Abstract
Background
Helicobacter pylori HP0958 protein (FlgZ) prevents the rapid turnover of RpoN (σ54), a transcription factor required for expression of several flagellar genes in H. pylori. FlgZ possesses a zinc-ribbon domain (DUF164) that contains two conserved CXXC motifs which coordinate a zinc ion and is thought to interact with nucleic acids or proteins. Two conserved cysteine residues in FlgZ (Cys-202 and Cys-223) were replaced with serine to assess their significance in FlgZ function. After confirming the importance of the CXXC motifs in the DUF164 domain of FlgZ, the distribution of DUF164 proteins and RpoN homologs in other bacteria was examined to determine if a correlation existed for the concurrence of the two proteins.
Results
Levels of RpoN were greatly reduced in H. pylori strains that expressed the FlgZC202S or FlgZC223S variants. The FlgZC202S variant, but not the FlgZC223S variant, accumulated at levels similar to the wild-type protein. DUF164 proteins are not universally distributed and appear to be absent in several major bacterial taxa, including Cyanobacteria as well as Alpha-, Beta- and Gammaproteobacteria. With the exception of the Actinobacteria, members of which generally lack RpoN, genes encoding DUF164 proteins and RpoN are frequently found in the same genome. Interestingly, many of the DUF164 proteins in Actinobacteria and Bacteroidetes lack most or even all of the conserved cysteine residues.
Conclusions
These findings suggest the importance of the zinc-ribbon domain of FlgZ in protecting RpoN from turnover. Since many bacteria that possess a DUF164 protein also contain RpoN, DUF164 proteins may have roles in RpoN protection or function in other bacteria.
Keywords: Helicobacter pylori, sigma54, RpoN, HP0958, FlgZ
Background
Helicobacter pylori is a member of the Epsilonproteobacteria that colonizes the human gastric mucosa where it causes a variety of gastrointestinal diseases, including acute gastritis, peptic and duodenal ulcers, B cell MALT lymphoma, and gastric adenocarcinoma [1-3]. Colonization of the gastric mucosa by H. pylori requires the bacterium to be motile [4,5], which is achieved through a cluster of polar sheathed flagella.
The bacterial flagellum is a complex structure consisting of three basic substructures - the basal body, hook and filament. Dozens of genes are required for flagellar biogenesis and the expression of these genes is regulated by a transcriptional hierarchy in which genes are expressed as their products are needed for assembly [6,7]. All three of the RNA polymerase sigma (σ) factors in H. pylori are involved in flagellar biogenesis. In general, genes needed early in flagellar assembly are under control of the primary σ factor RpoD (σ80), while RpoN (σ54) is responsible for transcription of genes needed midway through flagellar assembly and transcription of late flagellar genes is dependent on FliA (σ28) [8-13].
Transcriptional activation of the RpoN-dependent flagellar genes in H. pylori requires the response regulator FlgR and its cognate histidine kinase FlgS [12,14]. Although the signaling pathway that regulates expression of the RpoN flagellar regulon is poorly understood, transcriptional control of the regulon has been shown to be intimately associated with the flagellar protein export apparatus in H. pylori and its close relative Campylobacter jejuni [10,15-18]. Transcription of the H. pylori RpoN regulon also requires the putative RpoN chaperone and zinc-ribbon domain protein HP0958 [19,20], which we hereafter refer to as FlgZ (flagellar-associated zinc-ribbon domain protein).
FlgZ was originally identified as interacting with RpoN in a high-throughput screen of a yeast two-hybrid system [21]. Subsequently, FlgZ was shown to be required for motility in H. pylori [19,20] and to prevent the rapid turnover of RpoN - the half-life of RpoN is > 4 hours in wild type compared to ~30 minutes in the flgZ mutant [19]. FlgZ may have additional roles in H. pylori motility as it also interacts with the flagellar protein export apparatus protein FliH in the yeast two-hybrid system [21]. In addition, Douillard and co-workers showed that FlgZ binds flaA transcript (flaA encodes the major flagellin of H. pylori) and is needed for optimal production of FlaA [22]. These researchers proposed that FlgZ may work together with FliH to direct flaA transcripts to the basal body of the nascent flagellum to couple the translation and secretion of FlaA [22]. Although FlgZ may play a direct role in the expression and export of FlaA, this activity is not essential for motility since overproduction of RpoN suppresses the motility defect of the flgZ mutation [19].
FlgZ is a 254 amino acid polypeptide and a recently solved crystal structure of the protein revealed that it consists of a highly elongated, kinked coiled-coil hairpin domain (resides 1-170) followed by a zinc-ribbon domain (residues 174-238) [23]. FlgZ is a member of a family of proteins which contain a predicted zinc-ribbon domain of unknown function (PF02591; COG1579). Most members of this protein family, including FlgZ, contain the motif Y/F_X18-23_ CXXC_X18-26_CXXC, in which the conserved cysteine residues coordinate a zinc ion [23]. Zinc fingers function as interaction modules that bind DNA, RNA, proteins or small molecules [24-28]. Given the potential for FlgZ to act as both an RpoN chaperone and a RNA chaperone in a coupled translation-protein secretion process [19,22], it is unclear which of these functions is associated with the FlgZ zinc-ribbon domain.
To examine the issue of whether the zinc-ribbon domain of FlgZ is needed to protect RpoN from turnover in H. pylori, we changed two of the conserved cysteine residues in FlgZ (Cys-202 and Cys-223) to serine residues and examined the phenotypes of H. pylori strains expressing the FlgZ variants. Strains expressing either the FlgZC202S or FlgZC223S variant were non-motile and had reduced levels of RpoN, indicating the importance of these conserved cysteine residues in the function or stability of FlgZ. A search of bacterial genome sequences revealed that homologs of FlgZ (characterized by the presence of the DUF164) are widespread among bacterial species, but appear to be absent in several major taxonomic groups. In many taxonomic groups there is a statistically significant correlation between the presence of FlgZ homologs and RpoN, suggesting a role for FlgZ homologs in RpoN function in a diverse set of bacteria.
Results and discussion
Conserved cysteine residues in the zinc-ribbon domain of FlgZ are important for function and stability
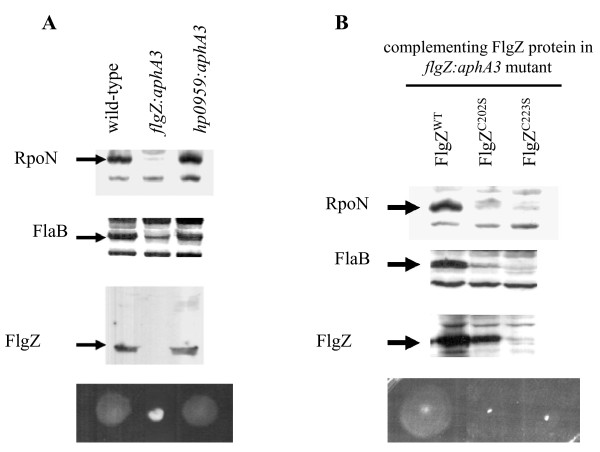
To determine if the conserved cysteine residues in the zinc-ribbon domain of FlgZ are critical for the function of the protein, mutations were introduced in each of the conserved CXXC motifs and the phenotypes of strains expressing the resulting FlgZ variants were examined. The conserved cysteine residues in H. pylori 26695 FlgZ are Cys-199, Cys-202, Cys-223 and Cys-226. Cys-202 and Cys-223 were changed to serine residues and the resulting FlgZ variants were expressed in a mutant strain of H. pylori 43504 in which flgZ (hp0958) was disrupted with a kanamycin-resistance cassette. As shown in Figure 1A and reported previously, disruption of flgZ in H. pylori resulted in a loss of motility and reduced RpoN levels [19]. Wild-type FlgZ expressed from the hp0405 locus in the flgZ mutant restored motility and supported wild-type levels of RpoN and FlaB (Figure 1B). FlaB is a minor flagellin whose expression is dependent on RpoN [10,12,29], and levels of this flagellin were reduced in the flgZ mutant. In contrast to wild-type FlgZ, the FlgZC202S and FlgZC223S variants failed to restore motility or support wild-type levels of RpoN and FlaB. Western blot analysis showed that the FlgZC202S variant was expressed at near wild-type levels in H. pylori. Taken together, these data suggest that the zinc-ribbon domain of FlgZ has a role in protecting RpoN from turnover. It is unclear if the FlgZC202S variant is defective in binding RpoN or in a subsequent step needed to protect RpoN from turnover.
Figure 1.
Effects of FlgZ variants on motility and RpoN stability. (A) Wild-type H. pylori strain ATCC 43504, flgZ:aphA3 and hp0959:aphA3 mutants were analyzed by western blotting for the proteins indicated in the top three panels. Construction of the mutant strains was described previously [19]. Approximately 1 × 108 cells were lysed and loaded in each lane. Following protein transfer, nitrocellulose membranes were probed with antiserum directed against RpoN, FlaB or FlgZ proteins that were fused to the maltose-binding protein (MBP) as described [19]. Results of motility assays are shown in the bottom panel. Each strain was inoculated into semisolid motility agar plates using a sterile toothpick and incubated 5 days at 37®C under microaerophilic conditions as described previously [19]. (B) Wild-type or mutant alleles of flgZ were introduced into the hp0405 locus of the H. pylori flgZ:aphA3 mutant as described previously [19]. The resulting strains were analyzed by western blotting for RpoN, FlaB and FlgZ (top three panels) and for motility (bottom panel) as described for panel A.
In contrast to the FlgZC202S variant, the FlgZC223S variant was not expressed stably in H. pylori. DNA sequencing of the flgZ allele encoding the FlgZC223S variant failed to reveal any additional mutations, suggesting that the serine substitution at Cys-223 decreases the stability of the protein. Caly and co-workers [23] reported that changing Cys-199 of FlgZ to alanine did not significantly alter the structure of the protein as assessed by circular dichroism. Taken together, these observations suggest substitutions in the second CXXC motif have a more profound effect on the structure of FlgZ than those in the first CXXC motif, while substitutions in either of the two CXXC motifs result in reduced RpoN levels and loss of motility.
Taxonomic distribution of proteins containing the DUF164
Having demonstrated the importance of at least one of the conserved cysteine residues within the DUF164 of FlgZ in protecting RpoN from turnover in H. pylori, we wished to determine if DUF164 proteins in other bacteria could have a role in RpoN function similar to that of FlgZ. To address this issue we investigated if the presence of a FlgZ homolog in a genome correlated with that of RpoN. For this purpose, we defined FlgZ homologs as proteins containing the DUF164 domain (PF02519), whereas all proteins belonging to the COG1508 according to the IMG annotations were considered RpoN homologs. We performed our analysis with a dataset restricted to finished genome sequences as well as the complete collection of both finished and draft sequences with the caveat that the apparent absence of either gene in a draft genome could be due to gaps in sequence coverage. The results of the analysis, which are summarized in Table 1 revealed several intriguing trends.
Table 1.
Taxonomic distribution of DUF164 proteins and RpoN in the domain Bacteria
| Taxonomic group | aNumber of genomes with both DUF164 protein and RpoN | aNumber of genomes with DUF164 protein only | aNumber of genomes with RpoN only | aNumber of genomes with neither DUF164 protein nor RpoN |
|---|---|---|---|---|
| Acidobacteria | 2 (2) | 0 (0) | 1 (1) | 0 (0) |
| Actinobacteria | 1 (2) | 73 (143) | 0 (0) | 16 (37) |
| Aquificae | 6 (8) | 0 (0) | 0 (0) | 0 (0) |
| Bacteroidetes | 24 (80) | 0 (2) | 0 (0) | 2 (4) |
| Chlamydiae | 15 (22) | 0 (0) | 0 (0) | 0 (0) |
| Chlorobi | 11 (12) | 0 (0) | 0 (0) | 0 (0) |
| Chloroflexi | 1 (2) | 3 (3) | 3 (5) | 6 (6) |
| Cyanobacteria | 0 (0) | 0 (0) | 0 (0) | 38 (51) |
| Deferribacteres | 0 (1) | 0 (0) | 0 (0) | 0 (0) |
| Dictyoglomi | 0 (0) | 2 (2) | 0 (0) | 0 (0) |
| Elusimicrobia | 0 (0) | 1 (1) | 0 (0) | 1 (1) |
| Fibrobacteres | 0 (0) | 0 (0) | 1 (2) | 0 (0) |
| Firmicutes | 17 (23) | 0 (1) | 76 (240) | 100 (243) |
| Clostridia | 17 (23) | 0 (1) | 25 (71) | 11 (55) |
| Clostridium | 12 (16) | 0 (1) | 10 (39) | 3 (12) |
| Fusobacteria | 0 (0) | 0 (0) | 0 (12) | 4 (8) |
| Gemmatimonadetes | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Lentisphaerae | 2 (2) | 0 (0) | 0 (0) | 0 (0) |
| Nitrospirae | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Planctomycetes | 3 (7) | 0 (0) | 0 (0) | 0 (0) |
| Alphaproteobacteria | 0 (0) | 0 (0) | 79 (122) | 42 (75) |
| Betaproteobacteria | 0 (0) | 0 (0) | 69 (141) | 3 (4) |
| Deltaproteobacteria | 32 (41) | 0 (1) | 1 (1) | 0 (0) |
| Epsilonproteobacteria | 22 (43) | 4 (5) | 0 (2) | 0 (1) |
| Gammaproteobacteria | 0 (0) | 0 (0) | 199 (368) | 44 (71) |
| Magnetococci | 0 (0) | 0 (0) | 1 (1) | 0 (0) |
| Zetaproteobacteria | 0 (0) | 0 (0) | 0 (1) | 0 (0) |
| Spirochaetes | 10 (27) | 2 (3) | 6 (6) | 0 (0) |
| Synergistetes | 0 (0) | 0 (0) | 1 (3) | 0 (1) |
| Tenericutes | 0 (0) | 0 (0) | 0 (0) | 25 (40) |
| Thermi | 0 (0) | 5 (8) | 0 (0) | 0 (0) |
| Thermotogae | 0 (0) | 0 (0) | 3 (3) | 8 (9) |
| TM7 | 0 (0) | 0 (0) | 0 (0) | 0 (1) |
| Verrucomicrobia | 3 (6) | 0 (0) | 0 (1) | 0 (0) |
| WWE1 | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Total | 150 (281) | 90 (169) | 440 (909) | 289 (552) |
aNumbers outside parentheses indicate results from analysis of finished genome sequence, while numbers within parentheses indicate results for both finished and unfinished genome sequences.
First, proteins characterized by the presence of the DUF164 are essentially restricted to the domain Bacteria as there are no representatives in the domain Archaea and only one in the domain Eukaryota. The sole eukaryotic protein is present in the sac fungus Phaeosphaeria nodorum SN15, and it differs substantially from FlgZ in both its size (571 amino acids in length versus 254 amino acids) and the presence of a SET domain (PF00856). Second, several bacterial taxonomic groups that are well represented in the JGI IMG database, such as Cyanobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Tenericutes, lack proteins that contain DUF164 (Table 1). Third, with only one exception, bacteria do not possess more than one DUF164 protein. The only exception we found in the database was Opitutus terrae PB90-1 (phylum Verrucomicrobia) which has two DUF164 proteins.
As shown in Table 1 DUF164 proteins are found in a broad range of taxonomic groups. While members of many of these groups, including Bacteroidetes, Chlamydiae, Chlorobi, Planctomycetes, Deltaproteobacteria, Epsilonproteobacteria, Spirochaetes, and Verrucomicrobia, often possess RpoN, members of Actinobacteria and Thermi typically lack RpoN. Indeed, homologs of rpoN are present in only two of 182 Actinobacteria genomes and none of the eight Thermi genomes in the JGI IMG database, indicating that the DUF164 proteins in these bacteria are obviously not involved in RpoN function.
We used the Fisher exact test to determine if there was a statistically significant association between DUF164 proteins and RpoN. The data from Table 1 were combined in a single 2-by-2 contingency table where the lines and columns contained the counts of genomes that possess and lack DUF164 proteins and RpoN, respectively. The online probability calculator from Vassar College (available at http://faculty.vassar.edu/lowry/VassarStats.html) was used to calculate the probabilities. One-tail probabilities obtained from the Fisher exact test close to zero (p < 0.05) indicate significant tendency of DUF164 and RpoN proteins to occur in the same genomes whereas probabilities close to 1 indicate a significant mutual avoidance of the two proteins. Analysis of the total finished genome sequences failed to reveal a significant tendency of DUF164 and RpoN to occur in the same genomes (p = 0.30). Analysis of a data set that included all of the finished genomes except the Actinobacteria genomes, however, indicated a significant tendency of DUF164 and RpoN proteins to occur in the same genomes (p < 10-13). A statistically significant association between DUF164 and RpoN proteins was also noted within the phylum Firmicutes (p < 10-5), where DUF164 proteins appear to be restricted to members of the class Clostridia (Table 1). The strong correlation between the presence of RpoN and DUF164 proteins in many bacterial genomes raises the possibility that, as with FlgZ, DUF164 proteins in these bacteria have a role in RpoN function.
Bacteria in taxonomic groups whose members generally possess both DUF164 and RpoN proteins often employ RpoN in unconventional ways. For example, H. pylori FlgR differs from most activators of σ54-RNA polymerase holoenzyme (σ54-holoenzyme) in that it lacks an enhancer-binding (i.e., DNA-binding) domain and activates transcription independently of an enhancer [29]. Many members of the Epsilonproteobacteria, Chlamydiae and Bacteroidetes groups similarly possess RpoN-dependent activators that lack enhancer-binding domains and presumably function as enhancer-independent activators [30-32]. Moreover, the RpoN-dependent activators Campylobacter jejuni FlgR (subphylum Epsilonproteobacteria) and Borrelia burgdorferi Rrp2 (phylum Spirochaetes) have been shown to activate transcription in an enhancer-independent manner [31,33]. Based on these comparisons, it is intriguing to speculate that FlgZ homologs may stimulate enhancer-independent transcriptional activation (which is presumably less efficient than enhancer-dependent transcription) in these bacteria. For instance, FlgZ homologs could facilitate interactions between RpoN and core RNA polymerase to increase the concentration of σ54-holoenzyme in the cell, resulting in increased promoter occupancy and allow for greater opportunity for productive interactions between the activator and σ54-holoenzyme.
Along a similar line of reasoning, competition among sigma factors for core RNA polymerase is an important factor in gene regulation [34]. If FlgZ and its homologs function as RpoN chaperones, such activity may allow RpoN to compete effectively with other sigma factors for binding RNA polymerase. This could be important for RpoN function in members of the Deltaproteobacteria which typically have exceptional numbers of RpoN-dependent activators (Myxococcus xanthus has ~50 different activators) and where RpoN is a significant global regulator in these bacteria [35,36].
Architectural features of DUF164 proteins
Divergence of DUF164 in different bacterial lineages was investigated using alignments of DUF164 proteins downloaded from the Sanger Institute website for PF02591 (http://pfam.sanger.ac.uk/family?acc=PF02591). Interestingly, many DUF164 proteins (43 out of 349 in the full dataset) lack one or more of the conserved cysteine residues. Most of these proteins are from bacteria belonging to either the phylum Actinobacteria or Bacteroidetes, as illustrated by Figure 2. In some cases, DUF164 proteins lack all four of the conserved cysteine residues (indicated by four stars in Figure 2). For example, DUF164 proteins from Flavobacteria (phylum Bacteroidetes) generally have the motif SXXS_X20_ DXXS instead of the two CXXC motifs. Given the absence of the CXXC motifs in these proteins, it is unclear if they bind zinc ions or have functions similar to that of FlgZ.
Figure 2.
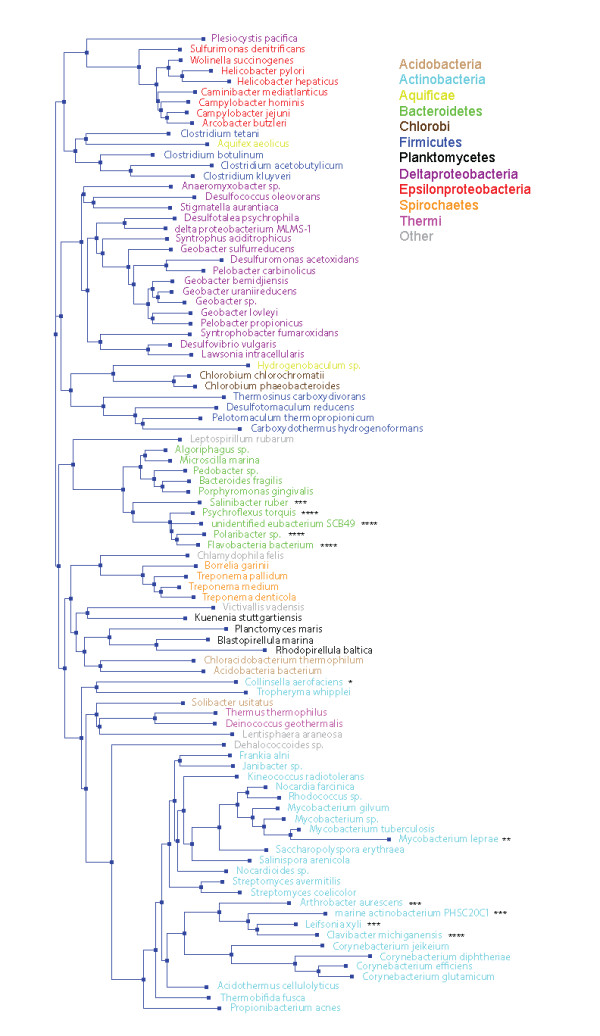
Phylogentic tree for DUF164 domain proteins. The figure presents a modified phylogenetic tree downloaded from Pfam (http://pfam.sanger.ac.uk/family?acc=PF02591). The tree was generated from an alignment of 93 seed DUF164 sequences using the FastTree algorithm [42]. Taxonomic groups for the bacterial species are color-coded. The numbers of stars indicate the number of cysteine residues in the conserved CXXC motifs that are replaced with other amino acid residues.
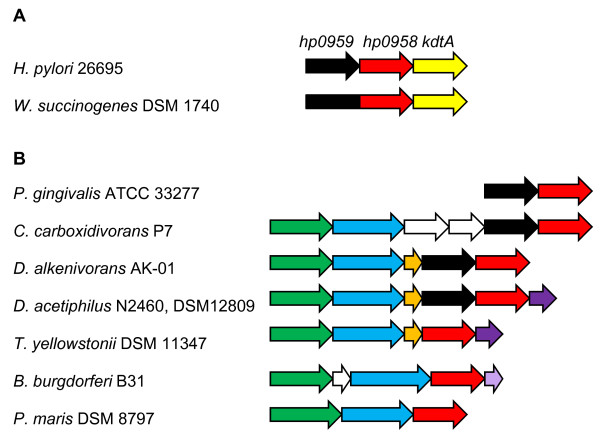
The domain organization for the 349 DUF164 proteins displayed in the Pfam database shows that the DUF164 domain is near the C-terminus in all the proteins in the database. All but three of the DUF164 proteins in the Pfam database are similar to FlgZ in that they possess a predicted coiled-coil domain near the N-terminus and lack other defined protein family domains. A notable exception to this architectural arrangement is the DUF164 protein from the Epsilonproteobacterium Wolinella succinogenes WS2117 which also contains a domain belonging to the NIF3 (NGG1p interacting factor 3) family (PF01784; COG0327). NIF3 interacts with the yeast transcriptional co-activator NGG1p, but the significance of this interaction is not known [37]. A gene encoding a NIF3 domain protein (hp0959) is located immediately upstream of flgZ in H. pylori (Figure 3A), suggesting a gene fusion event occurred in W. succinogenes. Gene fusion events are among the best predictors of protein-protein interactions [38]. HP0959, however, is not required for motility or RpoN stability in H. pylori (Figure 1A; and [19]), and we do not have any independent evidence regarding direct physical interaction between HP0959 and FlgZ. It may be that the DUF164 and NIF3 domains function together in some capacity which is unique to W. succinogenes. W. succinogenes appears to utilize RpoN to control expression of nitrogen-fixation (nif) genes in addition to flagellar genes. W. succinogenes also differs from H. pylori in that it possesses a second rpoN gene which appears to be part of an operon that includes several nif genes. It is possible that the NIF3 and DUF164 domains in W. succinogenes function together to modulate the activity or level of this second RpoN protein.
Figure 3.
Arrangement of genes around flgZ homologs in select, diverse bacteria. (A) The flgZ gene (red) in H. pylori 26695 is flanked by hp0959 (black) and kdtA (yellow). In W. succinogenes DSM 1740 the flgZ and hp0959 homologs (locus number WS2117) are fused. Arrows depicting genes are not drawn to scale. (B) The arrangements of genes surrounding flgZ homologs of bacteria from diverse taxonomic groups are indicated. Bacteria and the phylum or subphylum to which they belong (in parentheses) are: Porphyromonas gingivalis ATCC 33277 (Bacteroidetes); Clostridium carboxidivorans P7 (Firmicutes); Desulfatibacillum alkenivorans AK-01 (Deltaproteobacteria); Denitrovibrio acetiphilus N2460, DSM 12809 (Deferribacteres); Thermodesulfovibrio yellowstonii DSM 11347 (Nitrospirae); Borrelia burgdorferi B31 (Spirochaetes); and Planctomyces maris DSM 8797 (Planctomycetes). Genes whose products share similar functions are color coded as follows: DNA primase (dnaG) (green); σ70 family protein (blue); tRNA-Met (orange); and RNase H (rnhA) (dark purple). Unique genes are indicated by white arrows, except for the gene encoding the catalytic RNA of RNase P (light purple) which is located downstream of the flgZ homolog in B. burgdorferi B31. Locus numbers for the genes (left to right) are as follows: PGN_1989 and PGN_1990 (P. gingivalis); CcarbDRAFT_2245 through CcarbDRAFT_2240 (C. carboxidivorans); Dalk_3077 through Dalk_3074 (D. alkenivorans); DacetDRAFT_16020 through DacetDRAFT_16060 (D. acetiphilus); THEYE_A1028 through THEYE_A1032 (T. yellowstonii); BB0710 through BB0713 plus BB_R0040 (B. burgdorferi); PM8707T_20723 through PM8707T_20733 (P. maris).
Homologs of flgZ display shared synteny in taxonomically diverse bacteria
Since genes associated with the same cellular system or process are often co-localized in bacterial genomes, we used the JGI IMG ortholog neighborhood viewer tool to examine the gene neighborhoods surrounding flgZ orthologs to gain additional insight into their potential functions. As indicated previously, the gene upstream of H. pylori flgZ encodes a protein with a NIF3 domain. This gene arrangement is present in all of the Epsilonproteobacteria genomes in which a flgZ ortholog was unambiguously identified, as well as the genomes of several more distantly related bacteria, including members of Deltaproteobacteria, Bacteroidetes, Actinobacteria and several clostridia (Figure 3B). As discussed previously and shown in Figure 1A, the NIF3 domain protein is not required for motility or protection of RpoN from turnover in H. pylori. This does not rule out the possibility that the NIF3 domain protein functions with FlgZ in some other capacity in H. pylori or that the NIF3 and DUF164 domains might work together in other bacteria.
In many diverse bacterial species, genes encoding proteins containing DUF164 are closely associated with rpoD (encodes the primary sigma factor) and dnaG (encodes DNA primase) (Figure 3B). These bacteria included representative species of Spirochaetes (12 unique species), Planctomycetes (3 species), Firmicutes (7 species), Nitrospirae (1 species), Deferribacteres (1 species), and Deltaproteobacteria (8 species). The rpoD genes in these bacteria are located upstream of the gene encoding the DUF164 protein and may be part of the same operon. Given the close association of these genes it is possible that DUF164 proteins in these bacteria have roles in the function of the primary sigma factor.
Conclusions
DUF164 proteins are widespread among bacteria, but the functions of these proteins are virtually unknown. To the best of our knowledge H. pylori FlgZ is the only member for which functions have been ascribed. Results presented here suggest the importance of the DUF164 of FlgZ in protecting RpoN from turnover. Given the co-occurrence of DUF164 and RpoN proteins in many bacterial genomes, we anticipate that DUF164 proteins have a role in RpoN function in other bacteria besides H. pylori. It is clear, however, that DUF164 proteins play other roles in some bacteria, such as the Actinobacteria, which lack RpoN. It is not unusual for bacteria to recruit existing genomic features to adopt additional functions, and the analysis of taxonomical distribution of FlgZ and RpoN homologs suggests that this may have been the case in the evolution of FlgZ. It is possible that the ancestral FlgZ had another function, which has been retained in some present-day bacteria, and adopted a new role in RpoN protection and flagellar regulation in some lineages. However, the decay of the CXXC motifs in some Actinobacteria and other phyla may indicate an alternative scenario where FlgZ homologs in these bacteria adopted a new function that does not require zinc coordination.
Methods
Bacterial strains and media
Escherichia coli DH5α was used for routine cloning procedures and was grown in Luria-Bertani broth or agar medium at 37®C. H. pylori ATCC 43504 and its derivatives were grown on tryptic soy agar (TSA) supplemented with 5% horse serum and grown at 37®C under an atmosphere of 4% oxygen, 5% carbon dioxide, and 91% nitrogen. When required, medium was supplemented with 30 μg/ml chloramphenicol or kanamycin.
Construction and expression of FlgZ variants in H. pylori
Plasmid pEU39 Cm [39] is a suicide vector used to introduce DNA into the H. pylori hp0405 locus (encodes a NifS-like protein). The plasmid carries part of hp0405 that has been disrupted with a cassette containing a Campylobacter coli chloramphenicol transacetylase (cat) gene for selection of recombinants. A derivative of plasmid pEU39 Cm that carries a ~1.5-kb DNA fragment bearing flgZ (hp0958) and most of the upstream gene (hp0959) from H. pylori 26695 was constructed as described previously [19]. Mutations were introduced in flgZ carried on this plasmid using the QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) as per the supplier's instructions. Sequences for the mutagenic primers (Integrated DNA Technologies, Coralville, IA) were: 5'-CAGGCTTGTGGGGGTAGCTTTATTCGGTTGATGATAAG-3' and 5'-CTTATCATTCAACCGAATAAAGCTACCCCCACAAGCCTG-3' to change Cys-202 to serine; and 5'-CGAGTGGGGATATGATCACTAGCCCGTATTGCGGGCG-3' and 5'-CGCCCGCAATACGGGCTAGTGATCATATCCCCACTCG-3' to change Cys-223 to serine (sites where changes were introduced are underlined). The flgZ alleles were sequenced by the Georgia Genomics Facility at the University of Georgia to confirm that the correct mutations had been introduced and that no other mutations had been introduced inadvertently. Plasmids bearing the mutant flgZ alleles were introduced by natural transformation into a H. pylori ATCC 43504 containing a flgZ:aphA3 mutation [19]. Recombinants were selected on TSA supplemented with serum and chloramphenicol and the presence of the flgZ alleles in the hp0405 locus was confirmed by PCR.
Motility assays
Motility agar plates consisted of Mueller-Hinton broth supplemented with 5% horse serum and 0.35% agar. Sterile toothpicks were used to inoculate motility agar with H. pylori strains. Motility of H. pylori strains was scored after incubating plates at 37®C under an atmosphere of 4% oxygen, 5% carbon dioxide, and 91% nitrogen for 4 to 5 days.
Western blot analysis
Immunoblotting with primary antibodies directed against maltose binding protein (MBP) fusions to FlgZ, RpoN or FlaB was done as described previously [19,29]. Antibodies directed against MBP-RpoN were affinity purified prior to use as described [19]. Primary antibodies bound to proteins immobilized on the membranes were detected by enhanced chemiluminescence using peroxidase-coupled goat anti-rabbit antibody (MP Biomedicals, Aurora, ID).
Analysis of genome sequences
FlgZ and RpoN homologs were identified from the United States Department of Energy's JGI IMG and Wellcome Trust Sanger Institute Pfam databases [40,41]. Genes surrounding flgZ homologs were analyzed using the JGI ortholog neighborhood viewer tool. The taxonomical distribution of FlgZ and RpoN homologs was compiled from data downloaded from the IMG database using an in-house computer program.
List of abbreviations
FlgZ: flagellar-associated zinc-ribbon protein; NIF3: NGG1p interacting factor 3; JGI: Joint Genome Institute; IMG: Integrated Microbial Genomes; TSA: trypic soy agar; cat: chloramphenicol transacetylase; MBP: maltose-binding protein; aphA3: kanamycin-resistance gene
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LP constructed and characterized the H. pylori strains used in the study. JT participated in the taxonomic analysis. JM participated in the taxonomic analysis, performed the statistical analysis of the data, and helped draft the manuscript. TH conceived of the study, participated in its design, and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Lara E Pereira, Email: leperei@emory.edu.
Jennifer Tsang, Email: jtsang@uga.edu.
Jan Mrázek, Email: mrazek@uga.edu.
Timothy R Hoover, Email: trhoover@uga.edu.
Acknowledgements
We thank Anna Karls and Robert Scott for helpful discussions. This work was supported by award AI080923 to TH from the National Institutes of Health and award DBI-0950266 to JM from the National Science Foundation.
References
- Blaser MJ, Parsonnet J. Parasitism by the 'slow' bacterium Helicobacter pylori leads to altered gastric homeostasis and neoplasia. J Clin Invest. 1994;94:4–8. doi: 10.1172/JCI117336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pylori and gastroduodenal disease. Annu Rev Med. 1992;43:135–145. doi: 10.1146/annurev.me.43.020192.001031. [DOI] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- Eaton KA, Morgan DR, Krakowka S. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect Immun. 1989;57:1119–1125. doi: 10.1128/iai.57.4.1119-1125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton KA, Morgan DR, Krakowka S. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:(4):694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Chang JH, Chung SI, Yum JS. Molecular cloning and characterization of the Helicobacter pylori fliP gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leying H, Suerbaum S, Geis G, Haas R. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol Microbiol. 1992;6:2863–2874. doi: 10.1111/j.1365-2958.1992.tb01466.x. [DOI] [PubMed] [Google Scholar]
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S, Josenhans C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol. 2004;52(4):947–961. doi: 10.1111/j.1365-2958.2004.04006.x. [DOI] [PubMed] [Google Scholar]
- Porwollik S, Noonan B, O'Toole PW. Molecular characterization of a flagellar export locus of Helicobacter pylori. Infect Immun. 1999;67:2060–2070. doi: 10.1128/iai.67.5.2060-2070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohn G, Scarlato V. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol. 1999;181(2):593–599. doi: 10.1128/jb.181.2.593-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Frank R. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol. 2000;182:2068–2076. doi: 10.1128/JB.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan E, Dorrell N, Foynes S, Anyim M, Wren BW. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J Bacteriol. 2000;182:5274–5277. doi: 10.1128/JB.182.18.5274-5277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50(2):687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- Schmitz A, Josenhans C, Suerbaum S. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J Bacteriol. 1997;179:987–997. doi: 10.1128/jb.179.4.987-997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TG, Pereira L, Hoover TR. Helicobacter pylori FlhB processing-deficient variants affect flagellar assembly but not flagellar gene expression. Microbiology. 2009;155(Pt 4):1170–1180. doi: 10.1099/mic.0.022806-0. [DOI] [PubMed] [Google Scholar]
- Pereira L, Hoover TR. Stable accumulation of sigma54 in Helicobacter pylori requires the novel protein HP0958. J Bacteriol. 2005;187(13):4463–4469. doi: 10.1128/JB.187.13.4463-4469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KA, Karim N, Worku M, Moore SA, Penn CW, O'Toole PW. HP0958 is an essential motility gene in Helicobacter pylori. FEMS Microbiol Lett. 2005;248(1):47–55. doi: 10.1016/j.femsle.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Rain J-C, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V. et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Douillard FP, Ryan KA, Caly DL, Hinds J, Witney AA, Husain SE, O'Toole PW. Posttranscriptional regulation of flagellin synthesis in Helicobacter pylori by the RpoN chaperone HP0958. J Bacteriol. 2008;190(24):7975–7984. doi: 10.1128/JB.00879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly DL, O'Toole PW, Moore SA. The 2.2 A resolution structure of the HP0958 protein from Helicobacter pylori reveals a kinked antiparallel coiled coil hairpin domain and a highly conserved Zn-ribbon domain. J Mol Biol. 2010;403(3):405–419. doi: 10.1016/j.jmb.2010.08.051. [DOI] [PubMed] [Google Scholar]
- Bouhouche N, Syvanen M, Kado CI. The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol. 2000;8(2):77–81. doi: 10.1016/S0966-842X(99)01679-0. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8(3):209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Markus MA, Morris BJ. RBM4: a multifunctional RNA-binding protein. Int J Biochem Cell Biol. 2009;41(4):740–743. doi: 10.1016/j.biocel.2008.05.027. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Sunde M. Zinc fingers--folds for many occasions. IUBMB Life. 2002;54(6):351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- Brahmachary P, Dashti MG, Olson JW, Hoover TR. Helicobacter pylori FlgR is an enhancer-independent activator of sigma54-RNA polymerase holoenzyme. J Bacteriol. 2004;186(14):4535–4542. doi: 10.1128/JB.186.14.4535-4542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck LL, Smith TG, Hoover TR. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 2007;15(12):530–537. doi: 10.1016/j.tim.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Joslin SN, Hendrixson DR. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J Bacteriol. 2008;190(7):2422–2433. doi: 10.1128/JB.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo IS, Stephens RS. A developmentally regulated two-component signal transduction system in Chlamydia. J Biol Chem. 2003;278:17314–17319. doi: 10.1074/jbc.M212170200. [DOI] [PubMed] [Google Scholar]
- Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol Microbiol. 2007;65(2):277–293. doi: 10.1111/j.1365-2958.2007.05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54(4):855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J Bacteriol. 2003;185(20):6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C, Krushkal J, Ueki T, Puljic M, Sun J, Juarez K, Nunez C, Reguera G, DiDonato R, Postier B. et al. Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens. BMC Genomics. 2009;10:331. doi: 10.1186/1471-2164-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tascou S, Uedelhoven J, Dixkens C, Nayernia K, Engel W, Burfeind P. Isolation and characterization of a novel human gene, NIF3L1, and its mouse ortholog, Nif3l1, highly conserved from bacteria to mammals. Cytogenet Cell Genet. 2000;90(3-4):330–336. doi: 10.1159/000056799. [DOI] [PubMed] [Google Scholar]
- Sprinzak E, Altuvia Y, Margalit H. Characterization and prediction of protein-protein interactions within and between complexes. Proc Natl Acad Sci USA. 2006;103(40):14718–14723. doi: 10.1073/pnas.0603352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Anderson I, Lykidis A, Mavromatis K. et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38(Database issue):D382–390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]