Abstract
Pathogenic mycobacteria, including Mycobacterium tuberculosis and Mycobacterium bovis, cause significant morbidity and mortality worldwide. However, the vaccine strain Mycobacterium bovis BCG, unlike virulent strains, triggers extensive apoptosis of infected macrophages, a step necessary for the elicitation of robust protective immunity. We here demonstrate that M. bovis BCG triggers Toll-like receptor 2 (TLR2)-dependent microRNA-155 (miR-155) expression, which involves signaling cross talk among phosphatidylinositol 3-kinase (PI3K), protein kinase Cδ (PKCδ), and mitogen-activated protein kinases (MAPKs) and recruitment of NF-κB and c-ETS to miR-155 promoter. Genetic and signaling perturbations presented the evidence that miR-155 regulates PKA signaling by directly targeting a negative regulator of PKA, protein kinase inhibitor alpha (PKI-α). Enhanced activation of PKA signaling resulted in the generation of PKA C-α; phosphorylation of MSK1, cyclic AMP response element binding protein (CREB), and histone H3; and recruitment of phospho-CREB to the apoptotic gene promoters. The miR-155-triggered activation of caspase-3, BAK1, and cytochrome c translocation involved signaling integration of MAPKs and epigenetic or posttranslational modification of histones or CREB. Importantly, M. bovis BCG infection-induced apoptosis was severely compromised in macrophages derived from miR-155 knockout mice. Gain-of-function and loss-of-function studies validated the requirement of miR-155 for M. bovis BCG's ability to trigger apoptosis. Overall, M. bovis BCG-driven miR-155 dictates cell fate decisions of infected macrophages, strongly implicating a novel role for miR-155 in orchestrating cellular reprogramming during immune responses to mycobacterial infection.
INTRODUCTION
Pathogenic mycobacteria, including Mycobacterium tuberculosis, Mycobacterium bovis, etc., are the major cause of morbidity and mortality across the world, and in this regard, the fate of the infected host macrophages, significant effectors of innate immunity, could act as a rate-limiting step in ensuing immunity (29, 32, 47, 52, 55). Interestingly, avirulent pathogenic mycobacteria, including the vaccine strain M. bovis BCG, unlike virulent M. tuberculosis, cause extensive apoptosis of infected macrophages, which suggests a significant contribution of the apoptosis process to the initiation and subsequent amplification of innate as well as adaptive immune responses (33).
Among various cues that could lead to apoptosis of host cells, the initiation of the apoptotic machinery by posttranscriptional mechanisms assumes significant importance (37). Among posttranscriptional control mechanisms, microRNAs (miRNAs) are suggested to regulate several cellular processes, as they can regulate the expression profiles of 20 to 30% of genes in the human genome (2, 21). miRNAs are evolutionary conserved, non-protein-coding, single-stranded 20- to 22-nucleotide RNA molecules. miRNAs constitute an endogenous class of regulatory RNA molecules that regulate target mRNA either by translational repression or by degradation (2, 10, 18). The target mRNA silencing is brought about by loading mature miRNA onto the RNA-induced silencing complex (RISC), which results in the target mRNA's silencing through mRNA cleavage or translational repression (20, 35, 48). These attributes often act as key regulators of various cellular processes like cell proliferation (26, 69, 71), differentiation (56, 59), autophagy (22, 27), and apoptosis (12, 31, 58). Importantly, recent studies have revealed the central role of miRNAs in innate immune responses to pathogens and a variety of stimuli (30, 60–62). Though much about the miRNAs' synthesis and their mode of action is known, the molecular basis of the regulation and function of specific miRNAs and their roles during immunological processes such as apoptosis await further investigations.
In this perspective, various effectors of host immunity are known to be regulated by several miRNAs, and a prominent one among them, miRNA-155 (miR-155), often exhibits crucial roles during innate or adaptive immune responses (54, 64, 66). Significantly, miR-155 is known as a prototype multifunctional miRNA (17) and is recognized for its inducible expression in activated T cells, macrophages, and dendritic cells (DCs) (4, 67). Interestingly, miR-155 null mice exhibit a marked deficiency in various cell types, and ex vivo experiments demonstrate polarization of T cells toward a Th2 phenotype and severe compromise in the development of germinal centers and B-cell compartments or efficacy of humoral immunity in miR-155 null mice (54, 64). Recent reports have suggested novel roles for miR-155 in Th1 or Th17 differentiation during microbial infections as well as augmented expression of miR-155 in T cells during pathogenic microbial challenge (43, 49). Further, induced expression of miR-155 by lipopolysaccharide (LPS)-mediated Toll-like receptor 4 (TLR4) triggering resulted in modulation of interleukin-1 (IL-1) signaling events in human monocyte-derived DCs. Similarly, miR-155 triggered downregulation of PU.1, thus affecting the expression of DC-SIGN in human DCs (15). In regard to parameters associated with disease and immunity, miR-155 is implicated in autoimmune disorders like rheumatoid arthritis and acts as an important regulator of the oncogenic process by negatively regulating suppressor of cytokine signaling 1 (SOCS1) in breast cancer (28, 36, 40).
In view of the above observations, we set out to unravel the molecular mechanisms contributing toward M. bovis BCG-specific miR-155 expression, principally with regard to the role of miR-155 during M. bovis BCG-mediated apoptosis of macrophages. The present investigation demonstrates that M. bovis BCG-mediated TLR2 stimulation triggers augmented expression of miR-155 in macrophages. Significantly, induced expression of miR-155 involved integrated signaling cross talk among the members of phosphatidylinositol 3-kinase (PI3K), protein kinase Cδ (PKCδ), and mitogen-activated protein kinase (MAPK) pathways, which resulted in the participation of NF-κB and c-ETS in transcriptional activation of miR-155 promoter. Further, M. bovis BCG-triggered miR-155 modulated the expression of apoptosis effectors including PUMA, NOXA, BID, BIM, BAK1, and SMAC, thus leading to apoptosis of infected macrophages. Importantly, M. bovis BCG infection-induced apoptosis was severely compromised in macrophages derived from miR-155 knockout mice compared to wild-type (WT) mice. We present the evidence that miR-155 regulates protein kinase A (PKA) signaling by directly targeting a negative regulator of PKA, protein kinase inhibitor-alpha (PKI-α). Consequently, enhanced activation of PKA signaling directs the generation of PKA C-α, phosphorylation of MSK1, cyclic AMP response element binding protein (CREB), and histone H3. Importantly, miR-155-driven PKA signaling resulted in the activation of apoptotic effectors, active caspase-3, and BAK1 and the cytosolic translocation of cytochrome c, which involved signaling integration among members of MAPKs and epigenetic or posttranslational modification of histones or CREB. Thus, augmented PKA signaling by M. bovis BCG-driven miR-155 dictates cell fate decisions of infected macrophages, emphasizing a novel role for miR-155 in host immunity to mycobacterial infections.
MATERIALS AND METHODS
Cell culture, mice, and bacteria.
The RAW 264.7 mouse macrophage cell line was cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco-Invitrogen, United States) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich, St. Louis, MO), and the cultures were incubated at 37°C in 5% CO2. Macrophages were obtained from peritoneal exudates of C57BL/6 wild-type or TLR2−/− mice and were maintained in DMEM. Bone marrow cells were isolated from femur bones of C57BL/6 wild-type or miR-155−/− mice and cultured in DMEM with 10% FCS and 10% L929 cell-conditioning medium as a source of macrophage colony-stimulating factor (M-CSF) to differentiate into bone marrow-derived macrophages (BMDM). After 7 days, BMDM were used for different experiments. All studies involving mice were carried out after approval from the Institutional Ethics Committee for animal experimentation as well as from the Institutional Biosafety Committee. M. bovis BCG Pasteur 1173P2 was grown in Middlebrook 7H9 broth to log phase and then aliquoted, following which it was stored at −70°C. Representative vials were thawed, and the mycobacterial cells' viability was then assessed by plating on Middlebrook 7H10 agar plates. For all experiments macrophages were infected with M. bovis BCG at a multiplicity of infection (MOI) of 10.
Treatment with pharmacological reagents.
All pharmacological reagents were obtained from Calbiochem (San Diego, CA) and were reconstituted in sterile dimethyl sulfoxide (DMSO) (Sigma-Aldrich). In all experiments, the following inhibitors were used 60 min before experimental treatments: Myr-PKI-α (1 μM), H89 (10 μM), KT5720 (10 μM), LY294002 (50 μM), wortmannin (10 μM), rapamycin (100 nM), PKCα inhibitor (50 μM), PKCβ inhibitor (20 μM), PKCδ inhibitor (10 μM), PKCε inhibitor (50 μM), PKCζ inhibitor (5 μM), SB203580 (20 μM), U0126 (10 μM), SP600125 (50 μM), RAF1 inhibitor (2 μM), manumycin (10 μM), BAY 11-7082 (20 μM). DMSO (1%) was used as a vehicle control. A tested concentration of inhibitors was used after careful titration experiments assessing the viability of the macrophages by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay.
Reagents and antibodies.
General laboratory reagents were purchased from Sigma-Aldrich (St. Louis, MO) and Merck, Germany. Cell culture media were obtained from Gibco-Invitrogen. Fetal bovine serum was obtained from Sigma-Aldrich. Cell culture antibiotics were purchased from Sigma-Aldrich. Anti-PKI-α (H-55) and anti-BAK1 (H-211) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-MyD88 (D80F5), anti-cytochrome c, anti-caspase-3, anti-PKA C-α, anti-Ser376 phospho-MSK1, anti-Ser133 phospho-CREB, anti-Ser10 phospho-histone H3, anti-Thr202/Tyr204 phospho-ERK1/2, and anti-Thr180/Tyr182 phospho-p38 MAPK antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-proliferating cell nuclear antigen (PCNA) antibody was purchased from Calbiochem, and anti-β-actin antibody (AC-15) was procured from Sigma-Aldrich.
PKI-α 3′UTR-ΔmiR-155.
The miR-155 target site in the 3′ untranslated region (UTR) of PKI-α WT plasmid was mutated by nucleotide replacements by site-directed mutagenesis using the megaprimer inverse PCR method. The forward primer comprised the desired mutation, and respective reverse primers were used to generate the megaprimer. The megaprimer was used in turn to amplify the entire plasmid.
Luciferase assays.
For luciferase analysis, RAW 264.7 mouse macrophages were cultured in 60-mm dish plates containing 800 μl of DMEM (Gibco-Invitrogen). Each reporter vector (5 μg) plus 5 μg of β-galactosidase vector and/or 20 μM miRIDIAN miR-155 mimic (Dharmacon, Lafayette, CO) or 20 μM miR-155 inhibitor (Dharmacon) or 100 μM miR-155 small interfering RNAs (siRNAs) or respective control mimics or inhibitors or siRNAs (Ambion, Grand Island, NY) were then added to 300 μl of DMEM containing 5 μl of DharmaFECT 4 (Dharmacon), tubes were incubated for 30 min at room temperature, and the mixture was added to each plate. After 36 h of transfection, cells were lysed in reporter lysis buffer (Promega, USA) and assayed for luciferase activity. The results were normalized for transfection efficiencies by assay of β-galactosidase activity.
miR-155 detection.
For detection of miR-155 by real-time quantitative reverse transcriptase PCR (RT-PCR), total RNA was isolated from macrophages using the TRI reagent (Sigma-Aldrich) according to the manufacturer's protocol. Real-time quantitative RT-PCR for miR-155 was done using TaqMan miRNA assays (Applied Biosystems, Foster City, CA) as per the manufacturer's instructions. U6 snRNA was used for normalization.
miR-155 expression analysis in vivo.
For in vivo expression analysis of miR-155, 104 M. bovis BCG cells or phosphate-buffered saline (PBS) as vehicle control was injected intravenously into the tail vein of mice. Each in vivo experiment involved 6 to 8 animals per group. After 7 days of infection, mice were sacrificed and spleen and lymph nodes were collected. Alternatively, mice were infected intraperitoneally with 106 M. bovis BCG cells for 12 h. Total RNA was isolated from macrophages using the TRI reagent according to the manufacturer's instructions, and expression levels of miR-155 were analyzed by real-time quantitative RT-PCR.
Tuberculosis patients and healthy individuals.
The study population was comprised of healthy individuals (n = 6) and pulmonary tuberculosis patients (n = 11) reporting to the National Institute of Mental Health and Neurosciences, Bangalore, India. Radiological and clinical examinations were performed on each of the subjects under study to confirm active tuberculosis in pulmonary tuberculosis patients and to exclude individuals with active tuberculosis disease in the group of healthy individuals. The study subjects had given written consent, and the study was approved by the Institutional Bioethics Committee.
Real-time quantitative RT-PCR.
Macrophages were treated as described above, peripheral blood mononuclear cells (PBMCs) from healthy subjects and tuberculosis patients were isolated, and total RNA was isolated using TRI reagent in accordance with the manufacturer's instructions. A real-time quantitative RT-PCR was performed using the SYBR green PCR mixture (KAPA Biosystems, Woburn, MA) for quantification of Puma, Noxa, Bid, Bim, Bak1, Smac, and Gapdh, which was used as internal control. Primer sequences used in the study were as follows: Puma forward, 5′-AGCAGCACTTAGAGTCGCC-3′, Puma reverse, 5′-CCTGGGTAAGGGGAGGAGT-3′; Noxa forward, 5′-GCAGAGCTACCACCTGAGTTC-3′, Noxa reverse, 5′-CTTTTGCGACTTCCCAGGCA-3′; Bid forward, 5′-GCCGAGCACATCACAGACC-3′, Bid reverse, 5′-TGGCAATGTTGTGGATGATTTCT-3′; Bim forward, 5′-ATGGCAAAGCAACCTTCTGAT-3′, Bim reverse, 5′-GCTCTGTCTGTAGGGAGGTAGG-3′; Bak1 forward, 5′-GTTTTCCGCAGCTACGTTTTT-3′, Bak1 reverse, 5′-GCAGAGGTAAGGTGACCATCTC-3′; Smac forward, 5′-GCTGAGATGACTTCAAAACACCA-3′, Smac reverse, 5′-TGAATGTGATTCCTGGCGGTT-3′; Gapdh forward, 5′-GAGCCAAACGGGTCATCATCT-3′, Gapdh reverse, 5′-GAGGGGCCATCCACAGTCTT-3′.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were carried out following a protocol provided by Upstate Biotechnology (Lake Placid, NY) with modifications. Macrophages were transfected with miR-155 mimic or miR-155 inhibitor oligonucleotides or respective controls. After 48 h of transfection, macrophages were left uninfected or infected with M. bovis BCG for 8 h. The cells were fixed with 1.42% formaldehyde for 15 min at room temperature followed by inactivation of formaldehyde with 125 mM glycine. Nuclei were isolated from macrophages, and chromatin was sheared using a cup sonicator (Sonics and Materials, Inc., Bozeman, MT). Chromatin extracts containing DNA fragments with an average size of 500 bp were immunoprecipitated using anti-Ser133 phospho-CREB antibody. Purified DNA was analyzed by a real-time quantitative RT-PCR using the SYBR green PCR mixture. Regions with the phospho-CREB binding site in mouse Puma, Noxa, Bid, Bim, Bak1, and Smac promoters were amplified using the following primer pairs: Puma forward, 5′-GTGTGACCCAGTGAGCC-3′, Puma reverse, 5′-ACGCTCAGACTAACGGACT-3′; Noxa forward, 5′-GTCTCGAGACCTGCTCC-3′, Noxa reverse, 5′-CTCTCTGTTCAGGCGC-3′; Bid forward, 5′-GCATCTCTGATAGGGCCGAT-3′, Bid reverse, 5′-AGGAAGGTACACCCGGAAC-3′; Bim forward, 5′-TGTGCGCTCTTGTAGCG-3′, Bim reverse, 5′-GCCGTCCCAATCAATGTT-3′; Bak1 forward, 5′-GTTTTACTGTTGTCTGGCGG-3′, Bak1 reverse, 5′-GCCTACACAGTTGAATGATGG-3′; Smac forward, 5′-ATGGCCAACAACCAGCCA-3′, Smac reverse, 5′-GAGTCGTCGACCTCCGTC-3′; 28S rRNA forward, 5′-CTGGGTATAGGGGCGAAAGAC-3′, and 28S rRNA reverse, 5′-GGCCCCAAGACCTCTAATCAT-3′. All results were normalized either by respective input values or by amplification of 28S rRNA. All ChIP experiments were repeated at least three times.
Immunoblotting analysis.
Macrophages were harvested after treatment, washed with ice-cold PBS, and lysed in buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml each aprotinin, leupeptin, and pepstatin, 1 mM Na3VO4, 1 mM NaF). The total amount of protein was quantified, and equal amounts of proteins were resolved in 12% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). Membranes were blocked with 5% nonfat dried milk in Tris-buffered saline-Tween-20 (TBST) and probed with a primary antibody overnight at 4°C. After washings with TBST, membranes were probed with target-specific primary antibody followed by horseradish peroxidase-conjugated secondary antibody (Jackson Immunologicals, West Grove, PA). Blots were developed with an enhanced chemiluminescence detection system (Perkin Elmer Bioscience, Waltham, MA) in accordance with the manufacturer's protocol.
Nuclear and cytosolic subcellular fractionation.
Macrophages were harvested after respective experimental treatments and gently resuspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], and 0.5 mM PMSF). After incubation on ice for 15 min, cell membranes were disrupted with 10% NP-40. Cytosolic extract was separated from nuclei and mitochondria by centrifugation at 13,000 rpm for 15 min at 4°C. Nuclei were lysed with buffer C (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF), and nuclear protein extract was collected after centrifugation at 13,000 rpm for 20 min at 4°C. The nuclear and cytosolic fractions were resolved on denaturing polyacrylamide gel, and further processing was done as mentioned in “Immunoblotting analysis.”
Macrophage cell viability assay.
Macrophages were transfected with the miR-155-overexpressing construct (pPRIME-CMV-GFP-miR-155) or with the control vector (pPRIME-CMV-GFP) and/or infected with M. bovis BCG for 96 h. Macrophage cell viability was assayed by trypan blue staining. Briefly, trypan blue was mixed with treated or infected macrophages at a final concentration of 0.04%, the mixtures were incubated for 3 min, and the dye-excluding (viable) and the stained (nonviable) cells were counted microscopically.
Detection of apoptosis induction in macrophages.
Macrophage apoptosis upon M. bovis BCG infection was evaluated by augmented expression of proapoptotic genes, activation of caspase-3, cytosolic translocation of cytochrome c, annexin V staining, sub-G1 analysis, and DNA content analysis. Total cell lysate (for caspase-3 activation) or cytosolic fractions (for cytochrome c translocation) of macrophages that were either infected with M. bovis BCG or transfected with miRNA-155 mimics or miRNA-155 inhibitors were resolved onto SDS-PAGE gel and probed with respective antibodies to assess activation of caspase-3 or translocation of cytochrome c. For measurement of apoptosis using annexin V staining, the apoptotic population of macrophages was determined utilizing the annexin V-fluorescein isothiocyanate (FITC) kit (Miltenyi Biotech, United States) in accordance with the manufacturer's instructions. The data were analyzed by confocal microscopy. In an independent set of experiments, macrophages were stained with propidium iodine (PI), and the sub-G1 population and total DNA content were determined by flow cytometry and confocal microscopy, respectively.
Mycobacterial viability assay.
Macrophages were transfected with the miR-155 overexpressing construct (pPRIME-CMV-GFP-miR-155) or with the control vector (pPRIME-CMV-GFP) for 36 h, cells were infected with M. bovis BCG at a MOI of 10 for 24 h, and over the next 72 h cells were maintained in antibiotic-containing medium. Intracellular mycobacteria were harvested by lysing macrophages, and cell lysate was plated onto Middlebrook 7H10 agar plates. Total CFU were counted, and the CFU results from macrophages overexpressing miR-155 were compared with those from control macrophages.
Statistical analysis.
Levels of significance for comparison between samples were determined by the Student t test distribution. The data in the graphs are expressed as means ± standard errors (SE). Graphpad Prism 3.0 software (GraphPad software, La Jolla, CA) was used for all statistical analyses.
RESULTS
miR-155 is upregulated in response to M. bovis BCG infection.
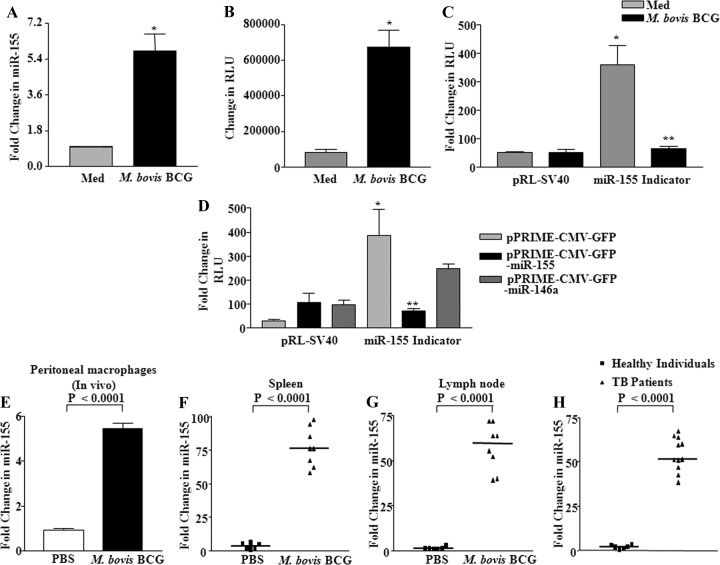
We first examined the expression levels of miR-155 triggered upon infection of macrophages with M. bovis BCG, and real-time quantitative RT-PCR demonstrated a significant increase in miR-155 expression (Fig. 1A). Our objective to study miR-155 derived from the following observations. The global expression analysis of miRNAs of macrophages infected with M. bovis BCG revealed upregulation of many miRNAs, including miR-155 (data not shown). In addition, reports have suggested that miRNA-155 could act as an initial immune sensor, which possibly modulates both innate and adaptive immune responses to a wide variety of infections or cancers (16, 19, 54, 64, 68). In addition to real-time quantitative RT-PCR, M. bovis BCG infection significantly enhanced the miR-155 promoter activity as shown in Fig. 1B. Further, experiments with specific miRNA indicator plasmids were utilized, in which miRNA binds to the complementary sequence fused to green fluorescent protein (GFP). As demonstrated in Fig. 1C, infection with M. bovis BCG clearly triggered induced selective expression of miR-155 in macrophages as evaluated by a marked inhibition of luciferase activity of the miR-155 indicator plasmid. However, luciferase activity remained unperturbed in controls. For additional validation, expression plasmids for miR-155 or miR-146a were cotransfected in RAW 264.7 macrophages with miR-155 indicator plasmid. Interestingly, GFP luciferase activity was markedly reduced only in the cells expressing miR-155, thus confirming the specificity and substantiating infection-induced expression of miR-155 (Fig. 1D).
Fig 1.
M. bovis BCG induces upregulation of miR-155 expression. (A) Mouse peritoneal macrophages were infected with M. bovis BCG, and expression of miR-155 was assayed by real-time quantitative RT-PCR. (B) miR-155 promoter activity was measured by transient transfection of RAW 264.7 macrophages with miR-155 promoter-luciferase construct prior to the addition of M. bovis BCG stimulation (data are means ± SE, n = 3). (C) Macrophages were transfected with miR-155 indicator or renilla plasmid (pRL-SV40) prior to M. bovis BCG infection. The luciferase activity of miR-155 indicator was assayed. Data are means ± SE, n = 3. (D) Macrophages were cotransfected with the control vector alone (pPRIME-CMV-GFP) or miR-155 (pPRIME-CMV-GFP-miR-155) or pPRIME-CMV-GFP-miR-146a overexpression plasmids along with the miR-155 indicator construct. The luciferase assay was performed 48 h posttransfection. Data are means ± SE, n = 3. (E) C57BL/6 mice were intraperitoneally infected with 106 M. bovis BCG bacilli for 12 h, while control mice were injected with PBS. Macrophages were harvested, and total RNA was isolated to estimate the total expression level of miR-155 (data are means ± SE; n = 8 for PBS-injected mice, and n = 8 for M. bovis BCG-infected mice). (F and G) In vivo expression of miR-155 was analyzed by injecting M. bovis BCG or PBS as vehicle control into the tail vein of mice. On the seventh day, mice were sacrificed and spleen (F) and lymph nodes (G) were removed for expression analysis of miR-155 (data are means ± SE; n = 6 for PBS-injected mice, and n = 8 for M. bovis BCG-infected mice). (H) Comparison of miR-155 expression profiles in PBMCs from pulmonary tuberculosis patients and from healthy individuals as assayed by real-time quantitative RT-PCR. (data are means ± SE; n = 6 for healthy individuals, and n = 11 for TB patients). Med, medium; *, P < 0.05 versus control; **, P < 0.05 versus miR-155 indicator control.
In order to bring relevance to the biology of mycobacterial infection in vivo, a suggested murine model to study systemic infection-triggered inflammation was utilized (11, 51). In this perspective, mice were infected with M. bovis BCG intraperitoneally or intravenously, and as shown in Fig. 1E, F, and G, M. bovis BCG triggered robust expression of miR-155 in vivo in peritoneal macrophages, spleen, and lymph nodes. Accordingly, PBMCs derived from pulmonary tuberculosis patients exhibited augmented expression of miR-155 compared to controls (Fig. 1H).
M. bovis BCG-triggered TLR2-PI3K-PKCδ-MAPK signaling regulates induced expression of miR-155.
TLR2 receptor engagement by M. bovis BCG elicits a gamut of cellular immune responses that comprise a wide spectrum of signaling cascades, including PI3K signaling (5, 8, 9, 24). PI3K signaling exerts significant effects on cell fate decisions by diligently regulating a wide range of cellular genes such as apoptotic or antiapoptotic genes, transcription factors, etc. Further, we and others have demonstrated the activation of PI3K signaling in host cells upon infection with pathogenic mycobacteria (6, 7, 44, 45). Here, we investigated whether M. bovis BCG-triggered activation of PI3K could regulate induced expression of miR-155. As shown in Fig. 2A and Fig. S1A in the supplemental material, disruption of TLR2 receptor triggering by utilization of macrophages from TLR2 null mice or overexpressed TLR2 dominant negative construct (TLR2 DN) abrogated M. bovis BCG's ability to induce miR-155 expression. Further, signaling perturbations either by dominant negative constructs or by pharmacological inhibitors to members of the PI3K pathway, LY294002, wortmannin (p85 PI3K), and rapamycin (mTOR), significantly attenuated M. bovis BCG-triggered miR-155 expression (Fig. 2B; see Fig. S1B in the supplemental material). Accordingly, M. bovis BCG infection triggered the phosphorylation of p85 subunit of PI3K, 4EBP1, and other important members of the PI3K pathway (see Fig. S1C in the supplemental material).
Fig 2.
M. bovis BCG-triggered activation of TLR2 signaling instructs macrophages to induce miR-155 expression. (A) WT or TLR2−/− macrophages were infected with M. bovis BCG or left uninfected. Expression of miR-155 was analyzed by real-time quantitative RT-PCR after 12 h of infection (n = 3). (B) Macrophages were pretreated with PI3K pathway inhibitors (LY294002, wortmannin, or rapamycin), and after M. bovis BCG infection miR-155 expression was analyzed by real-time quantitative RT-PCR. DMSO was used as vehicle control (n = 3). (C) Pretreatment of macrophages with specific pharmacological inhibitors of various PKC isoforms followed by real-time quantitative RT-PCR analysis of miR-155 expression upon M. bovis BCG infection (n = 3). (D) Real-time quantitative RT-PCR analysis of M. bovis BCG-triggered expression of miR-155 in macrophages that were pretreated with inhibitors U0126 (ERK1/2), SB203580 (p38), SP600125 (JNK1/2), manumycin (RAS), and RAF1 inhibitor (RAF1) (n = 3). (E) Macrophages were pretreated with Bay 11-7082 prior to M. bovis BCG infection, and expression levels of miR-155 were assayed by real-time quantitative RT-PCR (n = 3). (F, G, and H) Macrophages were transfected with WT miR-155 promoter-luciferase construct or mutant promoter-luciferase plasmids for NF-κB (F), c-ETS (G), and AP-1 (H) binding sites. Later, cells were left uninfected or infected with M. bovis BCG for 12 h followed by evaluation by luciferase reporter assay (n = 3). RLU, relative luciferase units; Med, medium; WT, wild type; TLR2−/−, TLR2 knockout; *, P < 0.05 versus control; **, P < 0.05 versus M. bovis BCG-infected miR-155 promoter-luciferase. For all panels, data are means ± SE.
Among many signaling kinases, PKC operates as regulatory kinase by modulating downstream signaling cascades, including activation of MAPK. Thus, PKC considerably modulates the transcriptome profile of the wide variety of immune cells; and significantly, regulatory functions within immune cells often require strong collaboration between PI3K and PKC activity (24, 45). In this perspective, pharmacological inhibition of PKC strongly inhibited M. bovis BCG-induced miR-155 expression (Fig. 2C). Importantly, inhibition of PKCδ activity among various isoforms either by specific pharmacological inhibitor or by overexpression of dominant negative mutants of PKCδ abolished M. bovis BCG infection-induced expression of miR-155 (Fig. 2C; see Fig. S1D in the supplemental material). Inhibition of PI3K activity strongly reduced the M. bovis BCG-mediated activation of PKCδ (Fig. S1E). In accordance with previous observations, inhibition of PKCδ reduced infection-triggered activation of ERK1/2 and p38 MAPKs (Fig. S1F). Similarly, inhibition of RAS, RAF1, and members of MAPKs, namely, ERK1/2 and p38 MAPKs, by means of either pharmacological inhibitors or dominant negative constructs significantly compromised M. bovis BCG-triggered miR-155 expression (Fig. 2D; see Fig. S1G in the supplemental material).
Several genes involved in immunity contain cis-acting consensus elements for various transcription factors, including NF-κB, and transcriptional expression often involves specific recruitment of transcription factors to the promoters of target genes. Notably, NF-κB among many transcription factors often acts as “gain control” for a wide range of signaling events in many cell types (41). In this regard, BAY 11-7082, a specific inhibitor of IκB-α phosphorylation, markedly diminished induced expression of miR-155 expression (Fig. 2E). We explored the roles for NF-κB, c-ETS, and AP-1 during M. bovis BCG-induced miR-155 expression in macrophages. In this context, we utilized miR-155 promoter constructs that lack binding sites for each of these transcription factors. As shown in Fig. 2F, G, and H, M. bovis BCG challenge fails to trigger activity of miR-155 promoter deletion constructs that lack binding sites for either NF-κB or c-ETS, but not AP-1. Prominently, interference in the PI3K, PKCδ, or MAPK activity reversed M. bovis BCG-mediated nuclear translocation of NF-κB (data not shown). These results strongly advocate for a multichotomous nature of M. bovis BCG-driven signaling, in which PI3K, PKCδ, MAPK, and NF-κB-c-ETS integrate to arbitrate induced miR-155 expression.
miR-155 is required for M. bovis BCG-mediated apoptosis of macrophages.
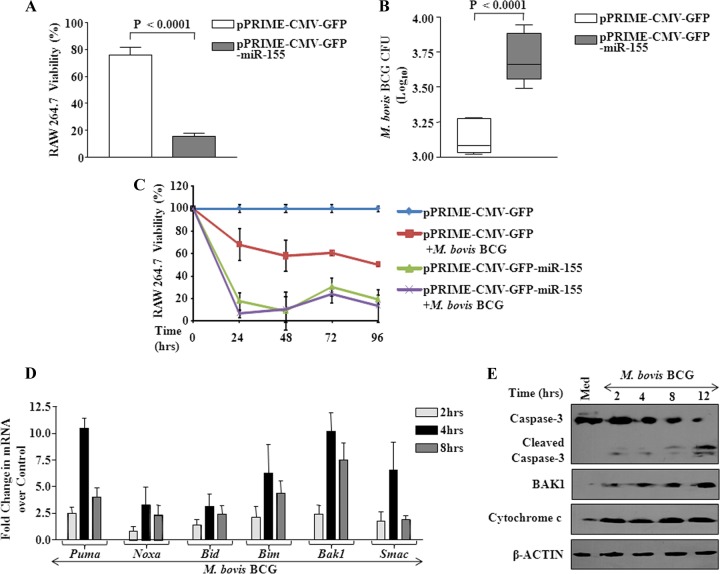
We investigated the effects of enforced expression of miR-155 in RAW 264.7 macrophages. As shown in Fig. 3A and B, ectopic expression of miR-155 resulted in loss of viability of macrophages with a concomitant increase in intracellular M. bovis BCG CFU compared to vector controls. Significantly, apoptosis of macrophages by enforced expression of miR-155 could be further augmented by infection with M. bovis BCG in a time-dependent manner (Fig. 3C). Validating the occurrence of apoptosis, investigation of kinetics and expression patterns of various proapoptotic genes demonstrated that M. bovis BCG triggered upregulation of transcript levels of PUMA, NOXA, BID, BIM, BAK1, and SMAC in macrophages as early as 4 h postinfection (Fig. 3D). One of the hallmarks of apoptosis is the loss of integrity of the mitochondrial outer membrane, a significant event resulting in the release of cytochrome c, which culminates in the activation of caspases. In this regard, infection of macrophages with M. bovis BCG triggered the release of cytochrome c in a time-dependent manner with a concomitant increase in BAK1 expression or caspase-3 processing, a necessary prerequisite for generation of active caspase-3 (Fig. 3E).
Fig 3.
Enforced expression of miR-155 augments macrophage apoptosis. (A) miR-155 was overexpressed in RAW 264.7 macrophages by transfecting pPRIME-CMV-GFP-miR-155 plasmid. Control cells were transfected with pPRIME-CMV-GFP. Macrophage viability was estimated by trypan blue staining (data are means ± SE, n = 12). (B) Macrophages were treated as described for panel A and infected with M. bovis BCG for 96 h. Intracellular survival of M. bovis BCG was assayed by estimating CFU for miR-155 overexpressed macrophages and for vector-transfected control macrophages. Data are means ± SE, for 6 separate experiments. (C) Kinetics of viability of macrophages transfected with miR-155 overexpression plasmid during infection with M. bovis BCG (data are means ± SE, n = 12). (D) Kinetics of proapoptotic gene expression over 8 h after M. bovis BCG infection of macrophages (data are means ± SE, n = 3). (E) Western blot analysis of cleavage of caspase-3, BAK1, and cytochrome c in macrophages with infection with M. bovis BCG for 6 h. β-Actin served as loading control, and blots are representative of 2 separate experiments.
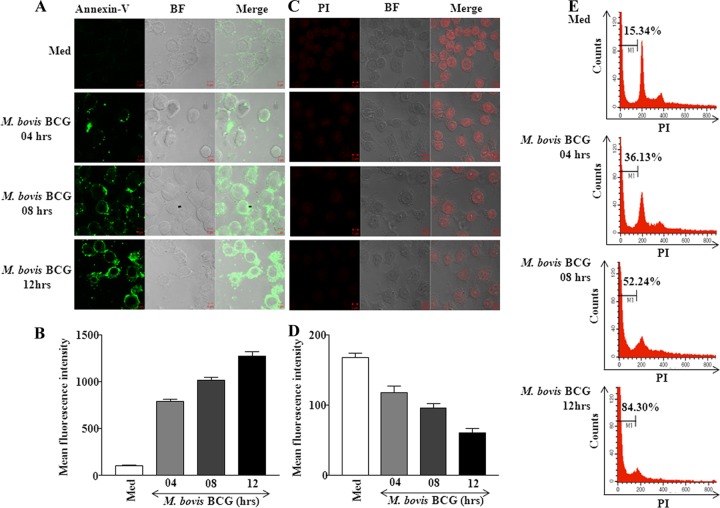
The activation of apoptosis effectors led to a significant increase in apoptosis as assayed by annexin V staining (Fig. 4A and B) and to loss of small DNA fragments as assayed for total DNA content using PI staining (Fig. 4C and D). Further, the proportion of the sub-G1 population in M. bovis BCG-infected macrophages increased in a time-dependent manner (Fig. 4E).
Fig 4.
M. bovis BCG triggers apoptosis of infected macrophages. (A) Macrophages were infected with M. bovis BCG for indicated times. After infection, cells were washed with PBS and stained with annexin V-FITC for 30 min followed by fixation in 3.7% paraformaldehyde. The cells were analyzed by confocal microscopy. The representative images are shown. Data are representative of 4 separate experiments. (B) Quantification of mean fluorescence intensities of panel A confocal images (data are means ± SE, n = 4). (C and D) After infection, macrophages were fixed with 70% ethanol to leak out apoptotic DNA fragments and total DNA content was visualized by confocal microscopy after staining cells with PI. Shown are a representative of 3 independent experiments (C) and quantification of the same (D). (E) Macrophages were infected with M. bovis BCG for indicated times, fixed in 70% ethanol, stained with PI, and analyzed by flow cytometry. A representative result of 3 experiments is shown. The sub-G1 subpopulation is indicated by M1, and the percentage of total cells undergoing apoptosis is indicated. Med, medium; BF, bright field; PI, propidium iodide.
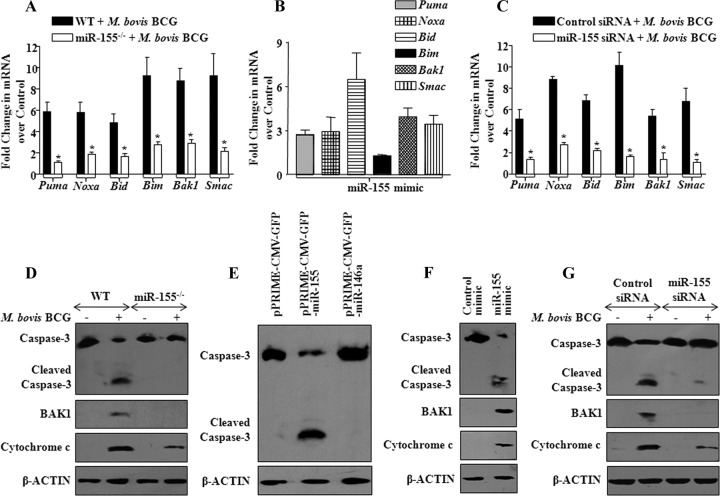
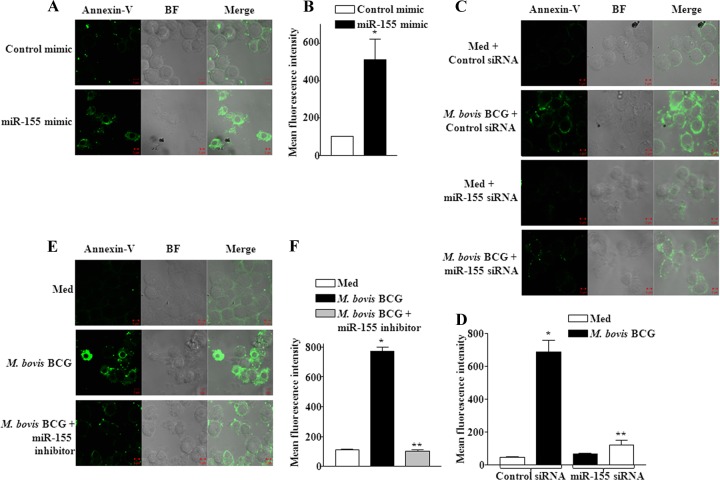
In order to further strengthen and validate our observations, we utilized miR-155 null macrophages. As shown in Fig. 5A and D, M. bovis BCG infection-induced apoptosis was severely compromised in bone marrow-derived macrophages (BMDM) derived from miR-155 knockout mice compared to the apoptosis in WT mice, as analyzed by the expression levels of proapoptotic genes as well as the reduced activation of caspase-3 and BAK1 expression and the release of cytochrome c. We performed gain-of-function and loss-of-function studies to validate the role of miR-155 during M. bovis BCG infection of macrophages. Significantly, enforced expression of miR-155 resulted in augmented expression of PUMA, NOXA, BID, BIM, BAK1, and SMAC transcripts in macrophages (Fig. 5B; see Fig. S2A and B in the supplemental material). Importantly, repression of M. bovis BCG-triggered miR-155 expression, either by miR-155 specific small interfering RNA (siRNA) or by miR-155 inhibitor (anti-miR-155 oligonucleotide), but not controls, considerably reduced the transcripts levels of PUMA, NOXA, BID, BIM, BAK1, and SMAC (Fig. 5C; see Fig. S2C, D, E and S3A in the supplemental material). Notably, ectopic expression of miR-155, but not miR-146a, triggered activation of apoptotic cascades in macrophages as evidenced by the generation of active caspase-3 (Fig. 5E and Fig. S2A). Further, enforced expression of miR-155 by miR-155 mimic validated the key role of miR-155 in activation of apoptosis (Fig. 5F and Fig. S2B). Accordingly, miR-155 siRNA or miR-155 inhibitor blocks M. bovis BCG-induced activation of apoptotic effectors (Fig. 5G and Fig. S3B) as well as apoptosis (Fig. 6A, B, C, D, E, and F), thus strongly assigning a considerable role for miR-155 in infection-triggered apoptosis of macrophages. The specificity of tested miR-155 inhibitor was confirmed by derepression of MyD88 protein, a known miR-155 target, as well as by decreased miR-155 promoter luciferase activity (see Fig. S3C and D in the supplemental material).
Fig 5.
miR-155 modulates effectors of apoptosis. (A) miR-155−/− or WT bone marrow-derived macrophages (BMDM) were infected with M. bovis BCG, and the expression profiles of proapoptotic genes were assayed using real-time quantitative RT-PCR. Data represent fold changes over uninfected cells. *, P < 0.05 versus WT M. bovis BCG-infected BMDM (data are means ± SE, n = 5). (B) The role of miR-155 in modulating expression of proapoptotic genes was validated by transfection of miR-155 in RAW 264.7 macrophages, and real-time quantitative RT-PCR was performed to monitor the changes in expression levels of proapoptotic genes (data are means ± SE, n = 3). (C) miR-155 activity was silenced using miR-155 siRNA prior to M. bovis BCG infection, and expression of proapoptotic genes was analyzed by real-time quantitative RT-PCR (data are means ± SE, n = 3). *, P < 0.05 versus control siRNA-transfected and M. bovis BCG-infected macrophages. (D) Activation of caspase-3, cytosolic translocation of cytochrome c, and expression of BAK1 in miR-155−/− or WT BMDM were assayed using Western blotting after infection with M. bovis BCG. A representative result of 3 experiments is shown. (E) miR-155 and miR-146a were overexpressed in macrophages, and activation of apoptosis was assayed by monitoring cleavage of caspase-3. β-Actin served as loading control, and blots are representative of 3 separate experiments. (F) miR-155 mimics were transfected into macrophages, and activation of apoptosis was monitored. (G) Macrophages were transfected with either control siRNA or miR-155 siRNA and then infected with M. bovis BCG. Cleaved caspase-3, BAK1, and cytochrome c protein levels were monitored by Western blotting. Western blots are representative of 3 independent experiments. Med, medium; WT, wild type; miR-155−/−, miR-155 knockout.
Fig 6.
miR-155 is an indispensable mediator of apoptosis. (A and B) Macrophages were transfected with either control mimics or miR-155 mimics, and induction of apoptosis was monitored by annexin V-FITC staining by confocal microscopy. The quantification of apoptosis cells was evaluated from 4 separate experiments (B). (C and D) miR-155 was knocked down using miR-155-specific siRNA, and induction of apoptosis was monitored as described for panel A and quantified. A representative result of 4 experiments is shown (D). (E and F) Macrophages were transfected with miR-155 inhibitor oligonucleotides prior to M. bovis BCG infection. The effects of M. bovis BCG infection and the role of miR-155 inhibitor on M. bovis BCG-induced apoptosis were analyzed by confocal microscopy. Quantitative analysis was performed (F). A representative result of 4 experiments is shown. Med, medium; BF, bright field.
miR-155 targets PKI-α, a negative regulator of PKA.
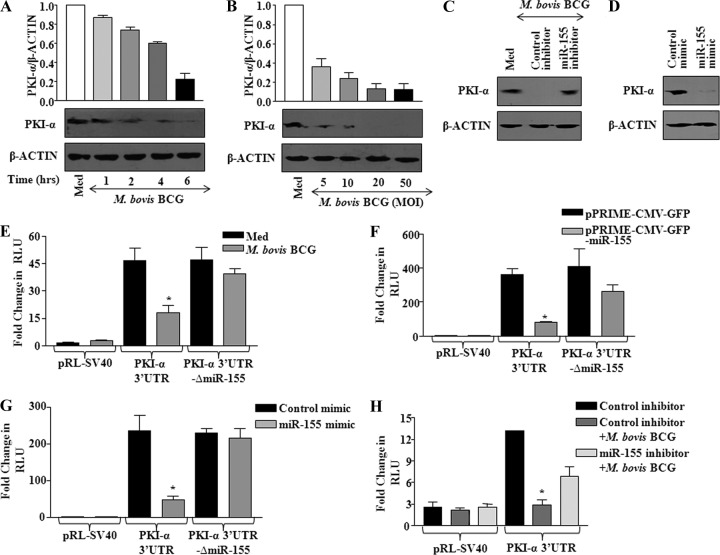
The activation of chronic inflammation during infection with pathogenic microbes often involves not only an unrestrained migration of immune cells including macrophages and a rapid enrichment of the proinflammatory cytokine milieu but also the rapid multiplication and dissemination of infected microbes by speedy apoptosis of infected macrophages. In this complex settings of bidirectional signaling between host and pathogen, activation of specific host signaling pathways that regulate expression of apoptotic or antiapoptotic genes will be a crucial determinant of cell fate decisions. Protein kinase A (PKA) is one of the rate-limiting signaling components that control multiple facets of immune responses, including DC maturation, antigen processing and presentation, T-cell proliferation, and effector functions, among others. Importantly, contributions to polarizing effects on macrophages by various stimuli are reported to involve the participation of miR-155. In this perspective, extensive bioinformatics analyses as well as a recent study by Fassi Fehri et al. (19) have suggested that protein kinase inhibitor-alpha (PKI-α) is a target for miR-155 and that miR-155 targets an 8-mer site located at positions 442 to 448 or 449 to 455 at the end of mouse or human, respectively, PKI-α 3′-UTR. Notably, PKI-α is a well-known endogenous inhibitor of PKA activity (14). Thus, we aimed to carry out an integrated investigation on the interaction of miR-155 and PKA signaling events. As a first step, we assessed the PKI-α protein expression levels in macrophages challenged with M. bovis BCG. Data depicted in Fig. 7A and B clearly demonstrate a time- and dose-dependent decrease of PKI-α expression upon infection with M. bovis BCG. Importantly, M. bovis BCG-mediated decrease in PKI-α protein could be reversed by ectopic expression of miR-155 inhibitor compared to control (Fig. 7C). Similarly, enforced expression of miR-155 mimic strongly reduced the basal expression level of PKI-α protein (Fig. 7D). Accordingly, M. bovis BCG infection or enforced expression of miR-155 by overexpression plasmid or mimics significantly reduced the luciferase activity of a PKI-α 3′UTR construct compared to that of a pRL-SV40 vector control. However, mutation of the miR-155 target site in 3′UTR of PKI-α WT plasmid abrogated M. bovis BCG's ability to reduce PKI-α 3′UTR activity, thus demonstrating the miR-155-dependent repression of PKI-α (Fig. 7E, F, and G). Consequently, the miR-155 inhibitor, but not the control oligonucleotide, nullified M. bovis BCG's ability to reduce the luciferase activity of the PKI-α 3′UTR construct (Fig. 7H).
Fig 7.
M. bovis BCG-induced miR-155 targets PKI-α. (A) Kinetics for PKI-α as assayed by Western blotting after M. bovis BCG infection of macrophages. The quantification of the same is shown (data are means ± SE, n = 3). (B) Macrophages were stimulated with addition of M. bovis BCG at various MOI (5, 10, 20, and 50) and assayed for PKI-α protein levels using Western blotting. Blots were quantified and represented as shown (data are means ± SE, n = 3). (C) M. bovis BCG-triggered miR-155 expression was silenced by transfecting macrophages with miR-155 inhibitor. PKI-α protein levels were monitored in uninfected and infected macrophages. A representative result of 3 experiments is shown. (D) Macrophages were transfected with miR-155 mimic or a control mimic, and expression of PKI-α protein was analyzed by Western blotting. A representative result of 3 experiments is shown. (E) PKI-α 3′UTR WT or PKI-α 3′UTR-ΔmiR-155 luciferase plasmids or vector control pRL-SV40 was transfected into macrophages prior to infection with M. bovis BCG. Luciferase activities were monitored by reporter luciferase assay (data are means ± SE, n = 3). (F) Macrophages were cotransfected with miR-155 overexpressing plasmid (pPRIME-CMV-GFP-miR-155) and PKI-α 3′UTR WT or PKI-α 3′UTR-ΔmiR-155 plasmids, and luciferase activities were measured. Data are means ± SE of 3 separate experiments. (G) miR-155 was overexpressed in macrophages by transfecting miR-155 mimic along with PKI-α 3′UTR WT or PKI-α 3′UTR-ΔmiR-155 luciferase plasmids, and the ability of miR-155 to target PKI-α 3′UTR was assayed by luciferase reporter assay. Data represent means ± SE, n = 3. (H) The effect of miR-155 on PKI-α was validated by cotransfection of PKI-α 3′UTR luciferase plasmid with miR-155 inhibitor prior to M. bovis BCG infection. Data are means ± SE of 3 separate experiments. RLU, relative luciferase units; Med, medium; *, P < 0.05 versus PKI-α 3′UTR luciferase control.
miR-155 regulates PKA signaling and macrophage apoptosis.
PKA is one of the effector kinases that regulate a wide array of cellular responses (63). cAMP binding to PKA results in the release of two catalytic subunits (C), and PKA C-α mediates activation of MAPKs in the cytosol or activation of CREB (cAMP response element binding protein) in the nucleus. Activation of MSK1 by MAPKs regulates induced expression of various immune genes through the involvement of phosphorylated CREB binding to CRE at promoters of target genes or chromatin modifications by phosphorylation at Ser10 of histone H3 (3, 53).
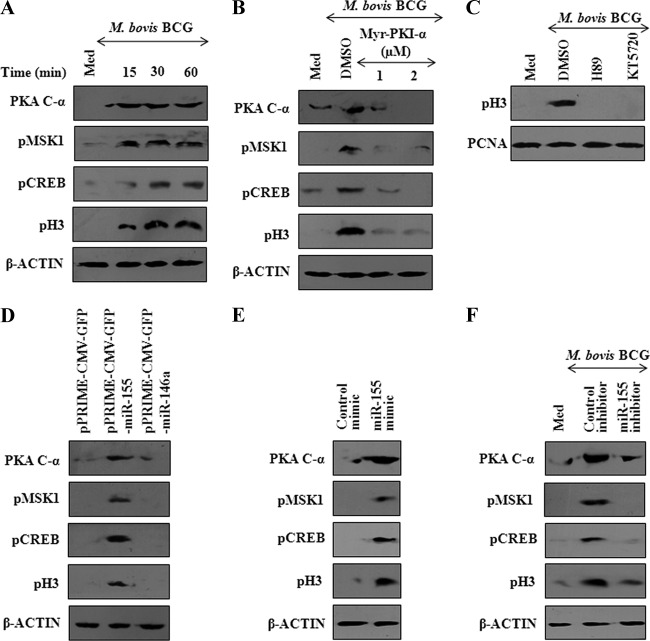
The PKA signaling is tightly regulated by PKI-α, a strong inhibitor of cAMP-dependent PKA activity, and PKI-α executes its inhibitory effects by high-affinity binding to free catalytic subunit as well as exporting nuclear catalytic subunit to the cytoplasm (13, 34). Interestingly, skeletal muscles derived from PKI-α null mice exhibited diminished levels of activation of transcription factor CREB (23). In this regard, we explored whether miR-155, by modulating PKI-α levels, regulates M. bovis BCG's ability to activate PKA signaling. As shown in Fig. 8A, infection of mouse peritoneal macrophages with M. bovis BCG induced the activation of PKA signaling as evidenced by the generation of PKA C-α and the phosphorylation of MSK1, CREB, and histone H3. Significantly, treatment of macrophages with Myr-PKI-α, an analogue of endogenous PKI-α, significantly diminished M. bovis BCG-mediated activation of PKA, MSK1, CREB, or histone H3 (Fig. 8B). Pharmacological inhibition of MSK1 (H89) or PKA (KT5720) activity markedly reduced M. bovis BCG-triggered histone H3 Ser10 phosphorylation (Fig. 8C). In order to further characterize the contribution of miR-155 in governing the expression of apoptotic genes, we investigated the histone H3 Ser10 phosphorylation upon enforced expression of miR-155 in macrophages. Accordingly, enforced expression of miR-155, but not miR-146a, resulted in generation of PKA C-α as well as activation of MSK1, CREB, or histone H3 (Fig. 8D and E and data not shown). On the contrary, inhibition of miR-155 expression by anti-miR-155 oligonucleotide inhibitor, but not by control oligonucleotide, resulted in a marked inhibition in the ability of M. bovis BCG to trigger PKA signaling (Fig. 8F).
Fig 8.
miR-155 activates PKA signaling. (A) Macrophages were stimulated by infection with M. bovis BCG, and the kinetics of PKA signaling activation was analyzed by Western blotting. Generation of PKA catalytic subunit α (PKA C-α) and phosphorylation of MSK1, CREB, and histone H3 are shown. Western blots are representative of 3 independent experiments. (B) Pretreatment of macrophages with myristoylated protein kinase A inhibitor peptide (Myr-PKI-α) prior to M. bovis BCG infection abrogates activation of PKA signaling. Western blots are representative of 3 independent experiments. (C) Macrophages were treated with H89 (MSK1 inhibitor) or KT5720 (PKA inhibitor) prior to M. bovis BCG stimulation. After 6 h of infection, cytosolic and nuclear fractions were separated and activation of histone H3 was assayed using Western blotting. Blots represent means of 3 separate experiments. (D) miR-155 or miR-146a was overexpressed in macrophages, and activation of PKA signaling was assayed by Western blotting. Western blots are representative of 3 independent experiments. (E) Macrophages were transfected with miR-155 mimic or control mimic, and activation of the PKA pathway was monitored as described for panel A. (F) M. bovis BCG-induced expression of miR-155 was nullified by transfecting miR-155 inhibitor prior to M. bovis BCG infection, and the regulation of the PKA pathway was monitored as described above. Western blots are representative of 3 independent experiments. β-Actin was used as loading control and DMSO as vehicle control. Med, medium.
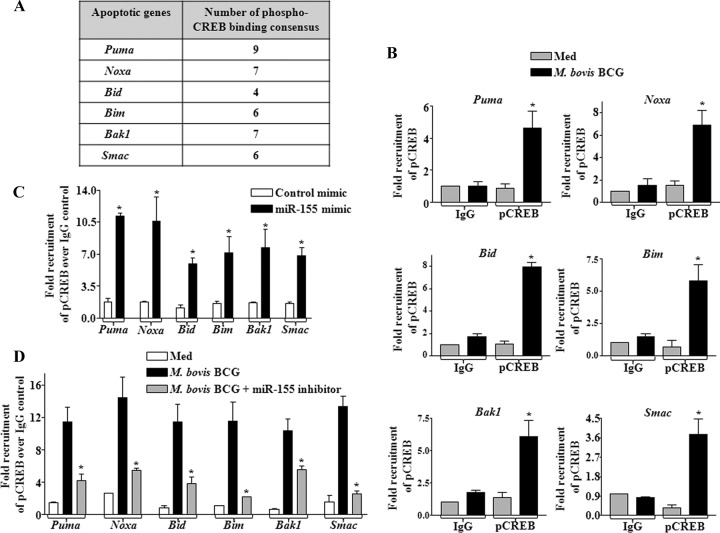
For further validation, we identified many sites for phospho-CREB binding consensus in 2-kb upstream nucleotide sequences of promoters of PUMA, NOXA, BID, BIM, BAK1, and SMAC genes (Fig. 9A). Accordingly, we carried out chromatin immunoprecipitation analysis, which revealed that infection with M. bovis BCG or enforced expression of miR-155 results in significant recruitment of phospho-CREB at the sites of selected apoptotic gene promoters in vivo (Fig. 9B and C). Corroborating this observation, miR-155 inhibitor treatment prior to M. bovis BCG infection blocked the recruitment of phospho-CREB to the promoters of PUMA, NOXA, BID, BIM, BAK1, and SMAC genes (Fig. 9D). These findings advocate miR-155 as a molecular switch that regulates M. bovis BCG-mediated macrophage apoptosis. These results also validate a significant role for miR-155 in regulation of PKA signaling during mycobacterial infection of macrophages.
Fig 9.
miR-155 recruits phospho-CREB to the promoters of apoptotic genes. (A) The 2-kb upstream nucleotide sequences of apoptotic genes Puma, Noxa, Bid, Bim, Bak1, and Smac were analyzed for phospho-CREB binding consensus sequences using the MatInspector program. (B) The chromatin immunoprecipitation (ChIP) assay was performed to analyze phospho-CREB (pCREB) recruitment to the promoters of proapoptotic genes upon M. bovis BCG stimulation of macrophages (data are means ± SE, n = 3; *, P < 0.05 versus IgG pulldown). (C) miR-155 was ectopically overexpressed in macrophages, and nuclear translocation of pCREB to the promoters of proapoptotic genes was observed by ChIP assay. Data are means ± SE, n = 3; *, P < 0.05 versus control mimic). (D) miR-155 inhibitor oligonucleotides were used to abolish M. bovis BCG-induced miR-155 activity, and the ChIP assay was done to analyze the total recruitment of pCREB to the promoters of proapoptotic genes as indicated (*, P < 0.05 versus M. bovis BCG). Data are means ± SE of 3 separate experiments.
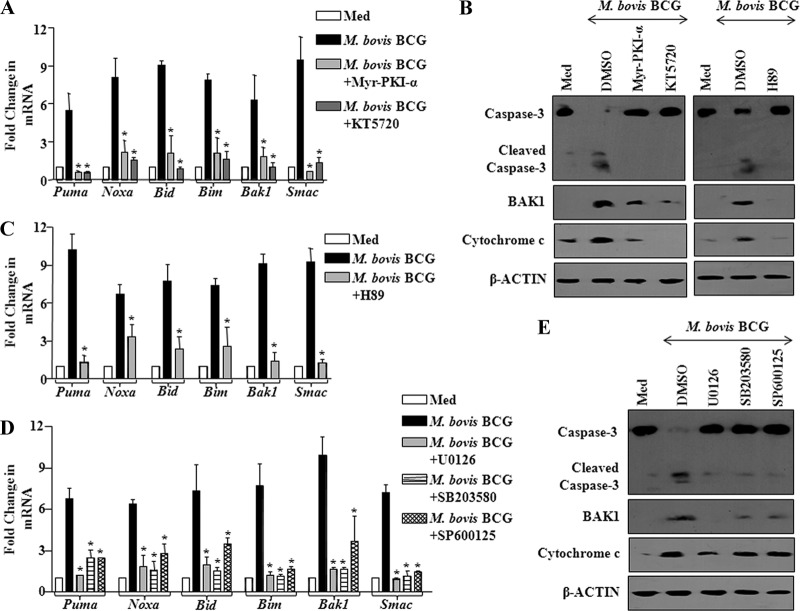
cAMP-driven PKA signaling is known to exert differential effects in effectuating either cellular survival or apoptosis of cells under various environmental cues. In this complex scenario, reports have indicated that PKA signaling could drive apoptosis of lymphoid cells that involve mitochondrion-mediated cascades, thus suggesting the interplay of multitudes of genes with a network of cellular pathways (42, 70). In this regard, M. bovis BCG-driven expression of apoptotic genes Puma, Noxa, Bid, Bim, Bak1, and Smac in macrophages could be effectively blocked by pharmacological inhibition of PKA (Myr-PKI-α or KT5720) (Fig. 10A). Accordingly, M. bovis BCG-triggered activation of apoptotic effectors, active caspase-3, BAK1 expression, or cytosolic translocation of cytochrome c could be abolished by inhibition of PKA or MSK1 (Fig. 10B). As described earlier, elevated levels of PKA signaling could differentially control the downstream effector kinases, including MAPKs, thus tightly regulating the epigenetic or posttranslational modification of histones or CREB (see Fig. S4A, B, C, D, and F in the supplemental material). In this regard, pharmacological inhibition of MSK1 (H89), ERK1/2 (U0126), p38 (SB203580), or JNK1/2 (SP600125) markedly reduced M. bovis BCG's ability to trigger apoptotic effectors PUMA, NOXA, BID, BIM, BAK1, and SMAC (Fig. 10C and D). Consequently, pharmacological inhibitor intervention identified involvement of MAPKs during M. bovis BCG-triggered activation of caspase-3, BAK1 expression, or cytosolic translocation of cytochrome c (Fig. 10E).
Fig 10.
M. bovis BCG-triggered miR-155 employs PKA signaling to modulate macrophage apoptosis. (A) Macrophages were treated with PKA specific inhibitors, Myr-PKI-α and KT5720, prior to M. bovis BCG stimulation. Changes in the proapoptotic gene expression profile were monitored by real-time quantitative RT-PCR analysis (data are means ± SE, n = 3). (B) Abrogation of PKA or MSK1 activity by pretreatment of macrophages with Myr-PKI-α or KT5720 or H89 prior to M. bovis BCG infection led to inhibition of caspase-3 cleavage and BAK1 and cytochrome c expression. DMSO served as vehicle control. Western blots are representative of 3 experiments. (C) Abrogation of MSK1 kinase activity by H89 inhibits M. bovis BCG-triggered expression of proapoptotic genes. Data represent means ± SE of 3 independent experiments. (D) Pretreatment of macrophages with a set of MAPK specific inhibitors prior to M. bovis BCG stimulation suppressed M. bovis BCG-triggered expression of proapoptotic genes in macrophages. Data are means ± SE of 3 separate experiments. (E) MAPKs play a crucial role in M. bovis BCG-induced apoptosis as analyzed by treating macrophages with pharmacological inhibitors of MAPKs and assaying cleavage of caspase-3 and protein expression levels of BAK1 and cytochrome c. Western blots are representative of 3 experiments. Med, medium; *, P < 0.05 versus M. bovis BCG stimulation.
DISCUSSION
In the present study, we demonstrate that M. bovis BCG infection-triggered expression of miR-155 in macrophages required the involvement of TLR2-mediated signaling events. The induced expression of miR-155 involved extensive signaling cohorts and cross talk among PI3K, PKCδ, and MAPK cascades. Our data support the evidence that miR-155 modulates PKA signaling, as PKI-α, a negative regulator of PKA signaling, is a direct target of posttranscriptional repression by miR-155. Prominently, miR-155-driven PKA signaling resulted in activation of apoptotic effectors, thus providing a molecular basis for M. bovis BCG's ability to induce apoptosis of macrophages.
The variable efficacy of the currently utilized vaccine M. bovis BCG has been attributed to various parameters associated with host-M. bovis BCG interactions. As mentioned, one such critical component is suggested to involve M. bovis BCG's ability to induce apoptosis of infected macrophages (57). Overexpression of miR-155 in dendritic cells increases apoptosis, and dendritic cells from miR-155 knockout mice were less apoptotic than those from WT mice (39, 65). M. bovis BCG-triggered caspase-dependent apoptosis during infection leads to the release of its antigens in the form of apoptotic blebs, which effectively culminates in activation of T cells by DC-mediated cross-priming (1, 46). Thus, apoptosis and cross-priming act as rate-limiting steps in the initiation of robust cellular immunity. In contrast, mycobacterium-mediated apoptosis of infected macrophages eventually leads to dissemination of the pathogen, tipping the balance of host immunity in favor of mycobacteria. These observations eloquently suggest a critical role for macrophage apoptosis in deciding the fate of mycobacterial infection.
In spite of noteworthy advancements, extensive understanding of the molecular mechanisms that govern M. bovis BCG-mediated apoptosis of host cells remains a challenge. Importantly, the contributory role, if any, of miRNA-mediated posttranscriptional repression of the cellular genes involved in host cells' apoptotic machinery is still unclear. For example, miR-155 is known to target suppressor of cytokine signaling 1 (SOCS1) in Treg cells, consequently regulating overall T-cell homeostasis. SOCS1 is implicated for its vital role in tumor necrosis factor alpha (TNF-α)- or reactive oxygen species (ROS)-triggered apoptosis in various cell types (25, 50). Further, evidence from studies on breast cancer cells implies that the single nucleotide mutation in the miR-155 binding site of the SOCS1 3′UTR resulted in significant amelioration of miR-155-mediated regulation of breast tumor cells (28).
Notably, in order to establish correlations with clinical manifestations during tuberculosis infection in vivo, we were able to detect significant expression of miR-155 levels in PBMCs of pulmonary tuberculosis patients. Accordingly, in vivo challenge with M. bovis BCG resulted in robust expression of miR-155 in peritoneal macrophages, spleen, or lymph nodes of infected mice. The in vivo augmented expression of miR-155 clearly signifies its role in influencing the pathophysiological attributes of tuberculosis. As shown, an elaborate signaling cross talk among the members of PI3K, PKCδ, and MAPK pathways is involved during infection-triggered expression of miR-155. It is well known that TLR2 receptor engagement by M. bovis BCG triggers an array of cellular reprogramming genes, which culminates in the apoptosis of infected macrophages (38). In this regard, perturbations leading to loss-of-function or gain-of-function experiments implied a critical role for miR-155 during M. bovis BCG-triggered apoptosis of macrophages by regulating apoptotic effectors PUMA, NOXA, BID, BIM, BAK1, and SMAC. Further, we show that miR-155 contributes to M. bovis BCG's ability to trigger apoptosis by directly targeting PKI-α, a negative regulator of PKA signaling in macrophages. This results in enhanced activation of PKA signaling, which acts as a prominent driving force in apoptosis of macrophages. Importantly, miR-155 effectively contributed to loss of viability of macrophages with a concomitant increase in M. bovis BCG CFU. Apoptosis of macrophages by enforced expression of miR-155 could be further potentiated by infection with M. bovis BCG. We also show that the induction of miR-155 results in epigenetic or posttranslational modification of histones or CREB, thus contributing to cellular reprogramming of various immune genes (Fig. 11).
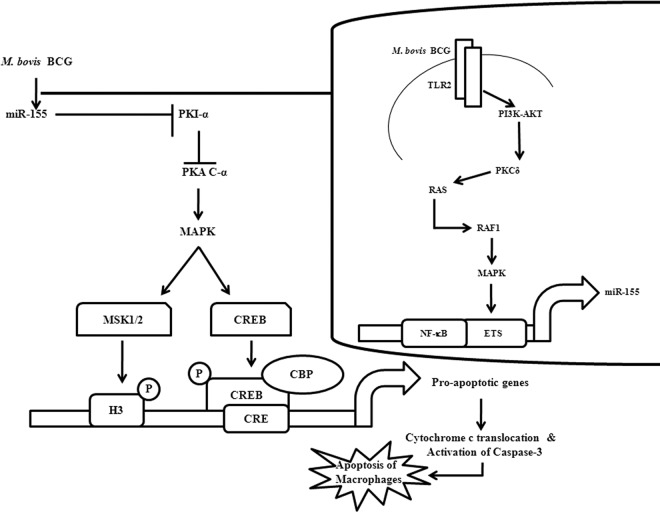
Fig 11.
Model depicting miR-155-mediated modulation of macrophage apoptosis. M. bovis BCG induces the upregulation of miR-155 by triggering the TLR2/PI3K pathway. PKCδ and RAS-RAF1-MAPKs play vital roles in M. bovis BCG-induced miR-155 expression through transcriptional factors NF-κB and c-ETS. Induced miR-155 targets PKI-α, a negative regulator of PKA signaling, resulting in the robust activation of the PKA signaling pathway. Overall, miR-155 induces the upregulation of proapoptotic genes by modulating PKA signaling, resulting in macrophage apoptosis.
Overall, our study demonstrates a new programming framework that orchestrates cellular signaling networks during M. bovis BCG-driven miR-155 expression, which acts as a crucial regulator of cell fate decisions of infected macrophages. These findings implicate a novel role for miR-155 in programming gene expressions during immune responses that regulate the pathogenesis of tuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Central Animal facility, IISc, for providing mice for experimentation. We thank Omana Joy, Kavya Ananthaswamy, and Puja Pai of the DBT-FACS facility and Samrajyam Nara of the MCB IISc confocal facility for their generous help and support. We acknowledge Kushagra Bansal, Sahana Holla, and Shambhuprasad Kotresh Togarsimalemath for critical comments and help. We sincerely thank Erik K. Flemington, Tulane University, New Orleans, LA, for miR-155 promoter luciferase plasmid, BICp-pGL3, BICp-pGL3-ΔNF-κB, BICp-pGL3-Δc-ETS, BICp-pGL3-ΔAP1, pPRIME-CMV-GFP, pPRIME-CMV-GFP-miR-155, and pPRIME-CMV-GFP-miR-146a constructs; and Thomas F. Meyer, Department of Molecular Biology, Max Planck Institute for Infection Biology, Berlin, for the kind gift of pRL-SV40 and PKI-α 3′UTR WT plasmids. We thank Elena Vigorito, Babraham Institute, Cambridge, United Kingdom, and Iain McInnes, University of Glasgow, Scotland, United Kingdom. The TLR2 DN cDNA construct was gifted by Douglas Golenbock, University of Massachusetts Medical School, Worcester, MA. We acknowledge Apurva Sarin, National Centre for Biological Sciences, Bangalore, India, and Kumaravel Somasundaram, Annapoorni Rangarajan, and Sathees Raghavan of Indian Institute of Science, Bangalore, India, for help during the study.
This study was supported by funds from the Department of Biotechnology (DBT), Department of Science and Technology (DST), Council for Scientific and Industrial Research (CSIR), and Indian Council of Medical Research (ICMR), Government of India.
Infrastructure support was received from ICMR (Center for Advanced Study in Molecular Medicine), DST (FIST), and UGC (special assistance) (K.N.B.).
A fellowship from CSIR (to D.S.G.) is acknowledged.
Footnotes
Published ahead of print 2 April 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albert ML, Sauter B, Bhardwaj N. 1998. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature 392:86–89 [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- 3.Aumo L, Rusten M, Mellgren G, Bakke M, Lewis AE. 2010. Functional roles of protein kinase A (PKA) and exchange protein directly activated by 3′,5′-cyclic adenosine 5′-monophosphate (cAMP) 2 (EPAC2) in cAMP-mediated actions in adrenocortical cells. Endocrinology 151:2151–2161 [DOI] [PubMed] [Google Scholar]
- 4.Banchereau J, Steinman RM. 1998. Dendritic cells and the control of immunity. Nature 392:245–252 [DOI] [PubMed] [Google Scholar]
- 5.Bansal K, et al. 2009. PIM2 induced COX-2 and MMP-9 expression in macrophages requires PI3K and Notch1 signaling. PLoS One 4:e4911 doi:10.1371/journal.pone.0004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bansal K, Narayana Y, Balaji KN. 2009. Inhibition of TNF-alpha-induced cyclooxygenase-2 expression by Mycobacterium bovis BCG in human alveolar epithelial A549 cells. Scand. J. Immunol. 69:11–19 [DOI] [PubMed] [Google Scholar]
- 7.Bansal K, Narayana Y, Patil SA, Balaji KN. 2009. M. bovis BCG induced expression of COX-2 involves nitric oxide-dependent and -independent signaling pathways. J. Leukoc. Biol. 85:804–816 [DOI] [PubMed] [Google Scholar]
- 8.Bansal K, et al. 2010. Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-kappaB signaling and drives Th2 immune responses. J. Biol. Chem. 285:36511–36522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bansal K, Trinath J, Chakravortty D, Patil SA, Balaji KN. 2011. Pathogen-specific TLR2 activation programs macrophages to induce Wnt-{beta}-Catenin signaling. J. Biol. Chem. 286:37032–37044 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 11.Be NA, et al. 2008. Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J. Infect. Dis. 198:1520–1528 [DOI] [PubMed] [Google Scholar]
- 12.Cimmino A, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 102:13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalton GD, Dewey WL. 2006. Protein kinase inhibitor peptide (PKI): a family of endogenous neuropeptides that modulate neuronal cAMP-dependent protein kinase function. Neuropeptides 40:23–34 [DOI] [PubMed] [Google Scholar]
- 14.de Lecea L, et al. 1998. Endogenous protein kinase A inhibitor (PKIalpha) modulates synaptic activity. J. Neurosci. Res. 53:269–278 [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Soto A, Puig-Kroger A, Vega MA, Corbi AL. 2005. PU.1 regulates the tissue-specific expression of dendritic cell-specific intercellular adhesion molecule (ICAM)-3-grabbing nonintegrin. J. Biol. Chem. 280:33123–33131 [DOI] [PubMed] [Google Scholar]
- 16.Donnem T, et al. 2011. Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J. Transl. Med. 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. 2009. miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta 1792:497–505 [DOI] [PubMed] [Google Scholar]
- 18.Farh KK, et al. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310:1817–1821 [DOI] [PubMed] [Google Scholar]
- 19.Fassi Fehri L, et al. 2010. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One 5:e9500 doi:10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazi F, et al. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123:819–831 [DOI] [PubMed] [Google Scholar]
- 21.Felekkis K, Touvana E, Stefanou C, Deltas C. 2010. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 14:236–240 [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel LB, et al. 2011. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 30:4628–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangolli EA, et al. 2000. Deficient gene expression in protein kinase inhibitor alpha null mutant mice. Mol. Cell. Biol. 20:3442–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghorpade DS, Kaveri SV, Bayry J, Balaji KN. 2011. Cooperative regulation of NOTCH1 protein-phosphatidylinositol 3-kinase (PI3K) signaling by NOD1, NOD2, and TLR2 receptors renders enhanced refractoriness to transforming growth factor-beta (TGF-beta)- or cytotoxic T-lymphocyte antigen 4 (CTLA-4)-mediated impairment of human dendritic cell maturation. J. Biol. Chem. 286:31347–31360 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.He Y, Zhang W, Zhang R, Zhang H, Min W. 2006. SOCS1 inhibits tumor necrosis factor-induced activation of ASK1-JNK inflammatory signaling by mediating ASK1 degradation. J. Biol. Chem. 281:5559–5566 [DOI] [PubMed] [Google Scholar]
- 26.Hwang HW, Mendell JT. 2006. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94:776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jegga AG, Schneider L, Ouyang X, Zhang J. 2011. Systems biology of the autophagy-lysosomal pathway. Autophagy 7:477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang S, et al. 2010. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70:3119–3127 [DOI] [PubMed] [Google Scholar]
- 29.Jo EK, Yang CS, Choi CH, Harding CV. 2007. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell Microbiol. 9:1087–1098 [DOI] [PubMed] [Google Scholar]
- 30.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 31.Jovanovic M, Hengartner MO. 2006. miRNAs and apoptosis: RNAs to die for. Oncogene 25:6176–6187 [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann SH. 2005. Robert Koch, the Nobel Prize, and the ongoing threat of tuberculosis. N. Engl. J. Med. 353:2423–2426 [DOI] [PubMed] [Google Scholar]
- 33.Keane J, Remold HG, Kornfeld H. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016–2020 [DOI] [PubMed] [Google Scholar]
- 34.Kumar P, Walsh DA. 2002. A dual-specificity isoform of the protein kinase inhibitor PKI produced by alternate gene splicing. Biochem. J. 362:533–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y, et al. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leng RX, Pan HF, Qin WZ, Chen GM, Ye DQ. 2011. Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev. 22:141–147 [DOI] [PubMed] [Google Scholar]
- 37.Li S, et al. 2004. Translation initiation factor 4E blocks endoplasmic reticulum-mediated apoptosis. J. Biol. Chem. 279:21312–21317 [DOI] [PubMed] [Google Scholar]
- 38.Lopez M, et al. 2003. The 19-kDa Mycobacterium tuberculosis protein induces macrophage apoptosis through Toll-like receptor-2. J. Immunol. 170:2409–2416 [DOI] [PubMed] [Google Scholar]
- 39.Lu C, et al. 2011. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 117:4293–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu LF, et al. 2009. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30:80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto S. 2011. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 21:116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muhl H, et al. 1996. Apoptosis is triggered by the cyclic AMP signalling pathway in renal mesangial cells. FEBS Lett. 382:271–275 [DOI] [PubMed] [Google Scholar]
- 43.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. 2011. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187:2213–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayana Y, Balaji KN. 2008. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J. Biol. Chem. 283:12501–12511 [DOI] [PubMed] [Google Scholar]
- 45.Narayana Y, et al. 2009. SOCS3 expression induced by PIM2 requires PKC and PI3K signaling. Mol. Immunol. 46:2947–2954 [DOI] [PubMed] [Google Scholar]
- 46.Nasser Eddine A, Kaufmann SH. 2005. Improved protection by recombinant BCG. Microbes Infect. 7:939–946 [DOI] [PubMed] [Google Scholar]
- 47.Nigou J, et al. 2002. Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes Infect. 4:945–953 [DOI] [PubMed] [Google Scholar]
- 48.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843 [DOI] [PubMed] [Google Scholar]
- 49.Oertli M, et al. 2011. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic gastritis and colitis. J. Immunol. 187:3578–3586 [DOI] [PubMed] [Google Scholar]
- 50.Oh J, Hur MW, Lee CE. 2009. SOCS1 protects protein tyrosine phosphatases by thioredoxin upregulation and attenuates Jaks to suppress ROS-mediated apoptosis. Oncogene 28:3145–3156 [DOI] [PubMed] [Google Scholar]
- 51.Ordway DJ, Orme IM. 2011. Animal models of mycobacteria infection. Curr. Protoc. Immunol. Chapter 19:Unit19.5 [DOI] [PubMed] [Google Scholar]
- 52.Pathak SK, et al. 2007. Direct extracellular interaction between the early secreted antigen ESAT-6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat. Immunol. 8:610–618 [DOI] [PubMed] [Google Scholar]
- 53.Pidoux G, Tasken K. 2010. Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J. Mol. Endocrinol. 44:271–284 [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez A, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schorey JS, Cooper AM. 2003. Macrophage signalling upon mycobacterial infection: the MAP kinases lead the way. Cell. Microbiol. 5:133–142 [DOI] [PubMed] [Google Scholar]
- 56.Shivdasani RA. 2006. MicroRNAs: regulators of gene expression and cell differentiation. Blood 108:3646–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skeiky YA, Sadoff JC. 2006. Advances in tuberculosis vaccine strategies. Nat. Rev. Microbiol. 4:469–476 [DOI] [PubMed] [Google Scholar]
- 58.Song G, Zhang Y, Wang L. 2009. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J. Biol. Chem. 284:31921–31927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song L, Tuan RS. 2006. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res. C Embryo Today 78:140–149 [DOI] [PubMed] [Google Scholar]
- 60.Sonkoly E, Stahle M, Pivarcsi A. 2008. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 18:131–140 [DOI] [PubMed] [Google Scholar]
- 61.Sonkoly E, et al. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2:e610 doi:10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taganov KD, Boldin MP, Baltimore D. 2007. MicroRNAs and immunity: tiny players in a big field. Immunity 26:133–137 [DOI] [PubMed] [Google Scholar]
- 63.Taylor SS, et al. 2004. PKA: a portrait of protein kinase dynamics. Biochim. Biophys. Acta 1697:259–269 [DOI] [PubMed] [Google Scholar]
- 64.Thai TH, et al. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608 [DOI] [PubMed] [Google Scholar]
- 65.Turner ML, Schnorfeil FM, Brocker T. 2011. MicroRNAs regulate dendritic cell differentiation and function. J. Immunol. 187:3911–3917 [DOI] [PubMed] [Google Scholar]
- 66.Vigorito E, et al. 2007. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallet MA, Sen P, Tisch R. 2005. Immunoregulation of dendritic cells. Clin. Med. Res. 3:166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao B, et al. 2009. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 200:916–925 [DOI] [PubMed] [Google Scholar]
- 69.Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. 2009. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem. Biophys. Res. Commun. 388:539–542 [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, et al. 2008. Gene expression signatures of cAMP/protein kinase A (PKA)-promoted, mitochondrial-dependent apoptosis. Comparative analysis of wild-type and cAMP-deathless S49 lymphoma cells.. J. Biol. Chem. 283:4304–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao C, et al. 2010. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U. S. A. 107:1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.