Abstract
The specific set of proteins bound at each genomic locus contributes decisively to regulatory processes and to the identity of a cell. Understanding of the function of a particular locus requires the knowledge of what factors interact with that locus and how the protein composition changes in different cell types or during the response to internal and external signals. Proteomic analysis of isolated chromatin segments (PICh) was developed as a tool to target, purify, and identify proteins associated with a defined locus and was shown to allow the purification of human telomeric chromatin. Here we have developed this method to identify proteins that interact with the Drosophila telomere-associated sequence (TAS) repeats. Several of the purified factors were validated as novel TAS-bound proteins by chromatin immunoprecipitation, and the Brahma complex was confirmed as a dominant modifier of telomeric position effect through the use of a genetic test. These results offer information on the efficacy of applying the PICh protocol to loci with sequence more complex than that found at human telomeres and identify proteins that bind to the TAS repeats, which might contribute to TAS biology and chromatin silencing.
INTRODUCTION
Proteomic analysis of isolated chromatin segments (PICh) was developed as an unbiased method of identifying proteins that physically interact with a specific locus in the genome (11), and this was done by using the telomeres of mammalian cells as a target. Telomeres are found in multiple copies of a simple repeat sequence and so do not offer the same challenge to the use of PICh as other genomic loci do. In this work, we applied PICh to the telomere-associated sequence (TAS) repeats of Drosophila to demonstrate the efficacy of the technology and to learn about the biology of these repeats.
TAS repeats are found in the subtelomeric region of chromosomes 2, 3, and X and nucleate a particular kind of heterochromatin which is responsible for the telomeric position effect (TPE; for a review, see reference 37). As also seen with pericentromeric heterochromatin-mediated position effect variegation (PEV), reporter genes inserted at the TAS repeats, or between the TAS repeats and the telomeric retrotransposon (HTT) arrays, variegate. The extent of gene silencing depends on the size, and hence the strength of the transcription activating effect, of the HTT array and the transcription-repressing activity of the TAS repeats (28). Interestingly, though, most modifiers of PEV [Mod(PEV)] have no effect on TPE; in fact, very few modifiers of TPE [Mod(TPE)] have been unambiguously described so far. Among these are the Polycomb group (PcG) genes, which in some studies have been proposed to act as dominant suppressors of TPE [Su(TPE)] and whose encoded proteins have been found to be located at the telomeric regions of polytene chromosomes (6). These findings indicate that TPE is a distinctive class of chromatin silencing which shares mechanistic features with both pericentromeric heterochromatin and PcG-mediated silencing of developmental regulators.
The extent to which PcG proteins and other reported Su(TPE) bind at TAS repeats and modify TPE, though, has become less clear since the finding of a high incidence of TPE-suppressing terminal deletions on chromosome 2L in public Drosophila mutant stocks (38, 46). This leads to a high rate of false-positive identifications of Su(TPE), in which the modifying activity is attributable to the 2L deletion, which eliminates the TAS repeats at that location and suppresses TPE in trans, rather than to the mutant gene being tested (38). The variability of results from genetic screenings for Mod(TPE) makes it difficult to advance hypotheses for the mechanisms working at TAS repeats. A possible way to understand these processes would be to identify which proteins can be found physically at the loci and then study them in more detail. We thus decided to use PICh to identify candidates for binding at TAS repeats.
The TAS repeats provide an excellent model for PICh development for several reasons. They are relatively large targets (∼45 kb/variant) with abundant repeated sequence, yet there are 30-fold fewer target sequences for a 25-nucleotide (nt) capture probe than in human telomeres. There are two families of TAS repeats, which provide not only different genomic locations to be targeted in parallel but also, due to differences in organization between the repeats, allows comparison of the efficiencies of PICh with different densities of target sequences. Finally, TAS repeats have reported functional differences between somatic tissues and the female germ line, where they function as Piwi-interacting-RNA-producing clusters (7, 55). The future extension of findings with cell lines into different physiological states will be informative of the role of TAS repeats in Drosophila chromatin regulation.
With only one validated locus targeted by PICh to date (human telomeres), we considered the various challenges of applying the method to other loci. Multiple factors have the potential to contribute to the success of an experiment like PICh, i.e., the relative abundance of the targeted sequence, the chromatin architecture of the locus, the density of the target sequence per DNA unit length, the design of the capture probe(s), and the balance between the stability of the cross-links between proteins and DNA and the efficiency of the capture probe invasion of the target DNA double strand. We have implemented pre-enrichment steps in the PICh protocol and introduced a series of filters to the identified proteins to rank the most likely candidate TAS proteins. With these modifications, we identified over 70 candidate proteins for direct binding to the two families of TAS repeats and validated 5 of these by chromatin immunoprecipitation (ChIP). We found that the majority of the proteins identified are not dominant Su(TPE), but the Brahma complex is a dominant Mod(TPE). These results suggest the existence of a distinctive mode of regulation at TAS repeats whereby chromatin silencing is less dependent on dose effects than in the case of PEV.
MATERIALS AND METHODS
Capture probe synthesis.
Dimethoxytrityl-protected locked nucleic acid T (DMT-LNA-T), DMT-LNA-ABz, and DMT-LNA-GDMF phosphoramidites were obtained from Exiqon; CPG oligonucleotide synthesis columns, Spacer 18, and desthiobiotinTEG phosphoramidites were obtained from Glen Research; dABz, dCBz, dT, and dGDMF phosphoramidites were obtained from Applied Biosystems. See the supplemental material for probe design considerations. Reagents were reconstituted into recommended concentrations with acetonitrile, and synthesis was done on an Expedite 8909 DNA synthesizer (Applied Biosystems) by using the recommended coupling conditions for each monomer. Capture probe was eluted from the resin with ammonium hydroxide and purified from a 15% acrylamide gel, and DMT was removed with 80% acetic acid prior to a final ethanol precipitation and resuspension in 0.1% TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0).
PICh.
Drosophila S3 and Kc cells were grown in suspension in CCM3 medium (HyClone) supplemented with Pen/Strep (Gibco) at 27°C and 100 rpm in 2.8-liter culture flasks to a density of 1 × 107 to 2 × 107/ml. For PICh experiments, typically, 1011 cells were centrifuged at 4,000 rpm at room temperature (RT) in a Beckman J6 centrifuge, washed with 400 ml 1× phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), centrifuged at 4,000 × g at RT, washed once with 5 packed cell volumes (pcv) of hypotonic buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl), resuspended with 3 pcv of hypotonic buffer, and swelled for 10 min on ice in two 100-ml Dounce homogenizers (Kontes); 37% formaldehyde (Fisher) was added to a final concentration of 3%, and the mixture was immediately homogenized with 15 strokes of a tight pestle and centrifuged for 10 min at RT at 5,000 × g. The supernatant was disposed of, and the pellet was resuspended with a total of 400 ml of cross-linking solution (3% formaldehyde, 1× PBS) and incubated for 30 min at RT on a shaking platform. The pellet was washed three times with PBS and then once with sucrose buffer (0.3 M sucrose; 10 mM HEPES, pH 7.9; 1% Triton X-100; 2 mM magnesium acetate), resuspended with 3 packed nuclear volumes (pnv) of sucrose buffer, and homogenized with 20 strokes of a tight pestle in an 100-ml homogenizer, and the pellet was kept after chromatin centrifugation at 5,000 × g for 10 min. Chromatin was washed once with RNase buffer (0.5% Triton X-100, 1× PBS) and resuspended with 5 pnv of RNase buffer, 0.01 pnv of RNase A (Sigma) was added, and the mixture was incubated for 5 h at RT on a rotating wheel and kept overnight at 4°C. The pellet was washed twice with 6 pnv of PBS and once with 6 pnv of LB3JD buffer (10 mM HEPES, pH 7.9; 0.1 M NaCl; 2 mM EDTA, pH 8.0; 1 mM EGTA, pH 8.0; 0.2% SDS; 0.1% Na-lauroylsarcosine), resuspended with 3 pnv of LB3JD, split into 5-ml aliquots in 15-ml polystyrene tubes, and sonicated on ice for a total processing time of 7 min of 15-s on pulses and 45-s off pulses in a Misonix sonicator with the power level set to 7.0 (39- to 42-W output). The aliquots were pooled and centrifuged at 25,000 × g for 1 h at RT, and chromatin was dialyzed against 30 volumes of buffer Y (5% glycerol; 20 mM HEPES, pH 7.9; 50 mM NaCl; 0.05% SDS; 0.05% Na-lauroylsarcosine; 0.02% Triton X-100; 1 mM EDTA, pH 8.0; 0.5 mM EGTA, pH 8.0) through a CE dialysis membrane with a molecular weight cutoff of 106 (Spectra/Por) for 4 h.
To preclear biotinylated molecules from the mixtures, chromatin was incubated for 5 min in a water bath at 60°C in 50-ml Falcon tubes, mixed regularly to transfer heat uniformly, and removed from the water bath. A 1:100 (vol/vol) Ultralink Plus streptavidin bead slurry (Thermo Scientific) was added, the mixture was incubated on a rotating wheel at RT for 2 h, and the flowthrough from an Econo-pac column (Bio-Rad) was collected.
For capture probe hybridization and purification, a 500-fold molar excess (to target copies) of capture probe was added to 10 mg of chromatin (as determined by A260) (in the case of TAS repeats, ∼3 nmol of capture probe/10 mg of chromatin). Capture probe hybridization was done with 15-ml polystyrene tubes as follows: 6 min at 70°C, 60 min at 37°C, 2.5 min at 60°C, 60 min at 37°C, 2.5 min at 60°C, and 120 min at 37°C. The mixture was transferred into 1.5-ml tubes and centrifuged for 15 min at the maximum speed at RT. The supernatant was transferred into a new 15-ml Falcon tube, 100 mM NaCl and 300 μl of MyOne magnetic streptavidin beads (Invitrogen) in LB3JD buffer were added, and the mixture was incubated on a rotating wheel at RT for 2 h. The beads were immobilized on a magnetic stand, washed seven times by gentle resuspension with 8 ml LB3JD buffer, transferred into a low-binding 1.5-ml tube, and washed twice for 5 min with 1 ml LB3JD buffer at 42°C at 1,000 rpm in a Thermomixer (Eppendorf) and then for 1 h at RT at 1,000 rpm. Elution was done for each sample with 1 ml elution buffer (12.5 mM biotin; 7.5 mM HEPES, pH 7.9; 75 mM NaCl; 1.5 mM EDTA, pH 8.0; 0.75 mM EGTA, pH 8.0; 0.15% SDS; 0.075% Na-lauroylsarcosine) at RT at 1,000 rpm for 2 h. Eluates were collected in a clean tube and centrifuged for 1 min to remove any magnetic beads that might have been carried over, and supernatants were transferred to new tubes. Proteins were precipitated by adding 100% cold trichloroacetic acid to a final concentration of 20%. Samples were incubated for 10 min on ice and centrifuged for 15 min at RT, and the supernatant was carefully removed. The pellet was washed twice with −20°C acetone by vortexing for a few seconds and centrifugation between washes. The pellets were briefly air dried and resuspended with 40 μl of cross-link reversal buffer (0.25 M Tris, pH 8.8; 2% SDS; 0.5 M β-mercaptoethanol); cross-links were reversed by sample incubation at 99°C for 25 min.
Samples were separated by SDS-PAGE (or stored at −20°C); gels were stained with the SilverQuest kit (Invitrogen) or with colloidal blue (Invitrogen) according to the manufacturer's instructions, and relevant regions of the gel were cut out of the TAS-specific and corresponding regions of the negative-control lanes and sent for analysis to the Taplin Mass Spectrometry Facility at the Harvard Medical School. Our decision to isolate large sections of the gel for mass spectrometry analysis was made to the detriment of deeper coverage but with the advantage of providing an overview of protein composition at a reduced cost. The clean appearance of the negative-control lane relative to the sample lanes indicates the extent to which we enriched for proteins with the specific capture probe. The number of peptides detected by mass spectrometry does not reflect the total amount of protein isolated by these two probes, given the mechanics of the liquid chromatography-tandem mass spectrometry apparatus, which discards a fraction of the injected peptides above its resolution power. For example, compare the number of bands detected by silver staining using specific versus nonspecific probes to the total number of proteins identified in each sample of the protein enrichment with the specific capture probes. This technical consideration means that more-dilute samples (such as the negative control) will have deeper coverage than more-concentrated samples (such as the specific purifications), thus leading to a reduction in the difference between the numbers of proteins identified in these samples. Increased coverage from the material enriched with a specific probe might be obtained by performing mass spectrometry analysis using greater numbers of smaller slices from the gel.
Bioinformatic analysis of candidate proteins.
The total proteins identified as associated with TAS repeats by mass spectrometry were filtered first by removing the ones identified in the negative-control PICh and then by removing proteins previously identified as the Drosophila TRAPome (44), which we called common contaminants. We then removed proteins for which only one peptide was detected, due to the lower confidence in the detection method. To sort the remaining candidates, we counted the peptides identified in each cell line for either TAS-L or TAS-R PICh experiments (PKc, PS3), and for each protein in the list, we determined two normalizing parameters. (i) The detectability score (D) represents a measure of the likelihood that a given protein will be detected in a mass spectrometry experiment. To calculate it, we input the amino acid sequence of the largest isoform (when multiple isoforms exist) of each protein identified in the PICh experiments into the Peptide Detectability Predictor (PDP) (52), and the number of peptides with a detectability score of >0.6 (on a scale of 0 to 1)/1,000 amino acids in the sequence was determined. To determine D, we assigned a value of 10 to the protein with the highest density of detectable peptides and normalized all of the other proteins on a scale of 0 to 10. (ii) The normalized gene expression score (EKc, ES3) is a proxy for the protein abundance in the respective cell lines. We used raw expression data for Kc and S3 cells from the ModENCODE project (9) and assigned a value of 10 to the highest gene expression level of all of the factors identified as candidates by PICh. The remaining factors' gene expression levels were normalized on a scale of 0 to 10. To calculate the confidence score (C), we used the following formula: C = (PKc + PS3)/{D + [(EKc + ES3)/2]}. The candidate TAS-L and TAS-R proteins were ranked according to this confidence score (see Tables 2 and 3; see also Tables S4 and S5 in the supplemental material).
Table 2.
TAS-L proteins ranked by confidence score
| Rank | Name | Domain or function | Modifier of silencinga |
Reference(s) and/or source | ||
|---|---|---|---|---|---|---|
| TPE | PEV | bwD | ||||
| 1 | XNP | Chromatin remodeler | − | + | + | This study, 20, 47 |
| 2 | Rrp1 | AP endonuclease | − | 38 | ||
| 3 | Dip3 | Myb/SANT-like domain, BESS motif | − | 38 | ||
| 4 | CG8289 | Chromodomain | ||||
| 5 | Bj1 | RCC1 superfamily | − | This study, 15 | ||
| 6 | borr | Chromosome passenger complex | − | This study | ||
| 7 | ial | Chromosome passenger complex | ||||
| 8 | Trl | GAGA factor, BTB/POZ domain | − | + | This study, 6, 17, 21, 38 | |
| 9 | CG8290 | − | 38 | |||
| 10 | CG3163 | myb/SANT-like domain | ||||
| 11 | smt3 | SUMO | − | 38 | ||
| 12 | crol | Zn finger | − | + | This study, 38, 47 | |
| 13 | CG4004 | Myb/SANT-like domain | ||||
| 14 | Orc2 | Origin recognition complex | − | + | 38, 42 | |
| 15 | CG33691 | |||||
| 16 | D1 | AT hook like | + | 3 | ||
| 17 | Mi-2 | Chromatin remodeler | + | 15 | ||
| 18 | Caf1 | WD40 repeats, histone binding | − | This study | ||
| 19 | GAG | GAG protein of Gypsy element | ||||
| 20 | Mcm2 | Minichromosome maintenance complex | − | 38 | ||
| 21 | CG1240 | SWIB/MDM2, Dek domains | − | This study | ||
| 22 | mu2 | |||||
| 23 | psq | BTB/POZ domain | − | + | 38, 47 | |
| 24 | Su(var)2-10 | Zn fingers, SAP and PINIT domains | ± | + | 15, 31, 38, 45 | |
| 25 | LBR | ICMT domain | − | 38 | ||
| 26 | fbl6 | F-box domain, leucine-rich repeats | − | 38 | ||
| 27 | Su(var)3-9 | Chromodomain, SET domain | ± | + | 15, 16 | |
| 28 | ran | GTPase | ||||
| 29 | CG1910 | − | 38 | |||
−, tested, no effect; +, tested, identified as modifier; ±, tested, modifier only in a subset of the studies or only some of the alleles.
Table 3.
TAS-R proteins ranked by confidence score
| Rank | Name | Domains/function | Modifier of silencinga |
Reference(s) and/or source | ||
|---|---|---|---|---|---|---|
| TPE | PEV | Others | ||||
| 1 | Rrp1 | AP endonuclease | − | 38 | ||
| 2 | Bj1 | RCC1 superfamily | − | This study, 15 | ||
| 3 | pont | AAA+ ATPase | ||||
| 4 | Trl | GAGA factor, BTB/POZ domain | − | + | This study, 6, 17, 21, 38 | |
| 5 | pita | Zn finger, C2H2 type | − | + | 38, 47 | |
| 6 | CG8289 | Chromodomain | ||||
| 7 | Mi-2 | Chromatin remodeler | + | 15 | ||
| 8 | crol | Zn finger | − | + | This study, 38, 47 | |
| 9 | D1 | AT hook like | + | 3 | ||
| 10 | XNP | Chromatin remodeler | − | + | + | This study, 20, 47 |
| 11 | LBR | ICMT domain | − | 38 | ||
| 12 | pzg | − | 38 | |||
| 13 | CG7946 | PWWP domain, LEDGF domain | − | 38 | ||
| 14 | mor | SANT, SWIRM, RSC8 domains | ± | This study, 6, 38 | ||
| 15 | RfC38 | AAA+ ATPase, RFC small subunit | − | 38 | ||
| 16 | Kdm2 | H3K4 demethylase | ||||
| 17 | CG1240 | SWIB/MDM2, Dek domains | − | This study | ||
| 18 | Dref | BED Zn finger | − | 38 | ||
| 19 | Chro | Chromodomain | − | 15 | ||
| 20 | HP1c | Chromodomain, chromo shadow | ± | 15, 38 | ||
| 21 | gp210 | − | 38 | |||
| 22 | smt3 | SUMO | − | 38 | ||
| 23 | crp | Helix-loop-helix domain | − | 38 | ||
| 24 | borr | Chromosome passenger complex | − | This study | ||
| 25 | Ubqn | Ubiquitin-like domain | ||||
| 26 | ran | GTPase | ||||
| 27 | Klp3A | Kinesin motor domain | ||||
| 28 | BEAF-32 | BESS motif, BED Zn finger | − | + | 15, 27 | |
| 29 | Caf1 | WD40 repeats, histone-binding | − | This study | ||
| 30 | Cp190 | BTB/POZ domain | ||||
| 31 | Cpr | NADPH-dependent flavin mononucleotide reductase | − | 38 | ||
| 32 | Parp | PARP, WGR, Zn finger, BRCT domains | ||||
| 33 | Adf1 | Myb/SANT-like domain | − | 38 | ||
| 34 | osa | ARID/BRIGHT DNA binding domain | ± | This study, 6, 38 | ||
| 35 | Dek | SAP domain | + | + | 32, 47 | |
| 36 | Orc1 | Origin recognition complex | − | 38 | ||
| 37 | HP1b | Chromodomain, chromo shadow | ||||
| 38 | Rpb5 | RNA polymerase II subunit | − | 38 | ||
| 39 | bocksbeutel | LEM domain | − | 38 | ||
| 40 | Nipped-B | Adherin | − | This study, 38 | ||
| 41 | ial | Chromosome passenger complex | ||||
| 42 | CG17078 | |||||
| 43 | GAG | GAG protein of Gypsy element | ||||
| 44 | CG1910 | − | 38 | |||
| 45 | ham | Zn finger | − | 38 | ||
| 46 | Dip3 | Myb/SANT-like domain, BESS motif | − | 38 | ||
| 47 | wds | WD40 domain | ||||
| 48 | zf30C | + | 38 | |||
| 49 | Snr1 | SNF5 superfamily | − | This study, 38 | ||
−, tested, no effect; +, tested, identified as modifier; ±, tested, modifier only in a subset of the studies or only some of the alleles.
Western blot assay.
Input chromatin at 0.1%, 0.03%, and 0.01% was separated along with 15% of the PICh-purified protein by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked for 1 h with 5% milk in PBS–1% Triton X-100 (PBST) and incubated overnight with the following dilutions of antibodies in PBST–3% milk at 4°C: rabbit antistromalin antibody at 1:2,000 (gift from Dale Dorsett), rabbit anti-SMC1 antibody at 1:2,000 (18), mouse anti-BEAF-32 antibody at 1:200 (5), rabbit antipontin antibody at 1:2,000, rabbit antireptin antibody at 1:2,000 (14), mouse antimodulo antibody at 1:1,000 (34), rabbit anti-Polycomb antibody at 1:2,000 (49), rabbit anti-dRING antibody at 1:1,000 (29), rabbit anti-Moira antibody at 1:2,000 (10), rabbit anti-Dsp1 antibody at 1:2,000 (39), rabbit anti-GAGAC-ter 581 antibody at 1:1,000 (4), rabbit anti-Woc antibody at 1:2,000 (43), rat anti-Row antibody at 1:1,000, rat anti-HP1c antibody at 1:1,000 (22), rabbit anti-Sle antibody at 1:2,000 (41), and rabbit anti-Gypsy/GAG antibody at 1:1,000 (50). Membranes were washed three times for 15 min each time with PBST and then incubated for 1 h with a 1:10,000 dilution in PBST of horseradish peroxidase-conjugated secondary anti-rabbit (GE), anti-mouse (GE), or anti-rat (Abcam) antibody. Membranes were washed three times for 15 min each time with PBST, incubated for 5 min with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), and exposed to Kodak BioMax film (Perkin-Elmer). Films were developed and digitalized, and images were processed using Adobe Photoshop (Adobe Systems).
ChIP.
Sg4 cells were grown in CCM3 medium supplemented with Pen/Strep. Flag-hemagglutinin (Flag-HA) tag expression vectors (pMK33-CFH-BD) (56) were cotransfected with pCoBlast (Invitrogen) using the FuGENE HD transfection reagent (Qiagen) and following the manufacturer's protocol. Blasticidin (Invivogen) at 25 μg/ml was added to the medium 2 days after transfection. At 24 h before cross-linking, transgene expression was induced by the addition of 1 mM CuSO4 and ChIP was performed with all of the transfected lines where transgene expression was detected by Western blot assay. ChIP experiments were performed essentially as described previously (33), with minor changes. All operations, unless otherwise noted, were at 4°C. Briefly, cells were cross-linked at a density of 5 × 106/ml for 10 min with 1.8% formaldehyde at RT, stopped with glycine at 0.125 M, washed with 1× PBS, resuspended, and incubated for 10 min in ChIP wash buffer A (10 mM HEPES, pH 7.6; 10 mM EDTA, pH 8.0; 0.5 mM EGTA, pH 8.0; 0.25% Triton X-100); this was repeated with ChIP wash buffer B (10 mM HEPES, pH 7.6; 100 mM NaCl; 1 mM EDTA, pH 8.0; 0.5 mM EGTA, pH 8.0). Nuclei were isolated by a brief incubation with 1% SDS in TE buffer and, after extensive washing with TE, resuspended with TE-phenylmethylsulfonyl fluoride (PMSF)-SDS (TE, 1 mM PMSF, 0.1% SDS) at a density of 1 × 108 cells/ml. Chromatin was solubilized using a Misonix sonicator to obtain a DNA length of 200 to 400 bp. Salt and detergent concentrations were corrected to 1% Triton X-100, 0.1% Na-deoxycholate (Na-DOC), and 140 mM NaCl, and the insoluble pellet was removed by centrifugation for 5 min at maximum speed in a microcentrifuge. Chromatin aliquots of 500 μl were precleared for 1 h with 20 μl of a protein A-Sepharose (PAS) slurry (Thermo Scientific) and incubated overnight with 1.5 μg of a rabbit polyclonal anti-HA antibody (Abcam) or with 1.5 μg of control rabbit IgG (Abcam). Antibody was captured with 30 μl of PAS for 3 h, beads were washed five times for 10 min each time with 1 ml of RIPA (140 mM NaCl; 10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0; 1% Triton X-100; 0.1% SDS; 0.1% DOC), one time with 1 ml of LiCl buffer (250 mM LiCl; 10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0; 0.5% NP-40; 0.5% DOC), and then twice with 1 ml of TE. Beads were then resuspended in 100 μl TE–50 μg/ml RNase A (Qiagen) and incubated for 30 min at 37°C, and then SDS at 0.5% and proteinase K (Roche) at 0.5 mg/ml were added and the mixture was incubated overnight at 37°C. The cross-links were reversed at 65°C for 6 h, DNA was extracted and ethanol precipitated, and the pellet was resuspended in 150 μl of double-distilled H2O. Quantitative PCR (qPCR) analysis was performed with the IQ SYBR green system (Bio-Rad) and the following primers: for TAS-R, TAS_ChIP7 (5′ GATGACAATGTAGTGAACGC 3′) and TAS_ChIP8 (5′ GCGCTCGACAGAATTTTCAT 3′); for TAS-L, TAS3L_ChIP1 (5′ TGACTGCCTCTCATTCTGTC3 ′) and TAS3L_ChIP2 (5′ TATCATCTCGTTCATCCGCC3 ′). The immunoprecipitated material was quantified against a calibration curve with dilutions of input DNA.
Mutant strains and genetic crosses.
Stocks were maintained and crosses were made on cornmeal molasses medium with dry yeast added to the surface at 25°C.
Mutants defective in candidate genes were chosen because they have a strong lethal or sterile phenotype or because they were described as null on FlyBase (53). One exception, XNPUY3132, is a gain-of-function mutation with no obvious phenotype. Stocks with a mutation of or a deficiency in a candidate gene and Mi(ET1) insertion stocks were obtained from the Bloomington Drosophila Stock Center. Depending on the affected chromosome, males from these stocks were crossed for four successive generations with either y w67c23; Sco/SM1 or y w67c23; Sb/TM6, Ubx females to remove extraneous Mod(TPE). New stocks were established after these backcrosses and tested for TPE by crossing with y w67c23; P(wvar)11-5 flies. Only stocks lacking a Mod(TPE) were used for further analysis. As noted previously (38), many of the stocks from the stock center carried Su(TPE) on chromosome 2. Thus, many of the chromosome 2 mutants were eliminated from further testing.
In tests for TPE using a white transgene inserted into a telomere, 3-day-old males were examined for eye color. Photographs were taken using a Nikon SMZ-U stereomicroscope with the diaphragm half open.
RESULTS
PICh optimization.
We have previously reported the development of PICh, a method to purify and identify proteins associated with a defined genomic location (11). The target sequences purified in that work (human telomeres) are highly abundant, and we were interested in determining whether less abundant genomic sequences can be isolated by this method. We chose to work with Drosophila cells because of the availability of multiple well-established lines that can be grown in large amounts, the lower complexity of the genome (∼20-fold smaller than the human genome), and the possibility of employing genetic assays to test candidates. As a target, we focused on the TAS repeats, a moderately repetitive group of genomic sequences.
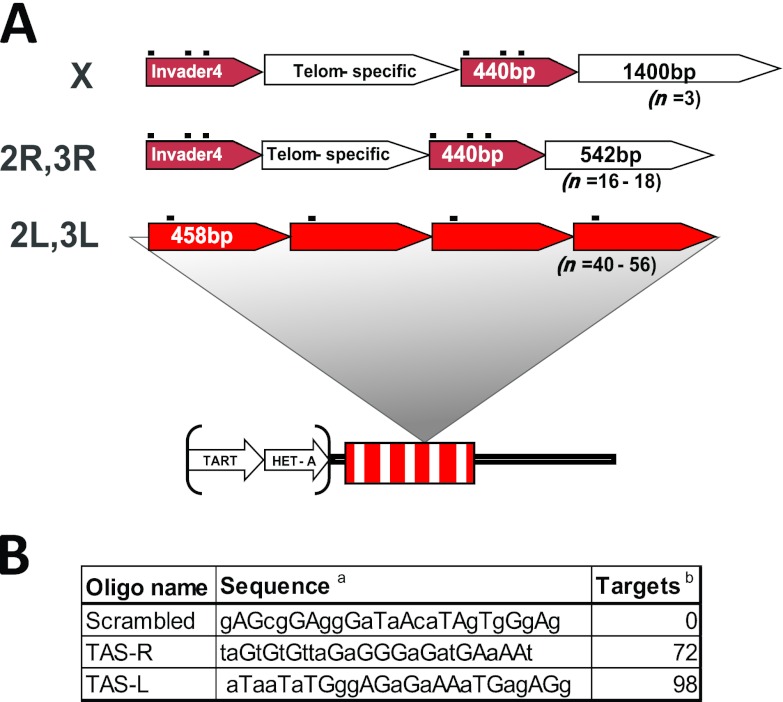
TAS repeats are subtelomeric satellite sequences which can be divided into at least two families: those found at the left ends of chromosomes 2 and 3 (2L and 3L; TAS-L) and those found at the XL, 2R, and 3R telomeres (TAS-R). The TAS-L family is composed of a unique canonical 458-bp sequence tandemly arranged in 40 to 60 repeat units per chromosome. The TAS-R family is characterized by the presence of two classes of intercalated subrepeats: a 440-bp unit derived from the 3′ untranslated region of the Invader 4 retroelement and a telomere-specific unit which differs between the XL TAS and the autosomes (Fig. 1A) (for a review, see reference 37). TAS-L and TAS-R do not share significant homology at the sequence level, so they offered two related targets for PICh that could be purified using distinct and nonoverlapping capture probes.
Fig 1.
Structures of the TAS repeats and sequences of PICh capture probes used in this study. (A) Schematic of the structure of TAS repeats (see the text for details): TAS repeats are adjacent to the telomeric retrotransposon arrays (TART, HET-A) and are organized as a single repeat unit in chromosomes 2L and 3L or as a combination of two repeat units, one of which is common to chromosomes 2R, 3R, and XL and the other of which differs between the autosomes and X. The black dots above the repeat blocks indicate the capture probe hybridization sequences. Telom, telomere. (B) Sequences of the capture probes used. a, homology region of the capture probes, not including the desthiobiotin and spacers (lowercase, DNA residues; uppercase, LNA). b, number of predicted targets in the haploid Drosophila genome.
We anticipated that the purification of chromatin and the identification of associated proteins from the TAS repeats would face several challenges compared to the purification of human telomeric chromatin. Compared to human telomeres, Drosophila TAS repeats comprise an equivalent percentage of the genome (∼0.02%, compared to 0.01 to 0.07% for human telomeres), but given their longer repeat sequence, the number of target positions for a given capture probe is considerably lower. Human telomeres are microsatellites composed of the simple TTAGGG repeat and stretching for lengths of ∼5 kb per chromosome end. They contain a large number of hybridization positions (a 6-nt sliding window along the chromosome ends), which significantly increases the opportunity for invasion by the capture probe compared to other, nonmicrosatellite sequences, where hybridization has to occur at a discrete position in the locus. Thus, even though the abundance of TAS repeats is similar to the abundance of human telomeres, the possible hybridization positions are ∼30-fold less abundant. Also, in choosing to target both TAS-L and TAS-R, we hoped to gain a better insight into the target constraints for PICh to work successfully. The TAS-L capture probe hybridizes to one position within the 458-bp repeat, whereas the TAS-R capture probe has three possible hybridization positions (one of which contains a single nucleotide change) within 300 bp (see Fig. S1 in the supplemental material), which we refer to throughout as a cluster of hybridization sites. Clustered hybridization sequences are expected to increase the likelihood that one homologous sequence in the cluster will be available for efficient hybridization to the probe, as factors such as nucleosome location or local protein binding sites might impact hybridization. The relative success of TAS-L versus TAS-R purification should show whether sequence abundance alone indicates a successful probe choice or whether the clustered target sequences are important for success.
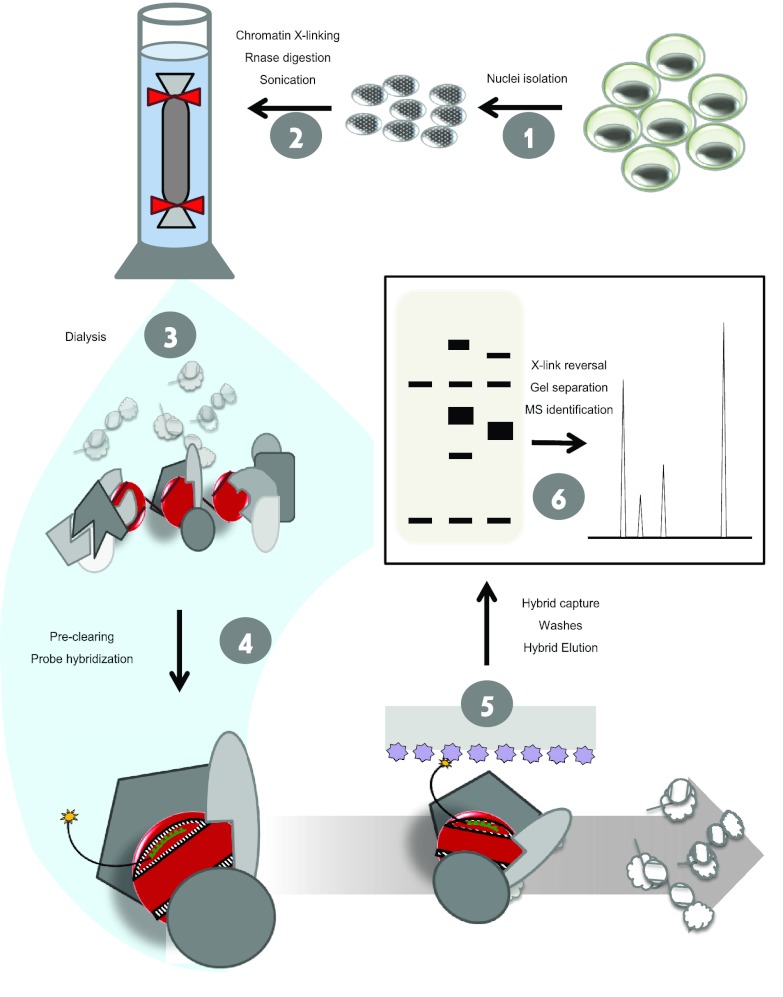
We altered the originally reported PICh protocol (Fig. 2) at specific steps to increase the efficiency of the method. One major consideration was the conditions used to prepare the sample for hybridization with the capture probe, as initial experiments indicated the importance of increasing the signal-to-noise ratio in this starting material. Attempts to purify TAS chromatin by the standard protocol yielded a large number of ribosomal proteins (data not shown). To combat this, we increased the RNase A incubation time (note that single-stranded nucleic acids will contribute disproportionately to the background signal because they are more easily available for spurious hybridization with the probe). Other abundant cytoplasmic proteins were also identified in the purified materials, so we isolated nuclei prior to cross-linking, rather than use whole cells, to limit contamination by cytoplasmic proteins. Finally, to remove non-cross-linked nucleic acids, proteins, and other components from the mixture, we dialyzed the solubilized chromatin through a membrane with a molecular weight cutoff of 106. With these combined steps, we achieved a chromatin sample that was enriched for cross-linked nuclear complexes of nucleic acids and protein with a reduced amount of contaminating materials.
Fig 2.
Optimized PICh protocol. Cell cultures are harvested and nuclei are isolated in step 1. Nuclei are cross-linked, RNA is digested, and chromatin is solubilized by sonication in step 2. Chromatin is dialyzed through a membrane with a molecular weight cutoff of 106 to obtain the substrate for hybridization in step 3. The desthiobiotinylated capture probe (shown as a black- and-green line with a yellow star representing desthiobiotin) is hybridized to the target DNA in a complex with the cross-linked associated proteins, including the histones (red) and nonnucleosomal chromatin proteins (gray) in step 4. The nucleoprotein complex is captured with streptavidin resin (lilac), and the nonassociated proteins and DNA (white outlined complexes) are washed away in step 5. The specific complexes are isolated, and the proteins are separated on a gel and subjected to mass spectrometric identification in step 6.
Purification of TAS repeats chromatin.
We designed PICh capture probes specific to the TAS-L and TAS-R families and used these to purify the chromatin from each version of TAS repeats. A scrambled capture probe with approximately the same base composition as the specific probes but no homologies in the Drosophila genome was used as a negative control (Fig. 1B). The starting biological materials for these experiments were the Kc167 (Kc) and S3 cell lines, which were chosen to obtain large quantities of homogeneous material, as well as to look at overlaps in the TAS protein composition in different biological contexts and thus allow the identification of candidate constitutive TAS proteins with higher confidence. The Kc and S3 cell lines display many similarities but have distinct embryonic origins, with Kc cells originating from young embryos and S3 cells originating from embryos on the verge of hatching (19), and distinct transcriptional profiles (9). By using these cell lines and capture probes that target the two different families of TAS repeats, we hoped to identify common themes and differences in the composition of the families of TAS repeats in different contexts.
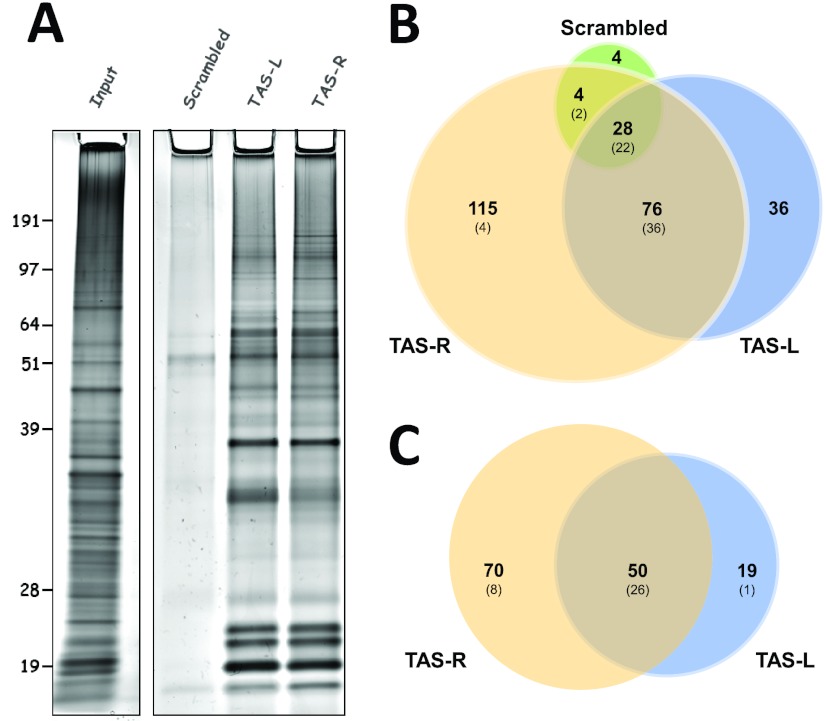
The PICh hybridization and capture protocol (11; see details in Materials and Methods) was used to purify TAS-associated proteins from the modified starting material described above. Samples purified using TAS-specific capture probes yielded a larger amount of total protein than did samples purified with the control (scrambled) capture probe, with a profile markedly different from that of the input protein, as determined by silver staining of the purified material (Fig. 3A). We cut the control, TAS-L, and TAS-R lanes of the protein gel into four slices and submitted each slice for mass spectrometry analysis. As expected, a much larger number of peptides was identified in the TAS-specific purifications, with ∼300 peptides for the control and 4- to 6.5-fold as many peptides for the specific purifications (see Table S1 in the supplemental material). PICh performed using the control capture probe yielded a mixture of highly abundant proteins, particularly histones, topoisomerase II, replication and transcription elongation factors, and heat shock proteins, most of which were also common to the TAS-specific purifications (Fig. 3B; Table 1). Many of these factors are likely to be present at TAS repeats but not specific to the locus. In our effort to identify TAS-specific factors, we removed all of these proteins from the list of candidates. We also removed proteins from the list of candidates that had only one peptide observed following mass spectrometry analysis (Table 1).
Fig 3.
TAS repeat chromatin purification. (A) Silver-stained gels with input Kc nuclear chromatin (left) and 20% of the protein isolated from Kc nuclear chromatin using the indicated capture probes (right). Molecular masses (kDa) are indicated on the left. (B) Overlap between the factors identified associated with the capture probes in Kc cells. The values in parentheses are the numbers of proteins in the respective sectors which are among the overall top 25% of proteins based on the absolute number of identified peptides. (C) Overlap between the factors identified associated with TAS-L and TAS-R capture probes after the removal of proteins identified in the negative control and common contaminants (see the text for details) using combined data from Kc and S3 cells. Values in parentheses are as in panel B.
Table 1.
Proteins filtered from the final TAS candidate lists
| Type | Proteinsg |
|---|---|
| Negative-control proteinsa | RpA-70, His H2B, Ef1α48D, gypsy/gag, Ote, Row, RfC3, Nop60B, mtSSB, Nopp140, CtBP, ZAM/gag, Transpac/gag, Gapdh1, Hsp70Aa, RpS2, Top1, CG11180, Dsp1, CG13295, RfC4, regucalcin, Taf6, CG3708, woc, ball, CG8142 |
| Negative-control proteins also listed as common contaminants | Top2, His H4, CG1516, dre4,d Act88F, His H2Avd, histone H1, histone H3, Lam, Hsc70-4, Gnf1, CG13096, Hsp27, Hsc70-5, stwl, Ssrp, CG6543, HP1d, Act42A, 14-3-3ζ, Trap1, Hrb87F, CG30122, retn, FK506-bp1, SF2, Fib |
| Common contaminantsb | LamC, SMC2, CG10576, Moe, tou, ATPsyn-β, glud, RpL6, Hsp60, mxc, CG12288, Hsp83, RpL8, l(2)03709, sqd, sesB, modd, RpS3A, CG2199, Ef2b, αTub84D, RpII140, βTub60D, blw, CG3680, Ca-P60A, kdn, Mcm3, msps, pAbp, l(3)72Ab, SMC1, CG6084, porin, scu, Hsp60C, ATPsyn-γ, WRNexo, Nup43, RpS8, ncd, Iswi, CG8677, gkt, eIF-4a, βTub56D, Cap, RpS6, Caf1-180, CG2118, SA, kise, l(3)mbt, su(Hw), Top3α, Rm62d,f, B52, Aly, lola, Incenp, slef, RpII215, dpa, HP5, pds5, lds, CG2982, BRWD3, E(bx), hang, RpL7A, mip130, Aldh, RpI135, Mcm6, Mcm5, nonA, CG42232, RpL13, CG4747, Nap1, CG5664, brme, DNApol-α180, Bap55, bor, CTPsyn, mus309d,e, Msh6, RpL10Ab, Ars2, ATPsyn-b, Mtor, lid, Klp61F, mus209d, Acon, RPA2, Kap-α3e, reptd |
| Proteins with only 1 peptide detectedc | GstD1, CG12592, CG15093, mre11, Dbp80, CG18292, CkIα, Orc4, CG33523, late, CG5703, cavd, CG7376, CG9797, CG9839, CG10139, Hrb27C, sxc, Acn, DnaJ-1, phr, l(1)G0004, CG12547, Caf1-105, Klp10A, lig3, Fancd2, CG17385, CG17896, Map60, exba, MAN1, grau, Hmu, Uch-L3, TfIIS, l(2)35Df, Nurf-38, SsRβ, Rab11, CG5857, DNApol-α73, Past1, JIL-1d, Elf, dalao, Rpd3d,e, pic, hay, Rcd1, RnrS, CG9135, CG9740, Tbp TH1, Transpac/pol, MBD-R2 |
Proteins identified in the scrambled capture oligonucleotide purification.
Proteins considered common contaminants because of their identification associated with various resins used in proteomic studies (44).
Proteins which were not identified in the negative control but for which only one peptide was identified in the specific PICh purifications, resulting in lower confidence.
Protein previously identified as Mod(PEV).
Protein previously identified as Mod(TPE).
Protein previously identified as Mod(bwD).
Boldface type indicates products of genes which have been associated with a chromatin-mediated silencing phenomenon, i.e., the genes under the d, e, and f index calls.
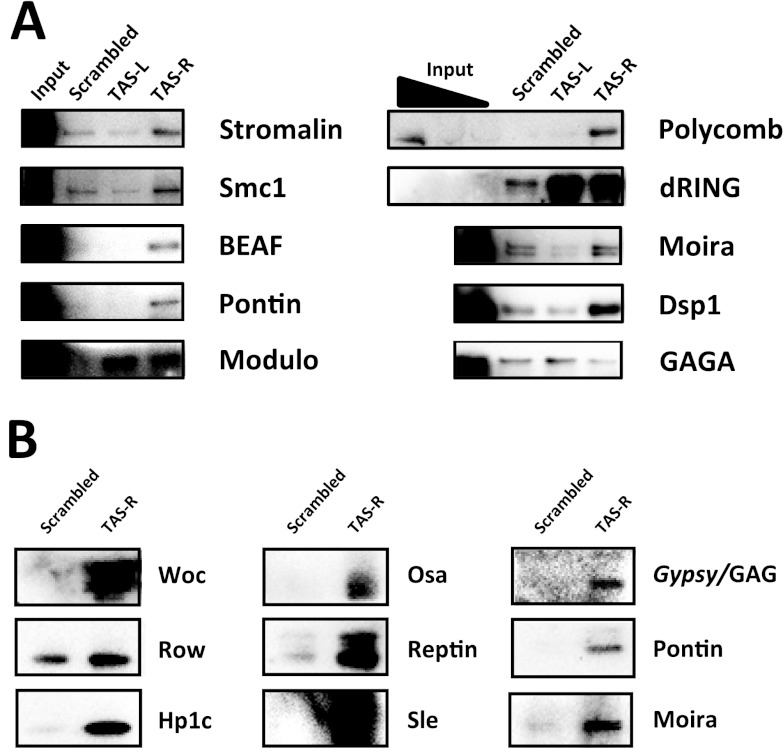
To confirm that the factors identified specifically in the TAS PICh experiments were indeed enriched in our purifications, we tested their abundance by Western blot assay (Fig. 4). Although high levels of enrichment of some factors, such as BEAF-32, pontin, or Osa, were obtained, other proteins, such as SMC1, stromalin, and Dsp1, showed little or no enrichment. When we searched for these proteins through a list of common contaminants in proteomic analysis (44), we found a good correlation between lack of enrichment by Western blot assay and the presence of the protein on the TRAPome list (44). This prompted us to filter the proteins found on the TRAPome list from our list of candidate TAS proteins (Table 1). After the subtraction of these common contaminants and low-confidence hits, the remaining proteins (Tables 2 and 3) showed GO terms anticipated to be associated with chromatin; among the top overrepresented terms were the cellular components “nucleus” and “chromosome” and the activities “DNA binding,” “chromatin binding,” and “regulation of gene expression” (see Table S2 in the supplemental material), consistent with the expected types of factors purified.
Fig 4.
Candidate TAS repeat factor enrichment levels by Western blot assay. (A) Candidate enrichment on PICh-purified protein from S3 cells. Lanes contain 15% of the purified protein and 0.01% input; for Polycomb and dRING, 0.03% and 0.1% input lanes are shown. (B) Negative control and TAS-R purifications from Kc cells. Lanes contain 15% of the purified protein.
The application of filters to these data sets is inexact, so caution must be used in evaluating whether any individual protein might be inappropriately filtered out (see Discussion). Subtraction of the proteins using the filters described above is expected to increase the likelihood that a remaining candidate is a true positive; however, some of the candidates removed might turn out to be bona fide TAS factors. Possible examples include HP1, previously shown to associate with the TAS region (24); Woc, a transcription factor associated with telomeres and mutants of which display telomeric fusion phenotypes (43); and Row, its partner protein (22). Woc and Row associate with Hp1c (1, 22), which was detected specifically at TAS-R in Kc cells (Table 3; see Table S1 in the supplemental material). Thus, though Woc and Row were seen in the control capture probe, they might be specifically enriched on TAS elements. Also, among the factors filtered out are some that have previously been associated with chromatin-silencing mechanisms (Table 1), which therefore might warrant further analysis to determine if they are true positives that should not have been filtered out.
The remaining proteins (Tables 2 and 3) were ranked by their likelihood of being genuine TAS-interacting proteins by several criteria. These were (i) the number of peptides detected, (ii) the relative expression levels of the genes in the cell line where peptides were identified (9), and (iii) a measure of the “detectability” of each protein by mass spectrometry. For the latter criterion, the sequence of each protein identified by PICh was analyzed using the PDP, an in silico tryptic digester (52), and the number of peptides with a “detectability score” of >0.6 by the PDP criteria was determined (1.0 is the maximum possible detectability score on this scale, and 0.6 was chosen arbitrarily). This value was then adjusted for the size of the protein by expressing it per 1,000 amino acids. The higher this value is, the more likely it is that the protein will be detected by mass spectrometry. Both the gene expression levels and the detectability score were normalized to a scale of 0 to 10, and the number of peptides identified was divided by the sum of these normalizing indexes. Highly expressing, highly detectable factors were thus brought to a lower confidence level than they would otherwise have had if the number of peptides identified were the only ranking factor. Tables 2 and 3 list the ranks of the TAS-L and TAS-R proteins, respectively, down to the lowest-ranking candidate tested by ChIP for TAS-R (Table 3, see below) and the corresponding candidates to the same confidence level for TAS-L (Table 2). For the complete lists, with the expression levels and detectability ranks, see Tables S4 and S5 in the supplemental material.
A distinct issue is the failure to detect bona fide interacting proteins (i.e., false negatives). Possible reasons to miss a bona fide interaction include the depth of proteomic coverage of the target loci by PICh, the extent to which proteins are difficult to identify due to low abundance, and poor detectability by mass spectrometry. One concern in our study was the complete absence of peptides for PcG proteins which have previously been reported to be localized at TAS repeats (2) and to be Mod(TPE) (6, 15), although the latter claim has been challenged for some of the genes (38). We looked for enrichment of Polycomb and dRING, two members of the PRC1 complex, in the purified proteins by Western blot assay. There is a clear enrichment of Polycomb in the TAS-R purification and also enrichment of dRING in both the TAS-L and TAS-R purifications (Fig. 4A). The false-negative results for these proteins are therefore due to a failure to detect them by mass spectrometry. This might be caused by a combination of low protein abundance (there is a very small amount of Polycomb transcript [9], as well as protein, as evidenced by the Western blot assay input signal [Fig. 3A]), shallower than the ideal depth of coverage of the purified material (due to the isolation of proteins from four sections of the gel, rather than the use of more discrete bands), and a low detectability of Polycomb and other PcG proteins by mass spectrometry. The Polycomb protein has 19.2% of the likely observable sequence coverage by mass spectrometry, compared to 81% of the observable sequence for topoisomerase II, a highly abundant protein in PICh experiments, according to the PeptideAtlas database (12). Using the criterion described above for peptide detectability, the density of highly detectable peptides for Polycomb is substantially lower than the average for detected proteins.
We conclude that proteins can be missed by PICh because of the depth of coverage by mass spectrometry and that the primary sequence of the protein and the resultant ability to detect its peptides through mass spectrometry might contribute to this issue. A straightforward way to alleviate this problem is to increase the amount of protein isolated and analyzed. PICh can also be used, as described above, with Western analysis as a detection method for proteins captured by a specific probe. This might prove useful in detecting additional members of a protein complex when one or more members have been identified by mass spectrometry or as a positive-control analysis method when optimizing PICh for a new sequence.
Validation of new TAS candidates.
Having assembled lists of potential TAS repeat factors, we next chose to validate a subset of these by examining their localization in chromatin. To that end, we transfected HA-tagged expression vectors into Sg4 cells and performed ChIP from the expressing lines. We picked factors from different positions in the candidate lists to test the likelihood of finding true-positive hits. We picked CG8289, a chromodomain protein ranked consistently high in all TAS purifications; Dip3 and Ial, found in both purifications but higher in the TAS-L rank (Tables 2 and 3); Klp3A, Zf30c, and Snr1, all specific to TAS-R purification, from further down in the ranking (Table 3); Polycomb, not identified by mass spectrometry but enriched at TAS-R (Fig. 4A); Row, identified in the negative control (Table 3; Fig. 4B) but, as discussed above, a plausible candidate TAS factor; and topoisomerase 3α, a factor common to all TAS purifications (see Table S1 in the supplemental material) but highly expressed protein and previously identified as a common contaminant in proteomic studies (44).
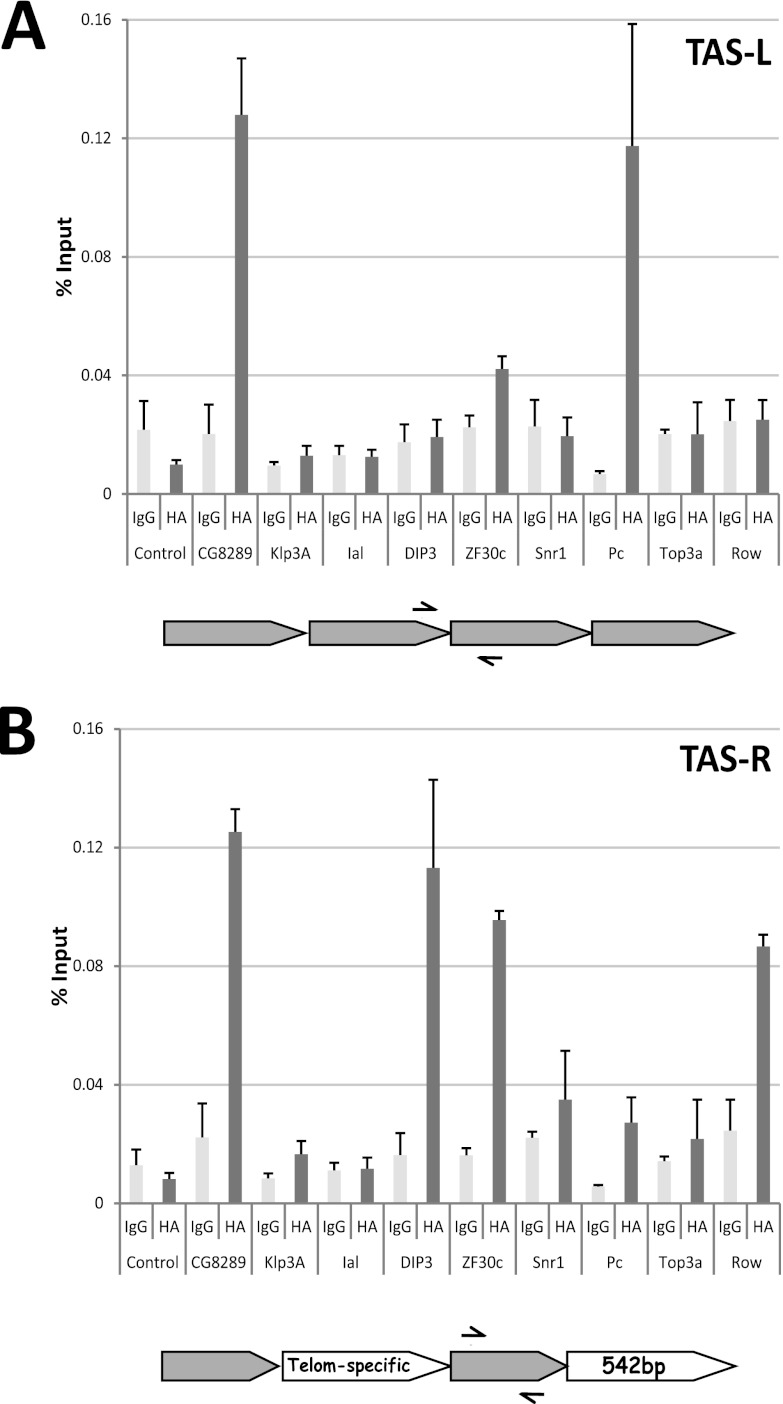
The tagged proteins were induced 24 h before formaldehyde cross-linking, and ChIP was performed with a polyclonal antibody for the HA tag. Binding at TAS repeats was determined by qPCR with primer pairs specific for TAS-L or TAS-R. The first observation was that the success rate for candidates as assessed by this approach was higher for TAS-R than for TAS-L (Fig. 5). This was not surprising, given the higher number of specific candidates identified (Table 3; see Table S1 in the supplemental material), and is consistent with the hypothesis that the PICh protocol works better with clustered capture probe hybridization sites. Second, the identification of proteins in the negative control should not be considered a disqualifying factor. An example of a protein that was found in the negative control yet shows binding by ChIP is Row: 4 total peptides were seen in the negative control versus 35 in the TAS-R PICh; similarly, for its partner Woc, 1 peptide was detected in the control versus 24 in the TAS-R PICh (see Table S1 in the supplemental material). These significant differences in abundance in the TAS-specific versus control purifications raised the possibility that these were true positives. Additionally, for Woc-Row, prior information was available that connected Row to binding at the TAS repeats. As discussed earlier, Row and Woc are known to be in a complex with HP1c (22), woc mutants have a telomere fusion phenotype, and all of these proteins have shown an enrichment on the TAS-R purifications, relative to the control and TAS-L (see Table S1 in the supplemental material), as well as enrichment by Western blot assay for Woc and HP1c (Fig. 3B). Consistently, when transfected into Sg4 cells, Row can be detected at TAS-R but not at TAS-L (Fig. 5). Top3α, on the other hand, as expected, was not detected at TAS repeats.
Fig 5.
ChIP analysis of candidate TAS protein binding at TAS repeats. Sg4 cells were transfected with expression vectors for HA-tagged versions of the indicated proteins, and ectopic expression of the transgenes was induced by the addition of CuSO4. At 24 h postinduction, cells were cross-linked and ChIP was performed. (A) ChIP, with primers specific for the TAS-L repeat, from cells transfected with expression vectors for the indicated tagged proteins. The two bars represent the average percentage of input DNA precipitated using a control IgG antiserum and an anti-HA polyclonal antibody, and the error bars are the standard errors from two or three independent transfections. CG8289 and Polycomb enrichments are significant (P < 0.05). (B) Same as panel A but with primers specific for the TAS-R repeat family. The Dip3 and Polycomb (P < 0.05) enrichments and the CG8289 and Row (P < 0.01) enrichments are statistically significantly different. A schematic of the respective TAS repeat structures and primers used (arrows) is below each graph. Pc, Polycomb; Telom, telomere.
The high-ranking candidate CG8289 was clearly detected at both TAS variants. CG8289 was identified in the PICh experiments at TAS-L in S3 and Kc cells and at TAS-R in S3 and Kc cells and embryos (see Table S3 in the supplemental material). Dip3, surprisingly, was not found at TAS-L but was detected at TAS-R, even though it ranked higher on the TAS-L list. Ial, the Drosophila homolog of the Aurora B kinase, which ranked high on the TAS-L list and lower on the TAS-R list, was not found at either of the TAS repeats by ChIP. Of the other TAS-R candidates tested, Klp3A was not confirmed, Zf30c was confirmed, and Snr1 was inconclusive (Fig. 5B). It is known that performing ChIP with components of ATP-dependent remodeling enzyme complexes such as Snr1 is complicated, presumably because of their transient interactions with DNA as they function (26). The Zf30c result is particularly interesting because its gene is deleted in a deficiency that has a dominant Su(TPE) phenotype (38). Given that it binds specifically at TAS-R and is associated with a Su(TPE), Zf30c is thus a good candidate as a chromatin regulation factor at TAS repeats.
Finally, Polycomb, found in the TAS-R purification by Western blot assay, is detected by ChIP on both TAS-L and TAS-R. Taken together, these results demonstrate a good rate of discovery for TAS-R candidates, with even proteins ranking at around number 50 proving to be present at the locus, and a lower efficiency for TAS-L, with only one of the top hits being confirmed by ChIP. This finding is consistent with the hypothesis that clustered target sequences might increase the efficiency of detection by PICh. As expected for a proteomic screening protocol, some nonspecific contaminants appear to have eluded the filters we employed and are therefore false positives. Further information concerning nonspecific proteins and success rates in purifying more loci are expected to help fine-tune filters and thereby to increase the level of bona fide protein identification by PICh.
The TrxG-Brahma complex is involved in the mechanism of TPE.
One use for a proteomic screen such as PICh is the identification of genes that can be tested using directed genetic protocols. This also serves as a form of validation, although not all mutations affecting bona fide interacting proteins will elicit a phenotype when tested by a specific genetic assay. Thus, such genetic assays cannot be used to remove candidates for interactions. The best-studied biological phenomenon associated with TAS repeats is the chromatin-silencing mechanism of TPE. The search for dominant Su(TPE) has produced few results. A deficiency screen for dominant Su(TPE) at the 2L telomere identified only a single gene, gpp, with mutations that could suppress TPE. Most of the suppressors identified in this screen were deficiencies for 2L TAS (38). Of the genes identified by PICh, we tested five that had not been identified in the deficiency screen: sle, CG8289, rept, cap, and smc1. None of the mutants tested showed suppression of TPE using the P(wvar)11-5 tester (23; data not shown).
As deficiencies of 2L TAS suppress TPE at a number of different chromosome ends (23), we next asked whether any of the factors identified by PICh could interfere with the TPE induced by Df(2L)M26 (here M26) (23) on P(w+)39C-62 (here C62), a mini-white insertion into 3R TAS (54). Flies with the C62 transgene alone exhibit an orange eye color due to TPE. In the presence of the M26 suppressor, silencing is lost and the eye color becomes red (Fig. 6). To look for dominant modifiers of this M26-C62 interaction, we crossed y w67c23; M26; C62 females with males mutant for putative TAS-binding factors identified by PICh. The males were either y w67c23; mutant/SM1 or y w67c23; mutant/TM6, Ubx, depending on the location of the candidate gene. Table 4 (see also Table S6 in the supplemental material) shows the genes tested and the results obtained. Whenever possible, we tested multiple alleles, including deficiencies, for candidate genes, although this was not possible in most cases. Two deficiency chromosomes, one for WRNexo and one for mor, showed a weak ability to suppress the suppression of TPE exhibited by M26. In these cases, the phenotype overlapped that of the wild type and these effects were not pursued further. The brm2 chromosome exhibited strong suppression of TPE suppression. It is well known that balancer chromosomes accumulate Mod(PEV) and Mod(TPE). We therefore repeated the assay of brm2 in the absence of the TM6 balancer by crossing y w67c23; brm2/TM6 males with y w67c23; Sb/TM6 females and then crossing the F1 y w67c23; brm2/Sb males with tester y w67c23; M26; C62 females. The result was essentially the same (Fig. 6).
Fig 6.
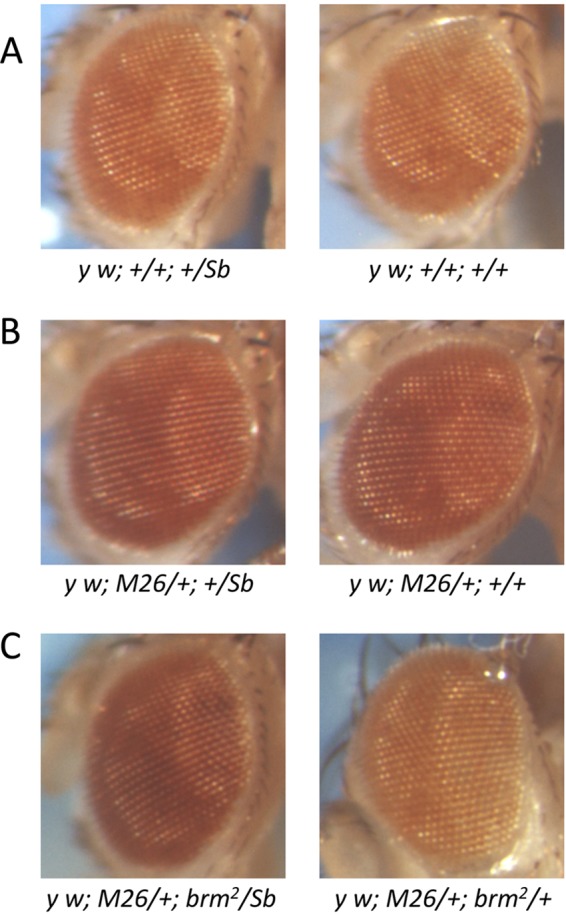
brm2 effects on Df(2L)M26 suppression of TPE. (A) Male progeny from a cross of y w67c23; +/Sb males and y w67c23; +; P(w+)39C-62 females. There are no Su(TPE) present in these males. Therefore, the mini-white insert in 3R TAS is repressed. (B) Male progeny from a cross of y w67c23; +/Sb males and y w67c23; Df(2L)D26; P(w+)39C-62 females. Df(2L)M26, a deficiency for 2L TAS and a Su(TPE). Therefore, the silencing of the mini-white insert imposed by 3R TAS is repressed and the mini-white gene exhibits increased expression. (C) Male progeny from a cross of y w67c23; brm2/Sb males and y w67c23; Df(2L)D26; P(w+)39C-62 females. Heterozygous Df(2L)M26/+; P(w+)39C-62/brm2 males exhibit silencing of the mini-white transgene similar to that seen when Df(2L)M26 is absent. Control Df(2L)M26/+; P(w+)39C-62/Sb sibling males, however, exhibit high expression of mini-white similar to that seen when silencing is suppressed.
Table 4.
Mod(TPE) screening results
| Gene | Allele | Modifier |
|---|---|---|
| Woc | Df(3R)BSC497 | No |
| Woc | Df(3R)BSC739 | Noa |
| Woc | Df(3R)D605 | Noa |
| WRNexo | Dr(3R)Cha7 | Weakb |
| Sle | 057 | No |
| Brm | 2 | Yes |
| SMC1 | exc46 | No |
| su(Hw) | 3 | No |
| Rept | 6945 | No |
| CG2199 | Df(3L)Ar14-8 | No |
| CG2199 | Df(3L)BSC289 | No |
| Snr1 | 01319 | No |
| Osa | 00090 | No |
| Mor | Df(3R)Po4 | Weakb |
| CAF1 | Df(3R)BSC471 | No |
| Borr | Df(2L)TE30Cb | No |
| Bj1 | Df(3L)XAS96 | No |
| XNP | 1 | No |
| XNP | UY3132 | Noa |
| CG1240 | Df(3L)BSC119 | No |
| Crol | 04418 | Noc |
| Crol | Df(2L)BSC243 | No |
| Trl | Df(3L)fz-M21 | Nod |
| Trl | Df(3L)XG3 | No |
A mutant was found, but it did not segregate with chromosome 3.
Efforts to map the modifier were unsuccessful because the phenotype overlaps that of the wild type.
A mutant was found, but it did not segregate with chromosome 2.
A mutant was found, but it was deemed to be a false positive because a chromosome bearing another allele of the same gene did not carry a mutant.
To further verify that Brahma plays a role in the mediation of TPE suppression, we mapped the genetic factor responsible for this suppression by meiotic recombination. The simple question is: does this genetic factor map close to brm? One easy approach is to use a dominant genetic marker close to brm and monitor the frequency of recombination. To do this, we used a Mi(3xP3-EGFP.ET1) element. There are 2,700 of these Mi(ET1) elements distributed randomly throughout the genome, which means that, on average, 1 of these elements is less than 1 map unit from any gene. They all carry a green fluorescent protein (GFP) marker. The Mi(ET1)sffMB06603 element is in chromosome region 72A5, 70 kb, and we estimate approximately 1 map unit, from brm. We collected Ubx male progeny from y w67c23; Mi(ET1)/brm2 females crossed with y w67c23; Sb/TM6, Ubx males, tested them for GFP expression, and crossed them with y w67c23; M26; C62 females to test for effects on M26 suppression of TPE. Of 216 males tested, 3 showed recombination between GFP and the suppressor of M26 suppression of TPE. This indicates that the genetic factor responsible for interference with M26 suppression is very close to brm and may, in fact, be the brm2 mutation.
Some of the mutants tested showed an interaction with the M26-C62 transgene combination (Table 4), but only the Mod(TPE) effect of the Brahma mutant brm2 (Fig. 6) mapped back to the mutant locus. A deficiency eliminating the Brahma complex Moira gene also showed an interaction in this assay, but the effect, though reproducible, was very subtle (see Table S6 in the supplemental material). We conclude that Brahma is involved in the regulation of TPE at TAS, consistent with our PICh analysis which identified several members of the Brahma complex as being physically associated with the TAS-R repeats (see Table S1 in the supplemental material), and “Brahma complex” as the GO term with the highest overrepresentation on the list of purified candidate TAS-R proteins (logarithm-of-odds score, 2.106; see Table S2 in the supplemental material).
DISCUSSION
We have isolated chromatin and identified proteins associated with the Drosophila TAS repeats present at the XL, 2R, 3R, 2L, and 3L telomeres. We validated the association of a subset of these factors by ChIP and analyzed their involvement in TPE. We have identified ∼70 factors not previously associated with the TAS repeats and have used that information in a directed genetic test to demonstrate a role for Brahma in TPE. We conclude that PICh works for target sequences less abundant than human telomeres and suggest that a close clustering of the capture probe targets might prove beneficial for PICh efficiency. These results represent a significant step in the direction of making PICh a universally applicable method while pointing out the difficulties involved in isolating complex loci using PICh and in appropriately filtering the resultant data to avoid false positives and false negatives.
Optimization of PICh technology.
The first application of PICh was in the purification of proteins associated with human telomeres (11). Expansion of PICh to less-abundant and more-dispersed targets than the TAS repeats reported here will likely require the use of a combination of probes capable of hybridizing to closely spaced sequences and would also benefit from further development of pre-enrichment strategies. Analysis of the resultant data will be made easier by the availability of an increasing number of public proteomic data sets of contaminating proteins and interaction networks, thus increasing the ability to filter the raw data and identify strong candidate proteins. Filtering out of possible false positives using lists of common contaminants should nevertheless be done with caution; these lists are under development, and some common contaminants might prove to be important at the locus examined.
We have identified strong candidate TAS-binding proteins using PICh, but a subset of these candidates will inevitably prove to be false positives. Given that PICh uses cross-linked material, a fraction of the false positives might be proteins that interact with a common contaminant and are thus carried over in the purification step. Protein-protein interaction data are increasingly comprehensive, and recent work has added to the list of protein complexes isolated from Drosophila (30). Nineteen of the proteins identified in TAS purifications have been reported to be part of a complex in which a common contaminant or a protein identified in the control PICh experiment was identified. This is merely an indicative number of false positives, as we have seen that not all of the proteins filtered by common-contaminant analysis—such as Brahma—are false positives. Likewise, such factors as Sle, Incenp, and Iswi, even though they have been removed from the final list of TAS candidates by virtue of their classification as common contaminants, deserve some attention, given their high enrichment in the TAS purifications (see Table S1 in the supplemental material). Careful analysis of all protein hits is thus required in order to minimize both the false-positive and false-negative candidates. The most decisive advance in the applicability of PICh will come from the production of other PICh data sets from different tissues and different organisms.
The chromatin at TAS repeats.
A key aspect of a developing methodology for performing proteomic screening, such as PICh, is to determine the fraction of proteins identified that are likely to be “true” interactors. These proteins can be determined retrospectively by determining the proteins identified that have previously been shown to have interactions by other analyses and can be determined prospectively by validating the association of novel candidates.
At least 17 (∼23%) of the 74 proteins identified as associated with the TAS-L/R purifications have previously been associated with some form of chromatin-mediated silencing or to have a telomere fusion phenotype when mutated. In contrast, such phenotypes have been described for only 8 to 12% of the factors in the negative control, common contaminants, and proteins with only one peptide identified. This implies enrichment for real TAS repeat-associated factors. Additionally, as noted above, some of the factors filtered from our final lists might be true TAS repeat factors. In all, 29 of the proteins detected (excluding the ones identified in the negative control) have been found to have a genetic interaction of some kind with heterochromatic silencing (Tables 1 to 3). Twelve of these have been associated with TPE, but the prevalence of terminal deficiencies on 2L is a serious problem for the interpretation of TPE suppression data (38, 46). Some of the putative Su(TPE) genes identified in numerous screenings have later been shown to have a second-site mutation at the tip of 2L which segregates with the TPE effect (38). Nevertheless, nine of the putative Su(TPE) genes found recently (15) have been identified in our experiments; others have not been identified in our experiments, including tefu (40) and gpp (48).
Some relatively widespread factors were identified at the TAS repeats by PICh, but the specificity of these associations is not clear, as they are expected to be located in many regions of the genome. These include the cohesin and condensin complex proteins modulo and Dsp1. Interestingly, these factors have been implicated in the regulation of the human subtelomeric D4Z4 repeats, which share structural similarities with the Drosophila TAS repeats (25, 57); likewise, cohesins have been implicated in heterochromatin formation at yeast subtelomeres (13). Also, Rm62, which is expressed ubiquitously and mutants of which interact genetically with dsp1 and pc, was detected on TAS-R in Kc cells only (see Table S1 in the supplemental material). Multiple Rm62 bands are found in polytene chromosomes, some colocalizing with Dsp1, notably in the telomeric regions, but a physical interaction with Dsp1 is seen only in young embryos and not in older embryos (35). Although Dsp1 has been discarded because one peptide was identified in the negative control, this suggests that Dsp1 and Rm62 might have a role together at TAS repeats and that there is specificity in PICh, as Kc cells are derived from young embryos and S3 cells are derived from old ones (19).
Finally, we have shown directly that CG8289, Dip3, Row, Zf30c, and Polycomb bind at least to one family of TAS repeats. These proteins have closer homologs in humans than in yeasts, mostly in the conserved chromodomain of CG8289 and zinc finger protein homology regions of Zf30c and Row. The lists of likely candidates for TAS-L and TAS-R factors that we have compiled can help to unravel the biological roles of TAS repeats and the mechanism of TPE.
Insights into the mechanism of TPE.
Drosophila assays for Mod(TPE) have proved to be prone to a high incidence of false-positive results (38). In addition to that, the occurrence of validated Mod(TPE) in genetic screenings is very rare, according to what one would expect based on the screenings for Mod(PEV) (16, 38). For these reasons, we used the candidates identified by PICh in functional tests of Drosophila. Of the 45 genes from our candidate lists (Tables 2 and 3) tested in this study or previously in a deficiency screen for dominant Mod(TPE) (38), mor, which is a weak modifier (not shown), and zf30c, which is eliminated in one of the deficiencies which act as Su(TPE) (38), have a level of functional validation in addition to the more thoroughly validated brm gene (Fig. 6). Brahma and Moira interact with each other as members of the SWI/SNF family Brahma chromatin-remodeling complex, emphasizing the role of this complex in TPE. There are three classes of explanations for why a low percentage of the proteins found by PICh are validated in this functional test. (i) Many of the proteins that bind to TAS repeats might be involved in functions that are unrelated to TPE, and/or (ii) PICh has been performed with cell lines which may have features different (harbor distinct sets of factors) from those of tissues in which TPE is assayed, and/or (iii) the mechanism by which proteins function in TPE is not dose dependent or involves redundant activities and thus does not create a dominant phenotype.
There is support for the latter possibility, in that screenings for dominant modifiers in other Drosophila-specific silencing models have had very low success rates. For example, screenings based upon the trans inactivation of bw+ by the bwD allele (51) and others based upon the trans suppression of PEV associated with a terminal chromosome deletion (16) have not identified any such modifiers. In the case of bwD, there was evidence that at least some modifiers are recessive (16). Comparable observations have been made for TPE, with loss of ATM causing a recessive suppressor phenotype (40), and for telomeric stability, with row mutants displaying a telomere fusion phenotype only when the protein is eliminated completely (22). While viable homozygous mutants for many genes in Drosophila are not readily available, one possible way to study genic modifiers is by looking at the effect of overexpression of such genes. Such a screen was conducted recently for Mod(bwD)s (47) and identified a plethora of new factors, some of which are identified here as candidate TAS-binding proteins (Tables 2 and 3) and most of which have no effect on PEV. These findings suggest that some silencing mechanisms follow rules different from those of the well-studied PEV model. It is possible that Mod(TPE), bwD, and TDA-PEV (16) are predominantly recessive. This would also be consistent with the findings on TPE in yeast, in which the study of heterochromatin, and notably of TPE, is done in the absence of the candidate modifier. Given that yeast is normally studied as a haploid, there is no distinction between dominant and recessive tests.
One reason why TPE effects are recessive might be the presence of specific DNA-binding factors which bring silencers preferentially to TAS. In yeast, RAP1 plays such a role, sequestering SIR proteins at telomeres (8, 36). If factors with a function similar to that of RAP1 exist in Drosophila, a fraction of the proteins involved in TPE will not necessarily show a phenotype when reduced by 50%, as their depletion will happen predominantly from loci for which their affinity is lower. Thus, one might expect traditional dominant Mod(TPE) screenings to yield a lower success rate than Mod(PEV) screenings. Indeed, that is what was observed historically and in this study.
The potential issue of the suitability of modifier screenings brings to light the usefulness of PICh. A more thorough analysis of these candidates might include the study of null mutants or the overexpression of such candidates when null mutants are not viable. The results obtained using PICh would help to direct such studies. In cases where limited information exists concerning the mechanism by which a given locus is regulated, knowing which factors bind to this locus will facilitate further genetic tests to validate function. Thus, in a genetically tractable organism, the application of PICh and genetic tests should synergize to more rapidly identify the full complement of proteins involved in a biological phenomenon.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Szostak lab (Massachusetts General Hospital) for help with oligonucleotide synthesis; D. Dorsett, A. Saurin, J. Pradel, M. Vidal, C. P. Verrijzer, D. Locker, C. Benyajati, M. Gatti, F. Azorín, C. Hama, A. Pelisson, and the Developmental Studies Hybridoma Bank for reagents; and Ross Tomaino for assistance with mass spectrometry.
Funding for this work was provided by the National Institutes of Health (GM43901) and by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health. J.M.A. was supported in part by a fellowship from the Fundação para a Ciência e Tecnologia (SFRH/BD/11800/2003).
Footnotes
Published ahead of print 9 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Abel J, Eskeland R, Raffa GD, Kremmer E, Imhof A. 2009. Drosophila HP1c is regulated by an auto-regulatory feedback loop through its binding partner Woc. PLoS One 4: e5089 doi:10.1371/journal.pone.0005089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. 2005. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J. Cell Sci. 118: 5465– 5477 [DOI] [PubMed] [Google Scholar]
- 3. Aulner N, et al. 2002. The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation. Mol. Cell. Biol. 22: 1218– 1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benyajati C, et al. 1997. Multiple isoforms of GAGA factor, a critical component of chromatin structure. Nucleic Acids Res. 25: 3345– 3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanton J, Gaszner M, Schedl P. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17: 664– 675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boivin A, Gally C, Netter S, Anxolabehere D, Ronsseray S. 2003. Telomeric associated sequences of Drosophila recruit Polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195– 208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brennecke J, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089– 1103 [DOI] [PubMed] [Google Scholar]
- 8. Buck SW, Shore D. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370– 384 [DOI] [PubMed] [Google Scholar]
- 9. Celniker SE, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927– 930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crosby MA, et al. 1999. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19: 1159– 1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Déjardin J, Kingston RE. 2009. Purification of proteins associated with specific genomic loci. Cell 136: 175– 186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deutsch EW, Lam H, Aebersold R. 2008. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9: 429– 434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dheur S, Saupe SJ, Genier S, Vazquez S, Javerzat J-P. 2011. Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol. Cell. Biol. 31: 1088– 1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diop SB, et al. 2008. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9: 260– 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doheny JG, Mottus R, Grigliatti TA. 2008. Telomeric position effect—a third silencing mechanism in eukaryotes. PLoS One 3: e3864 doi:10.1371/journal.pone.0003864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donaldson KM, Lui A, Karpen GH. 2002. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics 160: 995– 1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dorn R, et al. 1993. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics 133: 279– 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dorsett D, et al. 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743– 4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Echalier G. 1997. Drosophila cells in culture. Academic Press, Inc., New York, NY: [Google Scholar]
- 20. Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV. 2010. Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric beta-heterochromatin in vivo. J. Biol. Chem. 285: 15027– 15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farkas G, et al. 1994. The trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806– 808 [DOI] [PubMed] [Google Scholar]
- 22. Font-Burgada J, Rossell D, Auer H, Azorín F. 2008. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 22: 3007– 3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frydrychova RC, et al. 2007. Transcriptional activity of the telomeric retrotransposon HeT-A in Drosophila melanogaster is stimulated as a consequence of subterminal deficiencies at homologous and nonhomologous telomeres. Mol. Cell. Biol. 27: 4991– 5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frydrychova RC, Mason JM, Archer TK. 2008. HP1 is distributed within distinct chromatin domains at Drosophila telomeres. Genetics 180: 121– 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gabellini D, Green MR, Tupler R. 2002. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339– 348 [DOI] [PubMed] [Google Scholar]
- 26. Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19: 942– 954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilbert MK, Tan YY, Hart CM. 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173: 1365– 1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Golubovsky MD, Konev AY, Walter MF, Biessmann H, Mason JM. 2001. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics 158: 1111– 1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gorfinkiel N, et al. 2004. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 121: 449– 462 [DOI] [PubMed] [Google Scholar]
- 30. Guruharsha KG, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690– 703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hari KL, Cook KR, Karpen GH. 2001. The Drosophila Su(var)2–10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15: 1334– 1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kappes F, et al. 2011. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 25: 673– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kharchenko PV, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480– 485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krejci E, Garzino V, Mary C, Bennani N, Pradel J. 1989. Modulo, a new maternally expressed Drosophila gene encodes a DNA-binding protein with distinct acidic and basic regions. Nucleic Acids Res. 17: 8101– 8115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lamiable O, Rabhi M, Peronnet F, Locker D, Decoville M. 2010. Rm62, a DEAD-box RNA helicase, complexes with DSP1 in Drosophila embryos. Genesis 48: 244– 253 [DOI] [PubMed] [Google Scholar]
- 36. Marcand S, Buck SW, Moretti P, Gilson E, Shore D. 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 10: 1297– 1309 [DOI] [PubMed] [Google Scholar]
- 37. Mason JM, Frydrychova RC, Biessmann H. 2008. Drosophila telomeres: an exception providing new insights. Bioessays 30: 25– 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mason JM, Ransom J, Konev AY. 2004. A deficiency screen for dominant suppressors of telomeric silencing in Drosophila. Genetics 168: 1353– 1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mosrin-Huaman C, Canaple L, Locker D, Decoville M. 1998. DSP1 gene of Drosophila melanogaster encodes an HMG-domain protein that plays multiple roles in development. Dev. Genet. 23: 324– 334 [DOI] [PubMed] [Google Scholar]
- 40. Oikemus SR, et al. 2004. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 18: 1850– 1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orihara-Ono M, et al. 2005. The slender lobes gene, identified by retarded mushroom body development, is required for proper nucleolar organization in Drosophila. Dev. Biol. 281: 121– 133 [DOI] [PubMed] [Google Scholar]
- 42. Pak DTS, et al. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311– 323 [DOI] [PubMed] [Google Scholar]
- 43. Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. 2005. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol. Cell 20: 821– 831 [DOI] [PubMed] [Google Scholar]
- 44. Rees JS, et al. 2011. In vivo analysis of proteomes and interactomes using parallel affinity capture (iPAC) coupled to mass spectrometry. Mol. Cell. Proteomics 10 (6): M110.002386 doi:10.1074/mcp.M110.002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reuter G, Wolff I. 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182: 516– 519 [DOI] [PubMed] [Google Scholar]
- 46. Roegiers F, et al. 2009. Frequent unanticipated alleles of lethal giant larvae in Drosophila second chromosome stocks. Genetics 182: 407– 410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schneiderman JI, Goldstein S, Ahmad K. 2010. Perturbation analysis of heterochromatin-mediated gene silencing and somatic inheritance. PLoS Genet. 6: e1001095 doi:10.1371/journal.pgen.1001095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shanower GA, et al. 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169: 173– 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shao Z, et al. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37– 46 [DOI] [PubMed] [Google Scholar]
- 50. Syomin BV, Pelisson A, Ilyin YV, Bucheton A. 2004. Expression of the retrovirus gypsy Gag in Spodoptera frugiperda cell culture with the recombinant baculovirus. Dokl. Biochem. Biophys. 398: 310– 312 [DOI] [PubMed] [Google Scholar]
- 51. Talbert PB, LeCiel CD, Henikoff S. 1994. Modification of the Drosophila heterochromatic mutation brownDominant by linkage alterations. Genetics 136: 559– 571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang H, et al. 2006. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics 22: e481– e488 [DOI] [PubMed] [Google Scholar]
- 53. Tweedie S, et al. 2009. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 37: D555– D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wallrath LL, Elgin SC. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263– 1277 [DOI] [PubMed] [Google Scholar]
- 55. Yin H, Lin H. 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304– 308 [DOI] [PubMed] [Google Scholar]
- 56. Yu C, et al. 2011. Development of expression-ready constructs for generation of proteomic libraries. Methods Mol. Biol. 723: 257– 272 [DOI] [PubMed] [Google Scholar]
- 57. Zeng W, et al. 2009. Specific loss of histone H3 lysine 9 trimethylation and HP1γ/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 5: e1000559 doi:10.1371/journal.pgen.1000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.