at least several different RNA-binding proteins through different or overlapping sets of target mRNAs.
It is important to understand whether and how different RNA-binding proteins function coordinately to modulate a cellular event. We thus compared the target mRNAs of hnRNP Q1 with those of several other neuronal RNA-binding proteins retrieved from the literature (6, 12, 21). This analysis indicated that some hnRNP Q1 target mRNAs may also be targets for ZBP1 (34%), Staufen 1 (14%), Staufen 2 (34%), and FMRP (4%) (data not shown). Notably, several factors in the Rho-GTPase pathway appeared to be common targets of hnRNP Q1 and the Staufens. It would be interesting to examine whether hnRNP Q1 exists in mRNPs and granules containing any of those RNA-binding proteins and whether they coregulate the expression of certain targets in neurons.
Finally, it has been noted that various types of neuronal stimulation, including depolarization, can regulate filopodium formation (54). We observed that KCl treatment of neuronal cells could enhance the association of hnRNP Q1 with its target mRNAs (Fig. 5) and modify the distribution of hnRNP Q granules in neuronal processes (Fig. 1). Therefore, it would be interesting to know how environmental cues of neurite morphogenesis modulate the activity of hnRNP Q1 and even coordinate different RNA-binding proteins to achieve optimal neuronal functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank C.-K. James Shen, Sue Lin-Chao, and Jenn-Yah Yu for expression vectors and antibodies and the laboratory of Yi-Shuian Huang for providing rat embryos. We are grateful to Yi-Ping Hsueh and Yi-Shuian Huang (Academia Sinica, Taipei, Taiwan) for helpful suggestions during the course of this study and for critical reading of the manuscript.
This work was supported by grant NHRI-EX99-9737NI from the National Health Research Institutes of Taiwan.
Footnotes
Published ahead of print 9 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ambach A, et al. 2000. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur. J. Immunol. 30: 3422–3431 [DOI] [PubMed] [Google Scholar]
- 2. Anderson KD, et al. 2001. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 168: 250–258 [DOI] [PubMed] [Google Scholar]
- 3. Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. 2006. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol. Cell Neurosci. 32: 37–48 [DOI] [PubMed] [Google Scholar]
- 4. Bannai H, et al. 2004. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279: 53427–53434 [DOI] [PubMed] [Google Scholar]
- 5. Blanc V, et al. 2001. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J. Biol. Chem. 276: 10272–10283 [DOI] [PubMed] [Google Scholar]
- 6. Brown V, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487 [DOI] [PubMed] [Google Scholar]
- 7. Castets M, et al. 2005. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 14: 835–844 [DOI] [PubMed] [Google Scholar]
- 8. Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. 2008. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol. Cell. Biol. 28: 6929–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dent EW, et al. 2007. Filopodia are required for cortical neurite initiation. Nat. Cell Biol. 9: 1347–1359 [DOI] [PubMed] [Google Scholar]
- 10. Eom T, Antar LN, Singer RH, Bassell GJ. 2003. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23: 10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franklin JL, et al. 1999. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr. Biol. 9: 1448–1457 [DOI] [PubMed] [Google Scholar]
- 12. Furic L, Maher-Laporte M, DesGroseillers L. 2008. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 14: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gatto CL, Broadie K. 2009. The fragile X mental retardation protein in circadian rhythmicity and memory consolidation. Mol. Neurobiol. 39: 107–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glinka M, et al. 2010. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum. Mol. Genet. 19: 1951–1966 [DOI] [PubMed] [Google Scholar]
- 15. Goetze B, et al. 2006. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 172: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Higgs HN, Pollard TD. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70: 649–676 [DOI] [PubMed] [Google Scholar]
- 17. Holt CE, Bullock SL. 2009. Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hresko RC, Mueckler M. 2002. Identification of pp68 as the tyrosine-phosphorylated form of SYNCRIP/NSAP1. A cytoplasmic RNA-binding protein. J. Biol. Chem. 277: 25233–25238 [DOI] [PubMed] [Google Scholar]
- 19. Hresko RC, Mueckler M. 2000. A novel 68-kDa adipocyte protein phosphorylated on tyrosine in response to insulin and osmotic shock. J. Biol. Chem. 275: 18114–18120 [DOI] [PubMed] [Google Scholar]
- 20. Hufner K, et al. 2001. The verprolin-like central (vc) region of Wiskott-Aldrich syndrome protein induces Arp2/3 complex-dependent actin nucleation. J. Biol. Chem. 276: 35761–35767 [DOI] [PubMed] [Google Scholar]
- 21. Jønson L, et al. 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics 6: 798–811 [DOI] [PubMed] [Google Scholar]
- 22. Kabat JL, Barberan-Soler S, Zahler AM. 2009. HRP-2, the Caenorhabditis elegans homolog of mammalian heterogeneous nuclear ribonucleoproteins Q and R, is an alternative splicing factor that binds to UCUAUC splicing regulatory elements. J. Biol. Chem. 284: 28490–28497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525 [DOI] [PubMed] [Google Scholar]
- 24. Kim DY, Woo KC, Lee KH, Kim TD, Kim KT. 2010. hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res. 38: 7068–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurisu S, Takenawa T. 2009. The WASP and WAVE family proteins. Genome Biol. 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lai MC, Lee YH, Tarn WY. 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 19: 3847–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee A, et al. 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552 [DOI] [PubMed] [Google Scholar]
- 28. Liao G, Simone B, Liu G. 2011. Mis-localization of Arp2 mRNA impairs persistence of directional cell migration. Exp. Cell Res. 317: 812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Y, Szaro BG. 2011. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development 138: 3079–3090 [DOI] [PubMed] [Google Scholar]
- 30. Lowery LA, Van Vactor D. 2009. The trip of the tip: understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10: 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loya CM, Van Vactor D, Fulga TA. 2010. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 24: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu R, et al. 2004. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl. Acad. Sci. U. S. A. 101: 15201–15206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin KC, Zukin RS. 2006. RNA trafficking and local protein synthesis in dendrites: an overview. J. Neurosci. 26: 7131–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattila PK, Lappalainen P. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9: 446–454 [DOI] [PubMed] [Google Scholar]
- 35. Meyer G, Feldman EL. 2002. Signaling mechanisms that regulate actin-based motility processes in the nervous system. J. Neurochem. 83: 490–503 [DOI] [PubMed] [Google Scholar]
- 36. Miki H, Sasaki T, Takai Y, Takenawa T. 1998. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391: 93–96 [DOI] [PubMed] [Google Scholar]
- 37. Mingle LA, Bonamy G, Barroso M, Liao G, Liu G. 2009. LPA-induced mutually exclusive subcellular localization of active RhoA and Arp2 mRNA revealed by sequential FRET and FISH. Histochem. Cell Biol. 132: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mingle LA, et al. 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118: 2425–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mizutani A, Fukuda M, Ibata K, Shiraishi Y, Mikoshiba K. 2000. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 275: 9823–9831 [DOI] [PubMed] [Google Scholar]
- 40. Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20: 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan L, Zhang YQ, Woodruff E, Broadie K. 2004. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr. Biol. 14: 1863–1870 [DOI] [PubMed] [Google Scholar]
- 42. Paquette J, Tokuyasu T. 2010. EGAN: exploratory gene association networks. Bioinformatics 26: 285–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perycz M, Urbanska AS, Krawczyk PS, Parobczak K, Jaworski J. 2011. Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. J. Neurosci. 31: 5271–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. 2007. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One 2: e400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pollard TD. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36: 451–477 [DOI] [PubMed] [Google Scholar]
- 46. Reeve SP, et al. 2005. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr. Biol. 15: 1156–1163 [DOI] [PubMed] [Google Scholar]
- 47. Rohatgi R, et al. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97: 221–231 [DOI] [PubMed] [Google Scholar]
- 48. Rossoll W, et al. 2003. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sinnamon JR, Czaplinski K. 2011. mRNA trafficking and local translation: the Yin and Yang of regulating mRNA localization in neurons. Acta Biochim. Biophys. Sin. 43: 663–670 [DOI] [PubMed] [Google Scholar]
- 50. Vessey JP, et al. 2008. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 105: 16374–16379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang YP, Wang ZF, Zhang YC, Tian Q, Wang JZ. 2004. Effect of amyloid peptides on serum withdrawal-induced cell differentiation and cell viability. Cell Res. 14: 467–472 [DOI] [PubMed] [Google Scholar]
- 52. Weidensdorfer D, et al. 2009. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welshhans K, Bassell GJ. 2011. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 31: 9800–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu GY, Deisseroth K, Tsien RW. 2001. Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat. Neurosci. 4: 151–158 [DOI] [PubMed] [Google Scholar]
- 55. Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. 2008. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 314: 2618–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.