Abstract
Recent work identified the E3 ubiquitin ligase CRL4Cdt2 as mediating the timely degradation of Cdt1 during DNA replication and following DNA damage. In both cases, proliferating cell nuclear antigen (PCNA) loaded on chromatin mediates the CRL4Cdt2-dependent proteolysis of Cdt1. Here, we demonstrate that while replication factor C subunit 1 (RFC1)-RFC is required for Cdt1 degradation after UV irradiation during the nucleotide excision repair process, another RFC complex, Ctf18-RFC, which is known to be involved in the establishment of cohesion, has a key role in Cdt1 degradation in S phase. Cdt1 segments having only the degron, a specific sequence element in target protein for ubiquitination, for CRL4Cdt2 were stabilized during S phase in Ctf18-depleted cells. Additionally, endogenous Cdt1 was stabilized when both Skp2 and Ctf18 were depleted. Since a substantial amount of PCNA was detected on chromatin in Ctf18-depleted cells, Ctf18 is required in addition to loaded PCNA for Cdt1 degradation in S phase. Our data suggest that Ctf18 is involved in recruiting CRL4Cdt2 to PCNA foci during S phase. Ctf18-mediated Cdt1 proteolysis occurs independent of cohesion establishment, and depletion of Ctf18 potentiates rereplication. Our findings indicate that individual RFC complexes differentially control CRL4Cdt2-dependent proteolysis of Cdt1 during DNA replication and repair.
INTRODUCTION
Maintenance of genomic information depends on faithful replication during S phase and segregation of duplicated chromosomes during mitosis, which is critical for proper cell function and survival (39). During S phase, every segment of the chromosomal DNA must be replicated only once during the cell cycle. Recent studies revealed essential roles for ubiquitin-mediated proteolysis in ensuring that DNA replication occurs only once per cell cycle (5, 7). The DNA replication-licensing factor Cdt1 associates with the origin recognition complex (ORC), which is bound to replication origins and, in conjunction with Cdc6, loads the MCM2-7 (minichromosome maintenance subunits 2 to 7) complex onto the chromatin, thereby licensing DNA for an additional round of replication. Once DNA replication is initiated upon activation of the S-phase cyclin-dependent kinases (S-CDK), relicensing of any part of the replicated regions is prevented by strict regulation of the Cdt1 protein levels in mammalian cells (38). Although Cdt1 accumulates during G1 phase, it is degraded and maintained at low levels upon the initiation of DNA replication. S-CDK associates with Cdt1 through its Cy motif and phosphorylates Cdt1 to create a phosphorylated degron, a specific sequence element in target protein for ubiquitination, that is recognized by CRL1Skp2, also known as SCFSkp2 (28, 37, 45, 51). Subsequent studies demonstrated that the Cullin4 (Cul4)-containing E3 ligase Cul4-DDB1-Cdt2, known as CRL4Cdt2, plays a central role in Cdt1 degradation in cells of organisms from yeast to mammals, although CRL1Skp2 operates redundantly in mammalian cells (4, 17, 18, 20, 37, 44, 45). The WD40 repeat protein Cdt2 is the crucial substrate-recognizing subunit of CRL4 E3 ligase. Upon the initiation of DNA replication, Cdt1 associates with proliferating cell nuclear antigen (PCNA) on the chromatin through a PCNA-interacting motif (PIP box) and then is ubiquitinated by CRL4Cdt2, which comprises a feedback control to block licensing. When cells are exposed to DNA-damaging agents, such as UV, Cdt1 degradation is induced through the same PCNA-dependent CRL4Cdt2 pathway. Following local UV or laser irradiation, both Cdt1 and CRL4Cdt2 are rapidly recruited to the damaged sites, depending on the chromatin association of PCNA (19, 43). Detailed analyses using Xenopus laevis egg extracts demonstrated that either the initiation of replication or incubation with damage-containing DNA triggers PCNA loading on chromatin, the association of Cdt1 with PCNA through its PIP box, and the recruitment of Cdt2 (4, 15). Other proteins downregulated by the CRL4Cdt2 pathway include p21, Xic1, and Set8 in vertebrates, all of which contain the PIP box (1, 2, 9, 21, 24, 26, 36, 40, 52). These proteins share conserved amino acids within and downstream from the PIP box, creating a specialized degron for the CRL4Cdt2 pathway (15, 32).
PCNA forms a homotrimeric DNA sliding clamp, and its loading on the chromatin depends on the DNA-dependent ATPase complex called “replication factor C (RFC) complex.” There are four RFC complexes in eukaryotes; the canonical RFC complex, RFC1-RFC, comprises the largest subunit, RFC1 (also known as p140), and four smaller subunits (RFC2 to -5 [RFC2-5]), whereas the alternative RFC complexes, Ctf18-RFC, Elg1-RFC, and Rad17-RFC, all contain RFC2-5 but their largest subunits differ from RFC1 (25, 29). RFC1-RFC loads PCNA onto the primer/template junction, which then acts as a processivity factor for the replicative DNA polymerases DNA pol delta and epsilon (55). The three other RFC complexes are also involved in chromatin metabolism, such as DNA damage checkpoint responses (Rad17-RFC) (58), sister chromatid cohesion (Ctf18-RFC) (14, 27, 30), and the maintenance of genome stability (Elg1-RFC) (6, 22). Rad17-RFC is dedicated to a distinct heterotrimeric clamp, called Rad9-1-1, whereas both Ctf18-RFC and Elg1-RFC operate with PCNA. RFC complexes are required not only for PCNA loading but also have roles in assisting the recruitment of PCNA-interacting proteins, such as DNA polymerases (41, 42).
When cells are UV irradiated, DNA lesions are repaired by the versatile nucleotide excision repair (NER) pathway (10, 11). More than 20 polypeptides, including the 7 xeroderma pigmentosum (XP)-related proteins, are involved in NER dual incision, which removes damage-containing oligonucleotides. The resulting gap has a 3′-OH terminus that is structurally similar to the replication intermediates (12). Biochemical studies with the in vitro NER system revealed that gap-filling DNA synthesis depends on PCNA (34, 50).
In the present study, to elucidate the interplay between chromatin loading of PCNA and Cdt1 proteolysis, we examined the role of the different RFC complexes. After UV irradiation, PCNA loading by RFC1-RFC is important for the degradation of Cdt1. In contrast, we demonstrate that another RFC complex, Ctf18-RFC, is a key player in Cdt1 proteolysis to prevent rereplication during S phase. Although Ctf18-RFC is known to be involved in sister chromatid cohesion, Cdt1 degradation appears to proceed independently of the establishment of cohesion. Our findings provide novel insight into the control of Cdt1 proteolysis, which is closely coupled with DNA metabolism through the different RFC complexes.
MATERIALS AND METHODS
Cell culture.
HeLa cells, HeLa cells stably expressing Cdt1(1-151) Cy-9mycNLS (containing the N-terminal 151 amino acid sequence of Cdt1 and a mutated cyclin binding [Cy] motif that does not allow recognition by the cyclin A-CDK-dependent CRL1Skp2), Cdt1(1-28) 9mycNLS (containing only the N-terminal 28 amino acids of Cdt1 that serve as a degron for CRL4Cdt2), or Cy-mutated full-length Cdt1, HEK293 cells, HEK293 cells stably expressing Cdt2-FLAG, HEK293T cells, normal fibroblasts, and XPA-deficient XP2OSSV cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 5% CO2. HCT116 cell culture conditions were the same except that McCoy's medium was used. To synchronize cells in the early S phase, HeLa cells were blocked using the thymidine and aphidicolin block method. Proteasome inhibitor MG132 was used at 25 μM. UV-C (254-nm) irradiation of whole cells in dishes was performed at 20 to 100 J/m2 using a UV cross-linker (FS-800; Funakoshi). To analyze the DNA content, flow cytometry was performed as described previously. For synchronization of Cdt1(1-151) Cy in the early and middle S phase, cells were treated with 2 mM thymidine or 5 μg/ml aphidicolin for 18 h and released for 3 h.
Antibodies, Western blotting, and immunofluorescence.
For Western blotting, whole-cell lysates were prepared by lysing cell pellets directly in SDS-PAGE buffer. For immunofluorescence analysis, HeLa or HEK293T cells were fixed in 4% paraformaldehyde (Wako) for 10 min, permeabilized in 0.25% (vol/vol) Triton X-100 in phosphate-buffered saline (PBS), and stained with the antibodies indicated below as described previously. For double staining, Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 592-conjugated anti-rabbit antibodies were used as secondary antibodies with Hoechst 33258 to visualize DNA. The following primary antibodies were used: RFC1 (sc-20993; Santa Cruz), Ctf18 (H00063922-M01; Abnova), Elg1 (ab72111-100; Abcam), Rad17 (sc-5613; Santa Cruz), Smc3 (A300-060A; Bethyl), Cdt1 (38), Cdt2 (36), cyclin A (mouse, Ab-6, Neomarkers, and rabbit, H-432; Santa Cruz), Myc (sc-789 and sc-40; Santa Cruz), FLAG (F3165 and F7425, Sigma), PCNA (PC10; Santa Cruz), cyclobutane pyrimidine dimer (CPD) (TDM-2; Cosmo Bio), RCC1 (35), Orc2 (68636E; Becton Dickinson), and phospho-histone H3(Ser10) (06-570; Upstate). Protein levels were analyzed with ImageJ software.
RNA interference (RNAi) knockdown experiments.
The double-stranded RNAs were transfected at 100 μM using Oligofectamine (Invitrogen) or HiPerFect (Qiagen). Twenty-four hours after the first transfection, the second transfection was performed, and cells were cultured for two more days. The following small interfering RNAs (siRNAs) were made by Dharmacon: PCNA, CGGUGACACUCAGUAUGUC; Skp2, GCAUGUACAGGUGGCUGUU; and Cdt2, CCAGGAGGUGAUAAACUUU. The siRNA for luciferase (siLuc), known as GL2, was used as a control siRNA. siRNAs for RFC1 (product number HSS109188), Ctf18 (HSS127220), Elg1 (HSS129124), Rad17 (HSS109000), and SMC3 (HSS113498) were purchased from Invitrogen.
Chromatin fractionation.
Cell extracts were prepared using 0.1% Triton X-100 mCSK buffer {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.9, 100 mM NaCl, 300 mM sucrose, 0.1% (vol/vol) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 mM β-glycerophosphate, 1 mM Na3VO4, 10 mM NaF}. Approximately 5 × 105 cells were washed with ice-cold PBS and lysed with 0.1 ml of 0.1% Triton X-100 mCSK buffer for 15 min on ice. After centrifugation (15,000 rpm for 15 min at 4°C), the precipitate was washed with the same volume of ice-cold 0.1% Triton X-100 mCSK buffer and subsequently suspended in SDS sample buffer.
RESULTS
RFC1-RFC is required for UV-induced degradation of Cdt1.
To date, there are three known types of PCNA-loading RFC complexes, each of which has independent roles in different aspects of DNA metabolism. Based on this, together with our identification of small RFC subunits 2 and 3 in Cdt2 immunoprecipitates (unpublished data), we examined whether a certain type of RFC complex is involved in Cdt1 degradation through PCNA-CRL4Cdt2-mediated ubiquitination. To test this possibility, we knocked down the expression of the largest subunit of each of the four RFC complexes (RFC1, Ctf18, Elg1, and Rad17) individually with small interference RNAs (siRNAs) in 293T cells and examined Cdt1 levels after UV irradiation with immunofluorescence and immunoblot analyses (Fig. 1A; also see Fig. S1 and S2 in the supplemental material). We also included cells treated with siRNAs for PCNA and Cdt2. Flow cytometric analyses revealed that none of these siRNAs significantly affected cell cycle progression (see Fig. S1B). Since Cdt1 is present during G1 phase and is degraded upon the onset of S phase, while cyclin A is present from S phase through M phase, Cdt1-positive cells and cyclin A-positive cells are detected in a mutually exclusive fashion in asynchronously growing cells, as in the cells treated with control siRNA (Fig. 1A, Cont and UV−). These staining patterns were unaffected after transfection with any siRNA, because CRL1Skp2 is active to degrade Cdt1 in S phase. As previously reported, UV-induced Cdt1 degradation was blocked in siPCNA-transfected cells (Fig. 1A, UV+; also see Fig. S2 in the supplemental material). Suppression of Rad17, which is involved in loading of the Rad9-1-1 checkpoint clamp instead of PCNA, had no effect on Cdt1 degradation. On the other hand, among the three PCNA-loading RFC complexes, we found that UV-induced Cdt1 degradation was inhibited in RFC1-depleted cells but not in Ctf18- or Elg1-depleted cells. We obtained essentially the same results by immunoblot analysis using 293T, HeLa, and HCT116 cells (see Fig. S2).
Fig 1.
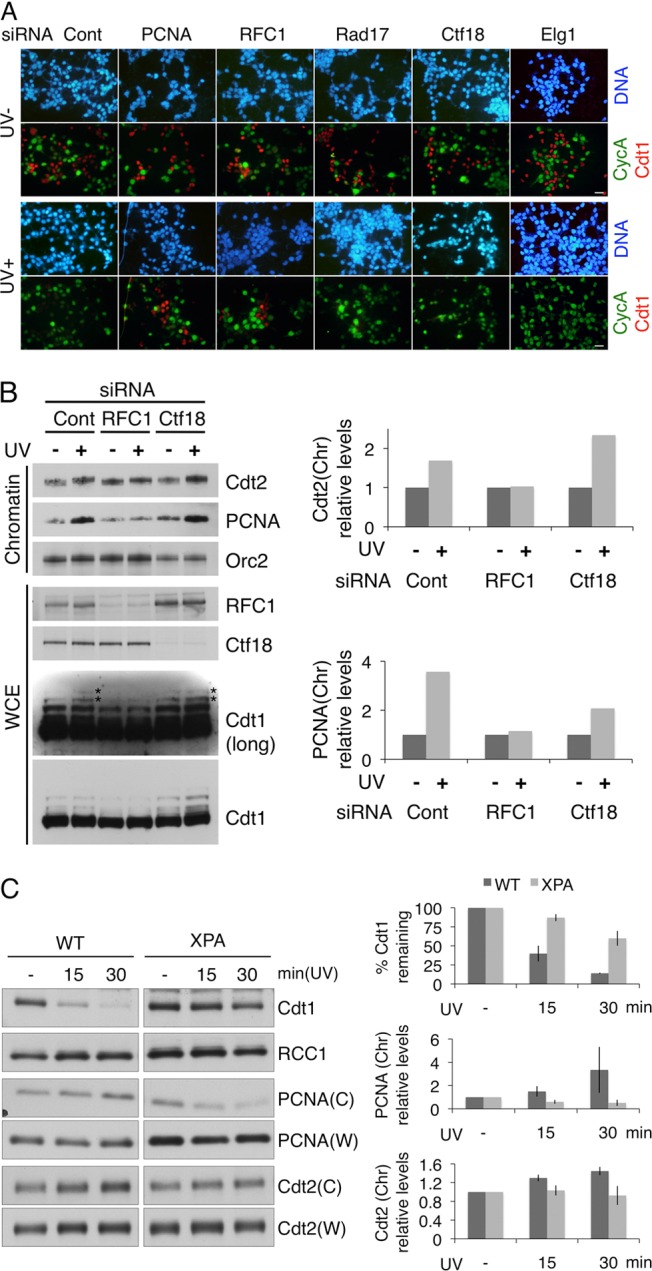
RFC1-RFC is required for Cdt1 degradation after UV irradiation during NER. (A) 293T cells were transfected with siRNAs and UV irradiated (UV+) or not (UV−). One hour later, cells were fixed and stained with antibodies for cyclin A (green) and Cdt1 (red) and with Hoechst stain. siRNA for luciferase was used as a control (Cont). Bars, 20 μm. (B) 293T cells were transfected with siRNAs together with MG132 1 h before half the culture was UV irradiated. After 1 h, whole-cell extract (WCE) and the chromatin-containing fraction were obtained (left). The polyubiquitinated form of Cdt1 is labeled with asterisks. Cdt2 and PCNA protein levels on chromatin (Chr) were quantified and normalized to the amount of Orc2 and are presented as values standardized to the amounts in non-UV-irradiated (−) cells, set as 1.0 (right). (C) Wild-type (WT) and XPA-deficient fibroblast cells (XPA) were UV irradiated at 20 J/m2. Protein levels at the indicated time points were analyzed. W and C stand for whole-cell extract and the chromatin-containing fraction, respectively. Protein levels were quantified and normalized to the amount of RCC1, a loading control. Relative Cdt1 protein levels in whole-cell extract after UV irradiation are presented as values standardized to the amount in non-UV-irradiated (−) cells, set as 100. PCNA and Cdt2 protein levels in the chromatin fraction (Chr) are presented as values standardized to the amount in non-UV-irradiated (−) cells, set as 1.0. The data represent the means ± standard deviations of two independent results.
Time course experiments also showed that Cdt1 degradation after UV irradiation was impaired in RFC1-depleted cells, as well as in Cdt2-depleted cells (see Fig. S3A in the supplemental material). Set8, another recently discovered target of CRL4Cdt2, was also stabilized in the RFC1-depleted cells but not when the expression of Ctf18 was suppressed. Simultaneous depletion of RFC1 and Ctf18 had no synergistic effect on either Cdt1 or Set8 degradation, indicating that RFC1-RFC is the unique RFC complex required for Cdt1 proteolysis after UV irradiation (see Fig. S3A).
We previously demonstrated that both Cdt1 and Cdt2 are recruited to the UV-damaged sites in a PCNA-dependent manner (19, 43). Therefore, we examined whether RFC1-RFC is required for the chromatin loading of PCNA and Cdt2 in response to UV irradiation. Cells were transfected with siRNAs for luciferase (control), RFC1, or Ctf18 and then incubated in the presence of the proteasome inhibitor MG132. After treatment with or without UV, various proteins in the whole-cell extracts or in the chromatin-bound fractions were examined. Unlike the control cells or siCtf18-transfected cells, the RFC1-depleted cells failed to accumulate PCNA or Cdt2 on chromatin after UV irradiation (Fig. 1B). Immunofluorescence analyses also showed that recruitment of PCNA and Cdt2 to the sites of DNA damage was defective in RFC1-depleted cells but not in Ctf18-depleted cells (see Fig. S3B in the supplemental material). In addition, the accumulation of polyubiquitinated forms of Cdt1 was also attenuated only in the cells treated with siRNA for RFC1 (Fig. 1B).
RFC1-RFC-dependent PCNA loading during the NER process is involved in Cdt1 degradation after UV irradiation.
In mammalian cells, UV-induced photolesions are repaired exclusively through the NER pathway, which involves the PCNA-dependent gap-filling DNA synthesis step. To determine whether Cdt1 degradation depends on NER, we examined XP2OSSV cells, which do not express the essential NER factor, XPA protein. Immunoblot analyses revealed that more than 80% of Cdt1 was degraded in normal cells within 30 min after UV irradiation, whereas degradation was obviously delayed in XPA-deficient cells (Fig. 1C). Both the PCNA and Cdt2 levels on the chromatin increased reciprocally after UV irradiation in normal cells but not in XPA-deficient cells. Local UV irradiation through micropore membrane filters also confirmed that recruitment of both PCNA and Cdt2 to the sites of DNA damage was abolished in XPA-deficient cells (see Fig. S4 in the supplemental material). These data indicate that, after UV irradiation, RFC1-RFC loading of PCNA to the single-stranded gaps created by the NER pathway is responsible for both the recruitment of CRL4Cdt2 and the ubiquitination of Cdt1.
Cdt1 proteolysis in the early S phase is mediated by Ctf18-RFC.
To determine whether the RFC1-RFC complex is involved in both modes of Cdt1 proteolysis (i.e., during S phase and after UV irradiation), cells were treated with siRNAs as described above, and the Cdt1 protein levels in S phase were examined. For this experiment, we took advantage of the HeLa cell line stably expressing Cdt1(1–151) Cy-9mycNLS (37), which contains the N-terminal 151-amino-acid sequence of Cdt1. Because the cyclin binding (Cy) motif is mutated in this construct, this form of Cdt1 is not recognized by cyclin A-CDK-dependent CRL1Skp2 but is degraded exclusively by CRL4Cdt2 both in S phase and after UV irradiation (37) (Fig. 2A, lanes 1 to 3). When cells transfected with each siRNA were incubated in the presence of thymidine to arrest in the early S phase, Cdt1(1–151) Cy was degraded in control cells (Fig. 2A, lane 3) but was stably present in the Cdt2- or PCNA-depleted cells (Fig. 2A, lanes 4 and 9). Unexpectedly, Cdt1(1–151) Cy was substantially stabilized in the Ctf18-depleted cells but not in the RFC1-depleted cells. Depletion of Rad17 or Elg1 did not affect the stability of the Cdt1 fragment. Similar results were obtained for the stability of the endogenous Set8 protein (Fig. 2A) and also when cells were arrested in S phase with aphidicolin treatment (see Fig. S5A in the supplemental material).
Fig 2.
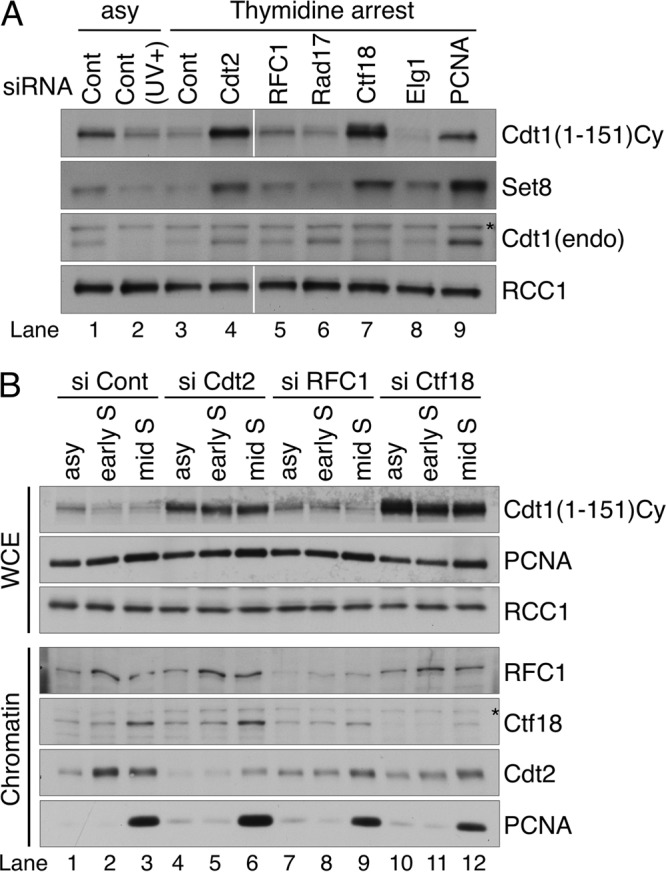
Ctf18 plays a major role in Cdt1 degradation in S phase. (A) HeLa cells expressing Cdt1(1–151) Cy were transfected with the indicated siRNAs and incubated with thymidine for 18 h to arrest cells in the early S phase. Whole-cell extract was prepared and blotted with anti-myc, Cdt1, Set8, and RCC1 antibodies. endo, endogenous. (B) Whole-cell extract and chromatin-containing extract prepared from asynchronously growing cells (asy) and from cells arrested with aphidicolin (early S) or released from aphidicolin arrest for 3 h (mid S) were transfected with the indicated siRNA. Protein levels were analyzed with indicated antibodies. RCC1 was used as a loading control. Asterisks on panels indicate nonspecific bands. Cont, control.
Ctf18-RFC has a key role in Cdt1 proteolysis during S phase.
To rule out the possibility that cell cycle arrest with DNA synthesis inhibitors itself adversely affected the stability of the Cdt1 protein, the cells were released from the aphidicolin block and the protein levels were analyzed 3 h later. At this time point, the majority of the cells were in mid-S phase regardless of the siRNA transfected, as revealed by flow cytometry, bromodeoxyuridine (BrdU) incorporation, and the presence of cyclin A (see Fig. S6 in the supplemental material). When compared among the aphidicolin-arrested (early-S-phase) cells, Cdt1(1–151) Cy was found to be maintained at higher levels in the Ctf18- or Cdt2-depleted cells than in the control or RFC1-depleted cells (Fig. 2B, compare lanes 2 and 8 with 5 and 11), the same as shown in Fig. 2A. At 3 h after release from aphidicolin arrest (mid-S phase), the levels remained high in the Ctf18- or Cdt2-depleted cells (Fig. 2B, lanes 6 and 12). Compared with its levels after the single knockdown of Ctf18, the levels of Cdt1(1–151) Cy did not increase after double knockdown of RFC1 and Ctf18 (see Fig. S5B in the supplemental material). Consistently, even in the asynchronously growing culture, Cdt1(1–151) Cy accumulated in the Ctf18-depleted cells (Fig. 2B, lane 10). We also examined the protein levels after depletion of the small subunit RFC4, which would result in disrupting the function of all RFC complexes. Although we could not block DNA replication completely after knocking down RFC4, Cdt1(1–151) Cy accumulated slightly compared with the level in control cells (see Fig. S5C in the supplemental material). The levels of Cdt1(1–151) Cy increased significantly after codepletion of Ctf18 but not of RFC1, even though DNA replication appeared to be more inhibited in cells doubly depleted of RFC1 and RFC4 than in cells depleted of Ctf18 and RFC4 (see Fig. S5C). Taken together, the above-described results indicate that Ctf18 acts as a major factor in Cdt1 degradation during S phase. An interesting observation is that although Cdt1(1–151) Cy was not degraded in the Ctf18-depleted cells at 3 h postrelease, a large amount of PCNA was detected on the chromatin, as observed in control or siRFC1-treated cells (Fig. 2B, lanes 3, 6, 9, and 12) (see below and Discussion).
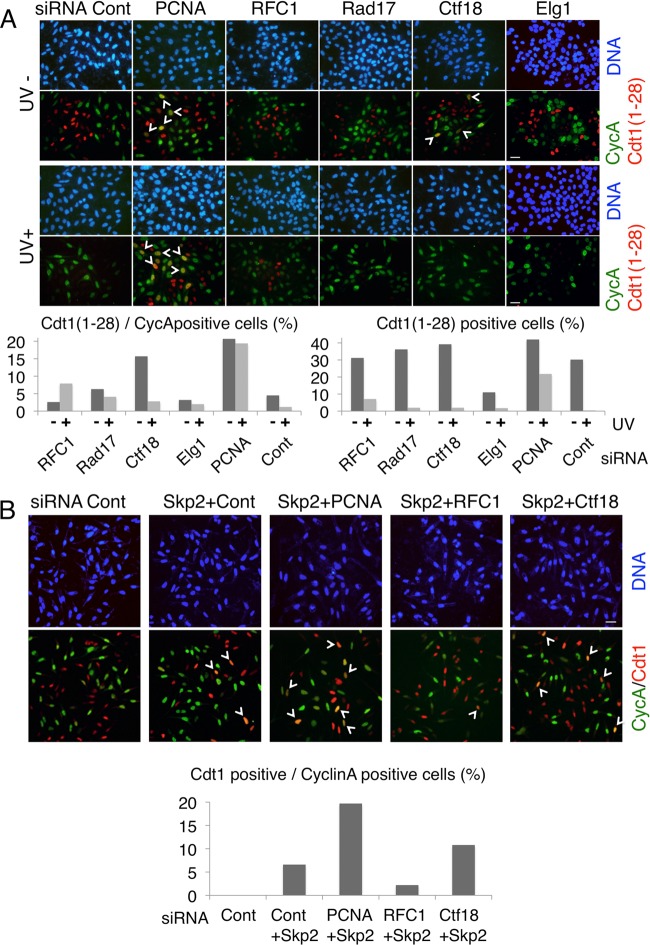
The key role of Ctf18 in CRL4Cdt2-mediated Cdt1 proteolysis during S phase was further confirmed by immunofluorescence analysis. We used the HeLa cell line stably expressing Cdt1(1–28) 9mycNLS, which contains only the N-terminal 28 amino acids of Cdt1 that serve as a degron for CRL4Cdt2. These cells were transfected with various siRNAs and stained simultaneously with anti-cyclin A (green signal) and anti-myc [Cdt1(1–28), red signal] antibodies (Fig. 3A, UV−). In control cells, cyclin A- and Cdt1(1–28)-positive cells were mutually exclusive, because Cdt1(1–28) was degraded in S-phase cells. Similar results were obtained with cells transfected with siRNA for RFC1, Rad17, or Elg1. In the PCNA- or Ctf18-knockdown cells, however, cell nuclei containing both signals (stained orange or yellow) were frequently observed, indicating that Cdt1(1–28) degradation in S phase was compromised in these cells (Fig. 3A, UV−, arrowheads and left graph).
Fig 3.
Ctf18 is required for Cdt1 degradation in S phase. (A) HeLa cells expressing Cdt1(1–28) and transfected with the indicated siRNA were UV irradiated (UV+) or not (UV−), and 1 h later, cells were fixed and costained with antibodies for cyclin A (CycA; green) and myc [Cdt1(1–28); red]. White arrowheads indicate nuclei that are positive for both cyclin A and Cdt1(1–28) and thus give yellow or orange signals. Bars, 20 μm. Cells stained with myc [Cdt1(1–28)] and cyclin A were counted. The frequencies of Cdt1(1–28)-positive cells among cyclin A-positive cells or in total cells are shown in graphs. (B) HeLa cells were transfected with the indicated combination of siRNAs and stained with antibodies for Cdt1 (red) and cyclin A (CycA; green). White arrowheads indicate doubly stained cells. Bar, 20 μm. Cdt1- and cyclin A-stained cells were counted, and the frequencies of Cdt1-positive cells among cyclin A-positive cells are shown in the graph (%). Cont, control.
Next, we examined whether endogenous Cdt1 is degraded during S phase in a Ctf18-dependent manner. Because endogenous Cdt1 is targeted by both CRL1Skp2 and CRL4Cdt2, CRL1Skp2 was selectively inactivated by the depletion of Skp2. HeLa cells were transfected with siSkp2 together with the siRNA for the control, PCNA, RFC1, or Ctf18 (Fig. 3B). In cells depleted of Skp2 alone or in combination with RFC1, only 2% to 6% of the cyclin A-positive cells were costained with anti-Cdt1, revealed by orange or yellow nuclear staining (Fig. 3B, arrowheads and graph). On the other hand, when PCNA or Ctf18 was knocked down together with Skp2, the doubly positive cells increased up to 10% to 20%, indicating that CRL4Cdt2 requires PCNA and its loader Ctf18-RFC for Cdt1 degradation during normal DNA replication in S phase.
RFC1 mediates Cdt1 proteolysis in both G1 and S phases after UV irradiation.
As described above, in G1 phase, Cdt1 was degraded after UV irradiation in an RFC1-dependent manner. Therefore, we examined whether Cdt1(1–28) that escaped degradation during S phase in the absence of Ctf18 was also resistant to UV irradiation. In the PCNA knockdown cells, Cdt1(1–28) remained stable in both G1 and S phases with or without UV irradiation (Fig. 3A). Consistent with the aforementioned results, Cdt1(1–28) present in G1-phase cells was not degraded after UV irradiation in the absence of RFC1. In contrast, both yellow and red signals were lost in the Ctf18 knockdown cells after UV treatment, indicating that Ctf18 is not necessary for the UV-induced degradation of Cdt1(1–28) (Fig. 3A, UV+ and right graph). When cells were simultaneously depleted of RFC1 and Ctf18, degradation of Cdt1(1–28) was prevented both in S phase and after UV irradiation, as in Cdt2-depleted cells, indicating that RFC1 is required for UV-induced degradation regardless of the cell cycle stage (G1 or S) (see Fig. S7 in the supplemental material). These results again indicate that Cdt1 proteolysis differentially utilizes the two RFC complexes, Ctf18-RFC during normal DNA replication in S phase and RFC1-RFC in response to UV irradiation.
Both RFC1 and Ctf18 associate with chromatin during S phase.
As mentioned above, although Cdt1(1–151) Cy was not degraded in the Ctf18-depleted cells at 3 h postrelease, a large amount of PCNA was detected on the chromatin (Fig. 2B). These results suggested that chromatin loading of PCNA alone is not sufficient but that Ctf18 is also required for CRL4Cdt2-mediated Cdt1 degradation during S phase. To evaluate the role of RFC complexes, we first examined the chromatin association of RFC1 and Ctf18 throughout the cell cycle. Synchronized HeLa cell cultures were obtained by thymidine-aphidicolin block and release. The progression of the cell cycle was monitored by flow cytometry and by the presence of Cdt1, cyclin A, and phosphorylated histone H3(Ser10). The levels of RFC1, Ctf18, and PCNA in the whole-cell extracts were almost constant throughout the cell cycle (Fig. 4A). Consistent with the notion that RFC1 and Ctf18 have a role in chromosomal events during S phase (RFC1 in replication fork progression and Ctf18 in the establishment and maintenance of chromosomal cohesion), the amounts of both proteins associated with chromatin increased as the cells underwent S phase (Fig. 4A). When the cells reentered G1 phase, however, only RFC1 was detected in the chromatin-bound fractions at levels similar to those in S phase under these conditions, although the levels of PCNA on the chromatin remained low once the cells completed DNA replication.
Fig 4.
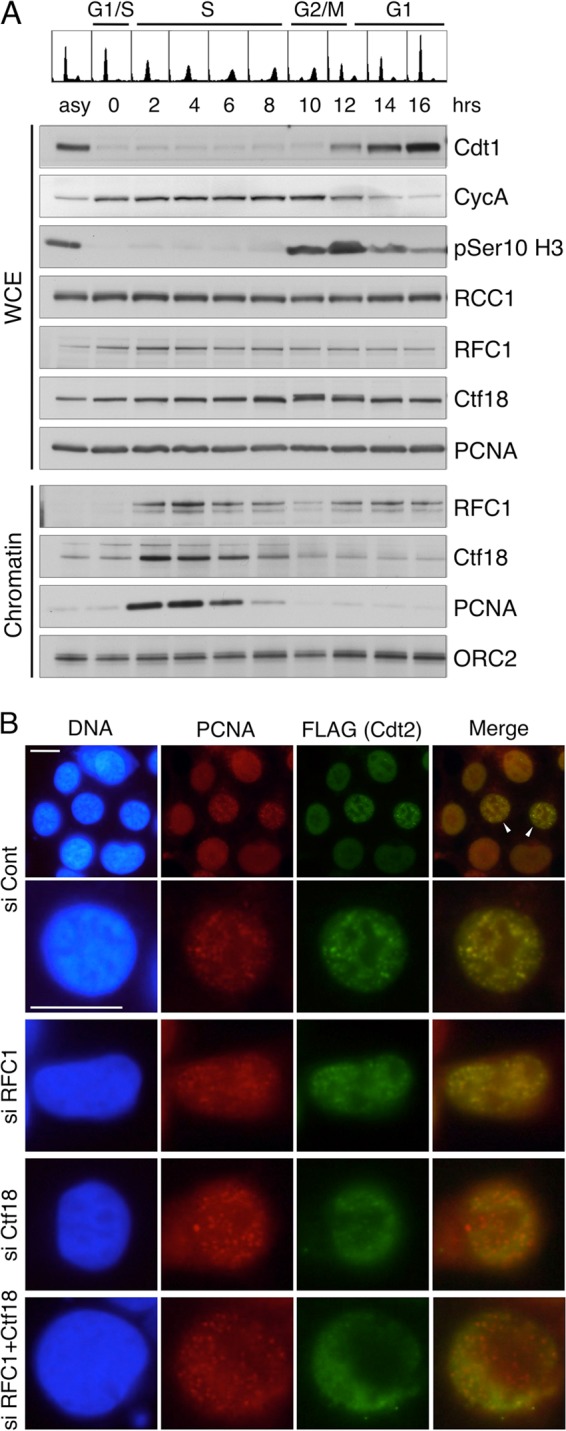
Ctf18 associates with chromatin and recruits Cdt2 to the PCNA foci during S phase. (A) HeLa cells synchronized at the early S phase by thymidine-aphidicolin block were released and collected at the indicated times. Whole-cell extract (WCE) and a chromatin-containing fraction were prepared, and proteins were analyzed. (B) 293 cells stably expressing Cdt2-FLAG were transfected with indicated siRNAs and stained for PCNA and Cdt2-FLAG. Bars, 10 μm. Cont, control.
Ctf18 is required for the recruitment of Cdt2 to replication foci containing PCNA.
Next, we examined the role of RFC proteins in the recruitment of Cdt2. When asynchronously growing 293 cells stably expressing Cdt2-FLAG were stained with PCNA, cells with foci were frequently observed (Fig. 4B, si Cont, arrowheads). These foci represent the sites of DNA replication (8). Costaining with PCNA and FLAG antibodies revealed that both proteins were well colocalized, indicating that CRL4Cdt2 is recruited to the chromatin sites where PCNA is loaded (Fig. 4B, si Cont). When cells had either RFC1 or Ctf18 knocked down, PCNA foci were detected in both cases and Cdt2 was colocalized with PCNA foci in RFC1-depleted cells. In contrast, the Cdt2 signal at the PCNA foci was highly reduced in Ctf18-depleted cells and also in Ctf18 and RFC1 double-knockdown cells. These results suggest that Ctf18-RFC is required for recruitment of CRL4Cdt2 to the PCNA loaded at DNA replication sites.
Based on the results described above, we examined whether two RFC complexes, Ctf18-RFC and RFC1-RFC, interacted with CRL4Cdt2. We used 293 cells stably expressing Cdt2-FLAG and 293 cells as a control. Asynchronously growing, UV-irradiated, or mid-S-phase-synchronized cells treated with MG132 were fixed with formaldehyde, cell extracts were used for immunoprecipitation with anti-FLAG antibodies, and Cdt2-bound proteins were examined. In addition to PCNA, both Ctf18 and RFC1 were detected in the precipitates from all lysates, but neither of them was detected in the control precipitates (see Fig. S8, lanes 10 to 12, in the supplemental material). The interaction of Cdt2 with PCNA and RFC1 but not with Ctf18 increased after UV irradiation, consistent with the role of these two loaders in Cdt1 degradation after DNA damage (see Fig. S8; compare lanes 10 and 11). In contrast, the interaction of Cdt2 with Ctf18 remained at the same level, though the interaction of Cdt2 with PCNA increased in S phase.
Ctf18-mediated Cdt1 proteolysis is independent of the establishment of cohesion but is required to block DNA rereplication.
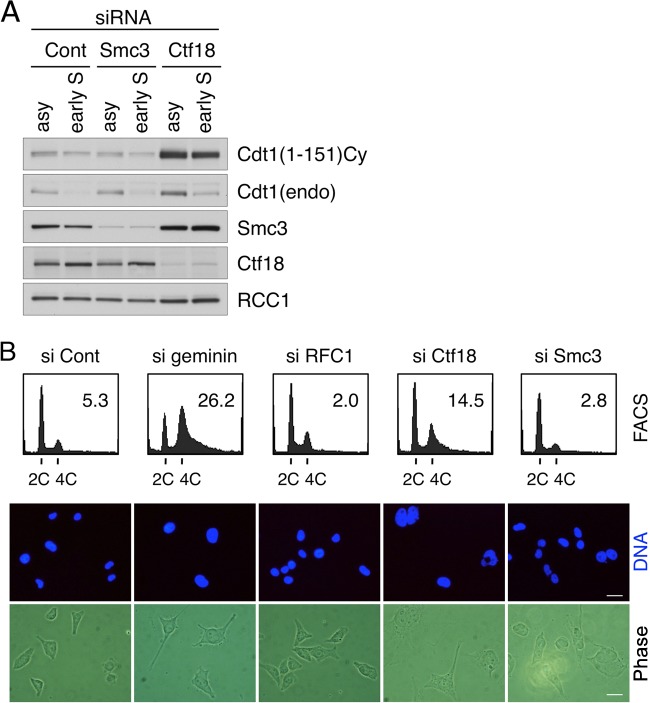
Previous reports indicated that Ctf18 is involved in the establishment of sister chromatid cohesion and in replication fork progression through interactions with the cohesin complex (3, 27, 53). Therefore, the cohesin protein or a Ctf18-dependent process of establishing cohesion might have been involved in the Cdt1 degradation. To address this possibility, Cdt1(1–151) Cy degradation was examined in cells depleted of the cohesin subunit Smc3. In cells arrested in early S phase, Cdt1(1–151) Cy was efficiently degraded, even when Smc3 expression was suppressed, compared with its degradation in Ctf18 knockdown cells (Fig. 5A). The Ctf18-RFC complex is composed of additional components, Dcc1 and Ctf8, which are also implicated in the establishment of cohesion. In contrast to the results for Ctf18, silencing of either of these components did not stabilize Cdt1(1–151) Cy (see Fig. S9A in the supplemental material). These results suggest that the function of Ctf18 in Cdt1 proteolysis is independent of the establishment of cohesion.
Fig 5.
Ctf18-RFC is involved in Cdt1 degradation independent of the establishment of cohesion and is required to block DNA rereplication. (A) HeLa cells expressing Cdt1(1–151) Cy were transfected with indicated siRNAs, and Cdt1 levels in asynchronously growing cells (asy) and cells arrested with aphidicolin for 18 h (early S) were analyzed. endo, endogenous. (B) HeLa cells stably expressing Cy-Cdt1 were transfected with the indicated siRNAs, and DNA content and nucleus size were analyzed by flow cytometry (fluorescence-activated cell sorter [FACS]) and DNA staining, respectively. Relative amounts of cells with a DNA content higher than 4C are shown in each histogram (%). Bars in lower panels are 20 μm. Cont, control; phase, phase contrast.
Previously, we and other groups reported that either Cdt1 overexpression or geminin knockdown leads to the overreplication of DNA (31, 37, 54, 57). Therefore, we examined the possible effects of Ctf18 knockdown on DNA replication. For these experiments, we used the HeLa cell line stably expressing Cy-mutated full-length Cdt1 (Cy-Cdt1) (37). This cell line showed extensive rereplication when geminin was depleted, as shown in Fig. 5B. In contrast to the RFC1 or Smc3 knockdown cells, whose cell cycle profiles were indistinguishable from those of the control cells, a significant population of the Ctf18 knockdown cells had a DNA content higher than 4C. Such cells were not detected in the Dcc1- or Ctf8-depleted Cy-Cdt1-expressing cells (see Fig. S9B in the supplemental material) or in the Ctf18-depleted wild-type Cdt1-expressing cells (see Fig. S10 in the supplemental material). In addition, nocodazole treatment did not prevent the appearance of such cells, suggesting that cells with higher DNA content were not due to mitotic failure (see Fig. S10). DNA staining also showed that the geminin- or Ctf18-depleted cells had larger nuclei than siRFC1, siSmc3, and control cells (Fig. 5B). These results suggest that compromising Ctf18 function for Cdt1 proteolysis led to DNA rereplication.
DISCUSSION
The DNA replication-licensing factor Cdt1 is degraded through the CRL4Cdt2-mediated ubiquitination pathway, which is dependent on PCNA-loaded chromatin. Here, we demonstrated distinct roles of two RFC complexes in Cdt1 proteolysis: RFC1-RFC is involved in the CRL4Cdt2 pathway triggered by UV irradiation and, unexpectedly, Ctf18-RFC plays a critical role in Cdt1 proteolysis during normal DNA replication in S phase.
In UV-irradiated cells, the NER pathway operated to repair the DNA photolesions. The damage-containing oligonucleotide was excised and the resulting gap filled in by DNA polymerases with the aid of PCNA. Consistent with the notion that the XPA protein is an essential component of the preincision NER protein complex, we found that XPA-deficient cells were defective in both the chromatin loading of PCNA and Cdt1 degradation in response to UV irradiation. Our RNAi experiments indicated that, among the known mammalian RFC complexes, RFC1-RFC is responsible for the UV-induced degradation of Cdt1. Our results suggest that other RFC complexes cannot substitute for RFC1-RFC in the gap-filling DNA repair synthesis associated with NER.
Contrary to our expectations, an alternative RFC complex, Ctf18-RFC, was identified as the major loader involved in Cdt1 degradation during S phase. In this phase, PCNA is loaded at the replication fork and interacts with numerous factors to orchestrate many aspects of the replication-linked processes and to regulate the proteins involved in cell cycle progression. One important role of PCNA is to target Cdt1 for CRL4Cdt2-mediated degradation on the chromatin, which serves as a negative feedback control to inactivate the initiator protein to inhibit rereplication. Importantly, such control is conserved in Escherichia coli, where it is known as RIDA (regulatory inactivation of DnaA) (23). The sliding clamp of E. coli DNA polymerase III (beta subunit) is loaded onto the DNA by the clamp loader gamma complex. In eukaryotes, the classical RFC complex containing RFC1 loads PCNA onto DNA during replication. Instead of RFC1-RFC, Ctf18-RFC was involved in Cdt1 degradation during S phase. Consistently, Ctf18, like RFC1, was detected on the chromatin during S phase. Ctf18-RFC, which is composed of Ctf18, Ctf8, Dcc1, and the core RFC subunits (RFC2-5), was originally identified as an alternative RFC complex required for sister chromatid cohesion in budding yeast (14, 30). EcoI acetyltransferase interacts with PCNA through its PCNA binding motif (PIP box) and acetylates Smc3 to establish cohesion (33, 46). Depletion of the cohesin subunit had no effect on Ctf18-mediated Cdt1 degradation, arguing against the notion that Cdt1 degradation is coupled with the establishment of cohesion. In addition, depletion of the small subunits of Ctf18-RFC, Ctf8 and Dcc1, which are required for cohesion, had no effect on Cdt1 degradation (see Fig. S9 in the supplemental material). We also found that, in Cy-Cdt1-expressing cells, depletion of Ctf18 resulted in the accumulation of cells with an abnormally high DNA content, a possible result of rereplication of DNA. Taken together, the present data demonstrate a novel role for Ctf18-RFC in preventing DNA replication-licensing after the onset of S phase by assisting the CRL4Cdt2-mediated proteolysis of Cdt1.
In the absence of Ctf18, Cdt1(1–151) Cy persisted during S phase despite the presence of a large amount of PCNA loaded onto the chromatin (Fig. 2B). This implies that the chromatin loading of PCNA is necessary but not sufficient for CRL4Cdt2-mediated proteolysis of Cdt1 during S phase. In addition to loaded PCNA, at least Ctf18 is required for Cdt1 degradation in S phase. We expect that in addition to its role as a PCNA loader, Ctf18 is required for recruiting CRL4Cdt2 to PCNA loaded on the chromatin. In accordance with this idea, we demonstrated that during S phase, Cdt2 localization at PCNA foci, which represent DNA replication sites, was dependent on Ctf18 but not on RFC1 (Fig. 4B). However, our data do not directly address whether the function of Ctf18 in Cdt2 recruitment is related to its role in PCNA loading. Then, how is Ctf18, rather than RFC1 involved during S phase? At the front of the leading-strand synthesis, a PCNA clamp slides continuously on DNA, while on lagging strands, PCNA must be loaded every time Okazaki fragment synthesis is initiated. In vitro analysis showed that both RFC1-RFC and Ctf18-RFC are capable of recognizing the 3′-OH end of a primer annealed to the template DNA strand and loading PCNA. One possibility is that each loader may not only load PCNA but also selectively recruit different factors that interact with PCNA during or after its loading; consequently, PCNA loaded by RFC1-RFC may be dedicated to replicative DNA polymerases for DNA synthesis, whereas PCNA loaded by Ctf18-RFC may function with CRL4Cdt2 to degrade Cdt1 and/or in other processes, such as the establishment of cohesion. This would be consistent with a previous report showing that, despite its ability to load PCNA, Ctf18-RFC cannot substitute for RFC1-RFC in the in vitro simian virus 40 (SV40) DNA replication system (49). Moreover, although both RFC1-RFC and Ctf18-RFC interact directly with translesion DNA polymerase η, only Ctf18-RFC stimulates its DNA synthesis (48). Given the abundance of interacting partners, it would be important to dictate properly selective functions to PCNA by, at least in part, the RFC complexes themselves. Any missorting of PCNA would induce a defect in DNA replication and DNA damage. This possibility would explain why PCNA loaded in the absence of Ctf18 could not participate in the CRL4Cdt2-dependent Cdt1 degradation pathway. On the other hand, it is also possible that subpopulations of PCNA loaded by different RFC complexes are marked differently for different aspects of DNA metabolism. However, we could not detect a difference in modifications, such as monoubiquitination and Tyr211 phosphorylation (56), on PCNA after depleting RFC1or Ctf18 (data not shown). In contrast to our results in human cells, RFC1 is involved in Cdt1 degradation during S phase in Saccharomyces pombe. We speculate that this difference may result from differences in the Cdt2 and Cdt1 structures. Mammalian Cdt2 has an extended C-terminal region compared with that of fission yeast Cdt2. This region might be involved in selective recruitment of CRL4Cdt2 to PCNA loaded by or linked with Ctf18 during S phase. Recently, it was reported that S. pombe Cdt1 has two PIP-like motifs (13), both of which are recognized for degradation, and this may render S. pombe Cdt1 able to be degraded, dependent on RFC1.
In contrast, RFC1-RFC is involved in Cdt1 degradation after UV irradiation in G1 phase. Interestingly, we found that Cdt1, which was stabilized in S phase in the absence of Ctf18, was degraded dependent on RFC1-RFC after UV irradiation (Fig. 3A, UV+; also see Fig. S7 in the supplemental material). Many reports support the notion that RFC1-RFC is recruited to the sites of DNA damage, where it recruits DNA pol delta for gap filling during NER. A recent report indicates that another replicative polymerase, pol epsilon, is recruited by Ctf18-RFC (41), raising the possibility that both RFC1-RFC and Ctf18-RFC are involved in UV-induced Cdt1 degradation. We found that the chromatin loading of Cdt2 and PCNA was defective in RFC1-depleted cells but not in Ctf18-depleted cells (Fig. 1B; also see Fig. S3B in the supplemental material), indicating that PCNA loading at the sites of DNA damage was performed mainly by RFC1-RFC. Interestingly, RFC1 but not Ctf18 was detected in the chromatin-containing fraction in G1 phase, as well as in S phase (Fig. 4A), suggesting that RFC1-RFC localizes more easily to the damaged sites, at least in G1 phase. In addition, we found that interaction of Cdt2 with RFC1 and PCNA but not with Ctf18 increased after UV irradiation (see Fig. S8 in the supplemental material). Given that Ctf18-RFC is also recruited to the damaged sites, this may occur only after PCNA is loaded by RFC1-RFC, perhaps, for example, especially to recruit DNA pol epsilon.
Among the PIP box-containing proteins, CRL4Cdt2 targets only selected ones. In mammalian cells, Cdt1, p21, and Set8 are reported targets of CRL4Cdt2. We and researchers in other laboratories have shown that targets of CRL4Cdt2 share a specialized degron created by the PIP box and additional residues within and downstream from the PIP box (15, 16, 32). In the present work, the degradation of Set8 in S phase and following UV irradiation was also dependent on the different loaders, as observed with Cdt1, suggesting that all the CRL4Cdt2 targets are regulated in the same way. In addition, it was reported that fission yeast Cdt1 contains dual PIP degrons that may be required for fine control of Cdt1 proteolysis (13) and that different ubiquitin-conjugating enzymes are involved in targeting different substrates for CRL4Cdt2 (47). Our finding that RFC complexes participate in distinguishing proteins for their interaction with PCNA may provide another layer of selection of target proteins and fine control for ubiquitination at the replication forks.
PCNA orchestrates many aspects of cellular functions associated with the faithful propagation of genomic and epigenetic information and the maintenance of genome integrity. The findings of this study demonstrate a novel link between RFC complexes and Cdt1 proteolysis. This implies that a sophisticated protein network coordinates the precise cell cycle progression, such as DNA replication, damage repair, and chromosomal cohesion, where the RFC complexes, together with PCNA, form a functional core. Further studies on the mechanisms underlying Cdt1 proteolysis mediated by PCNA and its loader RFC complexes will provide additional insight into the complexity of such a homeostatic control.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Z. Lygerou for corrections and critical reading of the manuscript.
This work was supported by Grants in Aid for Scientific Research, Scientific Research in a Priority Area, and the Global Center of Excellence Program from the Ministry of Education, Culture, Sport, Science, and Technology of Japan and by the Naito Foundation, the Hyogo Science and Technology Association, and the Uehara Memorial Foundation.
Footnotes
Published ahead of print 9 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abbas T, et al. 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abbas T, et al. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ansbach AB, et al. 2008. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol. Biol. Cell 19: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arias EE, Walter JC. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8: 84–90 [DOI] [PubMed] [Google Scholar]
- 5. Arias EE, Walter JC. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21: 497–518 [DOI] [PubMed] [Google Scholar]
- 6. Aroya SB, Kupiec M. 2005. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst.) 4: 409–417 [DOI] [PubMed] [Google Scholar]
- 7. Blow JJ, Dutta A. 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bravo R, Macdonald-Bravo H. 1987. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J. Cell Biol. 105: 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centore RC, et al. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dinant C, Houtsmuller AB, Vermeulen W. 2008. Chromatin structure and DNA damage repair. Epigenetics Chromatin 1: 9 doi:10.1186/1756-8935-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedberg EC, et al. 2006. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst.) 5: 986–996 [DOI] [PubMed] [Google Scholar]
- 12. Gillet LC, Scharer OD. 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106: 253–276 [DOI] [PubMed] [Google Scholar]
- 13. Guarino E, et al. 2011. Cdt1 proteolysis is promoted by dual PIP degrons and is modulated by PCNA ubiquitylation. Nucleic Acids Res. 39: 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanna JS, Kroll ES, Lundblad V, Spencer FA. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21: 3144–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Havens CG, Walter JC. 2009. Docking of a specialized PIP box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Havens CG, Walter JC. 2011. Mechanism of CRL4Cdt2, a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25: 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. 2006. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20: 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higa LA, et al. 2006. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- 19. Ishii T, et al. 2010. Proliferating cell nuclear antigen-dependent rapid recruitment of Cdt1 and CRL4Cdt2 at DNA-damaged sites after UV irradiation in HeLa cells. J. Biol. Chem. 285: 41993–42000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin J, Arias EE, Chen J, Harper JW, Walter JC. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23: 709–721 [DOI] [PubMed] [Google Scholar]
- 21. Jorgensen S, et al. 2011. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanellis P, Agyei R, Durocher D. 2003. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13: 1583–1595 [DOI] [PubMed] [Google Scholar]
- 23. Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94: 61–71 [DOI] [PubMed] [Google Scholar]
- 24. Kim DH, et al. 2010. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol. Cell. Biol. 30: 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim J, MacNeill SA. 2003. Genome stability: a new member of the RFC family. Curr. Biol. 13: R873–R875 [DOI] [PubMed] [Google Scholar]
- 26. Kim Y, Starostina NG, Kipreos ET. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22: 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lengronne A, et al. 2006. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- 28. Liu E, Li X, Yan F, Zhao Q, Wu X. 2004. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 279: 17283–17288 [DOI] [PubMed] [Google Scholar]
- 29. Majka J, Burgers PM. 2004. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 78: 227–260 [DOI] [PubMed] [Google Scholar]
- 30. Mayer ML, Gygi SP, Aebersold R, Hieter P. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7: 959–970 [DOI] [PubMed] [Google Scholar]
- 31. Melixetian M, et al. 2004. Loss of geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Michishita M, et al. 2011. Positively charged residues located downstream of PIP box, together with TD amino acids within PIP box, are important for CRL4(Cdt2)-mediated proteolysis. Genes Cells 16: 12–22 [DOI] [PubMed] [Google Scholar]
- 33. Moldovan GL, Pfander B, Jentsch S. 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- 34. Nichols AF, Sancar A. 1992. Purification of PCNA as a nucleotide excision repair protein. Nucleic Acids Res. 20: 2441–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishitani H, et al. 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 10: 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishitani H, et al. 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283: 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishitani H, et al. 2006. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. 2001. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276: 44905–44911 [DOI] [PubMed] [Google Scholar]
- 39. Nurse P. 1994. Ordering S phase and M phase in the cell cycle. Cell 79: 547–550 [DOI] [PubMed] [Google Scholar]
- 40. Oda H, et al. 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ogi T, et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37: 714–727 [DOI] [PubMed] [Google Scholar]
- 42. Overmeer RM, et al. 2010. Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30: 4828–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roukos V, et al. 2011. Dynamic recruitment of licensing factor Cdt1 to sites of DNA damage. J. Cell Sci. 124: 422–434 [DOI] [PubMed] [Google Scholar]
- 44. Sansam CL, et al. 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20: 3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Senga T, et al. 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281: 6246–6252 [DOI] [PubMed] [Google Scholar]
- 46. Sherwood R, Takahashi TS, Jallepalli PV. 2010. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev. 24: 2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shibata E, Abbas T, Huang X, Wohlschlegel JA, Dutta A. 2011. Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol. Cell. Biol. 31: 3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shiomi Y, Masutani C, Hanaoka F, Kimura H, Tsurimoto T. 2007. A second proliferating cell nuclear antigen loader complex, Ctf18-replication factor C, stimulates DNA polymerase eta activity. J. Biol. Chem. 282: 20906–20914 [DOI] [PubMed] [Google Scholar]
- 49. Shiomi Y, et al. 2004. The reconstituted human Chl12-RFC complex functions as a second PCNA loader. Genes Cells 9: 279–290 [DOI] [PubMed] [Google Scholar]
- 50. Shivji KK, Kenny MK, Wood RD. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell 69: 367–374 [DOI] [PubMed] [Google Scholar]
- 51. Sugimoto N, et al. 2004. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J. Biol. Chem. 279: 19691–19697 [DOI] [PubMed] [Google Scholar]
- 52. Tardat M, et al. 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- 53. Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vaziri C, et al. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell 11: 997–1008 [DOI] [PubMed] [Google Scholar]
- 55. Waga S, Stillman B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67: 721–751 [DOI] [PubMed] [Google Scholar]
- 56. Wang SC, et al. 2006. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8: 1359–1368 [DOI] [PubMed] [Google Scholar]
- 57. Zhu W, Chen Y, Dutta A. 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 24: 7140–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zou L, Cortez D, Elledge SJ. 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.